Abstract
The RecQL4 helicase is involved in the maintenance of genome integrity and DNA replication. Mutations in the human RecQL4 gene cause the Rothmund–Thomson, RAPADILINO and Baller–Gerold syndromes. Mouse models and experiments in human and Xenopus have proven the N-terminal part of RecQL4 to be vital for cell growth. We have identified the first 54 amino acids of RecQL4 (RecQL4_N54) as the minimum interaction region with human TopBP1. The solution structure of RecQL4_N54 was determined by heteronuclear liquid–state nuclear magnetic resonance (NMR) spectroscopy (PDB 2KMU; backbone root-mean-square deviation 0.73 Å). Despite low-sequence homology, the well-defined structure carries an overall helical fold similar to homeodomain DNA-binding proteins but lacks their archetypical, minor groove-binding N-terminal extension. Sequence comparison indicates that this N-terminal homeodomain-like fold is a common hallmark of metazoan RecQL4 and yeast Sld2 DNA replication initiation factors. RecQL4_N54 binds DNA without noticeable sequence specificity yet with apparent preference for branched over double-stranded (ds) or single-stranded (ss) DNA. NMR chemical shift perturbation observed upon titration with Y-shaped, ssDNA and dsDNA shows a major contribution of helix α3 to DNA binding, and additional arginine side chain interactions for the ss and Y-shaped DNA.
INTRODUCTION
RecQ-like DNA helicases form a ubiquitous protein family that plays a pivotal role in the maintenance of genome stability from bacteria to man (1). Five RecQ-like DNA helicases have been identified in the human cell, mutations of three of which are associated with autosomal, recessive human disorders that display various symptoms of genomic instability and premature ageing. Mutations in the RecQL2/BLM gene cause the Bloom’s syndrome, characterized by genomic instability and an increased risk of lymphomas and certain other types of cancer. Mutations in the RecQL3/WRN gene are implicated in the Werner syndrome, and individuals with this disease show symptoms of premature ageing including a high incidence of cancer. Mutations in the RecQL4 gene are associated with the Rothmund–Thomson (RTS), RAPADILINO and Baller–Gerold syndromes (1,2). RTS patients exhibit a heterogeneous clinical profile that includes physical and mental abnormalities, some features of premature ageing and an increased incidence of cancer, in particular osteosarcomas (2). RAPADILINO and Baller–Gerold patients also display developmental defects but less frequently cancer (3).
Numerous studies have indicated a role for Bloom protein (BLM), Werner Protein (WRN) and RecQL4 helicase at the crossroads of DNA replication, recombination and DNA damage response (1). The human RecQL4 gene contains 21 exons coding for a protein of 1208 amino acids (a.a.) (4). The corresponding protein lacks the RecQ carboxy-terminal and the helicase and RNaseD C-terminal domains present in other human RecQ helicases. Although initial studies suggested that RecQL4 lacks helicase activity, recent findings have unequivocally demonstrated a helicase activity for RecQL4 that is apparently masked by the strong annealing activity of the enzyme (5–8). Xu and Liu (6) proposed that the 400 amino-terminal a.a. may harbour a second helicase activity despite the absence of known nucleotide binding and helicase motifs.
Mouse models demonstrate that disruption of the RECQ4 gene at exons 5–8, encoding the amino-terminal region, results in early embryonic lethality (9). Conversely, mice with a disruption of the gene further downstream, leading to the deletion of the helicase domain are viable (10,11). This suggests that the amino-terminal region of RecQL4 executes an essential function in the cell. However, although the helicase domain is not required for viability, its loss causes a severe phenotype reminiscent of RTS (11).
The amino-terminus of RecQL4 possesses limited homology to the essential yeast replication factor Sld2 (12,13). In yeast, the binding of Sld2 to Dpb11 constitutes a critical regulatory step during initiation of DNA replication, and Xenopus laevis RecQL4 (XRecQL4) has also been reported to bind to XCut5, the frog orthologue of Dpb11 (13,14). The amino-terminal region of RecQL4 has also been reported to mediate interactions with several replication initiation factors (13,15). Importantly, it has been demonstrated that the corresponding region of the XRecQL4 is actively involved in replication initiation. Here, it is required for the loading of replicative DNA polymerases during the initiation of DNA replication (12,13).
Approximately 55 amino-terminal a.a. of human RecQL4 display the highest sequence homology to the N-terminus of Sld2 (12,13), and this region is conserved in metazoans. This led us to speculate that this part of the protein constitutes an autonomous subdomain of the RecQL4 N-terminal region. To address this assumption, we assessed binding of the human Dbp11 homologue TopBP1 and of DNA to the 54 residues of N-terminal RecQL4 (RecQL4_N54). In addition, we determined its solution structure via nuclear magnetic resonance (NMR) spectroscopy. In essence, RecQL4_N54 represents the minimal region for the interaction with TopBP1, binds Y-shaped, single-stranded (ss) and double-stranded (ds) DNA with no apparent sequence specificity and adopts a fold highly similar to homeodomains.
MATERIALS AND METHODS
Cloning, expression and purification of RecQL4_N54
A DNA insert corresponding to a.a. 1–54 of human RecQL4 was amplified by conventional polymerase chain reaction (PCR) with primers 5′-AACGGATCCATGGAGCGGCTGCGGGA-3′ and 5′-GACGAATTCTCACTGGCCCGTGGTACGCTTCA-3′, and a plasmid harbouring human RecQL4 cDNA as the template. The PCR product was inserted into the BamHI and EcoRI sites of vector pGEX1λT (GE Healthcare). The resulting vector pGEX-RecQL4_N54 encodes a glutathione S-transferase (GST) affinity purification tag followed by a thrombin digestion site and the 54 amino-terminal residues of human RecQL4 (Swiss-Prot entry O94761). Escherichia coli (BL21[DE3]) cells carrying the plasmid described above were grown at 37°C in 2 l Luria Bertani (LB) medium supplemented with 100 μg/ml of ampicillin for 3 h, after which the bacteria were collected by centrifugation. The cells were resuspended into 2 l of M9 labelling medium containing 4.0 g/l of 13C d-glucose and 1.0 g/l of 15N-NH4Cl (labelled compounds were purchased from EURISOTOP at 99% grade). The temperature was decreased to 20°C, and protein expression was induced by adding 1 mM isopropyl-β-d-thiogalactopyranoside. Cells were harvested 16 h after induction by centrifugation (5000g, 4°C, 10 min), washed with phosphate-buffered saline (PBS) and stored at −80°C after snap freezing. After thawing, bacterial cells were resuspended in 80 ml PBS, lysed by addition of 0.2 mg/ml lysozyme for 30 min on ice followed by 40 × 10 s pulses of ultrasound at 30% amplitude interrupted by 45 s incubations on ice utilizing a Branson W-450D Digital Sonifier. Triton X-100 was added to 3%, and after incubation at 4°C for 30 min followed by centrifugation at 30 000g at 4°C for 30 min, the supernatant was added to 0.7 ml/l culture glutathione Sepharose (GE Healthcare), and GST fusion proteins were allowed to bind for 30 min in the cold. The Sepharose was then washed with 3 × 5 ml cold PBS followed by release of RecQL4_N54 via digestion with 48 units of thrombin in 900 µl PBS for 16 h at 4°C. The supernatant of the thrombin digestion and 3 × 3 ml PBS washes were combined and incubated at 4°C for 30 min with 900 µl benzamidine Sepharose (GE Healthcare) to remove the thrombin and after separation of the Sepharose, applied to a 1 ml MonoS column (GE Healthcare) equilibrated with buffer A (20 mM potassium phosphate, pH 7.4, 1 mM EDTA and 1 mM DTT) using the ÄKTA purification system (GE Healthcare). RecQL4_N54 eluted from the column at ∼400 mM NaCl in a linear gradient of 0–2 M NaCl in buffer A. The fractions containing RecQL4_N54 were dialysed against 10 mM potassium phosphate. The protein was stable for several weeks at 4°C. For long-term storage, RecQL4 was snap frozen followed by lyophilization and storage at −20°C. Unlabelled protein samples were prepared as above, except that M9 medium was substituted with standard LB medium supplemented with ampicillin. As estimated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE), the purity of the protein was typically >90% with no prominent contaminants. Absorbance at 280 nm of guanidine hydrochloride denatured protein (εM = 8480/M/cm), and the Bradford assay was used for determining and adjusting protein concentration.
NMR spectroscopy
Samples were exchanged into in NMR buffer (20 mM d-Tris/HCl, pH 7.4, 100 mM NaCl, 10% D2O) using NAP25 columns and concentrated to 1.2 mM [U-15N]-RecQL4_N54 and 1.0 mM [U-13C,15N]-RecQL4_N54, respectively. Measurements in D2O were performed on lyophilized samples re-dissolved in D2O. NMR spectra were acquired at 20°C on Bruker Avance III spectrometers operated at 600 MHz with a cryoprobe and at 750 MHz. Data were processed with Topspin (Bruker Biospin) or NMRPipe (16) and analyzed with XEASY (17). The assignment of the 1H, 13C and 15N resonances was performed as described previously (18,19). Nuclear overhauser effect (NOE) constraints were taken from 3D 1H-1H-13C NOESY-heteronuclear single quantum coherence (HSQC) and 3D 1H-1H-15N NOESY-HSQC spectra (120 ms mixing time). For initial NOE peak picking, the program UNIO (20) was used. Upper limit distance constraints were classified according to their intensity in the NOESY spectra corresponding to distance limits of 2.8, 3.4, 4.6 and 5.6 Å. NOE intensities corresponding to fixed Hβ2-Hβ3, and aromatic ring distances were used for calibration.
Structure determination
The experimental distances of 1251 derived from NOE cross-peaks were used as upper limit constraints in CYANA (21). 52 JHNHα coupling constants extracted from a 3D HNHA experiment (22) and the NOE-derived distance constraints were subjected to a local conformational analysis using the FOUND module (23) and yielded 310 torsion angle restraints constraining 146 angles for the structure calculation. For secondary structure elements forming consistently in initial calculations, additionally 60 upper and lower limit H-bond constraints were introduced. The 20% of structures with the lowest CYANA target functions were subjected to energy minimization with OPAL (24). Figures were produced using MOLMOL (25). The quality of the structures was assessed by PROCHECK (26). The atomic coordinates and resonance assignments have been deposited in the Protein Data Bank (PDB code: 2KMU) and BioMagResBank (BMRB code: 16544), respectively.
DNA purification
The complementary 24-mer DNA oligonucleotides, LB2 and LB2-24R (Supplementary Table S2), were purchased from Eurofins and Purimex, dissolved in 10 mM Tris/HCl pH 8.0 and separately purified by high-performance liquid chromatography on a SP125/10 NUCLEOGEN DEAE 60–7 column (Macherey–Nagel) using a linear gradient of buffer A (20 mM sodium acetate, 20% acetonitrile, pH 5.5) to buffer B (20 mM sodium acetate, 20% acetonitrile, 1.5 M KCl, pH 5.5) within 60 min. The eluted oligonucleotides were precipitated by ethanol, redissolved in annealing buffer (10 mM Tris/HCl pH 7.5, 50 mM NaCl and 1 mM EDTA), heated up to 95°C for 5 min, cooled down to room temperature within 90 min using a PCR cycler and again precipitated by ethanol. After dialysis against water at 4°C overnight, the DNA was lyophilized and dissolved in an appropriate volume of NMR buffer.
DNA-binding analysis
DNA fragments derived from defined human origins of DNA replication were amplified from HeLa cell genomic DNA with the respective primer pairs as indicated in Supplementary Table S1. For electrophoretic mobility shift assay (EMSA) experiments, the fragments were re-amplified by preparative PCR followed by phenol/chloroform extraction. DNA fragments of various sizes were amplified as indicated above employing the primers detailed in Supplementary Table S2. LB2-24 dsDNA and LB2-24Y were prepared by hybridization of oligonucleotide LB24-F with LB2-24R and LB2-24RY, respectively. Binding of the indicated amounts of protein and DNA was performed for 10 min in binding buffer (20 mM HEPES–KOH, pH 7.5, 0.5 mM EDTA, 0.05% NP-40, 10% glycerol and 60 µg/ml BSA) on ice. The samples were resolved in a 15% native acrylamide gel in 0.5 × TBE buffer in the cold, and gels were stained with SYBR green EMSA gel stain (Invitrogen) and scanned with a Storm 860 Molecular Imager (GE Healthcare). For the detection of ssDNA, 5′-fluorescein-conjugated LB2-F was used.
DNA fragments for DNA bead-binding assays were prepared by preparative PCR using 5′-biotinylated primer bio-LB2 instead of LB2. The short fragment LB2_1-24 was prepared by hybridization of primers bio-LB2 and LB2-R24 as described. Two micrograms of RecQL4_N54 and 200 ng biotinylated DNA in 25 µl binding buffer were preincubated for 30 min on ice. Twenty-five microlitres of μMACS™ Streptavidin MicroBeads (Miltenyi Biotec) were immobilized in the magnetic column, equilibrated with 100 µl equilibration buffer for protein applications provided by the supplier followed by equilibration with 100 µl binding buffer. Then, the DNA-RecQL4_N54 binding reactions were passed through the column. The column was washed four times with 50 µl binding buffer, the column was removed from the magnet and the magnetic beads were recovered by rinsing three times with 50 µl binding buffer. Samples were supplemented with SDS loading buffer, denatured and resolved on a 15% SDS–PAGE followed by staining with SYPRO ruby protein gel stain (Invitrogen).
Yeast two-hybrid assay
DNA inserts corresponding to a.a. 1–675, 1–388, 1–115 and 1–54 of human RECQL4 were amplified by PCR using primers , forward: 5′-GCGAATTCATGGAGCGGCTGCGGGACGTG-3′ and corresponding reversed primers: (1–675) 5′-GCCGTCGACGTGCAGGTTGGTGGGAACTGG-3 ′, (1–388) 5′-GCCGTCGACCTCCCCTTTCTTCCGCCACTT-3 ′, (1–115) 5′-GCCGTCGACGCCTTTCAGATTGGCCTTGAG-3 ′, (1–54) 5′-GCCGTCGACCTGGCCCGTGGTACGCTTCAG-3′ and a plasmid harbouring human RECQL4 cDNA as the template. DNA insert corresponding to a.a. 54–675 was prepared using forward primer 5′-GCGAATTCGCCGGCGGCGGGCTCCGCAGCT-3′ and the reversed primer (1–675) (see above). The PCR fragments were digested with EcoRI–SalI and transferred into the GAL4-binding domain (BD) destination vector pGBKT7 (CLONTECH). A DNA insert corresponding to a.a. 1–780 of human TopBP1 (containing BRCT domains 0–5) was amplified by PCR with the primers BRCT0-2for: 5′-GAACTTCCATATGTCCAGAAATGACAAAGAACCG-3′ and BRCT0-5rev: 5′-GCGGTCGACTGCAGTATCTGAATTTAGATT-3′ using full-length TopBP1 cDNA construct KIAA 0259 (kindly provided by T. Nagase, Department of Human Gene Research, Kazusa DNA Research Institute, Chiba, Japan) as the template. The PCR fragment was digested with NdeI-SalI and transferred into the GAL4 activation domain (AD) vector pGADT7 (CLONTECH). The AD-TopBP1 (a.a. 793–1522) construct (containing BRCT domains 6–8) was described previously (27). For semiquantitative two-hybrid assays. Saccharomyces cerevisiae Y190 (CLONTECH) was transformed with corresponding pGBKT7- and pGADT7-constructs and β-galactosidase liquid culture assays using O-nitrophenyl β-d-galactopyranoside as substrate were performed as described previously (28). The β-galactosidase values represent the mean and standard deviations, respectively, of three independent experiments.
Purification of GST fusion proteins, IVT, GST pull-down
GST-tagged TopBP1 (a.a. 1–1522) and TopBP1 (a.a. 793–1522) were described previously (29), for GST-tagged TopBP1 (a.a. 1233–1264), ss oligonucleotides were annealed and inserted into the EcoRI/SalI digested expression vector pGEX-4T-1 (GE Healthcare). GST fusion proteins were purified as described previously (28). [35S]-methionine-labelled RECQL4 (a.a. 1–675) and [35S]-cysteine-labelled TopBP1 proteins were synthesized in vitro by coupled T7 RNA polymerase-mediated transcription and translation (IVT) in a reticulocyte lysate system as described by the manufacturer (Promega). DNA inserts corresponding to the a.a. 1233–1522 and 1264–1493 of TopBP1 were amplified by PCR with the following primers : topbp78forw: 5′-GCGAATTCCAAGCCCCCAGTATTGCCTTTCCA-3′ and topbp78rev: 5′-GCCGTCGACTTAGTGTACTCTAGGTCGTTTGAT-3′; topbp78neuforw: 5′-GCGAATTCTTAAAAAAACAGTACATATTTCAG-3′ and topbp78neurev: 5′-GCCGTCGACAATAAATGAAATAGCTTCTGG-3′, and the full-length TopBP1 cDNA construct KIAA 0259 as template. PCR products were digested with EcoRI/XhoI and transferred into the GAL4 AD vector pGADT7 (CLONTECH).
L-[35S]-methionine (specific activity: 1175 Ci/mmol) and [35S]-cysteine (specific activity: 1075 Ci/mmol) were obtained from Hartmann Analytic (Braunschweig, Germany). Glutathione–Sepharose beads (50 µl) bound by either GST-tagged TopBP1 and RECQL4 protein fragments or GST alone were washed with HB buffer (20 mM HEPES, 100 mM KCl, 5 mM MgCl2, 0.5 mM dithiothreitol and pH 7.4) and then resuspended in 50 µl HB buffer containing 0.5% Igepal CA–630. To each sample, 10 µl [35S]-methionine- or [35S]-cysteine-labelled interaction partner proteins were added. After incubation at 4°C under constant rotation for 2 h, the beads were extensively washed with HB buffer. Washed beads were boiled in SDS sample buffer. Bound proteins were separated by 10% SDS–PAGE and visualized by exposure on KODAK X-OMAT AR film.
Cell culture, co-immunoprecipitation and immunoblotting
HEK293T (ATCC-CRL-11268) cells were grown as monolayers in Roswell Park Memorial Institute medium supplemented with l-glutamine (PAA/GE Healthcare) and 10% heat-inactivated foetal calf serum at 37°C in an atmosphere of 5% CO2. Cells were co-transfected with a full-length expression construct of TopBP1 (a gift from J. Chen, University of Texas, Houston, USA) and pcDNA3-RecQL4 using FUGENE® HD transfection reagent (Roche) according to the manufacturer’s instructions. Co-transfected cells were harvested and lysed with a solution containing 50 mM Tris/HCl (pH 8.0), 150 mM NaCl, 1% Triton X-100 and a mixture of protease inhibitors (10 µg/ml leupeptin and aprotinin, 5 µg/ml pepstatin and 1 mM phenyl methyl sulphonyl fluorid (PMSF)). Lysates were cleared by centrifugation at 11 000 g at 4°C. TopBP1 polyclonal antibody (4 µg, Novus Biologicals), and anti-RecQL4 (a rabbit polyclonal antibody raised against recombinant a.a. 1–115 of human RecQL4) or pre-immune serum were incubated with 20 µl (bed volume) of protein G-Sepharose (GE Healthcare) and 1% BSA in lysis buffer overnight at 4°C. Beads were incubated with the same buffer containing 1.5 mg of precleared lysate for 2 h at 4°C and with a constant rotation speed. After extensive washing, co-immunoprecipitated proteins were eluted by boiling in SDS sample buffer at 95°C for 5 min from the beads.
Co-immunoprecipitated proteins were resolved by 10% SDS–PAGE and transferred to a nitrocellulose membrane (Whatman). Membranes were blocked in 5% skim milk for 45 min, washed thrice with Tris-buffered saline with Tween 20 (TBST) (10 mM Tris–HCl, pH 7.5, 150 mM NaCl and 0.5% Tween 20) and incubated with the primary antibodies: rabbit anti-TopBP1 (1:200; Novus Biologicals) and rabbit anti-RecQL4 (1:200) overnight at 4°C. After washing three times with TBST, the membranes were incubated with horseradish peroxidase–conjugated goat anti-(rabbit-IgG) secondary antibody (Dianova) for 1 h at room temperature. Protein bands were detected with ECL Prime Western Blotting Detection Reagent (GE Healthcare) as described by the manufacturer.
Surface plasmon resonance binding assay
Real-time analysis of the interaction between RecQL4_N54 and TopBP1 (a.a. 1233–1522) was performed on a Biacore 2000 system at 25°C, and data were processed with the evaluation software version 4.1.1 (GE Healthcare). Goat anti-GST antibody of 5100 response units (RU) were immobilized on flow cells 1 and 2 of a CM4 sensor chip by using the GST capture and amine coupling kits (GE Healthcare). GST-TopBP1 (a.a. 1233–1522) was injected on flow cell 2 at a flow rate of 10 µl/min until 830 RU had been bound. An equimolar amount of recombinant GST was captured on flow cell 1 (370 RU) for referencing. RecQL4_N54 was injected in running buffer (10 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA and 0.005% (v/v) surfactant P20) at concentrations from 3.13 to 200 µM. Sample injection and dissociation times were set to 60 s at a flow rate of 30 µl/min. Each injection was performed three times. Refractive index errors due to bulk effects were corrected with responses from reference flow cell 1 and by subtracting blank injections. Dissociation constants were calculated from the kinetic rate constants for GST-TopBP1 (a.a. 1233–1522)/RecQL4_N54 complex formation and dissociation derived from a 1:1 interaction model and from concentration-dependent steady-state binding of RecQL4_N54.
Quantitative real-time RecQL4_N54 DNA-binding assays
Surface plasmon resonance (SPR) binding experiments using 10 mM HEPES pH 7.4, 150 mM NaCl, 3 mM EDTA and 0.005% (v/v) surfactant P20 (HBS-EP150) as running and sample buffer were carried out on a Biacore T200 system at 25°C. DNA duplexes were produced by annealing complementary oligonucleotides using a 5-fold molar excess of the non-biotinylated oligonucleotide. The dsDNA (LB2-24 Y and LB2-24 ds) as well as the 5′-biotinylated LB2 24 ss oligonucleotide were injected on flow cells 2, 3 and 4 of a Neutravidin (Pierce)-coated CM3 sensor chip at a flow rate of 10 µl/min until about 44 or 22 RU had been bound, respectively (Supplementary Table S3). RecQL4_N54 samples triplicates were injected in HBS-EP150 at concentrations from 3.13 to 100 µM. Sample injection and dissociation times were set to 60 s at a flow rate of 30 µl/min. Refractive index errors due to bulk solvent effects were corrected with responses from non-coated flow cell 1 as well as subtracting blank injections. Dissociation constants were calculated from concentration-dependent steady-state responses of RecQL4_N54 binding.
Due to strong bulk effects and excessive non-specific binding of RecQL4_N54 to non-coated flow cell 1, dissociation constants could not be determined from SPR sensorgrams using a HEPES buffer that contained 50 mM NaCl (data not shown). Therefore, the experiments in 10 mM HEPES, pH 7.4, 50 mM NaCl, 3 mM EDTA and 0.005% (v/v) surfactant P20 (HBS-EP50) were conducted on an Octet RED96 bio-layer interferometry instrument (forteBio), operating at 25°C. Streptavidin (SA) dip and read biosensor tips with immobilized Y-shaped, dsDNA and ssDNA were exposed for 120 s to different concentrations of RecQL4_N54 (3.13–200 μM), respectively. Non-coated SA sensor tips were used for referencing. Dissociation constants were calculated from concentration-dependent steady-state binding of RecQL4_N54 (Figure 7D).
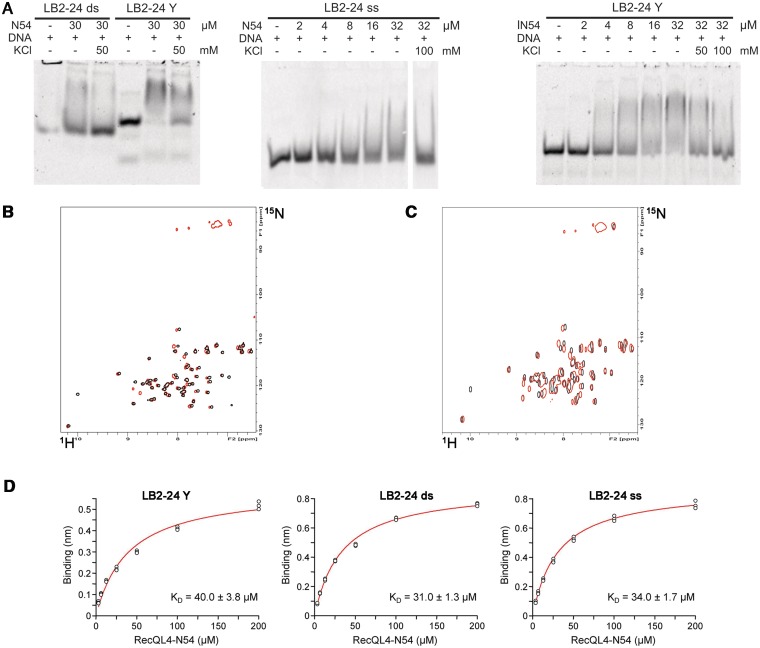
Figure 7.
RecQL4_N54 binding to Y-shaped DNA. (A) RecQL4_N54 prefers binding to Y-shaped DNA over ssDNA and dsDNA. Left panel: 30 µM RecQL4_N54 was preincubated in the presence of 2 µM of the indicated DNA and the DNA–protein complexes resolved by electrophoretic mobility gel shift assay. Middle panel: ssDNA binding—indicated amounts of RecQL4_N54 were preincubated with 1.3 µM oligomer LB2-F carrying a 5′-fluorescein, and the DNA–protein complexes resolved by electrophoretic mobility gel shift assay. Right panel: Binding to Y-shaped DNA—indicated amounts of RecQL4_N54 were preincubated with 2 µM LB2-derived Y-shaped DNA, and the DNA–protein complexes resolved by electrophoretic mobility gel shift assay. The DNA binding buffer was supplemented with KCl as indicated. Details for the DNA fragments are provided in Supplementary Table S1. (B) [1H,15N]–HSQC spectra (15N chemical shifts in F1 and 1H chemical shifts in F2) of RecQL4_N54 (200 µM) without (black contours) and with 3.5-fold excess (red contours) of Y-shaped DNA. (C) [1H,15N]-HSQC spectra of RecQL4_N54 (200 µM) without (black contours) and with 2-fold excess (red contours) of ssDNA. (D) Real-time binding analysis of RecQL4_N54 to Y-shaped, dsDNA and ssDNA. Fits of the biolayer interferometry steady-state binding of RecQL4_N54 using a HEPES buffer containing 50 mM NaCl.
RESULTS
The N-terminus of human RecQL4 interacts with TopBP1
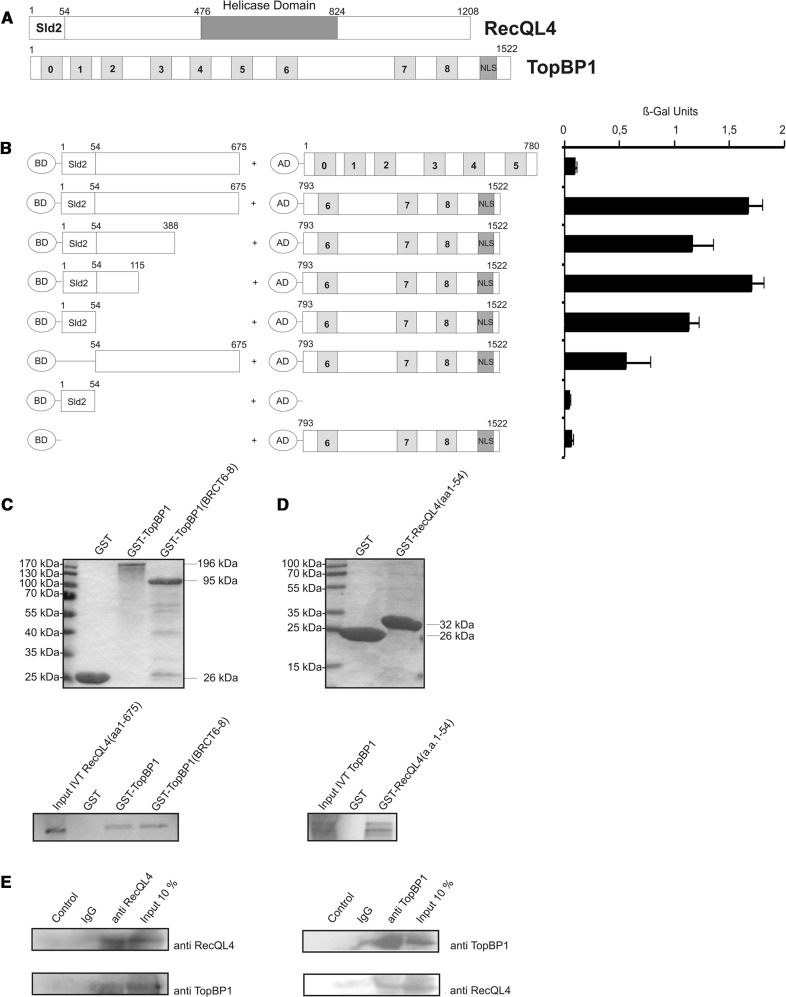
To assess the potential interaction between human RecQL4 and TopBP1 and to delineate the protein domains involved, we employed yeast two-hybrid analysis. An amino-terminal fragment corresponding to the first 675 a.a. of human RecQL4 was fused to the GAL4-BD. TopBP1 constructs covering BRCT domains 0–5 (a.a. 1–780) or BRCT domains 6–8 (a.a. 793–1522), respectively, were fused to the GAL4 AD (Figure 1A). After transformation of the constructs into S. cerevisiae reporter strain Y190, a β-galactosidase assay was performed. Significant transactivation was observed (Figure 1B) when the RecQL4 a.a. 1–675 construct was expressed in the presence of TopBP1-BRCT6-8, but not for TopBP1-BRCT0-5, suggesting a physical interaction between the N-terminus of the human RecQL4 and the BRCT6-8 domains of TopBP1. We then performed deletional mutagenesis to identify the TopBP1-interacting region of human RecQL4. This analysis indicated that a construct comprising the first 54 a.a. of human RecQL4 led to only moderately reduced β-galactosidase activity, similar to N388 and N115 fragments when compared with RecQL4 (a.a. 1–675). A RecQL4 construct lacking the first 54 a.a. supported β-galactosidase activation less significantly. This indicates that the 54 N-terminal residues of RecQL4 are important for the interaction with TopBP1-BRCT6-8, albeit contributions of more C-terminal positions are also apparent.
Figure 1.
The Sld2-homologous N-terminal domain of RecQL4 interacts directly with TopBP1. (A) Schematic depiction of human RecQL4 and TopBP1. (B) Specific interaction of the N-terminal domain of RecQL4 fused to the GAL4 DNA-BD with C-terminal part (a.a. 793–1522) of TopBP1 fused to the GAL4-AD in yeast. Interaction was semi-quantitatively assessed by β-galactosidase liquid culture activity assays. β-galactosidase values represent the mean and standard deviations of three independent experiments. (C) GST pull-down experiments performed with purified GST, GST–TopBP1 and GST–TopBP1 (a.a. 793–1522) and [35S]–methionine–labelled RecQL4 (a.a. 1–675) (lower panel). Coomassie brilliant blue R250-stained SDS–PAGE gels of the purified GST constructs are presented (upper panel). (D) GST pull-down experiments performed with purified GST and GST–RecQL4 (a.a. 1–54) and [35S]-cysteine-labelled full-length TopBP1. Coomassie brilliant blue R250-stained SDS–PAGE gels of the purified GST constructs are presented (upper panel). Input IVT: 10 µl of the in vitro translation product. (E) Reciprocal immunoprecipitation of RecQL4 and TopBP1. Cell extracts from human Hek293 cells transiently co-transfected with expression vectors for RecQL4 and ToPBP1 were subjected to immunoprecipitation using antibodies against RecQL4 and TopBP1 as described in ‘Material and Methods’. The precipitates were then analysed by SDS–PAGE and Western blot against the cognate proteins. Controls without antibody or with non-specific rabbit IgG served as negative control and 10% of the input served as an indicator for the (co-)immunoprecipitation efficiency.
In GST-pull-down assays, in vitro translated human RecQL4_1–675 bound to GST-TopBP1 full length and to GST-TopBP1-BRCT6-8 with comparable efficiencies, but not to GST alone (Figure 1C). Similarly, in vitro translated full-length TopBP1 bound to GST-RecQL4 a.a. 1–54 (RecQL4_N54, Figure 1D). TopBP1 did not bind to GST alone, confirming the specificity of the binding to the RecQL4 fragment. We next investigated whether an interaction between RecQL4 and TopBP1 can also be detected in cell extracts. Indeed, we were able to detect this interaction by reciprocal co-immunoprecipitations from HEK293T cell extracts expressing RecQL4 and TopBP1, using antibodies against RecQL4 and TopBP1 (Figure 1E).
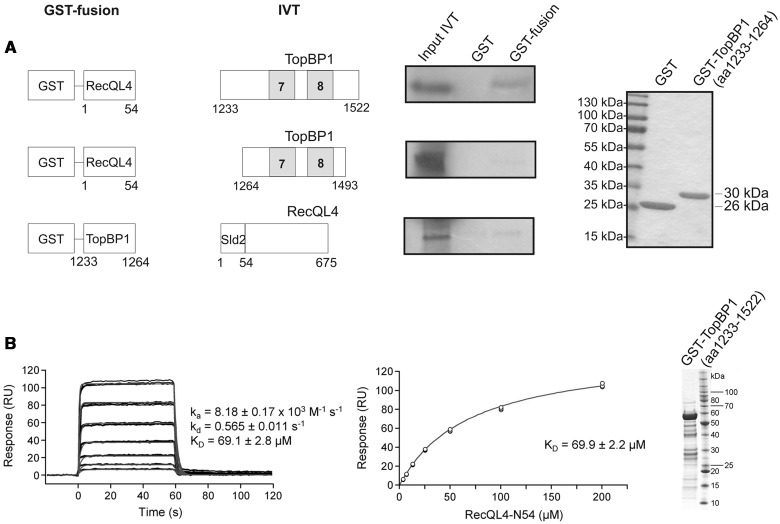
Subsequently, we used GST pull-down to further narrow down the RecQL4_N54-binding site of TopBP1 (Figure 2). A fragment comprising a.a. 1233–1522 (end) of human TopBP1 was efficiently bound by RecQL4_N54 (Figure 2A). In contrast, a construct limited to BRCT7/8 (a.a. 1264–1493) (30) was not bound. Similarly, the proline-rich region of TopBP1 preceding BRCT7/8 (a.a. 1233–1264) only pulled down minor amounts of RecQL4_1-675. Therefore, TopBP1 (a.a. 1233–1522) comprising BRCT7/8 together with the proline-rich region are required for the binding of RecQL4_N54. SPR was used to estimate the binding affinity of RecQL4_N54 to TopBP1 (a.a. 1233–1522). Using GST-TopBP1 (a.a. 1233-1522) immobilized via its GST moiety as bait, or GST alone as a control, and the RecQL4_N54 fragment as analyte, we observed specific binding (Figure 2B). A dissociation constant of ∼70 µM was determined from both the kinetic rates and the concentration-dependent steady-state binding. No binding of RecQL4_N54 to GST alone was observed.
Figure 2.
Characterization of the TopBP1 interaction with RecQL4. (A) Identification of the RecQL4_N54 BD of TopBP1 by GST pull-down. Pull-down experiments were performed as in Figure 1C–D with GST-fusion constructs as bait for in vitro translated proteins as indicated. Only TopBP1 (a.a. 1233–1522) shows efficient binding to GST-RecQL4_N54. Purified GST and GST-TopBP1 (a.a. 1233–1264) stained with Coomassie brilliant blue R250 are shown on the right. (B) Real-time in vitro SPR binding analysis of RecQL4_N54 to anti-GST antibody captured GST-TopBP1 (a.a. 1233–1522). Sensorgrams of 200, 100, 50, 25, 12.5, 6.25 and 3.13 µM RecQL4_N54 binding injected in triplicate (black lines) are shown overlaid with the best fit derived from a 1:1 interaction model (red lines) (left). Fit of the equilibrium data for TopBP1 (a.a. 1233–1522) binding (middle). The purified GST-TopBP1 (a.a. 1233–1593) used for the experiment stained with Coomassie brilliant blue R250 is shown on the right.
Taken together, a region comprising a.a. 1–54 of human RecQL4 appears to be both necessary and sufficient to bind the carboxy-terminal portion of TopBP1 including a.a. 1233–1522 of the latter. The results from yeast two-hybrid analysis and the moderate-binding constant further suggest additional contributions of regions proximal to RecQL4_N54 to the RecQL4-TopBP1 interaction.
The amino-terminal domain of human RecQL4
An in silico analysis (see Supplementary Information) of the minimal TopBP1-interacting region of human RecQL4 predicted a predominantly α-helical fold. This region was found (12,13) to align well with the yeast Sld2 homologues (Supplementary Figure S1A). Extensive homology searches show that this element is generally conserved in fungal Sld2 proteins and in the metazoan line where it fused N-terminal to a RecQ domain. Related sequences can also be sporadically found in other phylogenetic groups such as green algae, oomycetes or dinoflagellates. Interestingly, also the choanoflagellate Monosiga brevicollis possesses a predicted homologue containing an amino-terminal Sld2 homology region fused to a RecQ domain (Supplementary Figure S1), indicating that this arrangement may have been conserved even before diversion of the metazoan lineage. Extension of the alignment beyond this N-terminal homology region requires introduction of significant gaps into the alignment. The corresponding regions are predicted by the DISMETA server to be largely disordered in metazoan RecQL4 and fungal Sld2 sequences (data not shown). This suggested that the amino-terminus represents an autonomous structural element of the RecQL4/Sld2 family.
RecQL4_N54 could be expressed with high yields, both in complete and minimal medium as a GST fusion protein in E. coli, and purified to near homogeneity by a combination of affinity and ion exchange chromatography (Supplementary Figures S2A and B). The purified protein was analysed by CD spectroscopy (Supplementary Figure S2D). The far-UV CD spectrum of RecQL4_N54 exhibited double minima at 208 and 222 nm characteristic for significant α-helical content. The melting point Tm was found to be 331 K (58°C), based on the first-derivative analysis and the van’t Hoff plot of the CD signals at 192, 208 and 222 nm (Supplementary Figure S2E). We found that the temperature-induced melting was largely reversible (Supplementary Figure S2D). The CD analysis supports the view that RecQL4_N54 represents a stably folded domain. Gel filtration suggested that RecQL4_N54 exists as monomer in solution (Supplementary Figure S2C).
The NMR structure of RecQL4_N54
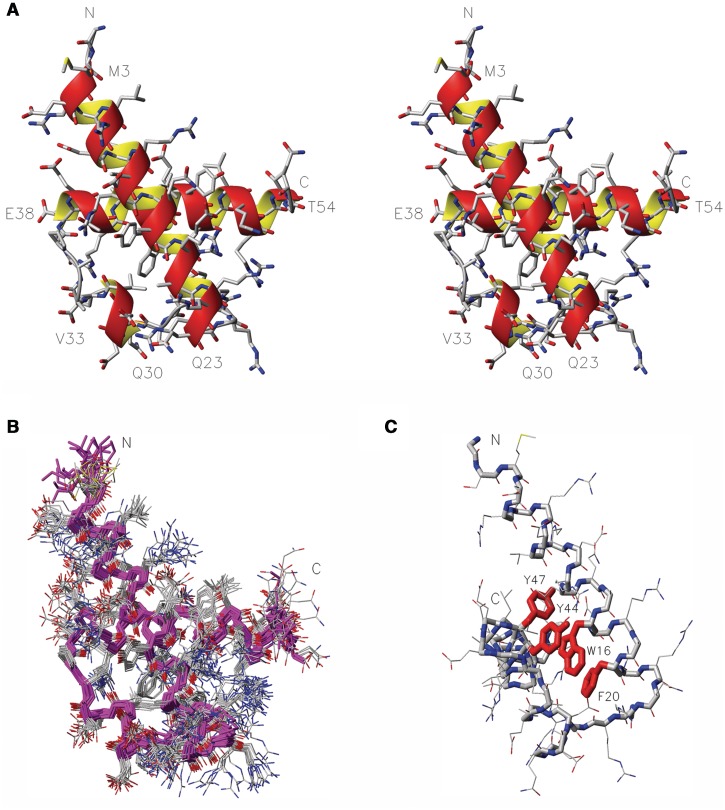
We determined the solution structure of RecQL4_N54 by heteronuclear NMR spectroscopy. The [1H,15N]-HSQC spectrum of RecQL4_N54 exhibited good spectral dispersion (Supplementary Figure S3) indicating a well-folded protein under NMR conditions. Resonance assignment was carried out by means of heteronuclear 3D experiments (e.g. HNCACB, CC(CO)NH, H(CCCO)NH and HCCH-COSY). The extent of proton assignment amounts to 99%. In addition, with the exception of the N′ and C′ resonances of residues Gly1 and Ser2, all heteroatoms accessible via standard heteronuclear NMR experiments including the aromatic resonances of proton-bearing carbons and nitrogen were assigned. The chemical shift assignments were used to predict the secondary structure composition of RecQL4_N54. The program CSI (31) predicted three α-helical elements. The RecQL4_N54 solution structure was calculated on the basis of 1251 nuclear Overhauser enhancement (NOE) constraints and 146 constrained torsion angles. Superimposition of the ensemble of 20 structures with the lowest CYANA target function is shown in Figure 3 together with a ribbon model of the lowest energy structure of RecQL4_N54. The refinement statistics is given in Table 1. RecQL4_N54 is composed of three α-helices connected by short loop elements. The first helix of RecQL4_N54 consists of 5.5 turns between residues Glu4 and Gln23. A six-residue loop connects the helix α1 to the short helix α2 comprising four residues (Gln30–Val33) and forming only one helical turn. The helix α2 connects to the last helical element via a four residue turn sterically not restricted owing to a central alanine dipeptide. The carboxy-terminal helix α3 starts after the Pro37. It comprises 4.5 turns formed by 17 a.a. (Glu38–Thr54). The aromatic side chains of Trp16, Phe20 and Tyr44 form a stacked arrangement as central element of the hydrophobic core (Figure 3C).
Figure 3.
NMR solution structure of RecQL4_N54. (A) Stereo view (side-by-side) of the structure closest to the mean. Heavy atom colouring: C (grey), N (blue), O (red) and S (yellow). Helices are indicated by an orange/yellow ribbon. Amino acid type and residue numbers for the start and end residue of the helical elements are annotated. (B) Superimposition of the 20 calculated structures with the lowest target function, backbone in magenta, heavy atom colouring as in A. (C) Aromatic core, the side chains of W16, F20, Y44 and Y47 are emphasized in red, aspect rotated with respect to (B) to display the core.
Table 1.
Structural statistics of RecQL4_N54
| NMR distance and dihedral constraints | |
| Distance constraints | 1251 |
| Hydrogen bond constraints | 60 |
| Total dihedral angle restraints | 310 |
| Structure statistics | |
| Violations, mean (SD) | |
| Target function (Å2) | 0.73 (0.14) |
| Distance constraints (Å) | 0.01 (0.0007) |
| Max. distance constraint violation (Å) | 0.21 (0.03) |
| Dihedral angle constraints (°) | 0.21 (0.0337) |
| Max. dihedral angle violation (°) | 2.14 (0.46) |
| AMBER physical energies (kcal/mol) after energy minimization | −831.30 (72.24) |
| Deviations from idealized geometry | |
| Bond lengths (Å) | 0.00562 (0.00019) |
| Bond angles (°) | 1.55700 (0.06497) |
| Mean global r.m.s.d. (Å) | |
| Backbone atoms (residues 3–56) | 0.73 (0.23) |
| Heavy atoms (residues 3–56) | 1.84 (0.28) |
RecQL4_N54 resembles a homeodomain
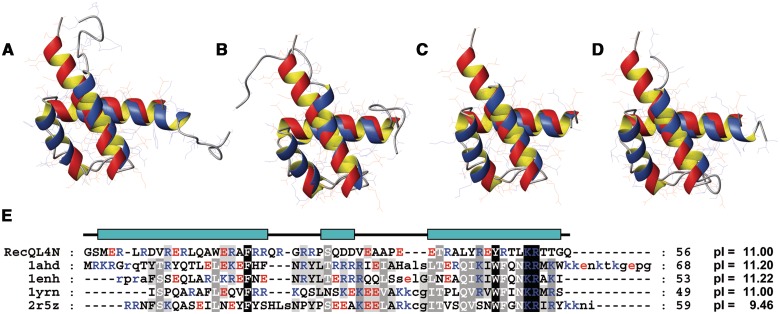
The overall topology of RecQL4_N54 is reminiscent of a helix-turn-helix motif. A search on the DALI server (32) using the lowest energy structure of RecQL4_N54 revealed a high structural similarity to homeodomains. The best-scoring hits represented predominantly homeodomains such as the ones of transcription factor Engrailed (PDB code: 1ENH) and of Mata1/Matα2 (1YRN). Furthermore, the structural alignment covered essentially the complete target domains in contrast to other sporadic DALI hits such as helix-turn-helix motifs of the chromosome partition protein MukB (3EUK) or the site-specific recombinase XerD (1AOP), where only a small fraction of the target sequence could be superimposed onto RecQL4_N54. A detailed comparison of RecQL4_N54 with representative homeodomains of Antennapedia [1AHD; backbone root-mean-square deviation (r.m.s.d.) 1.87 Å (33)], Engrailed [1ENH; 1.86 Å (34)], Mat a1/Mat α2 [1YRN; 1.76 Å (35)] and Scr/Exd [2R5Z; 1.79 Å (36)] yielded consistently an r.m.s.d. of ∼1.8 Å for the superimposition of the respective backbones (Figure 4A–D). Position and relative orientation of the three α-helices are conserved, but compared with classical homeodomains, the helix α2 is shortened by six residues. More importantly, the RecQL4_N54 helix α1 is extended by seven residues and thus lacks the amino-terminal non-helical extension typical for homeodomains, which is disordered in the absence of DNA. Upon DNA complex formation, this extension contributes to DNA-binding affinity and sequence specific contacts via minor groove interactions (37). The structure-based sequence alignment (Figure 4E) yields (only) two sequence identities (F20 and R52) and two conservative exchanges (Y47 and K51) between RecQL4_N54 and homeodomains (37,38). The remainder of RecQL4_N54, however, shares a high content of basic residues with homeodomains (Figure 4E).
Figure 4.
Superimposition of the NMR solution structure of RecQL4_N54 (orange ribbon) with homeodomains (blue ribbon). (A) Antennapedia homeodomain (PDB code 1AHD; backbone r.m.s.d. 1.87 Å), (B) Engrailed homeodomain (1ENH; 1.86 Å). (C) MATa1/MATα2 homeodomain (1YRN; 1.76 Å). (D) Homeodomain from the SCR/EXD complex (2R5Z; 1.79 Å). Homeodomains in (A), (C) and (D) are in the DNA-bound form, DNA not shown. (E) Structure-based sequence alignment of RecQL4_N54 against the depicted homeodomains. The RecQL4_N54 construct used for structure determination starts with two non-native residues derived from the GST tag after thrombin digest. Numbering of the a.a. positions refers to the construct and not the native RecQL4 sequence to maintain consistency with the related PDB (2KMU) and BMRB (16544) database entries. Therefore, the numbering of the residues is shifted by two compared to the native protein.
RecQL4_N54 binds double-stranded DNA
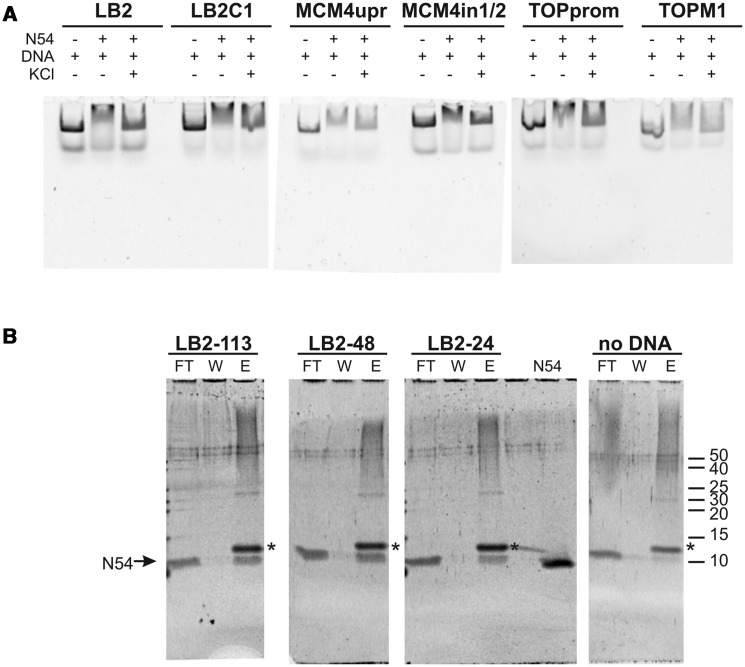
The structural homology to homeodomains led us to assess the DNA binding of RecQL4_N54. To this end, we performed EMSAs. Since RecQL4 has been implicated in the initiation of DNA replication and has been recently found to associate with several human origins of DNA replication (39), we initially tested the binding to dsDNA sequences derived from the human LB2, MCM4 and Topo1 origins of replication (40–42). RecQL4_N54 binds not only to the dsDNA fragment derived from the three well-defined origins but also to the sequences that are proximal to the respective origins (Figure 5A). This DNA binding appeared to be rather sensitive to salt, because the electrophoretic mobility shift was largely absent in the presence of 100 mM KCl. Also various other dsDNAs were shifted with similar efficiency (data not shown) indicating that RecQL4_N54 may bind dsDNA without apparent sequence specificity. To analyse this further, we performed EMSAs with different fragments derived from our original 232 bp fragment of the LB2 origin (Supplementary Figure S4). RecQL4_N54 bound all these fragments with similar efficiencies despite the absence of common recognisable conserved sequence elements. In conclusion, RecQL4_N54 exhibits no apparent sequence specificity.
Figure 5.
RecQL4_N54 binds dsDNA without apparent sequence preference. (A) Electrophoretic mobility gel shift assay were performed with RecQL4_N54 (2 µg/30 µM) and DNA fragments (200 ng) covering the sites of DNA replication initiation of three well-characterized human origins as well as three control fragments from proximal sites. Details on the DNA fragments are provided in Supplementary Table S1. The DNA binding buffer was supplemented with 100 mM KCl where indicated. (B) Binding of DNA fragments derived from the LB2 origin of replication (42) by RecQL4 was evaluated by a DNA bead binding assay. Biotinylated dsDNA was pre-incubated with RecQL4 following binding to magnetic SA beads immobilized to magnetic columns. The flow through (FT), the first wash (W) and the elution (E) fractions were analysed by SDS–PAGE. RecQL4_N54 is detected in the elution fractions of all samples containing DNA indicative for DNA binding irrespective of fragment length. The protein band corresponding to RecQL4_N54 is indicated by an arrow. The additional band in the elution (denoted by an asterisk) is SA released from the magnetic bead during sample preparation for SDS–PAGE. Input represents purified RecQL4_N54. The positions of the molecular weight markers (in kDa) are indicated on the right.
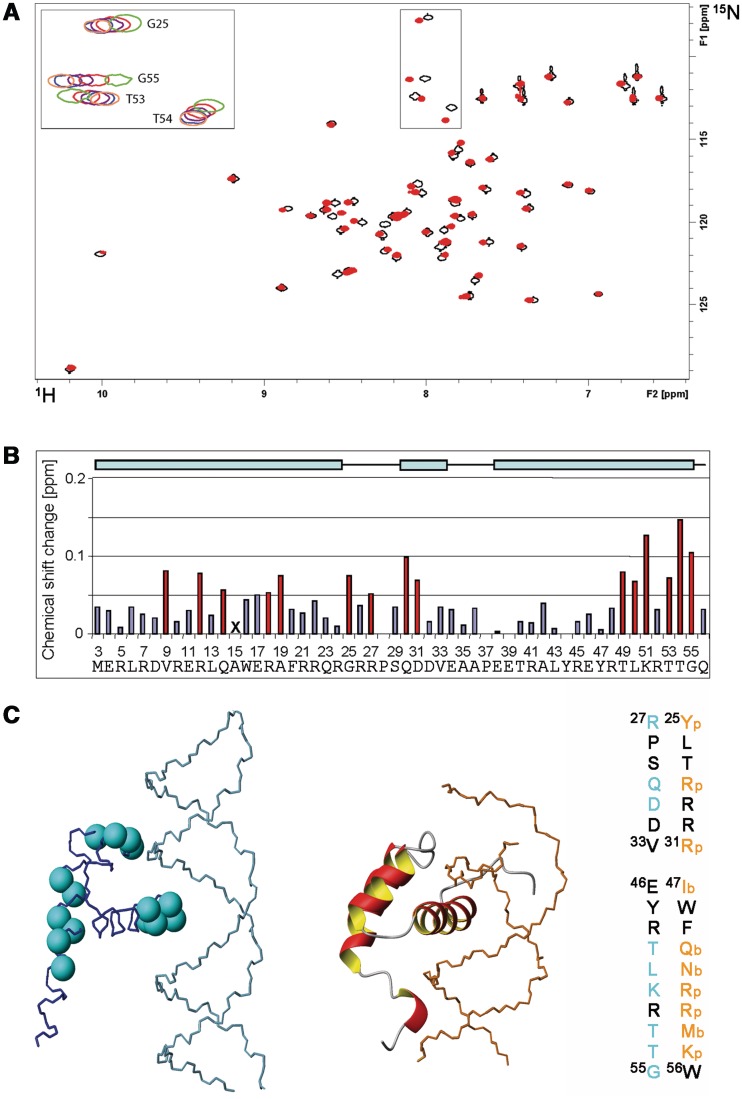
In general, various short dsDNA fragments (<50 bp) were shifted in EMSA experiments, albeit only poorly. However, an arbitrary shortening of the original 232 bp fragment from the LB2 origin down to 24 bp did not abolish DNA binding as detected by magnetic bead binding assays (Figure 5B). Collectively, the two binding assays suggest that RecQL4_N54 has also affinity to short dsDNA fragments, which is difficult to characterize under EMSA conditions. To confirm this and to narrow down the DNA-interaction region on RecQL_N54, an NMR-based mapping experiment (43) was carried out exemplary with the LB2-derived 24mer dsDNA. The dsDNA oligomer was added to the protein at a concentration of 170 µM in a stepwise manner so as to achieve a RecQL4_N54:DNA ratio of 1:0.5, 1:1, 1:2 and 1:3. Complex formation was monitored by acquisition of [1H,15N]- and [1H,13C]-HSQC spectra (Figure 6A and data not shown). An increasing DNA concentration led to incremental changes of the resonance positions for a defined subset of amide groups, and no significant spectral changes were observed above a 2-fold excess of DNA indicating the endpoint of titration (Figure 6A). The incremental chemical shift perturbation indicates binding in the fast exchange regime on the NMR time scale. Chemical shift perturbations were considered as significant when exceeding the 0.05 ppm average value for the combined proton/nitrogen chemical shift changes (44). Chemical shift perturbations for backbone amide resonances were observed for a number of residues throughout RecQL4_N54, but a cluster of C-terminal residues (Thr49, Lys51, Thr53 and Thr54) exhibited the most pronounced shift perturbation (Figure 6B), pointing towards a major contribution of helix α3 to DNA binding.
Figure 6.
Interaction of RecQL4_N54 with DNA. (A) [1H,15N]–HSQC spectrum (15N chemical shifts in F1 and 1H chemical shifts in F2) of RecQL4_N54 (170 µM) without (black contours) and with 3-fold excess (red contours) of dsDNA. The inset (upper left) depicts an overlay for selected cross-peaks (boxed in spectra) of spectra recorded with protein:DNA ratios of 1:0 (green contours), 1:0.5 (red), 1:1 (magenta), 1:2 (blue) and 1:3 (coral). Residue assignments for the inset are indicated, a fully assigned [1H,15N]–HSQC is given in the Supporting Information. (B) Chemical shift changes for each backbone amide group of RecQL4_N54 at 3-fold molar excess of the 24 mer dsDNA. The chemical shift change was determined for the 15N/1H cross-peak of the backbone amide group of each residue in the spectrum of free RecQL4_N54 to the nearest 15N/1H cross-peak in the spectrum of the RecQL4_N54:DNA complex using the relationship |Δδ(1H)| + |Δδ(15N)|/7 (45). Values above the average chemical shift perturbation of ∼0.05 ppm are depicted in red. The X for Ala15 indicates that the corresponding cross-peak was broadened beyond detection at 3-fold molar excess of dsDNA. The cyan bars on the top indicate the positions of the α-helices in RecQL4_N54. Note that the first native RecQL4 residue is M3. The numbering of the a.a. positions in RecQL4_N54 starts with two non-native residues derived from the expression construct to maintain consistency with the related PDB (2KMU) and BMRB (16544) database entries. Therefore, the numbering of the residues is shifted by two compared to the native protein. (C) Left panel: Cyan balls at the position of the nitrogen atom highlight the residues in RecQL4_N54 for which chemical shift perturbation above average was observed. Centre panel: For comparison, the Antennapedia homeodomain–DNA complex [PDB code 1AHD; (35)] is presented. Right panel: Sequence comparison between RecQL4_N54 and Antennapedia. Orange: Residues interacting with the phosphate/sugar backbone of the DNA (p) or contributing in base recognition interactions (b) in the Antennapedia homeodomain–DNA complex. Cyan: Residues experiencing significant chemical shift perturbation in RecQL4_N54.
RecQL4_N54 prefers Y-shaped DNA over single- and double-stranded DNA
We next addressed whether RecQL4_N54 binds other forms of DNA. The LB2-derived 24-mer dsDNA used for the NMR titration was only weakly shifted in EMSA experiments (Figure 7A), as also seen for other short DNA fragments. The upper strand of LB2-24 DNA, i.e. ssDNA, was shifted very poorly (Figure 7A, middle panel). This was also the case for ssDNAs of unrelated sequence (data not shown), indicating that RecQL4_N54 does not show a preference for ssDNA. Instead, a LB2-24-based, Y-shaped DNA consisting of 16 bp dsDNA with 8 nt ss overhangs (Supplementary Table S2; LB2_1-24 Y) was more efficiently shifted in EMSA experiments when compared with ssDNA and dsDNA (Figure 7A). Similarly, efficient binding was also found for other branched DNA structures such as Y-shaped DNAs of unrelated sequence, a DNA ‘bubble’, D- and R-loops and Holiday junctions (Supplementary Figure S5), suggesting that DNA binding is modulated by the presence of ds/ss junctions. Therefore, Y-shaped DNA was added to RecQL4_N54 (200 µM) in a stepwise fashion resulting in a molar ratio of 1:0.3, 1:0.7, 1:1, 1:1.5, 1:2 and 1:3.5 between protein and DNA, respectively. The perturbation of the amide group chemical shifts upon the addition of DNA was followed in [1H,15N]-HSQC experiments. Saturation of binding was achieved at 2-fold DNA excess (data not shown). Chemical shift changes upon addition of Y-shaped DNA were observed for the same subset of amide group resonances as for the dsDNA above. Interestingly, in the presence of Y-shaped DNA an additional set of resonances around 85 ppm (15N) becomes detectable (Figure 7B). This chemical shift range is indicative of Arg side chain guanidino groups, the protons of which become protected against exchange with the solvent upon DNA binding. These resonances are detectable neither for the free RecQL4_N54 nor for the dsDNA:RecQL4_N54 complex. Unfortunately, attempts to assign these resonances, using scalar coupling- and NOE-based experiments, turned out unsuccessful. We also performed comparable titrations with the upper strand of the 24-mer (Figure 7C). ssDNA protects the same two Arg side chain guanidino groups from solvent exchange as observed for Y-DNA, indicating that these are involved the interaction with ssDNA regions. In addition, chemical shift perturbation was observed for amide groups partly overlapping with those observed in the dsDNA titration experiment. Thus, the Y-DNA titration yields combined features of the ssDNA and dsDNA titration.
We also determined the apparent DNA-binding constants for ss-, ds- and Y-shaped DNA using biolayer interferometry. At conditions similar to those of the NMR experiments, we obtained comparable binding constants of 30–40 µM for all three DNA forms (Figure 7C and Supplementary Table S4). The repetition of the measurements by SPR at 150 mM NaCl also yielded similar binding constants of 130–160 µM for the three DNA species (Supplementary Figure S6 and Table S4). Yet, comparison of the calculated maximum binding capacities (Rmax calc.) and the experimentally determined maximum responses (Rmax) strongly suggests a different binding stoichiometry for Y-shaped DNA when compared with ss and ds DNA. Although for the former a 2:1 ratio for RecQL4_N54 binding was evident, an 1:1 binding was calculated for ds and ss DNA (Supplementary Table S4). Therefore, although the quantitative methods do not provide evidence for a higher affinity for Y-DNA, the EMSAs presented in Figure 7A and Supplementary Figure S5 together with the results from the NMR titrations, and the SPR experiments suggest that branched DNA structures may represent the preferred substrates.
DISCUSSION
Many DNA-binding proteins (e.g. repressor and homeotic proteins) exhibit helix-turn-helix-like motifs (45). Among these, homeodomains represent mainly eukaryotic sequence-specific DNA binding motifs and can also mediate protein–protein interactions [reviewed in (46)]. In metazoans, they play a fundamental role in development and evolution. Prominent representatives for this motif are the proteins encoded by the homeotic genes (37,47,48). Initially, the homeodomain was identified in Drosophila homeotic and segmentation proteins, and it was recognized that those proteins efficiently bind DNA through this structural motif (49,50). The homeodomain proteins elicit their function, e.g. by complex formation and specificity enhancer in promoter regions of their targets. Interestingly, a number of homeodomain containing proteins of the Hox family have been reported to be involved in several aspects of replication (51–56). Homeodomains have highly conserved sequences of ∼60 a.a. Generally, their structure displays a helix-loop-helix-turn-helix motif. The detailed protein:DNA interaction network was initially unravelled by NMR spectroscopy (57,58): Two α-helices (α1 and α3) mediate specific protein:DNA contacts and stabilize each other structurally. Interestingly, despite a low-sequence homology (Figure 4E, Supplementary Figure S1), the structure of RecQL4_N54 displays a high structural homology to homeodomain proteins, presented by superimpositions of various homeodomains with RecQL4_N54 (Figure 4). As in homeodomains, the RecQL4_N54 structure remains stable upon its removal from the remainder of the protein. We also observed strong fluorescence quenching of the invariant tryptophan residue buried in the native core of RecQL_N54 (data not shown), which is reminiscent of similar observations for homeodomains (59). The structure of RecQL4_N54 also resembles the canonical three helices, albeit helix α2 is considerably shortened down to one helical turn. This allows to accommodate the slightly elongated helix α3 in a position, which coincides with the helical arrangement of hitherto known homeodomains and to locate two sequence identities (T40 and R52) and three conservative exchanges (R45, Y47 and K51) in homologous positions. The chemical shift changes of several of these C-terminal residues (Figure 6B) suggest a dsDNA-binding mode similar to homeodomains, where essential sequence-specific major groove contacts are made by the C-terminal helix α3 (38). Despite the structural similarities, RecQL4_N54 displays also several features that distinguish it from canonical homeodomains. Most importantly, RecQL4_N54 lacks the disordered N-terminal extension archetypal for homeodomains in the free form. Instead of a flexible arm that establishes contacts to the minor groove upon DNA binding, helix α1 of RecQL4_N54 extends to the N-terminus. The moderate magnitude of chemical shift perturbation observed in the [1H,15N]- and [1H,13C]-HSQC spectra in the presence of the LB2 24mer dsDNA and Y-shaped DNA indicates negligible, if any, conformational changes in the structure of RecQL4_N54 upon DNA binding, suggesting that the N-terminus RecQL4_N54 constitutes a preformed DNA-binding module mediating interaction with DNA without sequence specificity as observed here. The significant structural homology to the DNA-bound form of the homeodomain homologues in 1AHD, 2R5Z and 1YRN (Figure 4) is consistent with this interpretation.
Most commonly, homeodomains bind to specific aTtA/tAaT DNA duplex motifs via a set of conserved a.a. (R3, R5, I47, Q50, N51 and M54) (37,60). Here, the arginines contact the minor groove via hydrogen bonds, while the uncharged residues in the helix α3 form hydrophobic interactions with, e.g. the thymine methyl groups of the major groove side. Taking into account the DNA-bound conformation of other homeodomains, the C-terminal threonines in RecQL4_N54 could act correspondingly to, e.g. the serine-45:adenine interaction in Antennapedia. For comparison (Figure 6C), a model of RecQL4_N54 bound to DNA is complemented by a backbone representation of the Antennapedia homeodomain–DNA complex (1AHD; 33). However, until now no sequence-specific DNA binding of RecQL4_N54 could be observed. Moreover, complex formation in the fast exchange regime on the NMR time scale suggested an affinity of 10 µM or above for the ds and Y-shaped DNA substrates. Consistent with this, a moderate binding affinity was determined by SPR and interferometry measurements. This is consistent with the function of RecQL4/Sld2 as general replication initiation and genome stability factor that has to act transiently and in a sequence-independent manner (1,12,13). It should be noted that homeodomains of several classes such as the Rhox, Lass or Satb proteins also appear to bind DNA without apparent sequence specificity (61). It is likely that other parts of the RecQL4 protein and interactions with other replication initiation factors are essential for the recruitment to origins of replication.
The RecQL4-TopBP1-interaction described here was not detected in a proteomic approach aimed to identify interaction partners after immunoprecipitation of a tagged full-length RecQL4 (15) but is in line with studies in Xenopus, where an (although larger) amino-terminal fragment was shown to interact with XCut5, the frog homologue of TopBP1. As for the Xenopus homologues (13), the interaction here did not depend on phosphorylation of the proteins. On the other hand, both human and Xenopus RecQL4 appear to be phosphorylated by S phase Cdks (13,15), and therefore, during replication, the interactions of RecQL4 are likely to be regulated by phosphorylation in vivo. Thangavel et al. (39) demonstrated that RecQL4 shows cell cycle-dependent association with origins of DNA replication, consistent with its reported role during the assembly of the replication initiation complex (12,13,62). This DNA association is likely to be modulated by interactions with MCM10, MCM2-7 (15,62) as well as with TopBP1 (Figure 1) or its Xenopus homologue xCut5 (13). As in Xenopus, there seems to be more than one interaction site between human RecQL4 and TopBP1. Although the DNA-binding constants of RecQL4 determined here appear to be in a similar range for ss, ds, and Y-shaped DNA, our EMSA and NMR analyses indicate a different binding mode for branched-type DNA. This suggests another role for RecQL4: the N-terminal domain could stabilize the emerging DNA ‘bubble’ during the initiation of DNA replication by binding at the ss–ds junction. A similar role was proposed for the budding yeast Sld2 (63) that contains an N-terminal domain homologous to RecQL4_N54. There, Sld2 was found to bind ss origin DNA and to stimulate DNA annealing. Although dsDNA was not bound by Sld2, the binding to, e.g. Y-shaped DNA was not tested. Conserved residues found in the N-termini of Sld2 and RecQL4 argue in favour of a similar function of both proteins. In Sld2, K44 and K50 are essential for yeast origin sequence binding (64). These residues correspond to R43 and K49 in human RecQL4 (Supplementary Figure S1). These positions are generally conserved in homeodomains and are located on helix α3 found important for DNA binding by our NMR experiments. Our preliminary mutational analysis indicates that K49N and R8Q/R43Q mutants do not bind dsDNA, but still interact with TopBP1 (data not shown). No annealing activity for RecQL4_N54 was detected here (data not shown), whereas a strong annealing activity for full-length human RecQL4 has been demonstrated (5,6). This indicates that additional DNA-binding sites of RecQL4 are important for its annealing activity. The role of structural transitions of DNA during initiation of replication probably needs more attention. It seems that the complex interaction framework of RecQL4 with other initiation factors and with the DNA at the initiation site may be required to spark origin firing.
Taken together, we demonstrate that the N-terminus of RecQL4 adopts a fold very similar to that of homeodomains and that this domain represents a DNA-binding module that is also involved in the interaction with human replication factor TopBP1. This study provides the first structural information for the Sld2/RecQL4 protein family. It should be highly rewarding to investigate the mechanistic interplay of the RecQL4_N54 domain and the remainder of the molecule and its interacting biomolecules to unravel a more complete understanding of the structure–function relationship for RecQL4. This should lead to a more in depth understanding of how DNA binding and annealing activities of RecQL4/Sld2 contribute to replication fork formation or to prevent a premature movement of the fork.
ACCESSION NUMBERS
PDB 2KMU.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–4, Supplementary Figures 1–5, Supplementary Methods and Supplementary References [65–67].
FUNDING
Deutsche Forschungsgemeinschaft [SFB 604/2-05/B02 to F.G. and F.H.]; The Fritz Lipmann Institute (FLI) and the Hans Knöll Institute (HKI) are members of the Science Association ‘Gottfried Wilhelm Leibniz’ (WGL) and are financially supported by the Federal Government of Germany and the State of Thuringia. Funding for open access charge: Leibniz Institute for Age Research – Fritz Lipmann Institute.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The skilful technical assistance of Claudia Franke and Sylke Fricke is gratefully acknowledged. The authors are grateful to T. Nagase, Kazusa DNA Research Institute, Chiba, Japan, and Y. Furuichi, AGENE Research Institute, Kamakura, Japan, for providing the cDNAs for human TopBP1 and RecQL4, respectively, and to J. Chen, University of Texas, Houston, USA, for making available the TopBP1 expression construct.
REFERENCES
- 1.Chu WK, Hickson ID. RecQ helicases: multifunctional genome caretakers. Nat. Rev. Cancer. 2009;9:644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- 2.Kellermayer R. The versatile RECQL4. Genet. Med. 2006;8:213–216. doi: 10.1097/01.gim.0000214457.58378.1a. [DOI] [PubMed] [Google Scholar]
- 3.Siitonen HA, Sotkasiira J, Biervliet M, Benmansour A, Capri Y, Cormier-Daire V, Crandall B, Hannula-Jouppi K, Hennekam R, Herzog D, et al. The mutation spectrum in RECQL4 diseases. Eur. J. Hum. Genet. 2009;17:151–158. doi: 10.1038/ejhg.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kitao S, Lindor NM, Shiratori M, Furuichi Y, Shimamoto A. Rothmund-thomson syndrome responsible gene, RECQL4: genomic structure and products. Genomics. 1999;61:268–276. doi: 10.1006/geno.1999.5959. [DOI] [PubMed] [Google Scholar]
- 5.Macris MA, Krejci L, Bussen W, Shimamoto A, Sung P. Biochemical characterization of the RECQ4 protein, mutated in Rothmund-Thomson syndrome. DNA Repair. 2006;5:172–180. doi: 10.1016/j.dnarep.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Xu X, Liu Y. Dual DNA unwinding activities of the Rothmund-Thomson syndrome protein, RECQ4. EMBO J. 2009;28:568–577. doi: 10.1038/emboj.2009.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capp C, Wu J, Hsieh TS. Drosophila RecQ4 has a 3'-5' DNA helicase activity that is essential for viability. J. Biol. Chem. 2009;284:30845–30852. doi: 10.1074/jbc.M109.008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki T, Kohno T, Ishimi Y. DNA helicase activity in purified human RECQL4 protein. J. Biochem. 2009;146:327–335. doi: 10.1093/jb/mvp074. [DOI] [PubMed] [Google Scholar]
- 9.Ichikawa K, Noda T, Furuichi Y. [Preparation of the gene targeted knockout mice for human premature aging diseases, Werner syndrome, and Rothmund-Thomson syndrome caused by the mutation of DNA helicases] Nippon Yakurigaku Zasshi. 2002;119:219–226. doi: 10.1254/fpj.119.219. [DOI] [PubMed] [Google Scholar]
- 10.Hoki Y, Araki R, Fujimori A, Ohhata T, Koseki H, Fukumura R, Nakamura M, Takahashi H, Noda Y, Kito S, et al. Growth retardation and skin abnormalities of the Recql4-deficient mouse. Hum. Mol. Genet. 2003;12:2293–2299. doi: 10.1093/hmg/ddg254. [DOI] [PubMed] [Google Scholar]
- 11.Mann MB, Hodges CA, Barnes E, Vogel H, Hassold TJ, Luo G. Defective sister-chromatid cohesion, aneuploidy and cancer predisposition in a mouse model of type II Rothmund-Thomson syndrome. Hum. Mol. Genet. 2005;14:813–825. doi: 10.1093/hmg/ddi075. [DOI] [PubMed] [Google Scholar]
- 12.Sangrithi MN, Bernal JA, Madine M, Philpott A, Lee J, Dunphy WG, Venkitaraman AR. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell. 2005;121:887–898. doi: 10.1016/j.cell.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 13.Matsuno K, Kumano M, Kubota Y, Hashimoto Y, Takisawa H. The N-terminal noncatalytic region of Xenopus RecQ4 is required for chromatin binding of DNA polymerase alpha in the initiation of DNA replication. Mol. Cell Biol. 2006;26:4843–4852. doi: 10.1128/MCB.02267-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- 15.Xu X, Rochette PJ, Feyissa EA, Su TV, Liu Y. MCM10 mediates RECQ4 association with MCM2-7 helicase complex during DNA replication. EMBO J. 2009;28:3005–3014. doi: 10.1038/emboj.2009.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 17.Bartels C, Xia T, Billeter M, Guntert P, Wuthrich K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 18.Schlott B, Wohnert J, Icke C, Hartmann M, Ramachandran R, Guhrs KH, Glusa E, Flemming J, Gorlach M, Grosse F, et al. Interaction of Kazal-type inhibitor domains with serine proteinases: biochemical and structural studies. J. Mol. Biol. 2002;318:533–546. doi: 10.1016/S0022-2836(02)00014-1. [DOI] [PubMed] [Google Scholar]
- 19.Ohlenschlager O, Ramachandran R, Flemming J, Guhrs KH, Schlott B, Brown LR. NMR secondary structure of the plasminogen activator protein staphylokinase. J. Biomol. NMR. 1997;9:273–286. doi: 10.1023/a:1018678925512. [DOI] [PubMed] [Google Scholar]
- 20.Herrmann T, Guntert P, Wuthrich K. Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J. Biomol. NMR. 2002;24:171–189. doi: 10.1023/a:1021614115432. [DOI] [PubMed] [Google Scholar]
- 21.Herrmann T, Guntert P, Wuthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 22.Vuister GW, Bax A. Quantitative J correlation: a new approach for measuring homonuclear three-bond J(HNHa) coupling constants in 15 N-enriched proteins. J. Am. Chem. Soc. 1993;115:7772–7777. [Google Scholar]
- 23.Guntert P, Billeter M, Ohlenschlager O, Brown LR, Wuthrich K. Conformational analysis of protein and nucleic acid fragments with the new grid search algorithm FOUND. J. Biomol. NMR. 1998;12:543–548. doi: 10.1023/A:1008391403193. [DOI] [PubMed] [Google Scholar]
- 24.Luginbuhl P, Guntert P, Billeter M, Wuthrich K. The new program OPAL for molecular dynamics simulations and energy refinements of biological macromolecules. J. Biomol. NMR. 1996;8:136–146. doi: 10.1007/BF00211160. [DOI] [PubMed] [Google Scholar]
- 25.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 26.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 27.Herold S, Wanzel M, Beuger V, Frohme C, Beul D, Hillukkala T, Syvaoja J, Saluz HP, Haenel F, Eilers M. Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol. Cell. 2002;10:509–521. doi: 10.1016/s1097-2765(02)00633-0. [DOI] [PubMed] [Google Scholar]
- 28.Borth N, Massier J, Franke C, Sachse K, Saluz HP, Hanel F. Chlamydial protease CT441 interacts with SRAP1 co-activator of estrogen receptor alpha and partially alleviates its co-activation activity. J. Steroid Biochem. Mol. Biol. 2010;119:89–95. doi: 10.1016/j.jsbmb.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 29.Wollmann Y, Schmidt U, Wieland GD, Zipfel PF, Saluz HP, Hanel F. The DNA topoisomerase IIbeta binding protein 1 (TopBP1) interacts with poly (ADP-ribose) polymerase (PARP-1) J. Cell Biochem. 2007;102:171–182. doi: 10.1002/jcb.21292. [DOI] [PubMed] [Google Scholar]
- 30.Leung CC, Gong Z, Chen J, Glover JN. Molecular basis of BACH1/FANCJ recognition by TopBP1 in DNA replication checkpoint control. J. Biol. Chem. 2011;286:4292–4301. doi: 10.1074/jbc.M110.189555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wishart DS, Sykes BD. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 32.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Billeter M, Qian YQ, Otting G, Muller M, Gehring W, Wuthrich K. Determination of the nuclear magnetic resonance solution structure of an Antennapedia homeodomain-DNA complex. J. Mol. Biol. 1993;234:1084–1093. doi: 10.1006/jmbi.1993.1661. [DOI] [PubMed] [Google Scholar]
- 34.Clarke ND, Kissinger CR, Desjarlais J, Gilliland GL, Pabo CO. Structural studies of the engrailed homeodomain. Protein Sci. 1994;3:1779–1787. doi: 10.1002/pro.5560031018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li T, Stark MR, Johnson AD, Wolberger C. Crystal structure of the MATa1/MAT alpha 2 homeodomain heterodimer bound to DNA. Science. 1995;270:262–269. doi: 10.1126/science.270.5234.262. [DOI] [PubMed] [Google Scholar]
- 36.Joshi R, Passner JM, Rohs R, Jain R, Sosinsky A, Crickmore MA, Jacob V, Aggarwal AK, Honig B, Mann RS. Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell. 2007;131:530–543. doi: 10.1016/j.cell.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gehring WJ, Qian YQ, Billeter M, Furukubo-Tokunaga K, Schier AF, Resendez-Perez D, Affolter M, Otting G, Wuthrich K. Homeodomain-DNA recognition. Cell. 1994;78:211–223. doi: 10.1016/0092-8674(94)90292-5. [DOI] [PubMed] [Google Scholar]
- 38.Billeter M, Guntert P, Luginbuhl P, Wuthrich K. Hydration and DNA recognition by homeodomains. Cell. 1996;85:1057–1065. doi: 10.1016/s0092-8674(00)81306-9. [DOI] [PubMed] [Google Scholar]
- 39.Thangavel S, Mendoza-Maldonado R, Tissino E, Sidorova JM, Yin J, Wang W, Monnat RJ, Jr, Falaschi A, Vindigni A. Human RECQ1 and RECQ4 helicases play distinct roles in DNA replication initiation. Mol. Cell Biol. 2010;30:1382–1396. doi: 10.1128/MCB.01290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biamonti G, Giacca M, Perini G, Contreas G, Zentilin L, Weighardt F, Guerra M, Della Valle G, Saccone S, Riva S, et al. The gene for a novel human lamin maps at a highly transcribed locus of chromosome 19 which replicates at the onset of S-phase. Mol. Cell Biol. 1992;12:3499–3506. doi: 10.1128/mcb.12.8.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Connelly MA, Zhang H, Kieleczawa J, Anderson CW. The promoters for human DNA-PKcs (PRKDC) and MCM4: divergently transcribed genes located at chromosome 8 band q11. Genomics. 1998;47:71–83. doi: 10.1006/geno.1997.5076. [DOI] [PubMed] [Google Scholar]
- 42.Keller C, Ladenburger EM, Kremer M, Knippers R. The origin recognition complex marks a replication origin in the human TOP1 gene promoter. J. Biol. Chem. 2002;277:31430–31440. doi: 10.1074/jbc.M202165200. [DOI] [PubMed] [Google Scholar]
- 43.Görlach M, Wittekind M, Beckman RA, Mueller L, Dreyfuss G. Interaction of the RNA-binding domain of the hnRNP C proteins with RNA. EMBO J. 1992;11:3289–3295. doi: 10.1002/j.1460-2075.1992.tb05407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williamson RA, Carr MD, Frenkiel TA, Feeney J, Freedman RB. Mapping the binding site for matrix metalloproteinase on the N-terminal domain of the tissue inhibitor of metalloproteinases-2 by NMR chemical shift perturbation. Biochemistry. 1997;36:13882–13889. doi: 10.1021/bi9712091. [DOI] [PubMed] [Google Scholar]
- 45.Brennan RG, Matthews BW. The helix-turn-helix DNA binding motif. J. Biol. Chem. 1989;264:1903–1906. [PubMed] [Google Scholar]
- 46.Wolberger C. Homeodomain interactions. Curr. Opin. Struct. Biol. 1996;6:62–68. doi: 10.1016/s0959-440x(96)80096-0. [DOI] [PubMed] [Google Scholar]
- 47.Gehring WJ, Muller M, Affolter M, Percival-Smith A, Billeter M, Qian YQ, Otting G, Wuthrich K. The structure of the homeodomain and its functional implications. Trends Genet. 1990;6:323–329. doi: 10.1016/0168-9525(90)90253-3. [DOI] [PubMed] [Google Scholar]
- 48.Gehring WJ. The homeobox in perspective. Trends Biochem. Sci. 1992;17:277–280. doi: 10.1016/0968-0004(92)90434-b. [DOI] [PubMed] [Google Scholar]
- 49.Hanes SD, Brent R. DNA specificity of the bicoid activator protein is determined by homeodomain recognition helix residue 9. Cell. 1989;57:1275–1283. doi: 10.1016/0092-8674(89)90063-9. [DOI] [PubMed] [Google Scholar]
- 50.Treisman J, Gonczy P, Vashishtha M, Harris E, Desplan C. A single amino acid can determine the DNA binding specificity of homeodomain proteins. Cell. 1989;59:553–562. doi: 10.1016/0092-8674(89)90038-x. [DOI] [PubMed] [Google Scholar]
- 51.Falaschi A, Abdurashidova G, Biamonti G. DNA replication, development and cancer: a homeotic connection? Crit. Rev. Biochem. Mol. Biol. 2010;45:14–22. doi: 10.3109/10409230903365608. [DOI] [PubMed] [Google Scholar]
- 52.Comelli L, Marchetti L, Arosio D, Riva S, Abdurashidova G, Beltram F, Falaschi A. The homeotic protein HOXC13 is a member of human DNA replication complexes. Cell Cycle. 2009;8:454–459. doi: 10.4161/cc.8.3.7649. [DOI] [PubMed] [Google Scholar]
- 53.de Stanchina E, Gabellini D, Norio P, Giacca M, Peverali FA, Riva S, Falaschi A, Biamonti G. Selection of homeotic proteins for binding to a human DNA replication origin. J. Mol. Biol. 2000;299:667–680. doi: 10.1006/jmbi.2000.3782. [DOI] [PubMed] [Google Scholar]
- 54.Salsi V, Ferrari S, Ferraresi R, Cossarizza A, Grande A, Zappavigna V. HOXD13 binds DNA replication origins to promote origin licensing and is inhibited by geminin. Mol. Cell Biol. 2009;29:5775–5788. doi: 10.1128/MCB.00509-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marchetti L, Comelli L, D'Innocenzo B, Puzzi L, Luin S, Arosio D, Calvello M, Mendoza-Maldonado R, Peverali F, Trovato F, et al. Homeotic proteins participate in the function of human-DNA replication origins. Nucleic Acids Res. 2010;38:8105–8119. doi: 10.1093/nar/gkq688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miotto B, Graba Y. Control of DNA replication: a new facet of Hox proteins? Bioessays. 2010;32:800–807. doi: 10.1002/bies.201000048. [DOI] [PubMed] [Google Scholar]
- 57.Otting G, Qian YQ, Billeter M, Muller M, Affolter M, Gehring WJ, Wuthrich K. Protein–DNA contacts in the structure of a homeodomain–DNA complex determined by nuclear magnetic resonance spectroscopy in solution. EMBO J. 1990;9:3085–3092. doi: 10.1002/j.1460-2075.1990.tb07505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Billeter M, Qian Y, Otting G, Muller M, Gehring WJ, Wuthrich K. Determination of the three-dimensional structure of the Antennapedia homeodomain from Drosophila in solution by 1H nuclear magnetic resonance spectroscopy. J. Mol. Biol. 1990;214:183–197. doi: 10.1016/0022-2836(90)90155-f. [DOI] [PubMed] [Google Scholar]
- 59.Nanda V, Brand L. Aromatic interactions in homeodomains contribute to the low quantum yield of a conserved, buried tryptophan. Proteins. 2000;40:112–125. doi: 10.1002/(sici)1097-0134(20000701)40:1<112::aid-prot130>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 60.Noyes MB, Christensen RG, Wakabayashi A, Stormo GD, Brodsky MH, Wolfe SA. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell. 2008;133:1277–1289. doi: 10.1016/j.cell.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Im JS, Ki SH, Farina A, Jung DS, Hurwitz J, Lee JK. Assembly of the Cdc45-Mcm2-7-GINS complex in human cells requires the Ctf4/And-1, RecQL4, and Mcm10 proteins. Proc. Natl. Acad. Sci. USA. 2009;106:15628–15632. doi: 10.1073/pnas.0908039106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kanter DM, Kaplan DL. Sld2 binds to origin single-stranded DNA and stimulates DNA annealing. Nucleic Acids Res. 2010;39:2580–2592. doi: 10.1093/nar/gkq1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bruck I, Kanter DM, Kaplan DL. Enabling association of the GINS protein tetramer with the mini chromosome maintenance (Mcm)2-7 protein complex by phosphorylated Sld2 protein and single-stranded origin DNA. J Biol. Chem. 2011;286:36414–36426. doi: 10.1074/jbc.M111.282822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nicholas KB, Nicholas HB, Jr, Deerfield DW., II GeneDoc: analysis and visualization of genetic variation. EMBNEW News. 1997;4:1–4. [Google Scholar]
- 67.Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32:W321–W326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.