Abstract
Different disease outcomes of Helicobacter pylori infection correlate with distinct inflammation patterns. These different inflammatory distributions may be initiated by differences in bacterial localization. One H. pylori property known to affect murine stomach localization is chemotaxis, the ability to move in response to chemical cues. In this report, we used nonchemotactic mutants (Che−) to analyze whether chemotaxis is required for initial colonization of particular stomach regions or for subsequent growth therein. We found that H. pylori behaves differently in the corpus, antrum, and corpus-antrum transition zone subregions of the stomach. This outcome suggests that these regions contain unique chemotactic signals. In the corpus, H. pylori utilizes chemotaxis for initial localization but not for subsequent growth. In contrast, in the antrum and the corpus-antrum transition zone, chemotaxis does not help initial colonization but does promote subsequent proliferation. To determine which chemoreceptor is responsible for the corpus-antrum phenotypes, we infected mice with strains lacking each chemoreceptor. Strains lacking TlpA, TlpB, or TlpC displayed only modest deviations from the wild-type phenotype, while strains lacking TlpD resembled the Che− mutant in their antral colonization defect and fared even worse than the Che− mutant in the corpus. Additional analysis showed that inflammation is worse in the antrum than in the corpus in both wild-type and Che− mutant infections. These results suggest that chemotaxis, specifically, that controlled by TlpD, is necessary for H. pylori to survive or grow in the environment of increased inflammation in the antrum.
INTRODUCTION
The gastric pathogen Helicobacter pylori chronically infects the stomachs of 50% of the world's population. Infection with H. pylori triggers gastric inflammation, also known as gastritis, which can lead to a wide range of disease outcomes, such as mild gastritis, gastric and peptic ulcers, gastric adenocarcinoma, and mucosa-associated lymphoid tissue (MALT) lymphoma (38). H. pylori-induced gastritis occurs most frequently in the antrum (A) (3, 26, 49), one of four distinct regions in the human stomach (19, 28). Each region is defined by the cell types that exist in the gastric glands. The two largest stomach regions are the antrum and the corpus (C), and a third region, the corpus-antrum (C-A) transition zone, separates them. Multiple lines of evidence suggest that H. pylori-triggered inflammation in each of these regions results in distinct disease outcomes. For example, gastritis that affects predominantly the antrum correlates with H. pylori-triggered duodenal ulcers (3, 56), a disease state that does not develop to gastric cancer (3, 45, 49). Gastritis that predominantly affects the corpus, in contrast, increases the risk of gastric ulcers and intestinal gastric cancer (3, 49), while gastritis throughout the stomach, so-called pangastritis, increases the risk for diffuse gastric cancer (49). These observations demonstrate that specific H. pylori diseases correlate with inflammation in specific stomach regions. The basis for these different patterns of inflammation is unclear, but one possible reason for the differential locations of diseased tissue is that different H. pylori strains preferentially colonize either the antrum or the corpus and in turn trigger elevated inflammation in that region. Thus, the localization of H. pylori within the stomach may be important for driving disease development.
Studies have shown that different H. pylori strains achieve higher bacterial colonization numbers in either the corpus or the antrum (1, 27, 33). For example, strain X47 has a strong preference for the corpus, and strain SS1 has an antral preference or colonizes the antrum and corpus equally, depending on the study (1, 27, 47). Three H. pylori characteristics have been found that influence stomach localization: (i) the presence of a cytotoxin-associated pathogenicity island, or cag-PAI (40); (ii) the presence of the ferric uptake regulator (Fur) (33), which is necessary for H. pylori adaptation to various conditions, including iron limitation (50), acid stress, and oxidative stress (6, 14); and (iii) the bacterial ability to be chemotactic, or to move in response to chemical cues (47).
Bacterial chemotaxis has been extensively studied in Escherichia coli and thus serves as a model for other bacteria (34, 52). Chemotactic responses initiate when chemoreceptors detect environmental cues and transmit information to CheA, a histidine kinase, via CheW, the coupling protein. In the absence of chemotaxis attractants, the ligand-free chemoreceptors activate the CheA histidine kinase, which in turn phosphorylates the response regulator CheY. Phosphorylated CheY (CheY-P) interacts with the flagellar motor to cause clockwise flagellar rotation and random bacterial movement. Conversely, when the chemoreceptors bind a chemoattractant ligand, the CheA histidine kinase is inactivated and CheY-P populations are reduced and replaced with nonphosphorylated CheY. Nonphosphorylated CheY does not interact with the flagellar motor, which in turn rotates in the default counterclockwise direction, and the bacteria swim in a straight line.
The chemotaxis system of H. pylori is similar to that of E. coli (29). H. pylori has the core signaling complex proteins CheW, CheA, and CheY, and disruption of any of these core proteins renders H. pylori nonchemotactic (5, 15, 37). H. pylori has three membrane-bound chemoreceptors, TlpA, TlpB, and TlpC, and one cytoplasmic chemoreceptor, TlpD (29). TlpA has been shown to sense arginine and bicarbonate (7), and TlpB has been shown to sense pH (10, 20) and autoinducer-2 (AI-2), the quorum-sensing molecule (39). TlpD is involved in energy taxis (44), and the ligands for TlpC are unknown.
Individual H. pylori strain SS1 chemoreceptor mutants colonize the mouse stomach to wild-type levels (2, 28) but fare less well in coinfections with the wild type. Specifically, mutants lacking TlpA or TlpC are outcompeted 25-to-50-fold by the wild type (2), while those lacking TlpB are not (31, 54). In comparison, nonchemotactic H. pylori SS1 mutants (Che−) that lack core-signaling components such as CheW, CheA, or CheY are outcompeted more than 1,000-fold. These Che− mutants additionally have colonization defects during the first 2 weeks of infection when they are the sole infecting strain (47). After 2 weeks, Che− mutants achieve wild-type colonization levels; however, they have a different distribution throughout the stomach. Unlike the wild type, Che− mutants have a striking inability to colonize the antrum, while they occupy the corpus at only modestly decreased levels compared to the wild type (47). Thus far, it is unknown why antral localization requires chemotaxis. Chemotaxis may be necessary to locate the antrum or to maintain colonization there, and one of the four chemoreceptors may be specifically responsible for the localization defects.
Since H. pylori infections are associated with antral gastritis (3, 26, 49), we found it interesting that bacterial chemotaxis both drives antral localization (47) and increases the inflammatory response to H. pylori (31, 41, 54). Therefore, we undertook this study to determine if chemotaxis is necessary for H. pylori to locate the antrum or to remain there after initial colonization and to determine which chemoreceptor is responsible for this localization phenotype.
MATERIALS AND METHODS
H. pylori strains and growth conditions.
H. pylori strain SS1 (27) and its isogenic mutants were used for all studies. SS1 was a gift of Jani O'Rourke (University of New South Wales) and was subjected to minimal laboratory passage before our experiments. SS1 ΔcheY::cat, ΔtlpA::cat, ΔtlpB::cat, ΔtlpC::cat, and ΔtlpD::cat were described previously (2, 31, 47, 54), and all have most of the target gene replaced with the Campylobacter cat gene. H. pylori was cultured on Columbia blood agar (Remel) with 5% difibrinated horse blood (Hemostat Laboratories, Davis, CA), 50 μg/ml cycloheximide, 10 μg/ml vancomycin, 5 μg/ml cefsulodin, 2.5 U/ml polymyxin B, and 0.2% β-cyclodextrin. Mouse stomach samples were plated on the same medium plus 5 μg/ml trimethoprim, 8 μg/ml amphotericin B, 10 μg/ml nalidixic acid, and 200 μg/ml bacitracin. To prepare H. pylori for mouse infection, we grew the bacteria with shaking in brucella broth (Becton, Dickinson and Company) with 10% fetal bovine serum (FBS; Gibco) (BB10). Cultures were incubated at 37°C with 7 to 10% O2, 10% CO2, and 80 to 83% N2, with chloramphenicol at 13 μg/ml to select for the various mutants as needed. For mouse infection, we inoculated animals orally intragastrically via a feeding needle (20 gauge by 1.5 in.) with a 500-μl inoculum containing ∼1 × 107 CFU/ml brucella broth-grown H. pylori.
Animal infections.
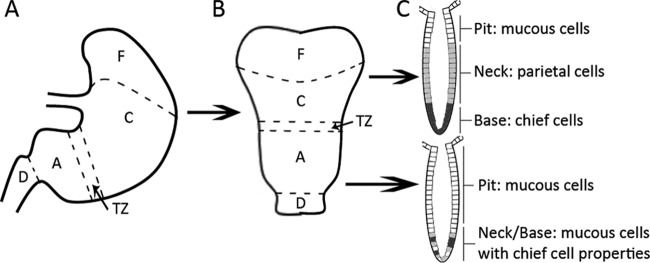
The University of California (U.C.) Santa Cruz Institutional Animal Care and Use Committee approved all animal protocols and experiments. Female C57Bl6/N mice (Helicobacter-free) (Taconic Laboratories, Germantown, NY) and female and male FVB/N mice (Helicobacter-free) (Charles River Laboratories) were housed at the animal facility of the U.C. Santa Cruz. Mice were between 6 and 8 weeks old at the time of H. pylori infection, and age-matched uninfected mice were included in all experiments. After the infection period, the animals were sacrificed via CO2 narcosis, and the stomach was dissected, opened along the lesser curvature, and divided into four parts—the corpus, the transition zone, the antrum, and the upper duodenum (Fig. 1). The nonglandular forestomach was identified as a yellow, thin tissue structure; it was removed from the stomach and not included in experimental analysis. The corpus is a brownish, thick-walled section of tissue, and the antrum is a paler, pinkish, thinner-walled section. The transition zone is an ∼1.0-mm-wide region demarcating these two sections. These regions were identified as described by Lee et al. (28). The tissue pieces were homogenized using a Bullet Blender (Next Advance) with 1.0-mm-diameter zirconium silicate beads and plated to determine the number of H. pylori CFU/gram of stomach.
Fig 1.
A diagram of the different physiological regions of the mouse stomach (28). (A) Side view of the stomach regions. (B) The stomach is opened up along the lesser curvature and divided down the middle along the longitudinal axis. (C) Representation of the different cell types found in the glands of the antrum and the corpus. F, forestomach; C, corpus; TZ, C-A transition zone; A, antrum; D, duodenum.
Histology and pathology.
After dissection, half of the stomach was placed in a histology cassette with a sponge (Fisher) and preserved in buffered formalin (Fisher). The tissue was embedded in paraffin, sectioned (5-μm slices), stained with hematoxylin and eosin, and evaluated in a blinded manner by a pathologist (J. E. Carter). Each stomach slice received a separate grade for each region of the stomach. Each slide was evaluated twice. Lymphocytic infiltration was scored as outlined by Eaton et al. (11), using scoring as follows: 0, no infiltrate; 1, mild, multifocal infiltration; 2, mild, widespread infiltration; 3, mild, widespread, and moderate multifocal infiltration; 4, moderate, widespread infiltration; and 5, moderate, widespread, and severe multifocal infiltration. Neutrophils were noted as either present or absent (data not shown). Gastric atrophy was assessed using the method of Rugge et al. (42) but was not present in any sample.
RESULTS
Chemotaxis is necessary for H. pylori to remain in the antrum after infection has established.
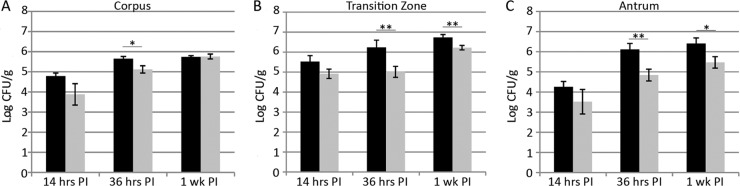
Terry et al. reported previously that Che− H. pylori bacteria are barely found in the murine antrum at 2 weeks postinoculation compared to wild-type H. pylori (47). This finding is consistent with chemotaxis either directing the bacteria to initially locate the antrum or allowing the bacteria to remain in the antrum after infection has been established. To determine at what point chemotaxis was needed in infection, we examined the numbers of wild-type and Che− H. pylori bacteria in the corpus, transition zone, and antrum at early time points (14 h) compared to later time points postinoculation. For our first time point, we wanted only to enumerate stably infecting bacteria, so we chose 14 h postinoculation, a time well after the 3 h required for the mouse stomach to empty from one feeding (46). At 14 h postinoculation, Che− bacteria did not have a substantial initial colonization defect in either the transition zone or the antrum, as indicated by numbers that did not differ significantly from those of the wild type (Fig. 2B and C). Interestingly, by 36 h postinoculation in these regions, the Che− H. pylori population had undergone only a modest increase in numbers compared to the substantial increase in wild-type numbers, and thus Che− colonization levels were significantly lower in the transition zone and the antrum. We observed that this phenotype remained at 1 week postinoculation (Fig. 2B and C), similar to what had been observed previously at 2 weeks (47). These results suggest that chemotaxis does not promote the initial antrum or transition zone seeding but is very important during the increase in H. pylori numbers that occurs from 14 h to 1 week postinoculation in these regions.
Fig 2.
Chemotaxis is required for antral colonization at time points after 14 h postinoculation (PI). Female FVB/N mice were infected with the SS1 wild type (black bars) or the isogenic Che− mutant (gray bars) for 14 h, 36 h, or 1 week. At each time point, stomachs were removed, and divided into the corpus (A), the C-A transition zone (B), and the antrum (C) and plated to determine H. pylori colonies. Error bars indicate standard error of the mean. n = 8 or 9 for 14 and 36 h, derived from two independent infections. n = 7 for 1 week, derived from one independent infection. *, P < 0.05; **, P < 0.01 (Student's t test). Data were also used for the doubling-time calculations in Table 1 and shown in Fig. 3.
In the corpus, Che− H. pylori colonization levels were somewhat lower at 14 h and remained so at 36 h, suggesting that chemotaxis plays a minor role in the initial seeding of this stomach region. Between 36 h and 1 week, numbers of both wild-type and Che− H. pylori bacteria increased such that they were nearly equal at 1 week postinoculation (Fig. 2A). Thus, it seems that chemotaxis is not needed for bacterial multiplication in the corpus.
We also examined the upper duodenum to identify if bacteria were washed out of the stomach into the duodenum. However, H. pylori colonies rarely grew from the duodenal samples, suggesting that the H. pylori bacteria were dead, unculturable, or present in the duodenum at numbers below our limit of detection (∼200 CFU per gram) (data not shown). We are therefore not able to estimate the contribution of bacterial loss due to flow out of the stomach.
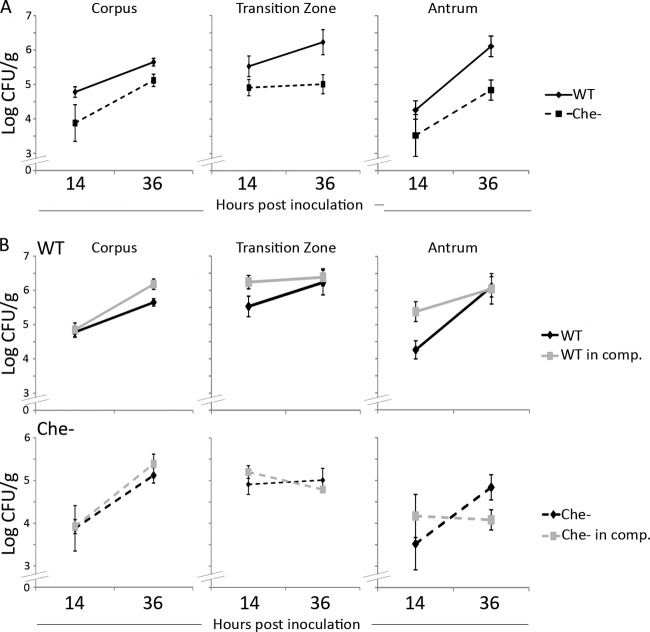
To estimate the doubling time of H. pylori colonies early in infection, when the bacterial numbers are increasing, we reasoned that we could use the equation for unrestricted growth. Early in infection, bacterial numbers are increasing due to multiplication but there is also presumably an unknown rate of removal of bacterial cells due to death and flow out of the stomach. Later in infection, in contrast, bacterial population growth rates are hypothesized to be similar to growth rates in a continuous-flow or chemostat culture although more complicated due to nonuniform nutrient distributions (16–18, 30). The chemotaxis mutants used here do not have in vitro growth defects, as measured in BB10 media for the Che−, the ΔtlpA, and the ΔtlpC mutants (2) and ΔtlpD mutants (S. M. Williams and K. M. Ottemann, personal communication). The ΔtlpB mutant behavior has not been directly measured, but it has no defect in an assay that depends on growth, the soft agar migration assay, suggesting that it does not have a growth defect (31). We found that the total population of wild-type H. pylori doubled approximately every 6.5 h in the corpus and 8 h in the transition zone. In the antrum, in contrast, wild-type H. pylori did considerably better, with the population doubling every 3 h (Fig. 3A; Table 1). Similar to that of the wild type, the population of the Che− mutant doubled approximately every 5.5 h in the corpus. However, in the antrum, the Che− strain population doubled approximately every 5 h, or at a rate nearly two times lower than that seen with the wild type in this region. Additionally, the Che− mutant population did not double in the transition zone over this time period (Fig. 3A; Table 1). These data further indicate that chemotaxis promotes growth or prevents H. pylori from washing out of the transition zone and antrum.
Fig 3.
Chemotaxis allows H. pylori to grow more quickly in the antrum and provides an advantage in a competition infection. (A) Female FVB/N mice were infected with the SS1 wild type (solid line) or the isogenic Che− mutant (dotted line) for 14 or 36 h. At each time point, stomachs were removed and divided into the corpus, the C-A transition zone, and the antrum and plated to determine the number of H. pylori. n = 8 or 9 for the wild type and the Che− mutant for all time points, derived from two independent infections. Data are also shown in Table 1 and Fig. 2. (B) Competition experiments were carried out by infecting female FVB/N mice with a 1:1 ratio of the wild type and the Che− mutant for 14 or 36 h. At each time point, stomachs were removed, divided, and plated as described for panel A. Black lines represent each strain in single infection, as described for panel A; gray lines indicate the strains in competition. n = 5 for the wild type and the Che− mutant for all time points, derived from one experiment. Error bars represent standard error of the mean.
Table 1.
Doubling times for H. pylori SS1 or its isogenic Che− (ΔCheY) mutant early after infectiona
| Infection category and strain | Doubling time (h) |
||
|---|---|---|---|
| Corpus | Transition zone | Antrum | |
| Single | |||
| WT | 6.46 | 7.87 | 3.00 |
| ΔCheY | 5.47 | 57.29 | 5.20 |
| Competition | |||
| WT | 5.28 | 49.49 | 10.55 |
| ΔCheY | 4.63 | No growth | No growth |
Doubling time = (22 h)/[(log CFU/gram36 h − log CFU/gram14 h)/log2]. Data are also shown in Fig. 3A.
Competition with wild-type H. pylori exacerbates the Che− H. pylori survival defect in the antrum.
We also determined how chemotaxis aids stomach region colonization under conditions where this process is more important—the competition infection—at the two earliest time points. For these experiments, we infected mice with a mixture of equal amounts of the wild type and the Che− mutant and assessed numbers of each strain at 14 and 36 h postinoculation as described above. Generally speaking, the numbers of both the wild type and the Che− mutant were not affected by the presence of the other strain in the corpus and transition zone at 14 or 36 h (Fig. 3B). One possibility is that the total bacterial numbers of both strains were below the threshold at which competition has an effect or, alternatively, that conditions that result in a competitive defect do not exist at those early infection times. The antrum represented a different situation. In this region, numbers of the Che− mutant did not increase (Fig. 3B). These results show that chemotaxis greatly facilitates bacterial survival or growth in this region, particularly when the wild type is included (Fig. 3B; Table 1).
Enhanced inflammatory responses may contribute to Che− antral localization defects.
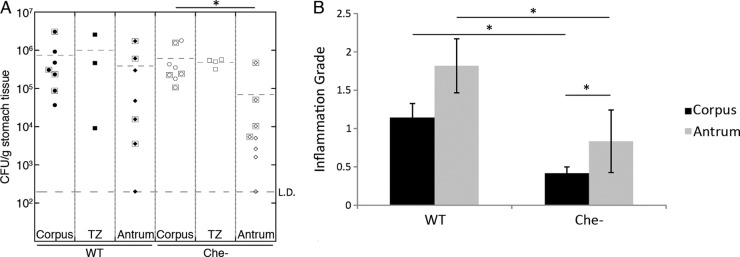
Several studies have reported that the majority of gastritis cases are antral dominant in humans (3, 26, 49). Since chemotaxis is necessary for H. pylori to remain in the antrum after infection has been established, we speculated that antral immune responses make this localization unfavorable for Che− H. pylori. Reports on the location of gastritis in mice suggest that there is increased inflammation in the corpus-antrum (C-A) transition zone and the antrum at 6 months postinoculation (27), and so we sought to examine if inflammation specifically in the antrum correlates with reduced Che− colonization. Inflammation is not detectable until after several months of H. pylori infection (12), so we examined inflammation and colonization levels in each physiologic region of stomachs from mice infected with wild-type or Che− H. pylori for 3 or 6 months. At these time points, we found that wild-type H. pylori induced more inflammation than Che− overall (Fig. 4B), as reported previously (54). Inflammation in the antrum was higher for both infection types compared to the corpus (Fig. 4B). Additionally, we noted that Che− strains continued to struggle to colonize the antrum (Fig. 4A). Thus, it appears that the antrum had significantly more inflammation than the corpus and was significantly more challenging for Che− H. pylori to colonize.
Fig 4.
The antrum harbors lower H. pylori colonization than other regions and more severe inflammation. (A) H. pylori colonization levels as CFU/gram of stomach from female C57BL/6N mice infected for 3 months (symbols surrounded by boxes) or 6 months (symbols without boxes) with H. pylori strain SS1 or its isogenic Che− mutant. n = 7 for the wild type, n = 8 for the Che− mutant. (B) Gastric inflammation tissue from mice infected for 6 months was analyzed by a pathologist and graded for inflammation. Error bars indicate standard error of the mean. Data derived from the same 6-month-infected mice as presented in panel A. L.D. = limit of detection. n = 5 for the wild type and the Che− mutant. *, P < 0.05 (Student's t test).
The chemoreceptor TlpD is required for growth and/or survival in the antrum.
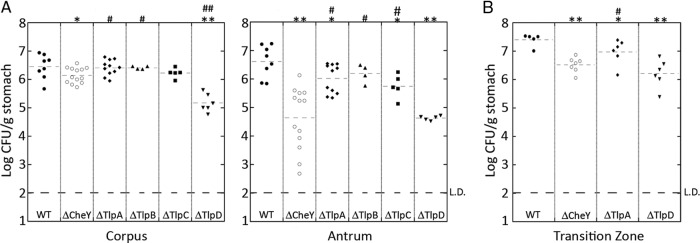
To help distinguish the signals required for colonization of the corpus versus the antrum, we next investigated whether individual chemoreceptor mutants had any distinct antrum or corpus colonization defects. To this end, we infected mice with SS1 ΔtlpA::cat, SS1 ΔtlpB::cat, SS1 ΔtlpC::cat, or SS1 ΔtlpD::cat and examined the colonization levels of each mutant 2 weeks postinoculation in the corpus and the antrum. As observed previously at this time point (47), the Che− mutant colonization levels in the antrum were significantly below that of the wild type (Fig. 5A). Loss of one receptor, TlpD, came the closest to phenocopying the antral colonization defect observed in the Che− mutant (Fig. 5A). In contrast, mutants lacking the TlpA, TlpB, or TlpC chemoreceptor had only slight defects in antral localization (Fig. 5A). To confirm these results, we repeated the experiment and included the transition zone. In this independent infection, the corpus and antrum colonization of TlpD again came closest to phenocopying the Che− antral colonization defect (Fig. 5A). Additionally, we observed that the Che− and TlpD mutants had significant defects in colonizing the transition zone (Fig. 5B). These findings suggested that the TlpD chemoreceptor is needed for H. pylori to flourish in the antrum and transition zone. Interestingly, we also observed that the ΔtlpD mutant had a significant colonization defect in the corpus (Fig. 5A), which was not observed with any other chemoreceptor mutant and was more significant than the slight colonization defect of the Che− strain in this region (Fig. 5A). We wanted to rule out the possibility that the tlpD mutant has a colonization defect that is general and not specific to the antrum. We therefore determined each strain's corpus/antrum ratio. If the tlpD mutant had a general defect, we would expect its corpus/antrum ratio to match that of the wild type. This analysis instead found that the tlpD mutant ratio was like that of the Che− mutant and not like that of the wild type (Table 2). Therefore, TlpD appears to be critical for specific H. pylori antral localization. Additionally, however, it seems that TlpD plays an important role in the corpus as well.
Fig 5.
The chemoreceptor TlpD is necessary for antral colonization. FVB/N mice were infected with the wild type or a Che−, TlpA−, TlpB−, TlpC−, or TlpD− mutant for 2 weeks, at which time the stomachs were removed, divided into (A) the corpus and antrum or (B) the corpus, antrum, and C-A transition zone, and plated to enumerate H. pylori colonies. n = between 4 and 13. L.D., limit of detection. In comparison to the wild type, *, P < 0.05; **, P < 0.0001. In comparison to the Che− mutant, #, P < 0.05; ##, P < 0.0001 (Student's t test).
Table 2.
Ratio of CFU/gram for the corpus versus the antruma
| Time PI and strain | Corpus/antrum ratio |
|
|---|---|---|
| Avg | Range | |
| 36 h | ||
| WT | 0.92 | 0.04–8.08 |
| ΔCheY | 1.05 | 0.29–18.0 |
| 2 wk | ||
| WT | 0.55 | 0.04–3.71 |
| ΔCheY | 12.0 | 2.08–1,117.74 |
| ΔTlpA | 1.90 | 0.57–15.62 |
| ΔTlpB | 1.39 | 0.75–4.96 |
| ΔTlpC | 2.30 | 1.75–6.76 |
| ΔTlpD | 5.19 | 1.10–507.35 |
| 6 mo | ||
| WT | 1.88 | 0.13–437.50 |
| ΔCheY | 8.96 | 3.32–2158.27 |
DISCUSSION
In this report, we provide evidence that bacterial chemotaxis promotes initial seeding in some stomach regions and the ability to rapidly multiply in others. Specifically, we found that H. pylori chemotaxis is needed to guide H. pylori to the corpus, but not to the antrum or transition zone, and that, once in the corpus, chemotaxis is not needed for H. pylori populations to increase. This finding suggests that nutrients are not limiting in the corpus, such that Che− mutants are able to obtain all that they need. The situation is different in the antrum and transition zone, where chemotaxis is required for the bacteria to achieve normal bacterial loads but not initial colonization. In these niches, Che− mutant numbers do not increase at the same rate as do those of the wild type. The main chemoreceptor that allows H. pylori to thrive in the antrum is TlpD, with the other chemoreceptors playing minor roles.
The mouse stomach has four distinct physiological regions: the forestomach, the corpus, the C-A transition zone, and the antrum (Fig. 1) (28). The gastric epithelium throughout the latter three regions is organized into repeating units called glands that invaginate from the surface. The glands in each stomach region are composed of different cells. The corpus glands contain zymogenic/digestive enzyme-secreting chief cells at the bottom and parietal/acid-producing cells, mucous neck cells, and mucous pit cells located progressively closer to luminal surface of the gland. There are also scattered but infrequent hormone-secreting enteroendocrine cells. The antrum similarly has scattered, infrequent enteroendocrine cells but, in contrast, has no parietal or chief cells. Instead, two types of mucous cells dominate these glands. One type is similar to the corpus mucous pit cells, while the other has properties that are a mixture of those of mucous neck cells and zymogenic cells (Fig. 1) (28, 36). Interestingly, mucin secreted by mucous neck cells inhibits H. pylori growth, while mucous pit cell mucin stimulates H. pylori growth (24). Accordingly, H. pylori has been found to occupy and replicate within the mucous pit cell mucin but not within the mucous neck cell mucin (23), suggesting that even within the distinctive stomach regions, there are some subareas more suitable for H. pylori than others.
The stomach regions described above present H. pylori with distinctive signals and challenges. For example, the cells in the antrum have a much higher turnover rate than those in the corpus, possibly increasing numbers of bacteria shed into the lumen (21). Also, mucous cells in both the corpus and the antrum secrete abundant superoxide anion (O2-) (48); however, the corpus may temper the levels of these oxygen radicals with the high parietal cell expression of free radical scavengers (35). Parietal cells also secrete concentrated HCl, which presumably results in more frequent exposure to acid in the corpus. The pH of the corpus, however, is not lower overall than that of the antrum, although it does display a broader range (8, 32, 51). Thus, the different regions of the stomach possess distinct physiological characteristics that likely present H. pylori with different chemotactic signals.
H. pylori-induced disease depends of the localization of inflammation: gastritis in the antrum predisposes patients to duodenal ulcer, while gastritis in the corpus predisposes patients to gastric ulcers and cancer (3, 49). While it is unknown why inflammation develops in different stomach regions, one explanation could be that inflammation follows H. pylori density and that different H. pylori strains have distinct preferences for the corpus or the antrum (1). Three bacterial genetic determinants have been found that influence corpus-antrum distribution: the cag-PAI, the Fur transcription factor, and chemotaxis. Loss of cagA delivery, by mutation of either cagA or cagY, creates strains that do better in the gerbil antrum and worse in the corpus than their parents (40). These strains also induce reduced inflammation in both regions compared to the wild type. H. pylori fur mutants, in contrast, are found in nearly wild-type numbers in the antrum at 1 day postinoculation but decrease in antral numbers at day 3 postinoculation (33). These phenotypes suggest that fur, like chemotaxis, is not needed for antral seeding but is needed for replication. Also, like the Che− mutant defect, the fur defect is less significant in the corpus. Fur regulates some chemotaxis genes, such as cheV1, possibly explaining the overlapping phenotypes (13, 33). Thus, these three bacterial properties affect corpus-antrum localization in different ways and suggest that normal H. pylori variations might influence stomach region density.
Our studies presented here demonstrate that chemotaxis does not help H. pylori locate the antrum but does help it achieve normal bacterial numbers in this region. This finding suggests that some parameter of the antrum is particularly difficult for H. pylori and that chemotaxis helps to overcome this challenge. We report here that one potentially challenging condition is the elevated inflammation found in the antrum, which we could detect at 6 months of infection. We did not, however, detect a correlation between inflammation grade and bacterial numbers, as also seen by Williams et al. (54). We did note, however, that there was greater variation in H. pylori numbers at later time points postinfection (Fig. 4A) than at earlier ones (Fig. 2 and 3). This difference may be a reflection of individual variation in the adaptive immune response to H. pylori, whereas the innate response, which drives earlier numbers, does not display as much variation. We do not yet know if early inflammatory events specifically in the antrum render this area inhospitable for growth in the first part of infection, but this possibility is consistent with our data.
Based on this information, we envision two possibilities: (i) chemotaxis allows H. pylori to locate a nutrient source necessary for growth and survival in the antrum or (ii) chemotaxis allows H. pylori to detect and avoid damaging compounds, such as those created by the immune response. Increased inflammation could amplify the necessity for chemotaxis in either situation, as inflammation is known to alter the nutrient status by chelating critical metals such as iron, zinc, and manganese (9, 25), and inflammation also increases production of reactive oxygen species (ROS) during the host's strong oxidative stress response (4). H. pylori appears to experience significant ROS exposure, as evidenced by the high cellular levels of enzymes such as SodB, catalase, AhpC, MdaB, and NapA (4, 22, 53).
To help us determine the nature of the chemotactic signal required for growth or survival in the antrum, we investigated which chemoreceptor was responsible for antral localization. Our studies determined that TlpA, TlpB, and TlpC each had only a moderate role in antral colonization. We were somewhat surprised that the acid-sensing TlpB did not play a more dominant role in antral localization, as one might have predicted that acid could repel H. pylori from the corpus to drive antral colonization (10). While pH gradients have been shown to impact the orientation of H. pylori in the mucus (43), our results suggest that pH sensing by TlpB does not contribute to H. pylori localization in the murine antrum. We do not know, however, if TlpB contributes to the initial seeding of the corpus.
TlpD appears to play the dominant role in the antral colonization of H. pylori, as well as in colonization in other regions. Interestingly, the tlpD mutant had a colonization defect in the corpus that was more severe than that seen with the completely nonchemotactic Che− mutant. Although we do not yet know why tlpD mutants experience such a corpus challenge, it is possible that the remaining chemoreceptors—TlpA, TlpB, and TlpC—attract H. pylori to an unsuitable niche, and normally, TlpD would act to at least partially repel the bacteria from a fatal interaction in this region.
TlpD is the only cytoplasmic chemoreceptor in H. pylori (44). Schweinitzer et al. demonstrated that TlpD senses cellular energy levels via the electron transport chain (44). Thus, TlpD may allow H. pylori to find optimal energy-generating conditions in the antrum and therefore promote colonization. One possibility is that increased antral inflammation alters the metabolically optimal area for H. pylori growth. For example, inflammation creates new respiratory electron acceptors in Salmonella infection through ROS-driven formation of tetrathionate from thiosulfate (55). In the case of H. pylori, it may be that inflammation limits the metabolically favorable region. Another possibility is that TlpD directs a repellent response to a deleterious component of the immune system, which allows H. pylori to escape and survive. This idea is consistent with our observed corpus phenotype in which tlpD mutants are even more defective than Che− mutants. One possible noxious compound is ROS. Our unpublished observations support the idea that wild-type H. pylori has a repellent response to ROS that is mediated by TlpD (Williams and Ottemann, personal communication).
In summary, we have shown that chemotaxis plays distinct roles in colonizing specific stomach regions, suggesting that there are subtle chemotactic cues that differ across this organ. Surprisingly, chemotaxis promotes initial seeding in some regions and growth in others. One of the four H. pylori chemoreceptors, TlpD, plays a dominant role in the growth phenotype. We hypothesize that antral success requires detection of a limited energy source or movement away from deleterious immune responses.
ACKNOWLEDGMENTS
We acknowledge Susan Williams and Marla Hill for experimental assistance.
The described project was supported by grant number AI050000 (to K.M.O.) from the National Institute of Allergy and Infectious Diseases (NIAID) at the National Institutes of Health.
This work is solely our responsibility and does not necessarily represent the official views of the NIH.
Footnotes
Published ahead of print 16 July 2012
REFERENCES
- 1. Akada JK. 2003. Helicobacter pylori tissue tropism: mouse-colonizing strains can target different gastric niches. Microbiology 149:1901–1909 [DOI] [PubMed] [Google Scholar]
- 2. Andermann TM, Chen Y-T, Ottemann KM. 2002. Two predicted chemoreceptors of Helicobacter pylori promote stomach infection. Infect. Immun. 70:5877–5881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Atherton JC. 2006. The pathogenesis of Helicobacter pylori-induced gastro-duodenal diseases. Annu. Rev. Pathol. 1:63–96 [DOI] [PubMed] [Google Scholar]
- 4. Baik S-C, Youn H-S, Chung M-H. 1996. Increased oxidative DNA damage in Helicobacter pylori-infected human gastric mucosa. Cancer Res. 56:1279–1282 [PubMed] [Google Scholar]
- 5. Beier D, Spohn G, Rappuoli R, Scarlato V. 1997. Identification and characterization of an operon of Helicobacter pylori that is involved in motility and stress adaptation. J. Bacteriol. 179:4676–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bijlsma JJE, et al. 2002. The Helicobacter pylori homologue of the ferric uptake regulator is involved in acid resistance. Infect. Immun. 70:606–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cerda O, Rivas A, Toledo H. 2003. Helicobacter pylori strain ATCC700392 encodes a methyl-accepting chemotaxis receptor protein (MCP) for arginine and sodium bicarbonate. FEMS Microbiol. Lett. 224:175–181 [DOI] [PubMed] [Google Scholar]
- 8. Cilluffo T, et al. 1990. Reproducibility of ambulatory gastric pH recordings in the corpus and antrum. Effect of food, time, and electrode position. Scand. J. Gastroenterol. 25:1076–1083 [DOI] [PubMed] [Google Scholar]
- 9. Corbin BD, et al. 2008. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science 319:962–965 [DOI] [PubMed] [Google Scholar]
- 10. Croxen MA, Sisson G, Melano R, Hoffman PS. 2006. The Helicobacter pylori chemotaxis receptor TlpB (HP0103) is required for pH taxis and for colonization of the gastric mucosa. J. Bacteriol. 188:2656–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eaton KA, Radin MJ, Krakowka S. 1995. An animal model of gastric ulcer due to bacterial gastritis in mice. Vet. Pathol. 32:489–497 [DOI] [PubMed] [Google Scholar]
- 12. Eaton KA, Danon SJ, Krakowka S, Weisbrode SE. 2007. A reproducible scoring system for quantification of histologic lesions of inflammatory disease in mouse gastric epithelium. Comp. Med. 57:57–65 [PubMed] [Google Scholar]
- 13. Ernst FD, et al. 2005. Transcriptional profiling of Helicobacter pylori Fur- and iron-regulated gene expression. Microbiology 151:533–546 [DOI] [PubMed] [Google Scholar]
- 14. Ernst FD, et al. 2005. Iron-responsive regulation of the Helicobacter pylori iron-cofactored superoxide dismutase SodB is mediated by Fur. J. Bacteriol. 187:3687–3692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Foynes S, et al. 2000. Helicobacter pylori possesses two CheY response regulators and a histidine kinase sensor, CheA, which are essential for chemotaxis and colonization of the gastric mucosa. Infect. Immun. 68:2016–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Freter R, Brickner H, Botney M, Cleven D, Aranki A. 1983. Mechanisms that control bacterial populations in continuous-flow culture models of mouse large intestinal flora. Infect. Immun. 39:676–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Freter R, Brickner H, Fekete J, Vickerman MM, Carey KE. 1983. Survival and implantation of Escherichia coli in the intestinal tract. Infect. Immun. 39:686–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Freter R, Stauffer E, Cleven D, Holdeman L, Moore R. 1983. Continuous-flow cultures as in vitro models of the ecology of large intestinal flora. Infect. Immun. 39:666–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Genta RM, Huberman RM, Graham DY. 1994. The gastric cardia in Helicobacter pylori infection. Hum. Pathol. 25:915–919 [DOI] [PubMed] [Google Scholar]
- 20. GoersSweeney E, et al. 2012. Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB. Structure 20:1177–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hansen O, Pedersen T, Larsen J. 1976. Cell proliferation kinetics in normal human gastric mucosa. Gastroenterology 70:1051–1054 [PubMed] [Google Scholar]
- 22. Harris A, et al. 2003. Catalase (KatA) and KatA-associated protein (KapA) are essential to persistent colonization in the Helicobacter pylori SS1 mouse model. Microbiology 149:665–672 [DOI] [PubMed] [Google Scholar]
- 23. Hidaka E, et al. 2001. Helicobacter pylori and two ultrastructurally distinct layers of gastric mucous cell mucins in the surface mucous gel layer. Gut 49:474–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kawakubo M, et al. 2004. Natural antibiotic function of a human gastric mucin against Helicobacter pylori infection. Science 305:1003–1006 [DOI] [PubMed] [Google Scholar]
- 25. Kehl-Fie TE, Skaar EP. 2010. Nutritional immunity beyond iron: a role for manganese and zinc. Curr. Opin. Chem. Biol. 14:218–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kuipers EJ. 1998. Review article: relationship between Helicobacter pylori, atrophic gastritis and gastric cancer. Aliment. Pharm. Ther. 12(Suppl 1):25–36 [DOI] [PubMed] [Google Scholar]
- 27. Lee A, et al. 1997. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology 112:1386–1397 [DOI] [PubMed] [Google Scholar]
- 28. Lee E, Trasler J, Dwivedi S, Leblond C. 1982. Division of the mouse gastric mucosa into zymogenic and mucous regions on the basis of gland features. Am. J. Anat. 164:187–207 [DOI] [PubMed] [Google Scholar]
- 29. Lertsethtakarn P, Ottemann KM, Hendrixson DR. 2011. Motility and chemotaxis in Campylobacter and Helicobacter. Annu. Rev. Microbiol. 65:389–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Licht TR, Christensen BB, Krogfelt KA, Molin S. 1999. Plasmid transfer in the animal intestine and other dynamic bacterial populations: the role of community structure and environment. Microbiology 145:2615–2622 [DOI] [PubMed] [Google Scholar]
- 31. McGee DJ, et al. 2005. Colonization and inflammation deficiencies in Mongolian gerbils infected by Helicobacter pylori chemotaxis mutants. Infect. Immun. 73:1820–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McLauchlan G, Fullarton GM, Crean GP, McColl KE. 1989. Comparison of gastric body and antral pH: a 24 hour ambulatory study in healthy volunteers. Gut 30:573–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Miles S, et al. 2010. Detailed in vivo analysis of the role of Helicobacter pylori Fur in colonization and disease. Infect. Immun. 78:3073–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Miller LD, Russell MH, Alexandre G. 2009. Diversity in bacterial chemotactic responses and niche adaptation. Adv. Appl. Microbiol. 66:53–75 [DOI] [PubMed] [Google Scholar]
- 35. Mills JC, et al. 2001. A molecular profile of the mouse gastric parietal cell with and without exposure to Helicobacter pylori. Proc. Natl. Acad. Sci. U. S. A. 98:13687–13692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mills JC, Shivdasani RA. 2011. Gastric epithelial stem cells. Gastroenterology 140:412–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pittman MS, Goodwin M, Kelly DJ. 2001. Chemotaxis in the human gastric pathogen Helicobacter pylori: different roles for CheW and the three CheV paralogues, and evidence for CheV2 phosphorylation. Microbiology 147:2493–2504 [DOI] [PubMed] [Google Scholar]
- 38. Polk DB, Peek RM. 2010. Helicobacter pylori: gastric cancer and beyond. Nat. Rev. Cancer 10:403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rader BA, et al. 2011. Helicobacter pylori perceives the quorum-sensing molecule AI-2 as a chemorepellent via the chemoreceptor TlpB. Microbiology 157:2445–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rieder G, Merchant JL, Haas R. 2005. Helicobacter pylori cag-type IV secretion system facilitates corpus colonization to induce precancerous conditions in Mongolian gerbils. Gastroenterology 128:1229–1242 [DOI] [PubMed] [Google Scholar]
- 41. Rolig AS, Carter JE, Ottemann KM. 2011. Bacterial chemotaxis modulates host cell apoptosis to establish a T-helper cell, type 17 (Th17)-dominant immune response in Helicobacter pylori infection. Proc. Natl. Acad. Sci. U. S. A. 108:19749–19754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rugge M, et al. 2002. Gastric mucosal atrophy: interobserver consistency using new criteria for classification and grading. Aliment. Pharm. Ther. 16:1249–1259 [DOI] [PubMed] [Google Scholar]
- 43. Schreiber S, et al. 2004. The spatial orientation of Helicobacter pylori in the gastric mucus. Proc. Natl. Acad. Sci. U. S. A. 101:5024–5029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schweinitzer T, et al. 2008. Functional characterization and mutagenesis of the proposed behavioral sensor TlpD of Helicobacter pylori. J. Bacteriol. 190:3244–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sipponen P, Hyvarinen H. 1993. Role of Helicobacter pylori in the pathogenesis of gastritis, peptic ulcer and gastric cancer. Scand. J. Gastroenterol. Suppl. 196:3–6 [DOI] [PubMed] [Google Scholar]
- 46. Symonds EL, Butler RN, Omari TI. 2000. Assessment of gastric emptying in the mouse using the [13C]-octanoic acid breath test. Clin. Exp. Pharmacol. Physiol. 27:671–675 [DOI] [PubMed] [Google Scholar]
- 47. Terry K, Williams SM, Connolly L, Ottemann KM. 2005. Chemotaxis plays multiple roles during Helicobacter pylori animal infection. Infect. Immun. 73:803–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Teshima S, Rokutan K, Nikawa T, Kishi K. 1998. Guinea pig gastric mucosal cells produce abundant superoxide anion through an NADPH oxidase-like system. Gastroenterology 115:1186–1196 [DOI] [PubMed] [Google Scholar]
- 49. Uemura N, et al. 2001. Helicobacter pylori infection and the development of gastric cancer. N. Engl. J. Med. 345:784–789 [DOI] [PubMed] [Google Scholar]
- 50. van Vliet AH, et al. 2002. The role of the ferric uptake regulator (Fur) in regulation of Helicobacter pylori iron uptake. Helicobacter 7:237–244 [DOI] [PubMed] [Google Scholar]
- 51. Verdú EF, et al. 1995. Effect of Helicobacter pylori status on intragastric pH during treatment with omeprazole. Gut 36:539–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wadhams GH, Armitage JP. 2004. Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell Biol. 5:1024–1037 [DOI] [PubMed] [Google Scholar]
- 53. Wang G, Alamuri P, Maier RJ. 2006. The diverse antioxidant systems of Helicobacter pylori. Mol. Microbiol. 61:847–860 [DOI] [PubMed] [Google Scholar]
- 54. Williams SM, et al. 2007. Helicobacter pylori chemotaxis modulates inflammation and bacterium-gastric epithelium interactions in infected mice. Infect. Immun. 75:3747–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Winter SE, et al. 2010. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467:426–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wroblewski LE, Peek RM, Wilson KT. 2010. Helicobacter pylori and gastric cancer: factors that modulate disease risk. Clin. Microbiol. Rev. 23:713–739 [DOI] [PMC free article] [PubMed] [Google Scholar]