Abstract
Programmed −1 ribosomal frameshifting is employed in the expression of a number of viral and cellular genes. In this process, the ribosome slips backwards by a single nucleotide and continues translation of an overlapping reading frame, generating a fusion protein. Frameshifting signals comprise a heptanucleotide slippery sequence, where the ribosome changes frame, and a stimulatory RNA structure, a stem–loop or RNA pseudoknot. Antisense oligonucleotides annealed appropriately 3′ of a slippery sequence have also shown activity in frameshifting, at least in vitro. Here we examined frameshifting at the U6A slippery sequence of the HIV gag/pol signal and found high levels of both −1 and −2 frameshifting with stem–loop, pseudoknot or antisense oligonucleotide stimulators. By examining −1 and −2 frameshifting outcomes on mRNAs with varying slippery sequence-stimulatory RNA spacing distances, we found that −2 frameshifting was optimal at a spacer length 1–2 nucleotides shorter than that optimal for −1 frameshifting with all stimulatory RNAs tested. We propose that the shorter spacer increases the tension on the mRNA such that when the tRNA detaches, it more readily enters the −2 frame on the U6A heptamer. We propose that mRNA tension is central to frameshifting, whether promoted by stem–loop, pseudoknot or antisense oligonucleotide stimulator.
INTRODUCTION
Accurate maintenance of the translational reading frame is essential in the production of functional proteins and spontaneous frameshifting occurs rarely, with an estimated frequency (in Escherichia coli) of 3 × 10−3 – 5 × 10−5 per codon (1). In some genes, however, mRNA elements are present that induce the ribosome to change reading frame at very high frequencies (reviewed in 2–4). These sites of programmed ribosomal frameshifting direct ribosomes into an overlapping open reading frame (ORF), generating a fusion protein containing the products of both upstream and downstream ORFs. Most widespread are sites of programmed −1 ribosomal frameshifting (–1 FS) where the ribosome slips back one nucleotide (nt) in the 5′-direction on the mRNA. Frameshifting in eukaryotes was first described as the mechanism by which the Gag-Pol polyprotein of the retrovirus Rous sarcoma virus (RSV) is expressed from overlapping gag and pol ORFs (5,6) and related signals have since been documented in many other viruses, including the clinically important human immunodeficiency virus types 1 and 2 (7) (HIV-1, HIV-2), human T-cell lymphotrophic virus types 1 and 2 (8,9) and the coronavirus responsible for severe acute respiratory syndrome (10). Frameshifting has also been increasingly recognized in conventional cellular genes of both prokaryotes and eukaryotes as well as in other replicating elements, such as insertion sequences and transposons.
The mRNA signal for −1 FS is composed of two elements, a slippery sequence with consensus X_XXY_YYZ (underlines denote zero frame; X can be any base, Y is A or U, Z is not G in eukaryotic systems) where the ribosome changes frame, and a downstream stimulatory RNA structure, a stem–loop or pseudoknot (reviewed in 3,4). Appropriate spacing (typically 5–8 nt) between slippery sequence and stimulatory RNA is also required for optimal −1 FS efficiency (11–13). There is considerable experimental support for the idea that ‘tandem-slippage’ of ribosome-bound peptidyl- and aminoacyl-tRNAs on the slippery sequence occurs upon encounter of the stimulatory RNA, with the tRNAs detaching from the zero frame codons (XXY_YYZ) and re-pairing in the −1 frame (XXX_YYY) (6,14). What actually drives tRNA movement in frameshifting is uncertain. There is accumulating evidence to suggest involvement of an intrinsic unwinding activity of the ribosome (15), with the stimulatory RNA exhibiting resistance to unwinding, perhaps by presenting an unusual topology. Failure to unwind the stimulatory RNA appropriately has been proposed to induce tension in the mRNA leading to uncoupling of the codon:anticodon complexes and realignment of the tRNAs in the −1 frame (16–29).
In recent years it has been discovered that efficient −1 FS can also be stimulated in some circumstances simply by annealing an RNA oligonucleotide downstream of a slippery sequence, at least in vitro (30–32). This was unexpected as mRNA-antisense oligonucleotide (AON) complexes appear to lack the structural features of naturally occurring stimulatory RNAs, such as stem–stem junctions, base triplexes or kinks, that have been associated with models implicating resistance to unwinding (reviewed in 3,4). In an attempt to gain insight into the mechanism of AON-induced −1 FS, we initiated a study to examine the effect on −1 FS of modulating the spacing distance between slippery sequence and annealed AON. During the initial in vitro translations carried out to validate the system, we were intrigued to observe ‘two’ frameshift products in the AON-stimulated frameshift assays. In this article, we describe our examination of the origin of these products. We show that in the experimental system employed, based on that developed by Howard and colleagues (30), both −1 ‘and’ −2 FS can occur efficiently on the HIV-1 slippery sequence (U6A) in response to bound AONs. Importantly, this effect is also seen when the AON stimulator is replaced by a stem–loop or pseudoknot stimulator. By examining −1 and −2 FS outcomes on mRNAs with varying slippery sequence-stimulatory RNA spacing distances, we found that the spacer-length optimum for maximal frameshifting is different depending upon the kind of stimulatory RNA employed, and that −2 FS is optimal at a spacer length 1–2 nt shorter than that optimal for −1 FS. We propose that the shorter spacer increases the tension on the mRNA such that when the tRNA detaches, it more readily enters the −2 frame on the U6A heptamer. These experiments provide the first observation of −2 FS on a eukaryotic viral heptameric slippery sequence and support the view that mRNA tension is central to the mechanism of frameshifting, not only with ‘traditional’ stem–loop or pseudoknot RNAs, but also with AON stimulators.
MATERIALS AND METHODS
Mutagenesis
Site-directed mutagenesis was performed using the Quikchange II site-directed mutagenesis kit (Stratagene) according to the manufacturer’s instructions.
Plasmids
Assessment of in vitro frameshift efficiencies employed plasmids derived from pFScass 5 (12). This vector contains the bacteriophage SP6 polymerase promoter driving expression of the influenza A/PR8/34 PB2 gene, with the minimal infectious bronchitis coronavirus (IBV) frameshifting signal (12) inserted at the Bgl II site at position 486 of the PB2 gene. We modified pFScass 5 by introducing a unique Mlu I site downstream of the inserted pseudoknot (creating plasmid pFScass 9), removed the entire IBV frameshifting signal using Bgl II and Mlu I and replaced it with a pair of complementary oligonucleotides encoding the AON-driven frameshifting signal utilized by Howard and colleagues (30), giving plasmid pFSHIV-AON (Figure 1). The frameshift signal in this plasmid comprises the HIV-1 slippery sequence U6A positioned 3 nt upstream of the binding site for a complementary AON. Frameshift assays in tissue culture cells employed derivatives of the dual luciferase reporter plasmid p2luc (33). The AON frameshifting signal present in pFSHIV-AON was cloned as a pair of complementary oligonucleotides into Bam HI and Sal I-cut p2luc such that expression of the downstream firefly luciferase gene required either a −1 (p2lucAON −1) or a −2 (p2lucAON −2) frameshift at the end of the upstream Renilla luciferase gene. To allow calculation of frameshifting efficiencies, two control plasmids (p2lucOinc and p2lucOP1inc) were prepared in which the two luciferases were aligned in-frame in order to obtain readings for normalization of data. Expression of the U6A slippery sequence-derived −2 FS product in transfected cells employed plasmid pFSeGFP-N2. This plasmid was prepared by subcloning a polymerase chain reaction (PCR)-generated DNA fragment encoding the U6A slippery sequence, a 7 nt spacer and a stimulatory stem–loop structure (see section ‘Results’) into XhoI/BamHI-cut pEGFP-N2 (Clontech, GenBank Accession number U57608). In this plasmid, the eGFP tag is expressed only in the −2 frame (following a frameshift event) and the natural start codon of eGFP was changed (to TCG) by site-directed mutagenesis to minimize leaky expression of eGFP. All plasmid sequences were confirmed by dideoxy sequencing.
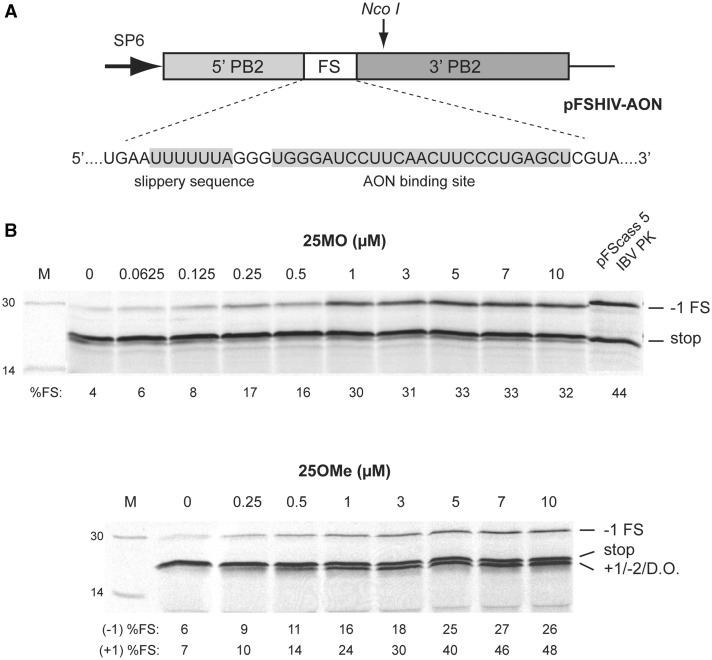
Figure 1.
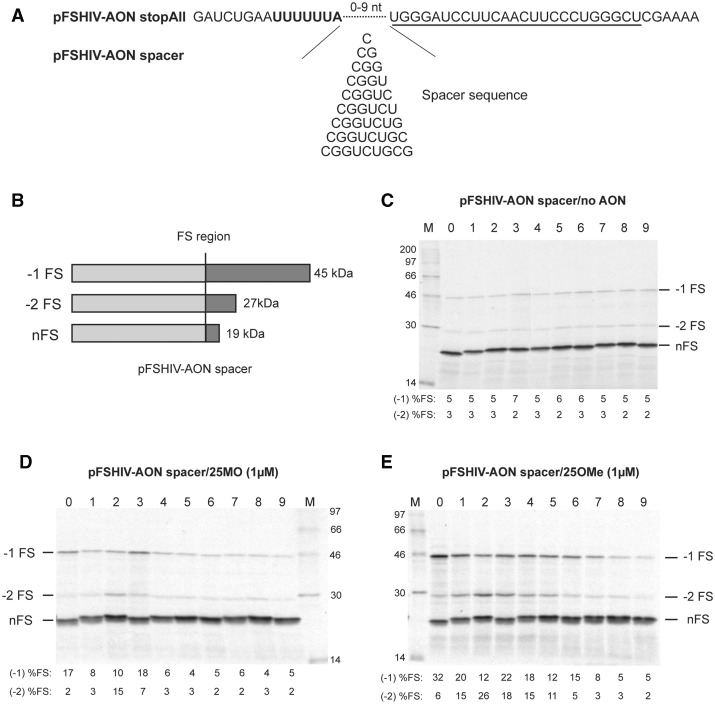
Antisense-oligonucleotide mediated frameshifting. (A) Plasmid pFSHIV-AON was prepared by cloning an AON-frameshift cassette (based on Ref. 30) into the influenza A PB2 gene within an SP6-based transcription vector. The cassette comprised the slippery sequence (U6A) of the HIV-1 frameshift signal, a 3 nt spacer (GGG) and a 25 nt stretch complementary to AONs 25MO or 25OMe. In in vitro translations in RRL, Nco I-linearized, SP6-derived mRNAs generate an 18 kDa non-frameshift product (stop) and a 28 kDa −1 frameshift product (−1 FS). (B) Messenger RNAs derived from Nco I-cut pFSHIV-AON were translated in RRL (at ∼50 µg/ml) in the presence of increasing quantities of 25MO or 25OMe. The products were resolved by 15% SDS–PAGE and visualized by autoradiography. In most lanes, an additional product is evident, thought to be derived from ribosomes that enter the +1/−2 frame (+1/−2) or fall off the template (or are permanently stalled) in the vicinity of the annealed AON (drop-off, d.o.). Control translations were also carried out using an mRNA derived from pFScass 5 (Ref. 12; pFScass 5/IBV PK) which contains the minimal IBV frameshift site. The frameshifting efficiency measured for each signal (to the nearest integer) is indicated below the relevant lanes (%FS; see section ‘Materials and Methods’) and takes into account the number of methionines present in each product (stop, 10; −1 FS, 10; +1/−2/d.o., 11). Due to the close migration of the +1/−2/d.o. product and the stop product in this experiment, the values represent our best efforts for quantification of each class of event. M; molecular size markers.
Oligonucleotides
The morpholino oligonucleotide 25MO (5′-AGCTCAGGGAAGTTGAAGGATCCCA-3′) was purchased from Gene Tools (Oregon, USA). The equivalent 2-O-Me oligonucleotide 25OMe (5′-AGCUCAGGGAAGUUGAAGGAUCCCA-3′), a truncated version (15OMe; 5′-AGUUGAAGGAUCCCA-3′) and an equivalent 15mer composed of RNA bases (15RNA) were from Thermo Scientific (Colorado, USA). The primary sequence of 25MO is identical to that of MOAB, described by Howard et al. (30).
In vitro transcription and translation
Frameshift reporter plasmids were linearized with Nco I or Bam H1 and capped run-off transcripts generated using SP6 RNA polymerase as described (12). Messenger RNAs were recovered by a single extraction with phenol/chloroform (1:1 vol/vol) followed by ethanol precipitation. Remaining unincorporated nucleotides were removed by gel filtration through a NucAway spin column (Ambion). The eluate was concentrated by ethanol precipitation, the mRNA resuspended in water, checked for integrity by agarose gel electrophoresis and quantified by spectrophotometry. Messenger RNAs were translated in nuclease-treated rabbit reticulocyte lysate (RRL; Promega) programmed with ∼50 µg/ml template mRNA. Typical reactions were of 10 µl volume and composed of 90% (vol/vol) RRL, 20 µM amino acids (lacking methionine) and 0.2 MBq [35S]-methionine. Reactions were incubated for 1 h at 30°C and stopped by the addition of an equal volume of 10 mM ethylenediaminetetraacetic acid (EDTA), 100 µg/ml RNase A followed by incubation at room temperature for 20 min. Samples were prepared for sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) by the addition of 10 vol of 2× Laemmli’s sample buffer (34), boiled for 3 min and resolved by SDS–PAGE. Dried gels were exposed to a Cyclone Plus Storage Phosphor Screen (PerkinElmer), the screen scanned using a Typhoon TRIO Variable Mode Imager (GE Healthcare) in storage phosphor autoradiography mode, and bands were quantified using ImageQuantTMTL software (GE Healthcare). Where used, AONs were added to translation reactions on ice at the same time as the mRNA, but were not pre-annealed to the mRNA. The calculations of frameshifting efficiency (%FS) took into account the differential methionine content of the various products and utilized two formulas. For a single frameshift event, %FS was calculated as 100{IFS1(MetS/MetFS1)/(IS + IFS1(MetS/MetFS1)} and where two frameshift events were noted, %FS = 100{IFS1(MetS/MetFS1)/[IS+IFS1(MetS/MetFS1)+IFS2(MetS/MetFS2)]}. In the formulae, the number of methionines in the stop, frameshift 1 and frameshift 2 products are denoted by MetS, MetFS1 and MetFS2, respectively; the densitometer values for the same products by IS, IFS1 and IFS2.
Frameshifting assays in tissue culture
COS 7 cells were maintained in Dulbecco’s modification of Eagle’s medium supplemented with 10% (vol/vol) fetal calf serum, 1% (vol/vol) penicillin/streptomycin and 1% (wt/vol) 25 mM glutamine. Plasmids were transfected using a commercial liposome method (Lipofectamine 2000, Invitrogen). 4 × 104 cells were seeded per well in 24-well plates and grown for 18–24 h until 80% confluency was reached. Transfection mixtures (containing plasmid DNA, serum-free medium [Optimem; Gibco-BRL] and Lipofectamine 2000) were set up as recommended by the manufacturer and added directly (dropwise) to the tissue culture cell growth medium. The cells were harvested 24 h post-transfection and reporter gene expression determined using a dual luciferase assay system kit (Promega). Each data point represents the mean value (± SEM) from six separate transfections.
Mass spectroscopy
Plasmid pFSeGFP-N2 expresses a fusion protein comprising N-terminal U6A-derived −2 FS peptide and a C-terminal eGFP tag. 293 T cells (7 × 106 per dish) were seeded onto 15 × 10 cm dishes and transfected with pFSeGFP-N2 (15 µg per dish) while in suspension using TransIT (Mirus Bio LLC). After 68 h, the proteasome inhibitor MG132 was added to the growth medium (to 20 μM), the cells were harvested 4 h later and lysed in 1 ml lysis buffer containing 50 mM Tris–HCl, pH 8, 5% glycerol, 0.5% IGEPAL CA-630, 200 mM NaCl, 1 mM DTT, 1 mM PMSF and 1 × complete protease inhibitor tablet (Roche). After clarification, the supernatant was mixed with 50 µl glutathione sepharose beads and incubated for 30 min at 4°C with rotation to pre-clear the lysate. Subsequently, the supernatant was incubated with 20 µl GFP-TRAP_A (ChromoTek) for 2 h at 4°C with rotation to bind GFP-tagged proteins. The beads were washed three times with wash buffer (10 mM Tris–HCl pH 8, 0.1% IGEPAL CA-630, 150 mM NaCl, 0.5 mM EDTA, 1 mM PMSF and 1× complete protease inhibitor tablet), transferred to an Ultrafree-MC spin column (0.22 µm; Millipore) and any remaining wash buffer removed by centrifugation. The beads were resuspended in 20 µl of 4× Laemmli’s buffer (250 mM Tris pH 6.8, 4% [wt/vol] SDS, 40% [vol/vol] glycerol, 10% [vol/vol] beta-mercaptoethanol, 20 μg/ml bromophenol blue), transferred to a fresh microcentrifuge tube and boiled for 10 min. The eluate was subjected to 12% SDS–PAGE, the proteins fixed with colloidal coomassie fix (45% [vol/vol] methanol, 1% [vol/vol] acetic acid) for 1 h, the gel stained with colloidal coomassie stain (17% [wt/vol] ammonium sulphate, 0.1% [wt/vol] coomassie G250, 0.5% [vol/vol] acetic acid and 34% methanol) overnight, washed with water and stored in 10% acetic acid solution. Band excision and mass spectroscopic (MS) analysis of the −2 FS product was carried out at the University of Cambridge, Department of Biochemistry Protein and Nucleic Acid Facility (PNAC). The excised band was subjected to reduction with 5 mM tris(2-carboxyethyl)phosphine and alkylation by addition of iodoacetamide (to 25 mM), followed by liquid removal and washes with 200 µl 100 mM ammonium bicarbonate with 50% acetonitrile. The gel pieces were dried in vacuo for 10 min and 25 µl of 100 mM ammonium bicarbonate containing 10 µg/ml modified trypsin (Promega) was added for trypsin digestion for 17 h at 32°C. Peptides were recovered and desalted using µC18 ZipTip (Millipore) and eluted to a maldi target plate using 2 µl alpha-cyano-4-hydroxycinnamic acid matrix (Sigma) in 50% acetonitrile, 0.1% trifluoroacetic acid. Peptide mass was determined using a Maldi micro MX MS (Waters) in reflectron mode and analysed with Masslynx software. For tandem ms/ms analysis (ESIMS/MS), desalted peptides in 70% methanol, 0.2% formic acid were delivered to a ThermoFinnigan LCQ Classic ion-trap MS using a static nanospray needle (Thermo Proxeon). Peptide masses of interest were manually selected for fragmentation using manufacturer-recommended settings. Fragment ions were matched to possible sequence interpretations using MS-Product (http://prospector.ucsf.edu/).
RESULTS
Identification of a novel product generated during translation of an AON-mediated ribosomal frameshifting signal
Our experimental system for studying AON-mediated frameshifting in vitro is based on that of Howard et al. (30) and is shown in Figure 1. We began by confirming that a 25 nt long morpholino (25MO) or 2-O-Me (25OMe) AON could stimulate–1 FS at the slippery sequence of HIV-1 (U6A) when bound 3 nt downstream on the mRNA. The frameshift reporter mRNA was transcribed from Nco I-cut pFSHIV-AON, a derivative of pFScass 5 (12; see section ‘Materials and Methods’), and translated in rabbit RRL in the presence of increasing concentrations of AON. As can be seen in Figure 1, both non-frameshifted (stop, 18 kDa) and −1 FS product (28 kDa) were seen, with the −1 FS efficiency peaking at ∼30%, a level similar to that measured for a control, pseudoknot-dependent frameshift signal (pFScass 5 IBV PK; 44% in this experiment). In the absence of added AON, the baseline −1 FS efficiency was 4–6%, indicating that the U6A heptamer is inherently slippery in RRL, as observed previously (12,30). In control translations with a non-targeting AON, or of mRNAs with mismatches in the AON binding site, very little frameshifting was seen, confirming the specificity of AON-mediated frameshifting (data not shown). Also evident in the 25OMe titration of Figure 1 was an unexpected product migrating just below the stop product. Based on the nucleotide sequence of the frameshift region and the position of termination codons in the different reading frames, this protein may correspond to ribosomes that have undergone a +1 (or −2) frameshift on the U6A heptamer. Alternatively, it could represent a peptide derived from ribosomes that had irreversibly stalled at the annealed AON, and subsequently dropped off the template (drop-off or d.o.). The unexpected product was also present, albeit to a lesser extent, in the 25MO titration. Given the possibility that it may represent an alternative frameshift product, we examined whether its generation was linked to the homopolymeric nature of the U6A slippery sequence by changing the slippery sequence of pFSHIV-AON to that present at the IBV (UUUAAAC) or simian retrovirus 1 (SRV-1) gag/pro (GGGAAAC) frameshift sites (Figure 2). With these mRNAs, little or no +1/−2/d.o. product was evident, indicating that its appearance is most likely linked to the U6A slippery sequence rather than to a compromise of elongation arising from the presence of a stably bound AON. Examining the AON titrations further (Figures 1 and 2), it can be seen that the plateau of −1 FS stimulation with 25MO was reached slightly earlier (at about 1 µM) than for 25OMe (3–5 µM). However, 25OMe was the more effective stimulator, promoting 49% −1 FS on UUUAAAC (c.f. 22% with 25MO), 42% on GGGAAAC (19% with 25MO) and, on the assumption that the novel product seen with U6A is an alternative frameshift product, a total of 74% frameshifting on U6A (33% with 25MO). The −1 FS efficiency engendered by 25OMe on the IBV and SRV-1 gag/pro slippery sequences (49% and 42%, respectively) was greater than that seen with that of HIV-1 (27%). However, it is likely that −1 FS on U6A would be greater if a proportion of ribosomes were not entering another frame.
Figure 2.
AON-mediated frameshifting on slippery sequences UUUAAAC and GGGAAAC. (A) Plasmids pFSIBV-AON and pFSSRV-AON were prepared by replacing the slippery sequence of pFSHIV-AON (U6A) by UUUAAAC or GGGAAAC respectively. (B) In vitro translations of pFSIBV-AON or pFSSRV-AON/Nco I mRNAs were carried out in the presence of increasing quantities of 25MO or 25OMe and the products analysed as in the legend to Figure 1. The non-frameshifted product (nFS), and those proteins arising from −1 or +1/−2 frameshifting or drop-off (d.o.) are indicated. Control mRNAs were also translated (pFScass5 IBV PK, HIV-AON/25OMe, HIV-AON/25MO) at the given AON concentrations (where relevant).
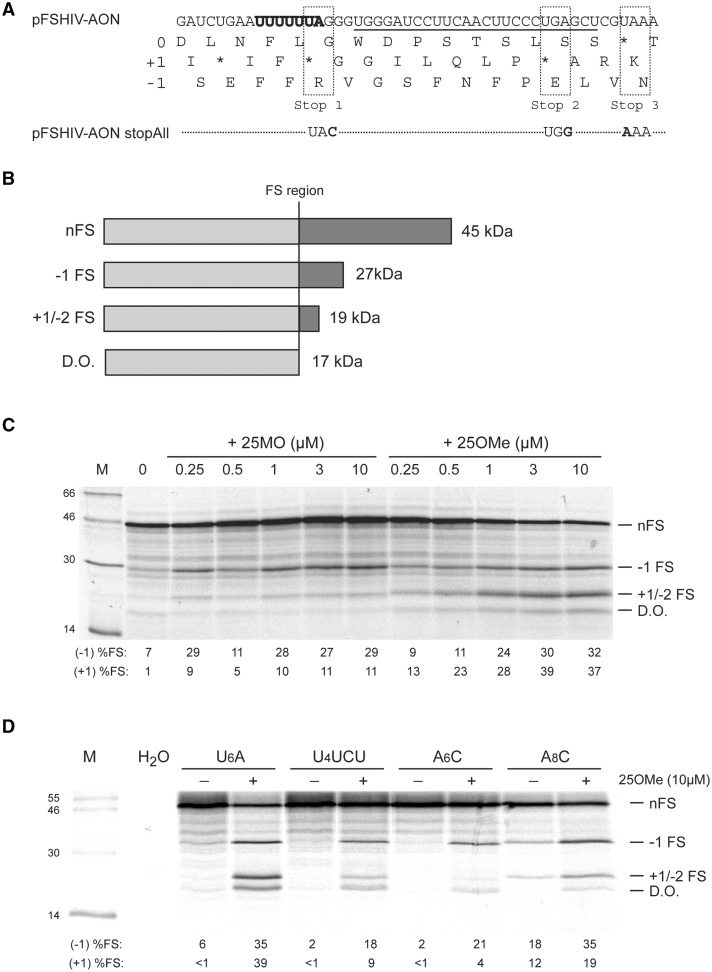
In an attempt to disentagle possible origins of the +1/−2/d.o. product, a variant of pFSHIV-AON was prepared (pFSHIV-AON stopAll) in which three local stop codons in the 0 and +1 reading frames were changed to sense codons such that the length of the ORFs downstream of the U6A stretch (in each of the three frames) would allow differentiation of a product of +1/−2 FS from that of a d.o. event based on size (Figure 3). This experiment revealed that the majority of the +1/−2/d.o. product appears to be derived from a +1/−2 FS, with a small proportion derived from a product whose size is consistent with prolonged (or permanent) stalling at the U6A heptamer (presumably) while the ribosome attempts to unwind the annealed oligonucleotide. Consistent with earlier work, with 25MO there was less +1/−2 FS product and little or no d.o. product, suggesting that morpholino oligonucleotides present less of a barrier to ribosomal elongation than their 2-O-Me counterparts. Within the context of pFSHIV-AON stopAll, three slippery sequence variants were prepared to address the question of whether 25OMe-stimulated +1/−2 FS was strictly dependent on a U-rich slippery sequence. As shown in Figure 3, with slippery sequence U4UCU, possessing an A-site codon sub-optimal for frameshifting, both −1 FS and +1/−2 FS were diminished (3- to 4-fold) suggesting a requirement for optimal A-site tRNA re-pairing in each case (see below). With slippery sequence A6C, efficient −1 FS was observed (21%) but the quantity of +1/−2 FS product was reduced (albeit detectable at ∼4%). We also tested A8C and were surprised to see relatively high expression of both −1 and +1/−2 FS products in the absence of added AON. The synthesis of these products could potentially be accounted for by transcriptional slippage on A8C by SP6 RNA polymerase during synthesis of the reporter RNAs (35). Alternatively, on this long homopolymeric A8C stretch, the ribosome can frequently lose frame even in the absence of a stimulatory element. A similar observation has been made for a U8 stretch (12). Significantly, in the presence of 25OMe, expression of the −1 FS product from the A8C mRNA rose to a level similar to that seen with the U6A mRNA and the +1/−2 FS product was also enhanced (to about half the value seen with U6A). Thus translational frameshifting is certainly taking place on this A-rich stretch and both −1 FS and +1/−2 FS products are synthesized.
Figure 3.
Confirmation of the +1/−2 FS event. (A) Nucleotide sequence and three-frame translation of the frameshift region of pFSHIV-AON. The slippery sequence is shown in bold, the binding site of 25MO/25OMe is underlined. The 0 and +1 frame stop codons (Stop 1–3) changed in pFSHIV-AON stopAll are boxed. (B) Diagrammatic representation of potential translation products of pFSHIV-AON stopAll and predicted molecular masses. (C) In vitro translation of pFSHIV-AON stopAll/Nco I mRNA was carried out in the presence of increasing quantities of 25MO or 25OMe and the products analysed as in the legend to Figure 1. The non-frameshifted product (nFS), and those proteins arising from −1 or +1/−2 frameshifting (−1 FS; +1/−2 FS), or from template drop-off at the site of the annealed oligonucleotide (d.o., see text) are indicted. The frameshifting efficiency measured for each signal (to the nearest integer) is indicated below the relevant lanes (%FS; see section ‘Materials and Methods’) and takes into account the number of methionines present in each product (nFS, 19; −1 FS, 10; +1/−2 FS, 11). (D) In vitro translation of slippery sequence variants of pFSHIV-AON stopAll. The wild-type Nco-I-derived mRNA and three slippery sequence variants were translated in RRL in the absence (−) or presence (+) of 10 µM 25OMe. Products were analysed and quantified as in the legend to Figure 1. In mutant (C)A6C, the bracketed C is included to indicate that this base was also changed from the wild-type context (which is (A)U6C) to avoid generation of an A7 stretch.
Stimulation of efficient −1 and −2 FS on the U6A heptamer in vitro and in transfected cells
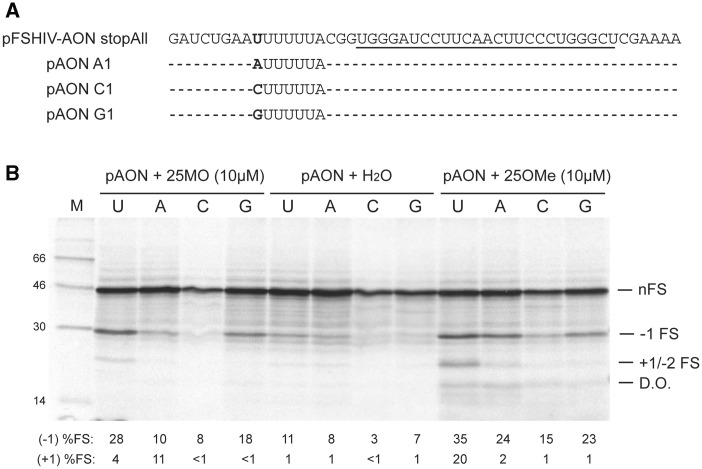
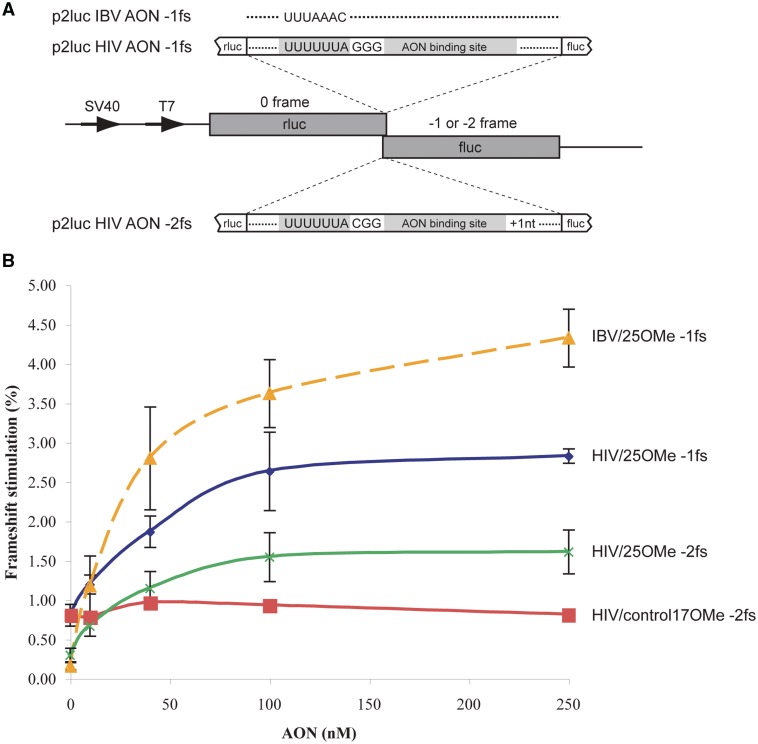
The reduction in +1/−2 FS frequency seen with the U4UCU heptamer (Figure 3) was thought-provoking in that it raised doubts as to whether a ‘traditional’ +1 FS event was occurring. In this mRNA, the A-site base changes (UUA to UCU) would not preclude forward movement of the P-site tRNA decoding the zero-frame UUU codon onto the overlapping +1 frame UUU codon, yet production of the +1/−2 product was diminished. To rule out that this was an effect of the identity of the A-site tRNA (tRNALeu versus tRNASer) on +1/−2 FS, we revisited the experiment, but changing the first base of the U6A heptamer (to A, C or U) in the context of pFSHIV-AON stopAll to probe P-site tRNA re-pairing in the −1 frame. From previous studies (6,12), we expected that tandem −1 slippage of P- and A-site tRNAs would be compromised to a greater or lesser extent, since the P-site codon would be sub-optimal for repairing in most cases, whereas a +1 movement of the P-site tRNA, as outlined above, would be unaffected. However, as shown in Figure 4, changing the first base to A, C or G effectively abolished the +1/−2 product. The effect on −1 FS was consistent with earlier work, with a reduction in all cases, especially with C at the first position. These data suggested strongly that the +1/−2 FS product results from −2 FS and this was confirmed by MS (for convenience, these data are presented at the end of the ‘Results’ section, since acquisition of sufficient trans-frame protein for MS analysis required additional knowledge obtained from experiments outlined in the following sections). The stimulation of both −1 and −2 FS on the U6A heptamer could also be engendered by an RNA-only oligonucleotide (15RNA) and a shorter 2-O-Me oligonucleotide (15OMe) (Supplementary Figure S1). Thus the capacity to stimulate both frameshift events is not specific to 25OMe. To confirm that AON-mediated −1 and −2 FS could also take place in a cellular context, the key elements of pFSHIV-AON were cloned into the dual luciferase frameshift reporter plasmid p2luc (33) such that the downstream firefly luciferase gene reported either −1 (p2lucHIV-AON −1 FS) or −2 (p2lucHIV-AON −2 FS) frameshifting, with the spacer length optimized in each case (−1 FS, 3 nt; −2 FS, 2 nt; see below). The relevant reporter plasmid together with increasing concentrations of 25OMe were transfected into COS 7 cells and luciferase activities measured 24 h later. Both −1 and −2 FS were detectable, with peak values of 2.6% (−1 FS) and 1.5% (−2 FS) and saturation at around ∼100 nM AON (Figure 5). A version of the −1 FS reporter plasmid with the IBV slippery sequence (p2lucIBV-AON) was also tested and showed similar levels of frameshifting, peaking at 4.5%. Although the absolute values are low, they are comparable to the frameshift efficiency of the natural HIV-1 signal in cells (∼5%; 36).
Figure 4.
Influence of the first base of the U6A slippery sequence on −1 and +1/−2 frameshifting. (A) The first base of the slippery sequence of pHIV-AON stopAll was changed to A, C or G. (B) In vitro translation of slippery sequence variants of pFSHIV-AON stopAll. The wild-type Nco-I-derived mRNA and three slippery sequence variants were translated in RRL following addition of 10 µM 25MO or 25OMe, or a water control. Products were analysed and quantified as in the legend to Figure 3.
Figure 5.
AON frameshift stimulation in mammalian cells. (A) Dual luciferase frameshift reporter plasmids were prepared in which downstream firefly luciferase (fluc) expression was dependent upon AON-mediated −1 frameshifting (p2luc IBV AON −1 FS, p2luc HIV AON −1 FS) or −2 frameshifting (p2luc HIV AON −2 FS). Plasmids were co-transfected with varying concentrations of 25OMe or a control 17OMe (lacking obvious complementarity to the target mRNA) using Lipofectamine 2000, with the concentration of AON calculated using the final overlay volume in the well. Frameshift efficiencies were determined by comparing luciferase activities to matched in-frame control constructs (see section ‘Materials and Methods’) co-transfected with 25OMe in separate dishes. (B) Measured frameshift efficiencies were plotted against AON concentration. Error bars represent the standard deviation of the mean from three experiments (control 17OMe titration was only done once).
Spacer-length dependence of programmed −1 or −2 ribosomal frameshifting on a U6A heptamer supports a role for mRNA tension in frameshifting
Based on the published literature, including our own studies of 80S ribosomes stalled at the IBV frameshift-stimulatory pseudoknot (22,37), we proposed a mechanical model of frameshifting in which a failure of intrinsic ribosomal helicase activity (15,28) to unwind efficiently the stimulatory RNA during the translocation step leads to the build up of tension in the mRNA and subsequently, breakage of codon:anticodon contacts and realignment of the tRNAs in the −1 reading frame. The validity of this model remains to be determined, but one prediction of it is that the magnitude of frameshifting should be influenced by relatively subtle changes to the length of the spacer separating the slippery sequence and stimulatory element. For the IBV frameshift signal, this is known to be the case; altering the natural spacing distance (6 nt) between pseudoknot and slippery sequence (UUUAAAC) by a single nucleotide either way has a 2-fold inhibitory effect on frameshifting, and a 5-fold reduction is seen when 2 nt are added to or removed from the spacer (12,37). Regarding AON-stimulated frameshifting, current evidence also supports a requirement for appropriate spacing. In the studies of Howard and colleagues (30), efficient frameshifting was seen with spacer lengths from −1 to 5 nt, with an optimum at 3 nt (as used in this study). Given the observation in the present work of AON-stimulated −1 and −2 FS, it was important to ascertain the optimal spacing distance for the two events. To do this, we modified pFSHIV-AON stopAll to generate plasmids (pFSHIV-AON spacer series) with spacers varying in length from 0 to 9 nt (Figure 6 panel A). In addition, for ease of comparison, a single base was added or removed from the mRNA downstream of the AON-binding region such that the size of the various translation products (stop, −1 FS, −2 FS) was maintained between constructs (Figure 6 panel B). In translations of the pFSHIV-AON spacer series, with added 25MO or 25OMe (at 1 µM in this experiment), we observed discrete peaks of frameshifting, with the overall pattern essentially the same, except that frameshifting was more efficient with 25OMe. The −1 FS product was evident across a broad range, spanning spacer lengths 0–7 nt, but with two optima, at 0 nt and at 3 nt, (particular noticeable in the 25MO titration). The −2 FS product had an optimal spacing of 2 nt and was evident over the range 1–5 nt with 25OMe, but more discrete with 25MO (2–3 nt). In the absence of added AON, both frameshift products were detectable although at low levels, and unsurprisingly, there were no spacer effects.
Figure 6.
The effect of slippery sequence-AON spacing on −1 and −2 frameshifting. (A) The spacer of pFSHIV-AON stopAll was changed from zero to nine nucleotides as indicated. In these plasmids, the sizes of the encoded frameshift products were normalised by appropriate deletion of bases downstream of the AON binding site. (B) Diagrammatic representation of potential translation products of pFSHIV-AON spacer mRNAs and predicted molecular masses. (C–E) Messenger RNAs derived from Nco I-cut pFSHIV-AON spacer variants were translated in RRL following addition of water (C), 1 µM 25MO (D) or 1 µM 25OMe (E). Products were analysed and quantified as in the legend to Figure 3. The numbers above each gel represent the spacer length. The frameshifting efficiency measured for each signal (to the nearest integer) is indicated below the relevant lanes (−1% FS; −2% FS) and takes into account the number of methionines present in each product (nFS, 10; −1 FS, 19; −2 FS, 11).
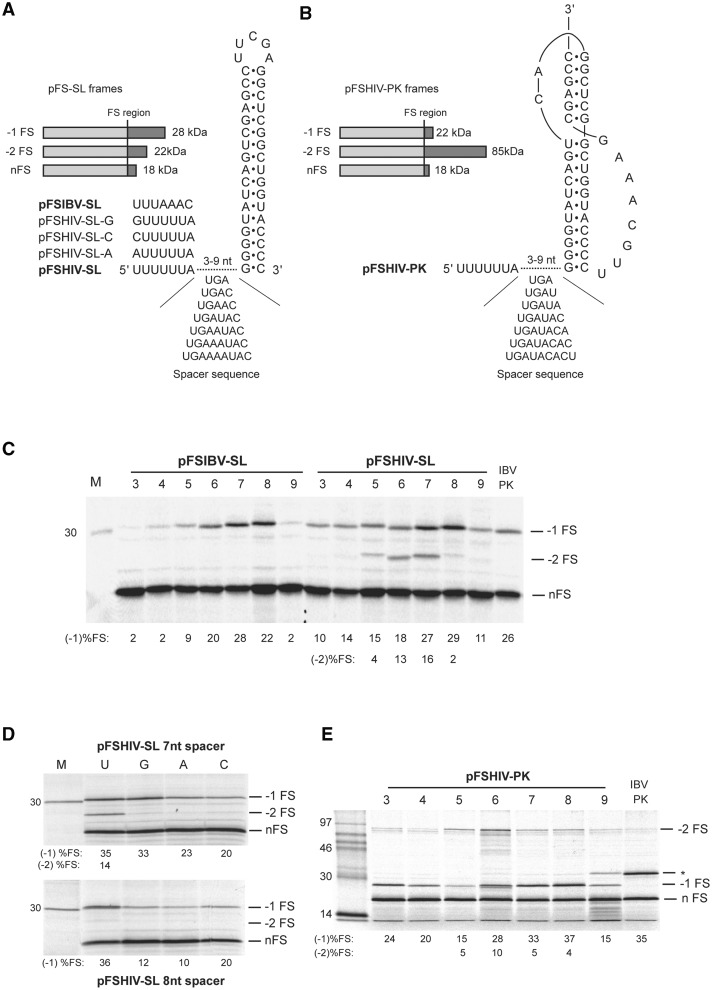
We went on to examine whether the −2 FS product could be engendered by cis-acting stimulatory RNAs and the spacing optima for such events. In these experiments, the AON-binding site was replaced by a pseudoknot (the minimal IBV pseudoknot; a functional version of the wild-type IBV pseudoknot with a shorter loop 3 [12]), generating plasmid pFSHIV-PK, or a stable stem–loop structure whose base composition was the same as the two stems of the minimal IBV pseudoknot, generating pFSHIV-SL (Figure 7). A variant of this plasmid with the IBV slippery sequence was also prepared (pFSIBV-SL). Subsequently, spacing variants of these plasmids were constructed (spacers of 3–9 nt) and the mRNAs translated in RRL. With pFSIBV-SL, only the −1 FS product was evident (consistent with the UUUAAAC slippery sequence being incompatible with −2 FS), with efficient −1 FS promoted over a narrow window of spacer length (6–8 nt), peaking at 7 nt. With pFSHIV-SL, both classes of frameshift product were seen. As with AON-mediated frameshifting, −1 FS on the U6A slippery sequence was observed at most spacer distances, peaking at 7–8 nt. The −2 FS product was more discrete, spanning spacers of 5–7 nt with a peak at 6–7 nt. Also shown in Figure 7 (panel D) are the translations of pFSHIV-SL-derived mRNA with spacers of 7 or 8 nt in which the first base of the slippery sequence was changed to G, A or C. Similar to the experiment of Figure 4, we found that disruption of this base inhibited both −1 and −2 FS. With the 8 nt spacer, only a trace of −2 FS is seen, as expected for this spacer length (c.f. Figure 7 panel C), and the inhibition of −1 FS was, if anything, more pronounced than with the 7 nt spacer. To account for the latter, we speculate that with the 7 nt spacer, a greater number of ribosomes have the capacity to frameshift (see Figure 7 panel C) and that partitioning occurs, with ribosomes able to enter either the −1 or −2 frame. With a 7 nt spacer, the block to −2 FS brought about by the slippery sequence changes may well direct ‘frameshift-competent’ ribosomes to partition more into the −1 frame. However, with an 8 nt spacer that does not promote −2 FS (see Figure 7 panel C), there are no additional ribosomes to partition, and the overall level of −1 FS is generally lower. The pattern of frameshifting observed with the stem−loop stimulator was also seen with the IBV pseudoknot (pFSHIV-PK). Again, −1 FS occurred across a broad range of spacer lengths, peaking at 6–8 nt, with the −2 FS product spanning a more discrete spacer range of 5–8 nt with a peak at 6 nt. Pseudoknot-induced −1 and −2 FS on U6A was also examined in constructs containing the SRV-1 gag/pro pseudoknot, one feature of which is a smaller stem 1 than the IBV pseudoknot (6 bp c.f. 11 bp). As shown in Supplementary Figure S2, a similar pattern of −1 and −2 FS was observed, with optimal spacing distances of 7–9 nt (−1 FS) and 6–7 nt (−2 FS).
Figure 7.
The effect of slippery sequence–stem–loop (SL) and slippery sequence-pseudoknot (PK) spacing on −1 and −2 FS. (A) The spacer of pFSHIV-SL and pFSIBV-SL was changed from three to nine nucleotides as indicated. In mRNAs derived from these plasmids, frameshifting is stimulated by a SL structure (whose length and base-pair composition is identical to the stacked stems of the minimal IBV PK; see text). Also shown is a diagrammatic representation of potential translation products of pFSHIV-SL mRNAs and predicted molecular masses. Note that zero frame ribosomes terminate at the stop-codon in the spacer in all cases. As previously, the sizes of the encoded frameshift products were normalized by appropriate deletion of bases downstream of the SL. Slippery sequence variants of pFSHIV-SL were also prepared in which the U6A sequence was changed to those indicated. (B) The spacer of pFSHIV-PK was changed from three to nine nucleotides as indicated. In these plasmids, frameshifting is stimulated by the minimal IBV PK (12). As with pFSHIV-SL and derivatives, zero frame ribosomes terminate at the stop-codon in the spacer in all cases, but note that the reading frames of frameshifted ribosomes differ. (C) Messenger RNAs derived from Nco I-cut pFSIBV-SL and pFSHIV-SL spacer variants were translated in RRL and products analysed and quantified as in the legend to Figure 3. The numbers above each gel represent the spacer length. The frameshifting efficiency measured for each signal (to the nearest integer) is indicated below the relevant lanes (−1% FS; −2% FS) and takes into account the number of methionines present in each product (nFS, 10; −1 FS, 10; −2 FS, 11). (D) Messenger RNAs derived from Nco I-cut pFSHIV-SL variants with 7 nt or 8 nt spacers were translated in RRL and products analysed and quantified as in panel C. Also tested were variants of the two plasmids in which the first position of the slippery sequence was changed to G, A or C. (E) Messenger RNAs derived from Bam HI-cut pFSHIV-PK spacer variants were translated in RRL and products analysed and quantified as in panel C. The numbers above each gel represent the spacer length. The frameshifting efficiency measured for each signal (to the nearest integer) is indicated below the relevant lanes (−1% FS; −2% FS) and takes into account the number of methionines present in each product (nFS, 10; −1 FS, 11; −2 FS, 35). The asterisk marks the position of the −1 FS product of pFS cass 5 control mRNA (IBV PK) and also an additional product seen in the translation of the pFSHIV-PK 9 nt spacer mRNA. The size of the latter product is consistent with one that would be synthesised following readthrough of the UGA codon the terminates the non-frameshifted product (present in the spacer; see text).
An interesting aspect of U6A-mediated frameshifting was the occurrence of −1 FS at shorter spacing distances (e.g. 3–5 nt in pFSHIV-SL), albeit at a lower efficiency (e.g. Figure 7 panel C). This could be a result of frameshifting one codon earlier on the mRNA, where the stimulatory RNA effect would be most apparent. At this position in pFSHIV-SL, the sequence G AAU UUU is present, which by chance is compatible (in principle) with tandem −1 FS. However, this sequence is also present in pFSIBV-SL, where −1 FS at spacers of 3–5 nt was less evident (Figure 7 panel C). An alternative explanation is that a single P-site tRNA slip is occuring on U6A (and to a lesser extent on UUUAAAC) at the shorter spacing distances, which is more favoured with P-site tRNAPhe (U UUU UUA) than tRNALeu (U UUA AAC). An unexpected observation was the appearance of a third ‘recoding’ product in translations of pFSHIV-PK with a 9 nt spacer. This product (asterisked in Figure 7 panel E) corresponds to readthrough of the UGA codon immediately downstream of the slippery sequence. Thus at an appropriate spacing distance, a frameshift-promoting pseudoknot can induce stop-codon readthrough. That this was observed at a spacing of 9 nt is consistent with the longer spacing requirements generally observed for those naturally occurring readthrough signals that have a stimulatory RNA component (38).
MS confirmation of the −2 FS event
In an attempt to determine the amino-acid sequence of the presumed −2 FS product, we carried out a large-scale in vitro translation of a tagged version of pFSHIV-AON with optimal (2 nt) spacer, but probably due to the low productivity of RRL, it proved impossible to isolate material of sufficient purity and yield for unambiguous N-terminal sequence determination by Edman degredation. A similar problem was encountered with a tissue culture expression system in which frameshifting was dependent upon co-transfection of 25OMe. However, it proved possible to purify sufficient +1/−2 product for MS analysis when a frameshift cassette of U6A, 7 nt spacer and stem–loop stimulator was expressed as an N-terminal fusion with eukaryotic green fluorescent protein (eGFP). This plasmid (pFSeGFP-N2, see section ‘Materials and Methods’) was transfected into 293 T cells and the −2 FS product purified by affinity chromatography (utilizing a GFP binding matrix), gel electrophoresis and band excision. Following digestion with trypsin, resultant peptides were analysed by MALDI mass fingerprinting and subsequent tandem mass spectrometry (ESIMS-MS). Peptides corresponding to 77% of the predicted fusion protein were identified and the sequence spanning the frameshift region was determined as LNFLYE, indicating −2 FS (Figure 8; raw data in Supplementary Figure S3). The peptide fingerprint data were scanned for other possible events, including the aforementioned P-site +1 FS (generating LNFYE), or sequential −1 ribosomal frameshifts in consecutive elongation cycles (with the first on G AAU UUU and the second on U UUU UUA; generating LNFFYE) but no matches were present. From the determined amino acid sequence, we predict that the −2 FS product is generated by tandem slippage of P- and A-site tRNAs. As the slip is −2, the P-site tRNA would re-pair on a codon that includes the base 5′ of the U6A heptamer (an A in these mRNAs) and it should be noted that the post-slippage contacts following this rearrangement are suboptimal, with the codon:anticodon complex (5′-AUU-3′/5′-GmAA-3′) mismatched at the first position. Altering the base 5′ of the slippery sequence to GU6A also did not inhibit −2 FS (Supplementary Figure S1). However, in −1 FS, it is known that there is tolerance for mismatches at the first position of the post-slip P-site complex (12; see also Figure 4) and this appears to hold true for −2 FS.
Figure 8.
Mass spectroscopy of the −2 FS product. Amino acid sequences of predicted −1 (A) and −2 frameshift products (B) that could be present at the N-terminus of the frameshift product derived from expression of pFSeGFP-N2 in tissue culture cells. Mass spectroscopy revealed only the sequence corresponding to −2 FS (see text and Supplementary Figure S3).
DISCUSSION
Studies of programmed −1 ribosomal frameshifting have focused predominantly on stem–loop and pseudoknot-dependent signals (reviewed in 3,4). From these investigations, a plausible model of frameshifting has emerged which posits a critical contribution of the stimulatory RNA in compromising the activity of the proposed intrinsic helicase activity of the 80S ribosome, located at the mRNA entry channel (15,28). A failure to unwind the stimulatory RNA appropriately during the elongation cycle would potentially compromise frame maintenance through the generation of tension in the mRNA, effectively pulling the mRNA in a 3′-direction while promoting breakage of the tRNA anticodon:codon interaction and realignment of the tRNA in the 5′-(−1) direction. In support of this model, ribosomes are known to pause at frameshift-stimulatory stem–loops and pseudoknots, indicating that such structures can act as barriers to elongation (20,21,37,39–43). In addition, cryo-EM reconstructions of 80S rabbit ribosomes stalled at the IBV frameshift-promoting pseudoknot have revealed a distorted tRNA in a hybrid A/P-like state, with the anticodon arm bent markedly towards the A-site of the ribosome (22,44,45). In these reconstructions, density that likely corresponds to the pseudoknot is observed at the mRNA entry channel close to the putative 80S helicase. These features are consistent with the model, with the pseudoknot resisting unwinding during eEF2-mediated translocation such that tension builds up in the mRNA, subsequently placing strain on the tRNA and resulting in the adoption of a bent conformation (22). In mechanical unwinding studies, a functional IBV-based pseudoknot has been shown to be a ‘brittle’ structure, with a shallow dependence of the unfolding rate on applied force and a slower unfolding rate than component hairpin structures (Green et al., 2008). This greater mechanical stability and kinetic insensitivity to force is consistent with a role in resistance to unwinding (23–25,29). Indeed, a number of pseudoknot features have been identified that could act in such resistance, such as the unusual topology of stems and loops, the geometry of the junction of the two stems and in some pseudoknots, base triplexes between loop 3 and stem 1 (reviewed in 3,4). While some or all of these features are readily identifiable in pseudoknot-stimulatory RNAs, the situation is not so clear-cut for stem–loop stimulatory RNAs. Nuclear magnetic resonance (NMR) analysis of the HIV-1 (46–48) and simian immunodeficiency virus (SIV) (49) structures has revealed a few features (inter-stem kink in HIV-1; stable loop in SIV-1) that may be relevant, but some other viral stem–loop stimulatory RNAs appear likely to have only regular A-form geometry.
The situation is even more convoluted when one considers AON-mediated frameshifting (30–32,50). The mRNA-oligonucleotide complexes would appear to lack unusual features like stem–stem junctions, triplexes or kinks and thus the mechanism by which they induce frameshifting is uncertain. It is known that the length of the AON can affect the efficiency of the process (32), thus the stability of the mRNA:AON duplex plays a role. However, while the specific chemical modifications present in 2-O-Me, MO, phosphorothioate (30) and locked nucleic acid (32) AONs can affect binding stability and target specificity, they are not fundamental for activity in frameshifting, as unmodified RNA oligonucleotides can also stimulate −1 FS, at least in vitro (30,31) and also −2 FS (Supplementary Figure S1). Recently, it has been shown that stem–loop structures can effectively substitute for RNA pseudoknots in some circumstances, with frameshift-stimulatory activity driven largely by the thermodynamic stability of the stem, but also influenced by loop size, composition and stem irregularities (51). The stem–loop employed here is also clearly capable of inducing frameshifting at certain spacer distances (Figure 7). With this in mind, it is likely the AON-mediated frameshifting is largely determined by stability and duplex length (32). Nevertheless, the encounter between ribosome and AON is clearly different from that of a cis-acting secondary structure. Firstly, the 80S helicase will encounter the 3′-hydroxyl of the annealed AON rather than a constrained duplex (or triplex in some pseudoknots). Secondly, the optimal spacing for frameshifting is quite different, being only 3 nt for AON-mediated −1 FS, but respectively, some 7 nt and 8 nt for optimal stem–loop or pseudoknot-mediated −1 FS. The simplest interpretation of these observations is that the putative 80S helicase unwinds several base-pairs of the annealed AON before its activity is compromised. Further work will be required to confirm a role for the helicase in frameshifting and to understand how it is compromised by what appears to be a regular mRNA:AON duplex. It may be that the presence of the free 3′-end of the AON is critical in this regard.
It is clear that 25OMe has a significant effect on ribosome progression, as evidenced by the appearance of a polypeptide corresponding to ribosomes stalled at the bound AON. This d.o. product accounted for up to 10% of the overall synthesis at high 25OMe or 15OMe concentration (10 µM). It has been speculated recently that ribosomal frameshifting frequencies have been generally underestimated as a failure to take into account ribosomes that have frameshifted yet failed to progress on the mRNA, which are often scored as non-frameshifted products (43). While the observation here of what appears to be AON-mediated drop-off of ribosomes lends support to the idea that mRNA structures can act as roadblocks to the elongating ribosome, such drop-off is far less apparent when ribosomes are challenged with natural frameshift-stimulatory RNAs (37). Indeed, in this study, the abundance of the d.o. product was greatly reduced when a 15mer unmodified RNA AON replaced 25OMe (Supplementary Figure S1). The relatively high proportion of ribosomes that appear to be stalled for an extended period probably reflects the very stable association of the OMe AONs with the mRNA template.
One of the unexpected outcomes of this work was the discovery of −2 FS on the U6A heptamer (and potentially—albeit to a lesser extent—on the A6C and A8C heptamers). Initially, we imagined that the protein in question originated by +1 FS, with the P-site tRNAPhe decoding UUU in the zero frame slipping forwards onto the overlapping +1 frame codon (also UUU) in a proportion of ribosomes stalled at the AON, stem–loop or pseudoknot, but this was ruled out experimentally. A −2 FS is consistent with the idea that mRNA tension promotes 5′-movement of the tRNAs in this context. Indeed, the spacing analysis of Figures 6 and 7 provides further support for this viewpoint. Irrespective of the nature of the stimulatory RNA, the spacing distance facilitating maximum −2 FS was consistently ∼1–2 nt less than that promoting −1 FS. Viewed simplistically, with the shorter spacer, the tension on the mRNA would be greater, increasing the likelihood of a −2 shift. While this hypothesis requires further substantiation, there is a precedent for the importance of mRNA tension, from studies of the +1 programmed frameshifting signal in the E. coli prfB gene encoding release factor 2. In this system, the interaction of a Shine-Dalgarno (SD)-like element in the mRNA with the anti-SD at the 3′-end of 16S rRNA is important in promoting efficient +1 FS at a 3′-recoding site (52). The effect of varying the spacing between SD sequence and P-site codon in the prf B system has been analysed by toeprinting (53). At a spacer length of 2 nt, noticeably shorter than that found naturally between SD and initiator AUG (5 nt; 54), 70S pre-translocation complexes could not be formed and instead, the tRNA added subsequently to fill the A-site moved spontaneously into the P-site, restoring the spacer to the natural length (5 nt). These data support a model in which formation of the SD–anti-SD helix in ribosomes stalled at the in-frame UGA codon of prfB generates tension on the mRNA that destabilizes codon:anticodon pairing in the P site and promotes slippage of the mRNA in the 5′-direction. It is plausible that the −1 and −2 FS we observe here originate in a similar manner, except that here, the tension pulls the mRNA in a 3′-direction, leading to −1 and −2 FS.
An interesting question is whether −2 FS is more widely exploited in virus or cellular gene expression. Only one example of a −2 FS signal has been documented to date, involved in the expression of tail assembly chaperone genes in bacteriophage Mu (55,56). Here, tandem −2 slippage occurs on a GG GGG CGA with the anticodon of the A-site tRNAArg (3′GCI5′, where I in the wobble position is inosine) forming a more stable post-slippage contact with the mRNA in the −2 frame rather than the −1 frame. Another potential −2 FS signal in Trichomonas vaginalis virus 1 has been suggested (57). In this virus, frameshifting most likely occurs on a conserved CC CUU UUU sequence, compatible with tandem −2 shifting. Examination of known viral −1 FS sites possessing a U6A or A6C slippery sequence, however, reveals that in most cases the spacing distances seem inappropriately long for efficient −2 FS. In HIV-1, where the stimulatory RNA forms immediately 3′ of a U6A slippery sequence (58), a stop codon in the −2 reading frame is present directly downstream of the U6A heptamer and appears to be present in all isolates of HIV-1 (59,60). Any ribosomes entering the −2 reading frame would terminate immediately, generating a truncated Gag polyprotein lacking viral proteins p1 and p6. As yet, there is no evidence to suggest that such a species is expressed in HIV-1 infected cells. Frameshifting in the expression of mammalian ornithine decarboxylase antizyme has remarkably been shown to be +1 in mammals and fission yeast, yet −2 in budding yeast (61,62). Precise details of the mechanism of the −2 FS remain to be elucidated, but as observed by Matsufuji and colleagues, lengthening the spacing between the antizyme shift site and its pseudoknot by three bases increased +1 FS in yeast at the expense of −2 FS, supporting a link between spacer length and frameshift direction (61). Intriguingly, replacing the 3′-stimulatory element that forms a component of the antizyme frameshifting signal with an annealed AON has been shown also to stimulate −1 FS when placed with zero spacing (50). However, this −1 product is not seen with the natural antizyme frameshift signal. Conclusive evidence of a functional −2 FS signal in virus or cellular genes is therefore still awaited.
In the past few years, AONs have been increasingly exploited as a tool to examine aspects of ribosomal frameshifting. The observation here of spacer-length-dependent −1 and −2 FS events supports the view that mRNA tension plays an important role in frameshifting and it will be interesting to see whether the structure of ribosomal complexes stalled at an AON–mRNA complex resemble those of ribosomes stalled at other frameshift-stimulatory elements (22,44,45).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–3.
FUNDING
Biotechnology and Biological Sciences Research Council U.K. [BB/G008205/1 to I.B. and BB/G00805/1 to R.J.C.G.]; Agency for Science, Technology and Research (A*STAR), Singapore (to Z.L.). Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Len Packman (Department of Biochemistry, University of Cambridge) for his assistance with the mass spectroscopy and associated methodology text.
REFERENCES
- 1.Parker J. Errors and alternatives in reading the universal genetic code. Microbiol. Rev. 1989;53:273–298. doi: 10.1128/mr.53.3.273-298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkins JF, Björk GR. A gripping tale of ribosomal frameshifting: extragenic suppressors of frameshift mutations spotlight P-site realignment. Microbiol. Mol. Biol. Rev. 2009;73:178–210. doi: 10.1128/MMBR.00010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giedroc DP, Cornish PV. Frameshifting RNA pseudoknots: structure and mechanism. Virus Res. 2009;139:193–208. doi: 10.1016/j.virusres.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brierley I, Gilbert RJC, Pennell S. Pseudoknot-dependent -1 ribosomal frameshifting: structures, mechanisms and models. In: Atkins JF, Gesteland RF, editors. Recoding: Expansion of Decoding Rules Enriches Gene Expression. Vol. Ch 7. Springer: 2010. pp. 149–174. [Google Scholar]
- 5.Jacks T, Varmus HE. Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting. Science. 1985;230:1237–1242. doi: 10.1126/science.2416054. [DOI] [PubMed] [Google Scholar]
- 6.Jacks T, Madhani HD, Masiarz FR, Varmus HE. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988a;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacks T, Power MD, Masiarz FR, Luciw PA, Barr PJ, Varmus HE. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988b;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 8.Nam SH, Copeland TD, Hatanaka M, Oroszlan S. Characterization of ribosomal frameshifting for expression of pol gene products of human T-cell leukemia virus type I. J. Virol. 1993;67:196–203. doi: 10.1128/jvi.67.1.196-203.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mador N, Panet A, Honigman A. Translation of gag, pro, and pol gene products of human T-cell leukemia virus type 2. J. Virol. 1989;63:2400–2404. doi: 10.1128/jvi.63.5.2400-2404.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thiel V, Ivanov KA, Putics A, Hertzig T, Schelle B, Bayer S, Weissbrich B, Snijder EJ, Rabenau H, Doerr HW, et al. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 11.Brierley I, Digard P, Inglis SC. Characterisation of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brierley I, Jenner AJ, Inglis SC. Mutational analysis of the ‘slippery-sequence’ component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 1992;227:463–479. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kollmus H, Honigman A, Panet A, Hauser H. The sequences of and distance between two cis-acting signals determine the efficiency of ribosomal frameshifting in human immunodeficiency virus type 1 and human T-cell leukemia virus type II in vivo. J. Virol. 1994;68:6087–6091. doi: 10.1128/jvi.68.9.6087-6091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baranov PV, Gesteland RF, Atkins JF. P-site tRNA is a crucial initiator of ribosomal frameshifting. RNA. 2004;10:221–230. doi: 10.1261/rna.5122604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takyar S, Hickerson RP, Noller HF. mRNA helicase activity of the ribosome. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 16.Brierley I, Rolley NJ, Jenner AJ, Inglis SC. Mutational analysis of the RNA pseudoknot component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 1991;220:889–902. doi: 10.1016/0022-2836(91)90361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wills NM, Gesteland RF, Atkins JF. Evidence that a downstream pseudoknot is required for translational read-through of the Moloney murine leukemia virus gag stop codon. Proc. Natl Acad. Sci. USA. 1991;88:6991–6995. doi: 10.1073/pnas.88.16.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 19.Yusupova GZ, Yusupov MM, Cate JH, Noller HF. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- 20.Plant EP, Jacobs KL, Harger JW, Meskauskas A, Jacobs JL, Baxter JL, Petrov AN, Dinman JD. The 9Å solution: How mRNA pseudoknots promote efficient programmed -1 ribosomal frameshifting. RNA. 2003;9:168–174. doi: 10.1261/rna.2132503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plant EP, Dinman JD. Torsional restraint: a new twist on frameshifting pseudoknots. Nucleic Acids Res. 2005;33:1825–1833. doi: 10.1093/nar/gki329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Namy O, Moran SJ, Stuart DI, Gilbert RJC, Brierley I. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature. 2006;441:244–247. doi: 10.1038/nature04735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen TM, Reihani SN, Oddershede LB, Sørensen MA. Correlation between mechanical strength of messenger RNA pseudoknots and ribosomal frameshifting. Proc. Natl Acad. Sci. USA. 2007;104:5830–5835. doi: 10.1073/pnas.0608668104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Green L, Kim CH, Bustamante C, Tinoco I., Jr Characterization of the mechanical unfolding of RNA pseudoknots. J. Mol. Biol. 2008;375:511–528. doi: 10.1016/j.jmb.2007.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen G, Chang KY, Chou MY, Bustamante C, Tinoco I., Jr Triplex structures in an RNA pseudoknot enhance mechanical stability and increase efficiency of -1 ribosomal frameshifting. Proc. Natl Acad. Sci. USA. 2009;106:12706–12711. doi: 10.1073/pnas.0905046106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazauric MH, Seol Y, Yoshizawa S, Visscher K, Fourmy D. Interaction of the HIV-1 frameshift signal with the ribosome. Nucleic Acids Res. 2009a;37:7654–7664. doi: 10.1093/nar/gkp779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mazauric MH, Leroy JL, Visscher K, Yoshizawa S, Fourmy D. Footprinting analysis of BWYV pseudoknot-ribosome complexes. RNA. 2009b;15:1775–1786. doi: 10.1261/rna.1385409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qu X, Wen JD, Lancaster L, Noller HF, Bustamante C, Tinoco I., Jr The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature. 2011;475:118–121. doi: 10.1038/nature10126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White KH, Orzechowski M, Fourmy D, Visscher K. Mechanical unfolding of the beet western yellow virus -1 frameshift signal. J. Am. Chem. Soc. 2011;133:9775–9782. doi: 10.1021/ja111281f. [DOI] [PubMed] [Google Scholar]
- 30.Howard MT, Gesteland RF, Atkins JF. Efficient stimulation of site-specific ribosome frameshifting by antisense oligonucleotides. RNA. 2004;10:1653–1661. doi: 10.1261/rna.7810204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olsthoorn RC, Laurs M, Sohet F, Hilbers CW, Heus HA, Pleij CW. Novel application of sRNA: stimulation of ribosomal frameshifting. RNA. 2004;10:702–703. doi: 10.1261/rna.7139704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu CH, Noteborn MH, Olsthoorn RC. Stimulation of ribosomal frameshifting by antisense LNA. Nucleic Acids Res. 2010;38:8277–8283. doi: 10.1093/nar/gkq650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, Atkins JF. A dual-luciferase reporter system for studying recoding signals. RNA. 1998;4:479–486. [PMC free article] [PubMed] [Google Scholar]
- 34.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 35.Anikin M, Molodtsov V, Temiakov D, McAllister WT. Transcript slippage and recoding. In: Atkins JF, Gesteland RF, editors. Recoding: Expansion of Decoding Rules Enriches Gene Expression. Vol. Ch 19. Springer: 2010. pp. 409–432. [Google Scholar]
- 36.Girnary R, King L, Robinson L, Elston R, Brierley I. Structure-function analysis of the ribosomal frameshifting signal of two human immunodeficiency virus type 1 isolates with increased resistance to viral protease inhibitors. J. Gen. Virol. 2007;88:226–235. doi: 10.1099/vir.0.82064-0. [DOI] [PubMed] [Google Scholar]
- 37.Kontos H, Napthine S, Brierley I. Ribosomal pausing at a frameshifter RNA pseudoknot is sensitive to reading phase but shows little correlation with frameshift efficiency. Mol. Cell. Biol. 2001;21:8657–8670. doi: 10.1128/MCB.21.24.8657-8670.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Firth AE, Wills NM, Gesteland RF, Atkins JF. Stimulation of stop codon readthrough: frequent presence of an extended 3′ RNA structural element. Nucleic Acids Res. 2011;39:6679–6691. doi: 10.1093/nar/gkr224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tu C, Tzeng T-H, Bruenn JA. Ribosomal movement impeded at a pseudoknot required for frameshifting. Proc. Natl Acad. Sci. USA. 1992;89:8636–8640. doi: 10.1073/pnas.89.18.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Somogyi P, Jenner AJ, Brierley I, Inglis SC. Ribosomal pausing during translation of an RNA pseudoknot. Mol. Cell. Biol. 1993;13:6931–6940. doi: 10.1128/mcb.13.11.6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lopinski JD, Dinman JD, Bruenn JA. Kinetics of ribosomal pausing during programmed -1 translational frameshifting. Mol. Cell. Biol. 2000;20:1095–1103. doi: 10.1128/mcb.20.4.1095-1103.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buchan JR, Stansfield I. Halting a cellular production line: responses to ribosomal pausing during translation. Biol. Cell. 2007;99:475–487. doi: 10.1042/BC20070037. [DOI] [PubMed] [Google Scholar]
- 43.Tholstrup J, Oddershede LB, Sørensen MA. mRNA pseudoknot structures can act as ribosomal roadblocks. Nucleic Acids Res. 2012;40:303–313. doi: 10.1093/nar/gkr686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moran SJ, Flanagan JF, 4th, Namy O, Stuart DI, Brierley I, Gilbert RJ. The mechanics of translocation: a molecular “spring-and-ratchet” system. Structure. 2008;16:664–672. doi: 10.1016/j.str.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flanagan JF, 4th, Namy O, Brierley I, Gilbert RJ. Direct observation of distinct A/P hybrid-state tRNAs in translocating ribosomes. Structure. 2010;18:257–264. doi: 10.1016/j.str.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Staple DW, Butcher SE. Solution structure of the HIV-1 frameshift inducing stem-loop RNA. Nucleic Acids Res. 2003;31:4326–4331. doi: 10.1093/nar/gkg654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gaudin C, Mazauric MH, Traïkia M, Guittet E, Yoshizawa S, Fourmy D. Structure of the RNA signal essential for translational frameshifting in HIV-1. J. Mol. Biol. 2005;349:1024–1035. doi: 10.1016/j.jmb.2005.04.045. [DOI] [PubMed] [Google Scholar]
- 48.Staple DW, Butcher SE. Solution structure and thermodynamic investigation of the HIV-1 frameshift inducing element. J. Mol. Biol. 2005;349:1011–1023. doi: 10.1016/j.jmb.2005.03.038. [DOI] [PubMed] [Google Scholar]
- 49.Marcheschi RJ, Staple DW, Butcher SE. Programmed ribosomal frameshifting in SIV is induced by a highly structured RNA stem-loop. J. Mol. Biol. 2007;373:652–663. doi: 10.1016/j.jmb.2007.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henderson CM, Anderson CB, Howard MT. Antisense-induced ribosomal frameshifting. Nucleic Acids Res. 2006;34:4302–4310. doi: 10.1093/nar/gkl531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu CH, Noteborn MH, Pleij CW, Olsthoorn RC. Stem-loop structures can effectively substitute for an RNA pseudoknot in -1 ribosomal frameshifting. Nucleic Acids Res. 2011;39:8952–8959. doi: 10.1093/nar/gkr579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weiss RB, Dunn DM, Dahlberg AE, Atkins JF, Gesteland RF. Reading frame switch caused by base-pair formation between the 3′ end of 16S rRNA and the mRNA during elongation of protein synthesis in Escherichia coli. EMBO J. 1988;7:1503–1507. doi: 10.1002/j.1460-2075.1988.tb02969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Devaraj A, Fredrick K. Short spacing between the Shine-Dalgarno sequence and P codon destabilizes codon-anticodon pairing in the P site to promote +1 programmed frameshifting. Mol. Microbiol. 2010;78:1500–1509. doi: 10.1111/j.1365-2958.2010.07421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen H, Bjerknes M, Kumar R, Jay E. Determination of the optimal aligned spacing between the Shine-Dalgarno sequence and the translation initiation codon of Escherichia coli mRNAs. Nucleic Acids Res. 1994;22:4953–4957. doi: 10.1093/nar/22.23.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu J, Hendrix RW, Duda RL. Conserved translational frameshift in dsDNA bacteriophage tail assembly genes. Mol. Cell. 2004;16:11–21. doi: 10.1016/j.molcel.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 56.Baranov PV, Fayet O, Hendrix RW, Atkins JF. Recoding in bacteriophages and bacterial IS elements. Trends Genet. 2006;22:174–181. doi: 10.1016/j.tig.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 57.Firth AE, Brierley I. Non-canonical translation in RNA viruses. J. Gen. Virol. 2012;93:1385–1409. doi: 10.1099/vir.0.042499-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dulude D, Théberge-Julien G, Brakier-Gingras L, Heveker N. Selection of peptides interfering with a ribosomal frameshift in the human immunodeficiency virus type 1. RNA. 2008;14:981–991. doi: 10.1261/rna.887008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Biswas P, Jiang X, Pacchia AL, Dougherty JP, Peltz SW. The human immunodeficiency virus type 1 ribosomal frameshifting site is an invariant sequence determinant and an important target for antiviral therapy. J. Virol. 2004;78:2082–2087. doi: 10.1128/JVI.78.4.2082-2087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Knops E, Brakier-Gingras L, Schülter E, Pfister H, Kaiser R, Verheyen J. Mutational patterns in the frameshift-regulating site of HIV-1 selected by protease inhibitors. Med. Microbiol. Immunol. 2012;201:213–218. doi: 10.1007/s00430-011-0224-z. [DOI] [PubMed] [Google Scholar]
- 61.Matsufuji S, Matsufuji T, Wills NM, Gesteland RF, Atkins JF. Reading two bases twice: mammalian antizyme frameshifting in yeast. EMBO J. 1996;15:1360–1370. [PMC free article] [PubMed] [Google Scholar]
- 62.Ivanov IP, Gesteland RF, Matsufuji S, Atkins JF. Programmed frameshifting in the synthesis of mammalian antizyme is +1 in mammals, predominantly +1 in fission yeast, but −2 in budding yeast. RNA. 1998;4:1230–1238. doi: 10.1017/s1355838298980864. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.