Abstract
Bioenergy is efficiently produced in the mitochondria by the respiratory system consisting of complexes I–V. In various organisms, complex I can be replaced by the alternative NADH-quinone oxidoreductase (NDH-2), which catalyzes the transfer of an electron from NADH via FAD to quinone, without proton pumping. The Ndi1 protein from Saccharomyces cerevisiae is a monotopic membrane protein, directed to the matrix. A number of studies have investigated the potential use of Ndi1 as a therapeutic agent against complex I disorders, and the NDH-2 enzymes have emerged as potential therapeutic targets for treatments against the causative agents of malaria and tuberculosis. Here we present the crystal structures of Ndi1 in its substrate-free, NAD+- and ubiquinone- (UQ2) complexed states. The structures reveal that Ndi1 is a peripheral membrane protein forming an intimate dimer, in which packing of the monomeric units within the dimer creates an amphiphilic membrane-anchor domain structure. Crucially, the structures of the Ndi1–NAD+ and Ndi1–UQ2 complexes show overlapping binding sites for the NAD+ and quinone substrates.
Keywords: alternative complex I, structural biology
Bioenergy is efficiently produced in the mitochondria by the respiratory system consisting of complexes I–V. The first of these complexes, NDH-1 (complex I, proton pumping NADH-Q oxidoreductase) serves as the major entry point into the respiratory chain, for electrons derived from metabolic processes. In various organisms, complex I can be replaced by the alternative NADH-quinone oxidoreductase (NDH-2), which catalyzes the transfer of an electron from NADH via FAD to quinone, without proton pumping (1). The NDH-2 enzymes are found in bacteria and the mitochondria of plants and fungi, but crucially, not in mammalian mitochondria. Plant and fungal mitochondria possess two types of NDH-2: one is directed to the matrix and catalyzes NADH oxidation in the matrix (designated the internal NADH dehydrogenase or Ndi), and the other faces the intermembrane space and oxidizes NADH in the cytoplasmic space (designated the external NADH dehydrogenase or Nde).
Complex I defects are associated with human diseases, including Leber’s hereditary optic neuropathy (LHON) and cancer (2). Neurodegenerative diseases (3), such as sporadic Parkinson’s disease and aging, have been linked to reactive oxygen species generation by complex I (4, 5). A number of studies have investigated the potential use of Ndi1 from Saccharomyces cerevisiae as a therapeutic agent against complex I disorders. For example, Ndi1 has been shown to rescue rodents from LHON symptoms and protect rat neurons from rotenone (a specific inhibitor of complex I) toxicity and Parkinsonian symptoms (2). Ndi1 can functionally replace complex I in both Caenorhabditis elegans (6) and cultured mammalian cells (7–9). Recently, the expression of yeast Ndi1 in Drosophila melanogaster mitochondria has been shown to confer protection against toxins (rotenone and paraquat) and lethal complex I knockdown (10). Importantly, Ndi1 has been shown to increase lifespan independently of the effects of diet.
The NDH-2 enzymes have also emerged as potential therapeutic targets for treatments against the causative agents of malaria (Plasmodium falciparum) (11–13) and tuberculosis (Mycobacterium tuberculosis) (14, 15). In P. falciparum, NADH:quinone oxidoreductase activity in the electron transport chain is carried out solely by NDH-2, with recent in vivo studies demonstrating the ability of high-affinity NDH-2 inhibitors to inhibit parasitic replication in tissue culture (16). Similarly, NDH-2 has been demonstrated to be critical in the life cycle of M. tuberculosis, where under hypoxic conditions the organism reverts to a nonreplicating drug-resistant state (17) in which NDH-2 is used directly for NAD+ recycling, thus indirectly for ATP synthesis (18). The inhibition of the NDH-2–dependent pathway has shown early promise for the development of antituberculosis drugs, targeting the persistent, nonreplicating form of the microbe (18).
Here we report the 3D structure of the Ndi1 from S. cerevisiae, in addition to the structures of Ndi1–NAD+ and Ndi1-ubiquinone-2 (UQ2) complexes. The structures show that Ndi1 homodimerizes to create an amphiphilic domain structure, which anchors the protein to the membrane. The structures of the Ndi1–NAD+ and Ndi1–UQ2 complexes show overlapping binding sites for the NAD+ and quinone substrates, which highlights Ndi1 as an example of the evolution of a membrane protein from an ancestral soluble protein.
Results
Overall Structure of Ndi1.
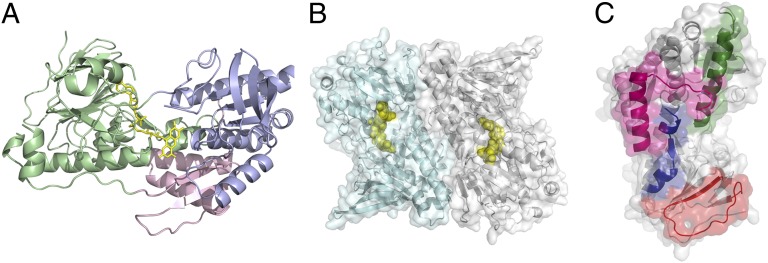
The crystal structure of Ndi1 was determined by a combination of multiple wavelength anomalous dispersion (MAD) and single isomorphous replacement with anomalous scattering (SIRAS) using Se-Met and mercury derivative crystals. The model refinement converged with residuals R = 23.4% and Rfree = 26.8% in the resolution range 40.0–2.7 Å (Tables S1 and S2). Each protomer of Ndi1 can be divided into three domains: (i) the FAD-binding domain (residues 43–178 and 342–442), (ii) the NADH-binding domain (residues 179–341), and (iii) the C-terminal, membrane-anchor domain (residues 443–513) (Fig. 1). The dinucleotide-binding domains are typical of the glutathione-reductase family of flavoenzymes, being composed of two Rossmann folds.
Fig. 1.
Crystal structure of Ndi1. (A) Cartoon representation of the Ndi1 monomer. The FAD-binding, NADH-binding, and membrane-anchor domains are colored green, lilac, and pink, respectively. The FAD (yellow) cofactor is represented as sticks. (B) The Ndi1 dimer. Both monomers are represented as cartoons, with molecule A in pale cyan and molecule B in gray. The FAD cofactors are shown as yellow spheres. Transparent surface representations of both monomers are also shown. The interaction surface between monomers is extensive at ∼9% of the surface area of each monomer. (C) Monomer B only from the representation in B is shown alone, with the regions of surface that interact with monomer A in the dimer highlighted: residues 103–116, blue; residues 145–165, red; residues 198–219 green; residues 489–513, pink.
There are two molecules in the asymmetric unit, which are packed as a dimer (Fig. 1). The interface between the Ndi1 monomers is extensive, with a buried surface area of ca. 1,920 Å2 per monomer, which is ∼9% of the solvent-accessible surface of each monomer and suggests that the homodimer is physiologically relevant (19). The observation of a dimeric Ndi1 structure is consistent with the biochemical analysis of the NDH-2 enzyme from Trypanosoma brucei [29% sequence identity with Ndi1 (20)] and with SDS/PAGE analysis of membranes harboring Ndi1, after chemical cross-linking (Fig. S1). Homooligomerization is a common feature among monotopic membrane protein structures (21). The homodimeric interface is composed of both electrostatic and hydrophobic interactions between monomers at the C-terminal membrane-anchor domain, including residues 489–513. There are also interactions between the inner tips of the molecules, which involve secondary structural elements not present in other flavoprotein dehydrogenases. These comprise residues 103–116 and 145–165 of the FAD-binding domain and residues 198–219 of the NADH-binding domain (Fig. 1).
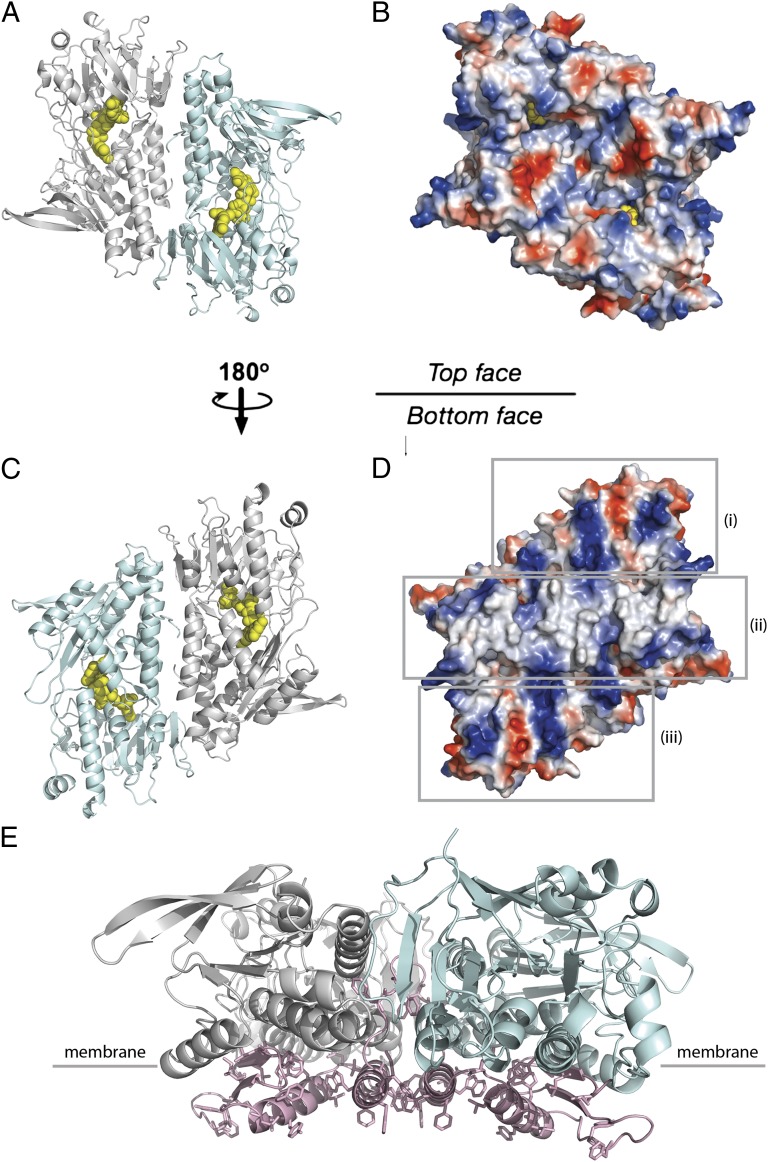
Membrane-Anchor Structure.
Fig. 2 shows the surface charge distribution on two flat sides of the Ndi1 dimer. On one side, there is a clear hydrophobic ridge bordered on the left and right by concentrated patches of positive charge. This is presumably the site where the protein inserts into the membrane, with the hydrophobic region buried and the phospholipid head groups interacting with the positively charged patches. In contrast, the face that presents to the matrix is predominantly charged. The membrane-anchor domain is composed of a contiguous 70-residue stretch at the C terminus of Ndi1 (residues 443–513 inclusive), with a three-stranded β-sheet connected to a helix-turn-helix structure. The C-terminal helix of this motif (residues 490–505) presents predominantly aromatic residues to the membrane (Fig. 2). In the dimer, the membrane-anchor domains of each monomer align to give the contiguous apolar ridge across the center of the complex that is seen in Fig. 2. The base of the ridge is the part of the protein most deeply inserted into the membrane and is ∼11 Å below the layer of the nucleotide-binding domains (Fig. 2). The twofold axis relating the monomers within the dimer is perpendicular to the membrane and maintains the complex on the surface of the membrane, allowing lipophilic substrate access from the membrane to the catalytic center of each molecule as discussed below.
Fig. 2.
Ndi1 surface electrostatic charge and hydrophobic membrane-anchor domain. The Ndi1 dimer is viewed from above (A and B) and below (C and D) the membrane plane. (A and C) Cartoon representations of the dimeric structure, with molecule A in pale cyan and molecule B in light gray. The FAD cofactors are highlighted as yellow spheres. (B and D) The electrostatic surface of the dimer, with surfaces of negative potential in red, positive in blue, and uncharged in white. The surface that faces the membrane (D) presents a hydrophobic ridge for membrane insertion [indicated (ii)], which is bordered by regions of positive charge [indicated (i) and (iii)]. (E) The Ndi1 dimer viewed parallel to the membrane. Molecule A is represented in pale cyan and molecule B in light gray. The membrane-anchor domains are highlighted in pale pink, with hydrophobic residues shown as sticks. The membrane anchor domains form part of the dimer interface and present a number of aromatic side chains for insertion into the membrane (the approximate position of the membrane surface is indicated).
FAD-Binding Site.
In the electron density, the FAD cofactor is clearly defined (Fig. S2), binding in an extended conformation, which is similar to that observed in the structures of a number of other FAD-dependent oxidoreductases [for example, sulfide:quinone oxidoreductase (SQR) from Aquifex aeolicus (22), rubredoxin reductase from Pseudomonas aeruginosa (23), glutathione reductase (24), and bacterial ferredoxin reductase (25)]. The adenosine group is anchored through interactions with the backbone nitrogen atoms of Ser-61 and Ala-129 and the backbone carbonyl atoms of Ala-129 and Val-176, and a hydrogen bond to the side chain of Arg-85. The pyrophosphate moiety participates in an extensive interaction network, making direct hydrogen-bonding interactions with the backbone nitrogen atoms of Trp-63, Gly-64, and Asp-383 and the side chain of a conserved Thr-91 (Fig. S2). Yang et al. (26) identified residues Pro-92 and Leu-93 as being positioned close to the FAD-binding site. These results are entirely consistent with the structure of Ndi1 as described here, which places residues Pro-92 and Thr-91 on the si-face of the FAD cofactor, with a hydrogen-bonding interaction between Thr-91 and the FAD.
The isoalloxazine ring of the FAD is positioned between the two Rossmann domains, which define two channels along the interdomain plane that lead to the active site (Fig. 3). The isoalloxazine ring is planar and makes direct hydrogen-bonding interactions with the backbone nitrogen atoms of Ala393 and Gln394, in addition to an interaction with residue Tyr-482 (Fig. S2).
Fig. 3.
Ndi1 substrate channels. Clipped electrostatic surface representation of the Ndi1 dimer, showing the FAD-binding pocket (FAD cofactor shown as sticks with yellow carbon atoms) and the hydrophilic (occupied by NAD+, with blue carbon atoms) and hydrophobic (occupied by UQ2, with pink carbon atoms) channels, within the Ndi1 monomer.
A channel approaches the FAD from the membrane insertion domain (Fig. 3). The channel is predominantly hydrophobic, lined by Trp-63, Phe-90, Leu-444, Leu-447, Leu-481, Tyr-482, Met-485, and Leu-487, but two polar residues also contribute (Fig. S3). The conserved residue Gln-394 is situated at the opening of the active site, and residue His-397 is located on the lower protein face.
Positions of the NAD+ and UQ2 Substrate Binding Sites Are Coincident.
To investigate the mode of substrate binding to Ndi1 and its reaction mechanism, two further structures were solved in complex with NAD+ and UQ2, respectively. The structure with NAD+ bound (Ndi1–NAD+; Tables S1 and S2) was refined to 2.9-Å resolution, and the structure with UQ2 bound (Ndi1–UQ2; Tables S1 and S2) to 3.0-Å resolution. Least-squared superpositions between the Ndi1, Ndi1–NAD+, and Ndi1–UQ2 models demonstrate that there are no gross conformational changes among the three structures (rmsds for the superpositions of 465 Cα atoms per monomer are 0.2 and 0.5 Å for the Ndi1/Ndi1–NAD+ and Ndi1/Ndi1–UQ2 superpositions, respectively).
The NAD+ binds in the second Rossmann domain in a predominantly positively charged cleft, with the nicotinamide ring approaching the re-face of the FAD (Fig. S4). With respect to the FAD, the NAD+ is less well defined in the electron density, although this is common for substrate complexes. The position of the ADP moiety is clear, with the ribose within hydrogen bonding distance of the side chain of Glu-272 and the adenine ring interacting with the backbone nitrogen groups of Ala-273 and Val-306. The exact position of the nicotinamide ring is more ambiguous, and the electron density suggests there may be multiple conformations. The nicotinamide ring stacks at a distance of 3.1 Å on the re-face of the isoalloxazine ring of the FAD cofactor (Fig. S4), which is suitable for hydride transfer, and is anchored by a hydrogen-bonding interaction with the backbone nitrogen atom of Gly-445. The Glu-272-NAD+ interaction, combined with selective pressure in the matrix [the concentration of NAD+ is 2- to 37-fold greater than that of NADP+ in plant mitochondria (27)], is presumably crucial in determining Ndi1 selectivity for NAD/H over NADP/H (the presence of a phosphate group at the 2′ position of the ribose ring would clash with the side chain of Glu-272 and preclude the hydrogen bond).
The position observed for the nicotinamide ring of NAD+ is also occupied by the head group of UQ2, which was clearly defined in a simulated annealing omit electron density map in the Ndi1–UQ2 structure (Fig. S5). In the refined model, the ring of the UQ2 molecule stacks ∼3.1 Å above the isoalloxazine ring of the FAD cofactor, and the tail has been modeled into the channel that approaches from the membrane. The UQ2 site is framed by a number of structural motifs, including the FAD cofactor, residues 237–240, 278–281, 391–394, and 445–447. In agreement with this, the residue 374–399 region was identified as the quinone binding domain by photoaffinity labeling experiments (28). Direct molecular contact with the UQ2 moiety is limited to a hydrogen-bonding contact between the UQ2 molecule and the peptide carbonyl group of Gly-445. Significantly, the positions of the head group of UQ2 and the nicotinamide of NAD superpose in the respective structures (Fig. 4). This suggests that the reaction may proceed by a ping-pong mechanism (Fig. S6).
Fig. 4.

UQ2- and NAD+-binding sites in the structure of Ndi1 are coincident. The structures of the Ndi1–NAD+ and Ndi1–UQ2 complexes are superposed. For simplicity, the polypeptide chain and the surface of only the Ndi1–NAD+ complex are represented in pale gray. The positions of the FAD cofactor (yellow carbon atoms), the NAD+ (blue carbon atoms), and UQ2 (pink carbon atoms) molecules are shown. In this superposition, the positions of the UQ2 moiety and the nicotinamide ring of the NAD+ molecule coincide.
Discussion
The Ndi1 structures presented here can be divided into three domains: FAD-, NAD+-binding, and membrane-anchor domains. These domains assemble within the structures, such that distinct water- and lipid-soluble substrate channels (from the matrix and from the membrane, respectively) converge adjacent to the FAD-binding site (Fig. 3). In fact, the Ndi1–NAD+ and Ndi1–UQ2 complex structures described in this work substantiate this analysis by showing that the NAD+- and UQ2-binding sites within the protein structure are coincident (Fig. 4), implying that the NAD+- and UQ2-binding events in the reaction mechanism of Ndi1 are mutually exclusive (Fig. S6). In addition, the observation of a single UQ2-binding site per monomer of Ndi1 is consistent with the most recent biochemical and kinetic analyses of the enzyme (26).
A number of studies analyzing the kinetic properties of the NDH-2 enzymes [from Yarrowia lipolytica (29), M. tuberculosis (14), and the Ndi1 from S. cerevisiae (30, 31)] have concluded that they operate by a ping-pong mechanism. However, a recent examination of the reaction mechanism of Ndi1 by site-directed mutagenesis, proteinase K digestion, and kinetic analyses concluded that the reaction proceeds through a ternary complex (26). The results of the mutagenesis experiments reported in this study agree well with the present structural analysis, placing residues Pro-92 and Leu-93 in close proximity to the FAD cofactor (Fig. S2B); however, the existence of a ternary complex, where Ndi1 is associated with NADH and quinone simultaneously, is inconsistent with the current structural data, which show coincident binding sites for NAD+ (but not NADH) and UQ2. The proteolytic digestion analyses reported by Yang et al. (26) proposed a conformational change in the structure of Ndi1, induced by NADH (but not NAD+), which is essential for the formation of a ternary complex. This analysis is not supported by the present structures, which show no molecular rearrangement on NAD+ or UQ2 binding. It should be noted that in contrast to the biochemical studies in solution, our structural data were obtained from crystals of the Ndi1–NAD+ and Ndi1–UQ2 complexes, using oxidized Ndi1 (but not the reduced Ndi1). The consolidation of our structural observations with recent work in solution must await further structural and biochemical analyses.
The existence of coincident NAD+- and UQ2-binding sites in the structures of Ndi1 is a similar scenario to that observed in the structural and functional analyses of the NAD(P)H-quinone oxidoreductase (NQOR) from rat (32). This enzyme, like Ndi1, is a FAD-containing protein, which catalyzes NAD(P)H-dependent two-electron reductions of soluble quinones. The crystal structures of two FAD-containing complexes of NQOR (one containing substrate NADP+ and the other with a duroquinone inhibitor) showed overlapping binding sites for the respective substrates. It is likely that the reaction mechanism of Ndi1 parallels that of NQOR. NQOR is, however, a soluble protein. In the case of Ndi1, it is important to note that the positions of binding for NAD+ from the matrix and ubiquinone from the membrane overlap in the reaction.
The structure of Ndi1 is comparable to that of another monotopic membrane protein, SQR from Aquifex aeolicus, a flavoprotein disulfide reductase [with 17% global sequence identity with Ndi1 (22)]. Like Ndi1, the structure of SQR includes two Rossmann domains, as well as a C-terminal membrane-anchor domain; however the protein is a trimer rather than a dimer. With the exclusion of the respective membrane-anchor domains, the two proteins are very similar, with an rmsd of 2.2 Å for 288 aligned Cα atoms. The membrane insertion domains in the two proteins each contain a three-stranded antiparallel β-sheet with two amphipathic helices and a C-terminal loop; however, they are topologically distinct. The membrane insertion domain of SQR occupies a central position below the Rossmann domains, as opposed to Ndi1, where it is situated predominantly below the NADH-binding domain.
SQR oxidizes sulphide ions with the electrons transferred through FAD to quinone. The sulphide ions bind at the re-face of the FAD at a similar position to that of NAD+ in Ndi1. Three extended loop regions occlude the space above the active site in SQR corresponding to the NAD+-binding site of Ndi1. The quinone binds at a distinct site. The head group binds 3.5 Å from the si-face of the FAD cofactor, with its tail group located in another subunit of the trimer. Quinone and sulfide (lipid- and water-soluble substrates, respectively) are therefore spatially distinct in these structures.
The convergence of the positions of the NAD+- and UQ2-binding sites within the structure of Ndi1 is perhaps the ultimate example of membrane protein evolution. Bracey et al. (21) reviewed the structural commonalities among monotopic membrane proteins (proteins that are membrane-bound but that do not cross the membrane) and observed significant structural homologies between monotopic membrane proteins and their soluble protein counterparts. They suggested that this may be a means of addressing enzymatic needs within membranes, without the constraints of typical polytopic membrane protein folds. The structure of Ndi1 fulfils this design idea, by not only bringing functionality to the membrane, but also by converging and adapting the substrate binding sites.
Materials and Methods
Expression, Purification, Crystallization, and Data Collection.
The mature Ndi1 enzyme was overexpressed and purified from Escherichia coli as previously described (31). A mutant Ndi1 in which residue glycine 192 was mutated to cysteine was used for the preparation of the mercury derivative. Crystallization trials were performed using the vapor diffusion technique. Crystals of the Ndi1–NAD+ and Ndi1–UQ2 complexes were prepared by co-crystallization. Further details on the crystallization conditions are given in SI Materials and Methods.
X-ray data were collected from frozen crystals at 100 K at the European Synchrotron Radiation Facility (ESRF; ID14-4, ID29, ID23-1, ID23-2), the Swiss Light Source (X06SA), and the Diamond Light Source (DLS, I-02), although the data used for the structure determination described here were collected at ESRF and DLS. Image data were processed using Denzo and Scalepack (33), HKL2000 (33), and the CCP4 program suite (34), or XDS (35) and SCALA (34) (Tables S1 and S2).
Structure Determination and Refinement.
The structure of Ndi1 was determined by a combination of MAD phasing, using data from crystals of the Se-Met protein, and SIRAS from a single Hg derivative. The atomic model was built with the programs O (36) and COOT (37) and refined using the programs REFMAC5 (38) [with TLS (39)] and CNS (40). Anisotropy-corrected data provided by the diffraction anisotropy server was used for density modification and refinement (41). During refinement, tight restraints between non–crystallographic-symmetry-related monomers were maintained. Water picking was carried out automatically with COOT (37), with consideration of conservative hydrogen-bonding criteria. The structures of the Ndi1–NAD+ and Ndi1–UQ2 complexes to 2.9 and 3.0 Å resolution, respectively, were solved by molecular replacement, using the Ndi1 structure as a search model. All figures were prepared with PyMOL. Solvent accessible surface area and interface area calculations were performed using the protein interfaces, surfaces, and assemblies service (PISA) at the European Bioinformatics Institute (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html) (42).
Supplementary Material
Acknowledgments
This work was funded by National Institutes of Health Grants R01GM033712 and R01EY020796 (to T.Y.) and supported by the Exploratory Research for Advanced Technology Human Receptor Crystallography Project from the Japan Science (to S.I.). The work was also partly funded by Biotechnology and Biological Sciences Research Council Grant BB/G023425/1 (to S.I.). Part of the work was performed in the Membrane Protein Laboratory funded by Wellcome Trust Grant 062164/ Z/00/Z (to S.I.) at the Diamond Light Source Limited. M.J.M. is supported by a La Trobe Institute for Molecular Science Senior Research Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 4G9K (Ndi1), 4GAP (Ndi1–NAD+), and 4GAV (Ndi1–UQ2)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1210059109/-/DCSupplemental.
References
- 1.Melo AM, Bandeiras TM, Teixeira M. New insights into type II NAD(P)H:quinone oxidoreductases. Microbiol Mol Biol Rev. 2004;68:603–616. doi: 10.1128/MMBR.68.4.603-616.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marella M, Seo BB, Thomas BB, Matsuno-Yagi A, Yagi T. Successful amelioration of mitochondrial optic neuropathy using the yeast NDI1 gene in a rat animal model. PLoS One. 2010;5:e11472. doi: 10.1371/journal.pone.0011472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schapira AH. Mitochondrial dysfunction in neurodegenerative disorders. Biochim Biophys Acta. 1998;1366:225–233. doi: 10.1016/s0005-2728(98)00115-7. [DOI] [PubMed] [Google Scholar]
- 4.Dawson TM, Dawson VL. Molecular pathways of neurodegeneration in Parkinson’s disease. Science. 2003;302:819–822. doi: 10.1126/science.1087753. [DOI] [PubMed] [Google Scholar]
- 5.Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 6.DeCorby A, Gásková D, Sayles LC, Lemire BD. Expression of Ndi1p, an alternative NADH:ubiquinone oxidoreductase, increases mitochondrial membrane potential in a C. elegans model of mitochondrial disease. Biochim Biophys Acta. 2007;1767:1157–1163. doi: 10.1016/j.bbabio.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 7.Bai Y, et al. Lack of complex I activity in human cells carrying a mutation in MtDNA-encoded ND4 subunit is corrected by the Saccharomyces cerevisiae NADH-quinone oxidoreductase (NDI1) gene. J Biol Chem. 2001;276:38808–38813. doi: 10.1074/jbc.M106363200. [DOI] [PubMed] [Google Scholar]
- 8.Seo BB, Wang J, Flotte TR, Yagi T, Matsuno-Yagi A. Use of the NADH-quinone oxidoreductase (NDI1) gene of Saccharomyces cerevisiae as a possible cure for complex I defects in human cells. J Biol Chem. 2000;275:37774–37778. doi: 10.1074/jbc.M007033200. [DOI] [PubMed] [Google Scholar]
- 9.Seo BB, et al. Molecular remedy of complex I defects: Rotenone-insensitive internal NADH-quinone oxidoreductase of Saccharomyces cerevisiae mitochondria restores the NADH oxidase activity of complex I-deficient mammalian cells. Proc Natl Acad Sci USA. 1998;95:9167–9171. doi: 10.1073/pnas.95.16.9167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanz A, et al. Expression of the yeast NADH dehydrogenase Ndi1 in Drosophila confers increased lifespan independently of dietary restriction. Proc Natl Acad Sci USA. 2010;107:9105–9110. doi: 10.1073/pnas.0911539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher N, Bray PG, Ward SA, Biagini GA. The malaria parasite type II NADH:quinone oxidoreductase: An alternative enzyme for an alternative lifestyle. Trends Parasitol. 2007;23:305–310. doi: 10.1016/j.pt.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 12.Dong CK, et al. Type II NADH dehydrogenase of the respiratory chain of Plasmodium falciparum and its inhibitors. Bioorg Med Chem Lett. 2009;19:972–975. doi: 10.1016/j.bmcl.2008.11.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biagini GA, et al. Generation of quinolone antimalarials targeting the Plasmodium falciparum mitochondrial respiratory chain for the treatment and prophylaxis of malaria. Proc Natl Acad Sci USA. 2012;109:8298–8303. doi: 10.1073/pnas.1205651109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yano T, Li LS, Weinstein E, Teh JS, Rubin H. Steady-state kinetics and inhibitory action of antitubercular phenothiazines on mycobacterium tuberculosis type-II NADH-menaquinone oxidoreductase (NDH-2) J Biol Chem. 2006;281:11456–11463. doi: 10.1074/jbc.M508844200. [DOI] [PubMed] [Google Scholar]
- 15.Teh JS, Yano T, Rubin H. Type II NADH: Menaquinone oxidoreductase of Mycobacterium tuberculosis. Infect Disord Drug Targets. 2007;7:169–181. doi: 10.2174/187152607781001781. [DOI] [PubMed] [Google Scholar]
- 16.Saleh A, Friesen J, Baumeister S, Gross U, Bohne W. Growth inhibition of Toxoplasma gondii and Plasmodium falciparum by nanomolar concentrations of 1-hydroxy-2-dodecyl-4(1H)quinolone, a high-affinity inhibitor of alternative (type II) NADH dehydrogenases. Antimicrob Agents Chemother. 2007;51:1217–1222. doi: 10.1128/AAC.00895-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wayne LG, Hayes LG. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun. 1996;64:2062–2069. doi: 10.1128/iai.64.6.2062-2069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rao SP, Alonso S, Rand L, Dick T, Pethe K. The protonmotive force is required for maintaining ATP homeostasis and viability of hypoxic, nonreplicating Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2008;105:11945–11950. doi: 10.1073/pnas.0711697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ponstingl H, Henrick K, Thornton JM. Discriminating between homodimeric and monomeric proteins in the crystalline state. Proteins. 2000;41:47–57. doi: 10.1002/1097-0134(20001001)41:1<47::aid-prot80>3.3.co;2-#. [DOI] [PubMed] [Google Scholar]
- 20.Fang J, Beattie DS. Novel FMN-containing rotenone-insensitive NADH dehydrogenase from Trypanosoma brucei mitochondria: Isolation and characterization. Biochemistry. 2002;41:3065–3072. doi: 10.1021/bi015989w. [DOI] [PubMed] [Google Scholar]
- 21.Bracey MH, Cravatt BF, Stevens RC. Structural commonalities among integral membrane enzymes. FEBS Lett. 2004;567:159–165. doi: 10.1016/j.febslet.2004.04.084. [DOI] [PubMed] [Google Scholar]
- 22.Marcia M, Ermler U, Peng G, Michel H. The structure of Aquifex aeolicus sulfide:quinone oxidoreductase, a basis to understand sulfide detoxification and respiration. Proc Natl Acad Sci USA. 2009;106:9625–9630. doi: 10.1073/pnas.0904165106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hagelueken G, et al. Crystal structure of the electron transfer complex rubredoxin rubredoxin reductase of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2007;104:12276–12281. doi: 10.1073/pnas.0702919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schulz GE, Schirmer RH, Sachsenheimer W, Pai EF. The structure of the flavoenzyme glutathione reductase. Nature. 1978;273:120–124. doi: 10.1038/273120a0. [DOI] [PubMed] [Google Scholar]
- 25.Senda T, et al. Crystal structure of NADH-dependent ferredoxin reductase component in biphenyl dioxygenase. J Mol Biol. 2000;304:397–410. doi: 10.1006/jmbi.2000.4200. [DOI] [PubMed] [Google Scholar]
- 26.Yang Y, et al. Reaction mechanism of single subunit NADH-ubiquinone oxidoreductase (Ndi1) from Saccharomyces cerevisiae: Evidence for a ternary complex mechanism. J Biol Chem. 2011;286:9287–9297. doi: 10.1074/jbc.M110.175547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agius SC, Rasmusson AG, Møller IM. NAD(P) turnover in plant mitochondria. Aust J Plant Physiol. 2001;28:461–470. [Google Scholar]
- 28.Murai M, et al. Characterization of the ubiquinone binding site in the alternative NADH-quinone oxidoreductase of Saccharomyces cerevisiae by photoaffinity labeling. Biochemistry. 2010;49:2973–2980. doi: 10.1021/bi100005j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eschemann A, Galkin A, Oettmeier W, Brandt U, Kerscher S. HDQ (1-hydroxy-2-dodecyl-4(1H)quinolone), a high affinity inhibitor for mitochondrial alternative NADH dehydrogenase: Evidence for a ping-pong mechanism. J Biol Chem. 2005;280:3138–3142. doi: 10.1074/jbc.M411217200. [DOI] [PubMed] [Google Scholar]
- 30.Velázquez I, Pardo JP. Kinetic characterization of the rotenone-insensitive internal NADH: Ubiquinone oxidoreductase of mitochondria from Saccharomyces cerevisiae. Arch Biochem Biophys. 2001;389:7–14. doi: 10.1006/abbi.2001.2293. [DOI] [PubMed] [Google Scholar]
- 31.Yamashita T, Nakamaru-Ogiso E, Miyoshi H, Matsuno-Yagi A, Yagi T. Roles of bound quinone in the single subunit NADH-quinone oxidoreductase (Ndi1) from Saccharomyces cerevisiae. J Biol Chem. 2007;282:6012–6020. doi: 10.1074/jbc.M610646200. [DOI] [PubMed] [Google Scholar]
- 32.Li R, Bianchet MA, Talalay P, Amzel LM. The three-dimensional structure of NAD(P)H:quinone reductase, a flavoprotein involved in cancer chemoprotection and chemotherapy: Mechanism of the two-electron reduction. Proc Natl Acad Sci USA. 1995;92:8846–8850. doi: 10.1073/pnas.92.19.8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 34.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 40.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 41.Strong M, et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.