Abstract
The majority of bacterial genes are located on the leading strand, and the percentage of such genes has a large variation across different bacteria. Although some explanations have been proposed, these are at most partial explanations as they cover only small percentages of the genes and do not even consider the ones biased toward the lagging strand. We have carried out a computational study on 725 bacterial genomes, aiming to elucidate other factors that may have influenced the strand location of genes in a bacterium. Our analyses suggest that (i) genes of some functional categories such as ribosome have higher preferences to be on the leading strands; (ii) genes of some functional categories such as transcription factor have higher preferences on the lagging strands; (iii) there is a balancing force that tends to keep genes from all moving to the leading and more efficient strand and (iv) the percentage of leading-strand genes in an bacterium can be accurately explained based on the numbers of genes in the functional categories outlined in (i) and (ii), genome size and gene density, indicating that these numbers implicitly contain the information about the percentage of genes on the leading versus lagging strand in a genome.
INTRODUCTION
It has been observed that the majority of bacterial genes tend to be located on the leading strand in a genome, and the percentage of such genes has a large variation across different bacteria, ranging from ∼45% to ∼90% (1,2). A number of studies have been carried out aiming to provide explanations for such observations. A key factor considered in these studies is the different mechanisms used by bacterial cells in replication of the leading and the lagging strands when cell replication and transcription occur simultaneously (3,4). Specifically, during chromosomal replication, deoxyribonucleic acid (DNA) and ribonucleic acid (RNA) polymerases move in the same direction on the leading strand but in opposite directions on the lagging strand, creating the possibility of head-on collisions between the two polymerases during transcription of some genes on the lagging strand, hence making the lagging strand the less efficient one between the two (1,4). In an earlier study, Brewer (3) suggested that bacterial cells may be under a selection pressure to have highly expressed genes reside on the leading strand. Rocha and Danchin (5,6) recently argued that it is really the essentiality instead of the needed expression levels of genes that may have driven certain genes to the leading strand. Although this interpretation seems to be correct, it provides only a partial answer as essential genes account for only a small portion of the whole gene set encoded in a bacterial genome, e.g. ∼10% in Escherichia coli (7,8) and ∼10% in Bacillus subtilis (9). Price et al. (10) observed that longer operons tend to be on the leading strand and suggested that there may be a selection pressure to have such an arrangement to avoid interruptions during transcription of such operons. Furthermore, Rocha (6,11) observed that the presence/absence of the DNA polymerase PolC in a genome is highly correlated with bacterial genomes having at least 70% of their genes on the leading strand or not. Hu et al. (12) proposed that replication-associated purine asymmetry may also contribute to the strand bias in a genome. In addition, Lin et al. (13) found that the essential genes on the leading strand are enriched in only a few of sub-categories of clusters of orthologous groups (14). Although this analysis provided useful insights of functional preference of genes to the leading and lagging strand, a larger analysis involving more genes and organisms is needed to ensure the generality of the observation. More importantly, the general issues of why the majority of bacterial genes tend to be located on the leading strands and why the percentage of leading strand genes has such a large variation across different organisms remain largely unanswered.
We present in this study a computational analysis of all the sequenced bacterial genomes aiming to provide a more general explanation to the above two observations. Our key findings are (i) genes of different functional categories have different level of tendency to be on the leading strand; (ii) genes of some functional categories such as transcription factor have higher preferences to be on the lagging strands; (iii) there is at least one balancing force that keeps genes from all moving to the leading strand during evolution, i.e. a more balanced genome facilitates a higher gene density in a genome and (iv) the percentage of leading-strand genes for a bacterium can be accurately explained in terms of genes in some functional categories outlined in (i) to (ii), genome size and gene density. On the basis of these findings, we believe that the percentage of genes on the leading versus lagging strand in a genome is the result of two sets of balancing forces, one that tends to drive genes of certain functional categories to the leading strands to make the bacteria more efficient in their responses to environmental changes and one that tends to keep the genome as compact as possible to stay energetically efficient when replicating and maintaining the genome.
MATERIALS AND METHODS
Data
The 725 bacterial genome sequences along with their predicted genes and functional annotations were retrieved from the NCBI FTP site (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/) as of 11 December 2010. The gene ontology (GO) annotations for these genomes were from the GOA Proteome Sets (v52) (15), and the GOslim definitions were downloaded from the Gene Ontology site (http://www.geneontology.org/GO_slims/goslim_generic.obo) (16). The microarray data for E. coli are downloaded from the M3D web site (http://m3d.bu.edu) (17).
High-level functional annotations of genes
GO (16) was used to define functional categories of gene products. Based on the GO annotation and GO hierarchy information, the Perl script map2slim (http://search.cpan.org/~cmungall/go-perl/scripts/map2slim) was used to the bacterial genomes for assignment of GOslim-based functional categories.
Determination of genes on leading and lagging strands
To determine whether a gene is on the leading versus lagging strand of a genome, the origin and the terminus of replication are needed. The origin of replication for each of the 725 bacterial genomes was retrieved from the DoriC database (18), which has been widely used in the comparative genomics analysis (19–21) and the origin prediction for newly sequenced genomes (22–28). The terminus of replication is thus calculated as the location of origin of replication plus half of the chromosome length. With these two positions, the leading and lagging strands are determined for each half of the chromosome according to a well-known fact that the leading strand always has more genes than the lagging strand does (11). For each bacterium, only the major chromosome is considered, and plasmids are excluded in this study.
Preference of functional categories on different strands
Given a GO functional category, an index x is calculated using the following formula:
where n0 is the number of leading-strand genes of this functional category, n1 is the number of lagging-strand genes of this category; x is calculated for all the GOslim functional categories, respectively, on the leading strand for all 725 genomes, so that for each category, there is a data set (A) of 725 values. In addition, the overall percentage of leading-strand genes (data set B) is obtained for each of the 725 genomes as well. For each GOslim functional category, a Wilcoxon rank sum test was performed to test whether the data sets A and B are from two distinct distributions. We also used the similar procedure to assess the preference of functional categories on the lagging strand. All the statistical analyses are conducted using the R statistical language (http://www.r-project.org).
Prediction of the percentage of leading-strand genes in a genome
Network training
A neural network model, with one hidden layer of 10 nodes, is used to predict the percentage of leading-strand genes in a genome using the total 57 inputs and then selected smaller numbers of 30, 25, 20, 15, 10 and 5 ones in this study. To reduce the possibility of the over-fitting problem, we used an early stopping technique, which divides the data into three subsets: training set used for computing gradient and updating the network weights and biases, validation set used for monitoring training process by its error rate and testing set used for assessing the neural network performance independently. When the network starts to over-fit the data, the error on the validation set begins to rise, and hence, the training process is stopped early. We used the default setting in the MATLAB Neural Network Toolbox, which arbitrarily divides the data into the three subsets, respectively: 507 (70%) for training set, 109 (15%) for validation set and 109 (15%) for testing set. The performance of a neural network is measured by mean squared error (MSE) and Pearson correlation score (R). The trained neural network can be downloaded from http://csbl.bmb.uga.edu/~xizeng/research/gene_strand_bias/.
Variable selection
Out of the initial set of 57 input variables, we have conducted a variable selection process based on the idea of mean impact value (MIV) (29). Based on the ranks of the MIVs of the input variables, input variables with insignificant MIV will be eliminated from the neural network model. MIVs are calculated as follows: vary the value of each input variable by increasing and decreasing 10% for all samples and get two outputs. Then subtract one from the other and obtain the impact change value [impact value (IV)] of the output due to the changes of the input variable values. Then the MIV is obtained by averaging the IVs across all trained networks: 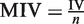 , where n is the times of network training.
, where n is the times of network training.
RESULTS
Characteristics of genes on leading strands
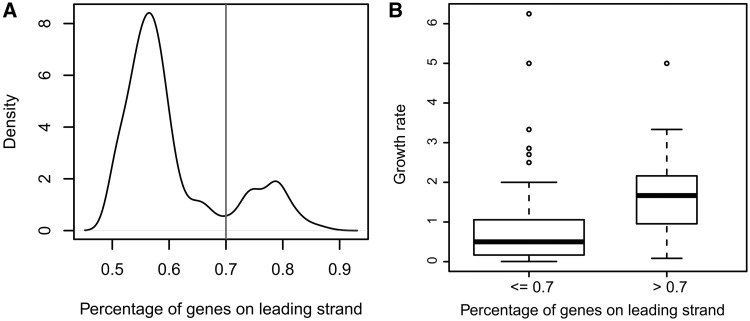
We have analyzed 725 sequenced eubacterial genomes for which origins of replication and GO-based annotations are available in terms of the strand biases of their protein-encoding genes, and archaea were excluded from our analysis as they may have multiple origins of replications. Figure 1A shows the percentage distribution of leading-strand genes across all the 725 genomes, ranging from 45% to ∼90%. This observation extends a previous observation made based on a few bacterial genomes. Across these 725 bacteria, the percentage of leading-strand genes does not show any correlation with genome sizes in terms of gene numbers (Supplementary Figure S1), whereas different phyla have substantially different averaged percentages of leading-strand genes (Supplementary Figure S2), which is consistent with a previous finding made on a smaller group of bacterial genomes (11,30).
Figure 1.
General characteristics for leading-strand genes. (A) Distribution of the number of bacteria with a specific percentage of genes on the leading strands; and (B) distribution of the percentages of leading-strand genes versus cell growth rate in the 104 bacterial genomes with growth rate data available.
We have also examined the relationship between leading strand bias and the growth rate for 104 of the 725 bacterial genomes, for which the doubling-time information is available (31). We found that bacterial genomes with high leading-strand bias (>70%) tend to have higher growth rates than those with low leading-strand bias (≤70%) measured by the Wilcoxon rank sum test with P value: 1.9 × 10−4, as shown in Figure 1B. This observation changes the previous conclusion that fast-growing bacteria have similar leading-strand bias to that of the slow-growing bacteria (11), which was made based on a substantially smaller number of genomes with known growth rates.
Functional categories whose genes have different preference to different strands
We have examined whether genes of different functional categories may have different level of preference to be on the leading versus the lagging strands across all bacteria. To do this, we checked 55 of the 127 GOslim functional categories (16) that have available gene assignments in at least 36 (5%) of the 725 genomes and have the number of genes with the median being between 5 and 500 (categories with >500 genes will be too general for our study) across all the genomes under consideration. For each of the 55 functional categories, we consider a functional category prefers a strand if genes in this category have a higher percentage than the average percentage of all genes on the strand across all genomes. The Wilcoxon rank sum test is used to assess the statistical significance of an observed preference measured using a P value. We found that 32 of the 55 categories prefer the leading strand with P value < 0.01, including genes related to ribosome, structural molecular activity, translation, RNA binding and cell cycle, with the detailed information presented in Table 1; and 11 categories prefer the lagging strand with P value < 0.01, including DNA-binding transcription factor activity, signal transducer activity and regulation of biological process, with details in Table 2. On average, 52% of the genes encoded in a bacterial genome are covered by the 43 (32 + 11) functional categories, and the detailed distribution of percentage across different bacterial genomes is given in Supplementary Table S1. Notably, transcription factor activity (GO:0003700) shows strong preference to the lagging strand. To confirm it, we have examined the set of all 271 annotated transcription factors in E. coli from the RegulonDB database (32) and found the same strand preference with P value 5.8 × 10−3 (Supplementary Table S2). One possible explanation is that transcription factors, particularly non-global transcription factors, are known to have low expression levels (33) and, hence, represent the last group of genes to move to the leading strand during evolution.
Table 1.
Preference of GOslim categories toward leading strands across 725 bacterial genomes
| GO branch | GO category | Preference |
|---|---|---|
| MF | GO:0005198 structural molecule activity | 7.28E-156 |
| MF | GO:0003723 RNA binding | 1.15E-107 |
| MF | GO:0008135 translation factor activity | 4.67E-78 |
| MF | GO:0005515 protein binding | 1.37E-32 |
| MF | GO:0003774 motor activity | 2.05E-24 |
| MF | GO:0000166 nucleotide binding | 5.93E-14 |
| MF | GO:0003676 nucleic acid binding | 1.04E-13 |
| MF | GO:0030234 enzyme regulator activity | 1.97E-08 |
| MF | GO:0016740 transferase activity | 9.62E-05 |
| BP | GO:0006412 translation | 6.77E-118 |
| BP | GO:0007049 cell cycle | 8.66E-72 |
| BP | GO:0019538 protein metabolic process | 7.64E-56 |
| BP | GO:0015031 protein transport | 7.60E-34 |
| BP | GO:0016043 cellular component organization | 1.81E-30 |
| BP | GO:0009605 response to external stimulus | 1.89E-20 |
| BP | GO:0007154 cell communication | 2.51E-18 |
| BP | GO:0006091 generation of precursor metabolites and energy | 1.11E-17 |
| BP | GO:0005975 carbohydrate metabolic process | 7.17E-16 |
| BP | GO:0019748 secondary metabolic process | 3.10E-15 |
| BP | GO:0006629 lipid metabolic process | 1.37E-11 |
| BP | GO:0009056 catabolic process | 1.51E-06 |
| BP | GO:0006519 cellular amino acid and derivative metabolic process | 2.41E-06 |
| BP | GO:0006811 ion transport | 5.37E-05 |
| BP | GO:0006950 response to stress | 3.26E-03 |
| CC | GO:0005840 ribosome | 1.97E-158 |
| CC | GO:0043226 organelle | 5.17E-116 |
| CC | GO:0005737 cytoplasm | 6.29E-56 |
| CC | GO:0005622 intracellular | 6.67E-41 |
| CC | GO:0005694 chromosome | 1.11E-26 |
| CC | GO:0043234 protein complex | 6.98E-26 |
| CC | GO:0005618 cell wall | 5.31E-11 |
| CC | GO:0005886 plasma membrane | 1.42E-06 |
The first column represents the three major GO categories: molecular function (MF), cellular component (CC) and biological process (BP).
Table 2.
Preference of GOslim categories toward lagging strands across 725 bacterial genomes
| GO branch | GO category | Preference |
|---|---|---|
| MF | GO:0003700 sequence specific DNA binding transcription factor activity | 3.09E-34 |
| MF | GO:0016209 antioxidant activity | 3.31E-11 |
| MF | GO:0003677 DNA binding | 3.41E-11 |
| MF | GO:0004871 signal transducer activity | 2.10E-06 |
| MF | GO:0004672 protein kinase activity | 9.34E-06 |
| MF | GO:0008233 peptidase activity | 1.08E-05 |
| BP | GO:0050789 regulation of biological process | 3.20E-15 |
| BP | GO:0019725 cellular homeostasis | 3.08E-11 |
| BP | GO:0006350 transcription | 2.94E-09 |
| BP | GO:0006464 protein modification process | 1.06E-06 |
| BP | GO:0007165 signal transduction | 6.31E-05 |
The first column represents the three major GO categories: molecular function (MF), cellular component (CC) and biological process (BP).
To check whether our analysis covers the observation that essential genes tend to be on the leading strands made by Rocha and Danchin (5), we created an artificial functional category ‘essential genes’ and applied our analysis to all the essential genes in 13 bacterial genomes in the DEG database (34), which has the most comprehensive annotated essential gene list. No surprise here as this category has a significant P value for preferring to be on the leading strand (Supplementary Figure S3), indicating that our explanation covers the observation made by Rocha and Danchin (5).
A balancing force: strand bias versus gene density
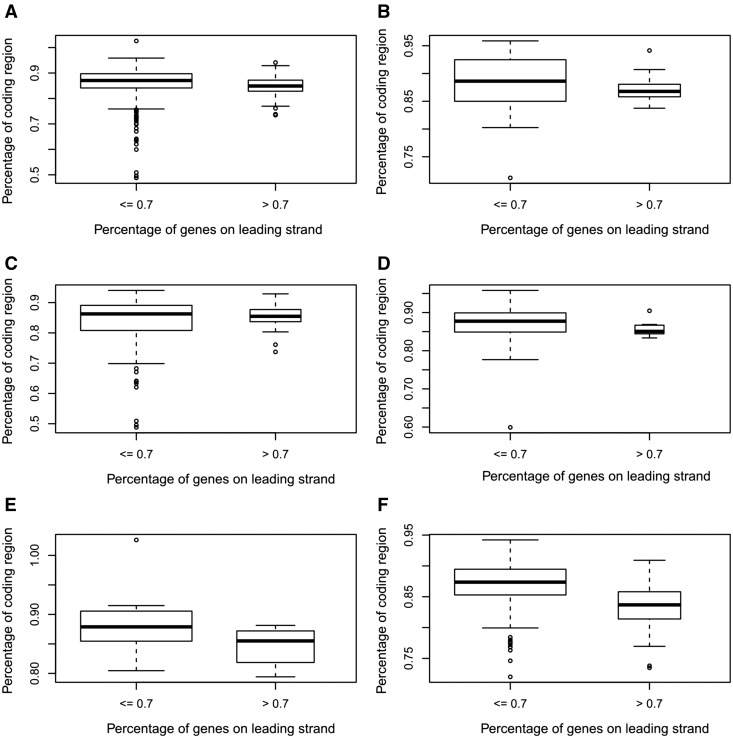
Our analysis suggests that there might be a selection pressure for a bacterium to have a more compact genome (i.e. a shorter genome without losing genes), particularly in a complex environment. To check this hypothesis, we have examined the percentages of coding regions in the two groups of bacteria, one containing all bacteria with at least 70% of the genes on the leading strands and one containing all the other 725 bacteria and checked their relationship with the living styles of the bacteria. Our analysis revealed that (i) the bacteria in the second group (with lower strand bias) tend to have higher percentages of coding regions than those in the first group, with a P value 1.8 × 10−8 based on the Wilcoxon rank sum test, as shown in Figure 2A and (ii) this tendency is more significant for bacteria living in complex environments, with P values ranging from 0.25 to 2.8 × 10−9, as shown in Figure 2B–F. One possible explanation is that there might be a selection pressure for bacteria living in nutrient-depleted environments to keep their genomes as compact as possible (without losing genes), and having a more balanced genome is one way to achieve this goal (a more balanced genome seems to allow a higher degree of overlap between regulatory regions of operons).
Figure 2.
Boxplots of the percentage of coding region versus the percentage of leading strand genes in a genome. (A) For all bacteria (P value of the Wilcoxon test: 1.1 × 10−8); (B) bacteria of specialized type with P value 0.22; (C) bacteria of host-associated type with P value 0.54; (D) bacteria of aquatic type with P value 0.065; (E) bacteria of terrestrial type with P value 0.0031 and (F) bacteria of multiple type with P value 1.9 × 10−9.
A model for interpreting the percentage of leading-strand genes
Our main hypothesis is that the percentage of leading-strand genes in a genome reflects the relationship between the key functionalities and the living environment of an organism. To check this hypothesis, we have examined the population of genes in each functional category encoded in each genome to see whether some of them can be used to predict the percentage of leading-strand genes.
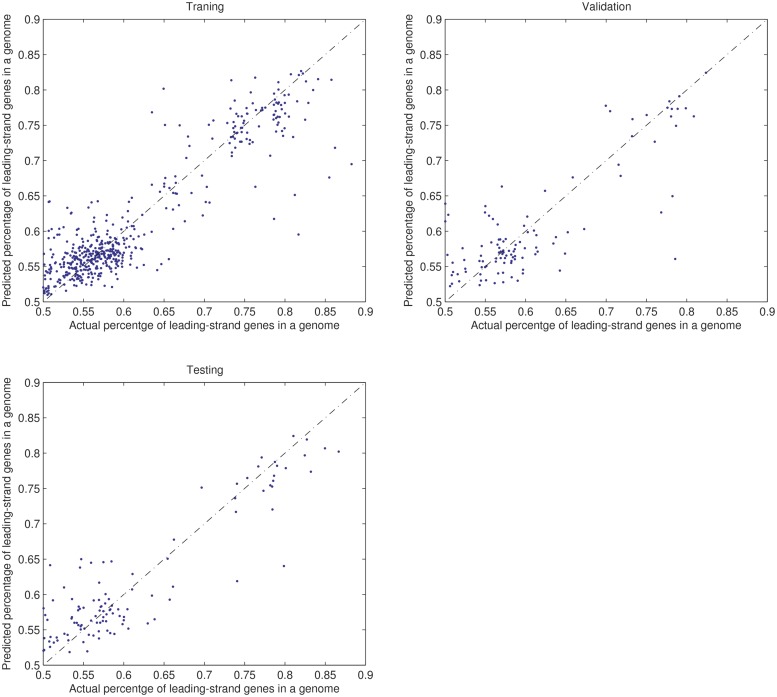
We trained 10 times a neural network with 57 input nodes, one node for each of the 55 functional categories, one node for gene density and one node for the genome size; one hidden layer of 10 nodes and one output node, where gene density is calculated as the percentage of non-coding region length against chromosome length. We split the 725 genomes into three sets: 507 (70%) as the training set, 109 (15%) as the validation set and 109 (15%) as the testing set. At the end of the training, the neural network has the following average performance results on the three data sets: MSE = 0.0015 and R (Pearson correlation score) = 0.91 between the desired and predicted values on the training set; MSE = 0.0023 and R = 0.85 on the validation set and MSE = 0.0021 and R = 0.87 on the testing set. Figure 3 shows the performance of a trained neural network on the different data sets.
Figure 3.
Performance in predicting the percentage of leading-strand genes in a genome by our trained neural network on the training, validation and testing set, respectively.
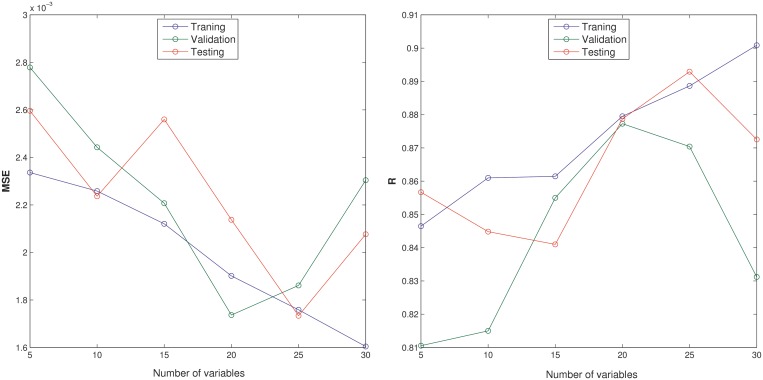
Using the variable selection procedure outlined in ‘Materials and Methods’ section, we have examined the IV of each input on the performance of each neural network trained by increasing or decreasing its value by 10% and used the averaged IV (MIV) for the 10 trained neural networks as a measure of the importance level of that variable. We have examined the performance of neural networks with smaller numbers of inputs with top MIV values: 30, 25, 20, 15, 10 and 5, as shown in Figure 4. Each network was trained three times. The networks with 25 inputs work best on the different data sets with the following average performance: MSE = 0.0018 and R = 0.89 on the training set; MSE = 0.0019 and R = 0.86 on the validation set and MSE = 0.0017 and R = 0.88 on the testing set. Specifically, these 25 inputs are listed in Table 3: cell cycle, iron transport, transport, response to stress, nucleobase, cellular homeostasis, translation and generation of precursor metabolites and energy under the biological process category; ribosome, cytoplasm, cell envelope and protein complex under the cellular component category; RNA binding, electron carrier activity, kinase activity, translation factor activity and structure molecule activity under the molecular function category; along with genome size and gene density. This prediction result is highly consistent with our above results shown in Figure 3.
Figure 4.
Evaluation of performance in the percentage of leading-strand genes in a genome with smaller numbers of inputs by our trained neural network.
Table 3.
Twenty-five selected inputs used in the neural network model
| Category | Variable | MIV |
|---|---|---|
| BP | GO:0007049 cell cycle | 0.012344 |
| BP | GO:0006811 ion transport | 0.005643 |
| BP | GO:0006629 lipid metabolic process | −0.00275 |
| BP | GO:0019748 secondary metabolic process | −0.00294 |
| BP | GO:0006810 transport | −0.00332 |
| BP | GO:0006950 response to stress | −0.00388 |
| BP | GO:0006139 nucleobase | −0.00402 |
| BP | GO:0019725 cellular homeostasis | −0.00466 |
| BP | GO:0006412 translation | −0.00825 |
| BP | GO:0006091 generation of precursor metabolites and energy | −0.01027 |
| CC | GO:0005840 ribosome | 0.009582 |
| CC | GO:0005737 cytoplasm | 0.007178 |
| CC | GO:0005622 intracellular | 0.003747 |
| CC | GO:0043226 organelle | 0.002657 |
| CC | GO:0030312 external encapsulating structure | −0.00234 |
| CC | GO:0030313 cell envelope | −0.00319 |
| CC | GO:0043234 protein complex | −0.0049 |
| MF | GO:0003723 RNA binding | 0.027956 |
| MF | GO:0009055 electron carrier activity | 0.003654 |
| MF | GO:0016301 kinase activity | 0.002421 |
| MF | GO:0030234 enzyme regulator activity | −0.00182 |
| MF | GO:0008135 translation factor activity | −0.00304 |
| MF | GO:0005198 structural molecule activity | −0.01745 |
| OT | Gene density | −0.00451 |
| OT | Genome size | −0.00515 |
Biological process (BP), cellular component (CC) and molecular function (MF) in the first column are the top-level categories in the gene ontology (GO) hierarchy; OT is for other variables that are not GO categories.
Using the selected 25 variables from each genome, we have constructed a new neural network with one hidden layer to predict the overall percentage of genes on the leading strand as follows:
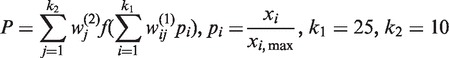 |
where P is the percentage of leading-strand genes in a genome, f is a hyperbolic tangent sigmoid transfer function, 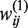 is the weight of the ith input to the jth node of the hidden layer and
is the weight of the ith input to the jth node of the hidden layer and 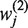 is the weight of the jth node in the hidden layer to the output node in the neural network model, k1 is the number of variables, k2 is the number of nodes of the hidden layer, pi is a scaling factor calculated as the ratio between the variable (xi) and the max value (xi,max) of this variable across all 725 bacterial genomes.
is the weight of the jth node in the hidden layer to the output node in the neural network model, k1 is the number of variables, k2 is the number of nodes of the hidden layer, pi is a scaling factor calculated as the ratio between the variable (xi) and the max value (xi,max) of this variable across all 725 bacterial genomes.
We speculate that the genes of certain functional categories need to be on the leading strands when living in certain environments to out-compete their competitors when food is limited and the competition is high; otherwise, the organism may keep the genes on the lagging strands as a more balanced genome may mean a more compact genome, which requires lower maintenance energy. A good example is that the chemotactic response of Pseudoalteromonas haloplanktis in exploiting ephemeral microscale nutrient patches is at least 10 times faster than that of E. coli (35), suggesting that P. haloplanktis may be genetically optimized for this particular capability. To check whether some genes are specifically located on the leading strand of the organism, we examined the strand distribution of genes across the 55 GOslim functional categories on P. haloplanktis and E. coli, and we found that genes of some functional categories are significantly enriched (with P value < 0.05) on the leading strand of P. haloplanktis than that of E. coli, including protein transport, DNA metabolic process, ion transport and signal transduction under the biological process category; organelle and intracellular under the cellular component; antioxidant activity and motor activity (Supplementary Table S3). This clearly makes sense as collectively having more genes related to motor activity, transporter activity and signal transduction on the leading strand may enable the bacteria to react much faster when the nutrients become available (36,37).
DISCUSSION
It has been observed that bacterial genomes have a large variation in terms of the percentage of their leading-strand genes, ranging from ∼45% to ∼90%. We have provided an explanation for the large variation of observed strand biases across 725 bacterial genomes, which extends substantially the previous explanations. Our key contributions through this study include that (i) the genes of certain functional categories that need to be on the leading strands of genomes, to enhance the survivability of the host; (ii) genes of some functional categories such as transcription factor have higher preference to be on the lagging strands; (iii) there is at least one balancing force that keeps genes from all moving to the more efficient leading strands during evolution, particularly in nutrient-depleted environments and (iv) the percentage of leading-strand genes for a bacterial genome can be well explained using the numbers of genes in 25 functional categories outlined in (i) to (ii), genome size and gene density. We anticipate that more sophisticated analyses could possibly lead to quantitative models relating the percentage of leading-strand genes in a bacterium to a few parameters, which reflect the relationships between the living environments of an organism and the ‘intended’ capabilities of the organism and the needs for its survival, giving rise to improved understanding about the rules that may determine which genes will be on the leading versus the lagging strand of a genome.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3 and Supplementary Figures 1–3.
FUNDING
Funding for open access charge: The National Science Foundation [DEB-0830024 and DBI-0542119] and the DOE BioEnergy Science Center [DE-PS02-06ER64304], which is supported by the Office of Biological and Environmental Research in the Department of Energy Office of Science.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank all the members of the CSBL Lab at the UGA, especially Dr. Victor Olman for discussion on statistical analyses and Hank Schwartz for discussion on transcription factor being enriched on lagging strand.
REFERENCES
- 1.Koonin EV. Evolution of genome architecture. Int. J. Biochem. Cell. Biol. 2009;41:298–306. doi: 10.1016/j.biocel.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zivanovic Y, Lopez P, Philippe H, Forterre P. Pyrococcus genome comparison evidences chromosome shuffling-driven evolution. Nucleic Acids Res. 2002;30:1902–1910. doi: 10.1093/nar/30.9.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brewer BJ. When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell. 1988;53:679–686. doi: 10.1016/0092-8674(88)90086-4. [DOI] [PubMed] [Google Scholar]
- 4.French S. Consequences of replication fork movement through transcription units in vivo. Science. 1992;258:1362–1365. doi: 10.1126/science.1455232. [DOI] [PubMed] [Google Scholar]
- 5.Rocha EP, Danchin A. Essentiality, not expressiveness, drives gene-strand bias in bacteria. Nat. Genet. 2003;34:377–378. doi: 10.1038/ng1209. [DOI] [PubMed] [Google Scholar]
- 6.Rocha EP. The replication-related organization of bacterial genomes. Microbiology. 2004;150:1609–1627. doi: 10.1099/mic.0.26974-0. [DOI] [PubMed] [Google Scholar]
- 7.Hashimoto M, Ichimura T, Mizoguchi H, Tanaka K, Fujimitsu K, Keyamura K, Ote T, Yamakawa T, Yamazaki Y, Mori H, et al. Cell size and nucleoid organization of engineered Escherichia coli cells with a reduced genome. Mol. Microbiol. 2005;55:137–149. doi: 10.1111/j.1365-2958.2004.04386.x. [DOI] [PubMed] [Google Scholar]
- 8.Kato J, Hashimoto M. Construction of consecutive deletions of the Escherichia coli chromosome. Mol. Syst. Biol. 2007;3:132. doi: 10.1038/msb4100174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, et al. Essential Bacillus subtilis genes. Proc. Natl Acad. Sci. USA. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Price MN, Alm EJ, Arkin AP. Interruptions in gene expression drive highly expressed operons to the leading strand of DNA replication. Nucleic Acids Res. 2005;33:3224–3234. doi: 10.1093/nar/gki638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rocha E. Is there a role for replication fork asymmetry in the distribution of genes in bacterial genomes? Trends Microbiol. 2002;10:393–395. doi: 10.1016/s0966-842x(02)02420-4. [DOI] [PubMed] [Google Scholar]
- 12.Hu J, Zhao X, Yu J. Replication-associated purine asymmetry may contribute to strand-biased gene distribution. Genomics. 2007;90:186–194. doi: 10.1016/j.ygeno.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Lin Y, Gao F, Zhang CT. Functionality of essential genes drives gene strand-bias in bacterial genomes. Biochem. Biophys. Res. Commun. 2010;396:472–476. doi: 10.1016/j.bbrc.2010.04.119. [DOI] [PubMed] [Google Scholar]
- 14.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barrell D, Dimmer E, Huntley RP, Binns D, O'Donovan C, Apweiler R. The GOA database in 2009—an integrated gene ontology annotation resource. Nucleic Acids Res. 2009;37:D396–D403. doi: 10.1093/nar/gkn803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faith JJ, Driscoll ME, Fusaro VA, Cosgrove EJ, Hayete B, Juhn FS, Schneider SJ, Gardner TS. Many microbe microarrays database: uniformly normalized affymetrix compendia with structured experimental metadata. Nucleic Acids Res. 2008;36:D866–D870. doi: 10.1093/nar/gkm815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao F, Zhang CT. Ori-Finder: a web-based system for finding oriCs in unannotated bacterial genomes. BMC Bioinformatics. 2008;9:79. doi: 10.1186/1471-2105-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vieira-Silva S, Rocha EP. The systemic imprint of growth and its uses in ecological (meta)genomics. PLoS Genet. 2010;6:e1000808. doi: 10.1371/journal.pgen.1000808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qu H, Wu H, Zhang T, Zhang Z, Hu S, Yu J. Nucleotide compositional asymmetry between the leading and lagging strands of eubacterial genomes. Res. Microbiol. 2010;161:838–846. doi: 10.1016/j.resmic.2010.09.015. [DOI] [PubMed] [Google Scholar]
- 21.Nakayama K, Kurokawa K, Fukuhara M, Urakami H, Yamamoto S, Yamazaki K, Ogura Y, Ooka T, Hayashi T. Genome comparison and phylogenetic analysis of Orientia tsutsugamushi strains. DNA Res. 2010;17:281–291. doi: 10.1093/dnares/dsq018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paul D, Bridges SM, Burgess SC, Dandass YS, Lawrence ML. Complete genome and comparative analysis of the chemolithoautotrophic bacterium Oligotropha carboxidovorans OM5. BMC Genomics. 2010;11:511. doi: 10.1186/1471-2164-11-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trost E, Gotker S, Schneider J, Schneiker-Bekel S, Szczepanowski R, Tilker A, Viehoever P, Arnold W, Bekel T, Blom J, et al. Complete genome sequence and lifestyle of black-pigmented Corynebacterium aurimucosum ATCC 700975 (formerly C. nigricans CN-1) isolated from a vaginal swab of a woman with spontaneous abortion. BMC Genomics. 2010;11:91. doi: 10.1186/1471-2164-11-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Vries SP, van Hijum SA, Schueler W, Riesbeck K, Hays JP, Hermans PW, Bootsma HJ. Genome analysis of Moraxella catarrhalis strain RH4, a human respiratory tract pathogen. J. Bacteriol. 2010;192:3574–3583. doi: 10.1128/JB.00121-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janssen PJ, Van Houdt R, Moors H, Monsieurs P, Morin N, Michaux A, Benotmane MA, Leys N, Vallaeys T, Lapidus A, et al. The complete genome sequence of Cupriavidus metallidurans strain CH34, a master survivalist in harsh and anthropogenic environments. PLoS One. 2010;5:e10433. doi: 10.1371/journal.pone.0010433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ran L, Larsson J, Vigil-Stenman T, Nylander JA, Ininbergs K, Zheng WW, Lapidus A, Lowry S, Haselkorn R, Bergman B. Genome erosion in a nitrogen-fixing vertically transmitted endosymbiotic multicellular cyanobacterium. PLoS One. 2010;5:e11486. doi: 10.1371/journal.pone.0011486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Falentin H, Deutsch SM, Jan G, Loux V, Thierry A, Parayre S, Maillard MB, Dherbecourt J, Cousin FJ, Jardin J, et al. The complete genome of Propionibacterium freudenreichii CIRM-BIA1, a hardy actinobacterium with food and probiotic applications. PLoS One. 2010;5:e11748. doi: 10.1371/journal.pone.0011748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Welsh EA, Liberton M, Stockel J, Loh T, Elvitigala T, Wang C, Wollam A, Fulton RS, Clifton SW, Jacobs JM, et al. The genome of Cyanothece 51142, a unicellular diazotrophic cyanobacterium important in the marine nitrogen cycle. Proc. Natl Acad. Sci. USA. 2008;105:15094–15099. doi: 10.1073/pnas.0805418105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dombi GW, Nandi P, Saxe JM, Ledgerwood AM, Lucas CE. Prediction of rib fracture injury outcome by an artificial neural network. J. Trauma. 1995;39:915–921. doi: 10.1097/00005373-199511000-00016. [DOI] [PubMed] [Google Scholar]
- 30.Worning P, Jensen LJ, Hallin PF, Staerfeldt HH, Ussery DW. Origin of replication in circular prokaryotic chromosomes. Environ. Microbiol. 2006;8:353–361. doi: 10.1111/j.1462-2920.2005.00917.x. [DOI] [PubMed] [Google Scholar]
- 31.Freilich S, Kreimer A, Meilijson I, Gophna U, Sharan R, Ruppin E. The large-scale organization of the bacterial network of ecological co-occurrence interactions. Nucleic Acids Res. 2010;38:3857–3868. doi: 10.1093/nar/gkq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gama-Castro S, Salgado H, Peralta-Gil M, Santos-Zavaleta A, Muniz-Rascado L, Solano-Lira H, Jimenez-Jacinto V, Weiss V, Garcia-Sotelo JS, Lopez-Fuentes A, et al. RegulonDB version 7.0: transcriptional regulation of Escherichia coli K-12 integrated within genetic sensory response units (Gensor Units) Nucleic Acids Res. 2011;39:D98–D105. doi: 10.1093/nar/gkq1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janga SC, Salgado H, Martinez-Antonio A. Transcriptional regulation shapes the organization of genes on bacterial chromosomes. Nucleic Acids Res. 2009;37:3680–3688. doi: 10.1093/nar/gkp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang R, Lin Y. DEG 5.0, a database of essential genes in both prokaryotes and eukaryotes. Nucleic Acids Res. 2009;37:D455–D458. doi: 10.1093/nar/gkn858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stocker R, Seymour JR, Samadani A, Hunt DE, Polz MF. Rapid chemotactic response enables marine bacteria to exploit ephemeral microscale nutrient patches. Proc. Natl Acad. Sci. USA. 2008;105:4209–4214. doi: 10.1073/pnas.0709765105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amos L, Klug A. Arrangement of subunits in flagellar microtubules. J. Cell Sci. 1974;14:523–549. doi: 10.1242/jcs.14.3.523. [DOI] [PubMed] [Google Scholar]
- 37.Wemmer KA, Marshall WF. Flagellar motility: all pull together. Curr. Biol. 2004;14:R992–R993. doi: 10.1016/j.cub.2004.11.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.