Abstract
Hsk1, Saccharomyces cerevisiae Cdc7-related kinase in Shizosaccharomyces pombe, is required for G1/S transition and its kinase activity is controlled by the regulatory subunit Dfp1/Him1. Analyses of a newly isolated temperature-sensitive mutant, hsk1-89, reveal that Hsk1 plays crucial roles in DNA replication checkpoint signaling and maintenance of proper chromatin structures during mitotic S phase through regulating the functions of Rad3 (ATM)-Cds1 and Rad21 (cohesin), respectively, in addition to expected essential roles for initiation of mitotic DNA replication through phosphorylating Cdc19 (Mcm2). Checkpoint defect in hsk1-89 is indicated by accumulation of cut cells at 30°C. hsk1-89 displays synthetic lethality in combination with rad3 deletion, indicating that survival of hsk1-89 depends on Rad3-dependent checkpoint pathway. Cds1 kinase activation, which normally occurs in response to early S phase arrest by nucleotide deprivation, is largely impaired in hsk1-89. Furthermore, Cds1-dependent hyperphosphorylation of Dfp1 in response to hydroxyurea arrest is eliminated in hsk1-89, suggesting that sufficient activation of Hsk1-Dfp1 kinase is required for S phase entry and replication checkpoint signaling. hsk1-89 displays apparent defect in mitosis at 37°C leading to accumulation of cells with near 2C DNA content and with aberrant nuclear structures. These phenotypes are similar to those of rad21-K1 and are significantly enhanced in a hsk1-89 rad21-K1 double mutant. Consistent with essential roles of Rad21 as a component for the cohesin complex, sister chromatid cohesion is partially impaired in hsk1-89, suggesting a possibility that infrequent origin firing of the mutant may affect the cohesin functions during S phase.
INTRODUCTION
DNA replication needs to be stringently regulated for cell growth and cell division to occur in a coordinated manner (Stillman, 1996). Initiation of DNA replication requires assembly of multiprotein complexes at chromosomal replication origins during late M to early G1 phase (Diffley et al., 1994; Newlon, 1997). This complex, termed prereplicative complex (preRC), includes ORC (Bell and Stillman, 1992), Cdc6 (Cocker et al., 1996), and MCM proteins (Tye, 1994; Kearsey et al., 1995; Chong et al., 1996; Donovan et al., 1997), which are conserved from yeasts to human. After cells pass Start, the preRC is activated and DNA synthesis is initiated at replication origins. This process is accompanied with dissociation of Cdc6 (Cocker et al., 1996; Tanaka et al., 1997) and of at least some components of the MCM complex from origins (Todorov et al., 1995; Aparicio et al., 1997; Kubota et al., 1997; Tanaka et al., 1997), resulting in a postreplicative complex (postRC), which is inactive until the next cell cycle. The firing of origins requires actions of at least two distinct serine/threonine kinases, namely, G1/S-specific CDK-Cyclin (Nasmyth, 1996; Stillman, 1996) and Cdc7-Dbf4 (Hartwell, 1971, 1973).
Saccharomyces cerevisiae CDC7 encodes a serine/threonine protein kinase required for the onset of DNA replication (Hollingsworth et al., 1992; Jackson et al., 1993). Genetic and biochemical evidence indicates that budding yeast Cdc7 triggers DNA replication during S phase by activating preRC assembled at each individual origin (Bousset and Diffley, 1998; Donaldson et al., 1998). For an active kinase, Cdc7, the catalytic subunit, requires Dbf4, the regulatory subunit, which is periodically expressed during the cell cycle (Chapman and Johnston, 1989; Jackson et al., 1993; Kitada et al., 1993; Cheng et al., 1999). Dbf4 also has been shown to interact with replication origins probably through other origin-binding proteins (Dowell et al., 1994).
Genes encoding Cdc7-related kinases have been isolated from the fission yeast, Xenopus, mouse, and human, indicative of the conserved functions of Cdc7-related kinases (Masai et al., 1995; Jiang and Hunter, 1997; Sato et al., 1997; Kim et al., 1998; Johnston et al., 1999; Masai et al., 1999). hsk1+ and dfp1+ (the same gene as him1+; Takeda et al., 1999) encode the catalytic and regulatory subunits, respectively, of the Cdc7-related kinase complex in the fission yeast. Both genes are essential for cell growth, and null mutants arrest with 1C DNA content (Masai et al., 1995; Brown and Kelly, 1998, 1999; Takeda et al., 1999). Hsk1 kinase activity is stimulated by association with Dfp1 and efficiently phosphorylates exogenous substrates as well as its own subunits in vitro. Whereas Hsk1 protein is expressed at a relatively constant level throughout the cell cycle, expression of Dfp1 protein is maximum at G1/S boundary through S phase and decreases at M/G1 boundary. Dfp1 is phosphorylated during S phase, probably reflecting autophosphorylation due to elevated kinase activity of Hsk1. Mutants of dfp1+ have been identified, which exhibit the defects in S/M checkpoint control induced by hydroxyurea (HU) as well as sensitivity to DNA damaging agents (Takeda et al., 1999). In keeping with this, dfp1+/him1+ is identical to rad35+. Among the proteins present in preRC, MCM2 has been suggested to be a major and conserved target of Cdc7 kinase (Lei et al., 1997; Sato et al., 1997; Brown and Kelly, 1998; Jiang et al., 1999; Kumagai et al., 1999; Takeda et al., 1999), although precise mechanisms of origin activation through Mcm phosphorylation are largely unknown.
To address the mechanism by which the Hsk1-Dfp1 kinase complex regulates G1/S transition and S phase progression and to explore any other physiological roles of this kinase in fission yeast, we have isolated and characterized a temperature-sensitive mutant of hsk1+. The isolated mutant hsk1-89 showed decreased intrinsic kinase activity and was defective in mitotic S phase. This mutant arrested its growth with 1C DNA content at 30°C and was not able to support Cdc18-induced overreplication, consistent with its essential role in initiation of DNA replication. Significant portion of the mutant cells entered mitosis with less than 1C DNA content, indicative of defect in replication checkpoint control. Unexpectedly, at 37°C, hsk1-89 cells arrested with near 2C DNA content and with abnormal nuclear structures and prematurely separated sister chromatids, and failed to enter mitosis. We have screened for the mutations that exhibit genetic interactions with hsk1-89, and identified cdc19-P1, rad3− and rad21-K1. We present here evidence showing that defects of hsk1-89 in initiation of S phase, checkpoint signaling, and maintenance of nuclear structures are linked to the functions of Cdc19, Rad3-Cds1, and Rad21 proteins, respectively.
MATERIALS AND METHODS
Yeast Strains, Media, and Genetics
Shizosaccharomyces pombe strains used in this study are summarized in Table 1. Cells were grown in rich (YE5S) or minimal (EMM) medium containing the required supplements. SPA or MEA was used for genetic crosses and sporulation. General genetic methods (Gutz et al., 1974; Moreno et al., 1991) and the procedure for transformation (Okazaki et al., 1990) were previously described. To induce expression from the nmt1 promoter (Maundrell, 1993), cells were grown to midexponential phase in EMM containing 10 μg/ml thiamine, washed three times with EMM, and then resuspended in fresh medium lacking thiamine at a density calculated to produce 1 × 107 cells/ml at the time of peak expression.
Table 1.
S. pombe strains used in this study
| Strain | Genotype | Derivations |
|---|---|---|
| JY319 | h− leu1-32 cdc24 | Our stock |
| JY741 | h− leu1-32 ura4-D18 ade6-M216 | Our stock |
| NT145 | h− leu1-32 | Our stock |
| NT63 | h− leu1-32 ura4-D18 ade6-M216 cdc2-33 | Our stock |
| NI65 | h− leu1-32 ura4 ade6-M216 cdc17 | Our stock |
| NI284 | h− leu1-32 ura4-D18 ade6-M216 cdc21 | T. Wang (Stanford University, Stanford, CA) |
| NI287 | h− leu1-32 ura4-D18 ade cdc18-K7 | T. Wang (Stanford University, Stanford, CA) |
| NI290 | h− cdc25-22 | T. Wang (Stanford University, Stanford, CA) |
| NI291 | h+ leu1-32 cdc19-P1 | Our stock |
| NI293 | h+ leu1-32 ura4-D18 ade6-M216 cdc22 | Our stock |
| NI319 | h− leu1-32 ura4-D18 ade6-M210 cdc19∷cdc19HA≫leu1+ | S. Forsburg (Salk Institute, USA) |
| NI322 | h− leu1-32 ura4-D18 ade6-M216 hsk1-89≫ura4+ | This study |
| NI355 | h− leu1-32 ura4-D18 nmt1-cdc18≫LEU2(int) | H. Nishitani (Kyushu University, Fukuoka, Japan) |
| NI358 | h+ leu1-32 ura4-D18 hsk1-89≫ura4+ | This study |
| NI359 | h− leu1-32 ura4-D18 hsk1-89≫ura4+ nmt1-cdc18≫LEU2(int) | This study |
| NI378 | h− leu1-32 ura4-D18 ade6-M216 cdc30 | P. Nurse (ICRF, Imperial Cancer Research Fund, London, UK) |
| NI392 | h− leu1-32 ura4-D18 ade6-M216 rad3∷ura4+ | Our stock |
| NI441 | h− leu1-32 ura4-D18 ade6-M216 rad21-K1≫ura4+ | Our stock |
| NI453 | h− leu1-32 ura4-D18 cds1∷ura4+ | K. Tanaka (University of Tokyo, Tokyo, Japan) |
| NI454 | h− leu1-32 cdc10-V50 | K. Tanaka (University Tokyo) |
| NI459 | h+ leu1-32 ura4-D18 ade6-M210 hsk1-89≫ura4+ chk1∷ura4+ | This study |
| NI462 | h+ leu1-32 ura4-D18 ade6-M210 hsk1-89≫ura4+ cdc22 | This study |
| NI464 | h− leu1-32 ura4-D18 ade6-M216 hsk1-89≫ura4+ cds1∷ura4+ | This study |
| NI465 | h+ leu1-32 ura4-D18 ade6 polα-ts13 | T. Wang (Stanford University) |
| NI466 | h+ leu1-32 ura4-D18 ade6 chkl∷ura4+ | This study |
| NI486 | h+ leu1-32 ura4-D18 hsk1-89≫ura4+ cdc19∷cdc19HA≫leu1+ | This study |
| NI496 | h+ leu1-32 ura4-D18 ade6-M210 hsk1-89≫ura4+ rad3∷ura4+ | This study |
| NI497 | h+ leu1-32 ura4-D18 ade6-M216 hsk1-89≫ura4+ rad21-K1≫ura4+ | This study |
| NI515 | h+ leu1-32 ade6-M210 hsk1-89≫ura4+ cdc19-P1 | This study |
| NI527 | h+ leu1-32 lys2+≫LacO his7+≫GFP-LacI-NLS | M. Yanagida (Kyoto University, Kyoto, Japan) |
| NI552 | h+ leu1-32 lys1+≫LacO his7+≫GFP-LacI-NLS hsk-89≫ura4+ | This study |
| NI562 | h+ leu1-32 lys1+≫LacO his7+≫GFP-LacI-NLS cdc25-22 | This study |
| NI563 | h+ leu1-32 lys1+≫LacO his7+≫GFP-LacI-NLS hsk-89≫ura4+ cdc25-22 | This study |
| NI581 | h− leu1-32 eso1-H17 | K. Tanaka (University of Tokyo) |
| NI625 | h+ ade6-M210 lysI+≫LacO his7+≫GFP-LacI-NLS cdc25-22 rad21-K1≫ura4+ | This study |
≫, marker gene (ura4+, leu1+ or LEU2) is integrated near the gene locus; int, indicates that the gene is integrated on chromosome.
Plasmid DNA and Polymerase Chain Reaction (PCR) Primers for Mutagenesis of hsk1+ Gene
pREP plasmids containing hsk1+ and dfp1+ were described previously (Masai et al., 1995; Takeda et al. 1999). DNA encoding hsk1-89 was amplified from total RNA prepared from hsk1-89 cells by reverse transcription-PCR and subcloned at NdeI-BamHI sites of pREP1, pREP41, or pREP81. For expression in insect cells, NdeI-BamHI fragment containing hsk1-89 was subcloned at SmaI-BglII site of pVL1393 after the NdeI site was blunt-ended by fill-in with the Klenow fragment. pUC19-Sp7GXR1 carrying the 4.6-kb XbaI-EcoRI hsk1+ genomic DNA (Masai et al., 1995) on pUC19 vector was used for subcloning of a 3.0-kb BamHI-EcoRI 3′-noncoding fragment by PCR as follows. PCR was conducted with a pair of primers, (hsk1-Bam-F; 5′-AATTTCGGATCCAGCAATCACTTAT-3′ and pUC-universal primer). A set of PCR primers (hsk1-Nde; 5′- AAACCGTTCATATGGCAGAGGCCC-3′ and hsk1-Bam-rev; 5′-GCTGGATCCGAAATTGCCAGCAAA-3′) was used for the mutagenesis of hsk1 open reading frame (ORF) (obtained as a NdeI-BamHI fragment; see below).
Isolation of Temperature-sensitive Mutant of hsk1+
The 1.5-kb NdeI-BamHI hsk1 cDNA fragment was amplified by PCR with the primer pair of hsk1-Nde and hsk1-Bam-rev in the presence of 0.2 mM MnCl2, the condition that significantly reduced the fidelity of nucleotide incorporation and induced misincorporation of nucleotides on the hsk1+ coding region. A mixture of mutated hsk1 fragments was cloned into pUC18. The BamHI-EcoRI 3′-noncoding fragment of hsk1+ into which a 1.8-kb ura4+ marker was inserted at the HindIII site (present at 0.8 kb downstream of the termination codon of hsk1+) was rejoined with mutated hsk1 (NdeI-BamHI) on pUC18. The resultant plasmids were digested with NdeI and EcoRI and the pool of mutated hsk1 fragments was directly introduced into ura4− haploid strain JY741. Stable Ura+ transformants were selected and the resultant 94 candidates were screened for temperature-sensitive phenotype on YE5S at 37°C. One strain (hsk1-89), which could not grow at 37°C, was selected, backcrossed with wild-type ura4− haploid strain, and used for further analysis.
Expression of Hsk1/Dfp1 Kinase Complex in Insect Cells and Purification of GST-MCM2N
Hsk1, Dfp1, and their derivatives were expressed on pVL1392 and pVL1393 plasmids in insect cells Sf9 was conducted as described previously (Takeda et al., 1999). Overexpression in Escherichia coli and purification of GST-SpMCM2N protein was performed as described (Ikeda et al., 1996; Takeda et al., 1999).
Antibodies
Anti-Hsk1 and anti-Dfp1 antibodies were prepared and used as described previously (Masai et al. 1995; Takeda et al., 1999). B-5-1-2 (Sigma, St. Louis, MO), and anti-hemagglutinin (HA) monoclonal antibody 12CA5 (Berkeley Antibody, Richmond, CA) were used to detect tubulin and HA-tagged fusion proteins, respectively.
Immunoprecipitation and Kinase Assays
One microgram of affinity-purified antibody or 10 μg of protein A-affinity column-purified antibody was added to the 200-μl extract (1 mg of protein), and protein–antibody complexes were collected onto protein A-Sepharose beads. After several washes of the beads with IP buffer (40 mM HEPES/KOH [pH 7.6], 100 mM potassium glutamate, 1 mM EGTA, 2 mM dithiothreitol, protease inhibitors [100 μg/ml TPCK, 0.1 μg/ml aprotinin, 0.5 μg/ml leupeptin, 0.1 μg/ml chymostatin, 1 μg/ml pepstatinA, and 10 μg/ml bacitracin], and 10% glycerol), they were run on SDS-PAGE for Western blotting or used for kinase assays. In vitro kinase assays were conducted as previously described (Takeda et al.; 1999) with a substrate protein indicated for each experiment. In-gel kinase assays were conducted as described previously (Geahlen et al., 1986; Waddell et al., 1995). Briefly, SDS-polyacrylamide gel (10%) was cast in the presence of 0.5 mg/ml myelin basic protein (Sigma) within the gel. Extracts (100 μg of protein) prepared by the boiling method were run on the gel. After electrophoresis, the gel was washed, denatured in 6 M guanidium hydrochloride, and renatured as described (Waddell et al., 1995). The gel was then equilibrated in the kinase buffer containing 40 mM HEPES/KOH (pH 7.6), 40 mM potassium glutamate, 5 mM magnesium acetate, 2 mM dithiothreitol, and 0.1 mM EGTA for 1 h at room temperature, and was incubated in the same kinase buffer containing 25 μM ATP and 100 μCi of [γ-32P]ATP for 30 min at room temperature, followed by extensive washing in 5% trichloroacetic acid and 1% sodium pyrophosphate. The gel was dried and autoradiographed.
Phosphatase Treatment
The immunoprecipitates, washed extensively with IP buffer, were resuspended in λ-phosphatase buffer containing 2 mM MnCl2 (New England BioLabs, Beverly, MA), and were divided into two equal aliquots. To one tube, 400 U of λ-phosphatase and 10 U of calf intestine alkaline phosphatase (New England BioLabs) were added, and both tubes were incubated for 1 h at 30°C.
Preparation of Extracts from Fission Yeast Cells and from Insect Cells
The whole cell extracts of fission yeast cells were prepared as follows. The harvested cells (from 50 ml of culture) were resuspended in 250 μl of water, boiled at 90°C for 5 min, and 300 μl of 2× concentrated Laemlli's SDS sample buffer containing 8 M urea and 500 μl of acid-washed glass beads was added. After vigorous vortex for 5 min, the samples were heated at 90°C for 5 min, sonicated for 1 min, and the supernatant was recovered by centrifugation. Concentrated extracts of fission yeast cells were prepared as previously described (Masai et al., 1995). Cell lysates were also prepared by vortexing the cells with glass beads in buffer containing 40 mM HEPES/KOH (pH 7.6), 1 mM EDTA, 0.5 M NaCl, 8 M urea, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride. Insect cells were infected with recombinant viruses at m.o.i. (multiplicity of infection) of 5, and after 3 d cells were harvested and extracts were prepared as described previously (Takeda et al., 1999).
Flow Cytometry, 4,6-Diamidino-2-phenylindole (DAPI) Staining, and Fluorescence Microscopy
Cells (5 × 106–1 × 107) were spun down, washed once with water, and then fixed in 70% ethanol. The fixed cells were treated with RNase A (0.1 mg/ml), stained with propidium iodide (2 μg/ml), and processed for flow cytometry by using Becton Dickinson FACScan, as described previously (Moreno et al., 1991). Fixed cells were stained with DAPI (0.1 μg/ml) (Takeda et al., 1999) and observed under fluorescence microscopy (OPTIPHOTO-2; Nikon, Tokyo, Japan). Detection of cen1-GFP signal(s) in the nucleus of S. pombe cells was conducted by using Realtime Digital Video Microscopy system (ARGUS-20; Hamamatsu Photonics, Hamamatsu City, Japan).
RESULTS
Isolation of a Temperature-sensitive Mutant of hsk1+
We previously reported that hsk1+ is essential for cell growth and is required for chromosomal DNA replication in S. pombe (Masai et al., 1995). To address physiological roles of hsk1+ in regulation of S phase as well as those at other phases in the cell cycle, we isolated a conditional lethal mutant of hsk1+. A 1.6-kb cDNA fragment containing the Hsk1 ORF was randomly mutagenized by PCR conducted in the presence of manganese and was rejoined with the 3′-noncoding region containing the S. pombe ura4+ as a selection marker. The linearized DNA containing the mutagenized ORF and ura4+ marker was directly introduced into a ura4− haploid strain to obtain alleles containing a mutagenized hsk1 and ura4+ gene in place of the chromosomal hsk1+. Among 94 stable Ura+ transformants selected at 25°C, one transformant (hsk1-89) could not grow at a high temperature (37°C). We confirmed that this temperature-sensitive (ts) phenotype was due to the mutations in the hsk1 gene by the following experiments. First, Southern analysis of the genomic DNA isolated from hsk1-89 mutant indicated that a ura4+ cassette was integrated at the expected site downstream of the hsk1 (our unpublished results). Second, sporulation of the heterozygous diploid cells constructed by a backcross of this mutant with the hsk1+ ura4− host strain produced no progeny, which segregated the ts and Ura+ phenotypes, indicating tight linkage of the hsk1 mutation with ura4+ marker (our unpublished results). Third, the ts phenotype was completely rescued by an ectopic plasmid, pREP81hsk1, which expressed hsk1+ under the control of the modified nmt1 promoter (Maundrell, 1993), but not with an empty vector (Table 2).
Table 2.
Complementation of hsk1-89 by plasmid carrying hsk1+ or dfp1+
| Strain | Plasmids | Growth
|
|
|---|---|---|---|
| 25°C | 37°C | ||
| wt (NT145) | pREP81 | +++ | +++ |
| hsk1-89 (NI358) | pREP81 | ++ | − |
| hsk1-89 | pREP81hsk1 | +++ | +++ |
| hsk1∷ura4+ | pREP81hsk1-89 | + | − |
| hsk1-89 | pREP41dfp1 | ++ | − |
| hsk1-89 | pREP1dfp1 | +++ | ++ |
Each transformant, grown at the permissive temperature (25°C) on a minimal plate, was replica-plated and was grown at the restrictive condition (37°C) for 5 d. Growth of each clone was examined and the extent of growth is expressed on semiquantitative scales of +++ (excellent growth equivalent to wild-type), ++ (fair growth), + (poor growth), and − (no growth).
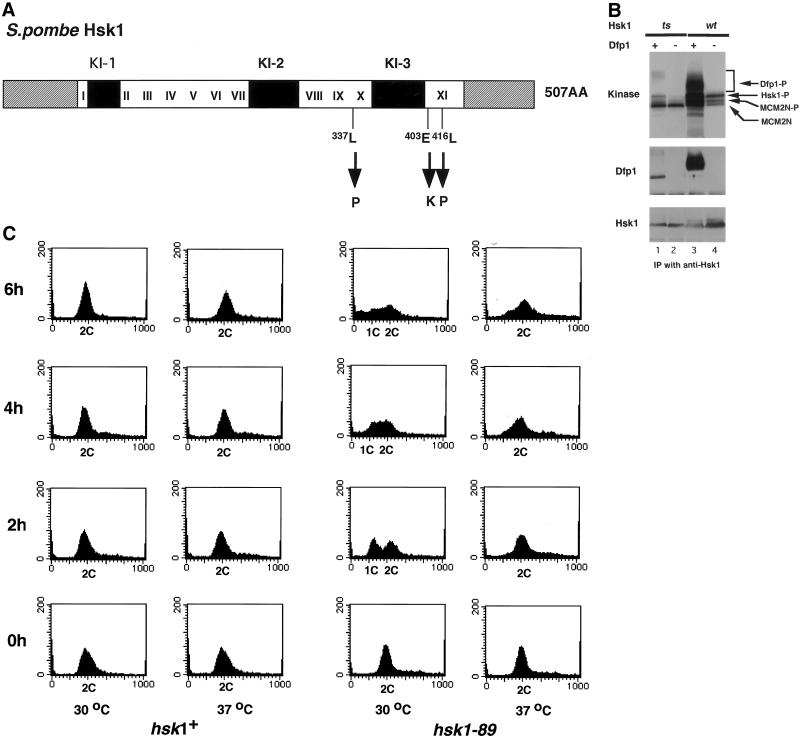
The hsk1-89 ORF was isolated from the ts mutant cDNA by reverse transcription-PCR and was subcloned into pREP81. The resulting pREP81hsk1ts rescued the lethal phenotype of hsk1 null strain only at 25°C but not at 37°C (Table 2), indicating that the amplified DNA fragment contained the ts mutation. Sequencing of the DNA fragment revealed that three amino acid residues (L337, E403, and L416) located in the C-terminal kinase domains X and XI were substituted in Hsk1-89 with proline, lysine, and proline, respectively (Figure 1A).
Figure 1.
Characterization of hsk1-89 strain and Hsk1-89 mutant protein. (A) Schematic drawing of the structure of Hsk1 protein and location of mutations in Hsk1-89. Amino acids, L337 (CTC), E403 (GAG), and L416 (CTC) were changed to P (CCC), K (AAG), and P (CCC), respectively, in Hsk1-89 mutant protein as indicated. White, black, and striped boxes indicate the conserved kinase domains (I-XI), nonconserved kinase inserts (KI-1, KI-2, and KI-3), and N-terminal and C-terminal tail regions, respectively. (B) Extracts were prepared from insect cells expressing Hsk1 (wild-type or mutant form) with (+) or without (−) Dfp1 protein as indicated in the figure, and immunoprecipitates prepared by anti-Hsk1 antibody were assayed in in vitro kinase reactions by using GST-SpMCM2N protein as a substrate (Takeda et al., 1999; top). The immunoprecipitates were also analyzed by Western blotting to detect Dfp1 and Hsk1 protein (middle and bottom). Hsk1-P, Dfp1-P, and MCM2N-P represent phosphorylated forms of Hsk1, Dfp1, and GST-SpMCM2N proteins, respectively, and MCM2N indicates the nonshifted (but phosphorylated) form of GST-MCM2N protein. (C) Wild-type (hsk1+) or hsk1-89 cells grown at 25°C were shifted to 30°C or 37°C and aliquots of cells were taken every 2 h and fixed. DNA contents of cells were analyzed by FACS.
Kinase Activity Is Reduced in hsk1-89 Mutant
The wild-type or mutant form of Hsk1 was expressed in insect cells singly or in combination with the regulatory subunit Dfp1 (Takeda et al., 1999). The immunoprecipitates prepared by anti-Hsk1 antibody were examined by Western blotting and kinase assays. The wild-type Hsk1 protein autophosphorylated in the absence of Dfp1 due to its intrinsic kinase activity (Figure 1B, lane 4; Takeda et al., 1999). This Hsk1 kinase activity was enhanced in the presence of Dfp1, resulting in a high level phosphorylation of both Hsk1 and Dfp1 (Figure 1B, lane 3; Takeda et al., 1999). Hyperphosphorylation of Dfp1 lead to appearance of mobility-shifted bands on SDS-PAGE (Figure 1B, lane 3). The phosphorylation of an exogenous substrate, GST-SpMCM2N containing the N-terminal 221 amino acids of S. pombe MCM2, was also greatly stimulated by the presence of Dfp1 (Figure 1B, lane 3; Takeda et al., 1999). In contrast, the mutant Hsk1 did not show any detectable level of autophosphorylation on its own, indicating that intrinsic kinase activity is impaired in hsk1-89. In the presence of Dfp1, only slight increase of autophosphorylation as well as Dfp1 phosphorylation was observed. Furthermore, the level of phosphorylation of the exogenous substrate was no more than the background level either in the presence or absence of Dfp1 (Figure 1B, lanes 1 and 2). Overexpression of Dfp1 partially suppressed growth defect of hsk1-89 at the restrictive temperature in a dose-dependent manner (Table 2), which is presumably due to increase of kinase activity of the mutant protein in the presence of excess amount of Dfp1 protein.
Defect of the hsk1-89 Mutant in Initiation of DNA Replication
The hsk1-89 mutant did not form colonies on a rich plate at temperatures >30°C. Fluorescence-activated cell sorter (FACS) analysis showed that 50% of hsk1-89 cells arrested with 1C DNA content at 2 h after shift-up to 30°C (Figure 1C), consistent with defect in G1/S transition, which was also observed in germinating spores carrying disrupted hsk1 (Masai et al., 1995). After prolonged incubation, the arrested cells committed to aberrant mitosis, resulting in generation of cut cells with <1C DNA content (Figures 1C and 3C, e) and their population reached 37% at 6 h after incubation at 30°C (Figure 3B, left).
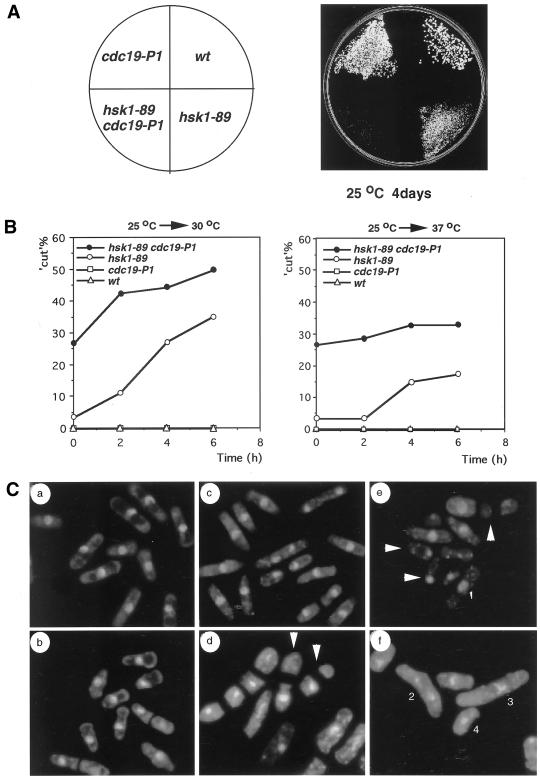
Figure 3.
Genetic interaction of hsk1-89 with cdc19-P1. (A) Growth of wild-type, cdc19-P1, hsk1–89, and hsk1-89 cdc19-P1 double mutant. Vegetatively growing cells of each strain were streaked on a rich plate and incubated at 25°C for 4 d. (B) Frequencies of cut cells in hsk1-89 and hsk1-89 cdc19-P1 after shift from 25 to 30°C (left) or to 37°C (right) were determined every 2 h. (C) Morphology of mutant cells stained with DAPI. (a) wild-type, 25°C, (b) cdc19-P1, 25°C, (c) hsk1-89, 25°C, (d) hsk1-89, cdc19-P1, 25°C, (e) hsk1-89, 6 h at 30°C, (f) hsk1-89, 6 h at 37°C. White arrows in d and e indicate cut cells. Abnormal chromosome structures (assigned by 1–4) in hsk1-89 at nonpermissive temperatures are indicated in e and f. Similar abnormal chromosomal morphologies of hsk1-89 are also shown in Figure 7B.
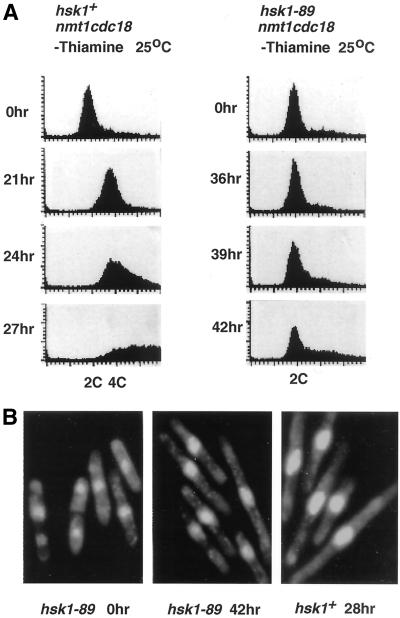
Fission yeast Cdc18, a homologue of S. cerevisiae Cdc6 (Cocker et al., 1996; Aparicio et al., 1997; Tanaka et al., 1997), is essential for the onset of S phase (Kelly et al., 1993) and is rapidly degraded after initiation of S phase by proteasome in a manner dependent on its phosphorylation by Cdc2 (Jallepalli et al., 1997; Lopez-Girona et al., 1998). Overexpression of Cdc18 protein in fission yeast inhibits G2/M transition and causes repeated rounds of DNA replication, resulting in swollen cells with polyploidy (Nishitani and Nurse, 1995). A haploid hsk1-89 or hsk1+ in which cdc18+ under the control of the nmt1 (Maundrell, 1993) was integrated into the chromosome was released from medium containing thiamine to that lacking thiamine at a permissive temperature to induce expression of Cdc18 protein. Changes in DNA contents were analyzed by flow cytometry. DNA content in the wild-type cells shifted from 2C to 4C at 21 h after induction and moved to even higher DNA content with concomitant appearance of elongated cells with larger nuclei, as previously reported (Figure 2, A and B; Nishitani and Nurse, 1995). In contrast, most cells stayed with 2C DNA content even at 36 h after induction in the hsk1-89 background (Figure 2A), although they started to elongate at 30 h due to G2 arrest caused by Cdc18 expression (our unpublished results). The DAPI staining of the hsk1-89 cells at 42 h and that of wild-type cells at 28 h after induction of Cdc18 indicated that the intensities of nuclear staining of hsk1-89 mutant cells were much less than that of the wild type (Figure 2B), consistent with the inhibition of overreplication in the mutant cells.
Figure 2.
Inhibition of Cdc18-induced overreplication in hsk1-89 mutant. (A) Cdc18 expression was induced from nmt1 promoter-driven cdc18+ integrated on the chromosome in hsk1+ or hsk1-89 at 25°C (Nishitani and Nurse, 1995). Aliquots were taken at the times indicated after shift to medium lacking thiamine and then analyzed by FACS. (B) hsk1-89 cells (center) at 42 h and hsk1+ cells (right) at 28 h after induction of Cdc18 protein were stained with DAPI and subjected to fluorescence microscopy to visualize nuclei. The extent of elongation of cell sizes is similar between the wild type and mutant. hsk1-89 cells (left) before induction of expression.
We have confirmed that Cdc18 protein can be overproduced in hsk1-89 carrying pREP3-cdc18 to the level similar to that observed in the wild-type cells containing the same plasmid (our unpublished results), indicating that expression and stability of Cdc18 protein is not affected by hsk1-89 mutation. FACS analysis indicated that most cells carrying hsk1-89 stayed with 2C DNA content at 28 h after induction under the same condition (our unpublished results), as was observed with the strain overexpressing Cdc18 from the chromosome. These results strongly suggest that Hsk1 kinase plays essential roles in initiation of DNA replication, consistent with its predicted crucial roles in activation of preRC generated by Cdc18 protein, and possibly in subsequent operation of replication checkpoint control (see later section).
Hsk1 Functions for Initiation of DNA Replication through Phosphorylating Cdc19
The hsk1-89 mutant grew at a permissive temperature more slowly than the wild type (Figure 3A) with a doubling time of 5 h. In release from G1 arrest, the transition through S to G2 phase was significantly delayed (our unpublished results), suggesting that S phase progression is partially defective even at a permissive temperature. To identify genetic interactions of Hsk1 with previously known cell cycle genes involved in G1/S transition, hsk1-89 was crossed with various cdc mutants (Table 3). Among them, hsk1-89 cdc19-P1 double mutant exhibited severe growth defect (Figure 3A and Table 3) and generated cut cells with higher frequency even at a permissive temperature (Figure 3B, left, and C, d). The frequency of cut cells reached ∼50% at 6 h after incubation at 30°C (Figure 3B, left).
Table 3.
Genetic interactions of hsk1-89 mutant with other mutations
| Additional mutation | Gene product | Functional point in cell cycle | Growth of double mutant at 25°C |
|---|---|---|---|
| cdc10-V50 | Transcription factor | late G1 | ++ |
| cdc18-K9 | MCM loading factor | G1/S | ++ |
| cdc19-P1 | MCM2 | G1/S | ± |
| cdc21 | MCM4 | G1/S | + |
| polα-ts13 | DNA polymerase I | early S | ++ |
| cdc22 | Ribonucleotide reductase | S | ++ |
| cdc24 | Reprication factor | S | ++ |
| cdc17 | DNA ligase | late S | ++ |
| cdc2-33 | Cyclin dept. kinase | G1/S and G2/M | ++ |
| cdc25 | Phosphatase | G2/M | ++ |
| rad3− | ATM homologue | Checkpoint | ± |
| cds1− | Checkpoint kinase | Checkpoint | + |
| chk1− | Checkpoint kinase | Checkpoint | ++ |
| rad21-K1 | Cohesin | Sister chromatid cohesion | ± |
| eso1-H17 | Ecol homologue | Establishment of cohesion | + |
Each double mutant cells were grown on YES plate at the permissive temperature (25°C) and growth was examined 4 d later. The extent of growth is expressed on semiquantitative scales of ++ (colony formation equivalent to hsk1-89 as shown in Table 1 and Figure 3A), + (less growth than hsk1-89; weak genetic interaction), ± (growth defect as shown in Figures 3A, 5B, or 7A; strong genetic interaction).
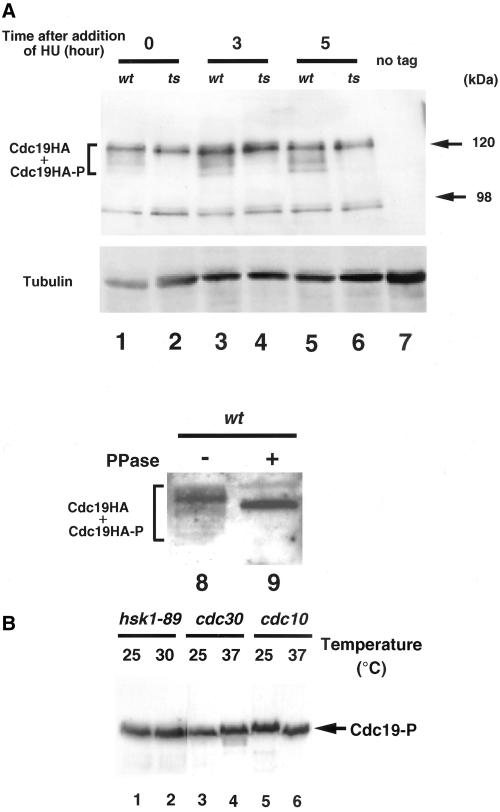
The above-mentioned results indicate that Hsk1 may play crucial roles in initiation of DNA replication of fission yeast by phosphorylating Mcm protein(s). We and others have reported that Mcm2 is specifically phosphorylated by Hsk1-Dfp1 kinase complex in vitro (Figure 1B; Brown and Kelly, 1998; Takeda et al., 1999). To examine whether Mcm2 is phosphorylated by Hsk1 in vivo, NI319 in which cdc19+ gene was replaced with HA-tagged cdc19+ (Sherman et al., 1998) was used to examine Cdc19/MCM2 protein in hsk1-89 cells. In extracts prepared from exponentially growing wild-type cells, ladders of bands that run faster on SDS-PAGE than the major bands were observed, whereas they were not detected in hsk1-89 grown under the same condition (Figure 4A, lanes 1 and 2). The intensities of these bands increased, when the extracts were prepared from the wild-type cells arrested with HU for 3 or 5 h (Figure 4A, lanes 3 and 5). Under the same conditions, these fast-migrating bands were nearly nondetectable in hsk1-89 (Figure 4A, lanes 4 and 6). These bands were not detected in nontagged strain (Figure 4A, lane 7), showing that they represent Cdc19/Mcm2 protein. To show that these extra bands are generated by phosphorylation, the immunoprecipitates with anti-HA antibody were pretreated by λ-phosphatas, and then were run on 8% SDS-PAGE. The fast migrating bands observed in the HU-treated wild-type extracts disappeared upon phosphatase treatment (Figure 4A, lane 9), indicating that they are indeed phosphorylated forms of Cdc19/Mcm2 protein. The phosphorylated forms of Cdc19/Mcm2 were also observed in HU-arrested cds1− cells at the same level as that in wild type (our unpublished results), indicating that Cds1 kinase, which is known to be activated in HU-treated cells, is not responsible for this phosphorylation. The mobility-shifted Cdc19/Mcm2 protein bands were observed also in a temperature-sensitive mutant of cdc30ts (encoding fission yeast Orc1) arrested at 37°C for 4 h but not in a cdc10ts mutant under the same condition (Figure 4B, lanes 3–6). Because cells are arrested at S phase in cdc30ts and at G1 in cdc10ts, this result is consistent with the prediction that Cdc19/Mcm2 protein is phosphorylated during S phase when Hsk1 kinase activity is high, but not in G1 phase when it is the lowest. When the hsk1-89 cells were arrested with 1C DNA content at 30°C for 2 h, the mobility-shifted forms of Cdc19/Mcm2 did not appear, consistent with the requirement of Hsk1 for S phase-specific phosphorylation of Cdc19/Mcm2 (Figure 4B, lanes 1 and 2). These results strongly indicate that Cdc19/Mcm2 protein is phosphorylated by Hsk1 in vivo during S phase and that this phosphorylation plays a critical role in initiation of DNA replication.
Figure 4.
Inability of Hsk1-89 to phosphorylate Cdc19/MCM2 protein in vivo. (A) Phosphorylation of Cdc19/MCM2 protein in vivo. Vegetatively growing (5 × 106 cells/ml) hsk1+ (NI319; lanes 1, 3, 5, 8, and 9) or hsk1-89 (NI486; lanes2, 4, 6, 10, and 11) carrying HA-tagged cdc19+ on the chromosome under its own promoter was arrested with 10 mM HU for 3 h (lanes 3 and 4) or for 5 h (lanes 5, 6, 8, and 9). Cell extracts were prepared by the regular glass bead method and run on an 8% SDS-PAGE. Cdc19 protein was immunoprecipitated by anti-HA antibody and was treated with λ-phosphatase before electrophoresis (lane 9) or untreated (lane 8). The extract from the wild-type (nontagged) cells (NT145) was run as a negative control (lane 7). Blotting was conducted by using anti-HA antibody. In lanes 8 and 9, the samples were run on a long gel (30 cm in length) at 500 V for 12 h at 4°C to maximize the separation of the phosphorylated forms. The lower panel for lanes 1–7 is the blotting with anti-tubulin antibody for loading control on a separate gel on which the same amount of each extract was run. (B) Phosphorylation of Mcm2 (Cdc19) in cdc mutants and hsk1-89 arrested at the nonpermissive temperature. hsk1-89 cells grown at 25°C (lane 1) were arrested at 30°C for 2 h (lane 2) and cdc30ts or cdc10ts cells vegetatively growing at 25°C (lanes 3 and 5) were arrested at 37°C for 4 h (lanes 4 and 6). Cell extracts were prepared and phosphorylation of Mcm2 (Cdc19) was analyzed as described in A, except that anti-Cdc19 antibody (a gift from Dr. S. Forsburg) was used.
Multiple bands for Cdc19/Mcm2 appear on SDS-PAGE, which was run under the condition to maximize the separation of phosphorylated forms (Figure 4A, lane 8). Upper bands migrating slowly on SDS-PAGE may also be phosphorylated forms of Cdc19/Mcm2. Because they can be detected also in hsk1-89, they may represent Cdc19/Mcm2 proteins phosphorylated by kinases other than Hsk1. We have recently shown that concerted action of Cdk is required for efficient phosphorylation of MCM complex by human Cdc7 in vitro (Masai et al., 2000). Phosphorylation of MCM by Cdk in vivo and in vitro and its effect on chromatin association and helicase activities have been reported (Hendrickson et al., 1996; Ishimi et al., 2000). It is likely that functions of MCM are under complex regulation of multiple sets of kinases during cell cycle.
Replication Checkpoint Signaling Is Defective in hsk1–89 Mutant
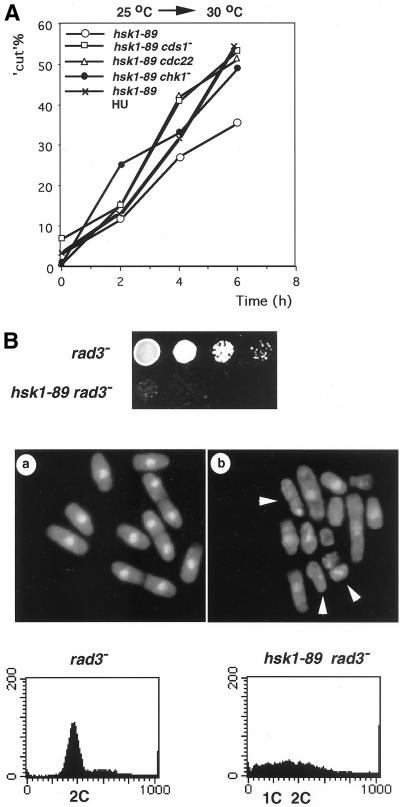
A number of gene products essential for the initiation of DNA replication (such as Cdc18, Cdt1, Orp1, Polα, and Rad4/Cut5) are known to be required also for induction of replication checkpoint signals, because loss or inactivation of these proteins causes an aberrant mitosis without DNA replication (Kelly et al., 1993; Saka and Yanagida, 1993; Hofmann and Beach, 1994; Saka at al., 1994; Grallert and Nurse, 1996; Bhaumik and Wang, 1998). Formation of cut cells in hsk1-89 at 30°C, which was also seen in hsk1 null mutants albeit with lesser frequency (Masai et al., 1995), is likely to be caused by defect in the same checkpoint control generated by the block in initiation of DNA synthesis. The cut cell formation was further enhanced in the cdc22 background or in the presence of 10 mM HU at 30°C (Figure 5A), suggesting that Hsk1 kinase activity may also play a role in DNA replication checkpoint control induced by nucleotide deprivation, which causes cells to arrest at early S phase.
Figure 5.
Defect of hsk1-89 in DNA replication checkpoint control. (A) Enhancement of cut cell formation in hsk1-89 by the presence of an additional mutation or by early S phase arrest with HU. Cells from strains indicated were grown at 25°C and were shifted to 30°C for 6 h (○, □, ▵, ●). hsk1-89 cells were shifted to 30°C in the presence of 10 mM HU (x; hsk1–89 HU). The number of cut cells was counted as described in Figure 3B. (B) Genetic interaction of hsk1-89 with rad3 deletion. rad3− cells were grown to 5 × 106 cells/ml at 25°C in YES liquid medium. hsk1-89 rad3− cells, which had doubling time of >6 h in YES at 25°C, were grown to 5 × 105 cells/ml, and then were concentrated to 5 × 106 cells/ml by centrifugation. Fivefold serial dilution of each cell suspension (5 × 106 cells/ml) was spotted onto a rich plate, which was incubated at the permissive temperature for 4 d (top). Cells were fixed, stained with DAPI, and examined under fluorescent microscopy (middle), or harvested and analyzed for DNA content by FACS (bottom). Arrowheads indicate examples of cut cells.
To investigate genetic interactions of hsk1+ with various checkpoint genes, double mutants between hsk1-89 and checkpoint mutants were constructed by genetic cross (Table 3). Most strikingly, hsk1-89 rad3− double mutant formed only microcolonies on a rich plate even at a permissive temperature (Figure 5B, top, and Table 3). Sixty percent of this double mutant displayed cut morphology (Figure 5B, middle), indicative of aberrant mitosis before completion of DNA replication. Consistent with this prediction, DNA content of the double mutant was highly heterogeneous, ranging from 2C to less than 1C (Figure 5B, bottom). These results also indicate that survival of hsk1-89 depends on checkpoint function of Rad3. In contrast, deletion of cds1+ or chk1+ did not significantly affect the viability of hsk1-89 at the permissive temperature (Table 3), although cut cell formation at 30°C was enhanced in both cds1− and chk1− backgrounds (Figure 5A). This suggests that these gene products contribute to the checkpoint control pathway, but that presence of either protein may be sufficient for maintaining the growth of hsk1-89 at the permissive temperature, consistent with the finding that Cds1 and Chk1 jointly enforce the replication checkpoint responses (Boddy et al., 1998; Zeng et al., 1998).
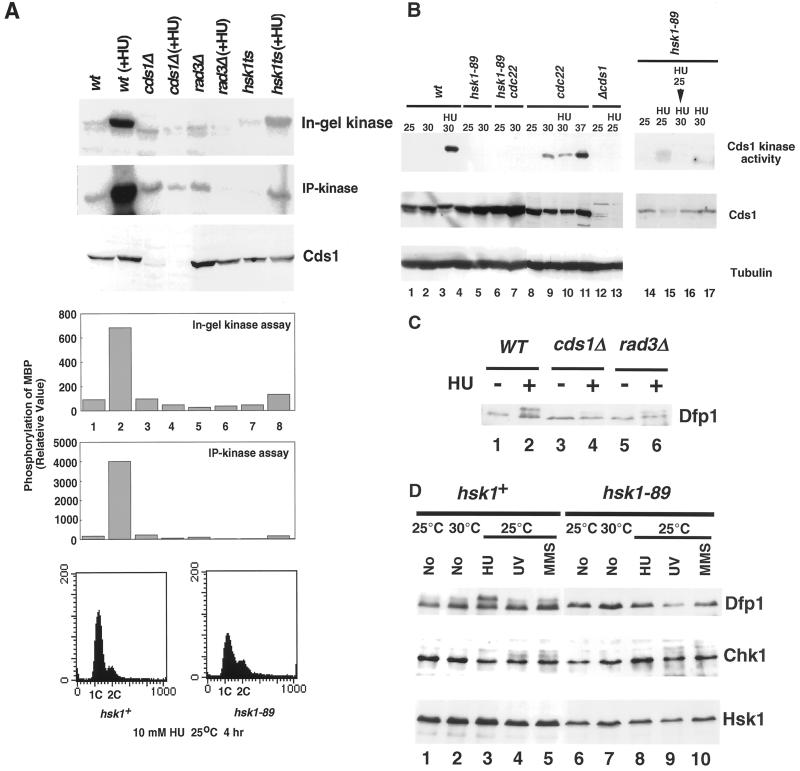
To investigate whether replication checkpoint signaling normally induced by early S phase arrest is defective in hsk1-89 mutant, Cds1 kinase activity, which is known to be specifically activated in response to early S phase arrest by nucleotide deprivation or in some S phase mutants (Lindsay et al., 1998), was measured by in-gel kinase assays by using the whole cell extract separated on a polyacrylamide gel polymerized in the presence of a substrate protein, myelin basic protein (MBP; Pellicioli et al., 1999). In this assay, ∼7-fold stimulation of Cds1 kinase activation was observed, when hsk1+ cells were arrested with HU at 25°C for 4 h (Figure 6A, top, lanes 1 and 2; Lindsay et al., 1998). Under this condition, 80% of cells arrested with 1C DNA content (Figure 6A, bottom). As expected, activation of Cds1 was not observed in cds1− as well as in rad3− cells (Figure 6A, top, lanes 3–6). In hsk1-89 cells, the basal level of Cds1 kinase activity was similar to that in the wild type, and only slightly increased after exposure to HU at the permissive temperature (Figure 6A, top, lanes 7 and 8). Seventy percent of hsk1-89 cells was arrested with 1C DNA content under this condition (Figure 6A, bottom). We also conducted standard in vitro kinase assays by using anti-Cds1 immunoprecipitates and MBP. Cds1 kinase activity in HU-treated hsk1-89 was >20-fold decreased compared with that of the wild-type cells (Figure 6A, middle panel and lower graph). Week but detectable activation of Cds1 kinase in hsk1-89 may be sufficient for maintaining replication checkpoint control induced by HU at the permissive temperature, because the frequency of cut cell formation in hsk1-89 at 10 h after treatment with HU at 25°C is low (<5%). However, when hsk1-89 cells arrested with HU at 25°C for 2 h were further incubated with HU at 30°C for 2 h, Cds1 kinase activity further decreased and small cut cells, similar to those observed in rad3− in the presence of HU, appeared with high frequency (Figure 6B, lanes 15 and 16; our unpublished results). Cds1 kinase is also activated when cells are arrested at early S phase by cdc22 mutation (Lindsay et al., 1998). In-gel kinase assays also detected Cds1 kinase activation in a cdc22 mutant at 30°C or 37°C (Figure 6B, lanes 9–11). The Cds1 kinase was not activated in hsk1-89 cdc22 double mutant at 30°C (Figure 6B, lanes 6 and 7), supporting our conclusion that Hsk1 is required for Cds1 kinase activation in response to early S phase arrest by nucleotide deprivation. These results indicate the possibility that Hsk1 plays an important role in Rad3-mediated Cds1 activation, which plays a central role in trigger of DNA replication checkpoint control.
Figure 6.
Defect of hsk1-89 in Cds1 kinase activation in response to nucleotide deprivation. (A) Cds1 kinase activity in various strains in the presence or absence of HU. (Top) Extracts were prepared by the boiling method (Takeda et al., 1999) from the strains indicated, which were grown at 25°C with or without 10 mM HU-treatment for 4 h, and in-gel kinase assays were conducted as described in MATERIALS AND METHODS. (Middle) Extracts were prepared from the same set of the strains at a low salt by glass beads disruption. Cds1 protein was immunoprecipitated by anti-Cds1 protein antibody from the extracts in the presence of 0.5 M NaCl, and the immunoprecipitates were used for in vitro kinase assays with 10 μg of MBP as a substrate. The reaction mixtures were run on 10% SDS-PAGE and phosphorylated MBP are shown. (Bottom) Extracts used for in-gel kinase assays were analyzed by Western blotting using anti-Cds1 antibody to detect Cds1 protein. In the upper graph, the extent of phosphorylation in in-gel kinase assay was quantified. In the middle graph, the extent of MBP phosphorylation in IP-kinase assay was quantified. The presence of approximately the same amount of Cds1 protein in the immunoprecipitates was confirmed (our unpublished results). Weak phosphorylation of MBP observed in cds1Δ and rad3Δ extracts is due to the presence of nonspecific kinases in the immunoprecipitates. In the bottom panel, DNA contents of HU-arrested hsk1+ or hsk1-89 cells were analyzed by FACS. (B) Cds1 kinase activity was measured by in-gel kinase assays in strains indicated, which were grown at the temperature shown (upper panel). Cells were arrested with 10 mM HU for 4 h (lanes 3, 10, 13. 15, 16, and 17). In lane 16, cells were grown at 25°C for 2 h and then shifted to 30°C for 2 h. Cds1 or α-tubulin protein was analyzed by immunoblotting (middle and lower panels). (C) HU-induced hyperphosphorylation of Dfp1 protein in various strains. Strains indicated were grown at 30°C. Ten millimolar HU was added to half of the cells and incubation was continued for another 4 h. Dfp1 protein was analyzed by Western blotting. (D) hsk1+ or hsk1-89 cells were grown at the temperature indicated without any treatment (No; lanes 1, 2, 6, and 7), treated with 10 mM HU for 4 h (HU; lanes 3 and 8), exposed to UV at 150 J/m2 followed by incubation in rich medium for 1 h (UV; lanes 4 and 9), or treated with 0.05% MMS for 4 h followed by incubation in rich medium for 1 h (MMS, lanes 5 and 10). Dfp1 protein (upper panel), Chk1 protein (middle panel; HA-tagged), or Hsk1 protein (lower panel) was detected by Western blotting. In C and D, extracts were prepared by the boiling method.
In contrast, Chk1 kinase activation, which occurs specifically during DNA damage checkpoint responses and is exemplified by the appearance of a mobility-shifted form of Chk1 protein, was normal in both hsk1+ and hsk1-89 cells (Figure 6D), indicating that Hsk1 is specifically required for DNA replication checkpoint responses. This is consistent with absence of cut cells after treatment of hsk1-89 cells with UV or methyl methane sulfonate (MMS) at a permissive temperature (our unpublished results).
Hyperphosphorylation of Dfp1 in HU Arrest Depends on Hsk1 Protein
We and others reported previously that Dfp1 protein is phosphorylated in a cell cycle-dependent manner. Mobility-shifted forms of Dfp1 appear during S through G2 phases (Brown and Kelly, 1999; Takeda et al., 1999). Dfp1 undergoes additional phosphorylation after early S phase arrest with HU and this phosphorylation depends on Cds1 kinase (Figure 6C, lanes 1–4; Brown and Kelly, 1999; Takeda et al., 1999). We found that this hyperphosphorylation did not occur in rad3− (Figure 6C, lanes 5 and 6), showing that HU-induced hyperphosphorylation of Dfp1 depends on the Rad3-Cds1 pathway.
We further examined whether these phosphorylations depend on Hsk1. In hsk1+ cells, mobility-shifted forms of Dfp1, which probably represent phosphorylated forms in G2 phase, were detected in asynchronous cell populations (Figure 6D, lanes 1 and 2). After arrested with HU, a distinct slow-migrating band appeared on SDS-PAGE (Figure 6D, lane 3). This hyperphosphorylated form of Dfp1 did not appear after treatment of the cells with UV or MMS (Figure 6D, lanes 4 and 5), indicating that it is specifically caused during DNA replication checkpoint responses but not during DNA damage checkpoint responses. In the hsk1-89 background, these shifted bands were not detected (Figure 6D, lanes 6–10), indicating that both cell cycle-regulated phosphorylation and HU-induced hyperphosphorylation require Hsk1. HU-induced hyperphosphorylation depends not only on Hsk1 but also on Rad3/Cds1 (Figure 6, C and D; Brown and Kelly, 1999). How can one reconcile these results? In the light of our finding that Cds1 activation after HU-induced arrest requires Hsk1 (Figure 6A), it would be an intriguing possibility that early S phase arrest, leading to transient activation of Hsk1 kinase, which in turn activates Cds1 kinase, which then causes hyperphosphorylation of Dfp1 protein in a feedback loop.
hsk1–89 Displays Apparent Defect in Mitosis, Which May Be Caused by Impaired Functions of Rad21 Cohesin
Incubation of hsk1-89 at 37°C resulted in appearance of cells with heterogeneous DNA content centered around 2C, and the majority of the cells did not pass through the first mitosis and only a small portion of the cells became 1C DNA after prolonged incubation (Figure 1C). Population of cut cells increased only up to 15% at 6 h at 37°C, and the high basal level of cut cells observed in cdc19-P1 hsk1-89 double mutant did not further increase upon shift to 37°C (Figure 3B, right). Strikingly, ∼30% of the population displayed condensed, scattered, or unequally segregated chromosomes at 6 h after shift to 37°C (Figure 3C, f). We did not observe any abnormal morphology of hsk1+ cells at 37°C (our unpublished results). The results indicate that hsk1-89 mutant exhibits apparent defect in mitosis at this temperature.
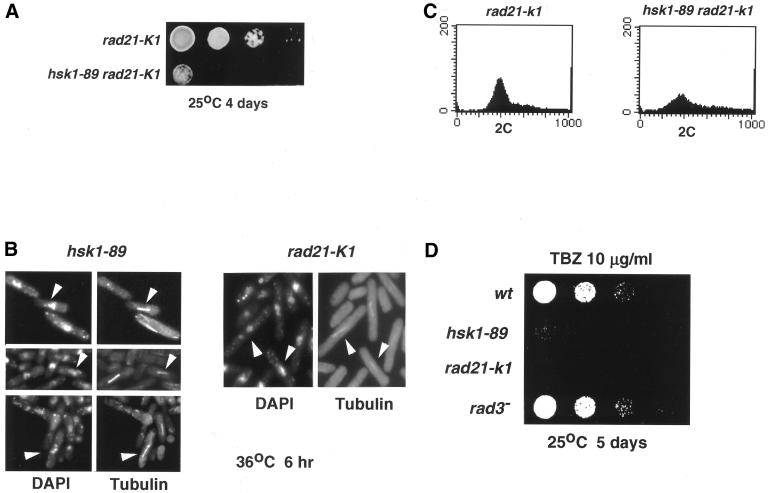
In survey of various mutants that show genetic interactions with hsk1-89, we discovered that a rad21-K1 hsk1-89 double mutant exhibits severe growth defect at a permissive temperature (Figure 7A and Table 3). Milder growth defect was also detected at the permissive temperature in double mutant between hsk1-89 and eso1-H17, which encodes mutant form of Eco1 (establishment of cohesion)/Ctf7 homologue in S. pombe (Tanaka et al., 2000; Table 3). The rad21-K1 mutant has been shown to be associated with pleiotropic defect such as uneven segregation of chromosomes, deficiency of DNA repair and loss of microtubule functions (Tatebayashi et al., 1998). At the permissive temperature, rad21-K1 hsk1-89 displayed various aberrant nuclear structures such as condensed chromosomes with short spindles and chromosomes stretched or unequally separated by elongated spindles, which are frequently observed in both hsk1-89 and rad21-K1 at 36–37°C (Figures 3C, f and 7B; Tatebayashi et al., 1998). The DNA content of this double mutant was similar to those of hsk1-89 at 37°C (Figures 1C and 7C). Moreover, like the rad21-K1 mutant (Tatebayashi et al., 1998), hsk1-89 was moderately sensitive to thiabendazole (TBZ), which is known to bind tubulin and inhibit its polymerization (Figure 7D), suggesting that hsk1-89 also affects microtubule functions. These results suggest that the mitotic defect in hsk1-89 may be caused by aberrant nuclear structures resulting from inefficient functions of Rad21 protein during S phase.
Figure 7.
Requirement of Hsk1 in maintenance of proper chromatin structures. (A) Genetic interaction between hsk1-89 and rad21-K1. Fivefold serial dilution of exponentially growing cultures indicated were spotted onto a rich plate, which was incubated at the permissive temperature. (B) Abnormal nuclear structures and tublin distributions in hsk1-89 (left) and rad21-K1 (right). Vegetatively growing cells were arrested at 36°C for 6 h and indirect immunofluorescence was conducted using anti-tublin. DAPI staining was conducted to visualize nuclei. Upper, middle, and lower panels of hsk1-89 represent aberrant nuclear structures such as condensed chromosomes with short spindles, abnormally stretched chromosomes, and those unequally separated by elongated spindles, respectively (indicated by white arrows). In this experiment, cells were shifted to 36°C to visualize the immunostained cells more clearly. Patterns of immunostaining and nuclear morphology at 36°C were very similar to those at 37°C (Figure 3C, f). (C) DNA content in rad21-K1 and hsk1-89 rad21-K1 double mutant. Cells grown at 25°C were harvested and analyzed for DNA content by FACS. (D) TBZ sensitivity of hsk1-89 mutant. Fivefold serial dilution of growing cells indicated were spotted onto a rich plate containing TBZ (10 μg/ml) and were incubated at the permissive temperature.
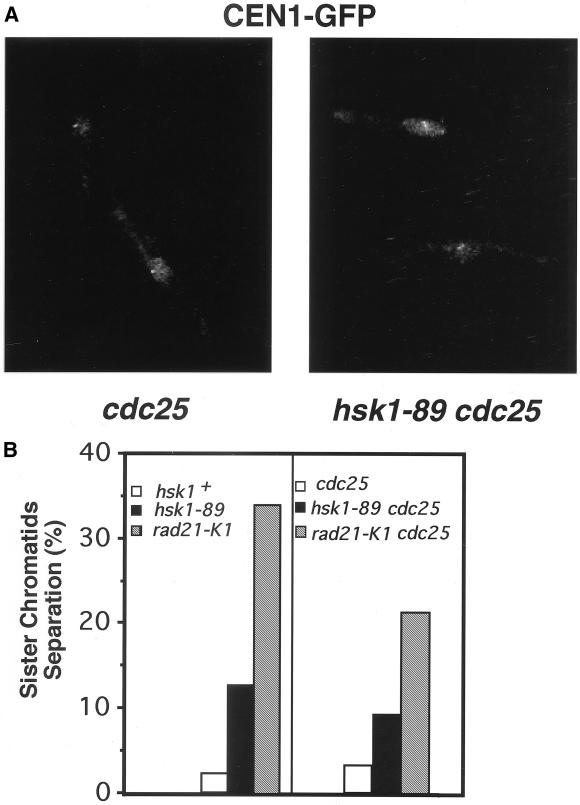
Scc1/Mcd1/Rhc21, the Rad21 homologue in S. cerevisiae, forms the complex called cohesin, which plays essential roles in sister chromatid cohesion during S phase through anaphase (Guacci et al., 1997; Michaelis et al., 1997). Therefore, sister chromatid cohesion may be defective in the hsk1-89 mutant cells at 36°C due to the impaired function of Rad21 protein. To examine this possibility, we constructed a hsk1-89 mutant strain carrying the cen1-GFP probe (NI552), in which behavior of centromeric DNA can be visualized with GFP-LacI-NLS, which binds to the clusters of the lacO (lac operator) sequences integrated near the cen1 (Nabeshima et al., 1998). When the exponentially growing hsk1+ cells (NI527) were shifted to 36°C for 5 h, 2.6% of cells in interphase or before mitosis exhibited split cen1-GFP signals in a single nucleus (Figure 8B, left). In contrast, untimely split cen1-GFP signals were detected in the nucleus of 12.7% of hsk1-89 cells (NI 552) under the same condition (Figure 8B, left). The premature separation of sister chromatids was observed in the nucleus of 34% of rad21-K1 mutant cells at the restrictive temperature (Figure 8B, left). To prevent cells from transition to anaphase when sister chromatids start to separate, we constructed rad21-K1 cdc25 or hsk1-89 cdc25 double mutant carrying the cen1-GFP probe to arrest the cells at G2/M transition. Whereas 3.9% of hsk1+ cdc25 cells (NI 562), which had been arrested at 36°C for 5 h, had the nucleus with the split signals (Figure 8, A and B, right), 22% of rad21-K1 cdc25 (NI625) cells treated in a similar manner exhibited sister chromatid separation in the nucleus. In hsk1-89 cdc25 cells (NI 563), sister chromatid separation was detected in 8.9% of the population (Figure 8, A and B, right). These results are consistent with partial defect in sister chromatid cohesion in the hsk1-89 mutant at 36°C.
Figure 8.
Premature sister chromatid separation in hsk1-89. Vegetatively growing hsk1+ or hsk1-89 cells carrying the GFP-LacI-NLS, which can bind to the lacO repeats integrated near the cen1 (Nabeshima et al., 1998), were arrested at 36°C for 5 h, fixed with methanol, and then subjected to fluorescence microscopy. (A) Nonsplit cen1-GFP signal in cdc25 cell (left) and a prematurely split cen1-GFP signals in hsk1-89 cdc25 cells (right). (B) Fractions of cells with split cen1-GFP signals are plotted for each strain indicated. The GFP signals in >100 nuclei for each cdc25+ strain (left), and those in >300 nuclei for each strain carrying cdc25 mutation (right) were examined. Populations of binucleate cells after cdc25 block (right) in hsk1+, hsk1–89, and rad21-K1 were 9.0, 2.4, and 3.9%, respectively, consistent with synchronistic arrest in G2/M boundary in these cdc25 strains under this condition. The above-mentioned experiments were conducted at 36°C for clearer visualization of cen1-GFP signals.
DISCUSSION
Cdc7-related kinases are evolutionary conserved from yeasts to human (Masai et al., 1995; Jiang and Hunter, 1997; Sato et al., 1997; Kim et al., 1998) and are expected to play key roles in initiation of DNA replication (Hartwell, 1971; Sclafani and Jackson, 1994). In this communication, we attempt to understand roles of Cdc7-related kinase in G1/S transition as well as its other important physiological functions by characterizing a newly isolated temperature-sensitive hsk1 mutant.
Essential Functions of Hsk1 for Initiation of S Phase
At 30°C, ∼50% of hsk1-89 cells arrest with 1C DNA content and eventually produce cut cells, indicating that the mutant is not able to enter S phase and shows replication checkpoint defect phenotype. The mutation also abrogated overreplication induced by a high-level expression of Cdc18, which is known to require Mcm and Orc functions as well (Kelly et al., 1993; Nishitani and Nurse, 1995; Nishitani, personal communication). Requirement of Hsk1 for Cdc18-induced overreplication lends further support for the idea that phosphorylation events by Hsk1 is strictly required for activation of preRC generated by Cdc18 and Mcm proteins, and clearly demonstrates that Hsk1 is essential for initiation of S phase.
It has been reported that the major substrate of Cdc7 and its related kinases in vitro is Mcm2 (Lei et al., 1997; Sato et al., 1997; Brown and Kelly, 1998; Kumagai et al., 1999; Takeda et al., 1999). We found severe growth defect of hsk1-89 in combination with cdc19-P1 encoding Cdc19/Mcm2 protein in fission yeast. Furthermore, we have detected in vivo phosphorylation of Cdc19/Mcm2 protein, causing its downward mobility-shift on SDS-PAGE, and this phosphorylation was not detected in hsk1-89 cells, consistent with very low kinase activity of the mutant protein in vitro. The level of this phosphorylation is specifically enhanced during S phase, but can be detected also during G2 phase, because it could be detected in exponentially growing culture where cells are mostly in G2 phase (Figure 4A, lane 1). It appears to be gone during G1 phase, because no mobility-shift was detected in cdc10 arrested cells. The timing of the appearance of the mobility-shifted forms of Cdc19 during cell cycle is largely consistent with the oscillation of Hsk1 kinase activity. This strongly indicates that Cdc19/Mcm2 is the physiologically important substrate of Hsk1. Only a portion of Cdc19/Mcm2 was detected as mobility-shifted phosphorylated forms even though cells were arrested in S phase (Figure 4A). This may indicate that only the Cdc19/Mcm2 protein associated with active replication origins may undergo S phase-specific phosphorylation. It is known that Mcm2 is present in excess over the number of replication origins. Similar S phase-specific phosphorylation of Mcm2 by Cdc7 kinase was previously reported in S. cerevisiae (Lei et al., 1997). Our results are in agreement with the notion that phosphorylation of Cdc19/Mcm2 protein by Hsk1 kinase is essential for the origin firing in the fission yeast.
Recent biochemical analysis indicated that helicase activity of human Mcm4-6-7 complex is inhibited by Mcm2 (Ishimi, 1997; Ishimi et al., 1998). It is of interest to determine whether phosphorylation of Mcm2 by Cdc7-related kinase affects its inhibitory effect on MCM helicase activity. Mapping of Hsk1-mediated phosphorylation site(s) of Cdc19/Mcm2 and genetic and biochemical analyses of the phosphorylation mutants will be needed to elucidate detailed molecular mechanisms of origin activation.
Hsk1 May Play Crucial Roles in the Rad3/Cds1-dependent DNA Replication Checkpoint Signaling Pathway
Significant fractions of the mutant cells, after being incubated at 30°C for 4 h, undergo premature mitosis, resulting in appearance of cut cells with less than 1C DNA content. This may represent uncoupling of S and M phases frequently observed in mutants defective in initiation of DNA replication such as cdc18, cdc21, cdt1, orp1, polα, and cut5 (Kelly et al., 1993; Saka and Yanagida, 1993; Hofmann and Beach, 1994; Saka et al., 1994; D'Urso et al., 1995; Grallert and Nurse, 1996; Maiorano et al., 1996). The aberrant mitosis at 30°C is enhanced by additional arrest at early S phase by a cdc22 mutation or by addition of HU. These results may indicate that checkpoint responses by nucleotide deprivation are also partially defective in hsk1-89. Similar defect in HU-induced replication checkpoint control was observed in mutants of dfp1+ encoding a regulatory subunit of Hsk1 kinase (Takeda et al., 1999).
We have discovered that activation of Cds1, a major mediator of replication checkpoint control downstream of Rad3, was severely impaired in the hsk1-89 mutant, indicating that Cds1 kinase activation requires Hsk1 kinase activity. This is consistent with the fact that Cds1 activation occurs only in S phase (Brondello et al., 1999), because Hsk1 kinase is active during S phase (Brown and Kelly, 1999; Takeda et al., 1999). Thus, Hsk1 kinase may play crucial roles in transmission of checkpoint signals from preRC or initiation complex to Rad3/Cds1 pathway. The cut cell formation of hsk1-89 was significantly enhanced by the presence of either chk1− or cds1− at 30°C or by that of rad3− at the permissive temperature. These results are consistent with the possibility that Hsk1 functions upstream of these checkpoint kinases or directly contributes to Cds1 activation in a pathway parallel to Rad3-Cds1. It is of interest whether Hsk1 modulates Rad3 and/or Cds1 kinase activity through direct phosphorylation.
We cannot entirely rule out the possibility that inefficient origin firing in hsk1-89 may somehow lead to insufficient generation of specific “DNA structures” even in the presence of HU, which are prerequisite for activation of Rad3-Cds1. However, we observed <20-fold reduction of the Cds1 kinase activity even in hsk-89 grown at 25°C (Figure 6A, second panel, lane 8). Inefficient origin firing of hsk1-89 at 25°C, if any, is not likely to explain this much reduction of Cds1 activation. We also did not observe any defect of Cds1 activation in response to HU in other mutants that may affect origin firing, such as cdc21(MCM4) and cdc30(orp1) (our unpublished results). Therefore, we favor the conclusion that Hsk1 is more specifically involved in Cds1 activation in response to nucleotide deprivation.
Combination of hsk1-89 and rad3− mutation results in severe growth retardation apparently caused by premature mitosis before completion of DNA replication. Mitosis may be coordinated with prolonged S phase of hsk1-89 cells through Rad3-mediated restraint of M phase. In the absence of Rad3 protein, mitosis may not be restrained due to complete defect in checkpoint control and premature mitosis would occur, resulting in cut cells with highly heterogeneous DNA content.
At this point, however, we cannot rule out the possibility that Rad3 protein is positively required for firing of origins in conjunction with Hsk1 and that enhanced defect in initiation in the double mutant leads to premature mitosis and generation of cut cells at a permissive temperature, as was observed in the hsk1-89 cdc19-P1 double mutant. In fact, rad3− has been reported to show synthetic defect with a number of initiation mutants in fission yeast.
In contrast to S phase checkpoint defect, DNA damage checkpoint, including phosphorylation of Chk1 protein, appears to be normal in hsk1-89, although the mutant cells restored growth more slowly than the wild type after exposure to UV or MMS (our unpublished results). We propose that Hsk1 kinase is required for recovery from DNA damage-induced arrest. Recently, Snaith et al. (2000) have reported another allele of hsk1(ts) and showed that Hsk1 is involved in DNA replication checkpoint control by being a target of Cds1 protein.
We show here that hyperphosphorylation of Dfp1 induced by HU arrest requires not only Cds1 (Figure 6; Brown and Kelly, 1999; Takeda et al., 1999) but also the wild-type level of Hsk1 kinase activity. In conjunction with the requirement of Hsk1 kinase activity for Cds1 activation, this result may indicate the presence of a Hsk1-Cds1 feedback loop mechanism, in which HU-induced S phase arrest activates Cds1 through Hsk1 and Cds1 in turn phosphorylates Dfp1 protein for feedback regulation. Alternatively, Hsk1-mediated prior phosphorylation of Dfp1 may be required for phosphorylation of the latter protein by Cds1.
Apparent Defect in Mitosis of hsk1-89 May Be Related to Malfunctions of Rad21 Cohesin
At 37°C, hsk1-89 cells arrested with heterogeneous DNA content centered around 2C and exhibited aberrant nuclear structures, including condensed, scattered, or unequally segregated chromosomes, indicative of mitotic defect of hsk1-89 mutant at this temperature. This may suggest that Hsk1 is required for the progression of mitosis per se. However, we think that possibility rather unlikely. When the mutant cells transiently arrested at M phase with nocodazole were temperature-shifted to 37°C for 1 h and released from metaphase arrest, ∼40% of the cells passed through mitosis (our unpublished results).
What, then, is causing this apparent mitotic defect? We have detected specific genetic interaction between hsk1+ and rad21+. Several rad21-K1-specific phenotypes such as aberrant chromatin structures and TBZ sensitivity were also observed in hsk1-89 mutant. Scc1/Mcd1, a S. cerevisiae homologue of Rad21, has recently been shown to form a complex called cohesin, which maintains sister chromatid cohesion during S phase through mitosis (Guacci et al., 1997; Michaelis et al., 1997). The sister chromatid cohesion is established during DNA replication by Eco1/Ctf7 protein (Skibbens et al., 1999; Toth et al., 1999). Rad21 is phoshorylated during S phase and undergoes proteolytic cleavage in anaphase (Tomonaga et al., 2000). We have shown that sister chromatid cohesion is partially impaired in hsk1-89 cells. The milder defect of sister chromatid cohesion in hsk1-89 compared with rad21-K1 may be due to the fact that a significant portion of the hsk1-89 cells still entered mitosis but arrested before the next S phase. After shift to 36°C, the majority of the cells arrested at S phase would not be able to establish sufficient sister chromatid cohesion due to defective origin firing, thus would be arrested with near 2C DNA content. In hsk1-89 arrested at 30°C, residual origin firing activity during S phase may be able to establish sister chromatid cohesion to the level sufficient for progression through mitosis. However, these cells may not be able to support initiation of the next S phase under the same condition. It is also possible that Hsk1 is more directly involved in activation of cohesin complex, including Rad21 protein during S phase and/or maintenance of sister chromatid cohesion in conjunction with Rad21. This function of Hsk1 may be specifically inactivated at 37°C not at 30°C. Synthetic growth defect of orp5 mutation on rad21 may also be consistent with requirement of proper origin firing for establishment of sister chromatid cohesion (Tanaka et al., personal communication). We speculate that Rad21-mediated sister chromatid cohesion depends on firing of origins, and infrequent origin firing would lead to insufficient establishment of sister chromatid cohesion, which would eventually result in defective mitosis.
Snaith et al. (2000) also reported aberrant chromosome structures in their hsk1(ts) cells. Disintegration of proper chromosome structures in hsk1 mutants may be a part of the reason that meiosis is completely blocked in these mutant strains (Forsburg et al. and Takeda et al., unpublished results).
In conclusion, we have generated a new ts mutant allele of hsk1+, and have shown that Hsk1 plays crucial roles in initiation and progression of S phase through phosphorylation of Cdc19/Mcm2 protein. We also showed novel genetic interaction between hsk1+ and rad3+ as well as that between hsk1+ and rad21+, and presented evidence for involvement of Hsk1 in Cds1 kinase activation for DNA replication checkpoint signaling and in maintenance of the chromosome structures during S phase, most likely through establishment of sister chromatids cohesion.
ACKNOWLEDGMENTS
We are grateful to Drs. K. Tanaka, H. Nishitani, S. Forsburg, P. Nurse, and M. Yanagida for gift of S. pombe strains. We thank Dr. H. Nisihitani for gift of anti-Cdc18, Dr. S. Forsburg for gift of anti-Cdc19 antibody, and Drs. Dominic Griffiths and Teresa Wang for gift of anti-Cds1 antibody. We also thank Etsuko Matsui for protein expression and extract preparations from insect cells, Hiromi Iiyama and Chika Taniyama for constructions of plasmid DNAs, and Drs. H. Okayama and Dr. Y. Watanabe for critical reading of the manuscript and our colleagues in the laboratory for valuable discussion and comments.
REFERENCES
- Aparicio OM, Weinstein DM, Bell SP. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45 during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- Bell S, Stillman B. Nucleotide-dependent recognition of chromosomal origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Bhaumik D, Wang TS-F. Mutational effect of fission yeast Polα on cell cycle events. Mol Biol Cell. 1998;9:2107–2123. doi: 10.1091/mbc.9.8.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy MN, Furnari B, Mondesert O, Russell P. Replication checkpoint enforced by kinase Cds1 and Chk1. Science. 1998;280:909–912. doi: 10.1126/science.280.5365.909. [DOI] [PubMed] [Google Scholar]
- Bousset K, Diffley JF. The Cdc7 protein kinase is required for origin firing during S phase. Genes Dev. 1998;12:480–490. doi: 10.1101/gad.12.4.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brondello J-M, Boddy MN, Furnari B, Russell P. Basis for the checkpoint signal specificity that regulates Chk1 and Cds1 protein kinases. Mol Cell Biol. 1999;6:4262–4269. doi: 10.1128/mcb.19.6.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GW, Kelly TJ. Purification of Hsk1, a minichromosome maintenance protein kinase from fission yeast. J Biol Chem. 1998;273:22083–22090. doi: 10.1074/jbc.273.34.22083. [DOI] [PubMed] [Google Scholar]
- Brown GW, Kelly TJ. Cell cycle regulation of Dfp1, an activator of the Hsk1 protein kinase. Proc Natl Acad Sci USA. 1999;96:8443–8448. doi: 10.1073/pnas.96.15.8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JW, Johnston LH. The yeast gene, DBF4, essential for entry into S phase is cell cycle regulated. Exp Cell Res. 1989;180:419–428. doi: 10.1016/0014-4827(89)90068-2. [DOI] [PubMed] [Google Scholar]
- Cheng L, Collyer T, Hardy CFJ. Cell cycle regulation of DNA replication initiator factor Dbf4. Mol Cell Biol. 1999;19:4270–4278. doi: 10.1128/mcb.19.6.4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong JP, Thommes P, Blow JJ. The role of MCM/P1 proteins in the licensing of DNA replication. Trends Biochem Sci. 1996;21:102–106. [PubMed] [Google Scholar]
- Cocker JH, Piatti S, Santocanale C, Nasmyth K, Diffley JFX. An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature. 1996;379:180–182. doi: 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- Diffley JFX, Cocker JH, Dowell SJ, Rowley A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell. 1994;78:303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- Donaldson AD, Fangman WL, Brewer BJ. Cdc7 is required throughout the yeast S phase to activate replication origins. Genes Dev. 1998;12:491–501. doi: 10.1101/gad.12.4.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan S, Harwood J, Drury LS, Diffley JFX. Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci USA. 1997;94:5611–5616. doi: 10.1073/pnas.94.11.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell SJ, Romanowski P, Diffley JFX. Interaction of Dbf4, the Cdc7 protein kinase regulatory subunit, with yeast replication origins in vivo. Science. 1994;265:1243–1246. doi: 10.1126/science.8066465. [DOI] [PubMed] [Google Scholar]
- D'Urso G, Grallert B, Nurse P. DNA polymerase alpha, a component of the replication initiation complex, is essential for the checkpoint coupling S phase to mitosis in fission yeast. J Cell Sci. 1995;108:3109–3118. doi: 10.1242/jcs.108.9.3109. [DOI] [PubMed] [Google Scholar]
- Geahlen RL, Anostario M, Jr, Low PS, Harrison ML. Detection of protein kinase activity in sodium dodecyl sulfate-polyacrylamide gels. Anal Biochem. 1986;153:153–158. doi: 10.1016/0003-2697(86)90074-6. [DOI] [PubMed] [Google Scholar]
- Grallert B, Nurse P. The ORC1 homolog orp1 in fission yeast plays a key role in regulating onset of S phase. Genes Dev. 1996;10:2644–2654. doi: 10.1101/gad.10.20.2644. [DOI] [PubMed] [Google Scholar]
- Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatids cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutz H, Heslot H, Leupold U, Lopreno N. Shizosaccharomyces pombe. In: King R C, editor. Handbook of Genetics I. New York: Plenum Press; 1974. pp. 395–446. [Google Scholar]
- Hartwell LH. Genetic control of the cell cycle in yeast II. Genes controlling DNA replication and its initiation. J Mol Biol. 1971;59:183–194. doi: 10.1016/0022-2836(71)90420-7. [DOI] [PubMed] [Google Scholar]
- Hartwell LH. Three additional genes required for deoxyribonucleic acid synthesis in Saccharomyces cerevisiae. J Bacteriol. 1973;115:966–974. doi: 10.1128/jb.115.3.966-974.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson M, Madine M, Dalton S, Gautier J. Phosphorylation of MCM4 by cdc2 protein kinase inhibits the activity of the minichromosome maintenance complex. Proc Natl Acad Sci USA. 1996;93:2223–12228. doi: 10.1073/pnas.93.22.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann JFS, Beach D. cdt1 is an essential target of the Cdc10/Sct1 transcription factor: requirement for DNA replication and inhibition of mitosis. EMBO J. 1994;13:425–434. doi: 10.1002/j.1460-2075.1994.tb06277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth RE, Jr, Ostroff RM, Klein MB, Niswander LA, Sclafani RA. Molecular genetic studies of the Cdc7 protein kinase and induced mutagenesis in yeast. Genetics. 1992;132:53–62. doi: 10.1093/genetics/132.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Arai K, Masai H. A fusion protein library: an improved method for rapid screening and characterization of DNA binding or interacting proteins. Gene. 1996;181:167–171. doi: 10.1016/s0378-1119(96)00497-0. [DOI] [PubMed] [Google Scholar]
- Ishimi Y. A DNA helicase activity is associated with an MCM4, -6 and -7 protein complex. J Biol Chem. 1997;272:24508–24513. doi: 10.1074/jbc.272.39.24508. [DOI] [PubMed] [Google Scholar]
- Ishimi Y, Komamura Y, You Z, Kimura H. Biochemical function of mouse minichromosome maintenance 2 protein. J Biol Chem. 1998;273:8369–8375. doi: 10.1074/jbc.273.14.8369. [DOI] [PubMed] [Google Scholar]
- Ishimi Y, Komamura-Kohno Y, You Z, Omori A, Kitagawa M. Inhibition of mcm4,6,7 helicase activity by phosphorylation with cyclin A/Cdk2. J Biol Chem. 2000;275:16235–16241. doi: 10.1074/jbc.M909040199. [DOI] [PubMed] [Google Scholar]
- Jackson AL, Pahl PMB, Harrison K, Rosamond J, Sclafani RA. Cell cycle regulation of the yeast Cdc7 protein kinase by association with the Dbf4 protein. Mol Cell Biol. 1993;13:2899–2908. doi: 10.1128/mcb.13.5.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jallepalli PV, Brown GW, Muzi-Falconi M, Tien D, Kelly TJ. Regulation of the replication initiator protein p65cdc18 by CDK phosphorylation. Genes Dev. 1997;11:2767–2779. doi: 10.1101/gad.11.21.2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Hunter T. Identification and characterization of a human protein kinase related to budding yeast Cdc7p. Proc Natl Acad Sci USA. 1997;94:14320–14325. doi: 10.1073/pnas.94.26.14320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, McDonald D, Hope TJ, Hunter T. Mammalian Cdc7-Dbf4 protein kinase complex is essential for initiation of DNA replication. EMBO J. 1999;18:5703–5713. doi: 10.1093/emboj/18.20.5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston LH, Masai H, Sugino A. First the CDKs, now DDKs. Trends Cell Biol. 1999;9:249–252. doi: 10.1016/s0962-8924(99)01586-x. [DOI] [PubMed] [Google Scholar]
- Kearsey SE, Maiorano D, Holmes EC, Todorov IT. The role of MCM proteins in the control of genome duplication. BioEssays. 1995;18:183–190. doi: 10.1002/bies.950180305. [DOI] [PubMed] [Google Scholar]
- Kelly TJ, Martin GS, Forsburg SL, Stephen RJ, Russo A, Nurse P. The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell. 1993;74:371–382. doi: 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- Kim JM, Sato N, Yamada M, Arai K, Masai H. Growth regulation of the expression of mouse cDNA and gene encoding a serine/threonine kinase related to Saccharomyces cerevisiae CDC7 essential for G1/S transition. J Biol Chem. 1998;273:23248–23257. doi: 10.1074/jbc.273.36.23248. [DOI] [PubMed] [Google Scholar]
- Kitada K, Johnson AL, Johnston LH, Sugino A. A multicopy suppressor gene of the Saccharomyces cerevisiae G1 cell cycle mutant gene dbf4 encodes a protein kinase and is identified as CDC5. Mol Cell Biol. 1993;13:4445–4457. doi: 10.1128/mcb.13.7.4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota Y, Mimura S, Nishimoto S, Masuda T, Nojima H, Takisawa H. Licensing of DNA replication by a multi-protein complex of MCM/P1 proteins in Xenopus eggs. EMBO J. 1997;16:3320–3331. doi: 10.1093/emboj/16.11.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai H, Sato N, Yamada M, Mahony D, Seghezzi W, Lees E, Arai K-I, Masai H , A novel growth- and cell cycle-regulated protein, ASK, activates human Cdc7-related kinase and is essential for G1/S transition in mammalian cells. Mol Cell Biol. 1999;19:5083–5095. doi: 10.1128/mcb.19.7.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Kawasaki Y, Young MR, Kihara M, Sugino A, Tye BK. Mcm2 is a target of regulation by Cdc7-Dbf4 during the initiation of DNA synthesis. Genes Dev. 1997;11:3365–3374. doi: 10.1101/gad.11.24.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay HD, Griffiths DJF, Edwards RJ, Christensen PU, Murray JM, Osman F, Walworth N, Carr AM. S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev. 1998;12:382–395. doi: 10.1101/gad.12.3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Girona A, Mondesert O, Leatherwood J, Russell P. Negative regulation of Cdc18 DNA replication protein by Cdc2. Mol Biol Cell. 1998;9:63–73. doi: 10.1091/mbc.9.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiorano D, van Assendelft GB, Kearsey SE. Fission yeast cdc21, a member of MCM protein family, is required for onset of S phase and is located in the nucleus throughout the cell cycle. EMBO J. 1996;15:861–872. [PMC free article] [PubMed] [Google Scholar]
- Masai H, Matsui E, You Z, Ishimi Y, Tamai K, Arai K-I. Human Cdc7-related kinase complex: in vitro phosphorylation of MCM by concerted actions of Cdks and Cdc7 and that of a critical threonine residue of Cdc7 by Cdks. J Biol Chem. 2000;275:29042–29052. doi: 10.1074/jbc.M002713200. [DOI] [PubMed] [Google Scholar]
- Masai H, Miyake T, Arai K-I. hsk1+, a Schizosaccharomyces pombe gene related to Saccharomyces cerevisiae CDC7, is required for chromosomal replication. EMBO J. 1995;14:3094–3104. doi: 10.1002/j.1460-2075.1995.tb07312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H, Sato N, Takeda T, Arai K-I. Cdc7 kinase complex as a molecular switch for DNA replication. Front Biosci. 1999;4:834–840. doi: 10.2741/masai. [DOI] [PubMed] [Google Scholar]
- Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Cohesin: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Moreno S, Clar A, Nurse P. Molecular genetic analysis of the fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Nabeshima K, Nakagawa T, Straight AF, Murray A, Chikashige Y, Yamashita M, Hiraoka Y, Yanagida M. Dynamics of centromeres during metaphase-anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol Biol Cell. 1998;9:3211–3225. doi: 10.1091/mbc.9.11.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. Viewpoints: putting the cell cycle in order. Science. 1996;274:1643–1645. doi: 10.1126/science.274.5293.1643. [DOI] [PubMed] [Google Scholar]
- Newlon CS. Putting it all together: building a prereplicative complex. Cell. 1997;91:717–720. doi: 10.1016/s0092-8674(00)80459-6. [DOI] [PubMed] [Google Scholar]
- Nishitani H, Nurse P. p65cdc18 plays a major role controlling the initiation of DNA replication in fission yeast. Cell. 1995;83:397–405. doi: 10.1016/0092-8674(95)90117-5. [DOI] [PubMed] [Google Scholar]
- Okazaki K, Okazaki N, Kume K, Jinno S, Tanaka K, Okayama H. High-frequency transformation method and library transducing vectors for cloning mammalian cDNAs by trans-complementation of Shizosaccharomyces pombe. Nucleic Acids Res. 1990;18:6485–6489. doi: 10.1093/nar/18.22.6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicioli A, Lucca C, Liberi G, Marini F, Lopes M, Plevani P, Romano A, Di Fiore PP, Foiani M. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 1999;18:6561–6572. doi: 10.1093/emboj/18.22.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka Y, Fantes P, Sutani T, Mclnerny C, Creanor J, Yanagida M. Fission yeast cut5 links nuclear chromatin and M phase regulator in the replication checkpoint control. EMBO J. 1994;13:5319–5329. doi: 10.1002/j.1460-2075.1994.tb06866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka Y, Yanagida M. Fission yeast cut5+, required for S phase onset and M phase restraint, is identical to the radiation-damage repair gene rad4+ Cell. 1993;74:383–393. doi: 10.1016/0092-8674(93)90428-s. [DOI] [PubMed] [Google Scholar]
- Sato N, Arai K-i, Masai H. Human and Xenopus cDNAs encoding budding yeast Cdc7-related kinases: in vitro phosphorylation of MCM subunits by a putative human homologue of Cdc7. EMBO J. 1997;16:4340–4351. doi: 10.1093/emboj/16.14.4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclafani RA, Jackson AL. Cdc7 protein kinase for DNA metabolism comes of age. Mol Microbiol. 1994;11:805–810. doi: 10.1111/j.1365-2958.1994.tb00358.x. [DOI] [PubMed] [Google Scholar]
- Sherman DA, Pasion SG, Forsburg SL. Multiple domains of fission yeast Cdc19p (MCM2) are required for its association with the core MCM complex. Mol Biol Cell. 1998;9:1833–1845. doi: 10.1091/mbc.9.7.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibbens RV, Corson LB, Koshland D, Hieter P. Ctf7p is essential for sister chromatids cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 1999;13:307–319. doi: 10.1101/gad.13.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snaith HA, Brown GW, Forsburg SL. Shizosaccharomyces pombe Hsk1p is a potential Cds1p target required for genome integrity. Mol Cell Biol. 2000;20:7922–7932. doi: 10.1128/mcb.20.21.7922-7932.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B. Cell cycle control of DNA replication. Science. 1996;274:1659–1664. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- Takeda T, Ogino K, Matsui E, Cho MK, Kumagai H, Miyake T, Arai K, Masai H. A fission yeast gene, him1+/dfp1+, encoding a regulatory subunit for Hsk1 kinase, plays essential roles in S phase initiation as well as in S phase checkpoint control and recovery from DNA damages. Mol Cell Biol. 1999;19:5535–5547. doi: 10.1128/mcb.19.8.5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Knapp D, Nasmyth K. Loading of an Mcm protein onto DNA-replication origins is regulated by Cdc6p and Cdks. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Yonekawa T, Kawasaki Y, Kai M, Furuya K, Iwasaki M, Murakami H, Yanagida M, Okayama H. Fission yeast Esol is required for establishing sister chromatid cohesion during S phase. Mol Cell Biol. 2000;20:3459–3469. doi: 10.1128/mcb.20.10.3459-3469.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebayashi K, Kato J, Ikeda H. Isolation of a Schizosaccharomyces pombe rad21ts mutant that is aberrant in chromosome segregation, microtuble function, DNA repair and sensitive to hydroxyurea: possible involvement of Rad21 in ubiquitin-mediated proteolysis. Genetics. 1998;148:49–57. doi: 10.1093/genetics/148.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todorov IT, Attaran A, Kearsey SE. BM28, a human member of the MCM2–3-5 family, is displaced from chromatin during DNA replication. J Cell Biol. 1995;129:1433–1445. doi: 10.1083/jcb.129.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomonaga T, Nagao K, Kawasaki Y, Furuya K, Murakami A, Morishita J, Yuasa T, Sutani T, Kearsey SE, Uhlman F, Nasmyth K, Yanagida M. Characterizaton of fission yeast cohesin: essential proteolysis of rad21 phosphorylated in the S phase. Genes Dev. 2000;14:2757–2770. doi: 10.1101/gad.832000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, Nasmyth K. Yeast cohesin complex requires a conserved protein, Eco1 (Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999;13:320–333. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye BK. The Mcm2–3-5 proteins: are they replication licensing factors? Trends Cell Biol. 1994;4:160–166. doi: 10.1016/0962-8924(94)90200-3. [DOI] [PubMed] [Google Scholar]
- Waddell TK, Fialkow L, Chan CK, Kishimoto TK, Downey GP. Signaling functions of L-selectin. Enhancement of tyrosine phosphorylation and activation of MAP kinase. J Biol Chem. 1995;270:15403–15411. doi: 10.1074/jbc.270.25.15403. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Forbes KC, Wu Z, Moreno S, Piwnica-Worms H, Enoch T. Replication checkpoint requires phosphorylation of the phosphatase Cdc25 by Cds1 or Chk1. Nature. 1998;395:507–510. doi: 10.1038/26766. [DOI] [PubMed] [Google Scholar]