Abstract
Dynamic actin filaments are a crucial component of clathrin-mediated endocytosis when endocytic proteins cannot supply enough energy for vesicle budding. Actin cytoskeleton is thought to provide force for membrane invagination or vesicle scission, but how this force is transmitted to the plasma membrane is not understood. Here we describe the molecular mechanism of plasma membrane–actin cytoskeleton coupling mediated by cooperative action of epsin Ent1 and the HIP1R homolog Sla2 in yeast Saccharomyces cerevisiae. Sla2 anchors Ent1 to a stable endocytic coat by an unforeseen interaction between Sla2’s ANTH and Ent1’s ENTH lipid-binding domains. The ANTH and ENTH domains bind each other in a ligand-dependent manner to provide critical anchoring of both proteins to the membrane. The C-terminal parts of Ent1 and Sla2 bind redundantly to actin filaments via a previously unknown phospho-regulated actin-binding domain in Ent1 and the THATCH domain in Sla2. By the synergistic binding to the membrane and redundant interaction with actin, Ent1 and Sla2 form an essential molecular linker that transmits the force generated by the actin cytoskeleton to the plasma membrane, leading to membrane invagination and vesicle budding.
Keywords: ANTH domain, membrane remodeling, PIP2
The dynamic actin cytoskeleton drives membrane shape changes in many key cellular processes such as morphogenesis, polarization, and vesicular trafficking. Although the idea was proposed long ago (1, 2), only recently was local actin polymerization confirmed to power clathrin-mediated endocytosis when the force provided by the endocytic proteins is not sufficient for vesicle budding (3–5). In mammalian cells the extra force from the actin cytoskeleton is needed under conditions that increase the energy requirements of membrane bending (5), whereas in yeast it is always required because of the high membrane tension created by the cellular turgor pressure (3).
Like the actin-driven membrane protrusion at the leading edge of migrating animal cells, the actin filaments at the endocytic sites are nucleated by the Arp2/3 complex. However, during endocytosis the membrane is pulled into the cell, not pushed out as in cell migration. Therefore, during endocytosis the plasma membrane must be coupled in some way to the actin cytoskeleton so that the force from the actin cytoskeleton can be transmitted to the membrane (2, 6). However, the molecular nature of this membrane–actin coupling is not known.
Potential candidates for mediating the membrane–actin coupling are the yeast protein Sla2 and its mammalian homolog HIP1R because (i) they can bind both the membrane and actin via their N-terminal ANTH domain and C-terminal THATCH/talin-like domain, respectively (7–10), and (ii) cells deficient in these proteins exhibit a specific endocytic block called an “uncoupling phenotype.” The knockout of the SLA2 gene in yeast (11) or siRNA-mediated down-regulation of HIP1R in HeLa cells (12) stops endocytic vesicle budding even though the endocytic coats are assembled on the plasma membrane and actin polymerizes continuously at these sites. This phenotype suggests that the actin cytoskeleton is not coupled to the membrane efficiently and therefore cannot drive its invagination. Paradoxically, endocytic vesicle formation is not dependent on the actin-binding activity of Sla2 (13, 14).
Importantly, a similar uncoupling phenotype has been observed recently in cells lacking endocytic epsins in yeast and in the cellular slime mold Dictyostelium discoideum, indicating that epsins may be involved in membrane–actin coupling (15, 16). However, epsins are canonical endocytic adaptors connecting the plasma membrane to the clathrin coat (17, 18) and are not known to bind to actin, thereby leaving their role in membrane–actin coupling unclear.
Here we describe the molecular mechanism by which epsin Ent1 and the HIP1R homolog Sla2 mediate membrane–actin coupling during endocytosis in the yeast Saccharomyces cerevisiae. Ent1 and Sla2 cooperate to bind to the plasma membrane and interact in a redundant manner with the actin cytoskeleton to transmit force from the cytoskeleton to the membrane.
Results
Functional Interplay of Sla2 and Ent1 at the Endocytic Site.
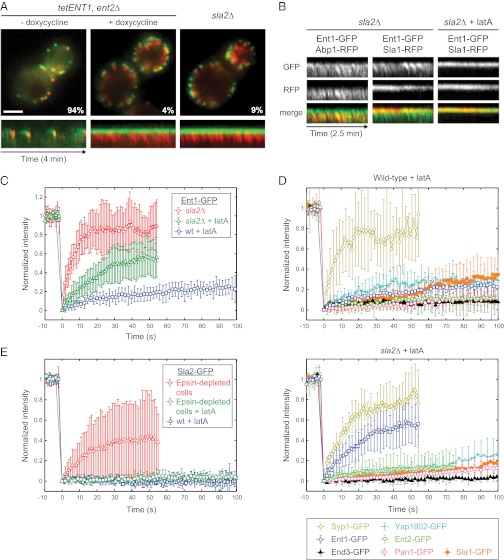
The uncoupling phenotype of SLA2 deletion was described as unique among deletion phenotypes of yeast endocytic factors (19). However, the yeast cells lacking both endocytic epsins Ent1 and Ent2 recently have been shown to exhibit an endocytic phenotype similar to sla2∆ (15). Because a single deletion of either the ENT1 or ENT2 gene does not cause any obvious phenotype, but the deletion of both genes was reported to be lethal (20), we constructed a yeast strain with the ENT2 gene deleted and the ENT1 gene regulated by a tetO promoter to study the effect of epsin depletion by live-cell imaging (Fig. S1 A and B). We found that endocytic coats (marked by the endocytic adaptor protein Sla1-GFP) were arrested in nonmotile endocytic patches on the plasma membrane, whereas dynamic actin tails (marked by the actin marker Abp1-RFP) were polymerized continuously from these sites, as in sla2∆ cells (Fig. 1A and Movie S1). In epsin-depleted and sla2∆ cells, only 4% and 9% of Sla1-GFP patches, respectively, showed normal internalization movement indicative of a successful endocytic event. These results suggest that, along with Sla2, the epsins may contribute to membrane–actin cytoskeleton coupling.
Fig. 1.
Sla2 stabilizes Ent1 at endocytic sites. (A) The uncoupling phenotype in epsin-depleted and sla2∆ cells. (Upper) Merged images of Sla1-GFP and Abp1-RFP in epsin-depleted (tetENT1, ent2∆) and sla2∆ strains are shown. The images of the epsin-depleted strain were taken before (− doxycycline) and after (+ doxycycline) the depletion of Ent1. The percentage of endocytic patches internalized during 4-min movies is shown for each strain. (Lower) Kymographs of Sla1-GFP and Abp1-RFP at endocytic patches of indicated strains. (Scale bar: 2 μm.) (B) Ent1 colocalizes with both the actin tails and the endocytic coat in sla2∆ cells. Kymographs of Ent1-GFP and Abp1-RFP or Ent1-GFP and Sla1-RFP at endocytic patches in sla2∆ cells treated or untreated with latA are shown as separate channels and merged images. (C) FRAP of Ent1-GFP at endocytic sites in latA-treated wild-type cells, latA-treated sla2∆ cells, and sla2∆ cells. The recovery curves represent the mean ± SD (n = 9–11 separate measurements). (D) FRAP analyses of the endocytic proteins Yap1802, Ent2, End3, Pan1, Sla1, and Syp1 at endocytic sites in latA-treated wild-type (Upper) and sla2∆ (Lower) cells. The curves represent the mean ± SD (n = 6–12). The recovery curves of Ent1-GFP photobleached under the same experimental conditions (from C) are shown for comparison. (E) FRAP of Sla2-GFP at endocytic sites in latA-treated wild-type cells, latA-treated epsin-depleted cells, and epsin-depleted cells. The recovery curves represent the mean ± SD (n = 5–12).
Epsins bind the membrane by an N-terminal ENTH domain (21–23), but they are not known to bind actin. However, Newpher et al. (24) reported that in sla2∆ cells Ent1-GFP exhibited an abnormal localization pattern that resembled the actin tails present in these mutant cells. To study this observation further, we expressed Ent1-GFP in an sla2∆ strain together with Abp1-RFP and Sla1-RFP markers, respectively. Interestingly, in sla2∆ cells Ent1-GFP partially localized to the actin tails marked by Abp1-RFP, whereas after inhibition of actin polymerization by latrunculin A (latA) Ent1-GFP localized only to the endocytic coat marked by Sla1-RFP (Fig. 1B and Movie S2). Therefore, Ent1 may interact not only with the membrane and coat proteins but also with the actin cytoskeleton of the endocytic site.
To understand better the possible interplay of Ent1 and Sla2 at the endocytic site, we analyzed fluorescence recovery after photobleaching (FRAP) of Ent1-GFP and Sla2-GFP at endocytic sites in wild-type cells and in sla2∆ or epsin-depleted cells, respectively. To perform FRAP experiments in wild-type cells, we arrested endocytosis with latA, which allows the assembly of the endocytic coat but blocks vesicle budding (11, 19, 24). Ent1-GFP recovery in latA-treated wild-type cells was very slow, indicating that Ent1 binds to the endocytic site in a stable manner (Fig. 1C and Movie S3). The recovery of Ent1-GFP in latA-treated sla2∆ cells was significantly faster (Fig. 1C and Movie S3). An even faster recovery of Ent1-GFP was observed in sla2∆ cells in the absence of latA (Fig. 1C and Movie S3). These results suggest that the binding of Ent1 to the endocytic site is stabilized by Sla2 and destabilized by F-actin.
To test if the effect of SLA2 deletion on Ent1 is specific, we performed FRAP experiments on other endocytic coat and early-module proteins (as classified in ref. 15). The coat proteins Yap1802, Ent2, End3, Pan1, and Sla1 showed very slow recoveries similar to the recovery of Ent1 in latA-treated wild-type cells whereas the early-module protein Syp1 recovered quickly (Fig. 1D). Importantly, for each of the proteins tested the recovery rates were similar in Sla2-expressing cells and sla2∆ cells, suggesting that the stabilizing effect of Sla2 is specific for Ent1 (Fig. 1D).
We then tested whether the localization and stability of Sla2 at the endocytic sites depends on the epsins. Contrary to Ent1-GFP behavior in sla2∆ cells, the localization and fluorescence recovery of Sla2 was not changed in epsin-depleted cells in comparison with wild-type cells (Fig. 1E and Fig. S1C), suggesting that Sla2 can bind tightly to the endocytic site independently of epsins. These data suggest that Sla2 helps tether Ent1 to a stable endocytic coat, whereas in the absence of Sla2 polymerizing actin may pull Ent1 off the membrane.
Sla2’s ANTH Domain Stabilizes Ent1’s ENTH Domain at the Endocytic Site.
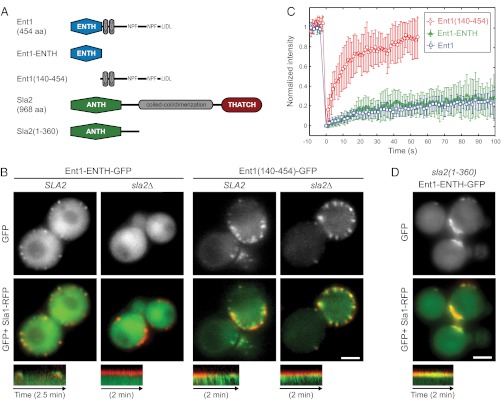
To understand how Ent1 interacts with Sla2 and the actin cytoskeleton, we expressed Ent1 in two parts: the N-terminal phosphatidylinositol (4,5)-bisphosphate [PI(4,5)P2]-binding ENTH domain (Ent1-ENTH; amino acids 1–154) and the C-terminal part [Ent1(140–454)] that binds other coat-associated proteins and clathrin (Fig. 2A) (18). We observed these fragments as GFP fusions in epsin-depleted cells. Ent1-ENTH-GFP localized to the endocytic coats marked by Sla1-RFP and rescued the endocytic defect of epsin depletion (Fig. 2B and Movie S4) (20). Ent1(140–454)-GFP failed to rescue the endocytic defect and colocalized with both the arrested coats and the actin tails (Fig. 2B and Movie S4). To assess the contributions of Ent1-ENTH and Ent1(140–454) to the stability of the Ent1 protein at the endocytic site, we performed FRAP with latA-treated epsin-depleted cells expressing either of the two Ent1 fragments (Fig. 2C and Movie S5). Ent1-ENTH appeared to be a very stable component of the endocytic site with exchange rates similar to those of the full-length Ent1. In contrast, Ent1(140–454) exhibited a very rapid exchange.
Fig. 2.
The Sla2 ANTH domain is required for stable binding of the Ent1 ENTH domain to the endocytic site. (A) The domain organization of Ent1 and Sla2 and the fragments used in this study. Protein domains and linear motifs for binding to EH-domain proteins (NPF) and clathrin (LIDL) are shown. (B) Localization of Ent1-ENTH-GFP or Ent1(140–454)-GFP together with Sla1-RFP in epsin-depleted cells in the presence or absence of Sla2. GFP and merged images are shown together with kymographs of Ent1 constructs and Sla1-RFP at endocytic patches. (C) FRAP of Ent1-ENTH-GFP and Ent1(140–454)-GFP at endocytic sites in latA-treated epsin-depleted cells. Curves represent the mean ± SD (n = 7–13 separate measurements). The Ent1-GFP FRAP in latA-treated wild-type cells (from Fig. 1C) is shown for comparison. (D) Sla2(1–360) stabilizes Ent1 ENTH at endocytic sites. Localization of Ent1-ENTH-GFP and Sla1-RFP in epsin-depleted sla2(1-360) cells (compare with B). GFP and merged images are shown together with the kymographs. All kymographs are oriented with the cell exterior at the top. (Scale bars: 2 μm in B and D.)
Next we observed both constructs in cells lacking Sla2. Interestingly, in epsin-depleted sla2∆ cells the membrane localization of Ent1-ENTH was lost completely, but the localization of Ent1(140–454) was not affected (Fig. 2B and Movie S4). These data suggest that the ENTH domain is necessary and sufficient for the Sla2-dependent tight binding of Ent1 to the endocytic sites and that the C-terminal part of Ent1 may interact with the actin cytoskeleton.
Because the N-terminal region of Sla2 that contains the ANTH domain [Sla2(1–360)] can support endocytosis in an otherwise wild-type strain (25) and localizes to endocytic sites independently of epsins (Fig. S1D), we tested if Sla2(1–360) is responsible for anchoring Ent1-ENTH to the endocytic site. Indeed, the expression of Sla2(1–360) was sufficient to recruit Ent1-ENTH to endocytic sites in epsin-depleted cells (Fig. 2D and Movie S4). However, these sites were nonmotile, suggesting that the membrane-binding domains of Ent1 and Sla2 are sufficient for localization of these proteins to the endocytic site but not for endocytosis. FRAP experiments confirmed that Sla2(1–360) is able to stabilize ENTH-GFP fully (Fig. S1E).
Ent1 ENTH and Sla2 ANTH Domains Interact to Bind the Membrane Cooperatively.
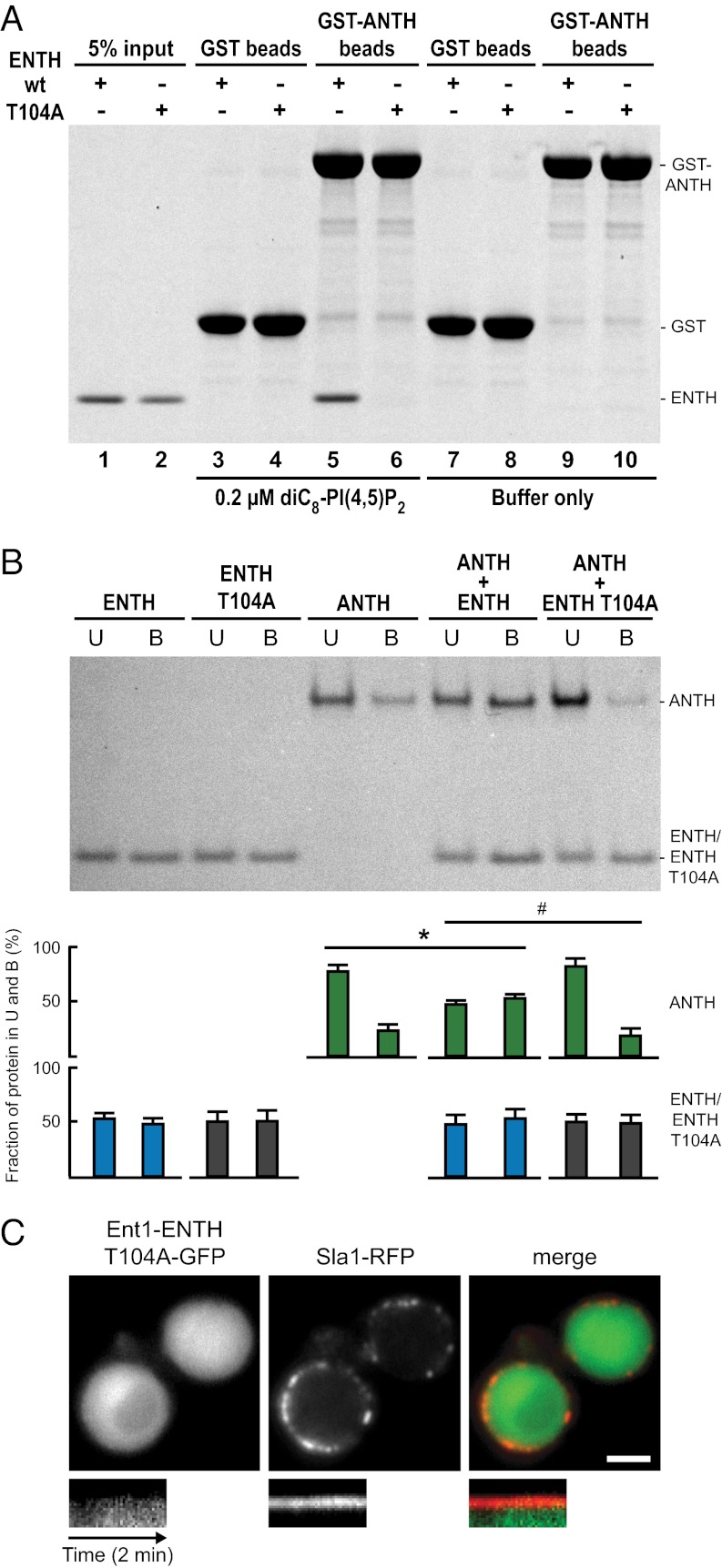
The localization and FRAP experiments suggested that Sla2 might anchor Ent1-ENTH to the endocytic site by a direct interaction between the ANTH and ENTH domains. To test this notion, we performed in vitro GST pull-down experiments with recombinant GST-ANTH and ENTH proteins. We also tested the possible interaction between the GST-ANTH domain and the ENTH domain carrying a T104A point mutation, which interferes with epsin function but is not involved in lipid binding (20, 26, 27). We did not detect any binding between GST-ANTH and ENTH or ENTH T104A (Fig. 3A, lanes 9 and 10). However, we also tested the interaction in the presence of dioctanoyl-PI(4,5)P2 [diC8-PI(4,5)P2], a water-soluble lipid ligand of the ANTH and ENTH domains (22, 28). Strikingly, in the presence of diC8-PI(4,5)P2 the ENTH domain interacted specifically with GST-ANTH, and this interaction was prevented by the T104A mutation in the ENTH domain (Fig. 3A, lanes 3–6). We obtained similar results when the ANTH domain was incubated with GST-ENTH or GST-ENTH T104A in the presence or absence of diC8-PI(4,5)P2 (Fig. S1F).
Fig. 3.
Ent1 ENTH and Sla2 ANTH domains interact with each other. (A) ENTH binds GST-ANTH in the presence of soluble lipid ligand diC8-PI(4,5)P2. Glutathione beads with GST-ANTH or GST alone were incubated with an equimolar amount of ENTH or ENTH T104A protein in the presence or absence of diC8-PI(4,5)P2 at 25 °C. Beads with bound proteins were analyzed by SDS/PAGE and Coomassie staining. A representative experiment from four independent pull-down assays is shown. (B) ENTH and ANTH bind cooperatively to PI(4,5)P2-containing liposomes in vitro. ENTH, ENTH T104A, and ANTH alone or in combination were incubated with PI(4,5)P2-containing liposomes. (Upper) Liposome-bound (B) and -unbound (U) protein fractions were solved by SDS/PAGE and measured densitometrically. (Lower) Bar graphs show the protein distribution between the fractions (% ± SD; n = 4 independent experiments). *P < 0.018 and #P < 0.021 (Student’s t test). (C) Localization of Ent1-ENTH T104A-GFP and Sla1-RFP in epsin-depleted cells. Separate channels and the merged image are shown together with the kymographs (compare with Fig. 2 B and D). Kymographs are oriented with the cell exterior at the top. (Scale bar: 2 μm.)
The dependence of the ENTH–ANTH interaction on the lipid analog suggests that the interaction between the ENTH and ANTH domains occurs after their binding to the membrane. We tested the functional effect of the ENTH–ANTH interaction on membrane binding using an in vitro liposome-binding assay. Both the ENTH and ANTH domains bound specifically to PI(4,5)P2-containing liposomes, and the T104A mutation did not alter ENTH binding (Fig. 3B). Remarkably, when the ANTH and ENTH domains were incubated together with liposomes, the binding of the ANTH domain to liposomes was increased significantly. When the ANTH domain was combined with the ENTH T104A mutant, no cooperative effect in liposome binding was observed (Fig. 3B). This result suggests that the binding of the Ent1 ENTH and the Sla2 ANTH domains to PI(4,5)P2 triggers a direct protein–protein interaction between these two domains, leading to cooperative membrane binding. The T104A mutation of ENTH abolishes the cooperative effect, possibly by interfering with the ENTH–ANTH interaction.
To test the role of the ENTH–ANTH interaction in vivo, we observed the ENTH T104A mutant as a GFP fusion in epsin-depleted cells. In contrast to the wild-type ENTH domain, ENTH T104A failed to localize to endocytic sites and did not rescue the endocytic defect of epsin depletion (Fig. 3C). This result suggests that Sla2 anchors Ent1 to the endocytic site by a direct, membrane-binding–induced interaction between the ANTH and ENTH domains.
In vivo Sla2 is bound tightly to the endocytic site independently of Ent1 and thus can stabilize Ent1 at the site. However, in our in vitro liposome-binding assay the Sla2 ANTH domain binds more weakly than the Ent1 ENTH domain (Fig. 3B), and therefore the effect of the cooperative action is seen as enhanced binding of the ANTH domain.
Identification of an F-Actin–Binding Domain in Epsin Ent1.
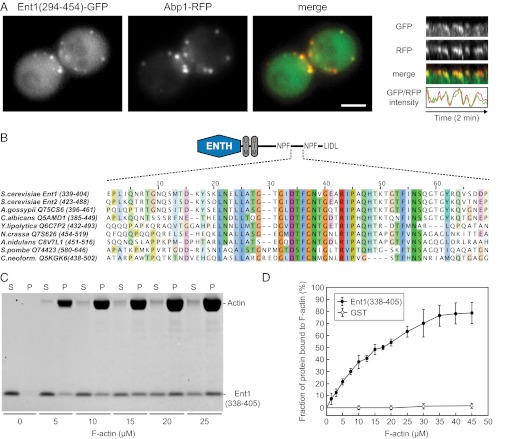
To map the interaction between the C-terminal part of Ent1 and the actin cytoskeleton, we observed different subfragments of Ent1(140–454) in epsin-depleted cells (Fig. S2A). Although the Ent1(140–294)-GFP fragment localized similarly to other coat proteins, the Ent1(294–454)-GFP fragment colocalized solely with the actin tails (Fig. S2 B and C). In addition, although the latA treatment did not affect the localization of the Ent1(140–294) fragment, it completely abolished the cortical localization of the Ent1(294–454) fragment (Fig. S2 B and C). When we expressed Ent1(294–454)-GFP in a wild-type strain, it localized to endocytic sites simultaneously with the actin marker Abp1-RFP (Fig. 4A and Movie S6), unlike the full-length Ent1 that localizes to the endocytic site significantly before actin (15). Sequence comparison revealed a conserved region (amino acids 338–405) in the central part of Ent1(294–454) (Fig. 4B). This fragment, Ent1(338–405), was sufficient for actin-tail localization in epsin-depleted cells (Fig. S2 B and C); therefore we refer to this region of Ent1 (amino acids 338–405) as the “actin cytoskeleton-binding” (ACB) domain. We then assessed the interaction of recombinant Ent1(338–405) with F-actin in vitro. Ent1(338–405) cosedimented with F-actin with a Kd of ∼17 μM (Fig. 4 C and D), indicating that Ent1 can interact with actin filaments directly. Although the affinity of the ACB–actin interaction is low, multiple Ent1 molecules and a dense F-actin network at the endocytic site may provide high avidity so that the interaction may be sufficiently strong for membrane–actin coupling while still allowing its dynamic regulation.
Fig. 4.
Identification of the Ent1 F-actin binding domain. (A) Colocalization of Ent1(294–454)-GFP with Abp1-RFP at endocytic sites. (Left) Separate channels and the merged image of an ABP1-RFP strain expressing Ent1(294–454)-GFP from a plasmid. (Right) Kymographs and fluorescence intensity profiles of Ent1(294–454)-GFP and Abp1-RFP at a single endocytic patch. Kymographs are oriented with the cell exterior at the top. (Scale bar: 2 μm.) (B) The multiple sequence alignment of the ACB region of S. cerevisiae Ent1, Ent2, and epsin proteins from indicated species. For each sequence the UniProt accession number and the amino acids aligned are indicated. (C) Ent1(338–405) binds to F-actin in the actin cosedimentation assay. Recombinant Ent1(338–405) protein (5 μM final concentration) was incubated with increasing amounts of F-actin. The fractions of unbound (S) and F-actin–bound protein (P) are shown. (D) Quantification of Ent1(338–405) binding to F-actin. The binding of Ent1(338–405) protein or GST control to F-actin was analyzed as in C. Data represent the mean ± SD (n = 3 independent experiments).
Ent2, the redundant paralog of Ent1, has both ENTH and ACB domains. The ENTH domain of Ent2 (Ent2-ENTH; amino acids 1–156) rescued the endocytic defect of epsin depletion (20) and localized to the endocytic coats similarly to Ent1’s ENTH (Fig. S3A). The Ent2-ENTH localization was dependent on the ANTH domain-containing fragment of Sla2 and was impaired by the T104A mutation, as was Ent1-ENTH (Fig. S3A), strongly suggesting that the ENTH domains of both Ent1 and Ent2 can interact with the Sla2 ANTH domain.
The C-terminal fragment of Ent2 containing the ACB domain [Ent2(421–609)] colocalized with the actin tails in epsin-depleted cells (Fig. S3 B and C). However, both the full-length Ent2-GFP in sla2Δ cells and the C-terminal part of Ent2 excluding the ENTH domain [Ent2(155–613)] in epsin-depleted cells localized strictly with coat proteins and not with the actin tails, in contrast to their Ent1 counterparts (Fig. S3 B and D). These results suggest that, although the Ent2 ACB domain has the ability to interact with the actin cytoskeleton, this interaction is suppressed in the full-length Ent2 protein.
Sla2 and Ent1 Interact Redundantly with the Actin Cytoskeleton.
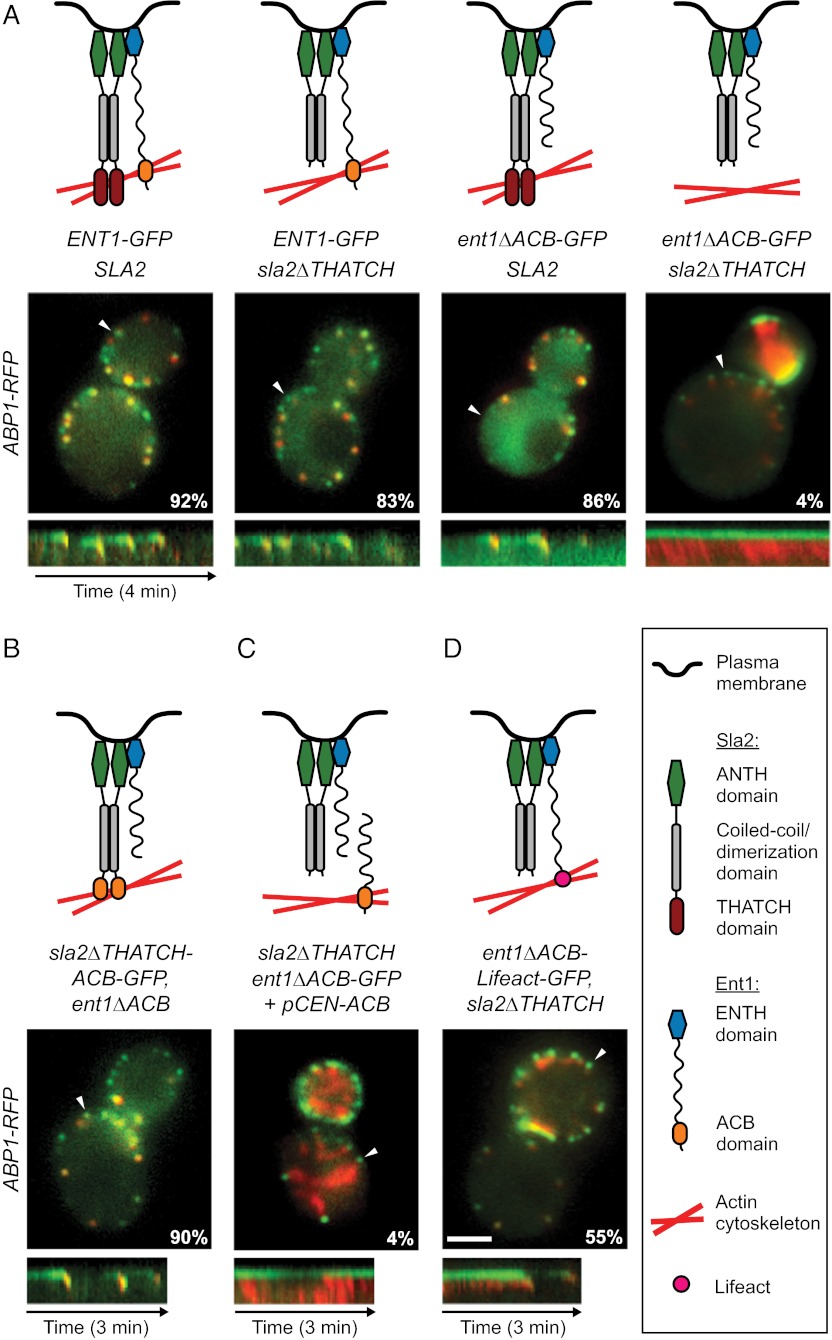
Although the actin cytoskeleton is essential for endocytic vesicle formation in yeast, neither the actin-binding THATCH domain of Sla2 nor the C-terminal part of Ent1 containing the ACB domain is required for this process (13, 14, 20). In fact, strains carrying individual deletions of the THATCH or the ACB domain (sla2∆THATCH and ent1∆ACB) exhibited normal endocytic dynamics with only a slightly decreased percentage of patches internalizing during a 4-min observation (Fig. 5A and Movie S7). We hypothesized that the ACB and the THATCH domains work redundantly to link the actin cytoskeleton to the membrane. Indeed, when the deletions of the THATCH and the ACB domains were combined, we observed the same uncoupling phenotype as in sla2∆ or epsin-depleted cells (Fig. 5A and Movie S7). Note that wild-type Ent2 protein was expressed normally in these cells, suggesting that Ent2 does not contribute significantly to membrane–actin coupling.
Fig. 5.
Ent1 ACB and Sla2 THATCH domains bind the actin cytoskeleton redundantly. (A) (Top) Schematic models of function of individual Ent1-Sla2 combinations. (Middle) Merged images of four strains expressing Sla2 or Sla2∆THATCH in combination with Ent1-GFP or Ent1∆ACB-GFP together with Abp1-RFP as an actin marker. The percentage of endocytic patches internalized during 4-min movies is shown for each strain. (Bottom) Kymographs of Ent1-GFP or Ent1∆ACB-GFP and Abp1-RFP from sites indicated by arrowheads in the merged images. (B) Sla2∆THATCH-ACB chimeric protein couples the membrane and the actin cytoskeleton. Sla2∆THATCH-ACB-GFP fusion protein was expressed in ent1∆ACB strain together with Abp1-RFP. The model of chimera function (Top), the merged image and percentage of internalized endocytic patches (Middle), and kymograph (Bottom) are shown as in A. (C) Expression of Ent1 ENTH and ACB domains in trans is not sufficient to couple the actin cytoskeleton to the membrane. Ent1(294–454) was expressed in ent1∆ACB-GFP sla2∆THATCH ABP1-RFP strain from a plasmid. The model of Ent1-Sla2 function (Top), the merged image and the percentage of internalized patches (Middle), and kymograph (Bottom) are shown as in A. (D) The Ent1 ACB domain can be replaced by the heterologous actin-binding domain Lifeact for membrane–actin coupling. The Ent1∆ACB-Lifeact-GFP fusion protein was expressed in the sla2∆THATCH strain together with Abp1-RFP. The model of chimera function (Top), the merged image and the percentage of internalized endocytic patches (Middle), and kymograph (Bottom) are shown as in A. (Scale bar: 2 μm.)
To test further the redundancy of the ACB and THATCH domains in membrane–actin coupling, we replaced the THATCH domain sequence of SLA2 gene with the ACB domain sequence of ENT1. We observed the chimeric Sla2∆THATCH-ACB-GFP protein together with Abp1-RFP or Sla1-RFP in an ent1∆ACB strain. The behavior of Sla2∆THATCH-ACB-GFP and both endocytic markers at the endocytic sites closely resembled the dynamics of Sla2, Abp1, and Sla1 proteins in the wild-type strain (Fig. 5B and Movie S7). This result strongly indicates that the Ent1 ACB domain can replace the Sla2 THATCH domain functionally and therefore that they share common biochemical functionality.
We then asked if the covalent link between the membrane-binding ENTH domain and the actin-binding ACB domain of Ent1 is required for membrane–actin coupling. We expressed the C-terminal fragment of Ent1 containing the ACB domain [Ent1(294–454)] in an sla2∆THATCH ent1∆ACB strain and analyzed the endocytic dynamics. The presence of the Ent1 ACB and ENTH domains as separate polypeptide chains was not sufficient to rescue the uncoupling phenotype of this strain (Fig. 5C and Movie S7).
Next we replaced the ACB sequence of the ENT1 gene with the sequence of a heterologous actin-binding domain, Lifeact, which binds actin but does not affect its polymerization or Arp2/3-mediated nucleation (29). The analysis of Sla1-RFP and Abp1-RFP dynamics showed that expression of the Ent1∆ACB-Lifeact-GFP fusion protein in an sla2∆THATCH strain rescued endocytosis to a great extent (Fig. 5D and Movie S7).
These data strongly suggest that Ent1 and Sla2 redundantly couple the actin cytoskeleton to the endocytic membrane via their THATCH and ACB domains, thereby transmitting the pulling force from the actin network to the membrane.
Phosphoregulation of the Ent1 ACB–Actin Cytoskeleton Interaction.
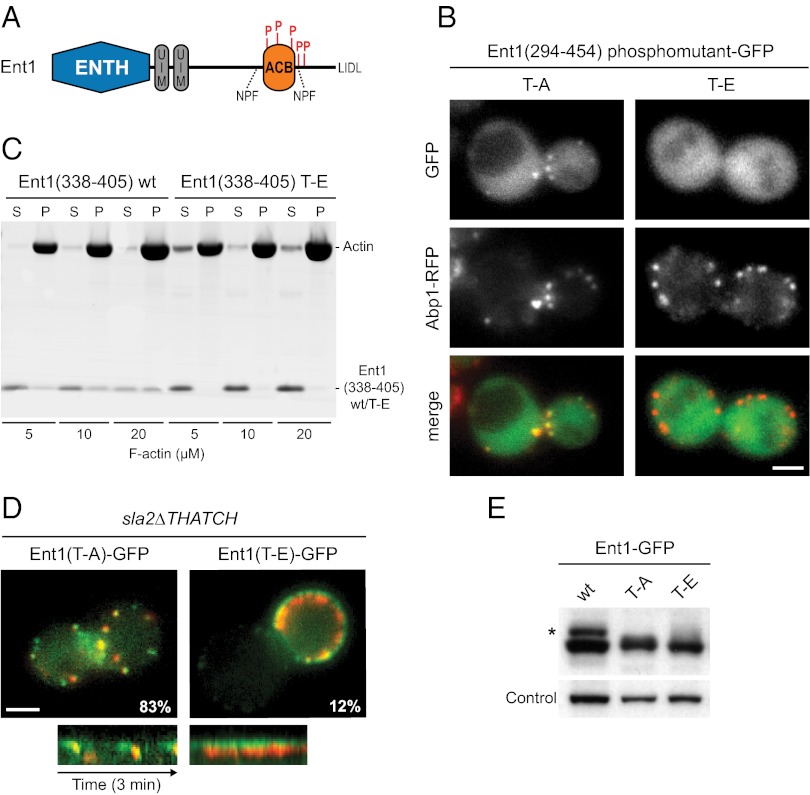
Ent1 is known to be phosphorylated on five threonines, T346, T364, T395, T415, and T427, by the Prk1 kinase (30, 31). Interestingly, these threonines are located within or in close proximity to the ACB domain (Fig. 6A). The Prk1 kinase (homologous to human AAK) regulates the disassembly of the endocytic coat by inhibiting interactions between endocytic proteins after vesicle scission (32). We therefore analyzed the possible role of Ent1 phosphorylation in the regulation of the Ent1–actin cytoskeleton interaction.
Fig. 6.
Phosphoregulation of the Ent1 ACB domain. (A) A schematic of the Ent1 domain organization with Prk1 phosphorylation sites indicated. (B) Localization of Ent1(294–454)-GFP phosphomutants. Separate channels and merged images of the ABP1-RFP strain expressing Ent1(294–454) T–A or Ent1(294–454) T–E GFP fusions from a plasmid are shown. (C) Comparison of wild-type Ent1(338–405) and Ent1(338–405) T–E mutant binding to F-actin in vitro. Recombinant proteins (5 μM final concentrations) were incubated with increasing amounts of F-actin. The fractions of unbound (S) and F-actin–bound protein (P) protein are shown. (D) The Ent1(T–E) phospho-mimicking mutant leads to the uncoupling phenotype. (Upper) Merged images of sla2∆THATCH strain expressing Abp1-RFP together with Ent1(T–A)-GFP or Ent1(T–E)-GFP, respectively. The percentage of Ent1-GFP patches internalized during 4-min movies is shown for each strain. (Lower) Kymographs of Ent1(T–A)-GFP or Ent1(T–E)-GFP together with Abp1-RFP at endocytic patches of sla2∆THATCH strain. Kymographs are oriented with the cell exterior at the top. (E) Expression levels of Ent1 constructs. Whole-cell lysates of strains expressing indicated Ent1 protein constructs were analyzed by immunoblotting with anti-HA antibody. The detection of Arc1 protein by anti-Arc1 antibody was used as a loading control. The asterisk indicates the phosphorylated fraction of wild-type Ent1 protein. (Scale bars: 2 μm in B and D.)
We first expressed the Ent1 C-terminal fragments with the Prk1 target threonines mutated to either alanines [Ent1(294–454) T–A] or glutamates [Ent1(294–454) T–E] as GFP fusions in wild-type cells. The nonphosphorylatable Ent1(294–454) T–A mutant colocalized with the actin patches marked by Abp1-RFP, whereas the phospho-mimicking mutant [Ent1(294–454) T–E showed only diffuse cytosolic localization (compare Figs. 6B and 4A). In a cosedimentation assay the F-actin binding of the phospho-mimicking mutant of the Ent1 ACB domain [Ent1(338–45) T–E] was severely reduced compared with the wild-type ACB domain (Fig. 6C). These results suggest that phosphorylation of Ent1 inhibits its interaction with endocytic actin filaments.
We then introduced the same T–A and T–E mutations into the chromosomal ENT1-GFP gene and observed the respective proteins in an sla2∆THATCH ABP1-RFP strain. Both Ent1 mutant proteins localized to endocytic sites. Although the endocytic patches were internalized normally in the Ent1 T–A mutant strain, the Ent1 T–E mutant strain showed a strong uncoupling phenotype (Fig. 6 D and E and Movie 8). This result suggests that the phosphorylation of Ent1 by Prk1 disrupts the coupling between Ent1 and the actin cytoskeleton, possibly facilitating the disassembly of the endocytic machinery after vesicle scission.
Discussion
In this study we show that Sla2 and Ent1 proteins couple the membrane and the actin cytoskeleton at the endocytic site. This coupling is essential for transmitting force from the actin cytoskeleton to the membrane to overcome the membrane tension that acts against membrane invagination (3, 5). Although Sla2 and its mammalian homolog HIP1R have both membrane- and actin-binding domains and have been implicated in membrane–actin coupling, the molecular mechanism was unknown previously (2, 6, 10). Importantly, the actin-binding activity of Sla2 has been shown to be unnecessary for endocytosis (13, 14). Epsins, on the other hand, are known to interact with the membrane and with other coat proteins, but no interaction with actin had been described. Our results provide a molecular mechanism for membrane–actin coupling mediated by the cooperative action of Sla2 and Ent1.
In both sla2∆ and epsin-depleted cells the endocytic coats remain nonmotile at the plasma membrane despite continuous actin polymerization at the endocytic sites. This phenotype suggests that in the absence of Sla2 or both epsins Ent1 and Ent2 the actin cytoskeleton is not properly coupled to the endocytic coat, and the mechanical force of actin polymerization cannot be used efficiently to invaginate the membrane.
Our FRAP experiments showed that Sla2, epsins, and other endocytic coat proteins are bound very stably to the endocytic site. This high stability likely is achieved by the assembly of the coat proteins into a stable lattice via multiple protein–protein interactions and may be required for maintaining the integrity of the coat under the forces from the actin cytoskeleton. We found that the stability of Ent1, but not the other coat proteins, depends critically on Sla2. Also, in the absence of Sla2 Ent1 exhibits a striking colocalization with the actin tails, hinting at an interaction between Ent1 and the actin cytoskeleton. Actin polymerization further increases the exchange rate of Ent1 in sla2∆ cells, suggesting that the flow of the actin network away from the membrane may pull Ent1 molecules off the membrane.
Ent1’s ENTH Domain and Sla2’s ANTH Domain Bind the Plasma Membrane Cooperatively.
Our experiments with fragments of Ent1 and Sla2 revealed that the interplay between these two proteins is mediated by their ENTH and ANTH domains. The membrane-binding ENTH domain is responsible for the stable binding of Ent1 to the endocytic site, whereas the C-terminal part of Ent1 interacts dynamically with the actin cytoskeleton and the endocytic coat. The localization of the ENTH domain to the endocytic site is fully dependent on Sla2 and specifically on its N-terminal fragment containing the ANTH domain.
These observations prompted us to test for a possible protein–protein interaction between the Ent1 ENTH and Sla2 ANTH domains using GST pull-down assays in vitro. Although no interaction could be detected between ligand-free ENTH and ANTH domains, specific binding between the Ent1 ENTH and Sla2 ANTH domains was observed readily in the presence of a soluble PI(4,5)P2 ligand. Importantly, when proteins were tested for their ability to bind to PI(4,5)P2-containing liposomes, the presence of ENTH led to a significant increase of the liposome-bound fraction of ANTH. These data strongly suggest that the Ent1 ENTH and Sla2 ANTH lipid-binding domains interact with each other upon membrane binding, thereby giving rise to a cooperative interaction with the membrane. The effects observed in vitro and in vivo appeared opposite: In vitro the ENTH domain supported the association of the otherwise weakly binding ANTH domain with liposomes, whereas in cells the ENTH domain required the ANTH domain for localization. This discrepancy is possibly caused by unknown factors present in vivo that strongly stabilize Sla2 at the endocytic site independently of epsins. Therefore in vivo the ANTH–ENTH interaction can be manifested as stabilization of the Ent1 protein at the endocytic site.
The behavior of the T104A mutant of Ent1’s ENTH domain further supports this notion. In vivo, Ent1-ENTH T104A could not localize to endocytic sites or complement the endocytic defect of epsin-depleted cells in the presence of Sla2. In vitro, the ENTH T104A domain did not enhance the binding of the Sla2 ANTH domain to liposomes, nor did it bind to ANTH in the presence of the PI(4,5)P2 ligand, although by itself it showed the same PI(4,5)P2-binding properties as wild-type ENTH. The conserved surface threonine 104, which lies outside the PI(4,5)P2-binding pocket of Ent1 ENTH and was shown to be important for epsin function (20, 26, 27), may be a part of an interaction surface between the Ent1 ENTH and Sla2 ANTH domains. Notably, the localization of the ENTH domain of Ent2 to the endocytic site also depends on the Sla2 ANTH domain and the identical threonine residue of Ent2 ENTH, suggesting that the ENTH domains of both Ent1 and Ent2 can interact similarly with Sla2 ANTH.
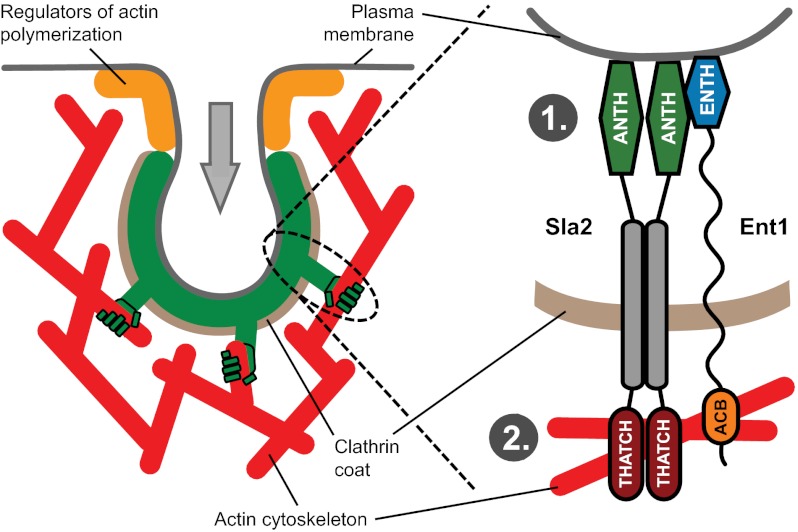
We propose that synergistic membrane binding by the ENTH and ANTH domains leads to tight anchoring of Ent1 and Sla2 to the membrane so that they can withstand pulling forces from the actin cytoskeleton. This synergy would explain why both the ENTH domain of Ent1 or Ent2 and the ANTH domain of Sla2 are required for endocytosis and why, although both Ent1 and Sla2 are capable of interacting with the membrane and actin, neither can perform the coupling function alone. A similar synergism has been suggested for different membrane-binding modules within a single protein (reviewed in ref. 33), but the cooperative effect between the ENTH and ANTH domains is an example of synergism between the membrane-binding domains of two different proteins.
Sla2 and HIP1R form dimers via their coiled-coil domains (14, 34), and the ENTH domain of mammalian epsin may oligomerize on liposomes in vitro (35). In addition, both proteins bind clathrin and other coat proteins (reviewed in ref. 10). These interactions may be important for proper transmission of the actin-generated force to the plasma membrane.
Interestingly, in Dictyostelium discoideum plasma membrane localization of the Sla2 homolog Hip1r depends on epsin’s ENTH domain (36). This dependence strongly suggests that the interaction between the lipid-binding domains of these two proteins is highly conserved. Because D. discoideum diverged early from the lineage leading to animals and fungi, it is likely that this ancient interplay between the epsin and Sla2/HIP1R proteins also is conserved in animals.
Sla2’s THATCH and Ent1’s ACB Domains Interact Redundantly with the Actin Cytoskeleton.
Our identification of the ACB F-actin–binding domain in Ent1 resolved the paradox that Sla2’s actin-binding THATCH domain is dispensable for membrane–actin coupling. Ent1’s ACB domain and Sla2’s THATCH domain act redundantly to couple the endocytic coat and the underlying membrane to the actin cytoskeleton. The deletion of the ACB domain of Ent1, similar to the deletion of the THATCH domain of Sla2, does not block endocytosis. However, the deletion of both domains causes a strong endocytic arrest and the formation of actin tails at the nonmotile endocytic sites. The ACB domain can be removed from Ent1 and fused to Sla2 in place of its THATCH domain without hampering endocytosis, showing that functionally the ACB domain can replace the THATCH domain’s role in interacting with the actin cytoskeleton. When the Ent1 protein was expressed as two polypeptides, physically separating the ENTH and ACB domains, endocytosis was blocked, suggesting that the membrane and actin cytoskeleton-binding domains must be linked physically. Finally, a nonrelated actin-binding module Lifeact (29) fused to Ent1 could support a significant level of endocytosis in the absence of both ACB and THATCH domains. This result further supports our hypothesis that the binding of Sla2 or Ent1 protein to the actin filament network at the endocytic site is critical for a successful endocytic invagination.
Ent1 was shown to be phosphorylated by Prk1 kinase on five sites in and near the ACB domain (30, 31). Prk1 has been implicated in the disassembly of the endocytic protein machinery after vesicle scission (32). Our results suggest that phosphorylation of Ent1 by Prk1 is not required for binding to actin but rather inhibits it. This notion is consistent with the proposed role of Prk1 kinase as a regulator of the disassembly of endocytic complexes, with Ent1 as its important target molecule (30).
Interestingly, although both Ent1 and Ent2 contain conserved ACB domains, only the Ent1 ACB domain can bind the actin cytoskeleton productively at the endocytic site. Our experiments indicated that the Ent2 ACB domain can interact with the actin cytoskeleton, but in the context of the full-length Ent2 protein it is not able to mediate this interaction. The appearance of two homologous epsins as a result of the whole-genome duplication in the ancestor of Saccharomyces cerevisiae may have allowed a functional diversification of Ent1 and Ent2 C-terminal parts. It is conceivable that the Ent1 ACB domain was fully dedicated to connecting the actin cytoskeleton to the endocytic coat, and the role of Ent2 was focused on mediating interactions with other coat proteins. Consistent with this hypothesis, Ent2 binds to clathrin with much higher affinity than Ent1 (37).
Taken together, our results show that Ent1 and Sla2 work together to couple the plasma membrane to the actin cytoskeleton at the endocytic site. The N-terminal Ent1 ENTH and Sla2 ANTH lipid-binding domains bind each other to cooperate in membrane binding. The C-terminal Ent1 ACB and Sla2 THATCH domains bind redundantly to actin filaments, providing a dynamic but robust coupling of the actin cytoskeleton to the endocytic coat. By this partly cooperative and partly redundant function, the Ent1–Sla2 linker transmits the pulling force from the polymerizing actin cytoskeleton to the plasma membrane, leading to membrane invagination and endocytic vesicle budding.
Materials and Methods
Strains and Plasmids.
Yeast strains and plasmid constructs are listed in Tables S1 and S2, respectively. Strains were grown at 25 °C in standard rich medium (yeast extract/peptone/dextrose; YPD) or synthetic defined (SD) medium with appropriate supplements. C-terminally tagged, deleted, and mutated alleles were generated by homologous recombination of the corresponding genes with PCR cassettes described previously (38), with the pMaM56-6 cassette (mCherry tagging), or with cassettes derived from pRS416-2xHA-meGFP (Table S2). The Lifeact coding sequence (29) was integrated together with the PCR cassette using a specific primer. To generate the epsin-depletion strains, the ENT1 promoter was replaced with the doxycycline-regulatable tetO2 promoter in the ent2Δ strain (and vice versa) using PCR cassette pCM224 (39). Strains expressing several fusion proteins and/or deletions were prepared by crossing. The expression levels of the protein constructs were assessed by immunoblotting with anti-FLAG or anti-HA antibodies. Depletion of epsin protein was achieved by the addition of 25 μg/mL doxycycline (Sigma) to the medium and incubation for 6–8 h at 25 °C.
Live-Cell Imaging.
For microscopy, yeast cells were grown in SD medium to an early logarithmic phase. Cells were attached to concanavalin A-coated coverslips (0.1 μg/mL) (Sigma) and observed at 25 °C. Where indicated, 200 μM latA (Enzo Life Sciences) was added 10 min before imaging. Two-color live-cell imaging was performed using an Olympus IX81 microscope equipped with a 100×/NA 1.45 PlanApo objective, an Orca-ER camera (Hamamatsu), and electronic shutters and filter wheels (Sutter Instrument) controlled by Metamorph software (Molecular Devices). The acquired image series were background subtracted and corrected for general photobleaching. Final image processing and analysis were done using ImageJ (http://rsb.info.nih.gov/ij/). The FRAP experiments were done with a custom-built set-up that focuses a 488-nm laser connected to an Olympus IX81 microscope (see SI Materials and Methods for details). The percentage of Sla1-GFP or Ent1-GFP patches exhibiting normal inward movement of at least 120 nm within 4-min movies was analyzed by a custom-written tracking program in MATLAB (Mathworks). Ninety to one hundred forty individual patches from 7–16 cells were counted for each strain/condition.
Liposome Flotation Assays.
Liposomes (20 μL) prepared by extrusion (see SI Materials and Methods for details) were incubated with 80 μL of a 2-μM solution of recombinant proteins (see SI Materials and Methods for details) in binding buffer [20 mM Hepes (pH 7.5) and 100 mM KCl] for 30 min at 25 °C. The suspension was mixed with 67 μL of 2.5-M sucrose in binding buffer, and a 125-μL aliquot was transferred into the 7 × 20 mm centrifuge tube (Beckman). The sample was overlaid by 100 μL of 0.75-M sucrose in binding buffer and 20 μL of binding buffer and was centrifuged at 435,000 × g for 30 min at 4 °C. The sucrose gradient then was fractioned into four 60-μL aliquots. The fractions were analyzed by SDS/PAGE using 4–12% Bis-Tris gel in MES buffer (Invitrogen) and Brilliant Blue G-Colloidal staining (Sigma). The amount of liposome-bound protein (top fraction, B) and unbound protein (bottom fraction, U) was quantified densitometrically using Kodak Image Station 4000MM PRO and ImageJ software.
GST Pull-Down and Actin Cosedimentation Assays.
Fifty microliters of glutathione beads (GE Healthcare) containing ca. 400 μg of GST-fusion protein (see SI Materials and Methods for details) were incubated with an equimolar amount of potential binding partner in 50 mM Hepes (pH 7.5), 150 mM KCl, and 1 mM DTT in the presence or absence of 0.2 μM diC8-PI(4,5)P2 (Echelon) for 30 min at 25 °C. Beads were washed briefly three times with 200 μL of buffer and then were incubated three times with 400 μL of buffer for 3 min. Finally, 5 μL of beads were resuspended in 45 μL of SDS/PAGE loading buffer, and bound proteins were analyzed by SDS/PAGE and Coomassie staining.
The actin cosedimentation assays were performed according to the manufacturer’s protocols (Cytoskeleton). See SI Materials and Methods for details.
Supplementary Material
Acknowledgments
We thank O. Gallego and K. Maeda for help with the liposome flotation assays; M. Knop and M. Meurer for plasmids; the European Molecular Biology Laboratory Protein Expression and Purification Facility for the recombinant proteins; and J. Ellenberg, A.-C. Gavin, C. Haering, M. Knop, and V. Sourjik for critical reading of the manuscript. M.S. is the recipient of Marie Curie Fellowship PIEF-GA-2009-237727.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 15092 (volume 109, number 38).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1207011109/-/DCSupplemental.
References
- 1.Perrais D, Merrifield CJ. Dynamics of endocytic vesicle creation. Dev Cell. 2005;9:581–592. doi: 10.1016/j.devcel.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Kaksonen M, Toret CP, Drubin DG. Harnessing actin dynamics for clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2006;7:404–414. doi: 10.1038/nrm1940. [DOI] [PubMed] [Google Scholar]
- 3.Aghamohammadzadeh S, Ayscough KR. Differential requirements for actin during yeast and mammalian endocytosis. Nat Cell Biol. 2009;11:1039–1042. doi: 10.1038/ncb1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferguson SM, et al. Coordinated actions of actin and BAR proteins upstream of dynamin at endocytic clathrin-coated pits. Dev Cell. 2009;17:811–822. doi: 10.1016/j.devcel.2009.11.005. erratum in Dev Cell (2010) 18:332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boulant S, Kural C, Zeeh JC, Ubelmann F, Kirchhausen T. Actin dynamics counteract membrane tension during clathrin-mediated endocytosis. Nat Cell Biol. 2011;13:1124–1131. doi: 10.1038/ncb2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galletta BJ, Mooren OL, Cooper JA. Actin dynamics and endocytosis in yeast and mammals. Curr Opin Biotechnol. 2010;21:604–610. doi: 10.1016/j.copbio.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCann RO, Craig SW. The I/LWEQ module: A conserved sequence that signifies F-actin binding in functionally diverse proteins from yeast to mammals. Proc Natl Acad Sci USA. 1997;94:5679–5684. doi: 10.1073/pnas.94.11.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun Y, Kaksonen M, Madden DT, Schekman R, Drubin DG. Interaction of Sla2p’s ANTH domain with PtdIns(4,5)P2 is important for actin-dependent endocytic internalization. Mol Biol Cell. 2005;16:717–730. doi: 10.1091/mbc.E04-08-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boettner DR, Friesen H, Andrews B, Lemmon SK. Clathrin light chain directs endocytosis by influencing the binding of the yeast Hip1R homologue, Sla2, to F-actin. Mol Biol Cell. 2011;22:3699–3714. doi: 10.1091/mbc.E11-07-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boettner DR, Chi RJ, Lemmon SK. Lessons from yeast for clathrin-mediated endocytosis. Nat Cell Biol. 2012;14:2–10. doi: 10.1038/ncb2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaksonen M, Sun Y, Drubin DG. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- 12.Engqvist-Goldstein AEY, et al. RNAi-mediated Hip1R silencing results in stable association between the endocytic machinery and the actin assembly machinery. Mol Biol Cell. 2004;15:1666–1679. doi: 10.1091/mbc.E03-09-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wesp A, et al. End4p/Sla2p interacts with actin-associated proteins for endocytosis in Saccharomyces cerevisiae. Mol Biol Cell. 1997;8:2291–2306. doi: 10.1091/mbc.8.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang S, Cope MJ, Drubin DG. Sla2p is associated with the yeast cortical actin cytoskeleton via redundant localization signals. Mol Biol Cell. 1999;10:2265–2283. doi: 10.1091/mbc.10.7.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carroll SY, et al. Analysis of yeast endocytic site formation and maturation through a regulatory transition point. Mol Biol Cell. 2012;23:657–668. doi: 10.1091/mbc.E11-02-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brady RJ, Damer CK, Heuser JE, O’Halloran TJ. Regulation of Hip1r by epsin controls the temporal and spatial coupling of actin filaments to clathrin-coated pits. J Cell Sci. 2010;123:3652–3661. doi: 10.1242/jcs.066852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen H, et al. Epsin is an EH-domain-binding protein implicated in clathrin-mediated endocytosis. Nature. 1998;394:793–797. doi: 10.1038/29555. [DOI] [PubMed] [Google Scholar]
- 18.Aguilar RC, Watson HA, Wendland B. The yeast Epsin Ent1 is recruited to membranes through multiple independent interactions. J Biol Chem. 2003;278:10737–10743. doi: 10.1074/jbc.M211622200. [DOI] [PubMed] [Google Scholar]
- 19.Kaksonen M, Toret CP, Drubin DG. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 2005;123:305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 20.Wendland B, Steece KE, Emr SD. Yeast epsins contain an essential N-terminal ENTH domain, bind clathrin and are required for endocytosis. EMBO J. 1999;18:4383–4393. doi: 10.1093/emboj/18.16.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itoh T, et al. Role of the ENTH domain in phosphatidylinositol-4,5-bisphosphate binding and endocytosis. Science. 2001;291:1047–1051. doi: 10.1126/science.291.5506.1047. [DOI] [PubMed] [Google Scholar]
- 22.Ford MGJ, et al. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- 23.De Camilli P, et al. The ENTH domain. FEBS Lett. 2002;513:11–18. doi: 10.1016/s0014-5793(01)03306-3. [DOI] [PubMed] [Google Scholar]
- 24.Newpher TM, Smith RP, Lemmon V, Lemmon SK. In vivo dynamics of clathrin and its adaptor-dependent recruitment to the actin-based endocytic machinery in yeast. Dev Cell. 2005;9:87–98. doi: 10.1016/j.devcel.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 25.Sun Y, Carroll S, Kaksonen M, Toshima JY, Drubin DG. PtdIns(4,5)P2 turnover is required for multiple stages during clathrin- and actin-dependent endocytic internalization. J Cell Biol. 2007;177:355–367. doi: 10.1083/jcb.200611011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aguilar RC, et al. Epsin N-terminal homology domains perform an essential function regulating Cdc42 through binding Cdc42 GTPase-activating proteins. Proc Natl Acad Sci USA. 2006;103:4116–4121. doi: 10.1073/pnas.0510513103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brady RJ, Wen Y, O’Halloran TJ. The ENTH and C-terminal domains of Dictyostelium epsin cooperate to regulate the dynamic interaction with clathrin-coated pits. J Cell Sci. 2008;121:3433–3444. doi: 10.1242/jcs.032573. [DOI] [PubMed] [Google Scholar]
- 28.Ford MGJ, et al. Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science. 2001;291:1051–1055. doi: 10.1126/science.291.5506.1051. [DOI] [PubMed] [Google Scholar]
- 29.Riedl J, et al. Lifeact: A versatile marker to visualize F-actin. Nat Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watson HA, Cope MJ, Groen AC, Drubin DG, Wendland B. In vivo role for actin-regulating kinases in endocytosis and yeast epsin phosphorylation. Mol Biol Cell. 2001;12:3668–3679. doi: 10.1091/mbc.12.11.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang B, Zeng G, Ng AYJ, Cai M. Identification of novel recognition motifs and regulatory targets for the yeast actin-regulating kinase Prk1p. Mol Biol Cell. 2003;14:4871–4884. doi: 10.1091/mbc.E03-06-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng G, Cai M. Prk1p. Int J Biochem Cell Biol. 2005;37:48–53. doi: 10.1016/j.biocel.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 33.Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 34.Wilbur JD, et al. Actin binding by Hip1 (huntingtin-interacting protein 1) and Hip1R (Hip1-related protein) is regulated by clathrin light chain. J Biol Chem. 2008;283:32870–32879. doi: 10.1074/jbc.M802863200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoon Y, et al. Molecular basis of the potent membrane-remodeling activity of the epsin 1 N-terminal homology domain. J Biol Chem. 2010;285:531–540. doi: 10.1074/jbc.M109.068015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Repass SL, Brady RJ, O’Halloran TJ. Dictyostelium Hip1r contributes to spore shape and requires epsin for phosphorylation and localization. J Cell Sci. 2007;120:3977–3988. doi: 10.1242/jcs.011213. [DOI] [PubMed] [Google Scholar]
- 37.Baggett JJ, D’Aquino KE, Wendland B. The Sla2p talin domain plays a role in endocytosis in Saccharomyces cerevisiae. Genetics. 2003;165:1661–1674. doi: 10.1093/genetics/165.4.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Janke C, et al. A versatile toolbox for PCR-based tagging of yeast genes: New fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- 39.Bellí G, Garí E, Aldea M, Herrero E. Functional analysis of yeast essential genes using a promoter-substitution cassette and the tetracycline-regulatable dual expression system. Yeast. 1998;14:1127–1138. doi: 10.1002/(SICI)1097-0061(19980915)14:12<1127::AID-YEA300>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]