Abstract
Protein posttranslational modifications (PTMs), particularly phosphorylation, dramatically expand the complexity of cellular regulatory networks. Although cysteine (Cys) in various proteins can be subject to multiple PTMs, its phosphorylation was previously considered a rare PTM with almost no regulatory role assigned. We report here that phosphorylation occurs to a reactive cysteine residue conserved in the staphylococcal accessary regulator A (SarA)/MarR family global transcriptional regulator A (MgrA) family of proteins, and is mediated by the eukaryotic-like kinase-phosphatase pair Stk1-Stp1 in Staphylococcus aureus. Cys-phosphorylation is crucial in regulating virulence determinant production and bacterial resistance to vancomycin. Cell wall-targeting antibiotics, such as vancomycin and ceftriaxone, inhibit the kinase activity of Stk1 and lead to decreased Cys-phosphorylation of SarA and MgrA. An in vivo mouse model of infection established that the absence of stp1, which results in elevated protein Cys-phosphorylation, significantly reduces staphylococcal virulence. Our data indicate that Cys-phosphorylation is a unique PTM that can play crucial roles in bacterial signaling and regulation.
Keywords: Ser/Thr kinase PknB, transcriptional regulation
Covalent posttranslational modification (PTM) of proteins greatly expands the coding capacity of prokaryotic/eukaryotic genomes, which leads to the production of much more diverse proteomes (1). Attaching different chemical groups, such as phosphate, acetate, lipids, and carbohydrates to amino acid residues in proteins allows these PTMs to fine-tune functions of proteins in response to various signaling events.
Among various PTMs, the reversible protein phosphorylation is the most widespread in signal transduction, which is a central process to the regulation of nearly every aspect of cell life, including growth, metabolism, motility, division, differentiation, organelle trafficking, and immunity, as well as learning and memory behaviors in higher organisms (2). In a typical phosphorylation event, a protein kinase transfers the γ-phosphate from ATP to specific amino acids of a protein to initiate signal transduction; in eukaryotes, these phosphorylated amino acids are usually Ser, Thr, and Tyr residues (3, 4). In contrast, His/Asp-based phospho-relay of two-component systems has been well recognized as a classic mode of signal transduction in the bacterial world (5). However, it wasn’t until recently that eukaryotic-like Ser/Thr/Tyr phosphorylation in bacteria started to receive significant attention (6–8). Particularly, in pathogenic bacteria such as Mycobacterium tuberculosis and Staphylococcus aureus, eukaryotic-like Ser/Thr kinases (and associated phosphatases) have often been shown to participate in virulence functions (5, 9–11). The kinase/phosphatase pairs are known as Stks/Stps in bacteria; the kinase Stk1 has been called as PknB or Stk, and the phosphatase Stp1 as Stp (9). Throughout this study, we will use Stk1 for the kinase and Stp1 for the phosphatase because these names are more common in the literature. Less common lysine and arginine phosphorylations have also been studied (12). For example, in Bacillus subtilis, a specific arginine kinase McsB has been isolated and shown to phosphorylate arginine residues of its binding partner CtsR (a stress-response transcriptional regulator) in vitro (13).
Cysteine is one of the more reactive amino acid side chains in proteins. The thiol group in cysteine plays a variety of roles in cellular processes such as enzymatic catalysis, metal binding, signal [reactive oxgen/nitrogen species (ROS/RNS)] sensing, and protein folding (disulfide formation). Thanks to its intrinsic reactivity, the Cys residue can be subject to a great number of PTMs that include alkylation (S-prenylation, S-methylation) (14, 15), acylation (S-palmitoylation), and oxidation (sulfenation, sulfination, S-nitrosylation, disulphide formation, and so forth) (14, 16, 17). The latter case enables Cys to serve as a regulatory switch on proteins that respond to redox change in their cellular environment (18–21). Despite these well-appreciated Cys modifications, Cys phosphorylation has long been regarded as a rare PTM. Cys phosphorylation was first discovered as enzymatic intermediates in the transport of carbohydrates by the bacterial phosphoenolpyruvate-dependent phosphotransferase system (22, 23), and later in the dephosphorylation of eukaryotic protein tyrosine phosphatase during catalysis (24). Because of its “rare occurrence” (likely a result of inappropriate assay conditions used in the past), the regulatory role of Cys-phosphorylation as a type of PTM remains elusive. Moreover, unlike conventional Ser/Thr/Tyr or His/Asp phosphorylation, no specific kinase/phosphatase pair has ever been recognized and designated for protein Cys-phosphorylation, not to mention its contribution to cellular signaling.
Here we report the identification of Cys-phosphorylation on a number of proteins, including several global transcriptional regulators in S. aureus. We demonstrate that Cys-phosphorylation, mediated by the sole eukaryotic-like Ser/Thr kinase/phosphatase (Stk1/Stp1) pair in S. aureus (25–27), significantly impacts the function of these proteins, thereby resulting in altered bacterial phenotypes.
Results
Cys-Phosphorylation of SarA/MgrA Family Proteins (SarA, MgrA, and SarZ) Mediated by Stk1-Stp1.
S. aureus, a major human pathogen that is the most common source of bacterial infections in the community and hospital, causes a wide variety of diseases, ranging from minor skin infections to life-threatening blood infections (28). The virulence of this organism is controlled by regulatory networks composed of a large array of regulatory proteins, such as the agr system and the staphylococcal accessary protein A (SarA)/MarR family global transcriptional regulator A (MgrA) family global regulatory proteins, which can respond to changing host microenvironments (29, 30). Recent studies have revealed that the sole and conserved Cys residue in SarA, MgrA, and SarZ (Fig. 1) acts as a redox switch to modulate the regulatory functions of these proteins (31–34). However, in some cases oxidation of Cys cannot fully account for the observed phenotypes, which led us to speculate that other PTMs might take place on these proteins to modulate their regulatory functions (35) (Fig. 1). Recent studies suggest that both MgrA and SarA could be subject to potential Ser/Thr phosphorylation mediated by a eukaryotic-like kinase Stk1 in S. aureus, and that the phosphorylation modulates the DNA-binding activity of these proteins (35, 36). Stk1-Stp1 is the only conserved pair of eukaryotic-like kinase-phosphatase in S. aureus. To investigate the phosphorylation mediated by Stk1-Stp1, we performed in vitro phosphorylation assays on SarA, MgrA, and SarZ using crude cell extract from S. aureus supplemented with ATP.
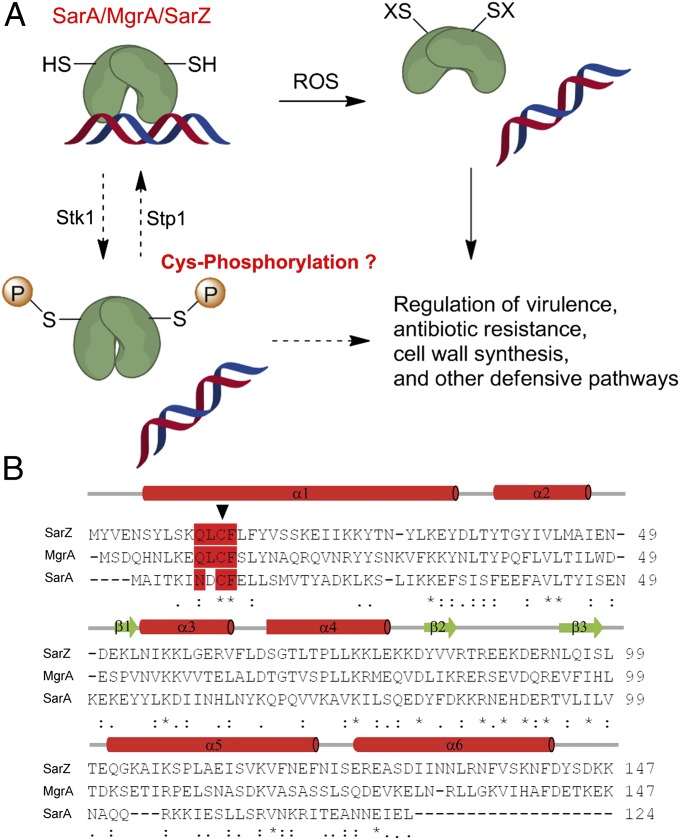
Fig. 1.
S. aureus SarA/MgrA family protein. (A) Model of gene regulation by the oxidation-sensing SarA/MgrA/SarZ. The N-terminal Cys residue, highly conserved in SarA/MgrA/SarZ, is subject to oxidation by ROSs, thus leading to dissociation from DNA. Phosphorylation of the same Cys residue mediated by Stk1-Stp1 might also modulate their target gene regulation. (B) Sequence alignment of SarZ, MgrA, and SarA. The Cys residue (indicated by an arrow) and the surrounding conserved residues are highlighted in red. The alignment was performed with ClustalW2 (61).
As shown in Fig. 2A, SarA, MgrA, and SarZ were indeed phosphorylated by cell extract isolated from the wild-type S. aureus strain Newman. Intriguingly, we noted that the observed phosphorylation occurred exclusively to the reduced forms of SarA, MgrA, and SarZ, but not to their oxidized forms (Fig. S1 A and B), which indicates that the phosphorylation of these proteins is highly redox-dependent. Subsequently, we tested mutant proteins with the conserved Cys residue mutated to Ser. No protein phosphorylation was observed for all three mutant proteins (Fig. 2A), indicating that the conserved reactive Cys is very likely the site of the observed phosphorylation.
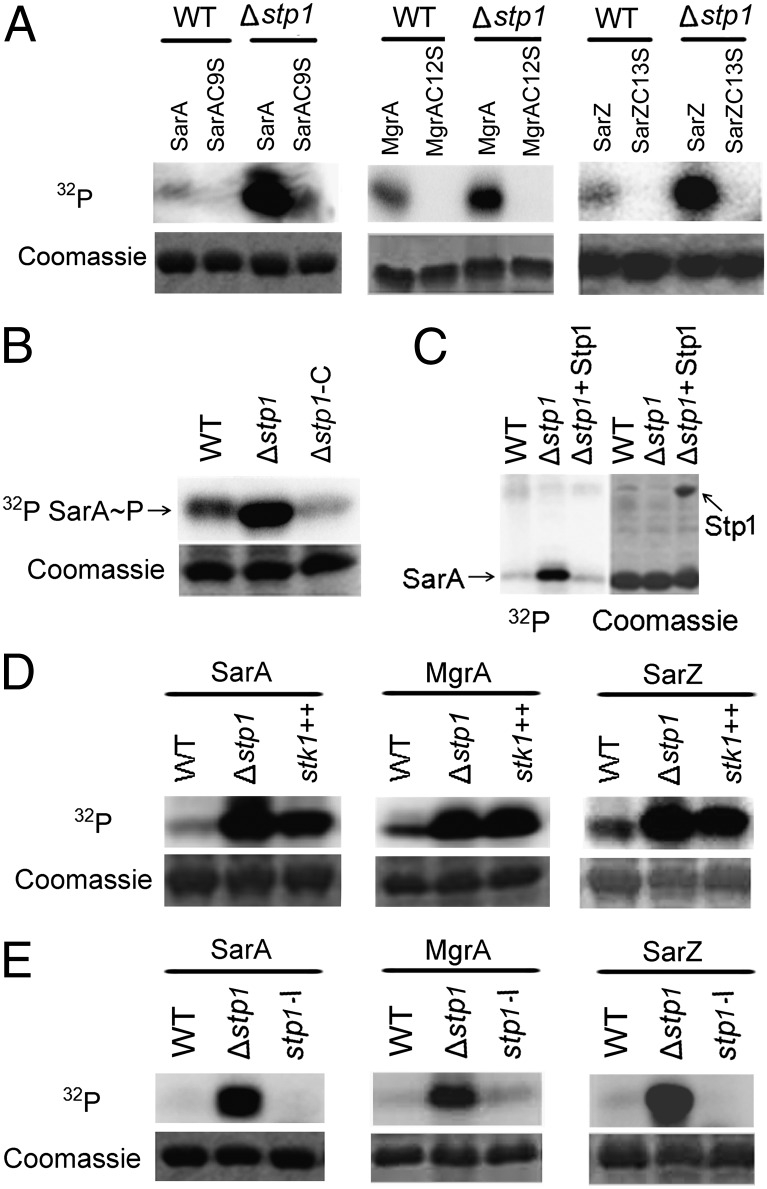
Fig. 2.
Phosphorylation of SarA family proteins (SarA, MgrA, and SarZ). (A) The Cys-to-Ser substitution abolished the phosphorylation of SarA, MgrA, and SarZ. WT, cell extract from the wild-type strain Newman; Δstp1, cell extract from Δstp1 strain (an in-frame deletion mutant of stp1). Protein bands stained with Coomassie blue after autoradiography were shown. (B) The phosphatase Stp1 dephosphorylates phospho-SarA (lane 3). The phospho-SarA was treated with Stp1 at 37 °C for 10 min before analysis. (C) The phosphorylation activity of cell extract from Δstp1 was restored by complementation with pYJ335::stp1 (lane 3). Δstp1-C, Δstp1/pYJ335::stp1. (D) Overexpression of Stk1 enhanced phosphorylation of SarA (lane 3). stk1++, cell extract from the wild-type strain carrying pYJ335::stk1, in which the expression of stk1 was induced by anhydrotetracycline (1 μg/mL). (E) Deactivation of stk1 abolished phosphorylation of SarA (lane 3). stp1-I, cell extract from the mutant with bursa aurealis transposon insertion in stp1 that deactivates both stp1 and stk1.
Because Stk1 and its associated phosphatase Stp1 constitute the sole Ser/Thr kinase-phosphatase pair in S. aureus, we tested their roles in the observed phosphorylation. An in-frame deletion mutant of the stp1 gene (Δstp1) was constructed. When cell extract from the Δstp1 mutant was tested, the phosphorylation levels of all three wild-type proteins were dramatically increased (Fig. 2A), yet the corresponding Cys-to-Ser mutants were not phosphorylated at all. Cell extract from the Δstp1 strain complemented with plasmid pYJ335::stp1 (Δstp1-C), expressing the wild-type Stp1, gave reduced protein phosphorylation compared with that of the Δstp1 mutant (Fig. 2B). Furthermore, the recombinant Stp1 expressed from Escherichia coli was able to remove the phosphate group of phosphorylated SarA (Fig. 2C), MgrA (Fig. S1C), and SarZ (Fig. S1D), supporting its phosphatase role. Consistently, overexpression of Stk1—the associated kinase of Stp1—in the wild-type background increased the phosphorylation level of all three proteins (Fig. 2D and Fig. S2A).
Surprisingly, when we performed the phosphorylation assays in the presence of high concentrations of DTT (10 mM), a widely used reducing agent in biochemical assays, we found that phosphorylation of both SarA and MgrA were almost abolished (Fig. S2B). In contrast, the phosphorylation of the kinase Stk1 itself, which is known to be a Ser/Thr type phosphorylation, was barely affected by the DTT treatment (Fig. S2B). Thus, the observed phosphorylation on the SarA/MgrA family proteins represents a unique type of modification chemically distinct from conventional Ser/Thr phosphorylation.
A bursa aurealis transposon insertion mutant of stp1 (stp1-I) (37) that deactivates both stp1 and its downstream cotranscribed stk1 (Fig. S3) was used for the in vitro phosphorylation assay. Cell extract from this mutant strain significantly diminished phosphorylation of all three proteins (Fig. 2E), indicating that Stk1 is essential for the observed protein phosphorylation. Collectively, our phosphorylation results suggest that phosphorylation occurs to the Cys residue conserved in the SarA/MgrA family proteins and that the sole Stp1-Stk1 pair in S. aureus mediates this Cys-phosphorylation.
Verification of Cys-Phosphorylation by LC-MS/MS.
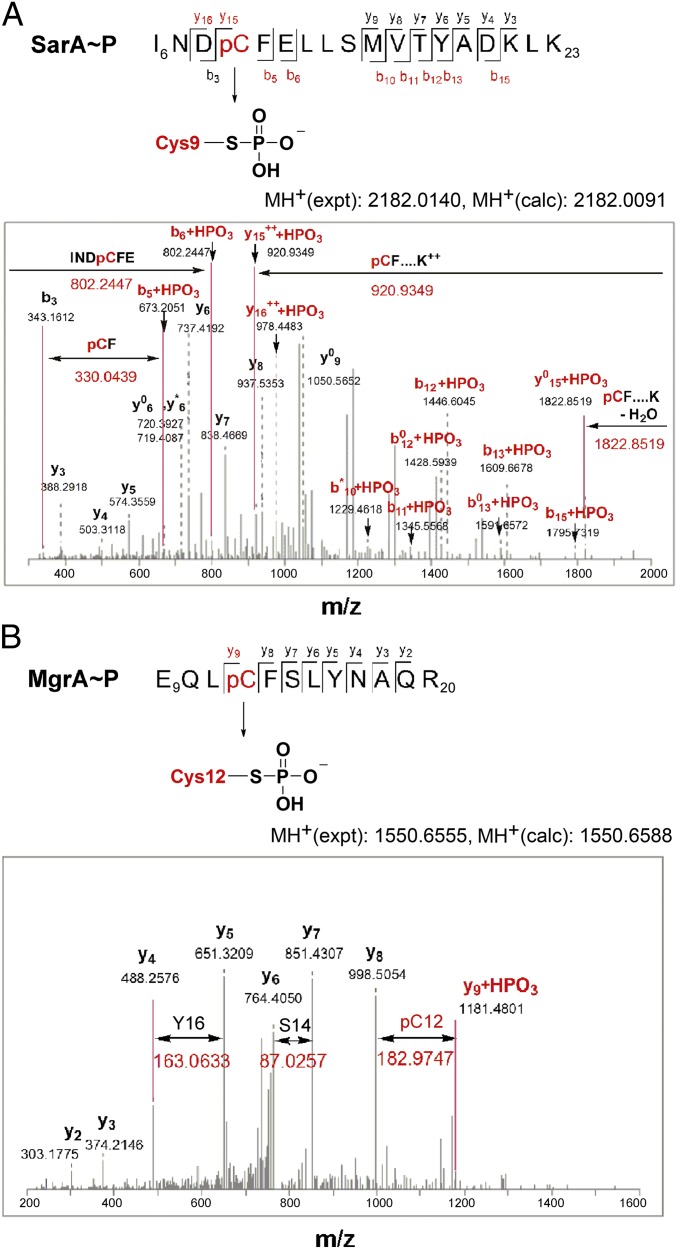
To further verify Cys-phosphorylation of the SarA/MgrA family proteins, we performed mass spectrometric characterization on the phosphorylated proteins. Despite the labile feature of phospho-Cys (38), we successfully identified the phospho-Cys modification in both phosphorylated SarA and phosphorylated MgrA (Fig. 3 and Fig. S4). In the case of SarA, after trypsin digestion, one phosphopeptide INDpCFELLSMVTYADKLK (observed m/z 2182.0140) was identified using the Mascot (v 2.3, MatrixScience) database search engine (Fig. 3A) (39). The detected y6 and y7 fragment ions showed that the phosphorylation was not on Thr-17 or Tyr-18, whereas fragment ions b5 and b6 indicated that the phosphorylation was on Cys-9 but not on Ser-14. In the case of MgrA, one phosphopeptide EQLpCFSLYNAQR (observed m/z 1550.6555) (Fig. 3B) was also identified. The y7 and y8 fragment ions were detected showing that neither Ser-14 nor Tyr-16 was phosphorylated in this sequence, and one y9 fragment ion showed that Cys-13 was phosphorylated (+80 Da mass shift). A mis-cleaved peptide from the same sequence GSHMNLKEQLpCFSLYNAQR (observed m/z 2318.0482) was also detected showing one b11 fragment ion confirming phosphorylation on Cys-13 (Fig. S4).
Fig. 3.
LC-MS/MS identification of Cys-phosphorylation of SarA and MgrA. (A) LC-MS/MS spectrum of the phosphopeptide I6NpCFELLSMVTYADKLK23 (observed m/z 2182.0140 Da corresponding to apo-peptide theoretical mass of 2102.0393 Da + 1 phosphate group 79.9747 Da) obtained after trypsin digestion of phospho-SarA. The b5 and b6 fragment ions corresponding to I6NDpCF and I6NDpCFE, respectively (observed m/z 673.2051 and 802.2247 corresponding to apo-fragment + 1 phosphate group 79.9437 Da) indicates the presence of phospho-Cys rather than on Ser or Thr. The phospho-Cys-9-Phe-10 (pCF) fragment is also highlighted by the mass difference of b5 and b3 fragment ions. (B) LC-MS/MS spectrum of the phosphopeptide E9QLpCFSLYNAQR20 (observed m/z 1550.6555 Da corresponding to apo-peptide experimental mass of 1470.6967 Da + 1 phosphate group 79.9588 Da) obtained after trypsin digestion of phospho-MgrA. The characteristic mass difference of the phospho-Cys-12 is highlighted. Fragment ions y5 and y7 indicate that phosphorylation is not on Ser-14 or Thr-16 and fragment y9 shows a mass shift of 80 Da from the phosphorylation of Cys-12. Individual fragments are labeled based on the b- or y-ion nomenclature. The phospho-fragments are colored red. Fragment ions arising from the neutral loss of water (−18 Da) are marked with a zero (0) and fragment ions with the loss of ammonia (−17 Da) are marked with an asterisk (*).
Cys-Phosphorylation of the SarA/MgrA Family Proteins Is Blocked by Oxidation and Alkylation.
Protein modifications are known to contribute to changes in cell physiology in response to particular signals. Pathogenic bacteria, such as S. aureus, require rapid response to a hostile environment during pathogenesis. It is well established that ROS serve beneficial roles for host defense, particularly when macrophages and neutrophils produce a burst of oxidants, such as H2O2, to kill invading microorganisms. The abrupt change of the redox status, on the other hand, is also used by the pathogen as a signal to adapt and evade the host defense. The reactive cysteine in the SarA/MgrA family proteins is known to be redox active and plays a significant role in responding to oxidative stress (40). As shown in Fig. S2 C and E, in the presence of either H2O2 or cumene hydrogen peroxide, the phosphorylation levels of both SarA and MgrA were greatly reduced, supporting the finding that oxidation blocks Cys-phosphorylation. Cys-oxidation in MgrA, SarZ, and SarA is critical for gene expression regulation in S. aureus. The same Cys residues can be phosphorylated, allowing the bacterium to perhaps balance the oxidation-sensing pathways through the same residues. Importantly, phosphorylation of the reactive Cys residues in these global regulatory proteins enables the bacterium to incorporate additional signaling pathways to affect/control existing gene regulation networks.
We also tested nucleophilic alkylators that can alkylate the reactive cysteine. Both iodoacetamide and maleimide significantly diminished the phosphorylation of SarA and MgrA (Fig. S2 D and F). In addition, we observed that the phosphorylated SarA is labile to iodine treatment but not to pyridine or hydroxylamine (HONH2) (Fig. S2G), consistent with a previous report that phospho-Cys displays unique liability toward iodine (I2) (24). All these data further confirm cysteine phosphorylation in these proteins.
Cys-Phosphorylation Modulates the DNA-Binding Ability of the SarA/MgrA Family Proteins.
To investigate whether Cys-phosphorylation modulates DNA-binding of SarA, we performed an EMSA with apo-SarA and phospho-SarA. Phospho-SarA was purified using phospho-protein purification kit (Qiagen) after the in vitro phosphorylation reaction. The hla promoter region contains a putative SarA-binding sequence (Fig. S5E) (41) and therefore was chosen to examine the effect of Cys-phosphorylation on the DNA-binding property of SarA. The observed binding of SarA to the hla promoter sequence is specific as SarA failed to show any binding toward the promoter region of SAV2033 as a control, which lacks the consensus sequence required for SarA binding (Fig. S5D). Compared with apo-SarA [Kd (SarA) = ∼32 nM], phospho-SarA displayed a reduced binding to the hla promoter region [Kd (SarA∼P) = ∼120 nM)] (Fig. S5A). The treatment of phospho-SarA with the phosphatase Stp1 restored the DNA-binding ability of SarA (Fig. S5B), indicating that Cys-phosphorylation accounts for the attenuation of the DNA-binding affinity of phospho-SarA. Attenuation of DNA binding by Cys-phosphorylation was also observed on MgrA [Kd (MgrA) = ∼102 nM compared with Kd (MgrA∼P) = ∼500 nM] and SarZ [Kd (SarZ) = ∼35 nM compared with. Kd (SarZ∼P) = ∼120 nM] (Fig. S6). Because glutamate (Glu) has been widely used as a phosphomimetic mutation (4), we further tested the DNA-binding properties of three mutant proteins, SarAC9E, MgrAC12E, and SarZC13E, using Glu as a mimic of phospho-Cys. Indeed, all Cys-to-Glu mutant proteins exhibited reduced binding affinities to their cognate DNA [Kd (SarAC9E) = ∼134 nM, Kd (MgrAC12E) = ∼480 nM, and Kd (SarZC13E) = ∼110 nM] (Figs. S5C and S6 B and D).
SarA, MgrA, and SarZ are dimeric proteins, in which the sole conserved Cys residue resides at the dimerization domain and is involved in hydrogen-bonding interactions with residues from the other monomer (31, 42, 43). To unveil how Cys-phosphorylation and the Cys-to-Glu substitution could impact DNA binding, we constructed initial structural models of wild-type Cys-phosphorylated SarZ and SarZC13E based on the crystal structure of SarZ in the Protein Data Bank (PDB ID: 3HSE) as described in SI Experimental Procedures. Molecular dynamics simulations were further performed on all three forms of SarZ to explore the detailed molecular basis. Time evolutions of centroid distance between chain A and B of dimeric SarZ from their initial conformation (t = 0) were recorded (Fig. S7B). As illustrated in Fig. S7A, the wild-type SarZ displayed very little conformational change during molecular dynamics (MD) simulation and the distance between chain A and B remained steady (Fig. S7B). However, a dramatic conformational change was observed in both SarZC13E and Cys-P-SarZ during MD simulation (Fig. S7B). Particularly for Cys-P-SarZ, the centroid distance between two monomers was sharply shortened by up to 0.75 Å after 40-ns MD simulation (Fig. S7B) and DNA binding domains of two chains almost clashed together (Fig. S7A), indicating that Cys-phosphorylation introduces a significant structural disturbance to SarZ dimerization. We attempted structural characterization of the Cys-to-Glu mutant proteins to validate the MD simulation result and probe the molecular basis underlying structural changes caused by Cys-phosphorylation. Although we were unable to grow good quality crystals of SarAC9E or MgrAC12E, we obtained the crystal structure of SarZC13E at 2.0 Å resolution (Fig. S8). This structure shows the presence of three strong hydrogen-bonding interactions between Glu-13 and two Tyr residues, Tyr-27 and Tyr-41, from the neighboring monomer (Fig. S8A). In contrast, only two weak hydrogen bonds exist in the wild-type SarZ (Fig. S8B). Consistent with the result from MD simulations, the enhanced hydrogen-bonding interactions between the two monomers, most likely existing in the Cys-phosphorylated protein, elicit a substantial conformational change in the DNA-binding domain of SarZ (Fig. S8C), with the DNA recognition helix α4 of SarZC13E displaced by 6 Å compared with that of the wild-type SarZ (Fig. S8D) (42). This observation suggests that Cys-phosphorylation of the SarA/MgrA family proteins induces changes at the dimeric interface, thereby modulating the DNA-binding properties of these proteins.
Phosphorylation of SarA Modulates Its Regulatory Function.
To probe the existence of Cys-phosphorylation in S. aureus, we expressed His6-SarA (Δstp1-sarA/pYJ335::sarA) in the stp1-deficient strain. The phosphorylated SarA could be directly detected using an antiphosphorylation antibody after purifying His6-SarA from the stp1-deficient strain with Ni-NTA beads (Fig. 4A, lane 1). In contrast, there was no detectable signal for His6-SarAC9S (with Cys-9 mutated to Ser) obtained from the stp1-deficient strain expressing Δstp1-sarA/pYJ335::sarAC9S (Fig. 4A, lane 2), indicating that the cysteine residue is the observed phosphorylation site of SarA in vivo.
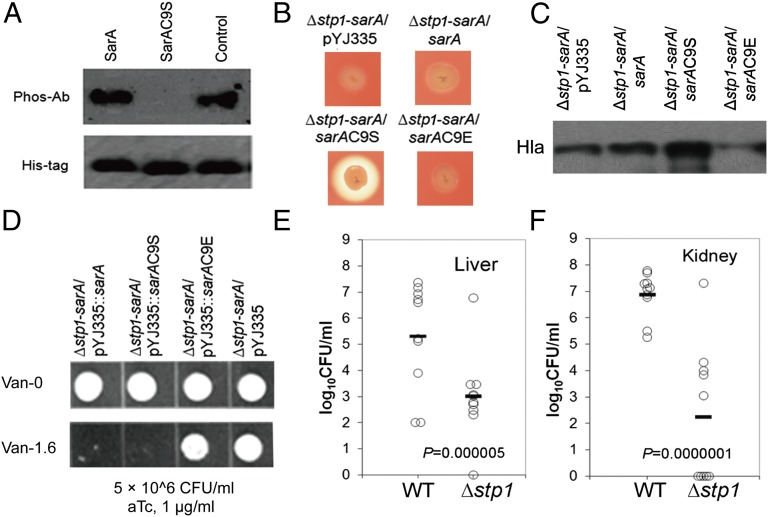
Fig. 4.
Cys-phosphorylation modulates bacterial virulence and antibiotic resistance. (A) Western blot analysis of phospho-SarA from cell extract. SarA, His6-SarA enriched with Ni-NTA beads from whole-cell extract of Δstp1-sarA/pYJ335::sarA; SarAC9S, His6-SarAC9S from Δstp1-sarA/pYJ335::sarAC9S; Control, the recombinant SarA expressed by E. coli after in vitro phosphorylation as a positive control. Phospho-Ab, antiphospho-Thr antibody; His-tag, InVision His-tag in-gel stain (Invitrogen). Δstp1-sarA, a double mutant with in-frame, unmarked stp1 deletion and bursa aurealis transposon insertion in sarA. (B) Hemolysis on the sheep blood agar. The strains tested were spotted on 5% (vol/vol) sheep blood agar plate. Zones of clearance indicate hemolysis. (C) Western blot analysis of the production of α-hemolysin (Hla) (see SI Experimental Procedures). (D) Vancomycin resistance assays. Aliquots (10 μL) of the diluted overnight cultures for each strain (5 × 106 CFU/mL) were spotted onto the tryptic soy agar (TSA) plates without (Upper) or with 1.6 μg/mL vancomycin (Lower). Both plates were supplemented with 1 μg/mL anhydrotetracycline (aTc) to induce the expression of SarA. (E and F) Effect of mutation of stp1 on the virulence of S. aureus in a mouse model of abscess formation. The wild-type strain Newman and the stp1 deletion mutant (Δstp1) were used to infect 10 mice each via retro-orbital injection. After 5 d, S. aureus colonization in murine liver (E) or kidney (F) was measured. Each circle represents one mouse. The horizontal black line represents the mean log10 CFU on the y axis. The statistical difference between mutant and wild-type strains was determined by Student t test (two-tailed). The limit of detection for organ infection is 100 CFU per organ.
SarA and Stp1 are positive regulators of α-hemolysin (hla) in S. aureus (44, 45). Consistent with previous results, we observed that either the Δstp1 or sarA mutant strain displayed reduced hemolysis compared with the wild-type strain, as indicated by zones of clearance on 5% (vol/vol) sheep blood agar (Fig. S9A). Double mutation of sarA and stp1 (Δstp1-sarA/pYJ335, a double mutant with in-frame, unmarked stp1 deletion and bursa aurealis transposon insertion in sarA) led to further attenuated hemolysis (Fig. S9A). The introduction of pYJ335-sarA into the ∆stp1-sarA double-mutant was unable to restore hemolysis (Fig. 4B). However, the introduction of pYJ335-sarAC9S into ∆stp1-sarA, with the expression of the Cys-to-Ser SarAC9S mutant protein, significantly enhanced hemolytic activity (Fig. 4B). We further observed that introduction of pYJ335-sarAC9E (a stable mimic of Cys-phosphorylated SarA) into Δstp1-sarA failed to enhance hemolysis (Fig. 4B). Western blot analysis showed that the production of α-hemolysin is very likely affected by phosphorylation of SarA (Fig. 4C and Fig. S9B). Specifically, the introduction of SarAC9S, a SarA mutant protein not subject to Cys-phosphorylation, significantly enhanced production of α-hemolysin, but wild-type SarA, as well as SarAC9E (a phosphomimetic mutant), failed to restore the α-hemolysin level in the Δstp1-sarA background presumably because of Cys-phosphorylation or Cys to Glu mutation of SarA (Fig. 4C and Fig. S9C). These results indicate that Cys-phosphorylation of SarA contributes to the attenuation of hemolysis observed for the Δstp1 strain (Fig. 4B and Fig. S9A), which is consistent with the weakened binding of the Cys-phosphorylated SarA to the hla promoter region observed by EMSA.
SarA also impacts the susceptibility/resistance of S. aureus to cell wall-targeting antibiotics. The sarA mutant strain in the Newman background displayed enhanced resistance to vancomycin (Fig. S9D), which could be complemented by expressing SarA using the plasmid pYJ335::sarA induced by anhydrotetracycline (aTc, 1 μg/mL) (Fig. S9D). In addition to the sarA mutant, both Δstp1 (Fig. S9D) and Δstp1-sarA mutant strains exhibited enhanced resistance to vancomycin compared with that of the wild-type Newman strain based on plate assays (Fig. 4D and Fig. S9D). We observed that aTc-induced expression of either wild-type SarA (Δstp1-sarA/pYJ335::sarA) or SarAC9S (Δstp1-sarA/pYJ335::sarAC9S), but not SarAC9E (Δstp1-sarA/pYJ335::sarAC9E), fully restored bacterial susceptibility to vancomycin. Collectively, these data support that phosphorylation of Cys-9 plays a critical role for the regulatory function of SarA in bacterial antibiotic resistance.
To further prove that protein Cys-phosphorylation is affected by cell wall-targeting antibiotics through Stk1, we examined the phosphorylation of SarA and MgrA mediated by Δstp1 cell extract in the absence or presence of antibiotics. As shown in Fig. S10, Cys-phosphorylation of both proteins was significantly inhibited by cell wall antibiotics vancomycin and ceftriaxone. A recent study in Streptococcus mutans suggested that carolactone, a biofilm inhibitor, acts by interfering with the function of S. mutans Stk1 (46). To determine whether the reduced Cys-phosphorylation was caused by the inhibition of S. aureus Stk1 by antibiotics, we tested the autophosphorylation of Stk1 in vitro in the absence or presence of various antibiotics including vancomycin, ceftriaxone, and erythromycin. Intriguingly, as shown in Fig. S10C, the phosphorylation of Stk1 was greatly inhibited by cell wall-acting antibiotics vancomycin and ceftriaxone, but not by erythromycin, a protein synthesis inhibitor. This observation agrees with the effects of antibiotics on Cys-phosphorylation of SarA (25). Taken together, these results suggest that Cys-phosphorylation of SarA plays important regulatory roles in bacterial virulence production and antibiotic resistance and that cell wall stress caused by antibiotic treatments might in turn serve one of the signals that affect Cys-phosphorylation in S. aureus.
Effect of Mutation of stp1 on the Virulence of S. aureus in a Mouse Model of Abscess Formation.
As global transcriptional regulators, SarA, MgrA, and SarZ are critical for bacterial virulence (31, 47–49). Given that deletion of stp1 inevitably accumulates Cys-phosphorylation among these proteins (SarA, MgrA, and SarZ), thereby partially impairing their DNA binding activity, we envision that deletion of stp1 might render S. aureus less virulent in a manner similar to the effects resulting from mutation of any of these transcriptional regulators. A mouse infection model of abscess formation was used to compare the infectivity of the Δstp1 deletion mutant with the wild-type Newman. As expected, the ∆stp1 strain was incompetent in establishing infection in the mouse. As shown in Fig. 4 E and F, bacterial loads in livers and kidneys of mice infected with ∆stp1 were dramatically reduced after 5 d. Compared with the Newman strain, the ∆stp1 strain showed a >2-log reduction (P = 5 × 10−6, t test) of virulence in livers of the infected mice (Fig. 4E). The virulence attenuation of ∆stp1 in kidneys of the infected mice was even more striking, exhibiting a >4-log reduction (P = 1 × 10−7, t test) of bacterial loads in comparison with wild-type Newman (Fig. 4F).
Discussion
Ser/Thr/Tyr phosphorylations are ubiquitous in eukaryotes (2, 50). These types of protein phosphorylations dominate eukaryotic signaling and regulation. Another common group of protein phosphorylation is the bacterial two-component system that uses His/Asp phosphorylations in signal relays in microbes (51, 52). His/Asp phosphorylations are transient and unstable to almost all enrichment and mass spectrometry detection procedures, thus making direct observation and characterization of these phosphorylations difficult and challenging (5). The presence of eukaryotic-like Ser/Thr kinase-phosphatase pairs in bacteria has fueled interests in elucidating their potential roles in bacterial signaling and regulation (53–55). The Stk1-Stp1 pair is conserved in Gram-positive bacteria and has been suggested to play global regulatory roles through Ser/Thr phosphorylation/dephosphorylation (6, 9). We show here that this kinase/phosphatase pair can phosphorylate Cys residues in different proteins in Gram-positive bacteria. Notably, the observed Cys-phosphorylation of global regulatory proteins exhibits significant regulatory functions, indicating the existence of a unique type of PTM that impacts biological signaling and regulation.
Despite the fact that cysteine is the most nucleophilic residue among natural amino acids and is known to be subject to multiple PTMs (56), Cys-phosphorylation was previously considered an unusual PTM with only a few known examples as catalytic intermediates in enzymatic reactions (24, 57, 58). We show here that the conserved Cys residue in the global transcriptional factors SarA, MgrA, and SarZ can be phosphorylated, which controls virulence and other properties of S. aureus. Previous in vitro kinase-mediated phosphorylation assays have indicated that the SarA/MgrA family proteins are subject to Ser/Thr phosphorylation (35, 36, 59). Our present study establishes Cys-phosphorylation as a main PTM occurring to these proteins. When the Cys residue was mutated to Ser, the mutant proteins could hardly be phosphorylated by cell extracts (Fig. 2A), indicating that the Cys residue is the major site of phosphorylation in the SarA/MgrA family proteins. The labile nature of Cys-phosphorylation under common experimental conditions might have contributed to previous lack of observation of this PTM on these regulatory proteins and other biological systems (38). To further support the observation that Cys-phosphorylation can be reversed by DTT, we used S-phosphocysteamine as a model substrate to test the stability of alkyl S-phosphate toward DTT under conditions similar to those used for the in vitro phosphorylation assay. As shown in Fig. S11, cysteamine, the dephosphorylated product, was observed after incubating S-phosphocysteamine with high concentrations of DTT (100–200 mM) and Mn2+ (200 mM). High concentrations of DTT above the physiological reduction range and excess of Mn2+ are required to reverse Cys-phosporylation, indicating stability of this PTM under physiological conditions. It is also possible that this type of modification could accumulate to significantly higher levels when bacteria encounter stresses such as ROS/RNS that shift the intracellular redox balance or through other signaling events that lead to activation of Stk1 or deactivation of Stp1.
Cys-phosphorylation could be more prevalent than previously thought and detected if appropriate experimental conditions are used. In fact, we found that Cys-phosphorylation also occurs to other global transcriptional regulators such as CymR (at the sole Cys-25 residue) (Fig. S12), a recently identified oxidation-sensing protein responsible for cysteine metabolism regulation in S. aureus (60). Given that all of the proteins we have shown to undergo Cys-phosphorylation are global regulators that control a broad spectrum of genes and properties in S. aureus, this study may suggest a previously unexplored paradigm of Cys-phosphorylation playing important roles in biological regulation in various organisms. We envision that certain kinases may have evolved to selectively phosphorylate cysteine over serine (or vice versa), even despite the near isostructures of these amino acids. The detailed molecular mechanism responsible for this Cys-phosphorylation event remains to be further elucidated.
Experimental Procedures
Detailed procedures are available in SI Experimental Procedures. See Table S1 for bacterial strains and plasmids used in this study and Table S2 for the list of primers used. Table S3 shows data collection and refinement statistics for the SarZC13E structure.
Supplementary Material
Acknowledgments
We thank Drs. O. Schneewind and D. Missiakas for providing transposon mutants and S. F. Reichard for editing the manuscript. This work was financially supported by National Institutes of Health National Institute of Allergy and Infectious Diseases Grant AI074658 (to C.H.); the Bairen Program of the Chinese Academy of Sciences (L.L.); a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Disease Award (to C.H.); and Shanghai Committee of Science and Technology Grant 10410703900 (to C.L.) and the 863 Program 2012AA020302 (to H.J.). F.S. is a Scholar of the Chicago Biomedical Consortium with support from The Searle Funds at The Chicago Community Trust.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205952109/-/DCSupplemental.
References
- 1.Walsh CT, Garneau-Tsodikova S, Gatto GJ., Jr Protein posttranslational modifications: The chemistry of proteome diversifications. Angew Chem Int Ed Engl. 2005;44:7342–7372. doi: 10.1002/anie.200501023. [DOI] [PubMed] [Google Scholar]
- 2.Cohen P. The origins of protein phosphorylation. Nat Cell Biol. 2002;4:E127–E130. doi: 10.1038/ncb0502-e127. [DOI] [PubMed] [Google Scholar]
- 3.Hunter T, Sefton BM. Transforming gene product of Rous sarcoma virus phosphorylates tyrosine. Proc Natl Acad Sci USA. 1980;77:1311–1315. doi: 10.1073/pnas.77.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tarrant MK, Cole PA. The chemical biology of protein phosphorylation. Annu Rev Biochem. 2009;78:797–825. doi: 10.1146/annurev.biochem.78.070907.103047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 6.Bakal CJ, Davies JE. No longer an exclusive club: Eukaryotic signalling domains in bacteria. Trends Cell Biol. 2000;10:32–38. doi: 10.1016/s0962-8924(99)01681-5. [DOI] [PubMed] [Google Scholar]
- 7.Kobir A, et al. Protein phosphorylation in bacterial signal transduction. Biochim Biophys Acta. 2011;1810:989–994. doi: 10.1016/j.bbagen.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 8.Muñoz-Dorado J, Inouye S, Inouye M. A gene encoding a protein serine/threonine kinase is required for normal development of M. xanthus, a gram-negative bacterium. Cell. 1991;67:995–1006. doi: 10.1016/0092-8674(91)90372-6. [DOI] [PubMed] [Google Scholar]
- 9.Ohlsen K, Donat S. The impact of serine/threonine phosphorylation in Staphylococcus aureus. Int J Med Microbiol. 2010;300:137–141. doi: 10.1016/j.ijmm.2009.08.016. [DOI] [PubMed] [Google Scholar]
- 10.Hegymegi-Barakonyi B, et al. Signalling inhibitors against Mycobacterium tuberculosis—Early days of a new therapeutic concept in tuberculosis. Curr Med Chem. 2008;15:2760–2770. doi: 10.2174/092986708786242886. [DOI] [PubMed] [Google Scholar]
- 11.Fernandez P, et al. The Ser/Thr protein kinase PknB is essential for sustaining mycobacterial growth. J Bacteriol. 2006;188:7778–7784. doi: 10.1128/JB.00963-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matthews HR. Protein kinases and phosphatases that act on histidine, lysine, or arginine residues in eukaryotic proteins: A possible regulator of the mitogen-activated protein kinase cascade. Pharmacol Ther. 1995;67:323–350. doi: 10.1016/0163-7258(95)00020-8. [DOI] [PubMed] [Google Scholar]
- 13.Fuhrmann J, et al. McsB is a protein arginine kinase that phosphorylates and inhibits the heat-shock regulator CtsR. Science. 2009;324:1323–1327. doi: 10.1126/science.1170088. [DOI] [PubMed] [Google Scholar]
- 14.Boal AK, et al. Structural basis for methyl transfer by a radical SAM enzyme. Science. 2011;332:1089–1092. doi: 10.1126/science.1205358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L, et al. Cysteine methylation disrupts ubiquitin-chain sensing in NF-κB activation. Nature. 2012;481:204–208. doi: 10.1038/nature10690. [DOI] [PubMed] [Google Scholar]
- 16.Reddie KG, Carroll KS. Expanding the functional diversity of proteins through cysteine oxidation. Curr Opin Chem Biol. 2008;12:746–754. doi: 10.1016/j.cbpa.2008.07.028. [DOI] [PubMed] [Google Scholar]
- 17.Nagahara N, Matsumura T, Okamoto R, Kajihara Y. Protein cysteine modifications: (1) Medical chemistry for proteomics. Curr Med Chem. 2009;16:4419–4444. doi: 10.2174/092986709789712880. [DOI] [PubMed] [Google Scholar]
- 18.Mongkolsuk S, Helmann JD. Regulation of inducible peroxide stress responses. Mol Microbiol. 2002;45:9–15. doi: 10.1046/j.1365-2958.2002.03015.x. [DOI] [PubMed] [Google Scholar]
- 19.Paget MS, Buttner MJ. Thiol-based regulatory switches. Annu Rev Genet. 2003;37:91–121. doi: 10.1146/annurev.genet.37.110801.142538. [DOI] [PubMed] [Google Scholar]
- 20.Antelmann H, Helmann JD. Thiol-based redox switches and gene regulation. Antioxid Redox Signal. 2011;14:1049–1063. doi: 10.1089/ars.2010.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brandes N, Schmitt S, Jakob U. Thiol-based redox switches in eukaryotic proteins. Antioxid Redox Signal. 2009;11:997–1014. doi: 10.1089/ars.2008.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meins M, et al. Cysteine phosphorylation of the glucose transporter of Escherichia coli. J Biol Chem. 1993;268:11604–11609. [PubMed] [Google Scholar]
- 23.Nuoffer C, Zanolari B, Erni B. Glucose permease of Escherichia coli. The effect of cysteine to serine mutations on the function, stability, and regulation of transport and phosphorylation. J Biol Chem. 1988;263:6647–6655. [PubMed] [Google Scholar]
- 24.Guan KL, Dixon JE. Evidence for protein-tyrosine-phosphatase catalysis proceeding via a cysteine-phosphate intermediate. J Biol Chem. 1991;266:17026–17030. [PubMed] [Google Scholar]
- 25.Beltramini AM, Mukhopadhyay CD, Pancholi V. Modulation of cell wall structure and antimicrobial susceptibility by a Staphylococcus aureus eukaryote-like serine/threonine kinase and phosphatase. Infect Immun. 2009;77:1406–1416. doi: 10.1128/IAI.01499-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Débarbouillé M, et al. Characterization of a serine/threonine kinase involved in virulence of Staphylococcus aureus. J Bacteriol. 2009;191:4070–4081. doi: 10.1128/JB.01813-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.von Dassow P, et al. Transcriptome analysis of functional differentiation between haploid and diploid cells of Emiliania huxleyi, a globally significant photosynthetic calcifying cell. Genome Biol. 2009;10:R114. doi: 10.1186/gb-2009-10-10-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 29.Novick RP. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol. 2003;48:1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- 30.Cheung AL, Nishina KA, Trotonda MP, Tamber S. The SarA protein family of Staphylococcus aureus. Int J Biochem Cell Biol. 2008;40:355–361. doi: 10.1016/j.biocel.2007.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen PR, et al. An oxidation-sensing mechanism is used by the global regulator MgrA in Staphylococcus aureus. Nat Chem Biol. 2006;2:591–595. doi: 10.1038/nchembio820. [DOI] [PubMed] [Google Scholar]
- 32.Chen PR, et al. A new oxidative sensing and regulation pathway mediated by the MgrA homologue SarZ in Staphylococcus aureus. Mol Microbiol. 2009;71:198–211. doi: 10.1111/j.1365-2958.2008.06518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujimoto DF, et al. Staphylococcus aureus SarA is a regulatory protein responsive to redox and pH that can support bacteriophage lambda integrase-mediated excision/recombination. Mol Microbiol. 2009;74:1445–1458. doi: 10.1111/j.1365-2958.2009.06942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ballal A, Manna AC. Control of thioredoxin reductase gene (trxB) transcription by SarA in Staphylococcus aureus. J Bacteriol. 2010;192:336–345. doi: 10.1128/JB.01202-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Truong-Bolduc QC, Hooper DC. Phosphorylation of MgrA and its effect on expression of the NorA and NorB efflux pumps of Staphylococcus aureus. J Bacteriol. 2010;192:2525–2534. doi: 10.1128/JB.00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Didier JP, Cozzone AJ, Duclos B. Phosphorylation of the virulence regulator SarA modulates its ability to bind DNA in Staphylococcus aureus. FEMS Microbiol Lett. 2010;306:30–36. doi: 10.1111/j.1574-6968.2010.01930.x. [DOI] [PubMed] [Google Scholar]
- 37.Bae T, et al. Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing. Proc Natl Acad Sci USA. 2004;101:12312–12317. doi: 10.1073/pnas.0404728101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Binkley F. Preparation and properties of S-phosphocysteine. J Biol Chem. 1952;195:283–285. [PubMed] [Google Scholar]
- 39.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 40.Chen PR, Brugarolas P, He C. Redox signaling in human pathogens. Antioxid Redox Signal. 2011;14:1107–1118. doi: 10.1089/ars.2010.3374. [DOI] [PubMed] [Google Scholar]
- 41.Chien Y, Manna AC, Projan SJ, Cheung AL. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J Biol Chem. 1999;274:37169–37176. doi: 10.1074/jbc.274.52.37169. [DOI] [PubMed] [Google Scholar]
- 42.Poor CB, Chen PR, Duguid E, Rice PA, He C. Crystal structures of the reduced, sulfenic acid, and mixed disulfide forms of SarZ, a redox active global regulator in Staphylococcus aureus. J Biol Chem. 2009;284:23517–23524. doi: 10.1074/jbc.M109.015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Y, et al. Structural and function analyses of the global regulatory protein SarA from Staphylococcus aureus. Proc Natl Acad Sci USA. 2006;103:2392–2397. doi: 10.1073/pnas.0510439103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burnside K, et al. Regulation of hemolysin expression and virulence of Staphylococcus aureus by a serine/threonine kinase and phosphatase. PLoS ONE. 2010;5:e11071. doi: 10.1371/journal.pone.0011071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chan PF, Foster SJ. Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J Bacteriol. 1998;180:6232–6241. doi: 10.1128/jb.180.23.6232-6241.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reck M, et al. The biofilm inhibitor carolacton disturbs membrane integrity and cell division of Streptococcus mutans through the serine/threonine protein kinase PknB. J Bacteriol. 2011;193:5692–5706. doi: 10.1128/JB.05424-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Booth MC, et al. Staphylococcal accessory regulator (sar) in conjunction with agr contributes to Staphylococcus aureus virulence in endophthalmitis. Infect Immun. 1997;65:1550–1556. doi: 10.1128/iai.65.4.1550-1556.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaito C, Morishita D, Matsumoto Y, Kurokawa K, Sekimizu K. Novel DNA binding protein SarZ contributes to virulence in Staphylococcus aureus. Mol Microbiol. 2006;62:1601–1617. doi: 10.1111/j.1365-2958.2006.05480.x. [DOI] [PubMed] [Google Scholar]
- 49.Luong TT, Dunman PM, Murphy E, Projan SJ, Lee CY. Transcription profiling of the mgrA Regulon in Staphylococcus aureus. J Bacteriol. 2006;188:1899–1910. doi: 10.1128/JB.188.5.1899-1910.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fiedler D, et al. Functional organization of the S. cerevisiae phosphorylation network. Cell. 2009;136:952–963. doi: 10.1016/j.cell.2008.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parkinson JS. Signal transduction schemes of bacteria. Cell. 1993;73:857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- 52.Casino P, Rubio V, Marina A. The mechanism of signal transduction by two-component systems. Curr Opin Struct Biol. 2010;20:763–771. doi: 10.1016/j.sbi.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 53.Mougous JD, Gifford CA, Ramsdell TL, Mekalanos JJ. Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat Cell Biol. 2007;9:797–803. doi: 10.1038/ncb1605. [DOI] [PubMed] [Google Scholar]
- 54.Molle V, Kremer L. Division and cell envelope regulation by Ser/Thr phosphorylation: Mycobacterium shows the way. Mol Microbiol. 2010;75:1064–1077. doi: 10.1111/j.1365-2958.2009.07041.x. [DOI] [PubMed] [Google Scholar]
- 55.Motley ST, Lory S. Functional characterization of a serine/threonine protein kinase of Pseudomonas aeruginosa. Infect Immun. 1999;67:5386–5394. doi: 10.1128/iai.67.10.5386-5394.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weerapana E, et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pannifer AD, Flint AJ, Tonks NK, Barford D. Visualization of the cysteinyl-phosphate intermediate of a protein-tyrosine phosphatase by X-ray crystallography. J Biol Chem. 1998;273:10454–10462. doi: 10.1074/jbc.273.17.10454. [DOI] [PubMed] [Google Scholar]
- 58.Postma PW, Lengeler JW, Jacobson GR. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Truong-Bolduc QC, Ding Y, Hooper DC. Posttranslational modification influences the effects of MgrA on norA expression in Staphylococcus aureus. J Bacteriol. 2008;190:7375–7381. doi: 10.1128/JB.01068-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ji Q, et al. Staphylococcus aureus CymR is a new thiol-based oxidation-sensing regulator of stress resistance and oxidative response. J Biol Chem. 2012;287:21102–21109. doi: 10.1074/jbc.M112.359737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chenna R, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.