Abstract
In Escherichia coli, tetracycline prevents translation. When subject to tetracycline, E. coli express TetA to pump it out by a mechanism that is sensitive, while fairly independent of cellular metabolism. We constructed a target gene, PtetA-mRFP1-96BS, with a 96 MS2-GFP binding site array in a single-copy BAC vector, whose expression is controlled by the tetA promoter. We measured the in vivo kinetics of production of individual RNA molecules of the target gene as a function of inducer concentration and temperature. From the distributions of intervals between transcription events, we find that RNA production by PtetA is a sub-Poissonian process. Next, we infer the number and duration of the prominent sequential steps in transcription initiation by maximum likelihood estimation. Under full induction and at optimal temperature, we observe three major steps. We find that the kinetics of RNA production under the control of PtetA, including number and duration of the steps, varies with induction strength and temperature. The results are supported by a set of logical pairwise Kolmogorov-Smirnov tests. We conclude that the expression of TetA is controlled by a sequential mechanism that is robust, whereas sensitive to external signals.
INTRODUCTION
Tetracycline is a polycyclic naphthacene carboxamide (1), isolated from Streptomyces genus of Actinobacteria (2), which prevents bacterial cell growth by binding to the 30 S ribosomal subunit. This binding interferes with the attachment of aminoacyl-tRNA to the mRNA-ribosome translation complex, inhibiting protein synthesis (3,4). In Escherichia coli, resistance to tetracycline is conferred by the extra-chromosomal Tn10 transposon encoded class B molecular determinants, forming the tet operon (5–8) (also named divergon (9) or regulon (10)). This operon consists of two structural genes, tetA and tetR. tetA is essential for tetracycline resistance (11), as it encodes for a membrane-targeted antiporter protein, TetA, responsible for active efflux of tetracycline, whereas tetR codes for TetR that regulates the tet operon.
In the absence of tetracycline, TetR binds to the operator sites of tetA and tetR, preventing their transcription (12). In the presence of tetracycline, TetR binds as a dimer to the biologically active tetracycline–Mg2+ complex, causing an allosteric conformational change in the repressor protein (13). This releases the repressor from the DNA, allowing RNA polymerase to bind and initiate transcription of tetA and tetR. The tet operon is thus a self-repressing system (12,14), capable of fast and efficient response to tetracycline. Its expression activity is largely independent of the metabolic state of the host cell, making it a preferential system to control recombinant gene expression (15) and to study mechanisms of gene expression (13). Studies of the β-galactosidase activity of the Tn10 tetR-tetA promoter have showed that PtetA is the strongest of the three promoters of this operon (16). It has thus been isolated, modified, and used in several studies (namely the tetR/O region) of gene expression (4), and its derivatives have been used in several synthetic circuits (17–23).
Studies suggest that gene expression kinetics in prokaryotes appears to be mainly controlled at the transcriptional level, particularly in initiation (24). In vitro measurements suggest that, in E. coli, transcription initiation is a sequential process (25). The first step, named ‘closed complex formation’, is the binding of the RNA polymerase enzyme to the promoter region and finding of the transcription start site (TSS). This is followed by isomerization, DNA unwinding and loading of the nucleotide strand (the ‘open complex formation’) (25). Afterwards, elongation of the RNA strand takes place (26), and once the termination site is reached, the single-stranded RNA molecule and the RNA polymerase are released (27). This sequential process (28) is subject to tight regulation that takes place at one or more stages.
Evidence suggests that, for most promoters in E. coli, the open complex formation is the main rate-determining step in initiation (29,30). However, the durations of other steps, namely the closed complex formation, isomerization, and promoter clearance are also sequence-dependent, differing widely between promoters and in different conditions (31). Environmental factors can also affect this kinetics (32,33). For example, apart from induction (34), factors such as temperature and pH can also affect the “rate-limiting” steps in initiation (30,35).
Changes in the kinetics of these steps allows the overall expression rate of a gene to be changed (8,25). For example, in vivo measurements of the expression of the synthetic promoter PLtetO-1 using the luciferase reporter system have shown that the rate of RNA production can change by 5000-fold with induction by anhydrotetracycline (aTc) (8). However, it is noted that this promoter was engineered with the aim of allowing tight regulation (wide range of induction) (8). Among the several tetracycline analogs, aTc was found to be an effective inducer even at very low concentrations (<50 ng/ml), as it binds to the TetR protein with very high affinity (∼35-fold higher than tetracycline) (36–38) and is less toxic to cells with a minimal inhibitory concentration of 4 µg/ml (39).
If transcription initiation is the rate-determining step in RNA production, and if it is composed of several sequential steps (30–32,40,41), provided that these are approximately exponentially distributed in duration, one would expect that transcript production is a sub-Poissonian process (40). However, recent measurements of in vivo cell-to-cell diversity in RNA numbers have exhibited higher diversity in RNA numbers than would be expected from a sub-Poissonian process of transcript production (42–44). In (44), it was hypothesized that this was because of either super-Poissonian RNA production or non-Poissonian RNA degradation. In (42,43), it was hypothesized that the cause is the existence of periods of activity and inactivity of the promoter. It is noted that measurements of cell-to-cell diversity in either RNA or protein numbers are affected by several processes other than transcription and translation. Specifically, the kinetics of degradation of RNA and proteins can be complex (45), that is, non-Poissonian, and there may be non-negligible differences in measurements using in vivo and in vitro techniques (24). Finally, another event that affects cell-to-cell diversity in RNA numbers is cell division, particularly because the intracellular environment of E. coli is not well-stirred (46). Even if the RNA molecules are partitioned in an unbiased fashion, the stochastic nature of this process will enhance the cell-to-cell diversity in the numbers of these molecules following division events (47,48). In addition, bacteria are known to partition unwanted protein aggregates in a biased fashion (49,50), including, for example, RNA tagged with MS2 coat protein fused with Green Fluorescent Protein (MS2-GFP) (51,52), and fluorescent proteins such as Tsr-Venus (53,54,55). This bias in partitioning ought to exacerbate cell-to-cell diversity in RNA and protein numbers when assessed by these methods. The degree of stochasticity in transcription thus needs to be assessed without the interference of subsequent events, by measuring transcript production one event at a time (40).
Here, using in vivo, single-molecule based techniques, we characterize the kinetics of initiation of PtetA (15,56), including its stochasticity and how it changes with induction strength and temperature. Namely, we assess the transcript production dynamics at the single event level. This is possible by a method recently developed to tag RNA in vivo in E. coli with MS2-GFP proteins, which allows individual transcription events to be detectable shortly after production (55,57), and the behaviour is similar to that of the unlabelled system (57).
We report measurements of time intervals between consecutive productions of RNA molecules under the control of PtetA when subject to several induction strengths and temperatures. From the distribution of these intervals, we analyse the dynamics of this promoter. We address the following questions. What is the in vivo kinetics of RNA production, one event at a time, under the control of PtetA, when fully induced? How does the in vivo kinetics of transcription change with induction? How noisy is this process? Finally, from the inference of number and durations of the rate-limiting steps, we address how the kinetics of the rate-limiting steps changes with temperature. In the end, we compare the kinetics of initiation of PtetA with that of the Plac/ara-1 promoter, also named Plar, which has been recently characterized (40) using the same methods.
MATERIALS AND METHODS
Chemicals
For routine cultures, the components of Luria-Bertani (LB) broth (Tryptone, Yeast extract and NaCl) were purchased from LabM (UK) and antibiotics from Sigma-Aldrich (USA). Phusion high-fidelity polymerase and other PCR reagents are from Finnzymes (Finland). Fermentas kits (Finland) for Plasmid isolation and PCR product extraction and purification were made as per the instructions provided. To perform qPCR, cells were fixed with RNAprotect bacteria reagent (Qiagen, USA). The Tris and EDTA for lysis buffer were purchased from Sigma-aldrich (USA) and lysozyme from Fermentas (USA). The total RNA extraction was done with RNeasy RNA purification kit (Qiagen, USA). DNase I, RNasefree for RNA purification, was purchased from Promega (USA). iScript Reverse Transcription Supermix for cDNA synthesis and iQ SYBR Green supermix for qPCr were purchased from Biorad (USA). Agarose for microscopic slide gel preparation and electrophoresis and isopropyl b-D-1-thiogalactopyranoside (IPTG) and aTc for induction of cells are from Sigma-Aldrich (USA). For staining DNA and RNA on gels, SYBR-Safe from Invitrogen (USA) was used.
Bacterial strain and growth conditions
The strain E. coli DH5α-PRO (identical to DH5α-Z1) (57) was used to clone and express the target and reporter genes. For overnight cultures, the strain from glycerol stock was inoculated in LB broth 10 g of tryptone, 5 g of yeast extract and 5 g of NaCl per litre, pH 7.0) (56) with appropriate antibiotics (100 μg/ml ampicillin and 35 μg/ml chloramphenicol) and incubated at 37°C with shaking (250 rpm).
Genetic constructs
We constructed the target gene PtetA-mRFP1-96BS with a 96 MS2-GFP binding site array in a single-copy BAC vector by restricting out the Plar promoter with BamH1 restriction endonuclease from a BAC clone carrying a target gene Plar-mRFP1-96BS (42) (a kind gift from Ido Golding, University of Illinois, IL), and replacing it with PtetA amplified from the pTetLux1 plasmid (56). The primers (Forward: 5′GGGATCCCTCACATGACCCGACAC 3′ and Reverse: 5′GGGATCCACTGCAATCGCGATAGC 3′) were designed to amplify the PtetA promoter with BamHI restriction site flanking regions. The amplicon and the BAC vector were subjected to BamH1 restriction digestion, followed with ligation of the amplified product into the BAC vector. Thus, we obtained a single copy F-based plasmid carrying the target region PtetA-mRFP1-96BS. This product was transferred into the competent E. coli strain DH5α–PRO host cells. The recombinants were selected with antibiotic screening and further confirmed with sequence analysis. The reporter molecules to visualize the target RNA were expressed from the pZS12MS2-GFP plasmid (55) (SC101 origin, 6–8 copies per cell, AmpR, PLlacO-1 promoter) cloned into the host strain, a kind gift from Philippe Cluzel, University of Chicago, IL. The tetR gene that encodes for a regulatory protein TetR, is integrated into the chromosome of E. coli strain DH5α–PRO, under the control of a strong promoter PN25, that ensures appropriate levels of repressor proteins for tight regulation and full induction, in spite of the residual binding affinity of tetR-aTc complex to DNA (8,55).
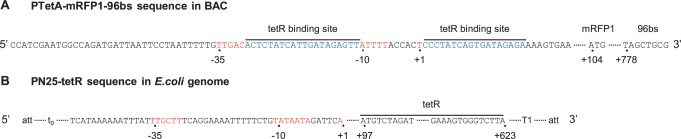
A detailed map of the tetA promoter sequence with the crucial elements, such as the TetR binding site, the −10/–35 regions, the TSS and the ribosome binding site region, as well as the beginning of the MS2-GFP binding region is shown in Figure 1. Note that there is no tetA gene in the genome of E. coli DH5α–PRO or in the genetic constructs that were transformed into the strain.
Figure 1.
(A) PtetA-mRFP1-96bs-BAC plasmid map: nucleotide sequence of the tet regulatory region on the BAC plasmid, beginning with PtetA (-35 and −10 consensus sequences), TSS (+1), palindromic patterns corresponding to two TetR binding sites (blue), mRFP1 start site at +104, and 96bs start site at +778. (B) PN25-tetR sequence integrated in the E. coli genome: PN25 controlled TetR protein coding gene and the lambda attP site. Terminators t0 and T1 prevent transcription from the integrated promoters into the neighbouring regions of the E. coli genome.
Induction of expression of the target gene and of the reporter gene
From the overnight culture, cells were inoculated into a fresh LB medium supplemented with antibiotics, with initial OD of 0.1 at 600 nm and incubated at a specific temperature (24°C or 37°C) to mid-logarithmic phase with 0.5 OD. To induce the production of MS2-GFP proteins, IPTG (1 mM) was added in the medium at 0.35 OD. The target mRNA from PtetA-mRFP1-96BS was then induced by adding aTc to the liquid culture. The target mRNA is rapidly tagged by the MS2-GFP proteins in the cytoplasm and can be detected as fluorescent spots soon after transcription occurs (57).
Quantitative PCR for mean mRNA quantification
The quantification of changes in the mean transcripts production rate of the target gene with induction strength and temperature, relative to a reference gene, were validated with qPCR. For the experimental samples, 10 ml of cells with 0.5 OD at 600 nm were induced with aTc (5–25 ng/ml) alone for 1 hour in liquid culture at a specific temperature (24°C or 37°C). Cells were then immediately fixed with RNAprotect bacteria reagent followed by enzymatic lysis with Tris-EDTA lysozyme buffer (pH 8.3). From the lysed cells, total RNA was isolated with RNeasy RNA purification kit. The total RNA was separated by electrophoresis through a 1% agarose gel and stained with SYBR®Safe DNA Gel Stain. The RNA was found intact with discreet bands for 16 S and 23 S ribosomal RNAs. To remove DNA contamination, RNA samples were treated with DNase I, RNase-free enzyme as per manufacturer’s instructions. The A 260 nm/280 nm ratio for the RNA samples assessed using GeneQuant pro UV/Vis Spectrophotometer (80-2114-98) were 2.0–2.1, indicating highly purified RNA, and the yield was estimated to be 0.4–0.5 µg/µl. cDNA was synthesized from 1 μg of RNA with iScript Reverse Transcription Supermix according to manufacturer’s instructions and stored at −20°C. The qPCR master mix contained iQ SYBR Green supermix with primers for the target and reference genes at a final concentration of 200 nM. We used three reference genes (16 S rRNA (42,58), 23 S rRNA (59) and dxs (60)) for internal reference, and similar patterns were observed in all cases. In the results section, we show the data relative to the 16 S rRNA reference gene, whereas in the supplement, we show the data relative to dxs (Supplementary Figure S2).
The primers for the target mRNA were (Forward: 5′ TACGAC GCCGAGGTCAAG 3′ and Reverse: 5′ TTGTGGGAGGTGATGTCCA 3′) to the region of mRFP1 (GenBank Accession Number: AF506027) (61) with amplicon length 90 bp and for the reference gene 16 S rRNA (EcoCyc Accession Number: EG30090) (Forward: 5′ CGTCAGCTCGTGTTGTGAA 3′ and Reverse: 5′ GGACCGCTGGCAACAAAG 3′), with amplicon length 74 bp (40), and primers were obtained from Thermo Scientific. The template for the reaction was 20 ng of cDNA with similar PCR efficiencies for both the target and reference genes, both greater than 95%. The thermal cycling protocol used was: 94°C for 15 s, 54°C for 30 s, and 72°C for 30 s up to 36 cycles, and in the end, one cycle of 94°C for 15 s. The fluorescence was read at the end of each cycle. These reactions were performed in three experiments, each with three replicates per condition with a final reaction volume of 50 µl. No-RT controls and no-template controls were used to crosscheck non-specific signals and contamination. The reaction was carried out in low-profile tube strips in a MiniOpticon Real time PCR system (Biorad). The Cq values generated by CFX ManagerTM Software were imported into Microsoft Excel, and the data were analysed following the Livak method (62) to obtain the fold changes in the target gene, normalized to the reference gene, and to calculate the standard error between experiments.
Fluorescent microplate reader measurement for mean protein levels
The mean fluorescence of the mRFP1 protein under the control of PtetA was measured with a Thermo Scientific* Fluoroskan Ascent Microplate Fluorometer. Cells at 0.5 OD600nm were induced with 15 ng/ml of aTc and incubated at a specific temperature (24°C or 37°C) for 1 hour with shaking. The optical density of induced and non-induced cells was measured after 1 hour. From that, 0.5 OD600nm of cells were taken, centrifuged and then re-suspended with fresh medium of 1:200 folds dilution. From this, 150 μl of cells were taken and placed on 96 well microplate and measured for relative fluorescence levels of mRFP1 protein with excitation (584 nm) and emission (607 nm) wavelengths (61). The cell density was kept identical in all wells of the plate for all measured conditions. We performed three independent experiments with three replicates for each condition.
Time-lapse microscopy
Cells were induced with IPTG and aTc as described earlier in the text. Five minutes after induction in liquid culture by aTc, cells were placed on a microscope slide between a coverslip and 1% LB-agarose gel with IPTG (1 mM) and aTc (15 ng/ml), to maintain full induction under the microscope. Cells were visualized in a Nikon Eclipse (TE2000-U, Nikon, Japan) inverted C1 confocal laser-scanning system with a 100× Apo TIRF (1.49 NA, oil) objective. The slide was kept in a temperature-controlled chamber, and the cells were focused within a few seconds under light microscope. Images were collected once per minute up to 1 hour under the fluorescence confocal microscope. Image acquisition began approximately 20 minutes after induction (including the 5 minutes induction in liquid culture). This interval is sufficient to reach a steady state level of induction (55,63). For image acquisition, we used Nikon software EZ-C1, under dark condition to minimize photolysis of aTc (photobleaching and nutrient depletion only become significant after 2 or more hours). GFP fluorescence was measured using a 488 nm argon ion laser (Melles-Griot) and a 515/30 nm detection filter. Images were acquired using medium pinhole, gain 130 and 1.68 µs pixel dwell. On the slide, the division time of the cells was approximately 40 minutes, likely because of the imaging.
We used a recent interacting multiple model filter based autofocus strategy (64). The method relies on the nature of the focal drift and exploits the interacting multiple model filter algorithm to predict the focal drift at time t based on the measurement at time t-1. It allows a drastic reduction of the number of required z-slices for focal drift correction, thus minimizing photo bleaching.
Cells and spots segmentation and the intensity jump-detection method
Detection of cells from the images is performed by a semi-automatic method (40). It consists of manually masking the regions that cells occupy during the imaging. For each image, the locations, dimensions and orientations of the cells within their masks were estimated by principal component analysis, assuming that the fluorescence inside the cell is uniformly distributed. This assumption is supported by measurements of the pixels intensities inside each cell, which were found to be fairly uniform (40). The segmentation of fluorescent spots (tagged RNAs) was performed by a kernel density estimation method (65) with a Gaussian kernel. An example of the results of the segmentation is shown in Supplementary Figure S1.
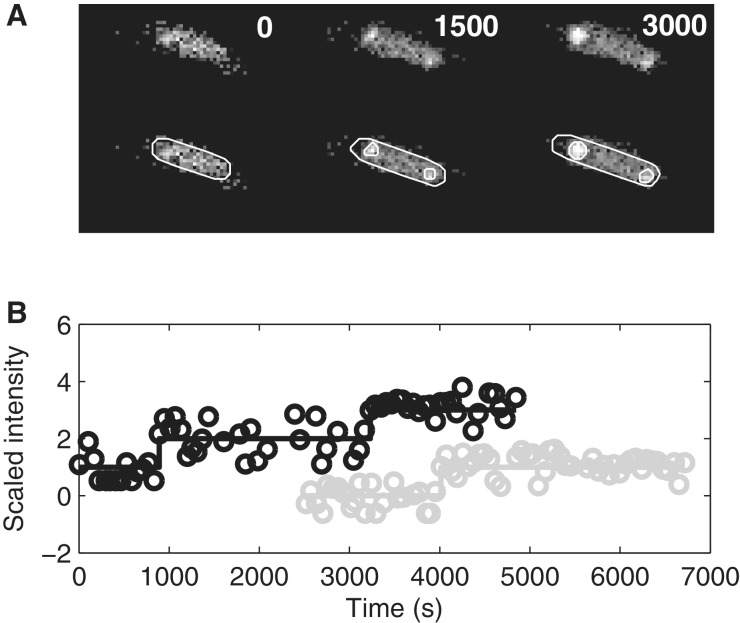
From these data, we compute the cell-background-subtracted total spot intensity time traces for each cell. Because the tagged RNAs do not degrade during the experiment (shown in the ‘Results’ section), this intensity should follow a monotonically increasing piecewise-constant function, where the jumps correspond to the appearance of novel mRNAs. This was verified by inspection (40). We fit such a function to the time trace by least squares, where the number of pieces in the function is determined by an F-test with a P value of 0.01, thus requiring higher order curves to fit significantly better to justify their use. Some intermediate results of this procedure, along with raw data, are shown in Figure 2. In Supplementary Data, we provide movies of two cells showing the time (in seconds) when each frame was obtained following induction of the target gene.
Figure 2.
Tagged RNAs in E. coli cells. (A) Unprocessed frames and segmented cells and RNA spots. The moments when images were taken are shown for each frame. (B) Examples of time series of scaled spot intensity levels from individual cells (circles) and the corresponding estimated RNA numbers (solid lines). The cell shown in (A) does not correspond to any of the traces in (B).
Inference of the number and duration of the sequential steps in transcription
From the distribution of intervals between productions of consecutive RNA molecules, we infer by maximum-likelihood the number and duration of the sequential steps in transcription initiation (40). We assume that the duration of each of the sequential steps follows an exponential distribution (40). Although the steps in initiation, such as the open complex formation, are likely not elementary (25), it was possible by this method to fit well the measured distributions in the case of Plar using a small number of steps that is consistent with the number of steps believed to be rate-limiting from in vitro studies (31,32). To support the results of the inference, we perform Kolmogorov-Smirnov tests to compare the measured distribution with the inferred one that best fits the data. This test is used to determine whether the inferred distribution does not fit the measured distribution.
The inference procedure assumes that the measured intervals between the productions of consecutive RNA molecules are not significantly affected by elongation. This relies on the fact that the mean duration of the intervals between consecutive transcription events is of the order of 600 s or higher, depending on induction strength and temperature (see Results section and in vitro studies (8)). On the other hand, elongation of the target gene was measured to take only tens of seconds (57). Possible sequence-dependent events, such as long transcriptional pauses, can also be ruled out as affecting significantly the measured distributions because, if existing, they would last only 10–100 s (e.g. 32 s half-life for the ops pause and 52 s for the his pause) (66). In addition, target RNA molecules become visible even while elongating (57), further diminishing possible effects of events in elongation in the measured distributions. In any case, although the elongation process may increase the variance of the distributions, it would not affect their means. Finally, the eventuality of possible premature terminations of transcription events can be ruled out in this case because they would generate distributions of intervals between transcription events with multiple peaks, centered on multiples of the mean interval between productions, which were not observed (see ‘Results’ section).
Here, we assume a d-steps model such that the durations of the d steps are exponentially distributed and are independent, with possibly different rates. The fit for each d-steps model is obtained by maximum likelihood estimation. The likelihoods are compared using likelihood ratio test, and the model with smallest d is selected that cannot be rejected at the significance level 0.01 in favour of a higher order model. This method was found to reliably distinguish the number of steps and the duration between any two steps when they differ by ∼25% or more in duration, from 200 intervals sampled from a stochastic model of gene expression with d exponentially distributed steps (40). Note that this method does not allow us to determine the temporal order of the sequential steps inferred. Only their number and durations can be assessed.
RESULTS
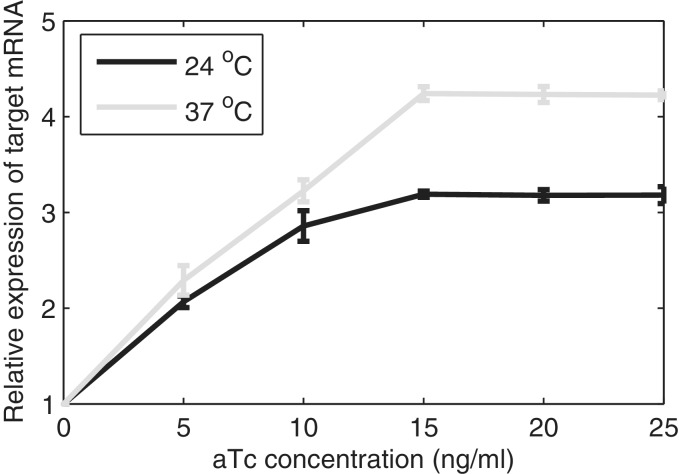
We study the in vivo kinetics of production of individual RNA molecules, as a function of inducer concentration and temperature, under the control of PtetA. First, we measured the relative mean RNA levels by qPCR as a function of induction and temperature. In Figure 3, we show the results using the 16 S rRNA gene as reference, whereas in supplement, we show the results using dxs as reference (Supplementary Figure S2), which are similar to 16 S rRNA. Previous studies suggest that the maximum induction is achieved with a concentration of aTc of 20 ng/ml or higher (8,56). The results (Figure 3 and Supplementary Figure S2) are in agreement, indicating that there is no significant increase in the rate of RNA production beyond 15 ng/ml of aTc. From these figures, we find that both the temperature and the inducer concentration significantly affect the rate of transcription under the control of PtetA.
Figure 3.
Relative expression level of target mRNA induced with different concentrations of aTc (ng/ml) at 24°C and 37°C, quantified by qPCR using the 16 S rRNA gene as reference. The standard deviation bars are from three independent experiments. In some cases, these bars’ lengths are too small to be visible.
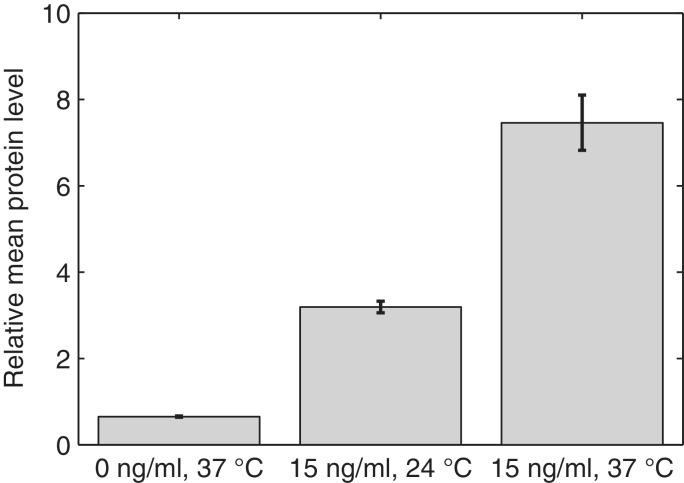
From here onwards, we focus on three conditions. Specifically, we measure gene expression in the absence of aTc at 37°C, and with 15 ng/ml aTc at both 37°C and 24°C so as to study how the kinetics changes with induction and temperature. First, we verify whether, for these conditions, the relative protein expression levels follow those of the RNA. Results of the measurements of relative fluorescence levels by microplate fluorometer are shown in Figure 4, and confirm that the protein levels follow the RNA levels. They also show similarity (same order of magnitude) to the measurements reported in (56) for this promoter using a luminescent reporter system, even though the cells in (56) were in late log-phase and had a 90 min induction period.
Figure 4.
Relative mean expression level of target proteins (mRFP1) estimated by microplate fluorometer in three conditions. The error bars are the standard deviation from the measurements in the different wells. For precise quantities, see Supplementary Table S1.
We next study the kinetics of RNA production in live, individual cells using the MS2-GFP tagging method (40,42). As described in the Methods section, the expression of the target gene is controlled by PtetA and is induced by aTc. The sequence of the target gene contains 96 binding sites for the MS2 coat protein. Because of these, the reporter proteins (MS2-GFP) can bind to the target RNA, producing a fluorescent spot that is detectable from fluorescence microscopy images.
Using this system, we first measured the cell-to-cell diversity in the number of tagged RNA molecules produced by individual cells over a certain period of time in all three conditions. In both the induced and non-induced cases, cells are placed under the confocal microscope 2 h following induction by IPTG. In the induced cases, the induction by aTc is done 1 h following the induction by IPTG.
From the images, we extracted the number of target RNA molecules in each cell, and calculated the mean and standard deviation of the number of RNA molecules in individual cells (Figure 5). We observe that the measured mean is in agreement with the measurements by qPCR (Figure 3), though the in vivo measurements have a slightly smaller relative increase with induction.
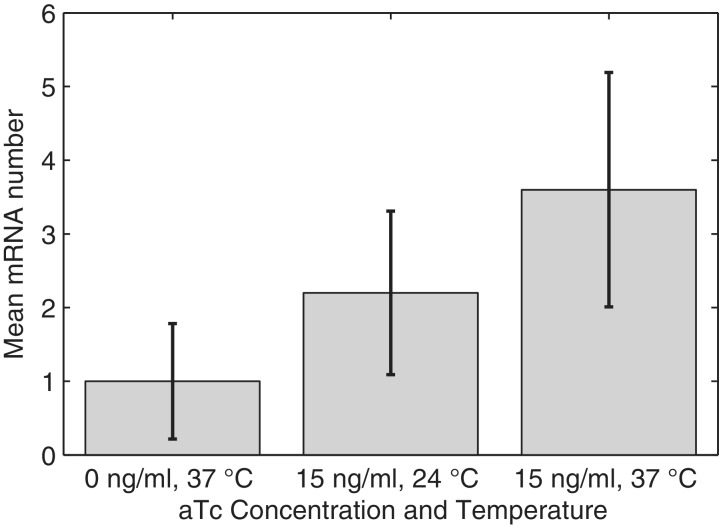
Figure 5.
Mean levels of the target mRNA relative to the non-induced condition at 37°C obtained from live cell imaging of the RNA tagged with MS2-GFP under the confocal microscope. Images taken 1 h after induction by aTc in three conditions. The error bars are the standard deviation of the number of RNA molecules in each cell. For precise quantities, see Supplementary Table S2.
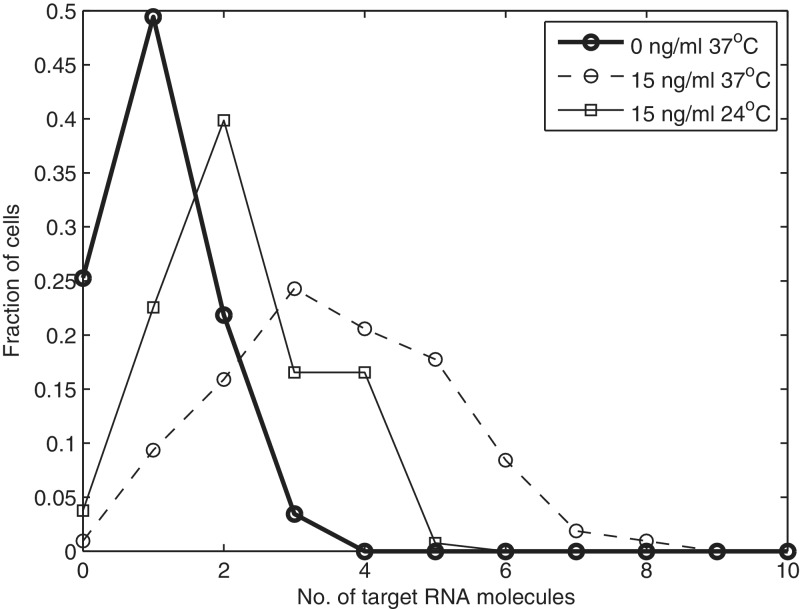
We also extracted the fraction of cells with a given number of RNA molecules (Figure 6). In the non-induced case (0 ng/ml aTc, 37°C), the variance of the distribution is 0.62, and the mean is 1.0. In the induced case, the variance is 2.5, and the mean is 3.6. Finally, for cells induced with 15 ng/ml aTc and incubated at 24°C, the variance of the distribution is 1.2, and the mean is 2.2. These values are of significance in that, in all cases, the variance is smaller than the mean (i.e. the Fano factor of the distribution is smaller than 1). If the process of RNA production was Poissonian, that is, if the intervals between consecutive productions were independent and followed an exponential distribution, the variance ought to be equal to the mean (i.e. Fano factor equal to 1). Because the tagged RNA molecules do not degrade, this can only be explained either by the dynamics of RNA production or of RNA partitioning in division (or the combination of these processes).
Figure 6.
Distribution of the fraction of cells with a given number of mRNA molecules, 1 h following induction by aTc (when induced) obtained from live cell imaging in three conditions.
To investigate this, we obtained distributions of intervals between productions of consecutive RNA molecules in individual cells in the three conditions (Figure 7). These distributions are not affected by the dynamics of RNA partitioning in division because we only count intervals between consecutive RNAs in single cells. In each case, several independent measurements were made, and the results were combined.
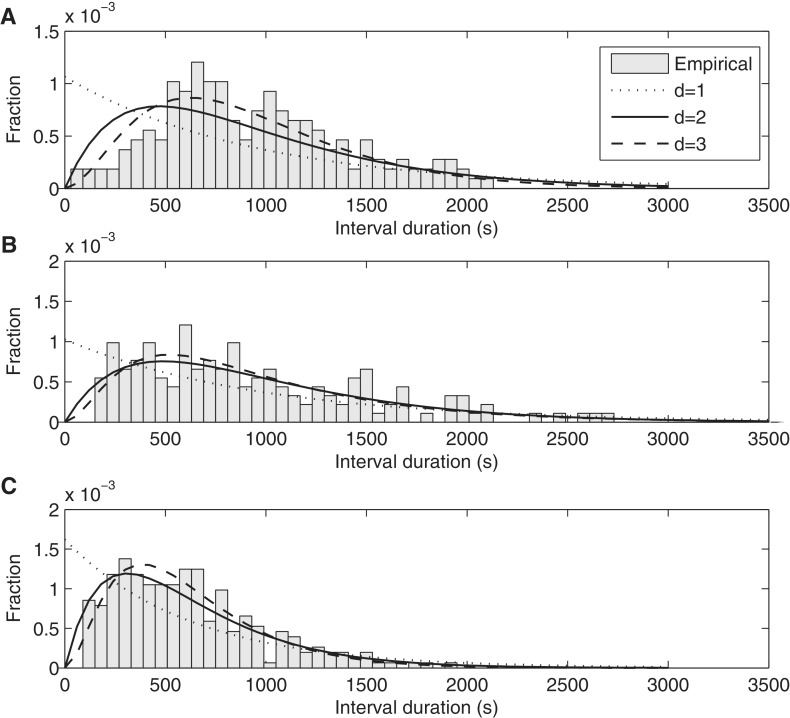
Figure 7.
Distributions of time intervals between productions of consecutive mRNA molecules in individual cells under the control of PtetA in conditions: (A) 0 ng/ml aTc at 37°C, obtained from 157 cells and 43 intervals (B) 15 ng/ml aTc at 24°C, obtained from 119 cells and 100 intervals, and (C) 15 ng/ml aTc at 37°C, obtained from 113 cells and 254 intervals. The probability density functions of inferred models of transcription initiation with differing number of rate-limiting steps are also shown in each plot. Note the different scales in the y-axis.
The number of cells observed in each condition is shown in Table 1 along with the number of intervals detected, as well as the square of the coefficient of variation (CV2, defined as the variance over the mean squared) of the intervals. It is noted that the mean duration of the intervals for the aTc 15 ng/ml (37°C) condition is in close agreement with in vitro measurements of the time it takes for a transcription initiation event to be completed once initiated (35).
Table 1.
Number of cells analysed, number of intervals between productions of consecutive RNA molecules detected in individual cells, mean interval duration, standard deviation of interval durations and CV2 of the interval durations for each condition
| Condition | aTc 0 ng/ml (37°C) | aTc 15 ng/ml (24°C) | aTc 15 ng/ml (37°C) |
|---|---|---|---|
| No. cells | 504 | 178 | 113 |
| No. intervals | 180 | 152 | 254 |
| Interval mean (s) | 939 | 974 | 617 |
| Interval std (s) | 459 | 676 | 367 |
| Interval CV2 | 0.21 | 0.48 | 0.35 |
From Figure 7, in all cases, the shapes of the distributions of intervals are not exponential-like. This implies that the process of RNA production under the control of PtetA is not Poissonian. Instead, because the standard deviations of the distributions are smaller than the means, resulting in a CV2 below 1 (Table 1), we can conclude that this process is sub-Poissonian. This explains the low values of cell-to-cell diversity in RNA numbers observed in cell populations (Figure 6).
From the comparison of the distributions A and C in Figure 7, we can assess the effects of induction in the dynamics of transcription under the control of PtetA. Note how distribution C, aside from having a smaller mean, is also slightly more exponential-like, explaining why the CV2 in RNA numbers is closer to 1 in case C than in case A (Table 1). From the comparison of the distributions B and C, we can assess the effects of lowering the temperature in the dynamics of transcription under the control of PtetA. When the temperature is reduced from 37°C (distribution C) to 24°C (distribution B), the mean interval between transcription events increases, and the shape of the distribution changes significantly. Namely, distribution B, corresponding to full induction at 24°C, is more exponential-like than distribution C.
These results rely on the intensity jump-detection method. This method assumes that the target RNA molecules are quickly bound by the MS2-GFP tagging proteins once transcribed (which was verified in (55)) and that, once that occurs, they do not degrade during the measurement. To validate this assumption of “immortality”, we studied the kinetics of degradation of these complexes. The MS2-GFP proteins used here are an assembly-defective mutant with the FG loop deleted (55,67). A recent study suggested that when using a target RNA with 48 binding sites for these MS2-GFP proteins, the resulting RNA-MS2-GFP complex may degrade during measurement sessions of a couple of hours (46). This would interfere, to some extent, with the intensity jump-detection method (see ‘Materials and Methods’ section). However, our target mRNA has 96 binding sites, and therefore the kinetics of degradation likely differs. We studied the degradation kinetics of the target RNA when bound the MS2-GFP and observed ∼120 RNA-MS2-GFP complexes produced in 50 cells for up to 2 h. We did not observe any degradation event of these complexes nor did we observe any significant loss of brightness in individual spots. We conclude that the additional MS2-GFP molecules provide sufficient stability to the complex, for us to consider them to be immortal for all practical purposes, implying that the total brightness of the spots within a single cell monotonically increases with time.
We next study the kinetics of the underlying processes responsible for shaping the interval distributions (Figure 7) and thus the observed cell-to-cell diversity in RNA numbers (Figure 6). For this, certain assumptions are necessary. First, we assume that the distributions are mainly shaped by the kinetics of transcription initiation. This relies on the following: in all distributions in Figure 7, the mean interval duration is higher than ∼500 s. Therefore, elongation is not expected to play a significant role in shaping the distributions because it lasts only tens of seconds (57). Also, as noted, the target RNA, when bound by MS2-GFP proteins, does not degrade during the measurements. Because of this, we assume that the distributions are shaped by transcription initiation, which includes steps such as the closed complex formation, isomerization and open complex formation (8,25–32,35,40,41,68).
We estimate by maximum likelihood the number and duration of the most prominent “rate-limiting” steps, that is, the ones that shape the distributions of intervals between transcription events (40). The results are shown in Figure 7 for each condition when assuming one, two and three steps. In Table 2, we show the log-likelihood values and the durations of the inferred steps for 1-step, 2-step and 3-step models, for each condition. The number of steps can be determined by using a likelihood-ratio test between pairs of models to reject a lower-degree model in favour of a higher-degree one (55). In Table 3, we show the results of the likelihood-ratio tests. For all distributions, the test rejects the 1-step (exponential) model in favour of the 2-step model (P values < 8.32 × 10−7). For distributions A and C, the 2-step model is also rejected in favour of the 3-step model (P values < 0.0019).
Table 2.
Log-likelihood and durations of the steps of the inferred models with d steps, for each condition
| aTc 0 ng/ml (37°C) |
aTc 15 ng/ml (24°C) |
aTc 15 ng/ml (37°C) |
||||
|---|---|---|---|---|---|---|
| d | Log-likelihood | Durations (s) | Log-likelihood | Durations (s) | Log-likelihood | Durations (s) |
| 1 | –1412 | 939 | –1198 | 975 | –1886 | 617 |
| 2 | –1369 | (470, 470) | –1174 | (487, 487) | –1836 | (309, 309) |
| 3 | –1356 | (313, 313, 313) | –1171 | (620, 240, 115) | –1828 | (254, 254, 109) |
There is no implied temporal order for the steps.
Table 3.
Likelihood-ratio test P values between pairs of models for each condition
| (d0, d1) | aTc 0 ng/ml (37°C) | aTc 15 ng/ml (24°C) | aTc 15 ng/ml (37°C) |
|---|---|---|---|
| (1, 2) | 0 | 3.02 × 10−12 | 0 |
| (2, 3) | 7.27 × 10−7 | 0.027 | 1.20 × 10−4 |
| (3, 4) | 0.342 | 0.268 | 0.370 |
The null model is the d0 step model (where d0 is 1, 2, or 3) while the alternative model is the d1 step model (where d1 = d0 + 1)
The time scales of the steps (for d = 2) are identical for all cases. As discussed in a previous work (40) this may be because of some unknown artefact of the inference method or be representative of the real kinetics of transcription initiation of this promoter. The method of inference was found to reliably distinguish the duration of each step when they differ by approximately 25% or more in duration, from 200 intervals sampled from a model of gene expression (40). For smaller differences, the solution can be biased toward inferring steps with identical durations, for unknown reasons. Nevertheless, given the number of intervals measured, it is possible to conclude that the steps do not differ by more than approximately 25%.
From Tables 2 and 3, provided that the assumed sequential model of transcription initiation (8,25–32,35,40,41,68) is correct, we conclude that, when not induced with aTc, transcription initiation controlled by PtetA has 3 rate-limiting steps, which are similar in duration (differing by less than 100–150 s between them). When fully induced, at 24°C, there are two dominant rate-limiting steps, similar in duration. Finally, at 37°C under full induction, there are three rate-limiting steps, two longer and similar in duration, and a (clearly) shorter third step. No significant improvement was obtained in the fit with more steps in any of the conditions.
We conclude that there are three rate-limiting steps in transcription initiation of PtetA. By lowering the temperature, two of the steps become longer in duration, whereas the third step remains unaltered and, because of its now relatively much shorter duration, it becomes barely detectable (P value of 0.0268, Table 3). Interestingly, the other two steps are not significantly affected by temperature (compare cases B and C in Table 2). Induction on the other hand affects the duration of the three steps (compare cases A and C in Table 2).
It is possible to provide additional support to these results, as well as to the assumption that each step follows an exponential distribution in duration, using a set of logical pairwise Kolmogorov-Smirnov (K–S) tests. One can compare the distributions of intervals between transcription events, for each condition, between the empirical cumulative distribution function of each case and the corresponding cumulative distribution function of the inferred models with d-steps, for all values of d. If the model accurately describes the measurements, the empirical and the inferred distributions should be indistinguishable by the K–S test. The comparisons (i.e. the P values) are shown in Table 4. Usually, for P values smaller than 0.01, it is concluded that the two distributions differ significantly.
Table 4.
P values from the Kolmogorov-Smirnov test between the empirical distribution and the various inferred models with d steps
| d | aTc 0 ng/ml (37°C) | aTc 15 ng/ml (24°C) | aTc 15 ng/ml (37°C) |
|---|---|---|---|
| 1 | 4.84 × 10−13 | 4.81 × 10−5 | 1.27 × 10−11 |
| 2 | 1.10 × 10−4 | 0.725 | 0.0485 |
| 3 | 0.0782 | 0.844 | 0.378 |
| 4 | 0.662 | 0.842 | 0.272 |
From Table 4, in case A (aTc 0 ng/ml, 37°C), the models with less than 3 steps do not accurately match the measured data. In case B (aTc 15 ng/ml, 24°C), only the 1-step model does not accurately match the data. This, as noted, is because of the increase in duration in two of the steps because of the lower temperature, rendering the effects of the third step much less significant in the overall distribution of intervals. Finally, in case C, we have the same result as in case A, that is, models with less than 3 steps do not accurately match the measured data. These results support the previous conclusions, using the likelihood ratio test, regarding the number of sequential steps that determine the shape of the distribution of intervals in each condition.
Finally, we tested whether during our in vivo measurements, the kinetics of production of RNA changed over time because of possible changes in the intracellular concentration of aTc. This could occur if degradation because of light sensitivity of intracellular aTc or its slow diffusion across the membrane were significant. That is, if either effect is significant, it should be possible to distinguish between the distributions of intervals obtained in the first 30 min and those obtained in the last 30 min of the hour-long measurements. We extracted these two sub-distributions from each induction condition and compared them with the Kolmogorov-Smirnov test. The Kolmogorov-Smirnov test was unable to differentiate the two distributions in any condition (all P values > 0.1), demonstrating that the measurements were done at an approximate steady state. In particular, for case A (aTc 0 ng/ml, 37°C), the P value was 0.32; for case B (aTc 15 ng/ml, 24°C); it was 0.16 and for case C (aTc 15 ng/ml, 37°C), it was 0.78.
DISCUSSION
Previous studies have shown that the kinetics of transcription initiation of PtetA is heavily dependent on induction and on environmental factors such as temperature (8,16,35,56). Other studies have shown that RNA production in bacteria is a stochastic (24,69) and a multi-step process (30), generally subject to complex regulatory mechanisms (8). Finally, recent studies have shown that the cell-to-cell diversity in RNA numbers, and likely protein numbers, can be significantly affected by processes other than gene expression (47,48,51). Therefore, the assessment of the kinetics of gene expression and regulation requires in vivo measurements of RNA production dynamics in individual cells, one event at a time, under various induction and environmental conditions (40).
The measured in vivo distributions of intervals between consecutive productions of RNA molecules in single cells are found to be sub-Poissonian, for the induction levels and temperatures tested. This was also observed in the case of Plar (40). The sub-Poissonian nature of the kinetics explains the low cell-to-cell diversity in RNA numbers observed at the cell population level. Relevantly, the distributions of intervals, including their mean and variance, are found to respond readily and discernibly to induction as well as temperature, revealing the plasticity of the expression mechanism of TetA. The plasticity appears to arise from the diversity of the changes in the durations of the various steps in response to differing induction levels and temperature.
Our results assume that the measurements are only affected by intrinsic noise sources in transcription. Downstream events, such as translation or RNA degradation, do not affect the results, as we detect RNA molecules as soon as these are produced and study only the time intervals between these events. It is necessary to discuss if other noise sources, aside intrinsic noise in transcription, could affect diversity in number of produced RNA molecules per cell. It may be that differences in the amounts of TetR and/or aTc in each cell could contribute to this diversity (i.e. be a significant source of extrinsic noise (70)). First, the strain used here over-expresses TetR (DH5αPRO produces constitutively around 7000 dimeric Tet repressors per cell during logarithmic growth (8)); thus, we expect the contribution of diversity in TetR numbers to the diversity in RNA numbers to be negligible. As for aTc, we found no evidence, at least at the population level, of varying rates of transcription initiation because of varying concentrations of aTc over time (e.g. because of depletion), as we found no difference in the kinetics of RNA production during the first and second half of the measurement period. Additionally, the tetA gene, responsible for expressing TetA, which confers tetracycline resistance (11) (by pumping it out of the cell) is not present in our strain. Finally, diversity in the numbers of either aTc or TetR in the cells is expected to only increase diversity, not decrease it (and thus cannot be the explanation for the observed sub-poissonian kinetics). Nevertheless, it is stressed that our measurements are not sufficient to predict the cell to-cell-diversity in number of RNA molecules under the control of PtetA as other factors, such as RNA degradation (45) and RNA partitioning in cell division (47) need to be considered.
In this regard, measurements by fluorescent in situ hybridization of RNA numbers under the control of 20 E. coli promoters (42,43) exhibited high values of cell-to-cell diversity. Another work reported Fano factors of mRNA numbers for 137 highly expressed genes in E. coli, ranging from 1 to 3, using a yellow fluorescent protein fusion library for E. coli (44). Although these results could be because of super-Poissonian transcript production from the strongly expressing promoters studied, other explanations cannot be ruled out. For example, it may be that although the production is sub-Poissonian, the observed diversity is a result of the subsequent contribution from complex RNA degradation mechanisms (45) and imperfect partitioning of RNA molecules in cell division ([47,48,51), among other things such as the biased segregation of unwanted substances (e.g. the fluorescent molecules used to tagged the RNA) (49,52). In any case, we note that our measured distributions of intervals between transcription events cannot be explained by models, such as the “on-off” model of transcription initiation (43,71), as this model entails super-Poissonian kinetics.
The kinetics of RNA production under the control of PtetA can be explained by a model of transcription initiation with successive “rate-limiting” steps, each of which is exponentially distributed in duration. From the inference of the number and duration of these steps in several conditions, we found that induction with aTc significantly changes the RNA production kinetics by reducing the duration of all rate-limiting steps, to various degrees. In particular, one of the steps becomes almost indiscernible. Meanwhile, lowering temperature under full induction by aTc increases the duration of two of the steps, but not of the third step, causing the kinetics to be well-fit by a two-step model under these conditions. Note that it is not possible to determine which steps [e.g. closed complex, open complex or isomerization (28)] are affected by aTc and temperature. Novel experimental techniques are necessary to perform this study in vivo. However, we can rule out TetR dissociation from the promoter as one of the rate-limiting steps. The complex TetR-promoter has a half-life of 12 s (37), which is a much faster process than the measured rate-limiting steps. Additionally, although TetR, when bound by aTc, may retain some ability to bind to the DNA [although this ability is reduced by approximately 9 orders of magnitude (72)], there is no evidence that this complex would have a longer half-life than when the DNA is bound by TetR alone.
A recent work used the methods used here to analyse the kinetics of Plar (40). The measurements were made at 24°C. It is of interest to compare them with our measurements regarding the response to induction. First, the kinetics of RNA production of Plar is also sub-Poissonian. Also, induction of Plar with IPTG and arabinose reduces the duration of the rate-limiting steps. The main differences between these two promoters are in the mean duration of the intervals at 24°C ([for Plar, this mean duration is ∼1500 s (40), whereas for PtetA, it is ∼1000 s]) and in the variability of the intervals. For Plar, the CV2 of the durations of these intervals is 0.70, whereas for PtetA, it is 0.52. From this, we conclude that the kinetics of transcription initiation of PtetA is less noisy than that of Plar. It is worthwhile to mention that the observations of the behaviour of PtetA (a native promoter) suggest that the sub-Poissonian kinetics of RNA production is not, for example, an artefact of the synthetic nature of Plar, and that it may be a common feature of the dynamics of transcript production in E. coli. Also, the results support several previous observations on the effect of temperature on the kinetics of transcription, but further show that the changes in kinetics are due, in part, to the alteration of the mean duration of the intermediate steps in transcription initiation.
PtetA controls the expression of TetA, which is responsible for the active efflux of tetracycline-Mg2+ complexes. This protein’s function justifies the need for such a stringent regulatory mechanism so as to ensure that TetA is present in the appropriate amount because both tetracycline and TetA (in high amounts) are harmful to the cell (12). We find that this control is achieved not only by the negative feedback mechanism of the tet operon (12) but also by a sub-Poissonian kinetics of transcription initiation. Relevantly, although robust (less noisy than a Poisson process), this system is nevertheless sensitive to external stimuli, such as tetracycline, and temperature because its behaviour discernibly changes with temperature and inducer concentration.
In the future, it will be of interest to further analyse how the dynamics of PtetA differs in other environmental conditions, such as in differing concentrations of hydrogen ions and metabolites. Such studies may provide insights on the plasticity of the kinetics of gene expression in bacteria and thus guide the engineering of synthetic genetic circuits with specific behavioural patterns.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1 and 2 and Supplementary Movies 1 and 2.
FUNDING
Academy of Finland [126803 to A.S.R.]; Finnish Funding Agency for Technology and Innovation (TEKES) [40284/08 to O.Y.-H.]; TUT President’s Doctoral Programme. Funding for open access charge: TEKES [40284/08 to O.Y.-H.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank F.G. Biddle for useful advice. A.-B. M. executed the qPCR measurements.
REFERENCES
- 1.McNaught AD, Wilkinson A. 2nd edn. Oxford: Blackwell Scientific Publications; 1997. IUPAC. Compendium of Chemical Terminology. (the “Gold Book”) [Google Scholar]
- 2.Duggar BM. Aureomycin: a product of the continuing search for new antibiotics. Ann. N. Y. Acad. Sci. 1948;51:177–181. doi: 10.1111/j.1749-6632.1948.tb27262.x. [DOI] [PubMed] [Google Scholar]
- 3.Connel SR, Tracz DM, Nierhaus KH, Taylor DE. Ribosomal protection proteins and their mechanism of tetracycline resistance. Antimicrob. Agents Chemother. 2003;47:3675–3681. doi: 10.1128/AAC.47.12.3675-3681.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schnappinger D, Hillen W. Tetracyclines: antibiotic action, uptake, and resistance mechanisms. Arch. Microbiol. 1996;165:359–369. doi: 10.1007/s002030050339. [DOI] [PubMed] [Google Scholar]
- 5.Hillen W, Scholimeier K, Gatz C. Control of expression of the Tn10-encoded tetracycline resistance operon II. Interaction of RNA polymerase and TET repressor with the tet operon regulatory region. J. Mol. Biol. 1984;172:185–201. doi: 10.1016/s0022-2836(84)80037-6. [DOI] [PubMed] [Google Scholar]
- 6.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nguyen TMN, Phan QG, Duong LP, Bertrand KP, Lenski RE. Effects of carriage and expression of the Tn10 tetracycline-operon on fitness of E. coli cells. Mol. Biol. Evol. 1989;6:213–225. doi: 10.1093/oxfordjournals.molbev.a040545. [DOI] [PubMed] [Google Scholar]
- 8.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lazarevic V, Margot P, Soldo B, Karamata D. Sequencing and analysis of the Bacillus subtilis lytRABC divergon: a regulatory unit encompassing the structural genes of the N-acetylmuramoyl-L-alanine amidase and its modifier. J. Gen. Microbiol. 1992;138:1949–1961. doi: 10.1099/00221287-138-9-1949. [DOI] [PubMed] [Google Scholar]
- 10.Collier GB, Mattson TL, Connaughton JF, Chirikjian JG. A novel Tn10 tetracycline regulon system controlling expression of the bacteriophage T3 gene encoding S-adenosyl-L-methionine hydrolase. Gene. 1994;148:75–80. doi: 10.1016/0378-1119(94)90236-4. [DOI] [PubMed] [Google Scholar]
- 11.Beck CF, Mutzel R, Barbe J, Müller W. A multifunctional gene (tetR) controls Tn10-encoded tetracycline resistance. J. Bacteriol. 1982;150:63–642. doi: 10.1128/jb.150.2.633-642.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hillen W, Berens C. Mechanisms underlying expression of Tnl0 encoded tetracycline resistance. Annu. Rev. Microbial. 1994;48:345–369. doi: 10.1146/annurev.mi.48.100194.002021. [DOI] [PubMed] [Google Scholar]
- 13.Orth P, Schnappinger D, Hillen W, Saenger W, Hinrichs W. Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system. Nat. Struct. Biol. 2000;7:215–219. doi: 10.1038/73324. [DOI] [PubMed] [Google Scholar]
- 14.Biliouris K, Daoutidis P, Kaznessis YN. Stochastic simulations of the tetracycline operon. BMC Syst. Biol. 2011;5:9. doi: 10.1186/1752-0509-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skerra A. Use of the tetracycline promoter for the tightly regulated production of a murine antibody fragment in Escherichia coli. Gene. 1994;151:131–135. doi: 10.1016/0378-1119(94)90643-2. [DOI] [PubMed] [Google Scholar]
- 16.Bertrand KP, Postle K, Wray LV, Jr, Reznikoff WS. Construction of a single-copy promoter vector and its use in analysis of regulation of the transposon Tn10 tetracycline resistance determinant. J. Bacteriol. 1984;158:910–919. doi: 10.1128/jb.158.3.910-919.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 18.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 19.Becskei A, Serrano L. Engineering stability in gene networks by autoregulation. Nature. 2000;405:590–593. doi: 10.1038/35014651. [DOI] [PubMed] [Google Scholar]
- 20.Becskei A, Séraphin B, Serrano L. Positive feedback in eukaryotic gene networks: cell differentiation by graded to binary response conversion. EMBO J. 2001;20:2528–2535. doi: 10.1093/emboj/20.10.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deans TL, Cantor CR, Collins JJ. A tunable genetic switch based on RNAi and repressor proteins for regulating gene expression in mammalian cells. Cell. 2007;130:363–372. doi: 10.1016/j.cell.2007.05.045. [DOI] [PubMed] [Google Scholar]
- 22.Nevozhay D, Adams RM, Murphy KF, Josic K, Balázsi G. Negative autoregulation linearizes the dose-response and suppresses the heterogeneity of gene expression. Proc. Natl. Acad. Sci. U.S.A. 2009;106:5123–5128. doi: 10.1073/pnas.0809901106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nandagopal N, Elowitz MB. Synthetic biology: integrated gene circuits. Science. 2011;333:1244–1248. doi: 10.1126/science.1207084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaern M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: from theories to phenotypes. Nat. Rev. Genet. 2005;6:451–464. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- 25.McClure WR. Mechanism and control of transcription initiation in prokaryotes. Annu. Rev. Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- 26.Uptain S, Kane M, Chamberlin M. Basic mechanisms of transcript elongation and its regulation. Annu Rev Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 27.Browning DF, Busby SJ. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2004;2:57–65. doi: 10.1038/nrmicro787. [DOI] [PubMed] [Google Scholar]
- 28.deHaseth PL, Zupancic ML, Record MT. RNA polymerase-promoter interactions: the comings and goings of RNA polymerase. J. Bacteriol. 1998;180:3019–3025. doi: 10.1128/jb.180.12.3019-3025.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nierman WC, Chamberlin MJ. Studies of RNA chain initiation by Escherichia coli RNA polymerase bound to T7 DNA. Direct analysis of the kinetics and extent of RNA chain initiation at T7 promoter A1. J. Biol. Chem. 1979;254:7921–7926. [PubMed] [Google Scholar]
- 30.McClure WR. Rate-limiting steps in RNA chain initiation. Proc. Natl. Acad. Sci. U.S.A. 1980;77:5634–5638. doi: 10.1073/pnas.77.10.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lutz R, Lozinski T, Ellinger T, Bujard H. Dissecting the functional program of Escherichia coli promoters: the combined mode of action of Lac repressor and AraC activator. Nucleic Acids Res. 2001;29:3873–3881. doi: 10.1093/nar/29.18.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buc H, McClure WR. Kinetics of open complex formation between Escherichia coli RNA polymerase and the lac UV5 promoter. Evidence for a sequential mechanism involving three steps. Biochemistry. 1985;24:2712–2723. doi: 10.1021/bi00332a018. [DOI] [PubMed] [Google Scholar]
- 33.Kandhavelu M, Lihavainen E, Muthukrishnan AB, Yli-Harja O, Ribeiro AS. Effects of Mg2+ on in vivo transcriptional dynamics of the lar Promoter. Biosystems. 2012;107:129–134. doi: 10.1016/j.biosystems.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 34.Smolander OP, Kandhavelu M, Mannerström H, Lihavainen E, Kalaichelvan S, Healy S, Yli-Harja O, Karp M, Ribeiro AS. Cell-to-cell diversity in protein levels of a gene driven by a tetracycline inducible promoter. BMC Mol. Biol. 2011;12:21. doi: 10.1186/1471-2199-12-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bertrand-Burggraf E, Lefèvre JF, Daune M. A new experimental approach for studying the association between RNA polymerase and the tet promoter of pBR322. Nucleic Acids Res. 1984;12:1697–1706. doi: 10.1093/nar/12.3.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Degenkolb J, Takahashi M, Ellested GA, Hillen W. Structural requirements of tetracycline-Tet repressor interaction: determination of equilibrium binding constants for tetracycline analogs with the Tet repressor. Antimicrob. Agents Chemother. 1991;35:1591–1595. doi: 10.1128/aac.35.8.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murphy KF, Adams RM, Wang X, Balázsi G, Collins JJ. Tuning and controlling gene expression noise in synthetic gene networks. Nucleic Acids Res. 2010;38:2712–2726. doi: 10.1093/nar/gkq091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schubert P, Pfleiderer K, Hillen W. Tet repressor residues indirectly recognizing anhydrotetracycline. Eur. J. Biochem. 2004;271:2144–2152. doi: 10.1111/j.1432-1033.2004.04130.x. [DOI] [PubMed] [Google Scholar]
- 39.Oliva B, Gordon G, McNicholas P, Ellestad G, Chopra I. Evidence that tetracycline analogs whose primary target is not the bacterial ribosome cause lysis of Escherichia coli. Antimicrob. Agents Chemother. 1992;36:913–919. doi: 10.1128/aac.36.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kandhavelu M, Mannerström H, Gupta A, Häkkinen A, Lloyd-Price J, Yli-Harja O, Ribeiro AS. In vivo kinetics of transcription initiation of the lar promoter in Escherichia coli. Evidence for a sequential mechanism with two rate limiting steps. BMC Sys. Biol. 2011;5:149. doi: 10.1186/1752-0509-5-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saecker RM, Record MT, deHaseth PL. Mechanism of bacterial transcription initiation: RNA polymerase - promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J. Mol. Biol. 2011;412:754–771. doi: 10.1016/j.jmb.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Golding I, Paulsson J, Zawilski SM, Cox EC. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123:1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 43.So LH, Ghosh A, Zong C, Sepúlveda LA, Segev R, Golding I. General properties of transcriptional time series in Escherichia coli. Nat. Genet. 2011;43:554–560. doi: 10.1038/ng.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yarchuk O, Jacques N, Guillerez J, Dreyfus M. Interdependence of translation, transcription and mrna. J. Mol. Biol. 1992;5:581–596. doi: 10.1016/0022-2836(92)90617-s. [DOI] [PubMed] [Google Scholar]
- 46.Llopis PM, Jackson AF, Sliusarenko O, Surovtsev I, Heinritz J, Emonetand T, Jacobs-Wagner C. Spatial organization of the flow of genetic information in bacteria. Nature. 2010;466:77–81. doi: 10.1038/nature09152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huh D, Paulsson J. Non-genetic heterogeneity from stochastic partitioning at cell division. Nat. Genet. 2010;43:95–100. doi: 10.1038/ng.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huh D, Paulsson J. Random partitioning of molecules at cell division. Proc. Natl Acad. Sci. USA. 2011;108:15004–15009. doi: 10.1073/pnas.1013171108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lindner AB, Madden R, Demarez A, Stewart EJ, Taddei F. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc. Natl Acad. Sci. USA. 2008;105:3076–3081. doi: 10.1073/pnas.0708931105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stewart E, Madden R, Paul G, Taddei F. Aging and death in an organism that reproduces by morphologically symmetric division. PLoS Biol. 2005;3:295–300. doi: 10.1371/journal.pbio.0030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lloyd-Price J, Lehtivaara M, Kandhavelu M, Chowdhury S, Muthukrishnan AB, Yli-Harja O, Ribeiro AS. Probabilistic RNA partitioning generates transient increases in the normalized variance of RNA numbers in synchronized populations of Escherichia coli. Mol. Biosyst. 2011;8:565–571. doi: 10.1039/c1mb05100h. [DOI] [PubMed] [Google Scholar]
- 52.Lloyd-Price J, Häkkinen A, Kandhavelu M, Marques IJ, Chowdhury S, Lihavainen E, Yli-Harja O, Ribeiro AS. Asymmetric disposal of individual protein aggregates in Escherichia coli, one aggregate at a time. J. Bact. 2012;194:1747–1752. doi: 10.1128/JB.06500-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu J, Xiao J, Ren X, Lao K, Xie XS. Probing gene expression in live cells. Science. 2006;311:1600–1603. doi: 10.1126/science.1119623. [DOI] [PubMed] [Google Scholar]
- 54.Xie XS, Choi PJ, Li GW, Lee NK, Lia G. Single-molecule approach to molecular biology in living bacterial cells. Annu. Rev. Biophys. 2008;37:417–444. doi: 10.1146/annurev.biophys.37.092607.174640. [DOI] [PubMed] [Google Scholar]
- 55.Le TT, Harlepp S, Guet CC, Dittmar K, Emonet T, Pan T, Cluzel P. Real-time RNA profiling within a single bacterium. Proc. Natl Acad. Sci. USA. 2005;102:9160–9164. doi: 10.1073/pnas.0503311102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Korpela M, Kurittu JS, Karvinen JT, Karp MT. A recombinant Escherichia coli sensor strain for the detection of tetracyclines. Anal. Chem. 1998;70:4457–4462. doi: 10.1021/ac980740e. [DOI] [PubMed] [Google Scholar]
- 57.Golding I, Cox E. RNA dynamics in live Escherichia coli cells. Proc. Natl Acad. Sci. USA. 2004;101:11310–11315. doi: 10.1073/pnas.0404443101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nevo-Dinur K, Nussbaum-Shochat A, Ben-Yehuda S, Amster-Choder O. Translation-independent localization of mRNA in Escherichia coli. Science. 2011;25:1081–1084. doi: 10.1126/science.1195691. [DOI] [PubMed] [Google Scholar]
- 59.Chini V, Foka A, Dimitracopoulos G, Spiliopoulou I. Absolute and relative real-time PCR in the quantification of tst gene expression among methicillin-resistant Staphylococcus aureus: evaluation by two mathematical models. Lett. Appl. Microbiol. 2007;45:479–484. doi: 10.1111/j.1472-765X.2007.02208.x. [DOI] [PubMed] [Google Scholar]
- 60.Lee C, Kim J, Shin SG, Hwang S. Absolute and relative QPCR quantification of plasmid copy number in Escherichia coli. J. Biotec. 2006;123:273–280. doi: 10.1016/j.jbiotec.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 61.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. A monomeric red fluorescent protein. Proc. Natl Acad. Sci. USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 63.Shales SW, Chopra I, Ball PR. Evidence for more than one mechanism of plasmid-determined tetracycline resistance in Escherichia coli. J. Gen. Microbiol. 1980;121:221–229. doi: 10.1099/00221287-121-1-221. [DOI] [PubMed] [Google Scholar]
- 64.Chowdhury S, Kandhavelu M, Yli-Harja O, Ribeiro AS. An interacting multiple model filter based autofocus strategy for confocal time-lapse microscopy. J. Microscopy. 2011;245:265–275. doi: 10.1111/j.1365-2818.2011.03568.x. [DOI] [PubMed] [Google Scholar]
- 65.Chen TB, Lu H, Lee YS, Lan HJ. Segmentation of cDNA microarray images by kernel density estimation. J. Biomed. Info. 2008;41:1021–1027. doi: 10.1016/j.jbi.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 66.Toulokhonov I, Landick R. The flap domain is required for pause RNA hairpin inhibition of catalysis by RNA polymerase and can modulate intrinsic termination. Mol. Cell. 2006;12:1125–1136. doi: 10.1016/s1097-2765(03)00439-8. [DOI] [PubMed] [Google Scholar]
- 67.Peabody DS. The RNA binding site of bacteriophage MS2 coat protein. EMBO J. 1993;12:595–600. doi: 10.1002/j.1460-2075.1993.tb05691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.China A, Tare P, Nagaraja V. Comparison of promoter-specific events during transcription initiation in mycobacteria. Microbiology. 2010;156:1942–1952. doi: 10.1099/mic.0.038620-0. [DOI] [PubMed] [Google Scholar]
- 69.Arkin A, Ross J, McAdams HM. Stochastic kinetic analysis of developmental pathway bifurcation in phage λ-infected Escherichia coli cells. Genetics. 1998;149:1633–1648. doi: 10.1093/genetics/149.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 71.Peccoud J, Ycart B. Markovian modelling of gene product synthesis. Theor. Popul. Biol. 1995;48:222–234. [Google Scholar]
- 72.Lederer T, Takahashi M, Hillen W. Thermodynamic analysis of tetracycline-mediated induction of Tet repressor by a quantitative methylation protection assay. Anal. Biochem. 1995;232:190–196. doi: 10.1006/abio.1995.0006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.