Abstract
The plant-specific, B3 domain-containing transcription factor ABSCISIC ACID INSENSITIVE3 (ABI3) is an essential component of the regulatory network controlling the development and maturation of the Arabidopsis thaliana seed. Genome-wide chromatin immunoprecipitation (ChIP-chip), transcriptome analysis, quantitative reverse transcriptase–polymerase chain reaction and a transient promoter activation assay have been combined to identify a set of 98 ABI3 target genes. Most of these presumptive ABI3 targets require the presence of abscisic acid for their activation and are specifically expressed during seed maturation. ABI3 target promoters are enriched for G-box-like and RY-like elements. The general occurrence of these cis motifs in non-ABI3 target promoters suggests the existence of as yet unidentified regulatory signals, some of which may be associated with epigenetic control. Several members of the ABI3 regulon are also regulated by other transcription factors, including the seed-specific, B3 domain-containing FUS3 and LEC2. The data strengthen and extend the notion that ABI3 is essential for the protection of embryonic structures from desiccation and raise pertinent questions regarding the specificity of promoter recognition.
INTRODUCTION
Seeds provide the basis for food, feed and bio-based economy. Their biological role is to nourish and protect the developing embryo and to facilitate the distribution of the progeny. The development of the seed involves the formation of both the embryo and the endosperm, followed by their maturation, their acquisition of first desiccation tolerance and then dormancy. These processes require a spatially and temporally well-regulated programme of gene expression. Transcription factors (TFs) such as LEAFY COTYLEDON1 and 2 (LEC1 and LEC2), ABSCISIC ACID INSENSITIVE3 (ABI3) and FUSCA3 (FUS3) are known to be key components of seed development (1–3 and references therein). LEC1, which is part of a CAAT-box binding complex (4), is required for both embryo development and seed maturation, while the B3 domain-containing ABI3, FUS3 and LEC2 TFs operate rather exclusively in seed maturation. All of these TFs are also important for a number of hormone-controlled processes (5). Furthermore, regulation of seed maturation includes post-transcriptional (6) and post-translational (7) processes as well as chromatin remodeling (8–11). The chromatin remodeling factor PICKLE was shown to repress the expression of the ABI3 gene (12), and the controlled degradation of the ABI3 protein is triggered by the ABI3-INTERACTING PROTEIN2 and mediated by the 26S proteasome pathway (7). In addition to the developmentally controlled transcription of ABI3 during seed development, its activity is controlled by alternative splicing (13).
Potential targets for ABI3, FUS3 and LEC2 include genes encoding seed storage proteins, oleosins and LEA proteins (14–17). The promoters associated with several of these genes contain the RY motif CATGCA, which acts as a binding site for the B3 domain (14,18–20). Both the dicotyledonous embryo and the monocotyledonous endosperm also exploit G-box-like (GBL) and CTTT-like motifs, which interact, respectively, with the TFs of the bZIP- and DOF class (3). The regulatory interaction between RY- and ABI3-like TFs also extends to non-seed plants such as ferns and mosses (21,22). ABI3-like proteins are important during the recovery phase from desiccation in the bryophyte Physcomitrella (23). Moreover, ABI3 was found to be active in dormant buds of poplar (24,25).
The ABI3 protein includes the three basic domains B1, B2 and B3, along with its N-terminal A1 domain. The latter is thought to be important for activation, while B1 and B2 may well be responsible for nuclear localization and interactions with other proteins (26–30). The B3 domain of ABI3 and its maize homologue VP1 is known to bind the RY motif of seed expressed genes (19,31).
The identification of ABI3 targets to date has been based on transcript analysis in either loss-of-function mutants (32) or over-expressing transgenics (16,33,34). These approaches have neither considered the genome in its entirety nor been able to distinguish between direct and indirect effects. This drawback can be overcome partially by using an inducible system for the TF (17,35). Chromatin immunoprecipitation (ChIP) methods (36,37) are well suited to the analysis of direct interactions between TFs and their target promoters and have been widely exploited to study regulatory networks in yeast (38–40). The extension of ChIP methodology to give genome-wide coverage (ChIP-chip) in plants has been described for a number of TFs (41–48). Herein, we report the application of genome-wide ChIP-chip and transcriptome analysis for the identification of a set of 98 genes forming a basic ABI3 regulon. Presumptive targets were further verified using quantitative reverse transcriptase–polymerase chain reaction (qRT–PCR) and/or transient promoter activation. The nature of the genes within the regulon emphasizes the importance of ABI3 during seed maturation. RYL and/or GBL motifs were found in almost all target promoters. However, the genome-wide frequency of such promoters raises intriguing questions concerning selective promoter activation by ABI3.
MATERIALS AND METHODS
Plant culture
Arabidopsis thaliana Col-0 was grown in soil under long day conditions (16 h light/8 h dark) at 22°C and 60% humidity for ChIP.
Transgenic 35S::ABI3:GR or 35S::FUS3:GR as well as Col-0 wild-type seedlings were grown on selective or pure MS medium, respectively, for 12 days. For induction, seedlings were transferred to liquid MS medium and treated with either 10 µM abscisic acid (ABA) and/or 30 µM dexamethasone (DEX) or the solvent ethanol as control. After 4 h (for array analysis) or 6 h (for qRT–PCR) shaking at 22°C, seedlings were frozen in liquid nitrogen.
Generation of transgenic lines
The ABI3 and FUS3 coding regions were amplified from cDNA and cloned through Gateway vector pDONR201 into pR1R2ΔGR (49) and used to generate transgenic plant lines (50) designated as 35 S::ABI3:GR and 35 S::FUS3:GR. Lines with single locus insertions were analyzed further. The primers used to amplify ABI3 and FUS3 coding regions (att sites in lower case) are
ABI3_for 5′-ggggacaagtttgtacaaaaaagcaggctATGAAAAGCTTGCATGTGGCGGCCA
ABI3_rev 5′-ggggaccactttgtacaagaaagctgggtGTTTAACAGTTTGAGAAGTTGGT
FUS3_for5′-ggggacaagtttgtacaaaaaagcaggctATGGTTGATGAAAATGTGGAAACCAATGCC
FUS3_rev 5′-ggggaccactttgtacaagaaagctgggtGGTAGAAGTCATCGAGAGAGATAT
RNA and cDNA
Total RNA of seedlings was prepared using the RNeasy Plant Mini Kit (Qiagen). RNA for qRT–PCR analysis of ABI3 expression during seed development was extracted using the hot borate method (51) from seeds dissected from the siliques at 5, 7, 9, 11, 13, 15, 17 and 19 days after flowering. One to three micrograms of total RNA isolated from seeds or ABI3::GR seedlings (see above) were treated with DNase I (Roche) and transcribed into cDNA (Revert AidTM H Minus First Strand cDNA Synthesis Kit; Fermentas).
Quantitative real-time PCR
Quantitatve real-time PCRs (40 cycles) were performed in triplicate in 384-well plates using SYBR Green Master Mix and a 7900HT real-time PCR system (Applied Biosystems). The used cycling conditions were as follows: initial denaturation step (95°C for 10 min), 40 cycles (95°C for 15 s; 60°C for 1 min) and final dissociation (95°C for 15 s; 60°C for 15 s; 95°C for 15 s). Data were processed using SDS 2.0 software (Applied Biosystems). Primers (Supplementary Table S3) were designed using Primer3 software and obtained from Metabion (Martinsried, Germany). Relative transcript levels were normalized to the expression of the reference gene EF1a (AT5G60390) in all seedling experiments and to UBQ 10 (AT4G05320) for ABI3 expression using primers described in Czechowski et al. (52).
Antibodies
To raise ABI3-specific antibodies, a fragment encoding amino acids 2–237 (omitting the B3 domain) of ABI3 (AT3G24650) served as antigen. The corresponding cDNA fragment was cloned into the expression plasmid pET23a (Novagen) and the His-tagged recombinant protein was purified from Escherichia coli BL21(DE3)LysS cells using affinity chromatography on Ni-agarose (Qiagen). ABI3-specific antibodies were raised in rabbits by injecting 800 µg antigen at 0, 28 and 38 days and affinity-purified on antigen-CNBr sepharose columns (GE healthcare). The antibody detects 0.1 ng ABI3 protein in western blots.
ChIP-chip and data analysis
Chromatin isolation and hybridization to SAP macroarray were done as described by Junker et al. (53). In brief, chromatin was isolated from Col-0 seeds (11–14 DAP) after cross-linking proteins and DNA with 1% formaldehyde for 20 min under vacuum and termination of the reaction with glycine (final concentration 125 mM). Seeds were crashed with a Potter homogenizer, filtered and the nuclei collected by centrifugation. After dissolving nuclear membranes with Triton X100 (1%), chromatin was collected by high-speed centrifugation and fragmented by ultra sonic treatment to an average size of 500 bp. An aliquot (0.5 µg) was saved to serve as input control. Ten micrograms of chromatin was combined with 2 µg anti-ABI3 antibody and the antibody–chromatin complex captured on protein A-coated magnetic beads (Invitrogen). The TF-DNA complex was eluted from beads using 1% sodium dodecyl sulfate (15 min at 65°C). Eluates and input chromatin were incubated overnight at 65°C to reverse the cross-links and DNA fragments were purified by QIAquick PCR Purification Kit (Qiagen). Chromatin fragments were blunted and ligated with adaptors oJW102 and oJW103 (jura.wi.mit.edu/young_public/hESregulation/ChIP.html). Adaptor-ligated fragments were amplified using oJW102 as primer. The SAP-promoter array (Systemic analysis of Arabidopsis Promoters (41)) was constructed by spotting 11 904 amplified promoter fragment on a set of two nylon membranes. Hybridization to this array was performed with [α-33P]dCTP labeled DNA fragments.
Five ABI3 ChIP-chip experiments using the SAP macroarray were performed, each consisting of a pair of hybridizations with amplified input chromatin and immuno-precipitated chromatin. Quantile normalization (54) was applied separately to the dataset of input and immuno-precipitated chromatin and log2 enrichment factors for most (>99.5%) of the spotted amplicons in all five experiments were calculated. A Hidden Markov model with scaled transition matrices (SHMM) (55) was applied to identify potential ABI3-bound regions. The 1125 top scoring amplicons were identified in each of the five experiments. Amplicons present in the top lists of at least four experiments were considered as putative target regions of ABI3.
Two independent biological experiments were performed using the Agilent Arabidopsis Genome Microarray Kit, Design Set 2x244K. Fragments of input and immuno-precipitated chromatin were amplified as above and labeled with Cy3-dUTP and Cy5-dUTP using the Agilent Genomic DNA labeling Kit (Agilent Technologies). Two color hybridization and signal extraction were done by ATLAS GmbH, Berlin, using standard procedures.
Log2 ratios of immuno-precipitated chromatin versus input chromatin of both experiments were quantile normalized (54) and a basic two-state Hidden Markov model (HMM) (55) was applied to identify potential ABI3-bound regions within −750 to +250 bp relative to the transcription start site (TSS). For each experiment, putative ABI3 target tiles were determined using state-posterior decoding (56), labeling a tile either as ABI3 target or non-target. 501 ABI3 target tiles occurring in both experiments were considered further.
Transcriptome analysis by Agilent 44K microarray hybridization
Two wild-type and two ABI3 expression experiments were performed using Agilent Arabidopsis V4 4x44K microarrays. Total RNA was isolated as described above. The microarrays were processed by ATLAS GmbH, Berlin. Measured signals were quantile normalized (54). Regulated candidates were identified using the Rank Product package of R (57,58) for two scenarios: (i) regulation by ABI3 alone, using DEX treated 35S::ABI3:GR seedlings as sample and mock-treated 35S::ABI3:GR as well as DEX-treated Col-0 seedlings as control and (ii) regulation by ABI3 in the presence of ABA, using ABA- and DEX-treated 35S::ABI3:GR seedlings as sample and ABA-treated 35S::ABI3:GR as well as ABA- and DEX-treated Col-0 samples as control (false discovery rate (FDR) max = 10%).
Transient promoter activation
Candidate ABI3 target promoters including non-translated leader sequence and a few N-terminal codons were amplified from genomic DNA and fused in-frame with the β-glucuronidase gene (GUS) of the plasmid pGUS1 (59). Primer sequences are given as Supplementary Material (Table S4). Constructs (10 µg) were transiently transformed into protoplasts of an embryogenic cell culture of Arabidopsis. To test promoter activation, an ABI3 expressing construct (20) was co-transformed. Transformed protoplasts were incubated at 21°C in the dark for 48 h without additional ABA. GUS activity was determined using Tropix GUS light-kit (Applied Biosystems) according to the manufacturer and measured in a Lumat LB9501 lumimeter (Berthold, Bad Wildbach, Germany). The ratio of activity with and without ABI3 was calculated. A ratio of larger than 4 is considered as activation.
Calculation of the seed maturation index
Expression values assigned to anatomical categories in Genevestigator version 2 (60,61) were used to characterize the expression of genes represented on the ATH1 Affymetrix chip. A maturation index (MI), ranging from 0 to 100, was calculated by dividing 100 times the expression value assigned to the seed category (E[seed]) with the sum of expression values of all tissue categories of the same or higher level (E[all]) (MI = 100 × E[seed]/∑E[all]). Characteristic values of the MI distribution are median = 3.44, 95% quantile = 9.2 and 99% quantile = 31.0. Genes with MI ≥ 31 are considered to be expressed in a seed-specific manner.
De novo motif discovery and motif search
Dispom (62) was used for de novo motif discovery. In analogy to the analysis of the ChIP-chip data, we used the promoter region from −750 to +250 bp relative to the most upstream annotated TSS of each gene for the analysis. As foreground dataset we used promoters of the ABI3 regulon, while we used 1000 randomly drawn promoters as background datasets. Default parameters of Dispom were applied, except that the initial motif length was set to 10 bases. After identification of a significantly (P ≤ 10−4) enriched motif, Dispom was run again with a new background dataset containing the motif(s) identified in previous run(s) with identical frequency as in the foreground dataset. Motif descriptions with respect to sequence, position, strand and sequence context within the promoter were used to predict binding sites in the most upstream promoters of all protein encoding genes in the Arabidopsis genome.
RESULTS
Identification of DNA fragments bound by ABI3
The use of ChIP-chip methodology allowed for the identification of DNA fragments interacting with ABI3. In the A. thaliana seed, ABI3 expression peaked around 11–14 days after pollination (DAP), as determined by qRT–PCR (Supplementary Figure S1) (61). Chromatin was isolated from wild-type seeds at this developmental stage and challenged with an anti-ABI3 polyclonal antibody raised against its N-terminal fragment (amino acids 2–237), excluding the B3 domain shared with FUS3 and LEC2. Cross-reactivity with FUS3, LEC2 and LEC1 has been tested in enzyme-linked immunosorbent assay and western blot (Supplementary Figures S2 and S3). The analytical ChIP quality of the antibody was demonstrated by its ability to enrich by up to 10-fold from immuno-precipitated chromatin, a promoter fragment of the ABI3 target gene AT2S3 (Supplementary Figure S4) (15,35). The identification of genomic DNA fragments binding with ABI3 was achieved using both the SAP-promoter macroarray (41) and the Agilent 2x244K genome tiling arrays. The SAP array covers about one-third of the intergenic regions of the genome. Theoretically, candidates identified with this platform should be also included in the whole genomic Agilent 2x244K array analysis. However, both datasets do not fully overlap, most likely due to the different type of arrays (spotted large intergenic regions versus 60 bp oligonucleotides). We therefore decided to exploit ChIP-chip candidates of both platforms. Five independent hybridizations (three biological and two technical replicates) were performed using the SAP array, producing a set of 102 promoter fragments which occurred within the 1125 top scoring fragments in at least four of the five replicates. A randomly chosen set of 1125 fragments would have resulted on average in the selection of a single interacting fragment. The criterion of at least four of five replicates was chosen to limit the number of potential false-positive predictions of top scoring fragments. For the five independent hybridizations on the SAP-promoter arrays, the intersection of the 1125 top scoring fragments for two hybridizations ranged from 199 to 318 potential target promoters between two hybridizations (average number of promoters: 223). A gene promoter was only identified as interacting candidate if the selected fragment is located between −750 and +250 bp relative to its putative TSS (as defined by mRNA features in the TAIR8 GFF file). Applying this criterion, the 102 SAP fragments identified a set of 121 gene promoters (Supplementary Table S1a). The enrichment of two of them (AT4G25140 and AT5G54740)—in addition to the napin AT2S promoter—was controlled by qRT–PCR (Supplementary Figure S4). For the Agilent array-based experiment, chromatin from two independent biological samples was used as the hybridization probe. The two experiments resulted in 2710 and 1830 oligonucleotides, respectively, and the 501 tiles enriched in both replicates were mapped to regions spanning −750 to +250 bp relative to putative TSSs, as above, resulting in the identification of 160 positive tiles (Supplementary Table S1b). A further 243 tiles located fully within coding sequences and the other 98 to non-genic regions. The 160 positive tiles identified as potential ABI3 targets could be assigned to 87 gene promoters, 31 of which were identical to members of the SAP set. Both platforms overlap and complement each other and both contributed to the identification of ABI3 target genes. The combined set of 177 putative ABI3 target sequences is detailed in Figure 1 and Supplementary Table S2.
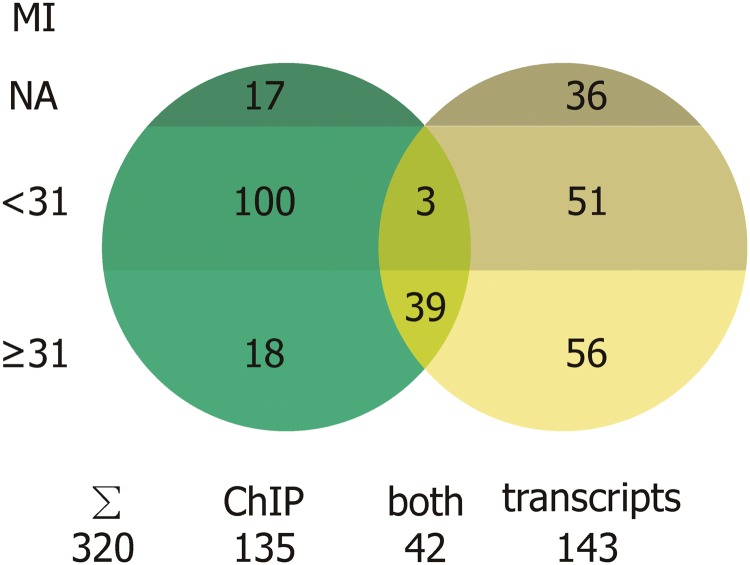
Figure 1.
Numbers of ABI3 target gene candidates identified by ChIP-chip (bound promoters) and transcriptome analysis (regulated genes). The candidate genes were subdivided according to their expression in seeds: Genes with seed MI > 31 exhibit a strong seed-specific expression (definition of MI see below); genes with MI < 31 are here considered as non-seed-specific genes; NA, MI not available. The overlapping 42 genes are called core regulon. ChIP-chip was performed with chromatin isolated from wild-type seeds; transcript analysis was performed with transgenic seedlings transformed with an ABI3-inducible gene construct.
Identification of ABI3-regulated genes
To exclude the interfering effects of other B3 domain TFs such as FUS3 and LEC2 with ABI3 in seeds, transcriptome analyses were performed in seedlings, exploiting an inducible ABI3 over-expression system in which ABI3 was fused with a glucocorticoid receptor domain sequence (GR) under the control of the CaMV 35S promoter (63). The ABI3::GR fusion protein remains in the cytosol until the GR ligand DEX is supplied, to initiate its translocation to the nucleus. Since ABI3-induced transcription in seedlings requires the presence of ABA (15,35,64), known to be elevated in seeds, four samples were compared in seedlings: untreated, ABA treated, DEX treated and treated with a combination of ABA and DEX. RNA was isolated after 4 h of incubation and used to probe an Agilent 4x44K microarray. Array data were analyzed using the RANK product method (57). An FDR of 10% was used as threshold for the selection of ABI3-responsive genes. This resulted in the identification of 185 ABI3-responsive gene products, of which 165 were up- and 20 down-regulated (Figure 1, Supplementary Table S1c).
Genes identified simultaneously by both ChIP-chip and transcript analysis
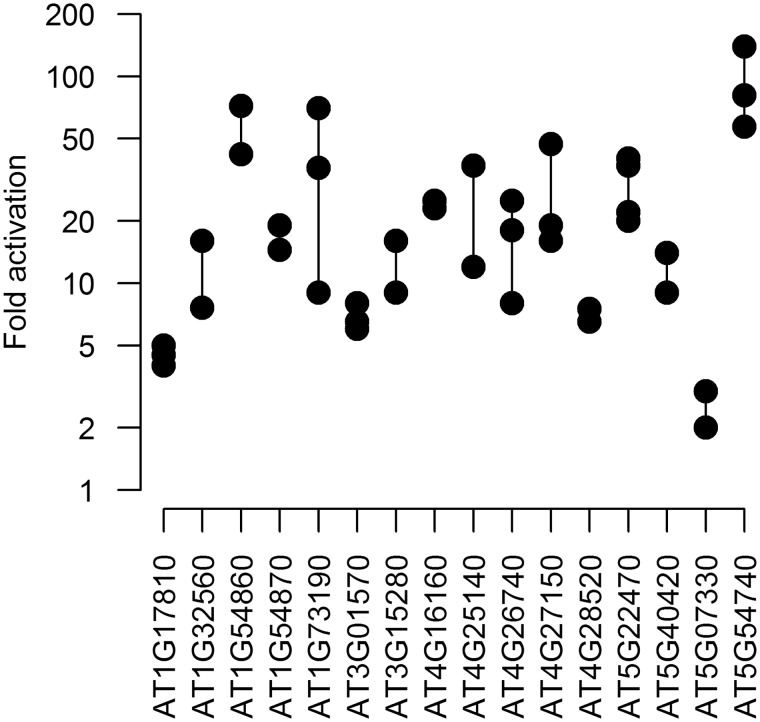
The ChIP-chip analysis identified 177 promoters binding to ABI3 and the transcriptome approach 185 regulated genes. The set of 42 genes (all up-regulated by ABI3) common to both sets (Figure 1 and Supplementary Table S2) was considered to be core of ABI3 target candidates. These 42 genes comprise 24 and 23% of genes identified by the ChIP-chip and transcriptome approach, respectively. A similar low extent of overlap between ChIP-chip and transcriptome analysis has been experienced in other studies (43,44,48). In total, 16 promoters of these 42 genes were transcriptionally fused to the uidA (GUS) reporter gene to allow the monitoring of promoter activation in a transient protoplast co-expression assay (19,65). Fifteen of them were activated by ABI3 by at least 4-fold and up to 85-fold (Figure 2 and Supplementary Tables S2 and S4). These results increased the level of confidence that the 42 candidate genes are genuine ABI3 targets.
Figure 2.
Promoter activation by ABI3 in transient assay. Protoplasts of an embryogenic Arabidopsis cell culture were transformed with promoter–reporter constructs and GUS activity was determined in the absence and presence of an ABI3 activator construct. Fold activation of individual experiments are given as dots.
Targets of ABI3 are specifically expressed in the seed
Since ABI3 is specifically expressed in the maturing seed, the expectation was that its targets would show a similar specificity in their expression. This expectation was evaluated by an analysis of the A. thaliana anatomy section of the Genevestigator database v2 (60,61), which is based on expression data recovered from a large number of microarray experiments. A seed MI was devised by dividing the expression level in the seed during developmental stages 6–10 by the sum of expression levels for all tissue categories. An MI of 31 corresponds to the 99% quantile, meaning that only 1% of the 20 041 genes assigned unambiguously on the ATH1 genes having an MI > 31, indicating strong seed specificity. An MI of 9.2 corresponds to the 95% quantile. The MI of ABI3 is 56. Of the 42 candidate ABI3 target genes, 39 recorded an MI > 31 (Figure 1 and Table 1) and all 42 had an MI > 9.2 (Supplementary Table S2).
Table 1.
MI contingency table and membership of the ABI3 core regulon
| MI |
Total |
||||||
|---|---|---|---|---|---|---|---|
| <31 |
≥31 |
||||||
| Number | % | Number | % | Number | % | ||
| ABI3 core regulon | Yes | 3 | 0.015 | 39 | 17.6 | 42 | 0.2 |
| No | 19 817 | 99.9 | 182 | 82.3 | 19 999 | 99.8 | |
| Total | 19 820 | 221 | 20 041 | ||||
All genes in the A. thaliana genome for which an MI was calculable were classified according their membership of the ABI3 core regulon and the size of their MI. MI > 31 were considered to indicate seed maturation specific expression.
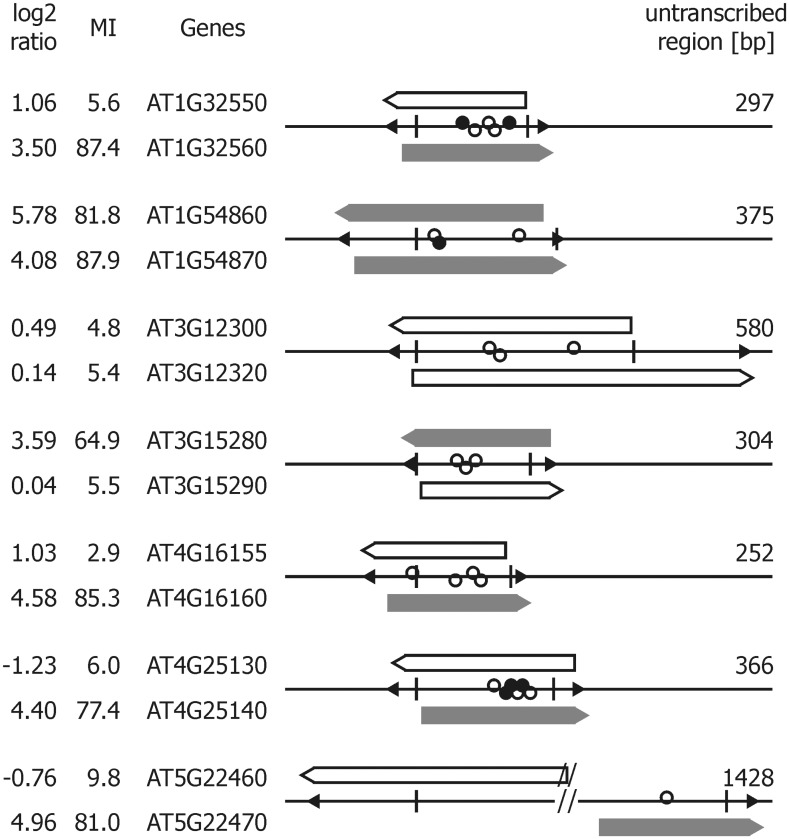
Several ABI3-bound intergenic regions are flanked by two genes in head to head constellation. If these regions are short (we used a criterion of 1500 bp, which is twice the average length of the used chromatin fragments), it might be doubtful which of both genes represents the true ABI3 target. To decide this question, seven such pairs with intergenic regions of 300–1400 bp in length have been analyzed further. The 5′ regions were cloned in both directions in front of the GUS reporter gene and their activation by ABI3 was determined using a transient promoter assay. If one of the flanking genes exhibited an MI > 31, it was this gene which became activated by ABI3, but not the other one. In one case, both flanking genes are seed specifically expressed with an MI > 31. Herein, both opposite promoters turned out to be activated by ABI3 in the transient assay (Figures 2 and 3 and Supplementary Table S4).
Figure 3.
ABI3-mediated activation of transcription of divergent, overlapping promoters. Intergenic regions of less than 1500 bp flanked by a pair of genes in opposite directions are depicted with their relevant features (filled triangles, translation start; vertical lines, proposed TSS; open circle, GBL motif; filled circles, RYL motif). Promoter regions cloned in front of GUS and assayed for activation by ABI3 in transiently transformed protoplasts of an embryogenic cell culture are shown above and below the intergenic regions (solid: ABI3 activated; open: not activated). Corresponding log2 transformed activation ratios as well as the MI and the gene ID are given on the left; the distance between the two proposed TSSs are noted on the right.
These results together confirmed that the expression of the ABI3 targets, like that of ABI3 itself, is highly seed specific.
Extended search for ABI3 targets
The strong correlation between the MI and being an ABI3 target prompted a search for additional targets among the 278 genes identified by only one of the ChIP-chip or the transcriptome analyses (Figure 1). Of these, 74 (18 from ChIP-chip and 56 from transcriptome analysis) recorded an MI > 31. The regulation by ABI3 of 69 of these was evaluated either by co-transformation with ABI3 in the promoter activation assay described above and/or were checked using qRT–PCR in transgenic seedlings expressing the inducible transgene fusion ABI3::GR (Table 2). Among the 69 genes tested, 56 were activated by ABI3 (Table 2 and Supplementary Table S2). Thus, the ABI3 regulon—as defined here—comprised 98 genes, consisting of the 42 targets present in both the ChIP-chip and transcriptome dataset, and the 56 with MI > 31 present in only one of the two datasets and evaluated by a second experimental approach (Table 3 and Supplementary Table S2).
Table 2.
The extended ABI3 regulon
| Genes | ChIP-chip | Transcript profiling | Total | % |
|---|---|---|---|---|
| MI ≥ 31 | 18 | 56 | 74 | 100 |
| Analyzed | 17 | 52 | 69 | 93 |
| ABI3 activated | 10 | 46 | 56 | 76 |
The regulation by ABI3 of genes associated with an MI > 31, either identified by ChIP-chip analysis or by genome-wide transcript profiling, was determined by qRT–PCR analysis of transgenic ABI3::GR plants or by transient promoter activation assays in transformed protoplasts. Genes induced by ABI3 constitute the extended ABI3 regulon.
Table 3.
Members of the ABI3 regulon, their putative function and the methods used for their validation
| Genes | Functions | Methods | Genes | Functions | Methods |
|---|---|---|---|---|---|
| AT4G27140 | SESA1, seed albumin1 | CT | AT5G18450 | AP2 domain TF | TP |
| AT4G27150 | SESA2, seed albumin2 | CTPA | AT5G07500 | PEI1, TF | CT |
| AT4G27160 | SESA3, seed albumin3 | CT | AT3G21370 | Beta glucosidase19 | CTP |
| AT5G54740 | SESA5, seed albumin5 | CTPA | AT5G01670 | Aldose reductase | CA |
| AT5G44120 | Cruciferin1 | CTP | AT2G28420 | Glyoxalase family | CPA |
| AT1G03880 | Cruciferin2 | CTP | AT1G12130 | Monooxgenase | TP |
| AT4G28520 | Cruciferin3 | CTA | AT1G54870 | Oxidoreductase | CTA |
| AT1G03890 | Cupin family | CT | AT3G05260 | Oxidoreductase | TP |
| AT3G22640 | Cupin family | CT | AT3G12203 | Carboxypeptidase | TP |
| AT4G36700 | Cupin family | TP | AT5G09640 | Serine carboxypeptidase | TP |
| AT2G28490 | Cupin family | CTP | AT3G54940 | Cystein proteinase, put. | TP |
| AT4G25140 | Oleosin1 | CTPA | AT2G31980 | Phytocystatin2 | TP |
| AT5G40420 | Oleosin2 | CTPA | AT4G10020 | Hydroxysteroid DH5 | TP |
| AT5G51210 | Oleosin3 | CTP | AT2G03520 | Ureide permease | TP |
| AT3G27660 | Oleosin4 | CP | AT5G22470 | Ribosyltransferase | CTPA |
| AT3G01570 | Oleosin family | CTA | AT5G62800 | Ubiquitin ligase, SINA | CA |
| AT2G25890 | Oleosin family | TP | AT3G61040 | Cytochrome P450 | TP |
| AT3G18570 | Oleosin family | TP | AT2G47770 | Benzodiazepine receptor | TP |
| AT1G32560 | LEA | CTPA | AT5G43770 | Proline-rich family | TP |
| AT2G21490 | LEA | CTP | AT2G23640 | Reticulan-like B13 | TP |
| AT3G15670 | LEA | CTP | AT3G21380 | Lectin family | CTP |
| AT5G44310 | LEA | CTP | AT5G55240 | caleosin-related | CT |
| AT4G21020 | LEA | CT | AT5G39720 | AIG2L, avirolence ind. | TP |
| AT5G06760 | LEA | CPA | AT1G67100 | LBD40, LOB domain 40 | TP |
| AT3G22490 | LEA | TPA | AT3G51810 | ATEM1, early methionin | TP |
| AT2G35300 | LEA | TP | AT2G36640 | ATECP63 | TP |
| AT2G42560 | LEA | TP | AT1G80090 | CBS family | TP |
| AT1G72100 | LEA | TP | AT5G01300 | ATPEBP | CTP |
| AT3G53040 | LEA | TP | AT1G04560 | AWPM19 family | CP |
| AT4G36600 | LEA | TP | AT1G54860 | Unknown | CTPA |
| AT2G18340 | LEA | TP | AT4G18920 | Unknown | CTP |
| AT1G73190 | Alpha-TIP | CTPA | AT5G45690 | Unknown | CTP |
| AT1G17810 | Beta-TIP | CTA | AT3G15280 | Unknown | CTA |
| AT5G07190 | ATS3, seed gene3 | TP | AT5G07330 | Unknown | CT |
| AT4G26740 | ATS1,seed gene1 | CTPA | AT1G02700 | Unknown | CT |
| AT4G09610 | GASA2 | CP | AT1G05510 | Unknown | CT |
| AT4G09600 | GASA3 | CA | AT1G27461 | Unknown | CT |
| AT2G42000 | Metallothionin-like | CTP | AT2G21820 | Unknown | CPA |
| AT2G23240 | Metallothionin-like | TP | AT5G04000 | Unknown | CA |
| AT2G15010 | Thionin | TP | AT4G27530 | Unknown | TP |
| AT2G38905 | Low temp., salt resp. | CT | AT2G19320 | Unknown | TP |
| AT4G39130 | Dehydrin family | TP | AT1G27990 | Unknown | TP |
| AT3G50980 | Dehydrin XERO1 | TP | AT1G65090 | Unknown | TP |
| AT1G48130 | ATPER, peroxiredoxin | CT | AT2G23110 | Unknown | TP |
| AT3G58450 | Stress protein | TP | AT5G16460 | Unknown | TP |
| AT4G25580 | Stress protein CAP160 | TP | AT1G29680 | Unknown | TP |
| AT3G56350 | Superoxide dismutase | TP | AT3G63040 | Unknown | TP |
| AT4G16160 | ATEOP16 | CTPA | AT1G47980 | Unknown | TP |
| AT4G26590 | ATOPT5 | CT | AT5G24130 | Unknown | TP |
Putative functions were extracted from TAIR8. Methods used C: ChIP-chip analysis, T: array-based transcriptome analysis, P: qRT–PCR and A: transient promoter activation assay.
Putative functions of ABI3 targets
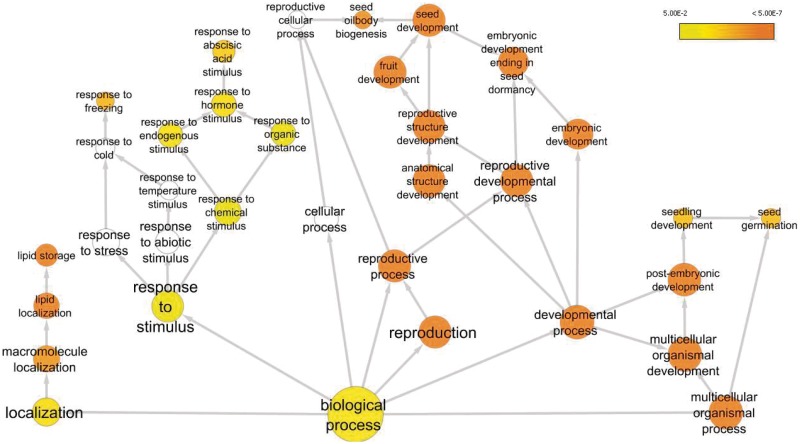
The functional characterization of the members of the ABI3 regulon was based on gene ontology (GO) terms (66). Embryo, seed and fruit development terms were over-represented, along with reproduction, development and lipid storage. Other represented functions were responses to ABA and freezing, germination and seedling development. Several genes previously implicated in seed-specific processes regulated by ABI3 and/or ABA were represented, including encoders of the 12S seed storage globulins (cupin superfamily) (67), 2S napin-like seed storage proteins (68), desiccation-related LEA proteins (69) and glycine-rich oleosins involved in seed lipid storage (70). Thus, the composition of the ABI3 regulon reflects the control of biological processes associated with seed maturation (Figure 4), although several genes encoding unknown proteins were also represented (Table 3).
Figure 4.
Members of the ABI3 regulon are involved in a range of biological processes. Over-representation for specific GO categories was assessed using the BiNGO plugin for Cytoscape. The color coding represents the significance of enrichment (P-value). The node size represents the number of ABI3 target genes in each particular category.
Sequence motifs in ABI3 target promoters
The definition of the ABI3 regulon provided an opportunity to identify ABI3-associated regulatory motifs. Promoter regions of −750 to +250 bp relative to the TSS (as used in the ChIP-chip analysis) of each of the 98 genes were subjected to an analysis by the Dispom algorithm (62). Two significantly over-represented motifs were discovered, namely GBL and RY element-like (RYL); the former resembled the well-characterized G-box-derived ABRE (ABA-responsive element) motif (ACGTG(T/G)C) (71,72) and the latter the RY/Sph motif CATGCA (20,73). Sequence logos of these motifs and their spatial distribution in the promoters of the ABI3 regulon genes are shown in Figure 5. The motifs are similar to regulatory elements described by Suzuki et al. (16,74) and were particularly abundant in the first 250 bp upstream of the TSS. RYL and GBL motif sequences within the promoters of all ChIP-chip and transcriptome candidate genes are given in Supplementary Table S2. The RY element is known to be bound by ABI3 (19), while motifs similar to GBL are implicated in the binding of bZIP-TFs, known to interact with ABI3 (26,29,30). Four classes of promoters were distinguished: those containing either both GBL and RYL, GBL only, RYL only or neither motif. The distribution of these classes within the regulon as well as in the whole genome classified according to the MI of the corresponding gene is listed in Table 4. Remarkably, within the class with both motifs present, most genes with a high MI turned out to be members of the ABI3 regulon (41 out of 49), whereas genes with low MI do not belong to the regulon (1 out of 182). This indicates that, in addition to motif content, higher order regulatory mechanisms need to be involved.
Figure 5.
Over-represented motifs and their position within the promoters of ABI3 regulon genes. GBL and RYL sequence motifs were detected in the region −750 to +250 bp relative to the TSS. Density refers to the frequency of motif occurrence in that region.
Table 4.
Motif content in the promoters of genes within the ABI3 regulon
| Promoter class | MI ≥ 31 |
MI < 31 |
||||
|---|---|---|---|---|---|---|
| Genome (g) | Regulon (r) | r/g (%) | Genome (g) | Regulon (r) | r/g (%) | |
| GBL and RYL | 49 | 41 | 84 | 182 | 1 | 0.5 |
| GBL | 64 | 35 | 55 | 1785 | 0 | 0 |
| RYL | 39 | 14 | 35 | 1449 | 1 | 0.07 |
| None | 70 | 5 | 7 | 16 399 | 1 | 0.006 |
| Total | 222 | 95 | 43 | 19 815 | 3 | 0.0015 |
Four promoter classes were recognized, according to the presence/absence of GBL and/or RYL motifs. Numbers of promoters of each class contained in the genome and the regulon are given. Promoters are further separated depending on their seed-specific activity as defined by the seed MI. MI > 31 relates to strong seed-specific promoter activity. Percentages of promoters of the regulon in relation to the whole genome content are calculated (r/g).
Genes of the ABI3 regulon are also regulated by other TFs
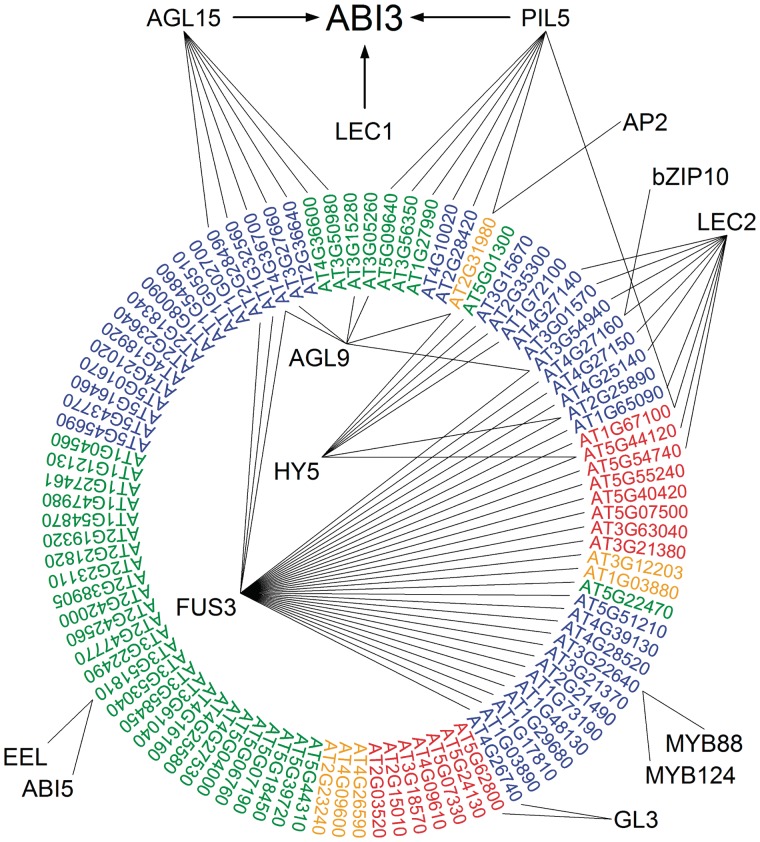
The seed-specific regulators ABI3, FUS3 and LEC2 share a B3 domain, which is required for the binding to RY elements (14,19,20). To analyze whether ABI3 and FUS3 regulate an overlapping set of targets, Arabidopsis seedlings transformed with a CaMV-35S promoter driven FUS3::GR construct were analyzed by qRT–PCR. The analysis of 92 genes of the ABI3 regulon was performed as described for the ABI3::GR transgenics (see above). A number of 24 ABI3 targets were found to be activated by FUS3 in an ABA-dependent manner, whereas 14 ABI3 targets were activated by FUS3 in an ABA-independent manner, only two of them respond to ABI3 in the absence of ABA (Supplementary Table S5). Target genes of FUS3 have also been described by Yamamoto et al. (17). Based on the criterion to be identified by both of these studies (this study and the study of Yamamoto), the overlapping 31 genes are considered to be true target genes of FUS3 (Figure 6 and Supplementary Table S5). Exploring the AGRIS ReIN database (http://arabidopsis.med.ohio-state.edu/), LEC2 as well as 11 more transcriptional regulators were found to also regulate genes of the ABI3 regulon (Figure 6 and Supplementary Table S5). The regulatory network shown in Figure 6 shows the overlap between the ABI3 regulon with the regulons of FUS3, LEC2 and the other TFs of different classes. All genes of the circle are targets of ABI3 as determined in this study. They are categorized according to the occurrence of regulatory motifs in the corresponding promoters (Supplementary Table S2). Target genes shared by ABI3, FUS3 and LEC2 are restricted to genes with RYL and GBL motifs or solely RYL motifs in their promoters. As expected, GBL motifs are dominantly present in promoters which are regulated by the bZIP factors bZIP10, ABI5, EEL and HY5. Factors AGL15, HY5 and PIL5 each share seven and AGL9 six target genes with ABI3 with low mutual overlap. An exception is the gene AT2G31980 encoding phytocystatin an inhibitor of cysteine protease, known to be a seed protein and related to abiotic stress (75). Besides by ABI3, this gene seems to be regulated by AGL9, HY5, PIF1 and AP2.
Figure 6.
The ABI3 regulon in the context of the AGRIS gene regulatory network. The network shows the overlap of the ABI3 regulon with the regulons of the B3 domain TFs FUS3 (AT3G26790) and LEC2 (AT1G28300) and other TF classes such as LEC1 (AT1G21970), HY5 (AT5G11260), AGL15 (AT5T13790), PIF1/PIL5 (AT2G20180), AP2 (AT4G36920), GL3 (AT5G41315), AGL9/SEP3 (AT1G24260), AtbZIP10 (AT4G02640), EEL (AT2G41070), ABI5 (AT2G36270), MYB88 (AT2G02820) and FLP/MYB124 (AT1G14350). Genes of the ABI3 regulon are arranged within a circle and categorized according to the occurrence of regulatory motifs in their corresponding promoters (RY+G-box: blue; RY: red; G-box: green and no motif: yellow). Additional information is given in Supplementary Table S5.
DISCUSSION
The development of the seed requires the spatial and temporal regulation of expression of many genes. TFs are central to the process of gene regulation. They operate by recognizing distinct DNA sequences, usually located upstream of the TSS of their target. A key step in the definition of the functional role of a particular TF remains the identification of its target(s), collectively referred to as a regulon. Herein, we have attempted to characterize the regulon recognized by ABI3, which is a key TF underpinning seed development. An extended combination of whole genome ChIP-chip and transcriptome experimental approaches with validation by qRT–PCR and transient promoter activation assay has led to an ABI3 regulon of at least 98 genes (Table 3). All these genes are strongly expressed in seeds, indicating their involvement in seed development. Most of the target promoters contain RYL and/or GBL motifs. However, the frequent genome-wide occurrence of these motifs in many other gene promoters raises questions about the specificity of recognition by ABI3.
Several ABI3 targets, in particular those encoding certain seed storage proteins, oleosins and LEA proteins (15,16,35,76–79) have been described before, whereas others have been identified newly in this study. Previous partially genome-wide approaches were taken to identify ABI3 target genes. These approaches exploited transgenic A. thaliana seedlings in which either ABI3 was constitutively expressed in the absence of exogenously supplied ABA (33) or in which VP1, the maize homologue of ABI3, was heterologously expressed in an abi3-6 mutant background (16). Based on arrays assaying some thousands of genes, the study of Nakashima et al. (33) allowed for the identification of 14 regulated genes, of which 10 are members of the ABI3 regulon. Suzuki et al. (16)—using an array containing 8000 genes—described 18 distinct response classes; two of these contain genes regulated exclusively by ABA, while in the remaining 16 classes, 114 genes were repressed and 150 genes were induced by VP1 (some in the presence and others in the absence of exogenously supplied ABA). Of the members of the ABI3 regulon, 42 were also identified by Suzuki et al. (16) (Supplementary Table S5). Due to the limited size of their array, not more than 48 overlapping genes could be expected. The 42 genes belong to only 3 of the 16 VP1-regulated classes and include exclusively VP1-up-regulated genes. No ABI3 repressed genes were identified in our study as being members of the regulon. Extrapolation from the 8000 genes being analyzed by Suzuki et al. (16) to the entire genome would predict a total population of around 800 VP1-regulated genes. The disparity between the size of the ABI3 regulon—as defined here with 98 genes—and the estimated number of VP1 up-regulated genes (around 800) may reflect important differences between the two experimental approaches, such as the use of a regulated expression of a homologous ABI3 gene versus the constitutive expression of the heterologous VP1 gene. Furthermore, differences concern the different cutoff values used in the transcriptome analysis as well as our additional ChIP-chip approach.
In large-scale experiments, there is a risk that false positives are included and true targets can be missed (false negatives). The ABI3 regulon, as defined here, certainly lacks a number of established true targets. One example is the gene encoding the TF HsfA9 (AT5G54070) (79). qRT–PCR successfully identified HsfA9 as an ABI3 target, but neither ChIP-chip nor transcriptome analysis picked it up. The low expression level of HsfA9 might be the reason that we failed to detect this gene in the transcriptome array analysis. Its promoter contains a CATGCATG motif at position −180 bp in front of the TSS. Nevertheless, it was not found in the ChIP-chip approach. A second example relates to the recently described SOMNUS gene (AT1G03790) (80), which was positive with respect to the transcriptome analysis, but did not exceed the activation threshold of 4-fold in the presence of ABA as detected by qRT–PCR. Within the promoter of SOMNUS (−750 to +250), neither a GBL nor an RYL motif could be detected. Both these genes encode TFs and are not included in the ABI3 regulon because their regulation by ABI3 could only be demonstrated by one experimental approach. Other putative target genes might have been missed due to the rather high MI threshold applied and by setting as a criterion that at least two experimental approaches were required to admit membership of the regulon. Furthermore, transcriptomics was done in seedlings with inducible ABI3 gene expression. Potential cooperating factors might be missing in seedlings, leading to a loss of true positives. To diminish the number of false positives, we applied a combination of experimental approaches using rather restrictive thresholds. Nevertheless, it cannot be excluded that genes which have been identified by transcriptome analysis and validated by qRT–PCR might be indirect targets since both approaches are based on transcript levels only.
Many of the proteins encoded by ABI3 targets appeared to be involved in protein and lipid storage and the acquisition of desiccation tolerance (Figure 4 and Table 3). Three TFs were also represented in the regulon; these were PEI1 (AT5G07500), an embryo-specific zinc finger, (81), AT1G67100 which encodes a plant-specific TF involved in the establishment of lateral organ boundaries (82) and AT5G18450 which encodes a member of the DREB subfamily of ERF/AP2 TFs, known to be involved in the response to drought stress (83). To et al. (84) suggested that ABI3 regulates the transcription of FUS3. Although FUS3 (AT3G26790) has been identified as candidate gene by the ChIP-chip SAP approach (Supplementary Table S1a), FUS3 induction could not be detected by qRT–PCR in ABI3 transgenic seedlings treated with DEX alone, as well as DEX and ABA (Supplementary Table S3). Also, no promoter activation was found in transient assay (Supplementary Table S4).
The Physcomitrella ABI3 homologue is required for desiccation tolerance (23), which suggests that ABI3 is part of an evolutionarily well-conserved regulatory network responsible for cellular drought tolerance, and the gene has been recruited in the context of seed development to regulate the desiccation processes required for the formation of a mature seed. The expression of 14 ABI3 regulon members was also noted in stamen and/or mature pollen (www.csbdb.mpimp-golm.mpg.de) which may reflect the shared physiological requirements of the seed and the pollen grain during their acquisition of desiccation tolerance.
Members of the ABI3 regulon are expected to contain common regulatory motifs in their promoters, and this assumption was made in identifying de novo potential TF binding motifs, GBL and RYL, in the promoter regions of genes specifically expressed in seeds. Similar motifs have been identified by Suzuki et al. (16) as well as based on a purely computational approach (85). Both motifs are abundant in 250 bp upstream of the TSS, as is the case for other regulatory signals (86). The proximity between the regulatory motifs and the TSS suggests a fairly direct interaction between the regulatory components and the basal transcription machinery. Some of the promoters of the genes belonging to the ABI3 regulon contain both GBL and RYL motifs, but others contain only one of these two types, which indicates that each of the motifs on its own can be sufficient to confer ABI3 dependency. ABI3 binds the RY motif through its B3 domain (19), but no direct interaction between ABI3 and GBL has to date been detected (31 and G. Mönke, unpublished data). The interaction between ABI3 and GBL may be mediated by GBL-bound bZIP-TFs, as described for ABI5 (30). Six of the ABI3 regulon members’ promoters lack both a GBL and an RYL motif; possibly due to the presence of only weakly conserved cis-elements or miss-annotation of TSSs. For instance, the upstream region of the cruciferin B gene (AT1G03880) contains two RYL elements far distant to the TSS. Otherwise, for three genes (AT2G31980, AT2G23240 and AT3G12203), it cannot be excluded that they represent indirect targets, because they have been selected by transcriptome analysis and qRT–PCR only (see above).
Most promoters of the ABI3 regulon genes require ABA for their activation in ABI3::GR transgenic seedlings (Supplementary Tables S1c and S3). This control process may include the phosphorylation-mediated activation of bZIP-TFs such as ABI5 (87,88) and/or the phosphorylation of ABI3 (89) following the ABA-repressed dephosphorylation of protein kinases (90,91). The role of ABA in overall gene regulation is still poorly understood by now. ABA might partially switch the developmental stage of the seedling toward seed development. Remarkably, in transient assay, the addition of ABA was not required for efficient activation, a point which is currently under study.
A genome-wide search for putative ABI3 target promoters using the features of the GBL and RYL motifs resulted in the identification of <3000 promoters (Table 4), a number which far exceeds the 98 functional ABI3 targets defined here. Large discrepancies between the numbers of functional target promoters and of in silico predicted promoters are commonly recorded (92), highlighting the existence of as yet unidentified features for transcriptional regulation, possibly of structural or epigenetic nature (93) and might reflect the existence of chromatin states recently described for the Arabidopis genome (94). Whether the discrepancy is due to selective binding or selective activation is an intriguing question. Among the ∼3000 A. thaliana promoters that contain GBL and/or RYL motifs, ABI3 binds preferentially to those where the relevant gene is specifically expressed in the seed (Table 5). Of 211 promoters in the Arabidopsis genome containing both motifs, 49 were associated with genes having an MI > 31 and 182 with MI < 31. As determined by ChIP-chip, ABI3 bound to 25 (51%) promoters of the MI > 31 group, but only 8 (4%) promoters of the MI < 31 group (Table 5 and Supplementary Table S2). Thus, the ChIP-chip results favor selective promoter binding and subsequent activation. Of the many promoters with an appropriate motif content only a small fraction is bound by ABI3 to become transcriptionally active. Therefore, our data do not support the other theoretical possibility that all motif containing promoters are bound by ABI3 and only a fraction becomes selectively activated. If selective promoter binding of ABI3 is a consequence of chromatin remodeling as described for the binding of PsABI3—the Pisum homologue—to the psp54 promoter (95) or if ABI3 binding mediates changes of chromatin structure as described for PvALF—the Phaseolus ABI3 homologue—and stabilizes its own interaction (9,96), is still a matter of debate.
Table 5.
Selective binding of ABI3 to promoters of seed-specific genes
| Promoter class | MI ≥ 31 |
MI < 31 |
P-value | ||||
|---|---|---|---|---|---|---|---|
| Genome (g) | ChIP-chip (ch) | ch/g (%) | Genome (g) | ChIP-chip (ch) | ch/g (%) | ||
| GBL and RYL | 49 | 25 | 51 | 182 | 8 | 4.4 | 1.6 × 10−13 |
| GBL | 64 | 18 | 28 | 1785 | 27 | 1.5 | 3.5 × 10−16 |
| RYL | 39 | 9 | 23 | 1449 | 3 | 0.2 | 4.6 × 10−13 |
| None | 70 | 4 | 5.7 | 16399 | 66 | 0.4 | 2.2 × 10−4 |
| Total | 222 | 56 | 25 | 19815 | 104 | 0.52 | 4.6 × 10−70 |
ABI3-bound promoters (ChIP-chip) were classified according to the presence/absence of GBL and/or RYL motifs and the seed-specific expression of the corresponding genes (MI). Comparison to the genome-wide occurrence of the promoters demonstrates specific binding of ABI3 to promoters of seed specifically expressed genes (MI > 31). Percentages and P-values are given.
ABI3, FUS3 and LEC2 belong to a class of seed-specific TFs characterized by a B3 DNA-binding domain, known to interact with RYL motifs. Nearly, one-third of the genes of the ABI3 regulon are also activated by FUS3, indicating a partial overlap between the target gene sets of both TFs (Figure 6 and Supplementary Table S5). The FUS3-mediated promoter activation seems to be less ABA-dependent than the ABI3-mediate regulation. Eight genes are regulated by all three B3 domain-containing TFs, with five of these genes involved in seed storage processes (Supplementary Table S5). This indicates that FUS3 and LEC2 can partially substitute for ABI3 in the regulation of seed storage processes, but probably not in processes related to the acquisition of desiccation tolerance. This notion is further supported by the partially overlapping phenotypes of corresponding fus3 and lec2 mutants (3,84). The presence of at least one RYL motif in the promoters of FUS3 and LEC2 co-regulated genes underlines the importance of this motif for the regulatory activity of these B3 domain-containing TFs (Figure 6 and Supplementary Tables S2 and S5). Several members of the ABI3 regulon are also regulated by other TFs (Figure 6 and Supplementary Table S5), possibly in the same or a different spatiotemporal context during seed development. Thus, PIL5 as ABI3 inhibits germination processes and cooperates with ABI3 in the regulation of SOMNUS (80). In accordance with its role during germination, HY5 negatively regulates the common targets and the two MADS box factors AGL9 and AGL15 are involved in embryogenesis, closely connected to seed development.
This study identifies a set of 98 genes being regulated by ABI3. The molecular functions of these ABI3 target genes support an essential role of ABI3 during seed maturation and acquisition of desiccation tolerance. The data suggest that the selective accessibility of target promoters for ABI3 is triggered by developmental switches toward embryogenesis and seed formation. It will be of great interest to further investigate the impact of ABA and higher order regulatory events to come up with a model for target gene prediction.
AVAILABILITY
Array data have been deposited at GEO:
http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GSE34043
GSE33951: Identification of target genes of ABI3 [Agilent 44k]
GSE34033: Identification of target genes of ABI3 [Agilent 2x244k]
GSE34041: Identification of target genes of ABI3 [SAP].
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–5 and Supplementary Figures 1–4.
FUNDING
Trilateral Project ARABIDO-SEED [BMBF grant 0313155 to A.J. and M.M.] coordinated by Prof. M. Caboche; Ministry of Culture of Saxony-Anhalt [XP3624HP/0606T to M.S. and J.K.]. Funding for open access charge: Leibniz-Institute of Plant Genetics and Crop Plant Research House hold budget.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We greatly appreciate stimulating discussions with colleagues within this project. B. Debreuq is acknowledged for the donation of vector plasmids. For excellent technical assistance, we thank Andrea Apelt, Annet Busching, Andreas Czihal, Ingelore Dommes, Sandra Drießlein, Dietlinde Fiedler, Susanne König, Elke Liemann, Elisabeth Nagel, Ingrid Pfort and Sabine Skiebe. We also thank Gunnar Huep, Gregor Mönke, Annegret Tewes, Svetlana Friedel and Prisca Viehöfer for their help with various techniques and constructs.
REFERENCES
- 1.Gutierrez L, Van Wuytswinkel O, Castelain M, Bellini C. Combined networks regulating seed maturation. Trends Plant Sci. 2007;12:294–300. doi: 10.1016/j.tplants.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 2.Santos-Mendoza M, Dubreucq B, Baud S, Parcy F, Caboche M, Lepiniec L. Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J. 2008;54:608–620. doi: 10.1111/j.1365-313X.2008.03461.x. [DOI] [PubMed] [Google Scholar]
- 3.Vicente-Carbajosa J, Carbonero P. Seed maturation: developing an intrusive phase to accomplish a quiescent state. Int. J. Dev. Biol. 2005;49:645–651. doi: 10.1387/ijdb.052046jc. [DOI] [PubMed] [Google Scholar]
- 4.Lotan T, Ohto M, Yee KM, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell. 1998;93:1195–1205. doi: 10.1016/s0092-8674(00)81463-4. [DOI] [PubMed] [Google Scholar]
- 5.Gazzarrini S, McCourt P. Cross-talk in plant hormone signalling: what Arabidopsis mutants are telling us. Ann. Bot. (Lond) 2003;91:605–612. doi: 10.1093/aob/mcg064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ng DW, Chandrasekharan MB, Hall TC. The 5′ UTR negatively regulates quantitative and spatial expression from the ABI3 promoter. Plant Mol. Biol. 2004;54:25–38. doi: 10.1023/B:PLAN.0000028767.06820.34. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X, Garreton V, Chua NH. The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev. 2005;19:1532–1543. doi: 10.1101/gad.1318705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berger N, Dubreucq B, Roudier F, Dubos C, Lepiniec L. Transcriptional regulation of Arabidopsis LEAFY COTYLEDON2 involves RLE, a cis-element that regulates trimethylation of histone H3 at lysine-27. Plant Cell. 2011;23:4065–4078. doi: 10.1105/tpc.111.087866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng DW, Chandrasekharan MB, Hall TC. Ordered histone modifications are associated with transcriptional poising and activation of the phaseolin promoter. Plant Cell. 2006;18:119–132. doi: 10.1105/tpc.105.037010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rider DS, Jr, Henderson JT, Jerome RE, Edenberg HJ, Romero-Severson J, Ogas J. Coordinate repression of regulators of embryonic identity by PICKLE during germination in Arabidopsis. Plant J. 2003;35:33–43. doi: 10.1046/j.1365-313x.2003.01783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki M, Wang HH, McCarty DR. Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiol. 2007;143:902–911. doi: 10.1104/pp.106.092320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perruc E, Kinoshita N, Lopez-Molina L. The role of chromatin-remodeling factor PKL in balancing osmotic stress responses during Arabidopsis seed germination. Plant J. 2007;52:927–936. doi: 10.1111/j.1365-313X.2007.03288.x. [DOI] [PubMed] [Google Scholar]
- 13.Sugliani M, Brambilla V, Clerkx EJ, Koornneef M, Soppe WJ. The conserved splicing factor SUA controls alternative splicing of the developmental regulator ABI3 in Arabidopsis. Plant Cell. 2010;22:1936–1946. doi: 10.1105/tpc.110.074674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braybrook SA, Stone SL, Park S, Bui AQ, Le BH, Fischer RL, Goldberg RB, Harada JJ. Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc. Natl Acad. Sci. USA. 2006;103:3468–3473. doi: 10.1073/pnas.0511331103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parcy F, Valon C, Raynal M, Gaubier-Comella P, Delseny M, Giraudat J. Regulation of gene expression programs during Arabidopsis seed development: roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell. 1994;6:1567–1582. doi: 10.1105/tpc.6.11.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki M, Ketterling MG, Li QB, McCarty DR. Viviparous1 alters global gene expression patterns through regulation of abscisic acid signaling. Plant Physiol. 2003;132:1664–1677. doi: 10.1104/pp.103.022475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamamoto A, Kagaya Y, Usui H, Hobo T, Takeda S, Hattori T. Diverse roles and mechanisms of gene regulation by the Arabidopsis seed maturation master regulator FUS3 revealed by microarray analysis. Plant Cell Physiol. 2010;51:2031–2046. doi: 10.1093/pcp/pcq162. [DOI] [PubMed] [Google Scholar]
- 18.Baumlein H, Nagy I, Villarroel R, Inze D, Wobus U. Cis-analysis of a seed protein gene promoter: the conservative RY repeat CATGCATG within the legumin box is essential for tissue-specific expression of a legumin gene. Plant J. 1992;2:233–239. [PubMed] [Google Scholar]
- 19.Monke G, Altschmied L, Tewes A, Reidt W, Mock HP, Baumlein H, Conrad U. Seed-specific transcription factors ABI3 and FUS3: molecular interaction with DNA. Planta. 2004;219:158–166. doi: 10.1007/s00425-004-1206-9. [DOI] [PubMed] [Google Scholar]
- 20.Reidt W, Wohlfarth T, Ellerström M, Czihal A, Tewes A, Ezcurra I, Rask L, Bäumlein H. Gene regulation during late embryogenesis: the RY motif of maturation-specific gene promoters is a direct target of the FUS3 gene product. Plant J. 2000;21:1–8. doi: 10.1046/j.1365-313x.2000.00686.x. [DOI] [PubMed] [Google Scholar]
- 21.Marella HH, Sakata Y, Quatrano RS. Characterization and functional analysis of ABSCISIC ACID INSENSITIVE3-like genes from Physcomitrella patens. Plant J. 2006;46:1032–1044. doi: 10.1111/j.1365-313X.2006.02764.x. [DOI] [PubMed] [Google Scholar]
- 22.Schallau A, Kakhovskaya I, Tewes A, Czihal A, Tiedemann J, Mohr M, Grosse I, Manteuffel R, Baumlein H. Phylogenetic footprints in fern spore- and seed-specific gene promoters. Plant J. 2008;53:414–424. doi: 10.1111/j.1365-313X.2007.03354.x. [DOI] [PubMed] [Google Scholar]
- 23.Khandelwal A, Cho SH, Marella H, Sakata Y, Perroud PF, Pan A, Quatrano RS. Role of ABA and ABI3 in desiccation tolerance. Science. 2010;327:546. doi: 10.1126/science.1183672. [DOI] [PubMed] [Google Scholar]
- 24.Rohde A, Prinsen E, De Rycke R, Engler G, Van Montagu M, Boerjan W. PtABI3 impinges on the growth and differentiation of embryonic leaves during bud set in poplar. Plant Cell. 2002;14:1885–1901. doi: 10.1105/tpc.003186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rohde A, Ruttink T, Hostyn V, Sterck L, Van Driessche K, Boerjan W. Gene expression during the induction, maintenance, and release of dormancy in apical buds of poplar. J. Exp. Bot. 2007;58:4047–4060. doi: 10.1093/jxb/erm261. [DOI] [PubMed] [Google Scholar]
- 26.Ezcurra I, Wycliffe P, Nehlin L, Ellerstrom M, Rask L. Transactivation of the Brassica napus napin promoter by ABI3 requires interaction of the conserved B2 and B3 domains of ABI3 with different cis-elements: B2 mediates activation through an ABRE, whereas B3 interacts with an RY/G-box. Plant J. 2000;24:57–66. doi: 10.1046/j.1365-313x.2000.00857.x. [DOI] [PubMed] [Google Scholar]
- 27.Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM. Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell. 1992;4:1251–1261. doi: 10.1105/tpc.4.10.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hill A, Nantel A, Rock CD, Quatrano RS. A conserved domain of the viviparous-1 gene product enhances the DNA binding activity of the bZIP protein EmBP-1 and other transcription factors. J. Biol. Chem. 1996;271:3366–3374. doi: 10.1074/jbc.271.7.3366. [DOI] [PubMed] [Google Scholar]
- 29.Lara P, Onate-Sanchez L, Abraham Z, Ferrandiz C, Diaz I, Carbonero P, Vicente-Carbajosa J. Synergistic activation of seed storage protein gene expression in Arabidopsis by ABI3 and two bZIPs related to OPAQUE2. J. Biol. Chem. 2003;278:21003–21011. doi: 10.1074/jbc.M210538200. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura S, Lynch TJ, Finkelstein RR. Physical interactions between ABA response loci of Arabidopsis. Plant J. 2001;26:627–635. doi: 10.1046/j.1365-313x.2001.01069.x. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki M, Kao CY, McCarty DR. The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell. 1997;9:799–807. doi: 10.1105/tpc.9.5.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finkelstein RR, Somerville CR. Three classes of abscisic acid (ABA)-insensitive mutations of Arabidopsis define genes that control overlapping subsets of ABA responses. Plant Physiol. 1990;94:1172–1179. doi: 10.1104/pp.94.3.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakashima K, Fujita Y, Katsura K, Maruyama K, Narusaka Y, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Transcriptional regulation of ABI3-and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol. Biol. 2006;60:51–68. doi: 10.1007/s11103-005-2418-5. [DOI] [PubMed] [Google Scholar]
- 34.Parcy F, Valon Ch, Kohara A, Misera S, Giraudat J. The ABSCISIC ACID-INSENSITIVE3, FUSCA3, and LEAFY COTYLEDON1 loci act in concert to control multiple aspects of Arabidopsis seed development. Plant Cell. 1997;9:1265–1277. doi: 10.1105/tpc.9.8.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kagaya Y, Toyoshima R, Okuda R, Usui H, Yamamoto A, Hattori T. LEAFY COTYLEDON1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE3. Plant Cell Physiol. 2005;46:399–406. doi: 10.1093/pcp/pci048. [DOI] [PubMed] [Google Scholar]
- 36.Collas P. The current state of chromatin immunoprecipitation. Mol. Biotechnol. 2010;45:87–100. doi: 10.1007/s12033-009-9239-8. [DOI] [PubMed] [Google Scholar]
- 37.Solomon MJ, Larsen PL, Varshavsky A. Mapping protein DNA interactions in vivo with formaldehyde—evidence that histone-H4 is retained on a highly transcribed gene. Cell. 1988;53:937–947. doi: 10.1016/s0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- 38.Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- 39.Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 40.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 41.Benhamed M, Martin-Magniette ML, Taconnat L, Bitton F, Servet C, De Clercq R, De Meyer B, Buysschaert C, Rombauts S, Villarroel R, et al. Genome-scale Arabidopsis promoter array identifies targets of the histone acetyltransferase GCN5. Plant J. 2008;56:493–504. doi: 10.1111/j.1365-313X.2008.03606.x. [DOI] [PubMed] [Google Scholar]
- 42.Kaufmann K, Muino JM, Jauregui R, Airoldi CA, Smaczniak C, Krajewski P, Angenent GC. Target genes of the MADS transcription factor SEPALLATA3: integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol. 2009;7:e1000090. doi: 10.1371/journal.pbio.1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao HY, Lee I, Deng X. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell. 2007;19:731–749. doi: 10.1105/tpc.106.047688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oh E, Kang H, Yamaguchi S, Park J, Lee D, Kamiya Y, Choi G. Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell. 2009;21:403–419. doi: 10.1105/tpc.108.064691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roccaro M, Somssich IE. Chromatin immunoprecipitation to identify global targets of WRKY transcription factor family members involved in plant immunity. Methods Mol. Biol. 2011;712:45–58. doi: 10.1007/978-1-61737-998-7_5. [DOI] [PubMed] [Google Scholar]
- 46.Tao Z, Shen L, Liu C, Liu L, Yan Y, Yu H. Genome-wide identification of SOC1 and SVP targets during the floral transition in Arabidopsis. Plant J. 2012;70:549–561. doi: 10.1111/j.1365-313X.2012.04919.x. [DOI] [PubMed] [Google Scholar]
- 47.Thibaud-Nissen F, Wu H, Richmond T, Redman JC, Johnson C, Green R, Arias J, Town CD. Development of Arabidopsis whole-genome microarrays and their application to the discovery of binding sites for the TGA2 transcription factor in salicylic acid-treated plants. Plant J. 2006;47:152–162. doi: 10.1111/j.1365-313X.2006.02770.x. [DOI] [PubMed] [Google Scholar]
- 48.Zheng Y, Ren N, Wang H, Stromberg AJ, Perry SE. Global identification of targets of the Arabidopsis MADS domain protein AGAMOUS-Like15. Plant Cell. 2009;21:2563–2577. doi: 10.1105/tpc.109.068890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004;39:366–380. doi: 10.1111/j.1365-313X.2004.02138.x. [DOI] [PubMed] [Google Scholar]
- 50.Bent A. Arabidopsis thaliana floral dip transformation method. Methods Mol. Biol. 2006;343:87–103. doi: 10.1385/1-59745-130-4:87. [DOI] [PubMed] [Google Scholar]
- 51.Wan CY, Wilkins TA. A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L) Anal. Biochem. 1994;223:7–12. doi: 10.1006/abio.1994.1538. [DOI] [PubMed] [Google Scholar]
- 52.Czechowski T, Bari RP, Stitt M, Scheible WR, Udvardi MK. Real-time RT–PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J. 2004;38:366–379. doi: 10.1111/j.1365-313X.2004.02051.x. [DOI] [PubMed] [Google Scholar]
- 53.Junker A, Monke G, Rutten T, Keilwagen J, Seifert M, Thi TM, Renou JP, Balzergue S, Viehover P, Hahnel U, et al. Elongation-related functions of LEAFY COTYLEDON1 during the development of Arabidopsis thaliana. Plant J. 2012;71:427–442. doi: 10.1111/j.1365-313X.2012.04999.x. [DOI] [PubMed] [Google Scholar]
- 54.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 55.Seifert M, Keilwagen J, Strickert M, Grosse I. Utilizing gene pair orientations for HMM-based analysis of promoter array ChIP-chip data. Bioinformatics. 2009;25:2118–2125. doi: 10.1093/bioinformatics/btp276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rabiner LR. A tutorial on Hidden Markov Models and selected applications in speech recognition. Proc. IEEE. 1989;77:257–283. [Google Scholar]
- 57.Breitling F, Herzyk P. Rank-based methods as a non-parametric alternative of the T-statistic for the analysis of biological microarray data. J. Bioinf. Comp. Biol. 2005;3:1178–1189. doi: 10.1142/s0219720005001442. [DOI] [PubMed] [Google Scholar]
- 58.Hong F, Breitling R, McEntee CW, Wittner BS, Nemhauser JL, Chory J. RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics. 2006;22:2825–2827. doi: 10.1093/bioinformatics/btl476. [DOI] [PubMed] [Google Scholar]
- 59.Stalberg K, Ellerstrom M, Josefsson LG, Rask L. Deletion analysis of a 2S seed storage protein promoter of Brassica napus in transgenic tobacco. Plant Mol. Biol. 1993;23:671–683. doi: 10.1007/BF00021523. [DOI] [PubMed] [Google Scholar]
- 60.Kleindt CK, Stracke R, Mehrtens F, Weisshaar B. Expression analysis of flavonoid biosynthesis genes during Arabidopsis thaliana silique and seed development with a primary focus on the proanthocyanidin biosynthetic pathway. BMC Res. Notes. 2010;3:255. doi: 10.1186/1756-0500-3-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Keilwagen J, Grau J, Paponov IA, Posch S, Strickert M, Grosse I. De-novo discovery of differentially abundant transcription factor binding sites including their positional preference. PLoS Comput. Biol. 2011;7:e1001070. doi: 10.1371/journal.pcbi.1001070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aoyama T, Chua NH. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 1997;11:605–612. doi: 10.1046/j.1365-313x.1997.11030605.x. [DOI] [PubMed] [Google Scholar]
- 64.Suzuki M, Kao CY, Cocciolone S, McCarty DR. Maize VP1 complements Arabidopsis abi3 and confers a novel ABA/auxin interaction in roots. Plant J. 2001;28:409–418. doi: 10.1046/j.1365-313x.2001.01165.x. [DOI] [PubMed] [Google Scholar]
- 65.Ivanov R, Tiedemann J, Czihal A, Schallau A, Diep lH, Mock HP, Claus B, Tewes A, Baumlein H. EFFECTOR OF TRANSCRIPTION2 is involved in xylem differentiation and includes a functional DNA single strand cutting domain 8. Dev. Biol. 2008;313:93–106. doi: 10.1016/j.ydbio.2007.09.061. [DOI] [PubMed] [Google Scholar]
- 66.Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S. AmiGO: online access to ontology and annotation data. Bioinformatics. 2011;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dunwell JM, Purvis A, Khuri S. Cupins: the most functionally diverse protein superfamily? Phytochemistry. 2004;65:7–17. doi: 10.1016/j.phytochem.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 68.Kreis M, Forde BG, Rahman S, Miflin BJ, Shewry PR. Molecular evolution of the seed storage proteins of barley, rye and wheat. J. Mol. Biol. 1985;183:499–502. doi: 10.1016/0022-2836(85)90017-8. [DOI] [PubMed] [Google Scholar]
- 69.Tunnacliffe A, Wise MJ. The continuing conundrum of the LEA proteins. Naturwissenschaften. 2007;94:791–812. doi: 10.1007/s00114-007-0254-y. [DOI] [PubMed] [Google Scholar]
- 70.Capuano F, Beaudoin F, Napier JA, Shewry PR. Properties and exploitation of oleosins. Biotechnol. Adv. 2007;25:203–206. doi: 10.1016/j.biotechadv.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 71.Guiltinan MJ, Marcotte WR, Jr, Quatrano RS. A plant leucine zipper protein that recognizes an abscisic acid response element. Science. 1990;250:267–271. doi: 10.1126/science.2145628. [DOI] [PubMed] [Google Scholar]
- 72.Hattori T, Totsuka M, Hobo T, Kagaya Y, Yamamoto-Toyoda A. Experimentally determined sequence requirement of ACGT-containing abscisic acid response element. Plant Cell Physiol. 2002;43:136–140. doi: 10.1093/pcp/pcf014. [DOI] [PubMed] [Google Scholar]
- 73.Dickinson CD, Evans RP, Nielsen NC. RY repeats are conserved in the 5′-flanking regions of legume seed-protein genes. Nucleic Acids Res. 1988;16:371. doi: 10.1093/nar/16.1.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Suzuki M, Ketterling MG, McCarty DR. Quantitative statistical analysis of cis-regulatory sequences in ABA/VP1- and CBF/DREB1-regulated genes of Arabidopsis. Plant Physiol. 2005;139:437–447. doi: 10.1104/pp.104.058412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hwang JE, Hong JK, Lim CJ, Chen H, Je J, Yang KA, Kim DY, Choi YJ, Lee SY, Lim CO. Distinct expression patterns of two Arabidopsis phytocystatin genes, AtCYS1 and AtCYS2, during development and abiotic stresses. Plant Cell Rep. 2010;29:905–915. doi: 10.1007/s00299-010-0876-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bies-Etheve N, Gaubier-Comella P, Debures A, Lasserre E, Jobet E, Raynal M, Cooke R, Delseny M. Inventory, evolution and expression profiling diversity of the LEA (late embryogenesis abundant) protein gene family in Arabidopsis thaliana. Plant Mol. Biol. 2008;67:107–124. doi: 10.1007/s11103-008-9304-x. [DOI] [PubMed] [Google Scholar]
- 77.Drea SC, Lao NT, Wolfe KH, Kavanagh TA. Gene duplication, exon gain and neofunctionalization of OEP16-related genes in land plants. Plant J. 2006;46:723–735. doi: 10.1111/j.1365-313X.2006.02741.x. [DOI] [PubMed] [Google Scholar]
- 78.Haslekas C, Grini PE, Nordgard SH, Thorstensen T, Viken MK, Nygaard V, Aalen RB. ABI3 mediates expression of the peroxiredoxin antioxidant AtPER1 gene and induction by oxidative stress. Plant Mol. Biol. 2003;53:313–326. doi: 10.1023/b:plan.0000006937.21343.2a. [DOI] [PubMed] [Google Scholar]
- 79.Kotak S, Vierling E, Baumlein H, Koskull-Doring P. A novel transcriptional cascade regulating expression of heat stress proteins during seed development of Arabidopsis. Plant Cell. 2007;19:182–195. doi: 10.1105/tpc.106.048165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Park J, Lee N, Kim W, Lim S, Choi G. ABI3 and PIL5 collaboratively activate the expression of SOMNUS by directly binding to its promoter in imbibed Arabidopsis seeds. Plant Cell. 2011;23:1404–1415. doi: 10.1105/tpc.110.080721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li Z, Thomas TL. PEI1, an embryo-specific zinc finger protein gene required for heart-stage embryo formation in Arabidopsis. Plant Cell. 1998;10:383–398. doi: 10.1105/tpc.10.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shuai B, Reynaga-Pena CG, Springer PS. The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 2002;129:747–761. doi: 10.1104/pp.010926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- 84.To A, Valon C, Savino G, Guilleminot J, Devic M, Giraudat J, Parcy F. A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell. 2006;18:1642–1651. doi: 10.1105/tpc.105.039925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Peng FY, Weselake RJ. Gene coexpression clusters and putative regulatory elements underlying seed storage reserve accumulation in Arabidopsis. BMC Genomics. 2011;12:286. doi: 10.1186/1471-2164-12-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Koudritsky M, Domany E. Positional distribution of human transcription factor binding sites. Nucleic Acids Res. 2008;36:6795–6805. doi: 10.1093/nar/gkn752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schutze K, Harter K, Chaban C. Post-translational regulation of plant bZIP factors. Trends Plant Sci. 2008;13:247–255. doi: 10.1016/j.tplants.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 88.Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl Acad. Sci. USA. 2000;97:11632–11637. doi: 10.1073/pnas.190309197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Guerriero G, Martin N, Golovko A, Sundstrom JF, Rask L, Ezcurra I. The RY/Sph element mediates transcriptional repression of maturation genes from late maturation to early seedling growth. New Phytol. 2009;184:552–565. doi: 10.1111/j.1469-8137.2009.02977.x. [DOI] [PubMed] [Google Scholar]
- 90.Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 91.Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pan Y, Tsai CJ, Ma B, Nussinov R. Mechanisms of transcription factor selectivity. Trends Genet. 2010;26:75–83. doi: 10.1016/j.tig.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bell O, Tiwari VK, Thoma NH, Schubeler D. Determinants and dynamics of genome accessibility. Nat. Rev. Genet. 2011;12:554–564. doi: 10.1038/nrg3017. [DOI] [PubMed] [Google Scholar]
- 94.Roudier F, Ahmed I, Berard C, Sarazin A, Mary-Huard T, Cortijo S, Bouyer D, Caillieux E, Duvernois-Berthet E, Al-Shikhley L, et al. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 2011;30:1928–1938. doi: 10.1038/emboj.2011.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gagete AP, Franco L, Isabel RM. The Pisum sativum psp54 gene requires ABI3 and a chromatin remodeller to switch from a poised to a transcriptionally active state. New Phytol. 2011;192:353–363. doi: 10.1111/j.1469-8137.2011.03818.x. [DOI] [PubMed] [Google Scholar]
- 96.Ng DW, Hall TC. PvALF and FUS3 activate expression from phaseolin promoter by different mechanisms. Plant Mol. Biol. 2008;66:233–244. doi: 10.1007/s11103-007-9265-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.