Abstract
Extensive synonymous codon modification of viral genomes appears to be an effective way of attenuating strains for use as live vaccines. An assumption of this method is that codon changes have individually small effects, such that codon-attenuated viruses will be slow to evolve back to high fitness (and thus to high virulence). The major capsid gene of the bacterial virus T7 was modified to have varying levels of suboptimal synonymous codons in different constructs, and fitnesses declined linearly with the number of changes. Adaptation of the most extreme design, with 182 codon changes, resulted in a slow fitness recovery by standards of previous experimental evolution with this virus, although fitness effects of substitutions were higher than expected from the average effect of an engineered codon modification. Molecular evolution during recovery was modest, and changes evolved both within the modified gene and outside it. Some changes within the modified gene evolved in parallel across replicates, but with no obvious explanation. Overall, the study supports the premise that codon-modified viruses recover fitness slowly, although the evolution is substantially more rapid than expected from the design principle.
Keywords: codon modification, vaccine design, fitness suppression, genome engineering
Introduction
Live vaccines typically provide better immunity than killed or subunit vaccines. Yet, the development of a safe, live vaccine from its pathogenic progenitor is a balancing act, requiring at least four properties. (1) The pathogen must be “attenuated,” growing well enough to effect immunity but not so well that it causes disease. (2) The attenuation must have a minimal effect on antigenic profile, so that vaccine-induced immunity is also directed against the wild-type pathogen. (3) The attenuated pathogen must not revert or evolve back to wild-type virulence when released in the host population. (4) it must be feasible to manufacture large quantities of the vaccine strain.
The focus of our study is the third requirement—avoiding reversion of the attenuated strain. Reversion is an evolutionary process. The very fact of attenuation usually means that the vaccine strain does not grow as well as the disease-causing strain within the host. Consequently, when a live vaccine strain starts the infection process within the vaccine recipient, selection will favor variants that grow faster and, by virtue of faster growth, these variants will possibly manifest higher virulence. If the vaccine strain can infect contacts of the vaccinated individual, the potential exists for chains of transmission, with selection for higher virulence and transmissibility spanning dozens to 100s of pathogen generations. If the vaccine strain evolves to high virulence, it can then initiate its own epidemics, as has been observed with the attenuated Sabin polio vaccine (Kew et al. 2005).
A new method for attenuating viruses for vaccines was proposed recently: engineer the viral genome with a large number of codon changes that do not change the protein sequence (Burns et al. 2006; Mueller et al. 2006). By deoptimizing codons—replacing wild-type codons with codons and codon combinations whose sequences impair replication and/or expression—it should be possible to lower viral fitness arbitrarily, satisfying property (1) above. And as the codon changes do not alter the protein sequence, the antigenicity should not differ from the wild-type virus [satisfying property (2)]. Finally, it has also been suggested that property (3) will be satisfied: codon changes tend to have individually small fitness effects, so many nucleotide changes will be required to restore wild-type fitness, itself requiring 100s or more generations (Burns et al. 2006; Mueller et al. 2006; Coleman et al. 2008).
Since this method was proposed, a handful of studies have tested properties of codon-modified viruses (Burns et al. 2006, 2009; Mueller et al. 2006, 2010; Coleman et al. 2008). The findings have been at least qualitatively consistent with expectations, and our purpose here is to examine the evolutionary biology of reversion more quantitatively than before. The study with the most extensive analysis of evolutionary reversion observed somewhat parallel fitness gains between some codon-attenuated poliovirus strains and their prototype parent in vitro, as if the fitness increase of codon-modified strains was due entirely to adaptation to culture conditions and not to reversing codon bias (Burns et al. 2006). Because the parental virus had not been previously adapted to the growth conditions, it is difficult to bound the magnitude of fitness gain due to codon reversion per se. Our main question is whether evolutionary reversion obeys the expectation of a slow rate of fitness increase when the initial virus is otherwise well adapted to growth conditions. In using a double-stranded DNA virus whose genome is highly modular and whose expression is relatively simple, our model system should be especially favorable to the assumptions underlying the needs of vaccine design by codon deoptimization.
Methods
Media and Strains
LB broth (10 g NaCl, 10 g Bacto Tryptone, 5 g Bacto Yeast extract per liter) was used for growth and plating. Plates contained 15 g/l Bacto Agar; when measuring plaque-forming units, cells and phage were mixed in 2–3 ml soft agar (7 g/l Bacto Agar) before overlaying on a plate.
IJ1133 [Escherichia coli K-12, F−ΔlacX74 thiΔ (mcrC-mrr)102::Tn10] was the host for all adaptations and assays. Escherichia coli K-12 JW5808 (BW25113 B::Kan) (Baba et al. 2006) was the host used when recombining T7 on gene 10 inserts in plasmids; this strain maintains a low copy number of the plasmid, reducing selection against possible deleterious effects of the cloned gene.
An isolate of T761 was used as the genomic backbone for this study. The population of T761 was derived from T7+ by adaptation to IJ1133 during 20 h of serial transfer under the same conditions as used here; final fitness was 41.9 doublings per hour from a starting fitness of 35.6 (Heineman et al. 2005). The benefit of using T761 over T7+ is that T761 is well adapted to the growth conditions, so we could study the effect of codon modification on adaptation per se. Compared with T7+, the population of T761 carried a deletion of the early region (bases 1257–2736) as well as six high-frequency point mutations, based on consensus Sanger sequences. As an intermediate step toward engineering codon changes into gene 10A, we selected an isolate of T761 deleted for the entire major/minor capsid gene 10; this isolate carried both the early and gene 10 deletions and eight point mutations (gene 9: 22392 G, gene 15: 29770 A, gene 16: 30861 C, 30945 G, 32860 A, 33023 G, gene 17: 34975 G, gene 17.5: 36389 G). This isolate was then used for recombination over the plasmids carrying the modified gene 10A sequences (which also had unmodified 10B sequences).
The capsid gene of T7 is translated in two forms: the major capsid protein has 345 amino acids (AA), whereas the minor form has the same initial 342 AA of the major capsid and then a 56 AA C-terminal extension. We limited our codon modifications to the gene 10A coding sequence (supplementary file S1, Supplementary Material online), changing the wild-type sequence to have progressively fewer preferred codons by the criteria of Zhou et al. (2009). These criteria assign a codon to the preferred class if it is overused in highly expressed genes of E. coli. As T7 infects E. coli and relies entirely on its host for translation, it seems justified to choose codons based on host use. To reduce the possibility of large fitness effects of individual codon changes, we avoided changing the first and last 14 codons, as those regions often have important regulatory functions.
We calculated the fraction of preferred codons, Fpr, as the fraction of preferred codons among all sites at which a distinction between preferred and nonpreferred codons could be made. This definition excluded from the calculation sites coding for Met, Trp, and Lys. Fpr in the engineered versions of 10A were 0.5, 0.3, 0.2, and 0.1, relative to a value of 0.68 for the wild type. The designs of different constructs were nested, so that constructs with higher levels of Fpr had all the changes of constructs with lesser Fpr (supplementary file S1, Supplementary Material online). In this way, a phenotypic comparison between, say, forms with 60 versus 90 codon changes can be attributed to the 30 codons that differ between them, rather than to the entire set of codon changes. Each codon-modified design of 10A was synthesized commercially (GeneScript) with flanking regions matching T7 exactly outside of this gene (through 10B) and cloned into plasmids; the plasmids were maintained at low or moderate copy number to minimize toxic effects. The insert sequence was verified and then used for recombination into the T761 deleted for gene 10. Sequences of the engineered 10A in the initial recombinant T7 were verified completely (partially in one case); no deviations from the expected sequences were noted. Each complete engineered phage is denoted as T7-10Apref, where the value of subscript pref refers to the fraction of optimal codons remaining. Plasmid pAR 436 (Studier and Rosenberg 1981), which carries the wild-type T7 gene 10 and flanking sequences, was used in one experiment to recombine wild-type gene 10 sequences back into T7-10A0.1.
Growth Conditions and Fitness Assays
All adaptation of transgenic phages used serial transfer throughout or in the final steps of the adaptation. Aliquots of cells were prepared from a culture of exponentially growing cells, stored at −80 °C, and thawed shortly before use. For serial transfers and fitness assays, cells were suspended in 10 ml LB in a 125-ml flask and grown with aeration (200 revolutions per minute) at 37 °C for 60 min before phage addition; cell density at this point was typically 0.5–1.0 × 108 cells per milliliter. After an appropriate amount of growth in one flask, a sample was transferred directly to another flask whose cells had grown for 1 h. During adaptations, the minimum number of phage transferred was 105, and before transfer to the next flask, cultures were sometimes allowed to progress to confluent lysis (clearing of the culture). During adaptation, transfer times and volumes were adjusted so that the culture either did not clear before transfer or cleared about the time of transfer. After transfer, a sample from each culture was stored, and the final culture from 1 day was used to seed the first culture when transfers were initiated on a subsequent day.
We conducted four adaptations with the most extreme construct, T7-10A0.1. Two short-term adaptations, denoted S1 and S2, each consisted of 20 h of serial transfer. A longer adaptation was carried out (L1) first by growth in a continuous flow system for 265 h and then by serial transfer for 11–12 h. This continuous flow system used a cell-only tube that fed unidirectionally into a tube with phage (Bull et al. 1997; Wichman et al. 2005). Volumes and flow rates were adjusted to maintain phage densities in the range of 106–108. Both tubes were immersed in a water bath to maintain a temperature of 37 °C and were aerated with filtered air into the liquid. The system was replaced every 2 days by inoculating sterile tubes with fresh cells and introducing phage from the most recent sample. As chemostats can select phage traits other than rapid growth (Bull et al. 2006), the final sample of this continuous culture adaptation (after 265 h) was coinfected with the initial T7-10A0.1 to allow recombination, and the recombinant pool was subjected to 11–12 h of serial transfer in flasks. This final phase of serial transfer specifically selects changes enhancing growth in the serial transfer environment because the recombination enables each mutation to be selected for its individual effect (Rokyta et al. 2002).
The fourth adaptation began with a plaque of T7-10A0.1 on a host carrying plasmid pAR436—the wild-type gene 10. The initial plating would have allowed recombination between the phage and the plasmid. With T7, the frequency of recombinants in a plaque grown on such a plasmid is ∼5%. Because the initial phage contained multiple mutations in gene 10, recombination could replace all or part of 10A with the plasmid sequence. Although the initial pool would have been mostly nonrecombinants with a minority of heterogeneous recombinants, the ensuing serial transfer, which was carried out for 6 h, would have selected the most fit genotypes from the recombinant mix. This evolved, recombinant phage is denoted T7-10A0.1R+.
Fitness is the growth rate of a phage population during serial transfer, represented as the number of doublings per hour. Doublings per hour is proportional to the ecologists’ intrinsic rate of increase (r), but values of doublings per hour are more easily interpreted. This measure assumes exponential growth, so the fitness assays strictly maintained low ratios of phage to cells throughout to avoid limiting growth from exhaustion of cells. The general approach to making a fitness measure is to grow the phage population through a series of serial transfers across flasks and compare the titers at two times. As it has recently been shown that fitness measured under these conditions can be sensitive to the timing of those samples (Bull et al. 2011), we standardized our fitness assays across all phages and replicates so that transfers were 20 min apart, and fitness was calculated from samples taken at 60 and 120 min since phage were added to the first culture.
Analysis of 454 Sequences
Sequences of most initial isolates used Sanger technology; sequences of evolved populations used the “454” methodology as provided by the Genomic Sequencing and Analysis Facility at UT Austin. The mapping software for 454 data was breseq (Barrick et al. 2009; http://barricklab.org/breseq), with some additional code written in C to calculate mutation frequency as a function of quality score. Base change frequencies at a site were calculated simply as the number of reads with a mutation divided by the total number of reads at that site, subject to the base read meeting a specific quality. Short indels were ignored because the 454 technology is known to produce a high rate of such errors in homopolymeric tracts. Indels are not expected to be favored within gene 10, and indeed, indels in 10 have not been seen to evolve in our many other studies (Bull and Molineux 2008). Base positions use the wild-type genome sequence as a reference, even though our phage T761 and its descendants lacked over 1.5 kb of the wild-type genome (GenBank V01146, Dunn and Studier 1983).
Results
Constructs: Design and Fitness
The major capsid gene 10 of wild-type phage T7, gp10, is the most highly expressed of the entire genome; 415 copies of the capsid protein are required for every virion, and over 100,000 copies of the protein are made in as little as 8 min (Molineux 2005). Two forms of gp10 are found in wild-type particles: gp10A is a 345 AA protein that constitutes ∼96% of the capsid protein found in virions; the minor species gp10B results from a programmed −1 translational shift in reading frame at codon 342 that results in the synthesis of a 398 AA product. Infective virions can be made using only gp10A or gp10B, and the biological function of the C-terminal extension of gp10B is unknown.
Thus, we reasoned that gene 10A should be a good candidate for fitness reduction by codon deoptimization. Each codon was assigned to a set of “preferred” or “nonpreferred” codons according to codon use in highly expressed genes of E. coli (Zhou et al. 2009). This set was then applied to T7 on the grounds that its main host is considered to be E. coli, that the engineered phages would be adapted to E. coli, and that the phage relies entirely on its host for translation.
In support of the premise that the fraction of preferred codon use (Fpr) is important to the expression of gene 10A, the Fpr in the wild-type sequence of T7 10A (0.68) places it in the upper 4% of E. coli genes. Expression levels of these E. coli genes are above the median, and the genes with the highest Fpr are the most highly expressed overall (expression levels in Keller et al. 2012; Fpr calculations done for the present analysis). Four constructs were designed that reduced the fraction of preferred codons in gene 10A to 50%, 30%, 20%, and 10% from the wild-type value of 68% (supplementary file S1, Supplementary Material online). The first and last 14 codons of gene 10A were kept unchanged to minimize effects on translation initiation and termination. In the most extreme deoptimization, just over half of the 10A codons were modified. We denote the resulting phages as T7-10Apref, where the value of pref is the level of preferred codon use in 10A.
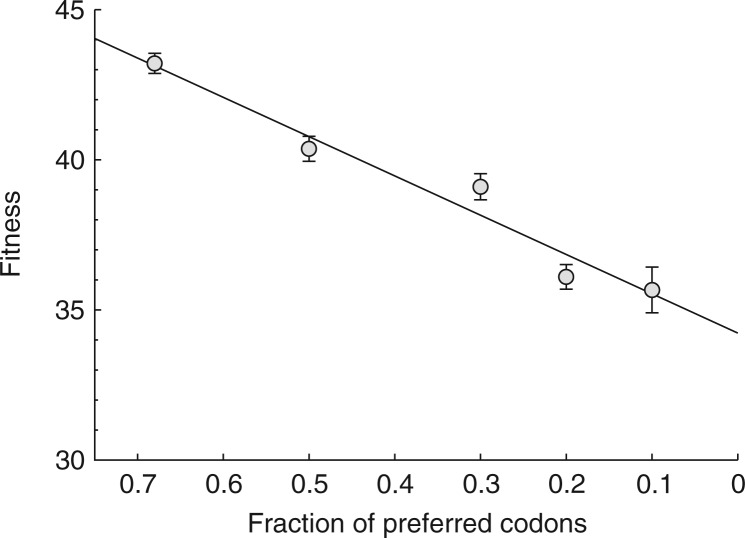
From the wild-type state of Fpr = 0.68, fitness declines linearly with the fraction of nonpreferred codons, although at least one of the points lies somewhat off the line (fig. 1). Linear declines in fitness components with codon deoptimization have also been observed in poliovirus (Burns et al. 2006). It is easily appreciated that the fit to linearity or to any other simple function provides an a priori predictability of how fitness will be reduced by a specific number of deoptimized codons. Indeed, most of the research on codon modification in poliovirus has been directed at identifying how to modify codons to achieve quantitative attenuation (Burns et al. 2006, 2009; Mueller et al. 2006; Coleman et al. 2008). A less obvious consequence of strict linearity is that it connotes equal (average) effects of different codon changes and the possibility that evolutionary reversion will be gradual.
FIG. 1.
Fitness declines approximately linearly with the level of codon deoptimization. The wild type is represented by the leftmost point, at 68% preferred codons in the major capsid gene, 10A. Four engineered constructs span 0.5–0.1 preferred codons in the gene. Fitness is measured in doublings per hour, a geometric measure (from left to right, mean fitnesses are 43.2, 40.4, 39.1, 36.1, and 35.7). The difference in fitness between the wild-type and most extreme engineered construct, T7-10A0.1, is 7.5.
Fitnesses of Adapted Viruses
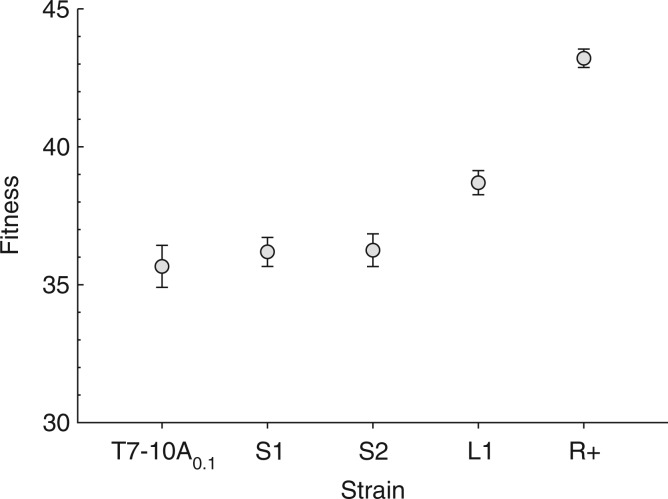
The most extreme construct, T7-10A0.1, was adapted to serial transfer in three independent replicates, one of them appreciably longer than the others (denoted L1 for the longer, S1 and S2 for the shorter ones). L1 began with 265 h of growth in a constant flow chemostat; the ending population was then recombined against the beginning population, and the recombinant pool was subjected to 11–12 h of serial transfer. This recombination-outgrowth phase selects those mutations evolved during continuous culture that are specifically beneficial in serial transfer (e.g., for comparison to S1 and S2), as it is known that phage propagation in chemostats can select a variety of phenotypes (Bull et al. 2006). As the lysis time of the adapted T7 is 13 min (Heineman et al. 2005), the total duration of L1 thus spanned 800–1,000 generations. Final fitness, measured on the population, was 38.7, an increase of three doublings per hour above that of its immediate ancestor, T710A0.1 (35.7, fig. 2); as will be noted below, none of the substitutions observed in this population were fixed, so the value of 38.7 is an average across genotypes. The two short-term adaptations, S1 and S2, were each carried out for 20 h (80–100 generations) of serial transfer. Neither the S1 nor S2 population exhibited a significant fitness increase over the ancestor; the observed increase was 0.5–0.6 doublings per hour in both lines.
FIG. 2.
Fitness of the initial T7-10A0.1 (leftmost, 35.7) and of four lines evolved from it (from S1 to R+, 36.2, 36.2, 38.7, and 43.2, respectively). The rightmost bar is for the phage T7-10A0.1R+ (labeled R+), created as a T7-10A0.1 recombined with a wild-type gene 10 plasmid and adapted for 6 h; it is used as the wild type in this paper and is also the leftmost point in figure 1.
To demonstrate the potential for rapid fitness recovery of T7-10A0.1 when many nucleotide reversions occur at once, the genetically modified virus was plated on a host with a plasmid containing the wild-type gene 10, creating a heterogeneous mix of the original phage plus partial and complete recombinants between the wild-type and the engineered gene 10. This pool was then adapted by serial transfer for 6 h. The fitness of the final population (T7-10A0.1R+) was considerably above that of the other adaptations (43.2) and was approximately the same value as observed for a T7 evolved specifically by serial transfer on the same host strain (Bull et al. 2011). Thus, we interpret this adaptation as having recovered pre-transgenic ancestral fitness; figures 1 and 2 use this evolved phage as the ancestral control. (Below we report that it attained the wild-type 10A sequence.)
Sequence Evolution
The initial T7-10A0.1 and the four populations evolved from it were subjected to 454 sequencing. The consensus sequence of gene 10A in the initial phage was identical to the design sequence, and the initial phage revealed no changes at frequencies of 10% or more. The consensus sequence of the evolved control phage T7-10A0.1R+ had fully reverted to the wild-type sequence, although a few residual design changes were present at or just under 10% frequency.
All adaptations had begun to accumulate nucleotide changes in the modified gene 10A. The two short adaptations (S1 and S2) had 3–4 changes each, but none reached even 50% frequency in the population of phages (table 1). The long-term adaptation (L1) had only two changes in 10A and was the only line to have changes in the minor capsid extension 10B. This portion of 10 had not been modified in the design. S2 and L1 also had changes outside of 10 that have no obvious biological import from the perspective of compensating for reduced expression of gene 10. Curiously, only four changes in these three adaptations were observed to reach or exceed 50% frequency; all were in L1, and two affected gene 10A.
Table 1.
Nucleotide Changes and Frequencies in Three Adapted Codon-Modified Lines.
| Line (frequencya) |
||||||
|---|---|---|---|---|---|---|
| Position | Base Change | AA Change | Gene (function) | S1 | S2 | L1 |
| 4303 | A →G | k 378 r | 1 (RNAP) | — | 0.115 | — |
| 3664 | A → C | n 165 t | 1 | — | — | 0.214 |
| 9158 | A → T | m 1 mb | 2.5 (ssDNA binding) | — | — | 0.389 |
| 14204 | G → A | g 93 d | 4.7 (DNA metabolism) | — | — | 0.196 |
| 14279 | T → C | i 118 t | 4.7 | — | — | 0.528 |
| 17766 | T → C | v 88 a | 6 (exonuclease) | — | 0.200 | — |
| 23129 | A → G | i 55 v | 10A (major capsid) | 0.438 | 0.114 | — |
| 23131 | A → C | i 55 i | 10A | 0.350 | 0.100 | 0.891 |
| 23425 | T → C | n 153 n | 10A | — | 0.188 | — |
| 23431 | T → C | n 155 n | 10A | — | 0.172 | — |
| 23434 | A → C | i 156 i | 10A | 0.111 | — | — |
| A → G | i 156 m | 10A | — | — | 0.500 | |
| 24088 | G → A | e 375 k | 10B (major capsid) | — | — | 0.720 |
| 24106 | G → A | a 361 t | 10B | — | — | 0.406 |
| 26984 | A → C | t 715 p | 12 (internal virion) | — | — | 0.429 |
| 36052 | A → C | s 477 r | 17 (tail fiber) | — | 0.158 | — |
| 38307 | T → C | l 313 s | 19 (terminase) | — | 0.125 | — |
NOTE.—A polymorphic, silent G → A change in gene 17 (base 35772, codon 383) was observed in all lines at a frequency of ∼0.4 relative to the wild-type base. This change was even observed in the initial T710A0.1 which was recently derived from a clonal isolate. We suspect that this change is a sequencing artefact and omit it from the table and our discussion of evolution in these lines.
To be reported here, the frequency of a nucleotide change had to reach 0.1 and be observed in at least two reads.
Change to a UUG start codon.
Across all three adaptations, three of the changes in 10A were silent reversions to the wild-type sequence, and one of those (base 23131, codon 55) was found in all three lines (tables 1 and 2). Parallel evolution was also evident at two other positions in 10A, although the nucleotide changes at 23434 were not the same in the two lines with evolution at this site. It is also noteworthy that all changes in 10A were confined either to codon 55 or codons 153–156.
Table 2.
Gene 10A Codons Experiencing Evolution.
| Codon Number | Designed Genome Codon | Wild-Type Codon | Evolved Codons |
|---|---|---|---|
| 55 | AUA | AUC | GUA, AUC |
| 153 | AAU | AAU | AAC |
| 155 | AAU | AAC | AAC |
| 156 | AUA | AUC | AUC, AUG |
Fitness Effects of Changes
With 182 codon changes, the fitness of T7-10A0.1 is 7.5 doublings per hour lower than that of wild type (180-fold fewer descendants per hour, hence only about 3-fold less per generation). The average effect per single nucleotide change is thus 0.04 doublings per hour. We could interpret this small value as supporting the premise motivating the use of codon modification for viral attenuation. However, a more important issue is whether the changes arising during evolutionary reversion of the engineered viruses also have small effects. The combined sequence results and fitnesses allow a qualitative determination of the latter quantity.
The summation of frequency changes in 10A for lines S1, S2, and L1 are 0.9, 0.57, and 1.4, respectively, and the summation of all genomic changes for those lines is 0.9, 1.2, and 4.3. Thus, the S1 and S2 adaptations resulted in less than one 10A substitution per genome on average and only slightly more than 1 in the much longer adaptation of L1. Estimated fitnesses of S1 and S2 are only slightly above that of T7-10A0.1—0.5–0.6 doublings per hour, the standard errors being compatible with no fitness difference up to a fitness increase of 2.7. The mean fitness of L1 was 3.2 doublings per hour above that of T7-10A0.1 (and possibly as high as 5.2). The higher fitness in L1 is not specifically attributable to changes in 10A, of course, because it accumulated several changes outside of the engineered gene, and those changes are unique to L1 and account for the majority of frequency change in this line. Only S1 experienced evolution limited to 10A, such that any fitness gain could be attributed to evolution within the transgene, and the fact that its fitness is not obviously higher than that of S2, despite the latter experiencing similar amounts of sequence evolution outside 10 as inside, is suggestive that the 10A changes are not contributing much. Thus, the fitness effects of evolution in the codon-modified gene after 20 h of adaptation (80–100 generations) appear to be less than 1 doubling per hour.
We can approach this question another way, asking how rapidly a mutation with a specific fitness effect reaches a particular frequency. Writing xt as the abundance of a genome with a particular change after t hours of exponential growth at a rate of Dx doublings per hour, we have:
| (1) |
Letting the frequency of the alternative genomic state be yt, we may calculate the relative change after t hours as:
| (2) |
This equation may be used to calculate the number of hours of transfer that will be required for a rare mutation with an arbitrary growth advantage to reach any ratio of abundance.
The relative gain of a rare mutation depends on the starting frequency, on its advantage, and on the time when finally sampled. For S1 and S2, the duration is 20 h and the starting frequency probably lies between 10−5 and 10−6. For these parameter values, the benefit of a mutation that reaches a frequency of 0.1–0.5 lies approximately in the range of 0.4–0.6 doublings hour. If the observed 10A mutations were this large and evolved to the observed frequencies, the predicted fitnesses would be similar to the observed fitness (the fitness—growth—of a polymorphic population is the sum of the growths of the different genotypes weighted by their initial frequencies). Thus, the S1 and S2 data are broadly compatible with fitnesses of ∼0.5 doublings per hour for the individual effects of the changes evolving in 10A.
This fitness effect is small on the scale of the large effect changes typically observed in experimental evolution of T7 (Bull and Molineux 2008). At the same time, an effect of 0.5–1.0 doublings per hour is 10- to 20-fold the average effect of a codon modification calculated for this system. Perhaps more telling, the long-term line L1 recovered 40% of the lost fitness (3 doublings per hour) with an average of 4.3 changes genome-wide across nine substitutions; fitness might thus be even higher in a genotype fixed for those mutations. Thus, the evolution proceeded much faster than if the fitness benefit of an adaptive change was the average effect of a codon reversion. It is of course plausible that these first adaptive changes would be the largest if adaptation was continued—that fitness would continue to improve but more slowly than observed in these early stages. It is also possible that fitness would plateau below the wild-type value.
Discussion
The magnitudes of fitness increase and the magnitudes of molecular evolution in this work are small by standards of other experimental evolution studies of T7 (Bull and Molineux 2008). At face value, this outcome supports the premise of codon modifications—that evolutionary reversion will be slow, whether of individual nucleotide changes or overall fitness. Yet, although this outcome is undeniable for this study, it need not generalize to other implementations of codon deoptimization, especially those in which fitness is depressed profoundly. Furthermore, secondary structure disruptions of the mRNA caused by codon changes may have a separate and large impact on fitness in translatability of an mRNA (Kudla et al. 2009; Gu et al. 2010; Zhou and Wilke 2011) or on replication and packaging of RNA viruses (Burns et al. 2009), and those effects can enable rapid fitness gains.
The fitness of T7 was initially reduced by 7.5 doublings per hour from our most extreme codon deoptimization, but this reduced fitness was 35 doublings per hour, which is still high. The absence of high-fitness reversions might then be due largely to the fact that the engineered fitness reduction was relatively small on a scale of total fitness; hence, the opportunities for large effect mutations were limited. However, the potential gain in fitness here is similar to that seen when adapting wild-type T7 to laboratory conditions. Specifically, Heineman et al. (2005) adapted wild-type T7 to the conditions used here, also for 20 h; initial fitness was 35.6, final fitness was 41.9, and 6 mutations were detected in the final consensus sequence. The opportunity for fitness improvement was virtually the same, but the fitness gain and sequence evolution was far more substantial than observed in the present study.
We thus cautiously suggest that an absence of high-frequency substitutions in lines adapted for 20 h is unusual and supports the premise that synonymous codon changes tend to have individually small effects by standards of T7 experimental evolution. Furthermore, the absence of fixed changes in the long-term line is also unusual and supports the view that evolution in response to codon modification may not be rapid even when the changes lie outside the modified portion of the genome. We emphasize, however, that our pre-engineered virus was well adapted to the growth conditions, and there may be rapid evolution when using a virus that, aside from codon changes, is also poorly adapted to the specific experimental growth conditions.
The high degree of parallel evolution despite the relatively small fitness effects further supports the thesis that adaptation of codon-attenuated viruses is slow. As there is no reason to suggest strong mutational biases being the cause of parallel evolution, we interpret the parallel evolution as evincing an absence of other mutations whose fitness effects are of equal or superior benefit.
The lack of fixed changes in the long-term line (L1) may seem unusual, given that the adaptation spanned 1,000 generations. However, most of the adaptation was carried out in a chemostat, and the chemostat population—which may indeed have carried fixed mutations—was subjected to a roughly 50% dilution and recombination with the starting phage and then serial transfer for 11–12 h before the population sequence was assayed. Any mutation fixed prior to this recombination step thus had 11–12 h to reach fixation again. Furthermore, any mutations beneficial in the chemostat but deleterious to serial transfer, which would have been selected against during this final phase, could have depressed the frequencies of beneficial mutations in linkage disequilibrium. Thus, we can merely conclude that any mutations observed in L1 were beneficial at some phase during the adaptation, and the absence of fixed mutations in L1 supports the view that none of the observed mutations were highly beneficial in serial transfer.
At least in poliovirus, there is debate about the specific mechanism by which codon changes reduce fitness. For example, one proposal is that suboptimal codon pairs are responsible (Coleman et al. 2008); another is that specific dinucleotides are responsible (Burns et al. 2006, 2009). Of course, any single design criterion will often overlap others. Nonetheless, the specific mechanism of inhibition should be reflected in the molecular bases of fitness recovery. In poliovirus, fitness recovery was associated with loss of several CpG dinucleotides, supporting their importance as the basis of fitness suppression (Burns et al. 2006)—CpG dinucleotides are specifically detrimental in eukaryotes as targets of methylation.
In our study, there were too few changes to support any strong inference about the basis of attenuation. Compensatory changes within the affected gene were confined to two small regions, perhaps reflecting an important regulatory role of those regions, but we have no basis for assigning importance to those regions (RNA folding algorithms do not suggest strong secondary structure of those regions). We did not observe any CpG losses in the codon-modified gene, but two of the adaptive changes listed in table 2 were losses of UpA dinucleotides (codons 156 and 155, the latter of which is followed by an A). UpA dinucleotides are potentially problematic in bacteria (Burns et al. 2009 and references therein), so this type of change is one to monitor in future studies of codon-modified bacteria and their viruses. We note, however, that codons 151–156 are unusually AU rich, even before deoptimization (151-AAA TAT AAT GAG AAC ATC-156).
Much of the molecular evolution here occurred outside the modified part of the genome, more so for the longest adaptation (L1). The two obvious explanations are (1) this evolution was not specific to the codon modifications and would have occurred in its absence or (2) this evolution was specific to the codon modifications. In previous work, we have used a specific recombination-based test to discriminate these alternative models, but that approach only works when the evolutionary changes are fixed in the adapted populations; here all the changes were at low to moderate frequencies. By using a genome that was well adapted to growth conditions, we have likely reduced the extent of changes under model (1). However, at least one of the changes observed in L1 (the 10B mutation at 24088) is likely not specific to the engineering, as it was observed in the original T761 population but was absent in our T7-10A0.1 and thus re-evolved in L1 (Heineman et al. 2005).
Model (2) is at least a plausible explanation for the compensatory changes outside of gene 10A. If fitness suppression by codon deoptimization results from reduced gene expression, then fitness may be partially recovered by large effect changes in other genes, accommodating the imbalance in abundances of different proteins. For example, deletion of the T7 ligase gene is lethal on a ligase-defective host, but compensatory evolution in other DNA metabolism genes (as well as in other genes) provides a remarkable fitness recovery (Rokyta et al. 2002; Harcombe et al. 2009); importantly, this fitness recovery does not (and cannot) involve restoration of normal ligase levels. Here, therefore, if the codon changes in 10A lower gp10A expression, changes in other genes might accommodate that lower expression and lead to higher fitness without any change in the codon-modified gene itself. It should thus be appreciated that the concentration of codon changes within the genome may have ramifications to evolutionary reversion. A design for codon deoptimization that avoids this problem is to distribute codon changes across multiple genes.
A corollary of model (2) is that codon superoptimization of a gene will also be deleterious. Thus, it is the balance of gene expression that underlies high fitness, and deviations in either direction will be deleterious. In principle, this prediction is easy to test, but any recoding of a gene to have high levels of preferred codon use runs the risk of disrupting some unknown property of the DNA or RNA that is deleterious for other reasons.
These adaptations are short term by evolutionary standards, and they leave open the question of whether continued growth under the same conditions would result in continued viral fitness improvement. From the perspective of vaccine development, continued improvement might lead to a new virulent virus. However, the potential for continued viral evolution need not pose a problem for vaccine use if the rate of fitness improvement is slow. If the vaccine strain is attenuated enough that anyone infected with it transmits to less than one new host on average (R0 < 1), the virus may invariably die out before it evolves a transmission rate high enough to be sustained (Antia et al. 2003).
Supplementary Material
Supplementary file S1 is available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank R. Springman and M. Schmerer for sequencing the initial recombinant phages; comments from two reviewers improved the manuscript. This work was supported by National Institute of Health GM 57756 and the Miescher Regents Professorship (to J.J.B.) and National Science Foundation Grant EF-0742373, National Science Foundation Cooperative Agreement No. DBI-0939454, and National Institute of Health R01 GM088344 (to C.O.W.).
References
- Antia R, Regoes RR, Koella JC, Bergstrom CT. The role of evolution in the emergence of infectious diseases. Nature. 2003;426:658–661. doi: 10.1038/nature02104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, Schneider D, Lenski RE, Kim JF. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461:1243–1247. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- Bull JJ, Badgett MR, Wichman HA, Huelsenbeck JP, Hillis DM, Gulati A, Ho C, Molineux IJ. Exceptional convergent evolution in a virus. Genetics. 1997;147:1497–1507. doi: 10.1093/genetics/147.4.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull JJ, Heineman RH, Wilke CO. The phenotype fitness map in phage experimental evolution. PLoS One. 2011;6:e27796. doi: 10.1371/journal.pone.0027796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull JJ, Millstein J, Orcutt J, Wichman HA. Evolutionary feedback mediated through population density, illustrated with viruses in chemostats. Am Nat. 2006;167:E39–E51. doi: 10.1086/499374. [DOI] [PubMed] [Google Scholar]
- Bull JJ, Molineux IJ. Predicting evolution from genomics: experimental evolution of bacteriophage T7. Heredity. 2008;100:453–463. doi: 10.1038/sj.hdy.6801087. [DOI] [PubMed] [Google Scholar]
- Burns CC, Campagnoli R, Shaw J, Vincent A, Jorba J, Kew O. Genetic inactivation of poliovirus infectivity by increasing the frequencies of CpG and UpA dinucleotides within and across synonymous capsid region codons. J Virol. 2009;83:9957–9969. doi: 10.1128/JVI.00508-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns CC, Shaw J, Campagnoli R, Jorba J, Vincent A, Quay J, Kew O. Modulation of poliovirus replicative fitness in HeLa cells by deoptimization of synonymous codon usage in the capsid region. J Virol. 2006;80:3259–3272. doi: 10.1128/JVI.80.7.3259-3272.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JR, Papamichail D, Skiena S, Futcher B, Wimmer E, Mueller S. Virus attenuation by genome-scale changes in codon pair bias. Science. 2008;320:1784–1787. doi: 10.1126/science.1155761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn JJ, Studier FW. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983;166:477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Gu W, Zhou T, Wilke CO. A universal trend of reduced mRNA stability near the translation-initiation site in prokaryotes and eukaryotes. PLoS Comput Biol. 2010;6:e1000664. doi: 10.1371/journal.pcbi.1000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcombe WR, Springman R, Bull JJ. Compensatory evolution for a gene deletion is not limited to its immediate functional network. BMC Evol Biol. 2009;9:106. doi: 10.1186/1471-2148-9-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heineman RH, Molineux IJ, Bull JJ. Evolutionary robustness of an optimal phenotype: re-evolution of lysis in a bacteriophage deleted for its lysin gene. J Mol Evol. 2005;61:181–191. doi: 10.1007/s00239-004-0304-4. [DOI] [PubMed] [Google Scholar]
- Keller TE, Mis SD, Jia KE, Wilke CO. Reduced mRNA secondary-structure stability near the start codon indicates functional genes in prokaryotes. Genome Biol Evol. 2012;4:80–88. doi: 10.1093/gbe/evr129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kew OM, Sutter RW, de Gourville EM, Dowdle WR, Pallansch MA. Vaccine-derived polioviruses and the endgame strategy for global polio eradication. Annu Rev Microbiol. 2005;59:587–635. doi: 10.1146/annurev.micro.58.030603.123625. [DOI] [PubMed] [Google Scholar]
- Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molineux IJ. T7 group. In: Calendar R, editor. The bacteriophages. Oxford: Oxford University Press; 2005. pp. 277–301. [Google Scholar]
- Mueller S, Coleman JR, Papamichail D, Ward CB, Nimnual A, Futcher B, Skiena S, Wimmer E. Live attenuated influenza virus vaccines by computer-aided rational design. Nat Biotechnol. 2010;28:723–726. doi: 10.1038/nbt.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S, Papamichail D, Coleman JR, Skiena S, Wimmer E. Reduction of the rate of poliovirus protein synthesis through large-scale codon deoptimization causes attenuation of viral virulence by lowering specific infectivity. J Virol. 2006;80:9687–9696. doi: 10.1128/JVI.00738-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokyta D, Badgett MR, Molineux IJ, Bull JJ. Experimental genomic evolution: extensive compensation for loss of DNA ligase activity in a virus. Mol Biol Evol. 2002;19:230–238. doi: 10.1093/oxfordjournals.molbev.a004076. [DOI] [PubMed] [Google Scholar]
- Studier FW, Rosenberg AH. Genetic and physical mapping of the late region of bacteriophage T7 DNA by use of cloned fragments of T7 DNA. J Mol Biol. 1981;153:503–525. doi: 10.1016/0022-2836(81)90405-8. [DOI] [PubMed] [Google Scholar]
- Wichman HA, Millstein J, Bull JJ. Adaptive molecular evolution for 13,000 phage generations: a possible arms race. Genetics. 2005;170:19–31. doi: 10.1534/genetics.104.034488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Weems M, Wilke CO. Translationally optimal codons associate with structurally sensitive sites in proteins. Mol Biol Evol. 2009;26:1571–1580. doi: 10.1093/molbev/msp070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Wilke CO. Reduced stability of mRNA secondary structure near the translation-initiation site in dsDNA viruses. BMC Evol Biol. 2011;11:59. doi: 10.1186/1471-2148-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.