Abstract
Candida albicans is the most frequently encountered human fungal pathogen, causing both superficial infections and life-threatening systemic diseases. Functional genomic studies performed in this organism have mainly used knock-out mutants and extensive collections of overexpression mutants are still lacking. Here, we report the development of a first generation C. albicans ORFeome, the improvement of overexpression systems and the construction of two new libraries of C. albicans strains overexpressing genes for components of signaling networks, in particular protein kinases, protein phosphatases and transcription factors. As a proof of concept, we screened these collections for genes whose overexpression impacts morphogenesis or growth rates in C. albicans. Our screens identified genes previously described for their role in these biological processes, demonstrating the functionality of our strategy, as well as genes that have not been previously associated to these processes. This article emphasizes the potential of systematic overexpression strategies to improve our knowledge of regulatory networks in C. albicans. The C. albicans plasmid and strain collections described here are available at the Fungal Genetics Stock Center. Their extension to a genome-wide scale will represent important resources for the C. albicans community.
Introduction
Candida albicans is a normal member of human natural cavities, especially of the gastrointestinal and urogenital tracts [1], [2]. In addition to its commensal activity and under specific conditions, this yeast becomes one of the major invasive fungal pathogen of humans, and can cause both mucosal and life-threatening disseminated infections [3], [4]. The significant mortality rate associated with candidiasis in immunocompromised patients drives the research efforts to improve our knowledge of C. albicans biology and pathogenesis [5].
During the last decade, progresses in gene inactivation methodologies have been the driving force to characterize C. albicans molecular processes [6], [7]. Several collections of heterozygous and homozygous knock-out (KO) mutants have been generated. These resources are now invaluable to study C. albicans regulatory networks, virulence, hyphal morphogenesis, biofilm formation, identify drug targets and evaluate the mode-of-action of antifungal compounds [8]–[20]. However, the use of KO mutants shows some limitations. First, since C. albicans is an obligate diploid organism with no known meiotic cycle, two rounds of gene disruption are required to produce each deletion mutant. The pioneering works for the construction of homozygous KO mutants in the C. albicans genome mentioned above were indeed tedious and time-consuming. Second, gene deletion approaches are not optimal in the case of functional redundancy or essential genes. To date, less than 25% of C. albicans genes have been functionally characterized, indicating that new approaches must be developed for the study of this pathogen.
The use of both systematic KO and overexpression (OE) approaches to investigate cellular processes has proven highly successful in the model yeast Saccharomyces cerevisiae. Genome-wide collections of S. cerevisiae OE strains have been assembled and used to perform large-scale functional analyses, leading to the identification of new signalling pathways, new targets and functions for transcription factors or protein kinases and, more largely, to improve our image of the functional landscape of the cell [21]–[27]. In contrast, OE strategies have not been exploited extensively in C. albicans. Fu et al. [28] established a collection of 26 heterozygous OE strains for genes encoding glycophosphatidylinositol-anchored (GPI) proteins. This study demonstrated the role in adherence for the product of the IFF4 gene, one of 11 members of the IFF genes family. More recently, Sahni et al. [29] constructed an OE library of 103 transcription factors which has been used in two independent screens, demonstrating a role for the Tec1 transcription factor in the response of white cells to pheromones [29] and identifying a critical function for the Brg1 transcription factor in C. albicans biofilm formation, filamentous growth and virulence [30]. Other studies have used OE of selected genes in order to test the relevance of regulatory networks and functional pathways inferred from gene expression studies [31]–[34]. Nevertheless, the collections of OE strains yet available are focused and despite the encouraging results obtained in the studies mentioned above, a flexible collection of OE plasmids encompassing the 6200 C. albicans genes and the corresponding C. albicans OE strains are still lacking.
Here, we constructed two collections of C. albicans OE strains, enriched for genes encoding protein kinases, protein phosphatases, transcription factors and other signalling proteins. These new resources took advantage of the highly efficient Gateway® technology to provide versatility to our system, leading to a first generation C. albicans ORFeome. To test the functionality and applications of our strategy, we performed screens for regulators of morphogenesis and growth rate in C. albicans, two research areas of crucial interest for the development of new antifungal strategies. Our results highlight the value of using gene OE as a complement to gene inactivation to both uncover gene function and reveal new regulators in C. albicans. They also pave the way for the development of genome-wide OE approaches for this major pathogen.
Results and Discussion
Development of Gateway Vectors for Overexpression in Candida albicans
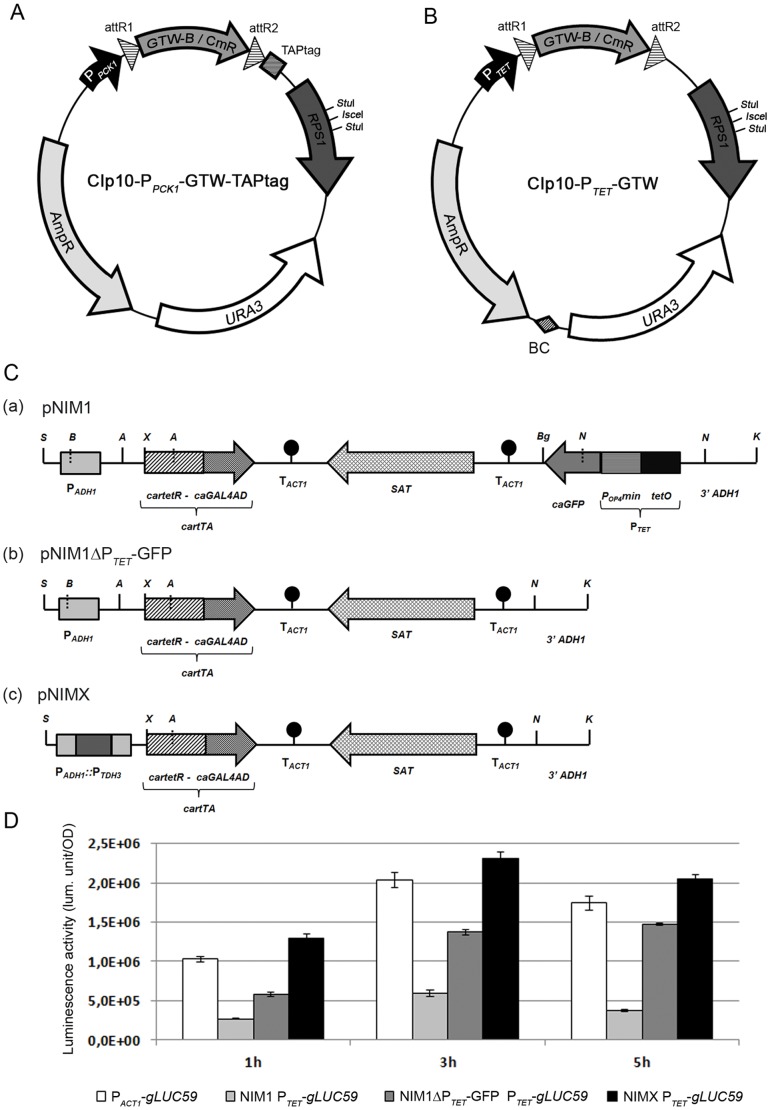
In order to develop a collection of C. albicans OE strains, we have taken advantage of the Gateway® methodology that enables recombination-mediated cloning of PCR-amplified ORFs into a donor vector and their subsequent recombination-mediated transfer into a variety of customized destination vectors [35]. We developed two conditional OE destination vectors named CIp10-PPCK1-GTW-TAPtag and CIp10-PTET-GTW (Fig. 1A and 1B respectively), both being derivatives of the C. albicans CIp10 integrative vector [36]. CIp10-PPCK1-GTW-TAPtag carries a Gateway® cassette flanked by the gluconeogenesis-induced C. albicans PCK1 promoter (PPCK1; [37]) and an in-frame sequence encoding a tag for tandem-affinity purification (TAPtag; Fig. 1B; [38]). Expression from PPCK1 is achieved in the presence of casamino acids and repressed in the presence of glucose. CIp10-PTET-GTW contains the TET promoter (PTET; [39]) that is activated in the presence of tetracycline derivatives. In contrast to CIp10-PPCK1-GTW-TAPtag, CIp10-PTET-GTW is not equipped with a TAPtag but with a unique barcode system (Fig. 1B and Materials and Methods for the barcoding procedure).
Figure 1. Gateway-adapted OE systems for C. albicans. Schematic maps of the CIp10-PPCK1-GTW-TAPtag (A) and CIp10-PTET-GTW (B) vectors.
The presence of attR recombination sites allows Gateway®-mediated cloning of ORFs in place of the GTW-B/CmR cassette. ORFs are expressed from the PCK1 promoter (PPCK1 - A) or the TET promoter (PTET - B) that are induced in gluconeogenic growth conditions or in the presence of tetracycline derivatives (doxycycline, anhydrotetracycline), respectively. In the first case, ORFs are fused to a TAPtag coding region, thus allowing production of proteins TAPtagged at their C-terminus. In the second case, each ORF is associated to a unique barcode (BC). Derivatives of CIp10-PPCK1-GTW-TAPtag and CIp10-PTET-GTW can be targeted to the C. albicans RPS1 locus when linearized with StuI or I-SceI and C. albicans transformants are selected for uridine prototrophy conferred by the URA3 gene. C. Schematic maps of the different transactivation cassettes used to promote expression from the TET promoter. The pNIMX cassette (c) is a derivative of pNIM1 (a; [39]). pNIMX was generated by deleting the PTET-GFP fusion in pNIM1, yielding pNIM1ΔPTET-GFP (b), and subsequently exchanging the PADH1 promoter upstream of the cartTA region by the TDH3 promoter (PTDH3). Relevant restriction sites are shown: A: AclI, B: BamHI, Bg: BglII, K: KpnI, N: NcoI, S: SacII, X: XbaI. D. The pNIMX transactivator cassette provides enhanced P TET -driven OE. C. albicans strains harbouring the CIp10-PTET-gLUC59 plasmid, with the gLUC59 luciferase reporter gene under the control of PTET, and either pNIM1, pNIM1ΔPTET-GFP or pNIMX (CEC1909, CEC2249 or CEC3083 respectively) were grown in YPD liquid medium supplemented with 50 µg.mL− 1 Dox. A C. albicans strain harbouring CIp10-PACT1-gLUC59 plasmid (CEC988) and expressing the gLUC59 reporter gene constitutively was used as a control and grown in the same conditions. Data represent luciferase specific activity detected from the different strains after 1, 5 and 8 h of growth in the presence of Dox. Assays were performed in duplicate and means and SD are shown.
Optimisation of Tetracycline-dependent Overexpression in Candida albicans
Expression from the PTET promoter requires a C. albicans-adapted reverse Tet-dependent transactivator (cartTA) that binds the tetO sequences in the PTET promoter in a tetracycline-dependent manner and drives transcription through the activation domain of the Gal4 protein of S. cerevisiae [39]. Different plasmids allowing expression of cartTA in C. albicans are available among which pNIM1 whereby cartTA is expressed from the promoter of the ADH1 gene and that harbours a PTET-GFP fusion (Fig. 1C–a; [39]). We reasoned that tetracycline-dependent OE of genes cloned downstream of PTET on CIp10-PTET-GTW plasmids might be enhanced by removing the PTET-GFP fusion from pNIM1 and expressing cartTA from a stronger promoter than that of the ADH1 gene. Therefore we produced two derivatives of the pNIM1 plasmid: pNIM1ΔPTET-GFP (Fig. 1C–b) lacks the PTET-GFP fusion; pNIMX (Fig. 1C–c) lacks this fusion and carries the cartTA coding region placed under the control of the strong and constitutive C. albicans TDH3 promoter (PTDH3; [40]. In order to test the relative efficiency of the pNIM1, pNIM1ΔPTET-GFP and pNIMX plamsids plasmids at driving OE from the PTET promoter, these plasmids were introduced in a C. albicans strain that harboured a fusion between PTET and the gLUC59 luciferase reporter gene [41]. Results presented in Fig. 1D showed that luciferase levels achieved from the strains harbouring pNIM1ΔPTET-GFP or pNIMX were respectively 3 or 5 times higher than those obtained in a C. albicans strain harbouring pNIM1. Noticeably, luciferase levels achieved from the strain transformed with pNIMX and the PTET-gLUC59 fusion were above those observed in a C. albicans strain harbouring a PACT1-gLUC59 fusion (Fig. 1D). Thus, C. albicans strains harbouring pNIMX were subsequently used to drive expression from the PTET promoter.
Validation of the OE-Gateway Vectors Developed for Candida albicans and Quantification of the OE Level
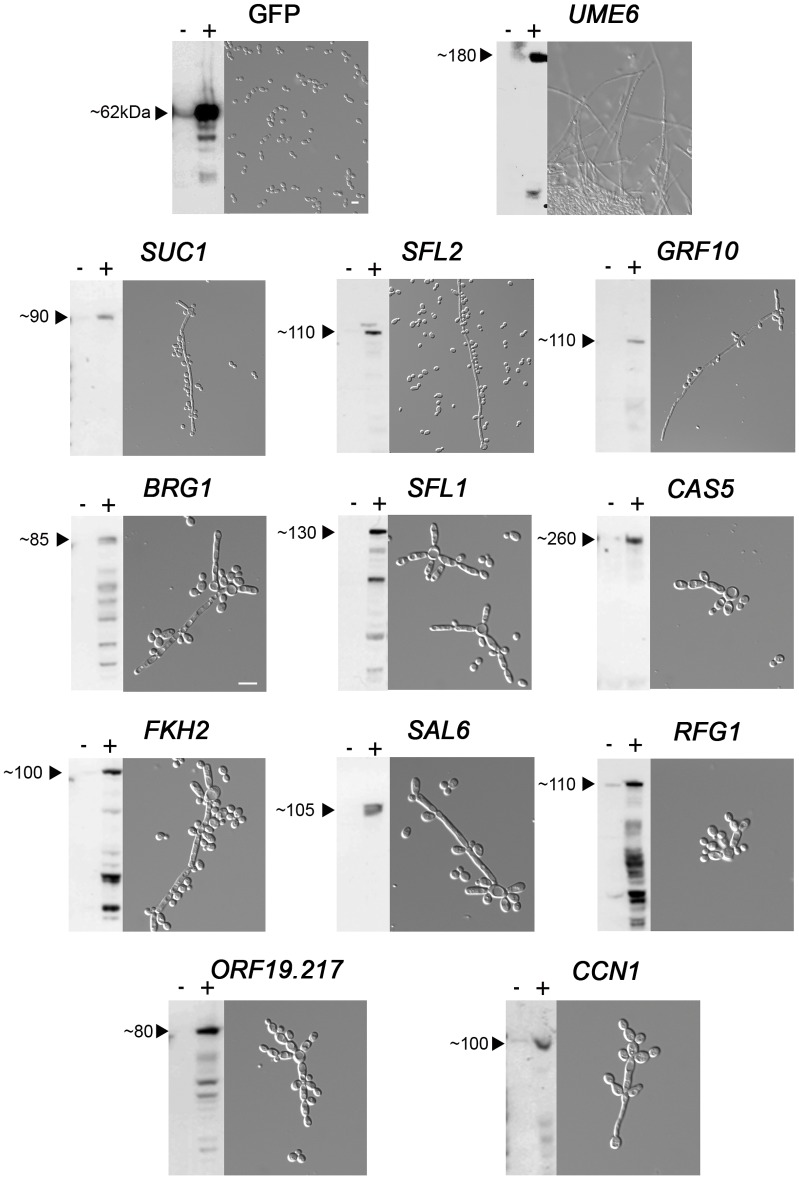
We verified that Gateway®-cloning of ORFs into both plasmids allowed efficient OE of proteins by transferring the GFP [42] and UME6 ORFs into these vectors. UME6 was selected as its OE has been shown to trigger hyphal formation [43], [44]. As shown in Fig. 2A, OE of UME6 resulted in the formation of hypha in conditions that do not normally trigger C. albicans morphogenesis. Production of TAP-tagged GFP and Ume6 proteins was also observed in strains harbouring derivatives of CIp10-PPCK1-GTW-TAPtag and grown under gluconeogenic conditions (Fig. 2B).
Figure 2. Functionality of the Gateway® OE systems. A. PPCK1-driven and PTET-driven OE of UME6 but not GFP triggers morphogenesis.
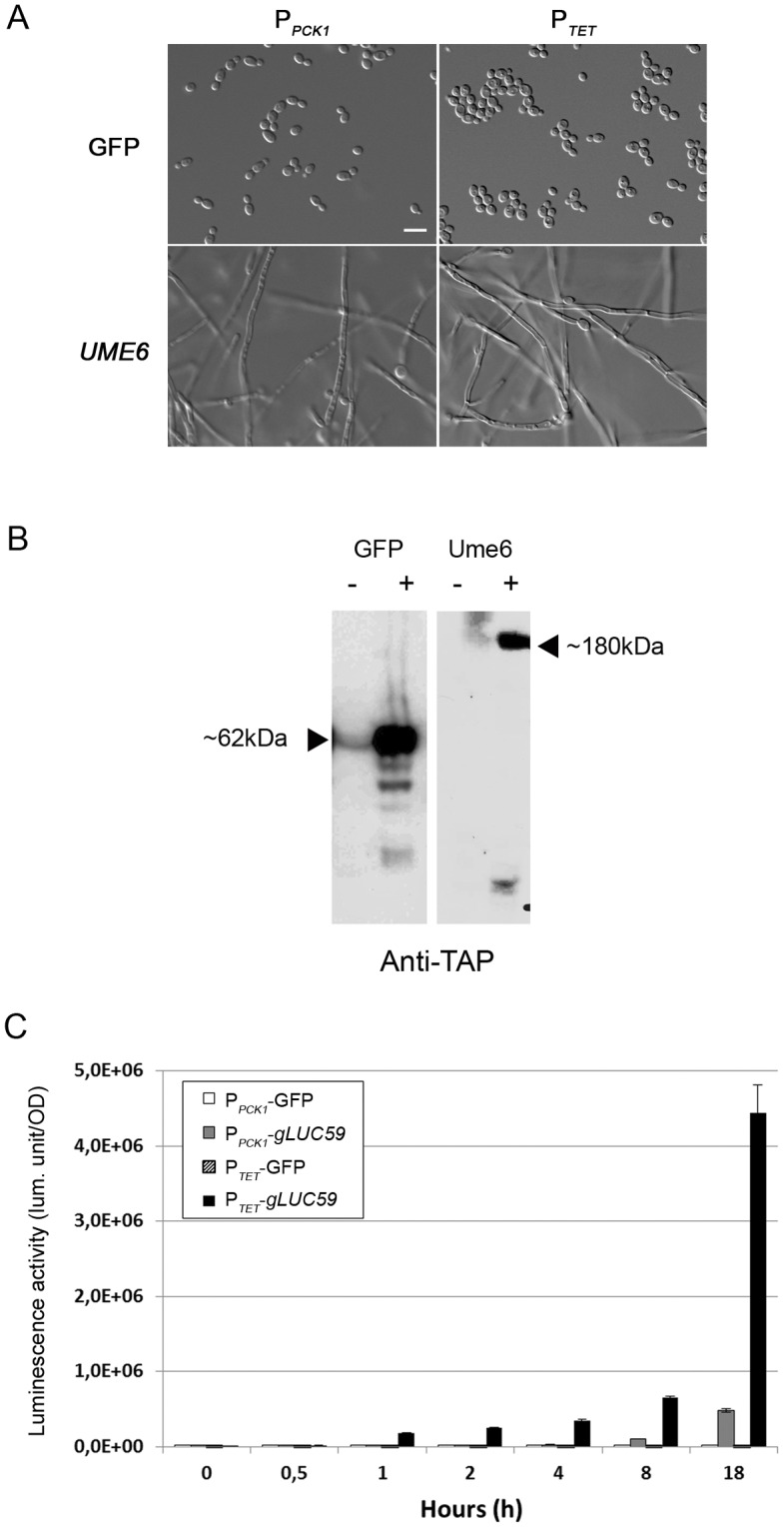
C. albicans strains with integrated CIp10-PPCK1-GTW-TAPtag or CIp10-PTET-GTW derivatives harbouring the GFP (CEC2407 or CEC2992, respectively) or UME6 (CEC1097 or CEC2994, respectively) ORFs were observed microscopically upon growth in gluconeogenic conditions or YPD supplemented with 50 µg.mL− 1 Dox at 30°C for 18 h. Scale bar = 5 µm. B. Production of TAPtagged proteins. C. albicans strains with integrated CIp10-PPCK1-GTW-TAPtag derivatives harbouring the GFP (CEC2407) or UME6 (CEC1097) ORFs were grown in SD (−) or YNB 2% casamino acids (+) for 6 h. Whole cell extracts were separated by SDS-PAGE and probed with a peroxidase-coupled antibody allowing the detection of TAPtagged proteins in gluconeogenic conditions only. Proteins of interest are indicated by an arrow along with their deduced size. C. Kinetics of expression from the PCK1 or TET promoters. C. albicans strains with integrated CIp10-PPCK1-GTW-TAPtag or CIp10-PTET-GTW derivatives harbouring the GFP (CEC2407 or CEC2992, respectively) or gLUC59 (CEC1906 or CEC3083, respectively) ORFs were grown in YNB 2% casamino acids or YPD supplemented with 3 µg.mL− 1 ATc for 18 h at 30°C. Data represent luciferase specific activity detected from the different strains at the indicated time points of growth under inducing conditions. Assays were performed in duplicate and means and SD are shown.
We also compared the strengths and expression kinetics of the PPCK1 and PTET promoters. Strains harbouring a PPCK1-gLUC59 fusion or a PTET-gLUC59 fusion and pNIMX were shifted to gluconeogenic conditions or grown in the presence of 3 µg.mL− 1 anhydrotetracycline (ATc), respectively, and luciferase activity was recorded at different time points following the shift. Results in Fig. 2C showed that an increase in luciferase activity was detectable after 1 h when using PTET while 4–8 h were needed to see such an increase when using PPCK1. Moreover, expression levels obtained from PTET were ca. 50 times those achieved from PPCK1 after 18 h of induction. Finally, a 460-fold and 24600-fold induction was observed with PPCK1 and PTET respectively after 18 h of induction. Thus, these two systems provide versatility in the levels and conditions of OE that can be advantageous when testing the effect of gene OE on a given phenotype.
Establishment of a Collection of Candida albicans OE Strains
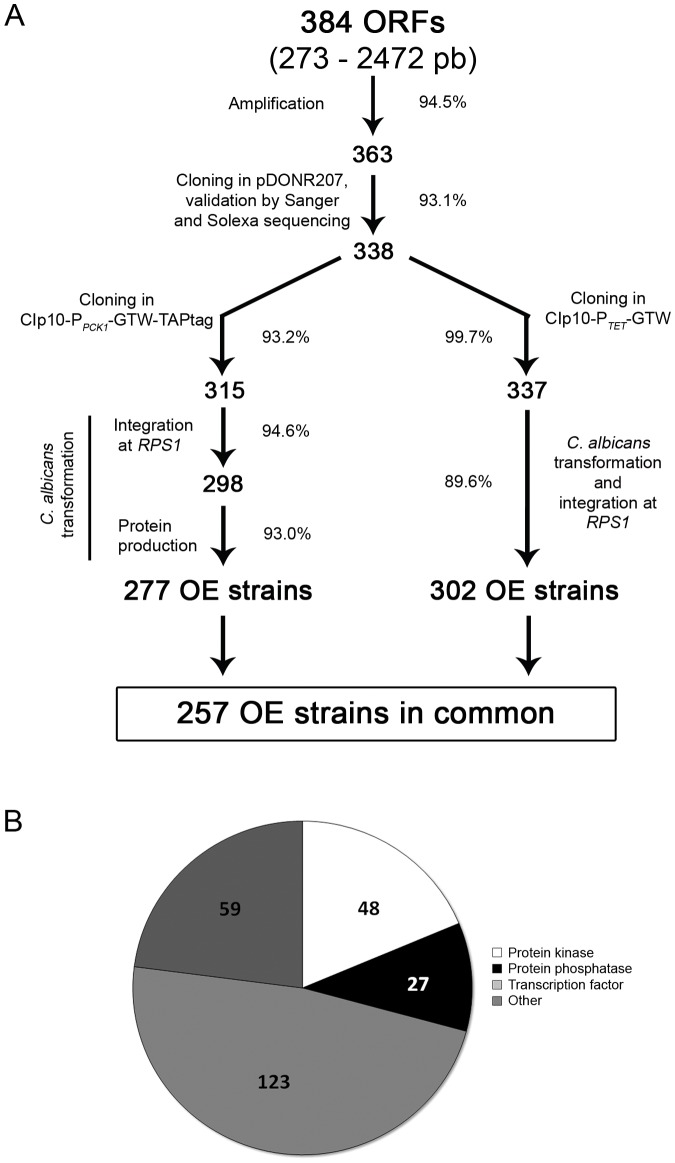
Based on these results, we generated two new collections of C. albicans OE strains. We focused our study on 384 C. albicans ORFs encoding 76 protein kinases (PKs), 36 protein phosphatases (PPs), 179 transcription factors (TFs) and 93 other proteins related to signalling. Corresponding PCR products from the start codon to the penultimate codon were cloned into the pDONR207 donor vector. Following Sanger and Illumina/Solexa sequence validation, a total of 338 (93.1%) derivatives of pDONR207 were obtained (Fig. 3A and Table S1).
Figure 3. Establishment of two collections of C. albicans OE strains. A. Schematic of the OE strain construction pipeline.
C. albicans ORFs were amplified from their start codon to their penultimate codon and cloned into the pDONR207 vector using Gateway®-mediated recombination. The resulting plasmids were analyzed individually by Sanger sequencing of the ORF-5′ and 3′ ends and in pools by Illumina/Solexa sequencing. Validated ORFs were transferred into CIp10-PPCK1-GTW-TAPtag or CIp10-PTET-GTW and the resulting plasmids were introduced at the RPS1 locus in C. albicans strain CEC161 or CEC2907, respectively. Production of a TAPtagged protein of the appropriate size was subsequently tested by Western-blot analysis of protein extracts of the C. albicans OE strains grown in YNB +2% casamino acids medium for 18 h in the case of the PPCK1-driven OE strains. For each step the success rate is indicated along with the number of validated plasmids or strains that have been obtained. B. Distribution of the 257 overexpressed ORFs overlapping both collections across functional categories.
ORFs cloned into pDONR207 were subsequently transferred into the CIp10-PPCK1-GTW-TAPtag and uniquely barcoded CIp10-PTET-GTW plasmids. A total of 315 CIp10-PPCK1-GTW-TAPtag derivatives and 337 CIp10-PTET-GTW derivatives were obtained (Fig. 3A; Table S1). These plasmids were subsequently introduced at the RPS1 locus in C. albicans wild-type strains CEC161 or CEC2907, respectively. Of the resulting 298 strains that harboured a CIp10-PPCK1-GTW-TAPtag derivative, 277 produced a TAP-tagged protein when grown in gluconeogenic conditions with approx. 15% showing relatively low levels of protein production (Fig. 3 and data not shown). Eventually, 277 C. albicans PPCK1-driven OE strains and 302 C. albicans PTET-driven OE strains were obtained, with 257 genes being represented in both sets of strains (Fig. 3A; Table S1). In summary, our procedure had more than 70% success rate in both cases, reflecting a near 90% success rate at each step. Results presented below focus on the phenotypes associated with the OE of those 257 genes for which PPCK1-driven and PTET-driven OE strains were available. These genes encode 48 PKs (44% of all annotated C. albicans PKs), 27 PPs (66% of all annotated C. albicans PPs), 123 TFs (51% of all annotated C. albicans TFs) and 59 other proteins related to signalling (Fig. 3B).
Screening for Genes Affecting Morphogenesis upon PPCK1-driven OE
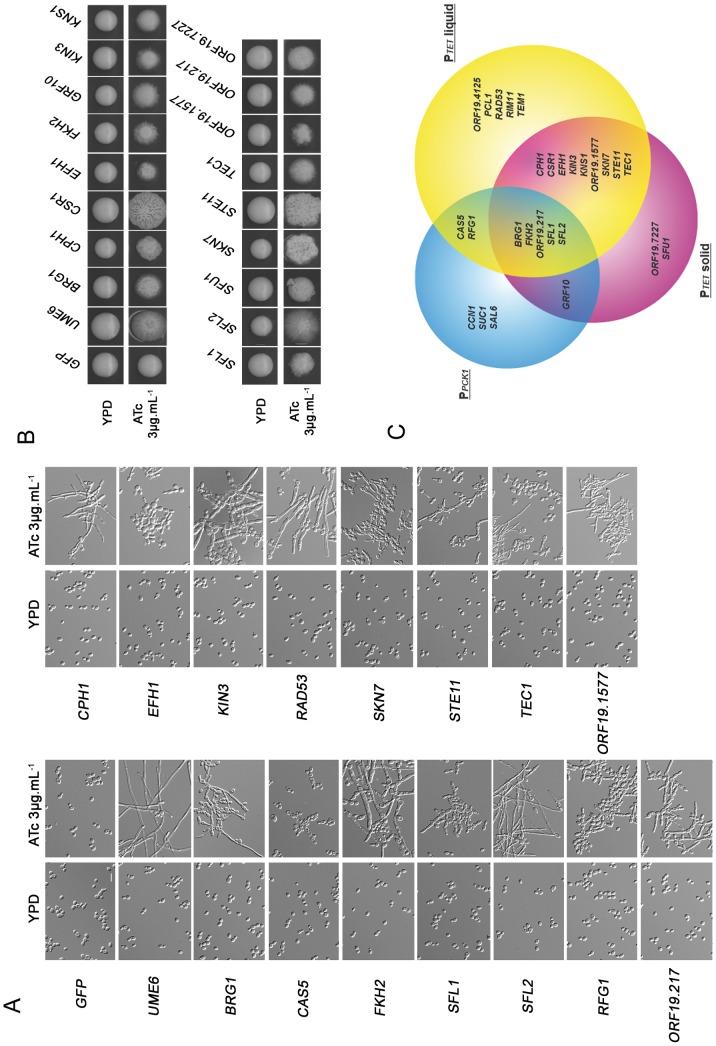
The ability of C. albicans to switch between yeast and hyphal forms is considered a major requirement for virulence and biofilm formation [11], [45]–[48]. Thus, we performed a screen to identify C. albicans genes whose PPCK1-driven OE triggers pseudohyphal or hyphal growth under conditions that normally promote yeast growth. As shown in Fig. 2A and 4, gluconeogenic conditions required for expression from PPCK1 are associated with growth in the yeast form only. Hence, the 257 C. albicans PPCK1-OE strains described above were grown individually in YNB 2% casamino acids at 30°C for 18 h and the cultures were observed microscopically. Eleven strains displayed pseudohyphal or hyphal growth in inducing conditions as shown in Fig. 4. The corresponding genes are listed in Table S2 and included 9 TFs, 1 PP and 1 PK regulatory subunit.
Figure 4. PPCK1-driven OE of 11 C. albicans genes triggers pseudohyphal or hyphal growth.
C. albicans strains with integrated CIp10-PPCK1-GTW-TAPtag derivatives harbouring ORFs for the indicated genes were grown in SD (−) or YNB 2% casamino acids (+) for 6 h (Western Blot) or in YNB 2% casamino acids for 18 h (microscopy). Whole cell extracts of uninduced and induced cultures were separated by SDS-PAGE and probed with a peroxidase-coupled antibody allowing the detection of TAPtagged proteins in gluconeogenic conditions only. Proteins of interest are indicated by an arrow along with their deduced size. 18 h induced cultures were observed microscopically and revealed OE-associated pseudo-filamentation or filamentation. Note that the 5 µm scale bar is different for photos of the two upper panels (GFP, UME6, SUC1, SFL2, and GRF10) and the rest of the figure.
Seven of these genes have been previously associated with morphogenesis including those encoding the Ccn1 G1 cyclin and the Cas5, Fkh2, Rfg1, Sfl1, Sfl2 and Brg1 transcription factors (Table S2). Indeed, inactivation of CCN1, CAS5, FKH2, SFL2 and BRG1 results in defects in filamentation [13], [17], [30], [34], [49]–[54]. On the other hand, Sfl1 and Rfg1 have been described as repressors of filamentation in C. albicans [10], [13], [55]–[58]. However, the role of RFG1 is not restricted to this function since its OE triggers pseudohyphal growth [59]. Noticeably, OE of SFL2 and BRG1 has been previously shown to trigger hyphal growth [30], [54], [60], [61]. In contrast, several genes identified in this screen were not known for their role in morphogenesis including those encoding the Sal6 phosphatase and the Suc1, Grf10, and Orf19.217 putative transcription factors (Table S2). Interestingly, we have observed that inactivation of GRF10 and ORF19.217 did not impair morphogenesis on a variety of hypha-inducing media despite the effect of their OE on morphogenesis (data not shown). These observations are concordant with results published by Homann et al. [13]. Thus, these results confirmed previous published data obtained with either KO or OE strategies and indicated that our OE approach could reveal genes with novel roles in C. albicans pseudohyphal or hyphal differentiation.
Screening for Genes Affecting Morphogenesis in Liquid Medium upon PTET-driven OE
Next, we performed a similar screen with the PTET-driven OE collection. Indeed, gene OE from PTET is highly advantageous since it can be used in any medium supplemented with a tetracycline derivative, while PPCK1-driven OE is strictly dependent upon gluconeogenic growth conditions. Moreover, we have observed that the level of OE is considerably higher with the PTET system (Fig. 2C). Thus, the 257 C. albicans PTET-driven OE strains were grown individually in liquid YPD supplemented with 3 µg.mL− 1 ATc at 30°C for 18 h and the cultures were observed microscopically. In these conditions, we observed that PTET-driven OE of 21 genes induced filamentation or pseudofilamentation (Fig. 5A and Table S2), among which 6 exhibited a weak phenotype (Fig. S2). This gene set included BRG1, SFL2, SFL1, RFG1, CAS5, FKH2 and ORF19.217 already identified in our screen of PPCK1-driven OE strains. In contrast, we did not observe filamentation upon PTET-driven OE of CAS5, GRF10, SAL6 and CCN1. Finally, PTET-driven OE of 14 additional genes triggered pseudofilamentation and/or filamentation (Fig. 5A, Fig. S2 and Table S2). These included TEC1, EFH1, CPH1, PCL1, RAD53, SKN7 and STE11 whose role in morphogenesis was previously uncovered using KO [17], [18], [30], [62]–[72] and OE mutants [30], [45], [65]–[67]. TEC1 and CPH1 are well-characterized regulators of morphogenesis. TEC1 encodes a TEA/ATTS transcription factor regulating hypha-specific genes as well as biofilm formation and pheromone signalling [11], [29], [64]. CPH1 encodes a transcription factor required for mating and hyphal growth on solid media and lies in the same Cek1-MAPK pathway than the Ste11 protein [62], [73], [74]. PCL1 encodes a cyclin homolog whose expression is induced upon filamentous growth [56]. It was also recently shown to be required for agar invasion at elevated temperature [72]. The Rad53 protein kinase is involved in DNA-replication and DNA-damage checkpoint pathways and its deletion is known to nearly completely abolish filamentous growth caused by genotoxic stresses [71]. SKN7 is predicted to encode a response regulator protein in a phosphorelay signal transduction pathway. Its deletion leads to a morphogenesis defect [68] but it has been mostly associated to a role in the response of C. albicans to oxidative stress and osmoregulation [13], [68]. Among the four remaining genes, RIM11 and KIN3 encode two previously characterized protein kinases that had not been associated with morphogenesis yet. Of notable interest, our screen uncovered two uncharacterised genes, ORF19.1577 and ORF19.4125, whose OE triggered filamentation. Heterozygous or homozygous mutants for these genes have been obtained [17], [18] but have not been associated to any relevant phenotype except for the heterozygous mutant ORF19.4125Δ that presents a reduced ability to invade agar compared to a wild-type strain [18].
Figure 5. PTET-driven OE screens confirm results obtained with the PPCK1 promoter and reveal the role of other C. albicans genes in morphogenesis. A. PTET-driven OE of 21 genes promotes pseudo-filamentation or filamentation in liquid media.
C. albicans strains with integrated CIp10-PTET-GTW derivatives harbouring ORFs for the indicated genes were grown in YPD or YPD supplemented with 3 µg.mL− 1 ATc for 18 h. Both cultures were observed microscopically and revealed OE-associated pseudofilamentation or filamentation. Genes whose PPCK1-driven OE results in pseudo-hyphal or hyphal growth are shown on the left panel whereas others genes are placed on the right panel. OE of 15 genes showing the strongest phenotypes are represented, the other 6 are shown in Fig. S2. Scale bar = 5 µm. B. P TET -driven OE of 17 genes promotes filamentation on solid media. Cultures of C. albicans wild-type strains with integrated CIp10-PTET-GTW derivatives harbouring ORFs for the indicated genes were spotted on YPD or YPD supplemented with 3 µg.mL− 1 ATc and were observed after 5 days of growth at 30°C. C. Overlap between the three morphogenesis screens. This venn diagram was obtained with the online software Gliffy (http://www.gliffy.com) and summarises results obtained in the screens performed with the PPCK1 and PTET promoters. Circle size is proportional to the number of genes identified.
Screening for Genes Affecting Morphogenesis in Solid Medium upon PTET-driven OE
We additionally performed a screen of our PTET-driven OE strain collection on YPD solid medium supplemented with 3 µg.mL− 1ATc and identified 17 genes whose OE triggered filamentation (Fig. 5B). Noticeably, three of these genes had not been identified in the screen performed in liquid conditions, namely SFU1, GRF10 and ORF19.7227. Moreover, the phenotype associated to the OE of CSR1 and KNS1 was relatively weak (Fig. S2). CSR1 (or ZAP1) encodes a zinc-finger TF involved in zinc homeostasis and in regulation of biofilm matrix production [33], [75]. It has been shown that deletion of CSR1/ZAP1 affects filamentous growth [10], [13], [17], [33], [76]. Similarly, a homozygous transposon insertion in ORF19.7227 that encodes a putative protein phosphatase inhibitor (PPI) decreases colony wrinkling but does not block true hyphal growth in liquid media [16], consistent with our observations. In contrast, the putative Ser/Thr PK Kns1 and the TF Sfu1 had not been previously associated to a function in filamentous growth. Indeed, KNS1 remains uncharacterized and SFU1 encodes a transcriptional repressor of iron-responsive genes [77]. Finally, we also noted that five of the 18 genes identified in the screen in liquid medium were not recovered in the screen on solid medium (PCL1, RFG1, CAS5, RIM11, ORF19.4125; Fig. 5A and B).
Taken together, the three screens performed using our PTET-driven and PPCK1-driven OE strain collections showed overlaps but specificities in the sets of genes that were identified based on the impact of their OE on morphogenesis (Fig. 5C), reemphasizing the interest of using versatile OE vectors. Moreover, all three screens revealed genes with previously unknown roles in morphogenesis, confirming the potential of an OE approach for C. albicans functional genomics.
Impact of PTET-driven OE on C. albicans Growth Rate
In the yeast S. cerevisiae, OE of up to 15% of the gene repertoire results in growth defects at the colony level. This is often due to pathway activation and can be used to identify targets of genes whose OE is toxic [23], [27]. Therefore, we assessed to what extent our PTET-driven OE system could trigger changes in C. albicans growth rate.
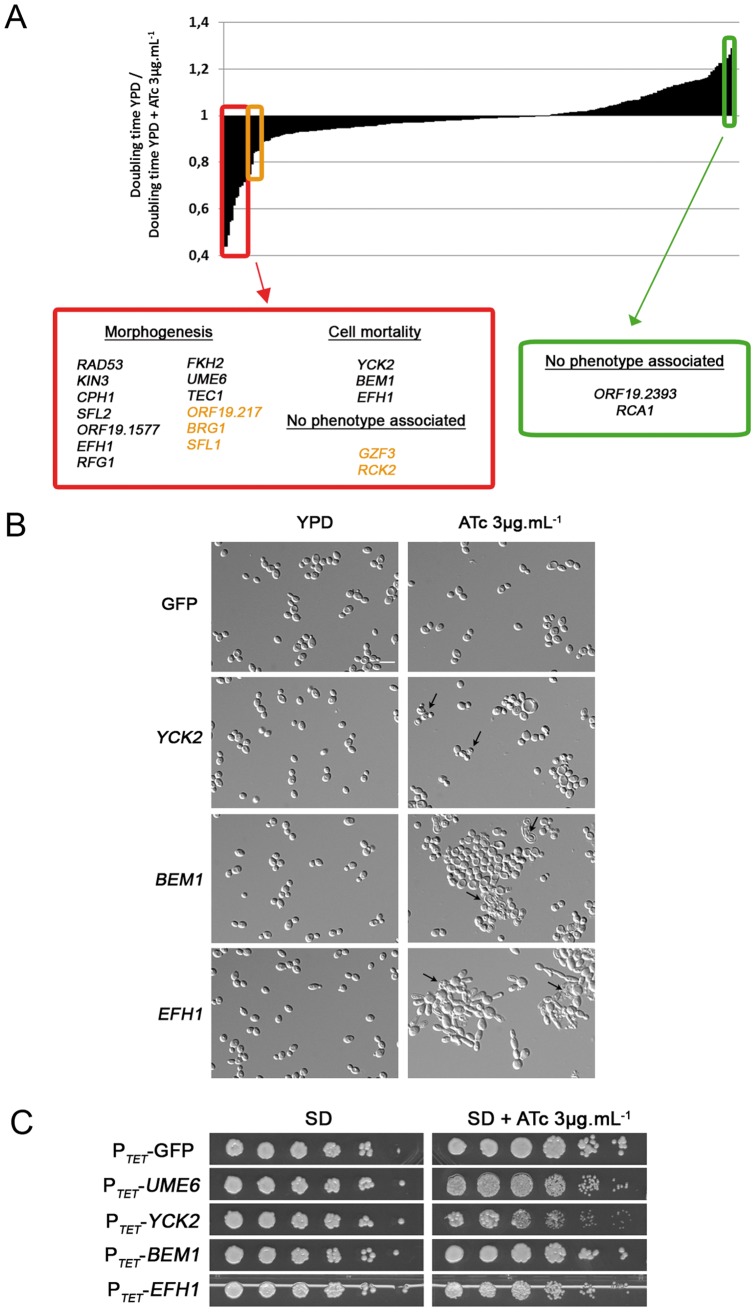
The 257 PTET-driven OE strains were surveyed during a growth kinetic in 96-well plates in the presence or absence of ATc. For each strain, a ratio equalliing to the doubling time monitored in non-inducing conditions (YPD) divided by the doubling time observed under inducing conditions (YPD supplemented with 3 µg.mL− 1 ATc) was calculated. This identified 17 genes whose OE decreased C. albicans growth rate (≥2 fold) and 2 genes whose OE increased C. albicans growth rate (>2 fold) (Fig. 6A). The latter two genes encode the bZIP domain-containing protein of the ATF/CREB family Rca1 and a putative TF of unknown function, Orf19.2393 (Fig. 6A). We did not observe other phenotypes associated with the OE of these genes in a wild-type strain (data not shown). The rca1ΔΔ mutant is viable but slow-growing and displays increased invasive growth [13], [17], consistent with our observations. This gene clearly plays important roles in C. albicans biology since it controls both the susceptibility to different antifungals [20] and carbonic anhydrase expression via the cAMP/PKA/Efg1 signalling pathway [78].
Figure 6. PTET-driven OE of 20 genes results in decreased or increased growth rate. A. Impact of OE on the growth rates of 257 OE strains.
Genes whose OE decreases or increases growth rate (≥2 fold and >2 fold, respectively) upon growth in liquid medium are listed in red or green boxes respectively. Genes are classified based on their OE phenotype from the more affected to the less affected. Genes in orange correspond to those whose OE decreased growth with a fold equal to 2. Data are mean and SD of 3 experiments. B. P TET -driven OE of YCK2 , BEM1 and EFH1 results in cell lysis. C. albicans strains with integrated CIp10-PTET-GTW derivatives harbouring the GFP, YCK2, BEM1 or EFH1 ORFs were grown in YPD or YPD supplemented with 3 µg.mL− 1 ATc for 18 h and observed microscopically. Cell lysis is indicated by arrows. Scale bar = 5 µm. C. P TET -driven OE of YCK2 and EFH1 reduces growth rate on solid medium. Serial dilutions of cultures of C. albicans strains with integrated CIp10-PTET-GTW derivatives harbouring the GFP, UME6, YCK2, BEM1 or EFH1 ORFs were spotted on SD or SD supplemented with 3 µg.mL− 1 ATc and observed after 3 days of growth at 30°C.
A majority of the genes whose OE resulted in decreased growth rate (13/17 genes or 76.4%) were among those whose OE triggered filamentation (Fig. 5A, 5B and 6A). Indeed, filamentation results in optical density readings that are not correlated with the actual growth rates. Nevertheless, we observed cell death when the BEM1, YCK2 or EFH1 genes were overexpressed (Fig. 6B). Decreased growth was also observed when strains overexpressing YCK2 or EFH1 were grown on solid medium (Fig. 6C). BEM1 has previously been shown to be essential in C. albicans [79], [80]. Despite the requirement of this gene for pseudohyphal and hyphal growth in S. cerevisiae and Yarrowia lipolytica, respectively [81], [82], the role of Bem1 in C. albicans morphogenesis remains unclear [79], [80]. None of the two other genes (YCK2 and EFH1) were previously associated with cell growth. In particular, YCK2 encodes a plasma membrane protein similar to the highly conserved serine/threonine casein kinase 1 (CK1) of S. cerevisiae and plays role in damaging oral epithelial cells and hyphal branching [83]. Thus, our results revealed genes involved in C. albicans fitness, including genes not previously described for such a role. However, if one excludes genes whose OE triggers morphogenesis, these genes represent a minor fraction of those we have tested. This is despite the fact that many of them have regulatory functions and suggests that C. albicans might be more robust than S. cerevisiae to the deleterious effects of gene OE and/or that our OE system is not sufficient to reveal such phenotypes.
Conclusions
In this study, we have reported the development of two collections of OE strains and the potential of OE screens in uncovering novel components of regulatory pathways in C. albicans. To date, OE screens have been rarely used for the identification of genes conferring specific phenotypes in C. albicans. Fu et al. [28] have established a collection of 26 C. albicans OE strains whereby genes encoding GPI-anchored proteins are overexpressed from a tetracycline-repressible promoter [84]. Sahni et al. [29] have constructed a collection of 107 C. albicans OE strains whereby genes encoding transcription factors are overexpressed from a tetracycline-inducible promoter [39]. These collections were developed using promoter replacement at the targeted gene through a split-marker strategy [28] or allelic exchange between an ADH1 allele and an OE plasmid obtained by restriction enzyme mediated cloning [39]. Therefore, these resources lack some of the versatility and reusability that is associated with the partial C. albicans ORFeome and collections of OE plasmids developed here. Indeed, our strategy was based on the highly efficient Gateway® recombinational cloning methodology [35], [85] that provided the possibility to shuttle ORFs between plasmids allowing gluconeogenesis- or tetracycline-inducible expression and production of tagged or untagged proteins. Moreover, because our OE plasmids used an integrative vector that is targeted highly efficiently to the C. albicans RPS1 locus, development of collections of OE strains in various genetic backgrounds is rather straightforward (AN, SBB and CE, unpublished data). Hence, our work has laid the ground for the establishment of a C. albicans ORFeome and a genome-wide collection of C. albicans OE strains, a collaborative project that is ongoing in our laboratory and that of C. Munro (University of Aberdeen; [86]).
Several of the genes that we have identified in our morphogenesis alteration screen were already known for their role in filamentation such as BRG1, CPH1, SFL2 and TEC1, thus validating our screen (Table S2). However, by comparison with previous studies, we noticed that OE of three genes present in our collection, namely RAS1, GPR1 and TPK2, and known to trigger morphogenesis upon OE [87]–[89] did not result in pseudohyphal or hyphal growth here. Indeed, we observed that amplification was associated with one mismatch every 1286 bp on average (see Materials and Methods for details) and certain genes harboured non-synonymous mutations (Table S1) that might have impacted their function. Moreover, we noticed some differences in the OE phenotypes according to the promoters we have used. This may be explained by 1) the difference of expression between the PCK1 and TET promoters, 2) the variable level of expression between clones of the same transformation due to tandem insertions and 3) the fact that some PPCK1-driven overexpressed proteins may not be fully functional because of the occurrence of the TAPtag at their carboxy-terminus. In conclusion, the TET promoter appears the most suitable system for large-scale OE screens for different reasons: 1) its induction is simpler and easier to control; 2) it provides a higher level of induction compared to the PCK1 promoter system; 3) it has been shown to function in animal models of C. albicans infections.
Among the 257 genes that we have tested, at least 137 have also been characterized by gene deletion and 34 (24.8%) were shown to be involved to some extent in morphogenesis [13], [17]. It is noteworthy that for a subset of the genes that we have identified based on the impact of their OE on morphogenesis the corresponding KO mutations did not impair morphogenesis (such as GRF10 and ORF19.217; [13] and data not shown). This highlights the complementarity between KO and OE screens and reinforces the potential of the latter in exploring regulatory networks [90].
In summary, this study provides an example of the potential of OE approaches in the investigation of C. albicans biology. Taken together, our results highlight the multiplicity of possible applications of our strategy such as a variety of phenotypic screens in C. albicans. In the short term, the development of diverse destination vectors and recipient strains will help to rapidly determine the function of a specific gene or to identify its partners, eg using suppressor screens or two-hybrid screens [26]. Moreover, the presence of barcodes in our PTET-driven OE strain collection will enable experiments in pools that will greatly facilitate large-scale experiments. Overall, the strain collections generated during this study and available through the Fungal Genetics Stock Center [91], as well as the OE strategy we developed, will allow the generation of a large number of relevant data for the whole Candida community.
Materials and Methods
Strains and Media
All C. albicans strains used in this study are listed in Table 1. Strains were grown at 30°C in YPD medium (1% yeast extract, 2% peptone, 2% glucose) or SD minimal medium [0.67% yeast nitrogen base (YNB; Difco) with 0.4 or 2% glucose] supplemented if necessary with arginine, histidine and uridine, at 20 mg.L− 1 and 2% agar for solid media. OE from PPCK1 was triggered in YNB plus 2% casamino acids liquid cultures at 30°C whereas OE from PTET was induced by the addition of 50 µg.mL− 1 doxycycline (Dox - Fluka) or 3 µg.mL− 1 anhydrotetracycline (ATc - Fisher Bioblock Scientific) in YPD at 30°C. ATc was preferred over Dox as this semi-synthetic tetracycline derivative has been described for its lower toxicity and its higher efficiency in the binding of the TetR repressor protein [92]. Furthermore, we have observed that 2 µg.mL− 1 ATc reproduced the effect of 50 µg.mL− 1 Dox, either on solid or in liquid medium and this concentration was not deleterious for growth or morphogenesis of C. albicans (Fig. S1 and data not shown). Dox- and ATc-containing cultures were maintained in the dark as these compounds are light sensitive.
Table 1. Strains used in this study.
| Strain name | Genotype | References |
| BWP17 | ura3Δ::λimm434/ura3Δ::λimm434 his1Δ::λimm434/his1Δ::λimm434 arg4Δ::λimm434/arg4Δ::λimm434 iro1Δ::λimm434/ iro1Δ::λimm433 | [101] |
| CEC161 | ura3Δ::λimm434/ura3Δ::λimm434 his1Δ::hisG/HIS1 arg4Δ::hisG/ARG4 | [102] |
| CEC955 | ura3Δ::λimm434/ura3Δ::λimm434 his1Δ::hisG/HIS1 arg4Δ::hisG/ARG4 ADH1/adh1::ADH1p-cartTA::SAT1::PTET-caGFP | This study |
| CEC988 | ura3Δ::λimm434/ura3Δ::λimm434 ARG4/arg4Δ::hisG HIS1/his1Δ::hisG RPS1/RPS1::CIp10-PACT1-gLUC59 | This study |
| CEC1097 | ura3Δ::λimm434/ura3Δ::λimm434 arg4Δ::hisG/ARG4 his1Δ::hisG/HIS1 RPS1/RPS1::CIp10-PPCK1-UME6-TAPtag | This study |
| CEC1906 | ura3Δ::λimm434/ura3Δ::λimm434 arg4Δ::hisG/ARG4 his1Δ::hisG/HIS1 RPS1/RPS1::CIp10-PPCK1-gLUC59-TAPtag | This study |
| CEC1909 | ura3Δ::λimm434/ura3Δ::λimm434 his1Δ::hisG/HIS1 arg4Δ::hisG/ARG4 ADH1/adh1::ADH1p-cartTA::SAT1::PTET-caGFP RPS1/ RPS1::PTET -gLUC59 | This study |
| CEC2175 | ura3Δ::λimm434/ura3Δ::λimm434 his1Δ::hisG/HIS1 arg4Δ::hisG/ARG4 ADH1/adh1:: PADH1-cartTA::SAT1 | This study |
| CEC2249 | ura3Δ::λimm434/ura3Δ::λimm434 his1Δ::hisG/HIS1 arg4Δ::hisG/ARG4 ADH1/adh1::ADH1p-cartTA::SAT1 RPS1/RPS1:: PTET -gLUC59 | This study |
| CEC2407 | ura3Δ::λimm434/ura3Δ::λimm434 his1Δ::hisG/HIS1 arg4Δ::hisG/ARG4 RPS1/RPS1::CIp10-PPCK1-GFP-TAPtag | This study |
| CEC2907 - CEC2908 | ura3Δ::λimm434/ura3Δ::λimm434 his1Δ::hisG/HIS1 arg4Δ::hisG/ARG4 ADH1/adh1::PTDH3-carTA::SAT1 | This study |
| CEC2992 | ura3Δ::λimm434/ura3Δ::λimm434 his1Δ::hisG/HIS1 arg4Δ::hisG/ARG4 ADH1/adh1::PTDH3-carTA::SAT1 RPS1/RPS1::CIp10-PTET-GFP | This study |
| CEC2994 | ura3Δ::λimm434/ura3Δ::λimm434 his1Δ::hisG/HIS1 arg4Δ::hisG/ARG4 ADH1/adh1::PTDH3-carTA::SAT1 RPS1/RPS1::CIp10-PTET-UME6 | This study |
| CEC3083 | ura3Δ::λimm434/ura3Δ::λimm434 his1Δ::hisG/HIS1 arg4Δ::hisG/ARG4 ADH1/adh1:: PADH1-cartTA::SAT1 RPS1/RPS1:: PTET -gLUC59 | This study |
Plasmids harbouring a Gateway® cassette were propagated in Escherichia coli strain TOP10 ccdBR (Invitrogen). Other plasmids were propagated in E. coli strain DH5α [93]. E. coli strains were grown in LB medium. Antibiotics were used at the following concentrations: ticarcillin, 50 µg.mL− 1; gentamycin, 10 µg.mL− 1; chloramphenicol, 15 µg.mL− 1.
Construction of a C. albicans Partial ORFeome
The detailed method for the cloning of C. albicans ORFs in the pDONR207 vector has been described [94]. Briefly, for each of the selected ORF, a forward primer including the attB1 site and the first 10 codons of the ORF and a reverse primer including the attB2 site and the last ten codons of the ORF were designed and synthesized at Pasteur-Genopole-Ile-de-France oligonucleotide synthesis platform (Table S1). ORFs were amplified from genomic DNA of C. albicans strain SC5314 [95] using Eppendorf Triple Master Taq polymerase and 30 cycles of amplification with elongation time varying from 1 to 3 min. according to the ORF size. The resulting PCR products were checked by agarose gel electrophoresis, ethanol precipitated and, following resuspension in Tris-EDTA (TE), mixed with the donor plasmid pDONR207 (Invitrogen), and subjected to a recombination reaction with Invitrogen Gateway® BP Clonase™. The recombination mixes were transformed into E. coli strain DH5αand one transformant per ORF was selected for further study. Plasmids were prepared using the Millipore™ MultiScreen™ HTS 96-well Filtration System and Millipore™ MultiScreen™ PLASMID. The cloned ORFs were sequenced from the 5′- and 3′-ends using Sanger sequencing. Moreover, a pool of the 347 plasmids was subjected to Illumina/Solexa sequencing in order to obtain full length sequencing of the ORFs. Sequencing reads were aligned to the ORF sequences available from the Candida Genome Database [96] using CLC Genomics Workbench version 4. Polymorphisms were compared to a database of SNPs obtained following Illumina/Solexa sequencing of C. albicans strain SC5314 (Diogo et al., manuscript in preparation). All plasmids with mutations causing a non-sense mutation or a frame-shift were excluded. Among the remaining 312 plasmids, we detected 494 synonymous and 405 non-synonymous mismatches. Among these, 362 synonymous and 209 non-synonymous mismatches had also been identified by Solexa/Illumina sequencing of strain SC5314 suggesting that they correspond to genuine SNPs. This indicated that our cloning procedure was responsible for 132 synonymous and 196 non-synonymous mutations in 422 kb insert sequences corresponding to one mutation at every 1286 bp. Information on these mutations is available in Table S1.
Construction of Gateway®-compatible C. albicans OE Vectors
The sequences of oligonucleotides used for cloning purposes are listed in Table S3. Two Gateway®-compatible vectors for conditional OE in C. albicans were constructed. First, oligonucleotides Nco-5′Sce and Nco-3′Sce were annealed and inserted into the NcoI site of the C. albicans CIp10 integrative vector [36]. This vector designated CIp10S is bearing the 18 bp I-SceI site that is not found in the C. albicans genome. Then, the C. albicans PCK1 promoter region (PPCK1) was amplified from C. albicans strain SC5314 genomic DNA using primers PRPKC1PR and TAPFUR. The TAPtag coding region was amplified using oligonucleotides TERPVUII and TAPFUF and plasmid pFA-TAP-URA3, a derivative of pFA-GFP-URA3 [97] where the PstI/AscI fragment harbouring the GFP coding region has been replaced by a PstI/AscI fragment carrying the TAPtag coding region amplified from plasmid pBS1479 [38] using oligonucleotides Tap1-PstI and Tap2-AscI that allow the addition of a (Gly-Ala)3 coding linker 5′ of the TAPtag coding sequence. Both PCR products were mixed and a fusion product was amplified using primers TERPVUII and PRPKC1PR and cloned into the TOPO-TA cloning vector (Invitrogen). The KpnI/PvuII PPCK1-TAPtag cassette was excised from the resulting plasmid and cloned into KpnI/EcoRV-digested CIp10S, yielding CIp10-PPCK1-TAPtag. The Gateway® RfB cassette was excised from pBS-RfB using EcoRV and cloned into EcoRV-digested CIp10-PPCK1-TAPtag, yielding CIp10-PPCK1-GTW-TAPtag. In order to construct the CIp10-PTET-GTW plasmid, a tetracycline-inducible promotor (PTET) was amplified from plasmid pTET25 [39] using oligonucleotides TETKpn and TetATGE5, and cloned into KpnI/EcoRV-digested CIp10-PPCK1-GTW-TAPtag, yielding CIp10S-PTET-TAPtag. This vector was amplified using Vect32 and Vect33 and the PCR product was digested with EcoRV and self-ligated yielding plasmid CIp10S-PTET that has three stop codons downstream of the EcoRV site. The EcoRV-digested Gateway® RfB cassette was cloned into EcoRV-digested CIp10S-PTET, yielding CIp10S-PTET-GTW. Subsequently, a derivative of CIp10S-PTET-GTW was constructed by StuI digestion and ligation of the annealed Vect30 and Vect31 oligonucleotides. This vector was designated CIp10-PTET-GTW. In this vector, the I-SceI site is closer to the RPS1 sequences that are used for integration at the C. albicans RPS1 locus than in the CIp10-PPCK1-GTW-TAPtag. Hence transformation efficiency and integration at the RPS1 locus are higher when using I-SceI-digested CIp10-PTET-GTW derivatives as compared to I-SceI-digested CIp10-PPCK1-GTW-TAPtag derivatives (data not shown). Yet, the use of StuI digestion to target derivatives of these plasmids at the RPS1 locus is still preferred.
A collection of CIp10-PTET-GTW derivatives was generated by the incorporation of specific molecular barcodes. We used the set of molecular barcodes previously designed for the construction of the S. cerevisiae deletion collections [98]. Briefly, these barcodes consist of a specific 20 bp sequence flanked by universal primer sequences (U1 and U2 or D1 and D2). These barcodes were amplified by PCR using genomic DNA prepared from a pool of the S. cerevisiae heterozygous deletion collection and primers Sac-U1 and Sac-U2 or Sac-D1 and Sac-D2. The resulting PCR products were digested with SacII and ligated into SacII-digested and dephosphorylated CIp10-PTET-GTW. Individual clones were recovered after E. coli TOP10 ccdBR transformation and the cloned barcodes were sequenced. Only plasmids with a tag showing a unique ID in the TAG4 yeast barcode array and without mismatch in the common primers U1-U2 or D1-D2 were kept. In total, 936 barcoded derivatives of CIp10-PTET-GTW were obtained.
Construction of C. albicans OE Strains
Detailed methods for the transfer of C. albicans ORFs from pDONR207 into the CIp10-PPCK1-GTW-TAPtag or barcoded CIp10-PTET-GTW plasmids as well as the integration of the resulting expression plasmids at the RPS1 locus have been described [94]. Briefly, an aliquot of each derivative of pDONR207 was mixed with 50 ng of one of the destination plasmids and subjected to a recombination reaction with Invitrogen Gateway® LR Clonase™. The recombination mixes were transformed into E. coli strain DH5αand one transformant was used for plasmid preparation as described above. EcoRV digestion was used to verify the cloning of the appropriate ORF. The expression plasmids bearing PPCK1 were digested by StuI (or I-SceI if necessary) and transformed into C. albicans strain CEC161 according to Walther and Wendland [99]. Transformants were selected for prototrophy and verified by PCR using primers CIpUL and CIpUR that yield a 1 kb product if integration of the OE plasmid has occurred at the RPS1 locus. Alternatively, the expression plasmids bearing PTET were transformed into C. albicans strain CEC2907 following StuI or I-SceI linearization. CEC2907 is a derivative strain of CEC161 transformed with pNIMX (Fig. 1B). pNIMX is a derivative of pNIM1 [39] that was modified in two steps. First pNIM1 (Fig. 1B.a) was digested by NcoI and BglII, treated to create blunt ends and self-ligated to reconstitute an NcoI site, yielding pNIM1ΔPTET-GFP (Fig. 1B.b). Next, the ADH1 promoter (PADH1) was replaced by the TDH3 promoter (PTDH3) as follows: a region of cartTA (containing the start codon) was excised from pNIM1ΔPTET-GFP using AclI and subcloned into ClaI-digested BLUESCRIPT-SK(-)-PTDH3. The resulting plasmid was linearized with XhoI, treated to create blunt ends and digested by XbaI. The XhoI(blunt)-XbaI fragment containing the PTDH3-cartTA fusion was then subcloned in pNIM1ΔPTET-GFP linearized with BamHI, treated to created blunt ends and digested by XbaI, yielding pNIMX (Fig. 1B.c), in which PTDH3 is inserted upstream of cartTA. Integration of pNIMX digested with KpnI and SacII at the ADH1 locus in strains CEC2907 was verified by PCR using primers NIM1_verif and ADH1_verif.
Analysis of TAPtagged Proteins by Western Blotting
A 20 mL culture in SD or YNB 2% casamino acids was inoculated at OD600 = 0.05 with a freshly grown colony. 10 ODs of exponentially growing cells were collected by centrifugation after 4–6 h of growth at 30°C, resuspended in lysis buffer (0.1 M NaOH, 0.5M EDTA, 2% SDS, 2% β-mercaptoethanol) and incubated 10 min at 90°C [100]. The lysate was neutralized with 5 µL 4M acetic acid, incubated 10 min at 90°C and 50 µL Loading buffer (0.25 M Tris-HCl pH 6.8, 50% glycerol, 0.05% bromophenol blue) were added. Proteins were separated on an Invitrogen 10% NuPage gel, transferred onto nitrocellulose and TAPtagged proteins were detected using peroxidase-coupled anti-peroxidase antibodies (Sigma) and an ECL kit (GE Healthcare).
Luciferase Assays
100 mL of YNB 2% casamino acids or YPD supplemented with 3 µg.mL− 1 ATc were inoculated with a freshly grown colony on SD 2% glucose resuspended in dH2O (in the case of PPCK1-driven OE strains) or an overnight culture in YPD at 30°C (in the case of PTET-driven OE strains). At each time point, a volume equivalent to 20 OD was centrifuged (2–5 min at 3500 rpm) and resuspended in 200 µL of R-luc buffer (NaCl 0.5 M, Na2HPO4 0.1 M pH 6.7, EDTA 1 mM). For luciferase assays, 100 µL of cells were mixed with 20 µL of 2 µM coelenterazine before luminescence (integration time: 1000 ms) and absorbance (wavelength: 610 nm) were measured using a microplate reader (TECAN Infinite 200). The final luminescence value is obtained by the following formula: Luminescence unit/Absorbance.
Spotting Assays
Strains in the PTET-driven OE collection were grown in 96-well plate in YPD (30 h; 30°C) and spotted on YPD plates supplemented or not with 3 µg.mL− 1 ATc using the RoTor robot (Singer Instrument). Alternatively, 5-fold serial dilutions of 3 mL overnight cultures in SD at 30°C were spotted on SD plates supplemented or not with 3 µg.mL− 1 ATc. In both cases, plates were grown at 30°C for 2–5 days and scanned with Epson perfection 4490. Spotting assays were performed in triplicate.
Growth Kinetics
Strains in the PTET-driven OE collection were grown in 96-well plates in YPD (30 h; 30°C) and inoculated at a final OD600 = 0.1 in 100 µL YPD supplemented or not with 3 µg.mL− 1 ATc. Growth at 30°C was monitored every 20 minutes using a microplate reader (TECAN Sunrise). Doubling-time (DT) was calculated by dividing by 2 the time between OD600 = 0.15 and OD600 = 0.6. Growth curves were performed in triplicate.
Microscopy and Image Analysis
Cells were observed with a Leica DM RXA microscope (Leica Microsystems). Images were captured with a Hamamatsu ORCA II-ER cooled CCD camera, using the Openlab software version 3.5.1 (Improvision Inc.), and then processed with Adobe Photoshop 10.0 software.
Supporting Information
Comparison of doxycycline (Dox) and anhydrotetracycline (ATc). A. 50 µg.mL−1 Dox or 2 µg.mL−1 ATc induce P TET to a similar extent. C. albicans strains with integrated CIp10-PTET-GTW derivatives harbouring the GFP or gLUC59 ORFs (CEC2992 or CEC3083, respectively) were grown in YPD supplemented with 50 µg. mL− 1 Dox or 2 µg.mL− 1 ATc for 18 h at 30°C. Data represent luciferase specific activity detected from the different strains at 0 and 18 h of growth under inducing conditions. Assays were performed in duplicate and means and SD are shown. B. Effects on morphogenesis are similar between 50 µg.mL−1 Dox and 2 µg.mL−1 ATc. C. albicans strain SC5314 and a strain overexpressing UME6 (CEC2994) were grown in YPD medium and spotted on YPD medium supplemented or not with tetracycline analog (50 µg.mL− 1 Dox or 2 µg.mL− 1 ATc). Pictures were taken after 5 days of growth at 30°C. C. ATc shows lower inhibition of C. albicans hyphal growth than Dox. C. albicans strain SC5314 was grown in YPD liquid medium supplemented or not with different concentrations of Dox or ATc for 18 h at 30°C and observed microscopically. Scale bar = 5 µm.
(TIF)
P TET -driven OE of 6 genes leads to a weak but significant phenotype in liquid media. C. albicans strains with integrated CIp10-PTET-GTW derivatives harbouring ORFs for the indicated genes were grown in YPD or YPD supplemented with 3 µg.mL− 1 ATc for 18 h. Both cultures were observed microscopically and revealed OE-associated pseudofilamentation or filamentation (germ tubes essentially). Scale bar = 5 µm.
(TIF)
Summary of the collections provided in this study.
(XLSX)
Candida albicans genes whose P PCK1 -driven or P TET -driven OE triggers pseudohyphal or hyphal growth.
(DOCX)
Oligonucleotides used in this study.
(DOCX)
Acknowledgments
We are grateful to other members of the Unité Biologie et Pathogénicité Fongiques for their constant support and numerous insights during the course of this project. We are grateful to C. Gouyette for oligonucleotide synthesis and C. Bouchier, L. Ma and S. Créno for sequencing of the plasmid clones. Thanks are due to C. Munro, A. Brown and G. Janbon insights in the manuscript and useful comments.
Funding Statement
This work has been supported by the European Commission (EURESFUN, LSHM-CT-2005-518199; Galar Fungail 2, MRTN-CT-2003-504148; FINSysB, PITN-GA-2008-214004), Agence Nationale de la Recherche (KANJI, ANR-08-MIE-033-01) and the Wellcome Trust (The Candida albicans ORFeome project, WT088858MA). Oligonucleotide synthesis and plasmid sequencing were supported by Pasteur-Génopole-Ile-de-France. AN was the recipient of a PhD fellowship from the DIM-MalInf Région Ile-de-France. VC was the recipient of a PhD fellowship of the European Commission (FINSysB, PITN-GA-2008-214004). SZ was the recipient from post-doctoral fellowships of the European Commission (FINSysB, PITN-GA-2008-214004) and Agence Nationale de la Recherche (KANJI, ANR-08-MIE-033-01). AF and ML were the recipients of post-doctoral fellowships of Institut Pasteur (Bourse Roux). DD was the recipient of a joined INRA/Institut Pasteur PhD fellowship. TR was the recipient of a post-doctoral fellowship from the European Commission (EURESFUN, LSHM-CT-2005-518199). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Odds FC, Webster CE, Mayuranathan P, Simmons PD (1988) Candida concentrations in the vagina and their association with signs and symptoms of vaginal candidosis. J Med Vet Mycol 26: 277–283. [DOI] [PubMed] [Google Scholar]
- 2.Calderone RA (2002) Taxonomy and biology of Candida. In Candida and Candididasis, ed. R. Calderone, 307–25. Washington DC: ASM Press.
- 3. Mavor AL, Thewes S, Hube B (2005) Systemic fungal infections caused by Candida species: epidemiology, infection process and virulence attributes. Curr Drug Targets 6: 863–874. [DOI] [PubMed] [Google Scholar]
- 4. Odds FC (2008) Secreted proteinases and Candida albicans virulence. Microbiology 154: 3245–3246. [DOI] [PubMed] [Google Scholar]
- 5. Pfaller MA, Diekema DJ (2007) Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev 20: 133–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hernday AD, Noble SM, Mitrovich QM, Johnson AD (2010) Genetics and molecular biology in Candida albicans . Methods Enzymol 470: 737–758. [DOI] [PubMed] [Google Scholar]
- 7. Noble SM, Johnson AD (2007) Genetics of Candida albicans, a diploid human fungal pathogen. Annu Rev Genet 41: 193–211. [DOI] [PubMed] [Google Scholar]
- 8. Davis DA, Bruno VM, Loza L, Filler SG, Mitchell AP (2002) Candida albicans Mds3p, a conserved regulator of pH responses and virulence identified through insertional mutagenesis. Genetics 162: 1573–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Roemer T, Jiang B, Davison J, Ketela T, Veillette K, et al. (2003) Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol Microbiol 50: 167–181. [DOI] [PubMed] [Google Scholar]
- 10. Uhl MA, Biery M, Craig N, Johnson AD (2003) Haploinsufficiency-based large-scale forward genetic analysis of filamentous growth in the diploid human fungal pathogen C.albicans . EMBO J 22: 2668–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nobile CJ, Mitchell AP (2005) Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr Biol 15: 1150–1155. [DOI] [PubMed] [Google Scholar]
- 12. Xu D, Jiang B, Ketela T, Lemieux S, Veillette K, et al. (2007) Genome-wide fitness test and mechanism-of-action studies of inhibitory compounds in Candida albicans . PLoS Pathog 3: e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Homann OR, Dea J, Noble SM, Johnson AD (2009) A phenotypic profile of the Candida albicans regulatory network. PLoS Genet 5: e1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Becker JM, Kauffman SJ, Hauser M, Huang L, Lin M, et al. (2010) Pathway analysis of Candida albicans survival and virulence determinants in a murine infection model. Proc Natl Acad Sci U S A 107: 22044–22049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Blankenship JR, Fanning S, Hamaker JJ, Mitchell AP (2010) An extensive circuitry for cell wall regulation in Candida albicans . PLoS Pathog 6: e1000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Epp E, Walther A, Lepine G, Leon Z, Mullick A, et al. (2010) Forward genetics in Candida albicans that reveals the Arp2/3 complex is required for hyphal formation, but not endocytosis. Mol Microbiol 75: 1182–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Noble SM, French S, Kohn LA, Chen V, Johnson AD (2010) Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet 42: 590–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Oh J, Fung E, Schlecht U, Davis RW, Giaever G, et al. (2010) Gene annotation and drug target discovery in Candida albicans with a tagged transposon mutant collection. PLoS Pathog 6: e1001140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bharucha N, Chabrier-Rosello Y, Xu T, Johnson C, Sobczynski S, et al. (2011) A large-scale complex haploinsufficiency-based genetic interaction screen in Candida albicans: analysis of the RAM network during morphogenesis. PLoS Genet 7: e1002058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vandeputte P, Pradervand S, Ischer F, Coste A, Ferrari S, et al. (2012) Identification and functional characterization of Rca1, a transcription factor involved in both antifungal susceptibility and host response in Candida albicans. Eukaryot Cell: 916–931. [DOI] [PMC free article] [PubMed]
- 21. Stevenson LF, Kennedy BK, Harlow E (2001) A large-scale overexpression screen in Saccharomyces cerevisiae identifies previously uncharacterized cell cycle genes. Proc Natl Acad Sci U S A 98: 3946–3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chua G, Morris QD, Sopko R, Robinson MD, Ryan O, et al. (2006) Identifying transcription factor functions and targets by phenotypic activation. Proc Natl Acad Sci U S A 103: 12045–12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sopko R, Huang D, Preston N, Chua G, Papp B, et al. (2006) Mapping pathways and phenotypes by systematic gene overexpression. Mol Cell 21: 319–330. [DOI] [PubMed] [Google Scholar]
- 24. Jin R, Dobry CJ, McCown PJ, Kumar A (2008) Large-scale analysis of yeast filamentous growth by systematic gene disruption and overexpression. Mol Biol Cell 19: 284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Magtanong L, Ho CH, Barker SL, Jiao W, Baryshnikova A, et al. (2011) Dosage suppression genetic interaction networks enhance functional wiring diagrams of the cell. Nat Biotechnol 29: 505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Prelich G (2012) Gene overexpression: uses, mechanisms, and interpretation. Genetics 190: 841–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gelperin DM, White MA, Wilkinson ML, Kon Y, Kung LA, et al. (2005) Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev 19: 2816–2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fu Y, Luo G, Spellberg BJ, Edwards JE Jr, Ibrahim AS (2008) Gene overexpression/suppression analysis of candidate virulence factors of Candida albicans . Eukaryot Cell 7: 483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sahni N, Yi S, Daniels KJ, Huang G, Srikantha T, et al. (2010) Tec1 mediates the pheromone response of the white phenotype of Candida albicans: insights into the evolution of new signal transduction pathways. PLoS Biol 8: e1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Du H, Guan G, Xie J, Sun Y, Tong Y, et al. (2012) Roles of Candida albicans Gat2, a GATA-type zinc finger transcription factor, in biofilm formation, filamentous growth and virulence. PLoS One 7: e29707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nobile CJ, Mitchell AP (2006) Genetics and genomics of Candida albicans biofilm formation. Cell Microbiol 8: 1382–1391. [DOI] [PubMed] [Google Scholar]
- 32. Nobile CJ, Nett JE, Andes DR, Mitchell AP (2006) Function of Candida albicans adhesin Hwp1 in biofilm formation. Eukaryot Cell 5: 1604–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nobile CJ, Nett JE, Hernday AD, Homann OR, Deneault JS, et al. (2009) Biofilm matrix regulation by Candida albicans Zap1. PLoS Biol 7: e1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, et al. (2012) A recently evolved transcriptional network controls biofilm development in Candida albicans . Cell 148: 126–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Walhout AJ, Temple GF, Brasch MA, Hartley JL, Lorson MA, et al. (2000) GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol 328: 575–592. [DOI] [PubMed] [Google Scholar]
- 36. Murad AM, Lee PR, Broadbent ID, Barelle CJ, Brown AJ (2000) CIp10, an efficient and convenient integrating vector for Candida albicans . Yeast 16: 325–327. [DOI] [PubMed] [Google Scholar]
- 37. Leuker CE, Sonneborn A, Delbruck S, Ernst JF (1997) Sequence and promoter regulation of the PCK1 gene encoding phosphoenolpyruvate carboxykinase of the fungal pathogen Candida albicans . Gene 192: 235–240. [DOI] [PubMed] [Google Scholar]
- 38. Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, et al. (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 17: 1030–1032. [DOI] [PubMed] [Google Scholar]
- 39. Park YN, Morschhauser J (2005) Tetracycline-inducible gene expression and gene deletion in Candida albicans . Eukaryot Cell 4: 1328–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Delgado ML, Gil ML, Gozalbo D (2003) Candida albicans TDH3 gene promotes secretion of internal invertase when expressed in Saccharomyces cerevisiae as a glyceraldehyde-3-phosphate dehydrogenase-invertase fusion protein. Yeast 20: 713–722. [DOI] [PubMed] [Google Scholar]
- 41. Enjalbert B, Rachini A, Vediyappan G, Pietrella D, Spaccapelo R, et al. (2009) A multifunctional, synthetic Gaussia princeps luciferase reporter for live imaging of Candida albicans infections. Infect Immun 77: 4847–4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schaub Y, Dunkler A, Walther A, Wendland J (2006) New pFA-cassettes for PCR-based gene manipulation in Candida albicans . J Basic Microbiol 46: 416–429. [DOI] [PubMed] [Google Scholar]
- 43. Carlisle PL, Banerjee M, Lazzell A, Monteagudo C, Lopez-Ribot JL, et al. (2009) Expression levels of a filament-specific transcriptional regulator are sufficient to determine Candida albicans morphology and virulence. Proc Natl Acad Sci U S A 106: 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zeidler U, Lettner T, Lassnig C, Muller M, Lajko R, et al. (2009) UME6 is a crucial downstream target of other transcriptional regulators of true hyphal development in Candida albicans . FEMS Yeast Res 9: 126–142. [DOI] [PubMed] [Google Scholar]
- 45. Huang H, Harcus D, Whiteway M (2008) Transcript profiling of a MAP kinase pathway in C. albicans . Microbiol Res 163: 380–393. [DOI] [PubMed] [Google Scholar]
- 46. Shapiro RS, Robbins N, Cowen LE (2011) Regulatory circuitry governing fungal development, drug resistance, and disease. Microbiol Mol Biol Rev 75: 213–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sudbery P, Gow N, Berman J (2004) The distinct morphogenic states of Candida albicans . Trends Microbiol 12: 317–324. [DOI] [PubMed] [Google Scholar]
- 48. Sudbery PE (2011) Growth of Candida albicans hyphae. Nat Rev Microbiol 9: 737–748. [DOI] [PubMed] [Google Scholar]
- 49. Loeb JD, Sepulveda-Becerra M, Hazan I, Liu H (1999) A G1 cyclin is necessary for maintenance of filamentous growth in Candida albicans . Mol Cell Biol 19: 4019–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bensen ES, Filler SG, Berman J (2002) A forkhead transcription factor is important for true hyphal as well as yeast morphogenesis in Candida albicans . Eukaryot Cell 1: 787–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sinha I, Wang YM, Philp R, Li CR, Yap WH, et al. (2007) Cyclin-dependent kinases control septin phosphorylation in Candida albicans hyphal development. Dev Cell 13: 421–432. [DOI] [PubMed] [Google Scholar]
- 52. Chamilos G, Nobile CJ, Bruno VM, Lewis RE, Mitchell AP, et al. (2009) Candida albicans Cas5, a regulator of cell wall integrity, is required for virulence in murine and toll mutant fly models. J Infect Dis 200: 152–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pukkila-Worley R, Peleg AY, Tampakakis E, Mylonakis E (2009) Candida albicans hyphal formation and virulence assessed using a Caenorhabditis elegans infection model. Eukaryot Cell 8: 1750–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Song W, Wang H, Chen J (2011) Candida albicans Sfl2, a temperature-induced transcriptional regulator, is required for virulence in a murine gastrointestinal infection model. FEMS Yeast Res 11: 209–222. [DOI] [PubMed] [Google Scholar]
- 55. Khalaf RA, Zitomer RS (2001) The DNA binding protein Rfg1 is a repressor of filamentation in Candida albicans . Genetics 157: 1503–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kadosh D, Johnson AD (2005) Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol Biol Cell 16: 2903–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bauer J, Wendland J (2007) Candida albicans Sfl1 suppresses flocculation and filamentation. Eukaryot Cell 6: 1736–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li Y, Su C, Mao X, Cao F, Chen J (2007) Roles of Candida albicans Sfl1 in hyphal development. Eukaryot Cell 6: 2112–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cleary IA, Mulabagal P, Reinhard SM, Yadev NP, Murdoch C, et al. (2010) Pseudohyphal regulation by the transcription factor Rfg1p in Candida albicans . Eukaryot Cell 9: 1363–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Spiering MJ, Moran GP, Chauvel M, Maccallum DM, Higgins J, et al. (2010) Comparative transcript profiling of Candida albicans and Candida dubliniensis identifies SFL2, a C. albicans gene required for virulence in a reconstituted epithelial infection model. Eukaryot Cell 9: 251–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lu Y, Su C, Liu H (2012) A GATA Transcription Factor Recruits Hda1 in Response to Reduced Tor1 Signaling to Establish a Hyphal Chromatin State in Candida albicans . PLoS Pathog 8: e1002663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Liu H, Kohler J, Fink GR (1994) Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266: 1723–1726. [DOI] [PubMed] [Google Scholar]
- 63. Csank C, Schroppel K, Leberer E, Harcus D, Mohamed O, et al. (1998) Roles of the Candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis. Infect Immun 66: 2713–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schweizer A, Rupp S, Taylor BN, Rollinghoff M, Schroppel K (2000) The TEA/ATTS transcription factor CaTec1p regulates hyphal development and virulence in Candida albicans . Mol Microbiol 38: 435–445. [DOI] [PubMed] [Google Scholar]
- 65. Lane S, Birse C, Zhou S, Matson R, Liu H (2001) DNA array studies demonstrate convergent regulation of virulence factors by Cph1, Cph2, and Efg1 in Candida albicans . J Biol Chem 276: 48988–48996. [DOI] [PubMed] [Google Scholar]
- 66. Lane S, Zhou S, Pan T, Dai Q, Liu H (2001) The basic helix-loop-helix transcription factor Cph2 regulates hyphal development in Candida albicans partly via TEC1 . Mol Cell Biol 21: 6418–6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Doedt T, Krishnamurthy S, Bockmuhl DP, Tebarth B, Stempel C, et al. (2004) APSES proteins regulate morphogenesis and metabolism in Candida albicans . Mol Biol Cell 15: 3167–3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Singh P, Chauhan N, Ghosh A, Dixon F, Calderone R (2004) SKN7 of Candida albicans: mutant construction and phenotype analysis. Infect Immun 72: 2390–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Staib P, Binder A, Kretschmar M, Nichterlein T, Schroppel K, et al. (2004) Tec1p-independent activation of a hypha-associated Candida albicans virulence gene during infection. Infect Immun 72: 2386–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bassilana M, Hopkins J, Arkowitz RA (2005) Regulation of the Cdc42/Cdc24 GTPase module during Candida albicans hyphal growth. Eukaryot Cell 4: 588–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Shi QM, Wang YM, Zheng XD, Lee RT, Wang Y (2007) Critical role of DNA checkpoints in mediating genotoxic-stress-induced filamentous growth in Candida albicans . Mol Biol Cell 18: 815–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Shapiro RS, Sellam A, Tebbji F, Whiteway M, Nantel A, et al. (2012) Pho85, Pcl1, and Hms1 signaling governs Candida albicans morphogenesis induced by high temperature or Hsp90 compromise. Curr Biol 22: 461–470. [DOI] [PubMed] [Google Scholar]
- 73. Kohler JR, Fink GR (1996) Candida albicans strains heterozygous and homozygous for mutations in mitogen-activated protein kinase signaling components have defects in hyphal development. Proc Natl Acad Sci U S A 93: 13223–13228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Leberer E, Wu C, Leeuw T, Fourest-Lieuvin A, Segall JE, et al. (1997) Functional characterization of the Cdc42p binding domain of yeast Ste20p protein kinase. EMBO J 16: 83–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ganguly S, Bishop AC, Xu W, Ghosh S, Nickerson KW, et al. (2011) Zap1 control of cell-cell signaling in Candida albicans biofilms. Eukaryot Cell 10: 1448–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kim MJ, Kil M, Jung JH, Kim J (2008) Roles of Zinc-responsive transcription factor Csr1 in filamentous growth of the pathogenic Yeast Candida albicans . J Microbiol Biotechnol 18: 242–247. [PubMed] [Google Scholar]
- 77. Lan CY, Rodarte G, Murillo LA, Jones T, Davis RW, et al. (2004) Regulatory networks affected by iron availability in Candida albicans . Mol Microbiol 53: 1451–1469. [DOI] [PubMed] [Google Scholar]
- 78. Cottier F, Raymond M, Kurzai O, Bolstad M, Leewattanapasuk W, et al. (2012) The bZIP transcription factor Rca1p is a central regulator of a novel CO(2) sensing pathway in yeast. PLoS Pathog 8: e1002485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Michel S, Ushinsky S, Klebl B, Leberer E, Thomas D, et al. (2002) Generation of conditional lethal Candida albicans mutants by inducible deletion of essential genes. Mol Microbiol 46: 269–280. [DOI] [PubMed] [Google Scholar]
- 80. Bassilana M, Blyth J, Arkowitz RA (2003) Cdc24, the GDP-GTP exchange factor for Cdc42, is required for invasive hyphal growth of Candida albicans . Eukaryot Cell 2: 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lorenz MC, Cutler NS, Heitman J (2000) Characterization of alcohol-induced filamentous growth in Saccharomyces cerevisiae . Mol Biol Cell 11: 183–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hurtado CA, Rachubinski RA (2002) Isolation and characterization of YlBEM1, a gene required for cell polarization and differentiation in the dimorphic yeast Yarrowia lipolytica . Eukaryot Cell 1: 526–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Park H, Liu Y, Solis N, Spotkov J, Hamaker J, et al. (2009) Transcriptional responses of Candida albicans to epithelial and endothelial cells. Eukaryot Cell 8: 1498–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Nakayama H, Mio T, Nagahashi S, Kokado M, Arisawa M, et al. (2000) Tetracycline-regulatable system to tightly control gene expression in the pathogenic fungus Candida albicans . Infect Immun 68: 6712–6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Rual JF, Hill DE, Vidal M (2004) ORFeome projects: gateway between genomics and omics. Curr Opin Chem Biol 8: 20–25. [DOI] [PubMed] [Google Scholar]
- 86.Legrand M, Munro CA, d'Enfert C (2011) Cool Tools 5: The Candida albicans ORFeome project. In: Calderone RA, Clancy CJ, editors. Candida and candidiasis, 2nd edition. Washington DC: ASM Press. 505–510.
- 87. Bockmuhl DP, Krishnamurthy S, Gerads M, Sonneborn A, Ernst JF (2001) Distinct and redundant roles of the two protein kinase A isoforms Tpk1p and Tpk2p in morphogenesis and growth of Candida albicans . Mol Microbiol 42: 1243–1257. [DOI] [PubMed] [Google Scholar]
- 88. Feng Q, Summers E, Guo B, Fink G (1999) Ras signaling is required for serum-induced hyphal differentiation in Candida albicans . J Bacteriol 181: 6339–6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Miwa T, Takagi Y, Shinozaki M, Yun CW, Schell WA, et al. (2004) Gpr1, a putative G-protein-coupled receptor, regulates morphogenesis and hypha formation in the pathogenic fungus Candida albicans . Eukaryot Cell 3: 919–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Yoshikawa K, Tanaka T, Ida Y, Furusawa C, Hirasawa T, et al. (2011) Comprehensive phenotypic analysis of single-gene deletion and overexpression strains of Saccharomyces cerevisiae . Yeast 28: 349–361. [DOI] [PubMed] [Google Scholar]
- 91. McCluskey K, Wiest A, Plamann M (2010) The Fungal Genetics Stock Center: a repository for 50 years of fungal genetics research. J Biosci 35: 119–126. [DOI] [PubMed] [Google Scholar]
- 92. Gossen M, Bujard H (1993) Anhydrotetracycline, a novel effector for tetracycline controlled gene expression systems in eukaryotic cells. Nucleic Acids Res 21: 4411–4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Taylor RG, Walker DC, McInnes RR (1993) E. coli host strains significantly affect the quality of small scale plasmid DNA preparations used for sequencing. Nucleic Acids Res 21: 1677–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Cabral V, Chauvel M, Firon A, Legrand M, Nesseir A, et al. (2012) Modular gene over-expression strategies for Candida albicans . Methods Mol Biol 845: 227–244. [DOI] [PubMed] [Google Scholar]
- 95. Gillum AM, Tsay EY, Kirsch DR (1984) Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet 198: 179–182. [DOI] [PubMed] [Google Scholar]
- 96. Arnaud MB, Costanzo MC, Skrzypek MS, Shah P, Binkley G, et al. (2007) Sequence resources at the Candida Genome Database. Nucleic Acids Res 35: D452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Gola S, Martin R, Walther A, Dunkler A, Wendland J (2003) New modules for PCR-based gene targeting in Candida albicans: rapid and efficient gene targeting using 100 bp of flanking homology region. Yeast 20: 1339–1347. [DOI] [PubMed] [Google Scholar]
- 98. Eason RG, Pourmand N, Tongprasit W, Herman ZS, Anthony K, et al. (2004) Characterization of synthetic DNA bar codes in Saccharomyces cerevisiae gene-deletion strains. Proc Natl Acad Sci U S A 101: 11046–11051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Walther A, Wendland J (2003) An improved transformation protocol for the human fungal pathogen Candida albicans . Curr Genet 42: 339–343. [DOI] [PubMed] [Google Scholar]
- 100. von der Haar T (2007) Optimized protein extraction for quantitative proteomics of yeasts. PLoS One 2: e1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Wilson RB, Davis D, Mitchell AP (1999) Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol 181: 1868–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Firon A, Aubert S, Iraqui I, Guadagnini S, Goyard S, et al. (2007) The SUN41 and SUN42 genes are essential for cell separation in Candida albicans . Mol Microbiol 66: 1256–1275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of doxycycline (Dox) and anhydrotetracycline (ATc). A. 50 µg.mL−1 Dox or 2 µg.mL−1 ATc induce P TET to a similar extent. C. albicans strains with integrated CIp10-PTET-GTW derivatives harbouring the GFP or gLUC59 ORFs (CEC2992 or CEC3083, respectively) were grown in YPD supplemented with 50 µg. mL− 1 Dox or 2 µg.mL− 1 ATc for 18 h at 30°C. Data represent luciferase specific activity detected from the different strains at 0 and 18 h of growth under inducing conditions. Assays were performed in duplicate and means and SD are shown. B. Effects on morphogenesis are similar between 50 µg.mL−1 Dox and 2 µg.mL−1 ATc. C. albicans strain SC5314 and a strain overexpressing UME6 (CEC2994) were grown in YPD medium and spotted on YPD medium supplemented or not with tetracycline analog (50 µg.mL− 1 Dox or 2 µg.mL− 1 ATc). Pictures were taken after 5 days of growth at 30°C. C. ATc shows lower inhibition of C. albicans hyphal growth than Dox. C. albicans strain SC5314 was grown in YPD liquid medium supplemented or not with different concentrations of Dox or ATc for 18 h at 30°C and observed microscopically. Scale bar = 5 µm.
(TIF)
P TET -driven OE of 6 genes leads to a weak but significant phenotype in liquid media. C. albicans strains with integrated CIp10-PTET-GTW derivatives harbouring ORFs for the indicated genes were grown in YPD or YPD supplemented with 3 µg.mL− 1 ATc for 18 h. Both cultures were observed microscopically and revealed OE-associated pseudofilamentation or filamentation (germ tubes essentially). Scale bar = 5 µm.
(TIF)
Summary of the collections provided in this study.
(XLSX)
Candida albicans genes whose P PCK1 -driven or P TET -driven OE triggers pseudohyphal or hyphal growth.
(DOCX)
Oligonucleotides used in this study.
(DOCX)