Abstract
The consolidation of long-term memory for sensitization and synaptic facilitation in Aplysia requires synthesis of new mRNA including the immediate early gene Aplysia CCAAT enhancer-binding protein (ApC/EBP). After the rapid induction of ApC/EBP expression in response to repeated treatments of 5-hydroxytryptamine (5-HT), ApC/EBP mRNA is temporarily expressed in sensory neurons of sensory-to-motor synapses. However, the molecular mechanism underlying the rapid degradation of ApC/EBP transcript is not known. Here, we cloned an AU-rich element (ARE)-binding protein, ApAUF1, which functions as a destabilizing factor for ApC/EBP mRNA. ApAUF1 was found to bind to the 3′ UTR of ApC/EBP mRNA that contains AREs and subsequently reduces the expression of ApC/EBP 3′ UTR-containing reporter genes. Moreover, overexpression of ApAUF1 inhibited the induction of ApC/EBP mRNA in sensory neurons and also impaired long-term facilitation of sensory-to-motor synapses by repetitive 5-HT treatments. These results provide evidence for a critical role of the posttranscriptional modification of ApC/EBP mRNA during the consolidation of synaptic plasticity.
Long-term facilitation (LTF) of sensory-to-motor synapses, induced by 5-hydroxytryptamine (5-HT), is a cellular mechanism underlying behavioral sensitization of withdrawal reflexes in Aplysia (1, 2). Long-term synaptic plasticity requires transcription of distinct sets of genes (3). In the early phase, the expression of immediate early genes, such as a transcription factor ApC/EBP, and a proteasome-associated enzyme ubiquitin C-terminal hydrolase are induced (4, 5). ApC/EBP is a transcription factor that activates transcription of the other late-response genes and, therefore, it is thought to be a molecular switch for the consolidation of LTF (6, 7). The expression of ApC/EBP is tightly regulated in a narrow time frame after 5-HT stimulation. ApC/EBP expression is first detected within 15 min after 5-HT treatment and decreases 4 h after the onset of stimulation (4). Interestingly, Yamamoto et al. showed that ApC/EBP protein is degraded through the ubiquitin-proteasome pathway (8). However, the molecular mechanisms underlying the rapid degradation of ApC/EBP transcripts are not clearly understood.
Gene expression can be regulated by multiple mechanisms such as transcriptional regulations and posttranscriptional modifications (9). One of the most important mechanisms of posttranscriptional gene regulation involves the regulation of mRNA stability (10–13). Many cytokine and proto-oncogene mRNAs have short half-lives, which is in part due to the presence of ARE(s) in 3′ untranslated regions (UTR), which function as destabilizing cis elements (14). AREs consist of one or more AUUUA motifs within a U-rich context, which associate with trans-acting factors to accelerate mRNA decay. It has been shown that ApC/EBP mRNA contains AREs in its 3′ UTR, and its stability can be increased by an ARE-binding protein, ELAV (15).
There are several ARE-binding proteins including AUF1, Hu proteins (ELAV family), and TTP. Among these proteins, AUF1 (hnRNP D) was originally identified as a destabilizing factor for c-Myc (16, 17). AUF1 exists as a family of four isoforms (p37, p40, p42, and p45) generated by alternative splicing of a single transcript in mammal, with each transcript having a different RNA binding specificity (18). Binding of AUF1 to ARE-containing mRNA leads to its association with additional factors such as the translation initiation factor eIF4G, which results in the degradation of AUF1 by the ubiquitin-proteasome pathway (19, 20).
In this study, we cloned an Aplysia homolog of AUF1 (ApAUF1) and demonstrated that ApAUF1 binds to the 3′ UTR of ApC/EBP mRNA and results in its degradation. Moreover, overexpression of ApAUF1 blocked the induction of ApC/EBP expression and the LTF induced by repetitive treatments of 5-HT.
Results
Cloning of ApAUF1 and Its Expression in Aplysia Sensory Neurons.
We identified an EST clone showing high homology to mammalian AUF1 (GenBank accession no. EY418173.1) in Aplysia kurodai EST database (www.seahare.org) (21). Further sequencing analysis revealed that the EST clone contains an ORF that encodes an AUF1-like protein (ApAUF1) of 323 amino acids (Fig. S1A). The ApAUF1 coding region shares 30–37% homology with mammalian AUF1 isoforms. ApAUF1 contains two RNA recognition motifs (RRMs), which are well-conserved among AUF1 homologs from other species, and a GY-rich domain, which are also found in mammalian AUF1 splicing variants (Fig. S1). In addition, at least four putative sites for serine phosphorylation, which are thought to play important roles in regulating protein activity, were recognized (NetPhos 2.0).
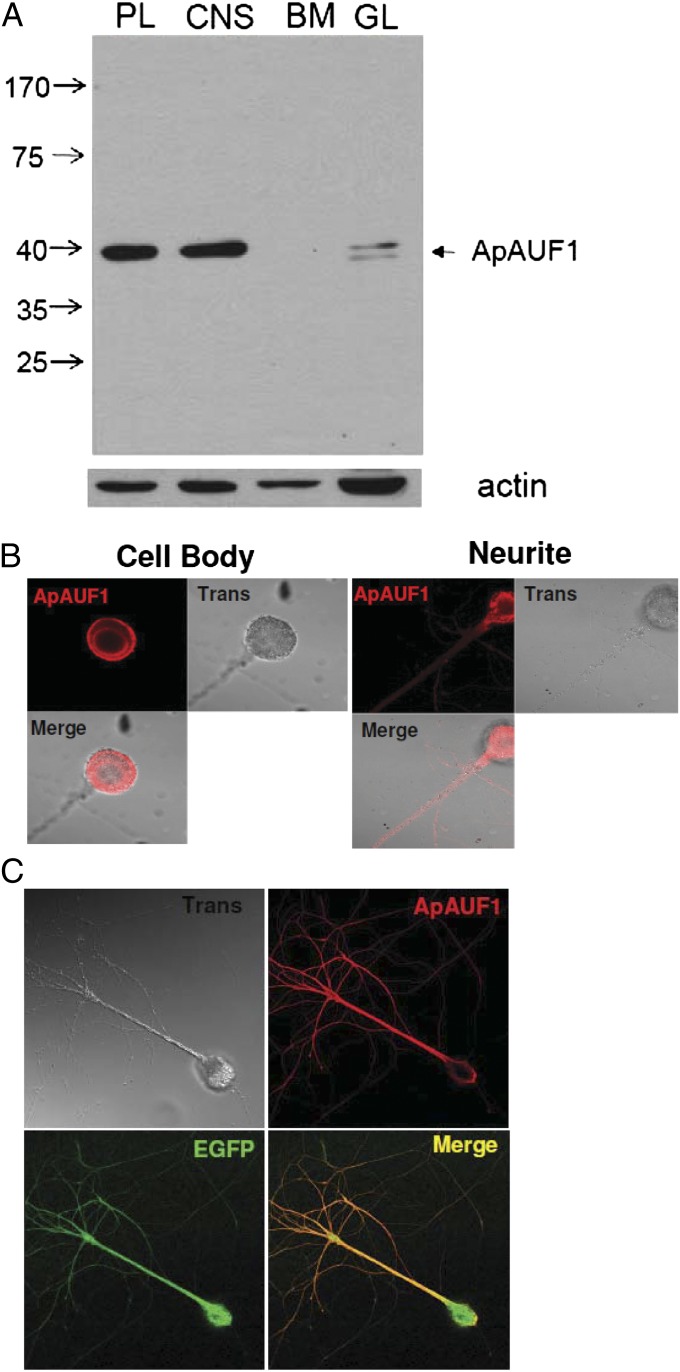
To examine the tissue distribution of ApAUF1, we generated an antibody against the full-length ApAUF1 protein. ApAUF1 is expressed in nervous system tissue including pleural ganglia but not in the buccal muscle (Fig. 1A). In the gill, two bands were detected, which suggests that two isoforms of AUF1 might be expressed. Using this antibody, we performed immunocytochemistry in cultured Aplysia sensory neurons. Endogenous ApAUF1 was localized mainly in the somatic region, near the plasma membrane and perinucleus region and also detected in the neurites, which is different from mammalian AUF1 proteins that are mainly localized in the nucleus (Fig. 1B). Consistently, overexpressed flag-ApAUF1 also showed a similar localization pattern in the sensory neurons (Fig. 1C).
Fig. 1.
Expression of ApAUF1 in the Aplysia nervous system. (A) Western blot analysis of multiple tissues of Aplysia. ApAUF1 is highly expressed in the nervous system of Aplysia. BM, buccal mass; CNS, central nervous system including abdominal ganglion and central ganglion; GL, gill; PL, pleural ganglion. Arrows indicate the molecular weights (kDa). (B) The subcellular localization of the endogenous ApAUF1 in sensory neurons. ApAUF1 is expressed mainly in the cytosol, specifically near the plasma membrane and nuclear membrane. ApAUF1 expression is also detected in the neurites. Trans, transmission image. (C) The subcellular localization of overexpressed flag-ApAUF1 in sensory neurons. Flag-ApAUF1 was detected by Flag antibody and GFP was coinjected as a marker. Overexpressed ApAUF1 shows similar localization pattern to the endogenous ApAUF1.
Association of ApAUF1 with ApC/EBP 3′UTR.
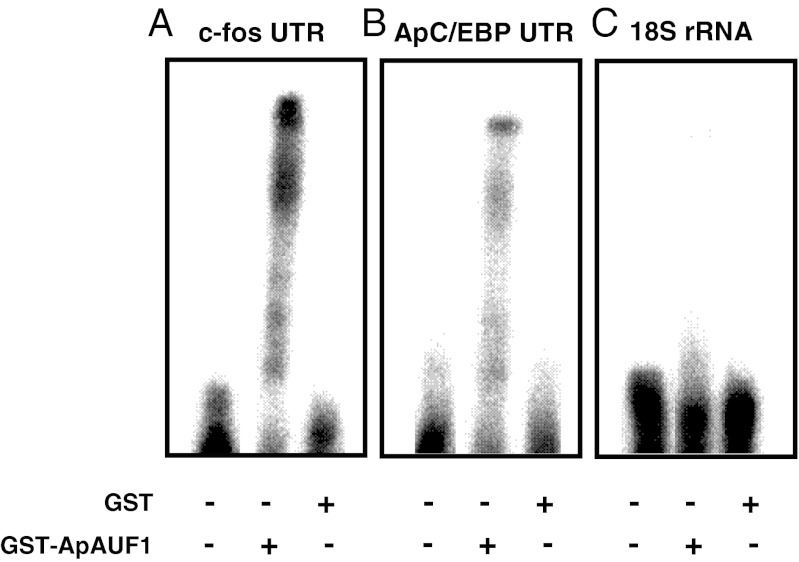
AUF1 is known to interact with many ARE-containing mRNAs that encode proto-oncogenes, nuclear transcription factors, and cytokines such as c-myc, c-fos, and TNF-α (14). To investigate whether ApAUF1 encodes a functional ARE-binding protein, we purified the recombinant ApAUF1 protein fused with GST and performed gel mobility shift assays by using the c-fos ARE (214 bp) as a target (22). α-32P-labeled c-fos ARE transcript was shifted by GST-ApAUF1, which suggests the formation of an RNA–protein complex (Fig. 2A). No band shift was detected when c-fos ARE transcript was incubated with GST alone. These results suggest that the cloned ApAUF1 encodes a functional ARE-binding protein.
Fig. 2.
Binding of ApAUF1 to the 3′ UTR of ApC/EBP mRNA. (A) In-gel mobility shift assays, a 32P-labeled riboprobe containing the ARE from c-fos mRNA was incubated with purified GST-ApAUF1 (500 nM) and resolved by native gel electrophoresis. Additional reactions were performed without protein (first lane) or with only GST protein (500 nM, last lane). RNA–protein complexes generated supershifted bands, indicating ApAUF1 encodes a functional ARE-binding protein. (B) Gel mobility shift assays of recombinant ApAUF1 binding to the ApCEBP 3′ UTR were performed in the same way as described in A and resulted in supershifted bands. The 3′ UTR of ApC/EBP mRNA (448 bp of AU-rich region) was labeled and used as a riboprobe. (C) Aplysia 18S rRNA (389 bp), which does not contain ARE sequences, was used as a probe to examine the binding specificity of ApAUF1. No specific interaction was observed between this negative control transcript and GST-ApAUF1.
Mouse C/EBPδ mRNA is highly unstable, and its 3′ UTR contains two putative AREs that interact with transacting factor(s), which are present in G0 growth arrested mammary epithelial cell lysates (23). Also, we showed that multiple copies of ARE pentamers (AUUUA), or extended pentamers (AUUUUA and AUUUUUA), exist in the 3′ UTR of ApC/EBP, and ApC/EBP is a binding target of an ARE-binding protein, ApELAV (15). It is also known that large numbers of mRNAs are shared targets of both ELAV and AUF1 (24). To examine whether ApAUF1 interacts with the putative ARE of ApC/EBP, we incubated recombinant ApAUF1 with the labeled 3′ UTR of ApC/EBP (448 bp), and we performed gel mobility shift assays. We found that the ApC/EBP 3′ UTR was shifted by purified ApAUF1 (Fig. 2B). ApAUF1 did not interact with Aplysia 18S rRNA (389 bp), which suggests that ApAUF1 specifically binds to AREs (Fig. 2C). These data indicate that the cloned ApAUF1 is an ARE-binding protein, which has ApC/EBP as a binding partner.
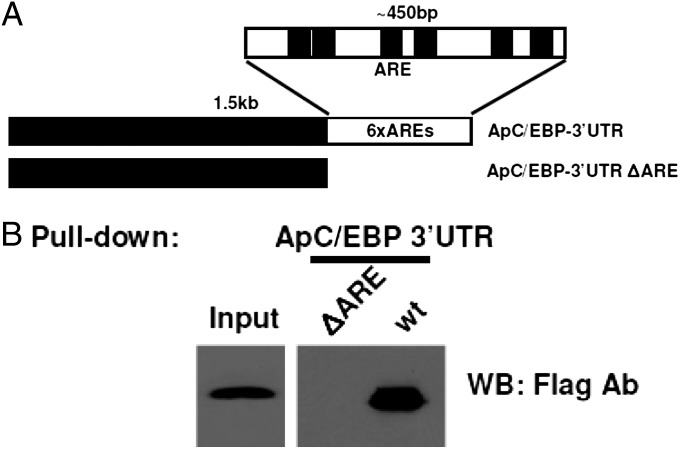
To examine whether ApAUF1 associates with ApC/EBP 3′ UTR through binding to AREs, we performed a RNA-protein pull-down assay by using a mutant RNA probe in which the putative AREs are deleted (Fig. 3A). After incubating biotin-labeled RNAs with protein lysates of HEK293T cells overexpressing flag-ApAUF1, RNA–protein complex were pulled down and analyzed in Western blot. Whereas the wild-type (WT) ApAUF1 bound to flag-ApAUF1, the ΔARE mutant RNA did not interact with flag-AUF1 (Fig. 3B), which demonstrates that the AREs are required for the interaction between ApAUF1 and ApC/EBP 3′ UTR.
Fig. 3.
ApAUF1 binds to the 3′ UTR of ApC/EBP through AREs. (A) Schematic structure of reporter RNAs. In ΔARE mutant, 448 bp containing putative six AREs in the 3′ UTR of ApC/EBP were deleted. (B) ApAUF1 interacts with intact 3′ UTR (wt), but not with ΔARE mutant RNA. In RNA-protein pull-down assay, biotin-labeled reporter RNA was incubated with lysate of HEK293T cells overexpressing 3x flag-ApAUF1. After pulling-down with avidin, RNA–protein complex was detected in Western blot by using an mFlag-m2 antibody.
Degradation of ApC/EBP ARE-Containing mRNA by ApAUF1.
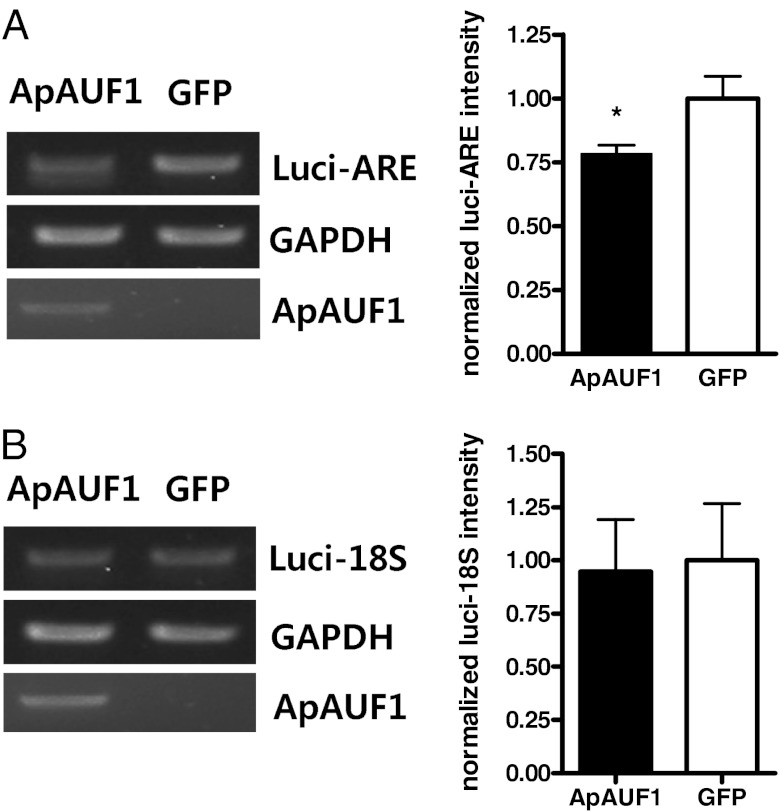
To determine whether ApAUF1 regulates the stability of mRNA, we measured mRNA levels of reporter genes after blocking new RNA synthesis. After overexpressing a reporter construct, which expresses firefly luciferase containing the ApC/EBP 3′ UTR (luci-ARE), with either ApAUF1 or EGFP in HEK293T cells, the cells were treated with actinomycin D for 6 h. RT-PCR analysis showed that ApAUF1 significantly reduced the level of ApC/EBP 3′ UTR-containing reporter mRNA, which suggests that ApAUF1 decreases the stability of ARE-containing mRNA (Fig. 4A; n = 5, unpaired two-tailed t test, *P < 0.05). Moreover, the reporter construct containing 18s rRNA in its 3′ UTR (luci-18S) was not affected by coexpression of ApAUF1, which suggests that ApAUF1 specifically acts on ARE-containing mRNAs (Fig. 4B). Altogether, our data suggest that ApAUF1 decreases ApC/EBP mRNA level by regulating the stability of mRNA through binding to the ARE in the 3′ UTR of ApC/EBP.
Fig. 4.
Destabilization of the reporter mRNA containing ApC/EBP ARE by ApAUF1. (A) The effect of ApAUF1 on the expression of luciferase mRNA containing the ApC/EBP ARE in its 3′ UTR (luci-ARE). HEK293T cells were transfected with luci-ARE and ApAUF1 or EGFP. After actinomycin D treatment (5 μg/mL, 6 h), total RNAs were extracted and subjected to RT-PCR. The mRNA level of luciferase-ARE, ApAUF1, and GAPDH was examined. The luciferase band intensities were normalized by using the intensity of GAPDH bands. ApAUF1 significantly reduced luci-ARE mRNA level compared with GFP group. n = 5 per group. *P < 0.05, t test. Error bars represent SEM. (B) The effect of ApAUF1 on the level of luciferase containing 18s rRNA (luci-18S) instead of the ApC/EBP ARE in its 3′ UTR. ApAUF1 has no effect on the expression of luci-18S. n = 5 per group.
Blockade of ApC/EBP Expression by Overexpression of ApAUF1.
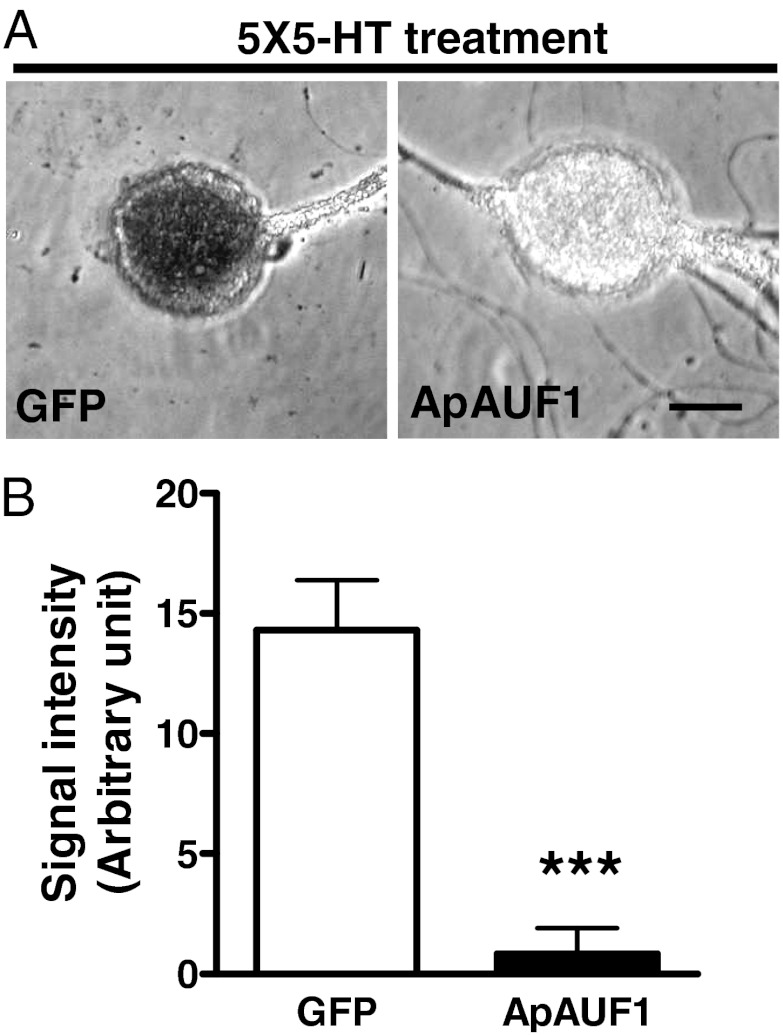
Given that ApAUF1 binds to the 3′ UTR of ApC/EBP in vitro and destabilizes reporter mRNA in HEK293 cells, we examined the effect of ApAUF1 overexpression on the induction of ApC/EBP mRNA in response to 5-HT treatment in Aplysia sensory neurons. Sensory neurons overexpressing ApAUF1 and the marker GFP gene were treated with five pulses of 5-HT (5× 5-HT), which induces ApC/EBP expression and LTF. Two hours after the onset of the 5-HT, ApC/EBP mRNA expression in the sensory neurons were examined by in situ hybridization. As a control, we overexpressed only GFP in sensory neurons without ApAUF1. The level of ApC/EBP mRNA was reduced more than 14-fold by ApAUF1 overexpression (n = 9) compared with control GFP group (n = 19) (t test, ***P < 0.001; Fig. 5). Using a sense probe against ApC/EBP mRNA as a negative control did not produce a detectible signal. This in situ hybridization result suggests that ApAUF1 suppresses the induction of ApC/EBP mRNAs in 5-HT–treated Aplysia sensory neurons.
Fig. 5.
Blockade of 5-HT–induced ApC/EBP induction by ApAUF1 overexpression. (A) Sensory neurons expressing ApAUF1 and GFP, or GFP alone, were stimulated with five pulses of 5-HT, and ApC/EBP mRNA was detected by in situ hybridization. Digoxigenin-labeled antisense riboprobes against ApC/EBP were used to detect ApC/EBP transcripts. Sensory cells expressing only GFP showed a strong signal for ApC/EBP when they were stimulated by 5-HT (Left). However, ApAUF1 overexpressing neurons exhibited less staining after 5-HT stimulation (Right). (Scale bar: 50 μm.) (B) ApC/EBP expression was quantified by measuring pixel intensity from the cell bodies. Signal intensities were significantly lower in ApAUF1 overexpressing neurons compared with GFP expressing control neurons (arbitrary unit of signal intensity, ApAUF1 + 5× 5-HT, 0.835 ± 1.065, n = 9; GFP + 5× 5-HT, 14.310 ± 2.076, n = 19; t test, ***P < 0.001).
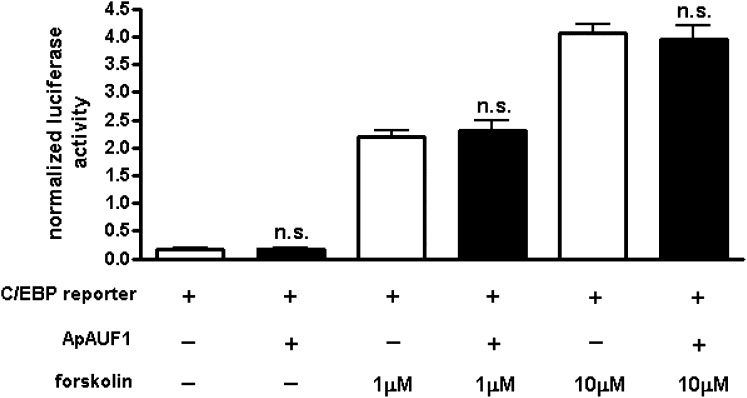
Although the mammalian AUF1 is a well-known posttranscriptional regulator of ARE-containing mRNAs, it is also known that AUF1 regulates transcription (25). To determine whether ApAUF1 represses the transcription of ApC/EBP mRNA, we examined the expression of the luciferase reporter gene containing ApC/EBP promoter in its 5′ region (C/EBP-luciferase). HEK293T cells were transfected with the C/EBP-luciferase reporter and ApAUF1, or GFP as a control. To stimulate C/EBP promoter-driven reporter expression, cells were treated with forskolin 24 h after transfection. Overexpression of ApAUF1 did not affect C/EBP promoter-driven expression of luciferase (Fig. 6). These results suggest that ApAUF1 does not negatively regulate the expression of endogenous ApC/EBP through a transcriptional mechanism, but by a posttranscriptional mechanism, such as mRNA degradation.
Fig. 6.
Effect of ApAUF1 on ApC/EBP promoter-driven gene expression. HEK293T cells were transfected with the luciferase reporter under control of the ApC/EBP promoter (C/EBP-luci) and ApAUF1, or EGFP. C/EBP promoter-mediated gene expression was stimulated by forskolin treatment, and the reporter gene expression was analyzed by using a luciferase assay. ApAUF1 overexpression had no significant effect on the transcription of ApC/EBP. Renilla luciferase was cotransfected and used for normalization. n = 6 per group. n.s., not significant, unpaired t test.
Blockade of Long-Term Facilitation by Overexpression of ApAUF1.
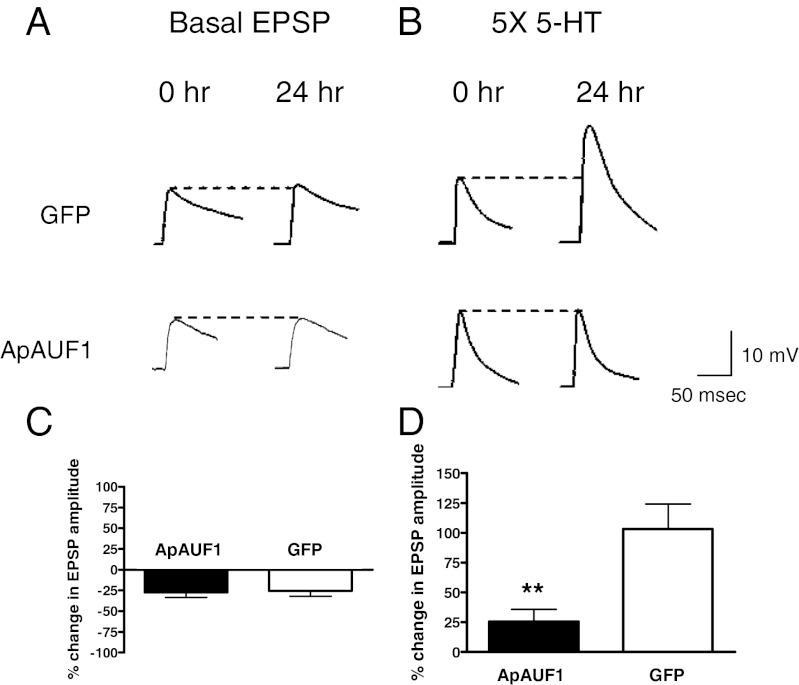
Inhibition of ApC/EBP expression through injection of antisense or double-stranded RNA (dsRNA) blocks LTF of Aplysia sensory-to-motor synapses, suggesting that ApC/EBP acts as a critical molecular switch for memory consolidation (4, 7). We observed the destabilizing effect of ApAUF1 on ApC/EBP mRNA (Fig. 5) and wanted to examine the effect of ApAUF1 overexpression on LTF induced by 5× 5-HT. First, we examined the effect of overexpression of ApAUF1 in sensory neuron on basal synaptic transmission in sensory-to-motor synapses by measuring EPSP amplitudes before and after overexpression of ApAUF1. Percentage changes in EPSP amplitude of ApAUF1 expressing synapses (−27.43 ± 5.92%, n = 20) were comparable to results obtained from control synapses expressing only GFP in sensory neurons (percent change in EPSP amplitude, −25.59 ± 6.36, n = 13; Fig. 7 A and C). Next, 5× 5-HT–induced EPSP amplitude changes were measured in sensory-to-motor synapses to examine the effect of AUF1 overexpression on LTF. Cultures injected with ApAUF1 showed decreased facilitation 24 h after five pulses of 5-HT (25.58 ± 10.22%, n = 18; Fig. 7 B and D). This facilitation was significantly different from the increase of EPSP amplitude measured in GFP-expressing synapses treated with 5× 5-HT (103.20 ± 20.98%, n = 13, t test, **P < 0.01; Fig. 7 B and D). This result shows that ApAUF1 overexpression represses LTF.
Fig. 7.
Impaired long-term facilitation by ApAUF1 overexpression. (A) Examples of EPSP recordings before (0 h) and after (24 h) overexpression of ApAUF1 + GFP (Lower) or GFP alone (Upper). Changes in EPSP amplitude were comparable between ApAUF1 expressing synapses and control synapses. (B) Examples of EPSP recordings before (0 h) and after (24 h) five pulses of 5-HT treatment. After confirming the presence of GFP expression in sensory neurons, five pulses of 5-HT were delivered to induce LTF. ApAUF1 overexpression blocked the 5-HT–induced increase in EPSP amplitude at 24 h. (C) Bar graph representing the effect of ApAUF1 overexpression on basal synaptic transmission. Basal synaptic transmission was not significantly different between the ApAUF1 and GFP control group (percent change in EPSP amplitude, −27.43 ± 5.92, n = 20 and −25.59 ± 6.37, n = 13, respectively; t test, P = 0.839). (D) Bar graph representing the effect of ApAUF1 overexpression on LTF. Overexpression of ApAUF1 in Aplysia sensory neurons of sensory-to-motor synapses significantly impaired 5-HT–induced LTF (percent change in EPSP amplitude, ApAUF1, 25.58 ± 10.22, n = 18 vs. GFP, 103.20 ± 20.98, n = 13; t test, **P < 0.01).
Discussion
In the present study, we cloned ApAUF1, which encodes a functional ARE-binding protein and acts as a destabilizing factor for the transcriptional activator ApC/EBP. Overexpression of ApAUF1 blocked the induction of ApC/EBP and LTF by 5-HT in Aplysia sensory-to-motor synapses.
ApC/EBP was identified as a downstream signaling molecule of CREB and was shown to be a positive transcriptional regulator in Aplysia sensory-to-motor synapses (4). In the rodent hippocampus, C/EBPβ and C/EBPδ were reported to be induced after inhibitory avoidance learning, suggesting that C/EBPs are well conserved components of the CREB-dependent signal pathway involved in memory consolidation (26). Transcriptional regulation of C/EBP expression has been extensively investigated (27, 28). Interestingly, ApC/EBP mRNA has a short lifetime (4, 8). Mammalian C/EBPδ mRNA is also known to be highly unstable, and it contains AREs in its 3′ UTR region (23). Yamamoto et al. suggested ubiquitin-proteasome–mediated degradation as a molecular mechanism underlying the rapid inactivation and degradation of ApC/EBP protein (8). This explanation is supported by the finding that Aplysia ubiquitin C-terminal hydrolase (Ap-uch) is induced by 5-HT treatment, and its expression reaches its peak 3.5 h after initiating 5-HT stimulation. This time course coincides with the time point when ApC/EBP activity begins to decrease (5). However, the mechanism regulating its rapid mRNA decay during memory consolidation is still unclear. Our results provide lines of evidence suggesting that an ARE-binding protein ApAUF1 is critically involved in the posttranscriptional degradation of ApC/EBP mRNA. First, ApAUF1 binds to 3′ UTR of ApC/EBP mRNA. Second, ApAUF1 decreases the stability of RNAs containing 3′ UTR of ApC/EBP mRNA. Third, ApAUF1 overexpression blocks the induction of ApC/EBP mRNA by 5-HT treatment. Finally, ApAUF1 overexpression impairs LTF by 5-HT treatment. Taken together, these results strongly suggest that ApAUF1 overexpression blocks LTF via degradation of ApC/EBP mRNA. Our data showing that ApAUF1 overexpression does not impair baseline synaptic transmission suggests that ApAUF1 may act on plasticity-related genes such as ApC/EBP. However, because it is possible that ApAUF1 can bind to other ARE-containing mRNAs besides ApC/EBP mRNA, the target profile of ApAUF1 needs to be further investigated. In addition, we cannot exclude the possibility that the overexpressed ApAUF1 impairs LTF via blocking other plasticity-related genes containing ARE. One caveat of this study is that we could not demonstrate the role of endogenous ApAUF1 in LTF. Because of the high expression level of the endogenous ApAUF1, multiple approaches such as injecting dsRNAs and anti-ApAUF1 antibody did not succeed in knocking down the expression of ApAUF1 in Aplysia sensory neurons. It is still possible that other RNA binding proteins are also involved in regulating the stability of ApC/EBP mRNA under the physiological condition.
Accordingly, we showed that the stability of ApC/EBP mRNA is positively regulated by another ARE-binding protein, ApELAV (15). ApAUF1 and ApELAV bind to the same AU-rich region of ApC/EBP and act as negative and positive regulators of ApC/EBP mRNA stability, respectively. It is of interest to study the molecular mechanisms involved in bidirectional modulation of ApC/EBP mRNA by these two opposing ARE-binding proteins. It is likely that the two ARE-binding proteins act dominantly at different time points to facilitate ApC/EBP mRNA stabilization and destabilization. Previous reports showed that mammalian ELAV-like proteins and AUF1 bind to common AU-rich target mRNAs concurrently and individually depending on their subcellular localization (24), developmental stage (29), or neuronal activity (30). For example, AUF1 and HuR bind to the ARE of α2 subunit of guanylyl cyclase mRNA in cerebellar granule cells. However, NMDA treatment decreased the level of nuclear AUF1 and, subsequently, increased the level of α2 guanylyl cyclase mRNA (30). Sequence analysis of ApAUF1 predicted at least four putative serine phosphorylation sites, suggesting that the activity of ApAUF1 may be regulated by protein kinases during synaptic plasticity, although this possibility remains to be investigated. Also, it is worthy to note that ApAUF1 shows the subcellular localization distinct from the mammalian AUF1 proteins (Fig. 1). It has been shown that nuclear import and export of mammalian AUF1 can be determined by the structure of C-terminal domains: Nuclear import is mediated by the uninterrupted C-terminal domain and nuclear export is facilitated by the presence of the amino acids encoded by exon 7 in the larger isoforms (31). Multiple alignments of ApAUF1 and mammalian isoforms suggest that the cloned ApAUF1 contains these additional sequences found in the larger isoforms p42 and p45, which interrupt its C-terminal domain and may facilitate the nuclear export of ApAUF1 (Fig. S2). Moreover, mammalian AUF1 isoforms show different localization patterns depending on the cell types (32). It may be due to the differences in interacting protein partners in different cell types, and ApAUF1 may also have specific binding proteins determining its subcellular localization in Aplysia neurons.
Evidence for the role of other RNA-binding proteins in the regulation of synaptic plasticity and memory has accumulated during the last decade (33, 34). The cytoplasmic polyadenylation element binding protein, CPEB, is one of the stabilizing components of the synaptic mark that regulates local protein synthesis and stabilizes the synapse-specific LTF in Aplysia (35–37). Syntaxin mRNA has been also reported to be a target of CPEB and Staufen, in Aplysia sensory neurons, and inhibiting the interaction between syntaxin mRNA and these binding proteins blocked LTF (38). In rodents, CPEB-1 knockout mice show deficits in some form of synaptic plasticity and extinction of hippocampus-dependent memory (39, 40). The expression of ELAV-like proteins, including HuD, which are mRNA stabilizing factors (10), are up-regulated in the CA1 region of the hippocampus after spatial memory formation (41–43). Moreover, transgenic mice overexpressing HuD have impaired fear conditioning and perform poorly in the Morris water maze test (44), showing the importance of regulated expression of RNA binding protein for proper memory processing.
Our findings provide a molecular mechanism for the decay of ApC/EBP mRNA and demonstrate the critical roles of posttranscriptional regulation of gene expression during the consolidation of synaptic plasticity. ApAUF1 may function as a checkpoint for the consolidation of LTF by clearing transcribed ApC/EBP mRNA. Malfunctioning of negative regulators such as ApAUF1 may result in the uncontrolled activation of positive regulators or the loss of temporal and spatial specificity of gene expression, which can disrupt normal memory consolidation.
Materials and Methods
Detailed methods are described in SI Materials and Methods.
RNA Gel Mobility Shift Assay.
The ApAUF1 GST-fusion protein purification and RNA gel mobility shift assay were performed as described (15). In the assay, 10, 100, or 500 nM purified proteins were added to the reaction. Because supershift of RNA probe was clearly detected only with 500 nM, the lanes loaded with 10 and 100 nM proteins were omitted in Fig. 2.
RNA-Protein Pull-Down Assay.
mRNA-protein pull-down assay was performed as described (45) with slight modifications.
In Situ Hybridization in Cultured Aplysia Sensory Neuron.
In situ hybridizations were performed as described by using an ApC/EBP mRNA specific probe (46). Signal intensity was measured by using Photoshop (Adobe).
Luciferase Assay.
HEK293T cells were transfected with 1 μg of the firefly luciferase reporter, 200 ng of ApAUF1, and 200 ng of renilla luciferase for normalization. Twenty-four hours after transfection, we treated the cells with 1 μM or 10 μM forskolin (Sigma), or DMSO, and we performed luciferase assays by using Dual Glo luciferase assay systems (Promega).
Sensory-to-Motor Coculture, 5-HT Treatment, and Electrophysiology.
Sensory-to-motor neuron coculture of A. kurodai, 5-HT treatment, and electrophysiological recording of basal synaptic transmission and 5 × 5-HT-induced long-term facilitation were conducted as described by Lee et al (7).
Statistics.
For pair-wise comparisons, Student’s t tests were used and P < 0.05 was considered to be significant. Data are represented as mean ± SEM.
Supplementary Material
Acknowledgments
This work was supported by the Creative Research Initiative Program of the Korean Ministry of Science and Technology and World Class University program; S.-L.C. and J.S. are supported by Brain Korea 21 fellowship; D.-J.J. was supported by Kyungpook National University Research Fund; and K.L. is supported by Basic Science Research Program Grant 2011-0028240 of the Ministry of Education, Science, and Technology.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
1Y.-S.L., S.-L.C., and H.J. contributed equally to this work.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1116224109/-/DCSupplemental.
References
- 1.Kandel ER. The molecular biology of memory storage: A dialogue between genes and synapses. Science. 2001;294:1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- 2.Bailey CH, Bartsch D, Kandel ER. Toward a molecular definition of long-term memory storage. Proc Natl Acad Sci USA. 1996;93:13445–13452. doi: 10.1073/pnas.93.24.13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee YS, Bailey CH, Kandel ER, Kaang BK. Transcriptional regulation of long-term memory in the marine snail Aplysia. Mol Brain. 2008;1:3. doi: 10.1186/1756-6606-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alberini CM, Ghirardi M, Metz R, Kandel ER. C/EBP is an immediate-early gene required for the consolidation of long-term facilitation in Aplysia. Cell. 1994;76:1099–1114. doi: 10.1016/0092-8674(94)90386-7. [DOI] [PubMed] [Google Scholar]
- 5.Hegde AN, et al. Ubiquitin C-terminal hydrolase is an immediate-early gene essential for long-term facilitation in Aplysia. Cell. 1997;89:115–126. doi: 10.1016/s0092-8674(00)80188-9. [DOI] [PubMed] [Google Scholar]
- 6.Alberini CM, Ghirardi M, Huang YY, Nguyen PV, Kandel ER. A molecular switch for the consolidation of long-term memory: cAMP-inducible gene expression. Ann N Y Acad Sci. 1995;758:261–286. doi: 10.1111/j.1749-6632.1995.tb24833.x. [DOI] [PubMed] [Google Scholar]
- 7.Lee JA, et al. Overexpression of and RNA interference with the CCAAT enhancer-binding protein on long-term facilitation of Aplysia sensory to motor synapses. Learn Mem. 2001;8:220–226. doi: 10.1101/lm.40201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamamoto N, Hegde AN, Chain DG, Schwartz JH. Activation and degradation of the transcription factor C/EBP during long-term facilitation in Aplysia. J Neurochem. 1999;73:2415–2423. doi: 10.1046/j.1471-4159.1999.0732415.x. [DOI] [PubMed] [Google Scholar]
- 9.McCarthy JE, Kollmus H. Cytoplasmic mRNA-protein interactions in eukaryotic gene expression. Trends Biochem Sci. 1995;20:191–197. doi: 10.1016/s0968-0004(00)89006-4. [DOI] [PubMed] [Google Scholar]
- 10.Guhaniyogi J, Brewer G. Regulation of mRNA stability in mammalian cells. Gene. 2001;265:11–23. doi: 10.1016/s0378-1119(01)00350-x. [DOI] [PubMed] [Google Scholar]
- 11.Bevilacqua A, Ceriani MC, Capaccioli S, Nicolin A. Post-transcriptional regulation of gene expression by degradation of messenger RNAs. J Cell Physiol. 2003;195:356–372. doi: 10.1002/jcp.10272. [DOI] [PubMed] [Google Scholar]
- 12.Mitchell P, Tollervey D. mRNA stability in eukaryotes. Curr Opin Genet Dev. 2000;10:193–198. doi: 10.1016/s0959-437x(00)00063-0. [DOI] [PubMed] [Google Scholar]
- 13.Bolognani F, Perrone-Bizzozero NI. RNA-protein interactions and control of mRNA stability in neurons. J Neurosci Res. 2008;86:481–489. doi: 10.1002/jnr.21473. [DOI] [PubMed] [Google Scholar]
- 14.Chen CY, Shyu AB. AU-rich elements: Characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 15.Yim SJ, et al. Regulation of ApC/EBP mRNA by the Aplysia AU-rich element-binding protein, ApELAV, and its effects on 5-hydroxytryptamine-induced long-term facilitation. J Neurochem. 2006;98:420–429. doi: 10.1111/j.1471-4159.2006.03887.x. [DOI] [PubMed] [Google Scholar]
- 16.Zhang W, et al. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol Cell Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeMaria CT, Brewer G. AUF1 binding affinity to A+U-rich elements correlates with rapid mRNA degradation. J Biol Chem. 1996;271:12179–12184. doi: 10.1074/jbc.271.21.12179. [DOI] [PubMed] [Google Scholar]
- 18.Wagner BJ, DeMaria CT, Sun Y, Wilson GM, Brewer G. Structure and genomic organization of the human AUF1 gene: Alternative pre-mRNA splicing generates four protein isoforms. Genomics. 1998;48:195–202. doi: 10.1006/geno.1997.5142. [DOI] [PubMed] [Google Scholar]
- 19.Laroia G, Cuesta R, Brewer G, Schneider RJ. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science. 1999;284:499–502. doi: 10.1126/science.284.5413.499. [DOI] [PubMed] [Google Scholar]
- 20.Laroia G, Sarkar B, Schneider RJ. Ubiquitin-dependent mechanism regulates rapid turnover of AU-rich cytokine mRNAs. Proc Natl Acad Sci USA. 2002;99:1842–1846. doi: 10.1073/pnas.042575699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee YS, et al. Transcriptome analysis and identification of regulators for long-term plasticity in Aplysia kurodai. Proc Natl Acad Sci USA. 2008;105:18602–18607. doi: 10.1073/pnas.0808893105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma WJ, Cheng S, Campbell C, Wright A, Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J Biol Chem. 1996;271:8144–8151. doi: 10.1074/jbc.271.14.8144. [DOI] [PubMed] [Google Scholar]
- 23.Dearth LR, DeWille J. An AU-rich element in the 3′ untranslated region of the C/EBP delta mRNA is important for protein binding during G0 growth arrest. Biochem Biophys Res Commun. 2003;304:344–350. doi: 10.1016/s0006-291x(03)00597-7. [DOI] [PubMed] [Google Scholar]
- 24.Lal A, et al. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004;23:3092–3102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dobi A, et al. AUF1 is expressed in the developing brain, binds to AT-rich double-stranded DNA, and regulates enkephalin gene expression. J Biol Chem. 2006;281:28889–28900. doi: 10.1074/jbc.M511858200. [DOI] [PubMed] [Google Scholar]
- 26.Taubenfeld SM, et al. Fornix-dependent induction of hippocampal CCAAT enhancer-binding protein [beta] and [delta] Co-localizes with phosphorylated cAMP response element-binding protein and accompanies long-term memory consolidation. J Neurosci. 2001;21:84–91. doi: 10.1523/JNEUROSCI.21-01-00084.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montarolo PG, Kandel ER, Schacher S. Long-term heterosynaptic inhibition in Aplysia. Nature. 1988;333:171–174. doi: 10.1038/333171a0. [DOI] [PubMed] [Google Scholar]
- 28.Guan Z, et al. Integration of long-term-memory-related synaptic plasticity involves bidirectional regulation of gene expression and chromatin structure. Cell. 2002;111:483–493. doi: 10.1016/s0092-8674(02)01074-7. [DOI] [PubMed] [Google Scholar]
- 29.Hambardzumyan D, et al. AUF1 and Hu proteins in the developing rat brain: Implication in the proliferation and differentiation of neural progenitors. J Neurosci Res. 2009;87:1296–1309. doi: 10.1002/jnr.21957. [DOI] [PubMed] [Google Scholar]
- 30.Jurado S, et al. NMDA induces post-transcriptional regulation of alpha2-guanylyl-cyclase-subunit expression in cerebellar granule cells. J Cell Sci. 2006;119:1622–1631. doi: 10.1242/jcs.02867. [DOI] [PubMed] [Google Scholar]
- 31.Sarkar B, Lu JY, Schneider RJ. Nuclear import and export functions in the different isoforms of the AUF1/heterogeneous nuclear ribonucleoprotein protein family. J Biol Chem. 2003;278:20700–20707. doi: 10.1074/jbc.M301176200. [DOI] [PubMed] [Google Scholar]
- 32.Arao Y, Kuriyama R, Kayama F, Kato S. A nuclear matrix-associated factor, SAF-B, interacts with specific isoforms of AUF1/hnRNP D. Arch Biochem Biophys. 2000;380:228–236. doi: 10.1006/abbi.2000.1938. [DOI] [PubMed] [Google Scholar]
- 33.Martin KC, Barad M, Kandel ER. Local protein synthesis and its role in synapse-specific plasticity. Curr Opin Neurobiol. 2000;10:587–592. doi: 10.1016/s0959-4388(00)00128-8. [DOI] [PubMed] [Google Scholar]
- 34.Schuman EM. mRNA trafficking and local protein synthesis at the synapse. Neuron. 1999;23:645–648. doi: 10.1016/s0896-6273(01)80023-4. [DOI] [PubMed] [Google Scholar]
- 35.Si K, et al. A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in aplysia. Cell. 2003;115:893–904. doi: 10.1016/s0092-8674(03)01021-3. [DOI] [PubMed] [Google Scholar]
- 36.Miniaci MC, et al. Sustained CPEB-dependent local protein synthesis is required to stabilize synaptic growth for persistence of long-term facilitation in Aplysia. Neuron. 2008;59:1024–1036. doi: 10.1016/j.neuron.2008.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Si K, Choi YB, White-Grindley E, Majumdar A, Kandel ER. Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell. 2010;140:421–435. doi: 10.1016/j.cell.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 38.Liu J, Hu JY, Wu F, Schwartz JH, Schacher S. Two mRNA-binding proteins regulate the distribution of syntaxin mRNA in Aplysia sensory neurons. J Neurosci. 2006;26:5204–5214. doi: 10.1523/JNEUROSCI.4917-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alarcon JM, et al. Selective modulation of some forms of schaffer collateral-CA1 synaptic plasticity in mice with a disruption of the CPEB-1 gene. Learn Mem. 2004;11:318–327. doi: 10.1101/lm.72704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berger-Sweeney J, Zearfoss NR, Richter JD. Reduced extinction of hippocampal-dependent memories in CPEB knockout mice. Learn Mem. 2006;13:4–7. doi: 10.1101/lm.73706. [DOI] [PubMed] [Google Scholar]
- 41.Pascale A, et al. Increase of the RNA-binding protein HuD and posttranscriptional up-regulation of the GAP-43 gene during spatial memory. Proc Natl Acad Sci USA. 2004;101:1217–1222. doi: 10.1073/pnas.0307674100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bolognani F, Merhege MA, Twiss J, Perrone-Bizzozero NI. Dendritic localization of the RNA-binding protein HuD in hippocampal neurons: Association with polysomes and upregulation during contextual learning. Neurosci Lett. 2004;371:152–157. doi: 10.1016/j.neulet.2004.08.074. [DOI] [PubMed] [Google Scholar]
- 43.Quattrone A, et al. Posttranscriptional regulation of gene expression in learning by the neuronal ELAV-like mRNA-stabilizing proteins. Proc Natl Acad Sci USA. 2001;98:11668–11673. doi: 10.1073/pnas.191388398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bolognani F, et al. Associative and spatial learning and memory deficits in transgenic mice overexpressing the RNA-binding protein HuD. Neurobiol Learn Mem. 2007;87:635–643. doi: 10.1016/j.nlm.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 45.Mastushita-Sakai T, White-Grindley E, Samuelson J, Seidel C, Si K. Drosophila Orb2 targets genes involved in neuronal growth, synapse formation, and protein turnover. Proc Natl Acad Sci USA. 2010;107:11987–11992. doi: 10.1073/pnas.1004433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim H, et al. A nucleolar protein ApLLP induces ApC/EBP expression required for long-term synaptic facilitation in aplysia neurons. Neuron. 2006;49:707–718. doi: 10.1016/j.neuron.2006.01.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.