Abstract
The ability to maintain intracellular concentrations of toxic reactive oxygen species (ROS) within safe limits is essential for all aerobic life forms. In bacteria, as well as other organisms, ROS are produced during the normal course of aerobic metabolism, necessitating the constitutive expression of ROS scavenging systems. However, bacteria can also experience transient high-level exposure to ROS derived either from external sources, such as the host defense response, or as a secondary effect of other seemingly unrelated environmental stresses. Consequently, transcriptional regulators have evolved to sense the levels of ROS and coordinate the appropriate oxidative stress response. Three well-studied examples of these are the peroxide responsive regulators OxyR, PerR, and OhrR. OxyR and PerR are sensors of primarily H2O2, while OhrR senses organic peroxide (ROOH) and sodium hypochlorite (NaOCl). OxyR and OhrR sense oxidants by means of the reversible oxidation of specific cysteine residues. In contrast, PerR senses H2O2 via the Fe-catalyzed oxidation of histidine residues. These transcription regulators also influence complex biological phenomena, such as biofilm formation, the evasion of host immune responses, and antibiotic resistance via the direct regulation of specific proteins.
INTRODUCTION
An effective oxidative stress defense response is a required item in the basic survival kit of all aerobic organisms as well as those anaerobes that exist in environments subject to transient exposures to oxygen. This is due to molecular oxygen's ability to accept electrons from cellular redox components to form toxic reactive oxygen species (ROS) (51). One such product is superoxide anion (O2•−), which results from a one-electron reduction of O2. Another, resulting from a two-electron reduction, is hydrogen peroxide (H2O2) (51). Both O2•− and H2O2 have the ability to oxidize exposed iron sulfur clusters, as has been observed for certain dehydratases (50), while H2O2 is known to oxidize Fe2+ in proteins containing mononuclear iron centers (100). This not only inactivates the enzymes but also results in the release of free ferric iron (Fe3+), which is converted to ferrous iron (Fe2+) in the intracellular reducing environment (51). This is significant since Fe2+ can reduce H2O2 to form hydroxyl radicals (•OH) in the Fenton reaction as follows: Fe2+ + H2O2 → Fe3+ + •OH + OH−.
Hydroxyl radicals are highly potent oxidants of cellular macromolecules that react at a diffusion-limited rate (51). Thus, the increased intracellular iron levels resulting from oxidative damage to a subset of iron-containing proteins can lead to increased •OH-mediated cellular damage, particularly to DNA (55). O2•−, H2O2, and •OH are all capable of causing a variety of oxidative lesions in proteins, DNA, and lipids (51). An important aspect of some of these secondary reactions is the production of a variety of organic peroxides that can also mediate further oxidative damage (97).
In addition to endogenous sources, bacteria also encounter ROS from external sources. Macrophages actively produce large amounts of O2•− and nitric oxide (NO•) in order to kill invading pathogens (92). The host defense response in plants is similar and also includes the synthesis of organic peroxides (62), while some microorganisms excrete ROS to inhibit the growth of competitors (42). Exposure to redox cycling compounds, such as the herbicide paraquat, or naturally occurring quinones can also serve as a source of ROS. These molecules can participate in cyclic reactions in which they transfer an electron from cellular electron donors, such as NADH oxidases, to O2 to generate O2•− (22).
Bacteria maintain a basal level of protective enzymes to detoxify O2•− and H2O2, thereby keeping their concentrations within safe limits (95). These include superoxide dismutases that use a metal center to catalyze the dismutation of O2•− to H2O2 and catalases that usually employ a heme cofactor to convert H2O2 to O2 and H2O (50). Peroxiredoxins, a class of peroxidases, function to reduce H2O2 and organic peroxides (ROOH) to either H2O or H2O and the corresponding alcohol (ROH) via the peroxide-mediated oxidation of cysteine thiols that are subsequently reduced using cellular electron donors to regenerate the active enzyme (88, 94).
When ROS levels exceed safe limits, bacteria have the ability to mount an inducible response, resulting in increased expression of ROS detoxification enzymes along with additional protective systems that repair oxidative damage, protect vulnerable enzymes from inactivation, and control the levels of free Fe2+ (87). For example, oxidative stress increases the demand for reducing equivalents, necessitating the induction of metabolic pathways to increase the reductant supply (32). The expression of iron-binding proteins, like Dps, is often induced in response to oxidative stress to reduce the levels of free Fe in order to prevent production of •OH via the Fenton reaction (12). Strategies are also employed to protect enzymes that contain vulnerable Fe-containing centers from oxidation by increasing the uptake of Mn2+, which is able to replace Fe2+ at some active sites, rendering them resistant to oxidation (50). In some cases, Fe-containing enzymes are replaced with analogs that do not require Fe (70).
The regulation of the expression of genes involved in the bacterial oxidative stress defense response is complex and often under the control of regulators that can directly sense the levels of specific ROS and activate or derepress target gene transcription. This review summarizes work concerning the mechanisms of oxidant sensing and transcriptional regulation by OxyR, PerR, and OhrR, three well-studied peroxide responsive regulators that have the ability to distinguish between ROOH and H2O2 (75). Two distinct mechanisms for peroxide sensing that utilize either the oxidation of cysteine residues (OxyR and OhrR) or the metal-catalyzed oxidation of histidine residues (PerR) have evolved in these regulators. In each case, oxidative modification of the regulator alters its DNA binding properties.
OxyR
OxyR is an H2O2-sensing transcriptional regulator of the LysR family (93) that is generally found in Gram-negative bacteria but is also known to occur in a few Gram-positive bacteria (78, 83). Like other regulators of this family, it contains a conserved N-terminal helix-turn-helix DNA binding domain, a central coinducer recognition and response domain which senses the regulatory signal, and a C-terminal domain that functions in multimerization and activation (59–61, 93, 115). OxyR functions primarily as a global regulator of the peroxide stress response that maintains intracellular H2O2 levels within safe limits (1) and also plays a role in the cellular response to thiol depletion (2). The OxyR regulon of Escherichia coli is comprised of over 20 genes, including genes involved in H2O2 detoxification (katE, ahpCF), heme biosynthesis (hemH), reductant supply (grxA, gor, trxC), thiol-disulfide isomerization (dsbG), Fe-S center repair (sufA-E, sufS), iron binding (yaaA), repression of iron import systems (fur), and manganese import (71, 124, 125). OxyR also upregulates the expression of OxyS, a small regulatory RNA that integrates peroxide stress with general stress pathways (34, 122). Although there are significant differences, the OxyR regulons of other organisms tend to include similar classes of genes (39, 82, 96, 104, 116).
While OxyR is primarily thought of as a transcriptional activator under oxidizing conditions that acts through direct interaction with the RNA polymerase α subunit (61, 103, 106), OxyR can function as either a repressor or activator under both oxidizing and reducing conditions (19, 45, 49, 96, 108, 121).
In E. coli, tetrameric OxyR binds to the 5′ promoter-operator regions of target genes at a conserved sequence motif (61, 108, 123) (Fig. 1). The oxidized (OxyRox) and reduced (OxyRred) forms of the protein adopt different conformational states that can sometimes result in changes in the DNA binding contacts, with the reduced form of OxyR contacting two pairs of major grooves separated by one helical turn while the DNA binding contacts of the oxidized form shift to contact four consecutive major grooves (108). These redox-dependent changes also alter its affinity for target promoters and can affect promoter conformation and, thereby, contacts to RNA polymerase (103, 107, 108).
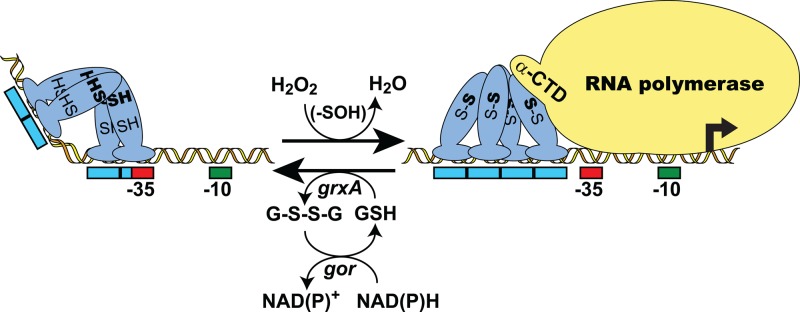
Fig 1.
Mechanism of transcription activation via intramolecular disulfide formation in 2-Cys OxyR. A model for H2O2-dependent, OxyR-mediated transcription activation of a target gene in E. coli is shown. Activation begins with the oxidation of the sensing cysteine (SH) residue of OxyR to sulfenic acid (-SOH), followed by the rapid formation of an intramolecular disulfide bond with the resolving cysteine (SH). The resulting conformational change often causes a shift in the DNase I footprint and can also affect DNA binding affinity and promoter conformation as well as render OxyR capable of interacting with RNA polymerase. Transcription activation involves the direct interaction of OxyR with the alpha subunit C-terminal domain (α-CTD) of RNA polymerase. Oxidized OxyR is reduced, using reduced glutathione (GSH) as the electron donor, via the glutaredoxin (grxA)/glutathione reductase (gor) system, with reducing equivalents ultimately supplied by NAD(P)H. Red and green boxes indicate RNA polymerase σ70 −35 and −10 promoter elements, respectively. Blue boxes indicate OxyR DNA binding contacts. Activation can also occur via oxidative modification of the sensing cysteine alone.
Sensing of H2O2 occurs via direct oxidation of OxyR at a specific “sensing” cysteine residue (Fig. 1). In E. coli, in which the normal intracellular H2O2 level is ∼20 nM, OxyR is present in its reduced form (95, 123). Rapid oxidation of OxyRred occurs when intracellular H2O2 levels reach ∼100 nM, which is well below the level at which growth is inhibited (∼2 μM) (33, 95). OxyRox is slowly reduced via glutaredoxin 1 using electrons supplied by reduced glutathione (2, 105, 123) (Fig. 1). This provides feedback regulation of the system, since expression of grxA (glutaredoxin 1) and gor (glutathione reductase) is induced during oxidative stress as part of the OxyR regulon (105, 123, 125). This dependence on reduction by GrxA also renders OxyR sensitive to the thiol-disulfide redox status (glutathione disulfide [GSSG]/reduced glutathione [GSH] ratio) of the cell (2, 123). Recent evidence indicates that a similar reductive recycling of OxyR occurs in Pseudomonas aeruginosa, which utilizes a thioredoxin/thioredoxin reductase system that is part of the OxyR regulon (116).
The exact mechanism(s) of oxidant sensing by OxyR is still a subject of active investigation and has generated some debate (44, 84). Two basic models have been proposed. The first, supported by clear evidence, indicates that H2O2-mediated activation of E. coli OxyR constitutes a simple on/off switch that occurs through the formation of a specific disulfide bond between the conserved cysteine residues C199 and C208 (65, 105, 123). Activation involves a two-step oxidation that begins when H2O2 reacts with a thiolate ion of the sensing cysteine, C199, to form a sulfenic acid (C199-SOH) (Fig. 1). C199-SOH rapidly reacts with C208-SH to form an intramolecular disulfide bond (65). This induces structural changes in the regulatory domain (amino acids 80 to 305) (18, 65) that result in altered associations between the subunits within the tetramer, leading to altered DNA binding properties and allowing productive interaction between OxyR and RNA polymerase (18, 106, 108).
The second “molecular code” hypothesis suggests that modification of C199 alone is sufficient to activate OxyR in the absence of disulfide bond formation and that the type of modification determines the regulatory outcome (56). In this case, a more selective regulatory response in which distinct subsets of OxyR target genes are differentially regulated depending on the particular oxidized form of OxyR involved is expected. Initially, this was based primarily on in vitro studies indicating that oxidative modification of only C199 was sufficient for activation, since hydroxylated Cys199-SOH, S-nitrosylated (Cys199-SNO), and S-glutathionylated (Cys199-S-S-G) forms of OxyR were able to activate transcription, and that the different modifications elicited distinct changes in the circular dichroism (CD) spectra of OxyR-DNA complexes (40, 56). The idea was subsequently reinforced by the observation that Cys199-thiol-esterification of OxyR resulted in activation of target genes both in vitro and in vivo (38). Until recently, there were little available data to adequately evaluate the molecular code model's prediction of modification-specific patterns of gene activation. In fact, microarray studies had indicated that OxyR did not play a significant role in gene activation in response to the S-nitrosylating agents nitrosylated glutathione (GSNO) and sodium nitrite under aerobic conditions (80). However, a recent analysis in E. coli has shown that S-nitrosylation of proteins occurs naturally during anaerobic respiratory growth on nitrate, resulting in the nitrosylation of OxyR at C199 that causes the activation of a set of genes that is distinct from those upregulated in response to H2O2 (98). Thus, it appears that in the case of E. coli OxyR, both intramolecular disulfide bond formation and modifications to Cys199 alone can elicit distinct OxyR-mediated regulatory outcomes.
Several recent studies also indicate that there is diversity in the activation mechanisms of OxyR orthologs in other organisms. For example, in vivo studies of the E. coli-like OxyR from Pseudomonas aeruginosa, involving free thiol labeling of OxyR in extracts of wild-type and C199/C208 single and double mutants exposed to H2O2, detected several oxidized forms of OxyR in addition to the one containing a disulfide bond between C199 and C208 (45). The resulting suggestion that Pseudomonas OxyR has a different redox cycle than the E. coli protein has been supported by a recent mutagenesis study indicating that a third cysteine residue (Cys296) may be involved in peroxide sensing in P. aeruginosa (4). This cysteine is not conserved in the E. coli protein and has so far been found only in a small group within the betaproteobacteria as well as in several Pseudomonas species (4).
Finally, a second OxyR structural class, containing only one cysteine, has been identified in Deinococcus radiodurans (13, 120). Two novel OxyR proteins (DrOxyR and DrOxyR2) are found in this organism, with each containing one essential sensing cysteine residue that roughly corresponds to C208 in the E. coli protein. DrOxyR and DrOxyR2 are 31% and 28% identical to E. coli OxyR, respectively, and contain conserved residues that have been implicated in DNA binding (R4, L32 S33, R50), activation (D142, R273), and multimerization (A233) (60, 115). However, residues in the E. coli protein that are predicted to play a role in sulfenic acid formation at C199 and subsequent disulfide bond formation with C208 (H198, R201, R266, T238) (59, 61) are absent. The differences in the sensing mechanisms between the 1-Cys and 2-Cys OxyR proteins have yet to be explored. However, DrOxyR is able to complement an E. coli OxyR mutant, thus indicating functional similarity, while in vitro chemical modification studies have shown that H2O2-dependent activation of DrOxyR involves the formation of a cysteine sulfenic acid at the essential sensing cysteine (13).
PerR
PerR functions as a peroxide responsive repressor and is a member of the Fur family of small, dimeric, metal-responsive transcriptional regulators (1). PerR is a global regulator that responds primarily to H2O2. It substitutes for OxyR in many Gram-positive bacteria (75). However, its distribution is not limited to Gram-positive bacteria (69, 78, 91, 112), and in several cases, PerR is found along with OxyR (36, 37, 111, 118). In Bacillus subtilis, PerR regulates itself (30) and other genes, including those involved in oxidative stress defense (katA, ahpCF, mrgA) (10, 14), metal homeostasis (zosA, fur, hemAXCDBL) (14, 30, 31), and surfactant production (srfA) (41), in response to peroxide exposure. As with OxyR, PerR homologs in other organisms tend to regulate similar classes of genes (7, 8, 36, 48, 79, 91, 117).
Each PerR monomer contains a binding site for a structural Zn2+ as well as a regulatory site that, in B. subtilis, binds either Fe2+ or Mn2+ (46, 52) (Fig. 2A). Repression is achieved when PerR, containing metals bound at both sites, binds DNA at a consensus Per-box (29) that either overlaps part of the promoter or is immediately downstream from it (36, 46). PerR-mediated positive regulation has also been observed in at least two cases and appears to involve PerR binding to distant upstream sites; however, the mechanism of activation has not been investigated (7, 41).
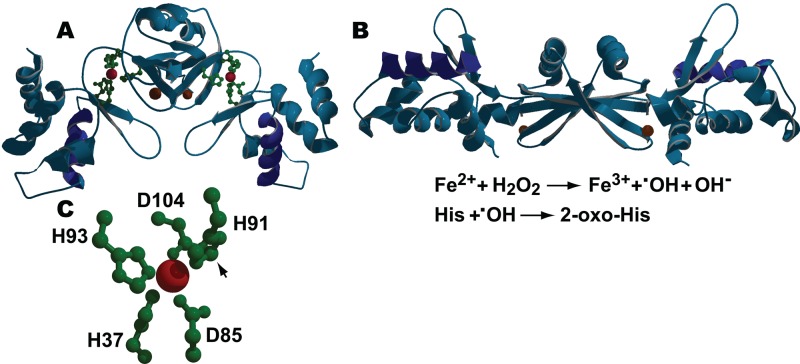
Fig 2.
H2O2-mediated inactivation of PerR. (A) The ribbon structure of the carbon backbone of reduced B. subtilis PerR-Zn2+-Mn2+, showing the side chains of the amino acids coordinating the regulatory metal (green) and the DNA binding helices (dark blue), is shown. The structural and regulatory metals, Zn2+ and Mn2+, are shown as brown and red spheres, respectively. In its reduced state, PerR-Zn2+-Fe2+/Mn2+ binds to sites overlapping target promoter/operators and blocks transcription. (B) The ribbon structure of oxidized PerR-Zn2+-Mn2+ is shown. The steps in the Fenton-mediated oxidation of His residues in PerR-Zn2+-Fe2+ by H2O2 are shown below the panel. Exposure to H2O2 results in the iron-catalyzed production of OH•, followed by the oxidation of the 2-carbon of the imidazole ring of one of two histidine residues (H37 and H91) that participate in coordinating the bound Fe2+ in each monomer. While H37 is the preferential target for oxidation, oxidation of either histidine to 2-oxo-histidine results in the disruption of normal Fe coordination, resulting in the destabilization of the DNA binding domain of the monomer and leading to the release of oxidized PerR from the DNA. (C) Close-up view of the Mn2+-containing regulatory metal binding site in reduced PerR that shows the relative positions of the metal-coordinating side groups of H37, H91, H93, D85, and D104 (positions of the coordinating side groups roughly correspond to those on the right side of panel A). The arrow indicates the 2-carbon of the imidazole group of H91, which is the site of oxidation in H91 and H37. In all cases, DNA binding helices are shown in dark blue. The Swiss Protein Data Bank (PDB) identifications (IDs) for reduced and oxidized PerR are 3F8N and 2RGV, respectively.
While coordination of either Fe2+ or Mn2+ at the regulatory site is required for DNA binding of the PerR dimer (46), the Fe2+-containing form (PerR-Zn2+-Fe2+) is thought to be responsible for H2O2 sensing in vivo. This is based on the observations that PerR-Zn2+-Fe2+ is >103-fold more sensitive to oxidation than PerR-Zn2+-Mn2+ (67) and that Fe2+ has a higher affinity (∼30-fold) for binding to the regulatory site than Mn2+. The exact physiological relevance of Mn2+ binding at the regulatory site is not fully understood, but Mn2+ can compete with Fe2+ for binding to PerR in vivo, thus altering the relative sensitivity of genes in the PerR regulon to H2O2 induction (14, 26, 66). Not only does Mn compete with Fe, but in some cases, Mn, not Fe, appears to function as the corepressor (30). Mn2+ may also serve to protect PerR from oxidative inactivation while still allowing PerR-mediated derepression of target genes in response to Fe2+ released due to oxidative damage (66).
The oxidation-dependent disruption of proper iron coordination at the regulatory site is the basis of oxidant sensing by PerR (Fig. 2). Accumulated evidence indicates that Fe2+ (or Mn2+) bound at the regulatory site is coordinated by three histidine (H37, H91, H93) and two aspartate (D104, D85) residues. These constitute a four-sided pyramid with alternating histidine (H37 and H91) and aspartate residues at the corners of the base and a His (H93) positioned at the apex (52) (Fig. 2C). Binding of the regulatory metal locks the dimer in a caliper-like configuration that stabilizes the positions of the N-terminal DNA binding domains of the monomers such that they can interact with the DNA (52) (compare Fig. 2A and B). H2O2-mediated inactivation of PerR involves the reduction of H2O2 by Fe2+ at the regulatory site to form the hydroxyl radical (•OH) (Fig. 2C). This highly reactive species oxidizes H37 and, to a lesser degree, H91 to 2-oxo-histidine, resulting in the disruption of normal metal coordination (67, 110). This causes an opening of the caliper configuration that negates DNA binding, a result which is seen in the crystal structures of demetalated, nonoxidized apo-PerR-Zn2+ as well as oxidized PerR-Zn2+-Fe2+ (52, 109, 110) (Fig. 2B). The region that allows access to Fe2+ bound at the regulatory site is rich in hydrophilic residues, thus explaining PerR's preference for H2O2 (52). The fact that there is no known physiological system to repair 2-oxo-histidine has led to the proposal that oxidized PerR is probably not recycled (67).
A second distinct mechanism of PerR inactivation occurs via exposure to nitrosylating agents such as nitrous oxide (NO). Exposure of B. subtilis to either an NO bolus or the NO generator Na-nitroprusside results in PerR-mediated induction of the PerR regulon during growth under aerobic and anaerobic conditions (77). The observation that anaerobic growth in the presence of Mn2+ reduces NO-induced expression of the PerR regulon while high iron levels enhance it suggests that induction is mediated by PerR-Zn2+-Fe2+. And it has been proposed that NO-mediated inactivation of PerR may be due to nitrosylation of bound Fe2+ (77).
The structural site that binds Zn2+ does not appear to play a role in oxidant sensing. The structural Zn2+ is coordinated by the cysteines C96, C99, C136, and C139 (67). In both PerR-Zn2+-Fe2+ and PerR-Zn2+-Mn2+, these cysteines have been shown to be highly resistant to oxidation with either H2O2 or diamide at levels higher than would be encountered in vivo (109).
OhrR
OhrR is a transcriptional repressor belonging to the MarR family of regulators and senses both organic peroxides and NaOCl (17). Generally, in their reduced form, OhrR dimers bind cooperatively to conserved, AT-rich, inverted-repeat sequences that overlap target gene promoters via winged-helix DNA binding domains (20, 28, 47, 76, 83) (Fig. 3A). OhrR is found in both Gram-positive and Gram-negative bacteria (1, 86) and can coreside with either OxyR (85) or PerR (27) or both (83). The first examples of OhrR to be characterized were shown to regulate themselves along with a gene encoding an organic peroxide-specific peroxiredoxin, Ohr (20, 27, 76, 83). Other orthologs that regulate a larger and more diverse set of target genes have since been characterized (16, 63, 72).
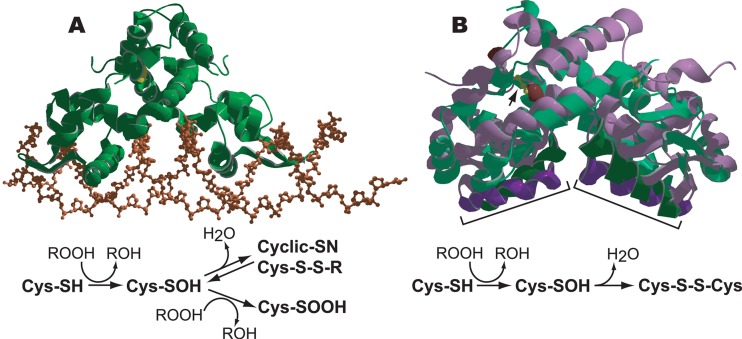
Fig 3.
Organic hydroperoxide (ROOH)-mediated derepression of OhrR-regulated promoters. (A) The structure of reduced 1-Cys B. subtilis OhrR (green ribbon) bound to the ohrA operator (DNA sugar phosphate backbone in brown) is shown. The redox cycle of B. subtilis OhrR is depicted below. Reduced 1-Cys OhrR binds to the target promoter/operator through the interaction of winged helix-turn-helix DNA binding domains with the DNA major groove, thereby blocking transcription. In the presence of organic hydroperoxide (ROOH), the single sensing cysteine in reduced OhrR, C15 (yellow), is oxidized to cysteine sulfenic acid (Cys-SOH). The Cys-SOH derivative remains bound to the promoter and must undergo one of several further modifications to induce the conformational change necessary to release the repressor. Cys-SOH can react either with a reduced cellular thiol to form a mixed thiol (Cys-S-S-R) or with the amino group of a neighboring amino acid residue to form a cyclic amide (Cys-SN). Both Cys-S-S-R and Cys-SN are recycled in vivo via reduction. Derepression can also occur through further oxidation of the Cys-SOH, for example, to cysteine sulfinic acid (Cys-SOOH). Overoxidized derivatives of OhrR are likely degraded. (B) An overlay of the ribbon structures of the reduced (purple) and oxidized (green) forms of the 2-Cys X. campestris OhrR showing the peroxide-induced shift in the positions of the DNA binding helices (dark purple and dark green, indicated by brackets). The redox-active cysteines in reduced OhrR are colored brown, while the disulfide bonds in the oxidized form are rendered in yellow (also indicated by an arrow in the right-hand monomer). The redox cycle of a 2-Cys OhrR is depicted below. The sensing cysteine (C22) of reduced 2-Cys OhrR bound to a target promoter/operator is oxidized in the presence of ROOH to cysteine sulfenic acid (Cys-SOH). The sensing Cys-SOH rapidly reacts with a second “resolving” cysteine (C127) residue to form an intersubunit disulfide bond that induces a conformational change that repositions the DNA binding helices and releases OhrR from the promoter. The oxidized disulfide bond-containing form of 2-Cys OhrR is likely actively rereduced in vivo. The PDB IDs for reduced B. subtilis OhrR proteins bound to the ohrA operator and oxidized and reduced X. campestris OhrR proteins are 1Z9C, 2PFB, and 2PEX, respectively.
Organic peroxide- or sodium hypochlorite (NaOCl)-mediated inactivation of OhrR, leading to derepression of OhrR-repressed genes, occurs via the oxidation of a sensing cysteine that undergoes subsequent disulfide formation (either intersubunit or mixed). This usually renders OhrR incapable of binding DNA, leading to derepression of target genes (1, 28, 85, 86). One exception to this general scheme occurs in Streptomyces coelicolor, in which reduced OhrR binds cooperatively to multiple sites overlapping the divergently transcribed promoters of ohrR and ohrA. Upon oxidation, OhrRox remains bound to a single high-affinity binding site within the ohrR-ohrA intergenic region that is upstream of the ohrR promoter and overlaps the ohrA −35 sequence. Interestingly, OhrRox bound at this site activates ohrR transcription but does not interfere with the transcription of ohrA (83).
The preferred organic peroxide inducer for OhrR can vary between organisms. For example, OhrR proteins of B. subtilis and Xanthomonas campestris are more sensitive to complex organic peroxides, such as linoleic acid hydroperoxide, while Agrobacterium tumefaciens OhrR preferentially senses less-complex organic peroxides like cumene hydroperoxide (57, 85, 102).
OhrR is now recognized to be a member of an oxidant-responsive subgroup of regulators of the MarR family that share a peroxide-sensing mechanism. Other members of the group include Pseudomonas aeruginosa OspR (63) and MgrA and SarZ in Staphylococcus aureus (54, 73). All sense peroxides or other oxidants through the initial oxidation of a sensing cysteine residue (15, 16, 63). However, OspR, MgrA, and SarZ differ from OhrR in that each appears to play a much more global regulatory role.
Like OhrR, Pseudomonas aeruginosa OspR appears to preferentially sense organic peroxides and mediates peroxide resistance through the regulation of a glutathione peroxidase gene (PA2826) while also directly regulating genes involved in quorum sensing and tyrosine metabolism. Alterations in OspR activity also affect β-lactam resistance, pigment production, and virulence in a murine acute pneumonia model (63).
MgrA and SarZ are global regulators in S. aureus that use peroxide as a signal for an adaptive shift to growth inside a host. MgrA is known to regulate, directly or indirectly, over 340 genes, including those encoding a variety of virulence factors as well as several antibiotic resistance determinants (72). SarZ, a close homolog of MgrA, also acts as a global regulator, controlling at least 80 genes involved in virulence, peroxide and antibiotic resistance, and the metabolic shift to anaerobic growth (16).
OhrR and its orthologs fall into one of two structural classes depending on the number of cysteines that participate in peroxide sensing (Fig. 3). Members of the first class, known as the 1-Cys class, which also includes OspR, MgrA, and SarZ, contain a single N-terminal sensing cysteine (C15 in B. subtilis OhrR) that is conserved in all OhrR proteins (47, 86). A second 2-Cys class contains an additional cysteine residue (C127 in X. campestris OhrR) in the C-terminal region that is also involved in peroxide sensing (86).
The current model for the mechanism of organic peroxide sensing in OhrR/MgrA family members is similar in several respects to peroxide sensing in OxyR. In both 1-Cys and 2-Cys OhrR proteins, the initial step in organic peroxide-mediated derepression involves the oxidation of the N-terminal sensing cysteine to a sulfenic acid (C-SOH) (28, 86) (Fig. 3). Organic peroxide specificity is due to a hydrophobic region lining the access channel to the sensing cysteine (47). This OhrR sulfenic acid derivative is still capable of DNA binding (68). At least one of the oxidized cysteines in the dimer must undergo additional modification in order for derepression to occur (25). This second stage of the sensing mechanism is where the 1-Cys and 2-Cys classes differ. Studies of B. subtilis OhrR have determined that inactivation of a 1-Cys OhrR can occur in several ways (Fig. 3A). The C15 sulfenic acid can either rapidly form a mixed disulfide with a cellular free thiol (i.e., bacillithiol, a recently identified 398-Da thiol) or react with the amino group of an adjacent amino acid residue to form a cyclic sulfenamide (17, 68). Mixed disulfide and protein sulfenamide formation is reversible since DNA binding activity of these derivatives of OhrR was able to be recovered in vitro by reduction in the presence of dithiothreitol (DTT) (68, 102). Irreversible inactivation of OhrR results when the C15 sulfenic acid is further oxidized to either a sulfinic (C15-SOOH) or sulfonic (C15-SO3H) acid, a process which occurs either in cells exposed to highly efficient inducers, like linoleic acid hydroperoxide, or in organic peroxide-exposed cells already undergoing disulfide stress (68).
The 2-Cys OhrR class, exemplified by X. campestris OhrR, contains an N-terminal sensing cysteine, C22, and two additional C-terminal cysteines, C127 and C131 (86). Only C22 and C127 are essential for regulator function (85, 86). Initial oxidation of the sensing C22 to sulfenic acid is followed by rapid formation of an intermolecular disulfide bond with C127 of the other subunit in the dimer (Fig. 3B). It is the formation of this intermolecular disulfide bond, not the oxidation of C22 to sulfenic acid, which renders the protein incapable of DNA binding, since amino acid substitutions at C127 in OhrR result in a protein that binds DNA in both its reduced and oxidized forms (3, 86). Therefore, disulfide bond formation in 2-Cys OhrR is functionally analogous to mixed disulfide formation in 1-Cys OhrR. Disulfide bond formation or mixed disulfide formation likely also serves to protect OhrR from overoxidation, which permanently inactivates the repressor (3, 68). The presence of a reduction system for oxidized OhrR containing either intermolecular or mixed disulfide bonds has been suggested but has not been identified (68). Possible candidates include systems utilizing dihydrolipoamide which have recently been shown to be involved in the reductive recycling of the organic peroxide-specific peroxiredoxins Ohr and OsmC in Xylella fastidiosa (24), while in B. subtilis and other Firmicutes, the bacilliredoxins, made up of several putative thiol-disulfide reductases (17, 43), are an attractive possibility.
The crystal structures of reduced unbound X. campestris OhrR and reduced B. subtilis OhrR bound to its operator sequence are similar (Fig. 3A and B), and conserved residues that appear to perform similar functions in both proteins have been identified (47, 81). Oxidation and disulfide bond formation in X. campestris OhrR induce dramatic structural changes, resulting in a 28° rotation of the winged helix-turn-helix DNA binding domains to positions that are incompatible with DNA binding (81) (Fig. 3B). No crystal structure is available for a 1-Cys OhrR containing a mixed disulfide; however, some insight into possible structural changes can be gained from the known structures of the reduced and oxidized forms of SarZ. C13-SOH in oxidized SarZ retains several of the normal hydrogen bonding interactions found in the reduced form. Thus, oxidation to Cys sulfenic acid has only a small effect on the positions of the DNA binding helices. This is consistent with observations that the sulfenic acid derivatives of OhrR and SarZ retain DNA binding activity (83, 89). Mixed thiol formation (C13-S-S-R) in SarZ disrupts these interactions, resulting in a conformational shift that increases the spacing between the DNA binding helices and negates DNA binding (89). The structural rearrangement observed in the mixed thiol derivative of SarZ is less pronounced than that seen in the oxidized disulfide-containing X. campestris OhrR, which displays a much greater degree of rotation between the subunits in the dimer (Fig. 3B). It will be interesting to see if the structural changes that occur upon mixed thiol formation in 1-Cys OhrR are similar to those in the dithiol derivative of the 2-Cys class or if they more closely resemble those seen in SarZ. In this regard, it is interesting to note that a 1-Cys B. subtilis OhrR can be converted to a functional 2-Cys OhrR by changing either of two Q residues (G120 or Q124), at positions corresponding to C127 and C131 in the X. campestris OhrR, to C so that the structural changes leading to derepression may be similar for the 1-Cys and 2-Cys OhrR proteins (101).
ROLE OF OxyR, PerR, AND OhrR IN VIRULENCE
ROS are an important component of the host immune response (62, 92), and it is becoming clear that peroxide is an important signal governing the expression of virulence genes. This is perhaps best illustrated by the peroxide-sensing regulators MgrA and SarZ in S. aureus, which have evolved to orchestrate the adaptive shift necessary to promote growth within a host in response to peroxide (5, 53). Although they are viewed primarily as modulators of the oxidative stress response, OxyR (64, 104), PerR (35, 48, 91, 117), and OhrR (3) have all been shown to be required for virulence in at least one experimental system. In the case of OxyR and PerR, their effect on virulence is not solely related to the regulation of oxidative stress defense genes. OxyR induces the expression of virulence factors that allow pathogens to evade host innate immunity (74, 90), while PerR has been shown to directly regulate an extracellular virulence factor (mitogen factor 3) in Streptococcus pyogenes (117). Other clinically relevant processes, such as biofilm formation (99, 119) and the oxidation state-independent regulation of several epigenetically controlled promoters determining phase variation (9, 113, 114), are also regulated by OxyR.
CONCLUSION
Much is known concerning the basic structural, mechanistic, and regulatory details of the peroxide-sensing regulators that are the subject of this review; however, many questions remain. Clearly, OxyR and its orthologs have evolved to sense a variety of oxidants in addition to H2O2 and have assigned a role for OxyR as a sensor of nitrosative stress during anaerobic growth. There are at least two distinct mechanisms of oxidant sensing, involving oxidative modification of a sensing cysteine alone (13, 98) or in concert with a second disulfide bonding partner (13), that are known in OxyR. There are also indications of the potential involvement of a third cysteine in peroxide sensing (4). The LysR-type regulator BenM, which controls genes involved in benzoate catabolism, is known to sense two coinducers (cis,cis-muconate and benzoate) that bind to distinct sites on the protein (23). Binding of each ligand singly activates transcription, while simultaneous binding of both coinducers results in a synergistic increase in target gene transcription (11) that is not observed for all BenM target promoters (21). It will be interesting to learn how oxidant-specific structural changes in OxyR affect promoter binding affinity and other OxyR-target promoter interactions as well as the ability of OxyR to interact with regulatory partners. Our knowledge of the full spectrum of oxidants that are sensed by these regulators is likely to be incomplete. For example, OhrR has only recently been shown to sense hypochlorite (17), while its natural organic peroxide inducers remain unidentified.
Finally, ROS are an important signal to pathogens, indicating contact with the host. There is a growing realization of the role of peroxide-sensing regulators in adapting to this environmental shift through the control of processes such as virulence factor expression (74, 90) and biofilm formation (99, 119). Furthermore, the lethality of some antibiotics is due, at least in part, to increased production of ROS (58), and it has been demonstrated that the levels of oxidative stress protective enzymes can affect antibiotic resistance levels (6). Therefore, a detailed understanding of the mechanisms and physiological functions of oxidative stress responsive regulators will further our understanding of bacterial adaptation to environmental changes in general and is likely to shed new light on aspects of many clinically important processes.
ACKNOWLEDGMENTS
We apologize to those investigators whose work we did not cite due to space limitations. We thank Piyapol Munpiyamit for help with figure preparation.
Research funding to S.M. and J.M.D. was provided by grants from the Chulabhorn Research Institute, Mahidol University National Science and Technology Development Agency (NSTDA), and the Center of Excellence for Environmental Health, Toxicology, and Management of Chemicals.
Footnotes
Published ahead of print 13 July 2012
REFERENCES
- 1. Antelmann H, Helmann JD. 2011. Thiol-based redox switches and gene regulation. Antioxid. Redox Signal. 14: 1049–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aslund F, Zheng M, Beckwith J, Storz G. 1999. Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol-disulfide status. Proc. Natl. Acad. Sci. U. S. A. 96: 6161–6165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Atichartpongkul S, Fuangthong M, Vattanaviboon P, Mongkolsuk S. 2010. Analyses of the regulatory mechanism and physiological roles of Pseudomonas aeruginosa OhrR, a transcription regulator and a sensor of organic hydroperoxides. J. Bacteriol. 192: 2093–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bae HW, Cho YH. 2012. Mutational analysis of Pseudomonas aeruginosa OxyR to define the regions required for peroxide resistance and acute virulence. Res. Microbiol. 163: 55–63 [DOI] [PubMed] [Google Scholar]
- 5. Ballal A, Ray B, Manna AC. 2009. sarZ, a sarA family gene, is transcriptionally activated by MgrA and is involved in the regulation of genes encoding exoproteins in Staphylococcus aureus. J. Bacteriol. 191: 1656–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bizzini A, Zhao C, Auffray Y, Hartke A. 2009. The Enterococcus faecalis superoxide dismutase is essential for its tolerance to vancomycin and penicillin. J. Antimicrob. Chemother. 64: 1196–1202 [DOI] [PubMed] [Google Scholar]
- 7. Brenot A, King KY, Caparon MG. 2005. The PerR regulon in peroxide resistance and virulence of Streptococcus pyogenes. Mol. Microbiol. 55: 221–234 [DOI] [PubMed] [Google Scholar]
- 8. Brenot A, Weston BF, Caparon MG. 2007. A PerR-regulated metal transporter (PmtA) is an interface between oxidative stress and metal homeostasis in Streptococcus pyogenes. Mol. Microbiol. 63: 1185–1196 [DOI] [PubMed] [Google Scholar]
- 9. Broadbent SE, Davies MR, van der Woude MW. 2010. Phase variation controls expression of Salmonella lipopolysaccharide modification genes by a DNA methylation-dependent mechanism. Mol. Microbiol. 77: 337–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bsat N, Herbig AF, Cassillas-Martinez L, Setlow P, Helmann JD. 1998. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol. Microbiol. 29: 189–198 [DOI] [PubMed] [Google Scholar]
- 11. Bundy BM, Collier LS, Hoover TR, Neidle EL. 2002. Synergistic transcriptional activation by one regulatory protein in response to two metabolites. Proc. Natl. Acad. Sci. U. S. A. 99: 7693–7698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Calhoun LN, Kwon YM. 2011. Structure, function and regulation of the DNA-binding protein Dps and its role in acid and oxidative stress resistance in Escherichia coli: a review. J. Appl. Microbiol. 110: 375–386 [DOI] [PubMed] [Google Scholar]
- 13. Chen H, et al. 2008. A novel OxyR sensor and regulator of hydrogen peroxide stress with one cysteine residue in Deinococcus radiodurans. PLoS One 3: e1602 doi:10.1371/journal.pone.0001602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen L, Keramati L, Helmann JD. 1995. Coordinate regulation of Bacillus subtilis peroxide stress genes by hydrogen peroxide and metal ions. Proc. Natl. Acad. Sci. U. S. A. 92: 8190–8194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen PR, et al. 2006. An oxidation-sensing mechanism is used by the global regulator MgrA in Staphylococcus aureus. Nat. Chem. Biol. 2: 591–595 [DOI] [PubMed] [Google Scholar]
- 16. Chen PR, et al. 2009. A new oxidative sensing and regulation pathway mediated by the MgrA homologue SarZ in Staphylococcus aureus. Mol. Microbiol. 71: 198–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chi BK, et al. 2011. S-bacillithiolation protects against hypochlorite stress in Bacillus subtilis as revealed by transcriptomics and redox proteomics. Mol. Cell. Proteomics 10: M111.009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Choi H-J, et al. 2001. Structural basis of the redox switch in the OxyR transcription factor. Cell 105: 103–113 [DOI] [PubMed] [Google Scholar]
- 19. Christman MF, Storz G, Ames BM. 1989. OxyR, a positive regulator of hydrogen peroxide-inducible genes in Escherichia coli and Salmonella typhimurium, is homologous to a family of bacterial regulatory proteins. Proc. Natl. Acad. Sci. U. S. A. 86: 3484–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chuchue T, et al. 2006. ohrR and ohr are the primary sensor/regulator and protective genes against organic hydroperoxide stress in Agrobacterium tumefaciens. J. Bacteriol. 188: 842–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Clark TJ, Momany C, Neidle EL. 2002. The benPK operon, proposed to play a role in transport, is part of a regulon for benzoate catabolism in Acinetobacter sp. strain ADP1. Microbiology 148: 1213–1223 [DOI] [PubMed] [Google Scholar]
- 22. Cohen GM, d'Arcy Doherty M. 1987. Free radical mediated cell toxicity by redox cycling chemicals. Br. J. Cancer Suppl. 8: 46–52 [PMC free article] [PubMed] [Google Scholar]
- 23. Craven SH, et al. 2009. Inducer responses of BenM, a LysR-type transcriptional regulator from Acinetobacter baylyi ADP1. Mol. Microbiol. 72: 881–894 [DOI] [PubMed] [Google Scholar]
- 24. Cussiol JR, Alegria TGP, Szweda LI, Netto LES. 2010. Ohr (organic hydroperoxide resistance protein) possesses a previously undescribed activity, lipoyl-dependent peroxidase. J. Biol. Chem. 285: 21943–21950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Eiamphungporn W, Soonsanga S, Lee J- W, Helmann JD. 2009. Oxidation of a single active site suffices for the functional inactivation of the dimeric Bacillus subtilis OhrR repressor in vitro. Nucleic Acids Res. 37: 1174–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Faulkner MJ, Ma Z, Fuangthong M, Helmann JD. 2012. Derepression of the Bacillus subtilis PerR peroxide stress response leads to iron deficiency. J. Bacteriol. 194: 1226–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fuangthong M, Atichartpongkul S, Mongkolsuk S, Helmann JD. 2001. OhrR is a repressor of ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtilis. J. Bacteriol. 183: 4134–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fuangthong M, Helmann JD. 2002. The OhrR repressor senses organic hydroperoxides by reversible formation of a cysteine-sulfenic acid derivative. Proc. Natl. Acad. Sci. U. S. A. 99: 6690–6695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fuangthong M, Helmann JD. 2003. Recognition of DNA by three ferric uptake regulator (Fur) homologs in Bacillus subtilis. J. Bacteriol. 185: 6348–6357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fuangthong M, Herbig AF, Bsat N, Helmann JD. 2002. Regulation of the Bacillus subtilis fur and perR genes by PerR: not all members of the PerR regulon are peroxide inducible. J. Bacteriol. 184: 3276–3286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gaballa A, Helmann JD. 2002. A peroxide-induced zinc uptake system plays an important role in protection against oxidative stress in Bacillus subtilis. Mol. Microbiol. 45: 997–1005 [DOI] [PubMed] [Google Scholar]
- 32. Giro M, Carrillo N, Krapp AR. 2006. Glucose-6-phosphate dehydrogenase and ferredoxin-NADP(H) reductase contribute to damage repair during the soxRS response of Escherichia coli. Microbiology 152: 1119–1128 [DOI] [PubMed] [Google Scholar]
- 33. Gonzalez-Flecha B, Demple B. 1997. Homeostatic regulation of intracellular hydrogen peroxide concentration in aerobically growing Escherichia coli. J. Bacteriol. 179: 382–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gonzalez-Flecha B, Demple B. 1999. Role for the oxyS gene in regulation of intracellular hydrogen peroxide in Escherichia coli. J. Bacteriol. 181: 3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gryllos I, et al. 2008. PerR confers phagocytic killing resistance and allows pharyngeal colonization by group A Streptococcus. PLoS Pathog. 4: e1000145 doi:10.1371/journal.ppat.1000145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hahn J-S, Oh S-Y, Chater KF, Cho Y-H, Roe J-H. 2000. H2O2-sensitive fur-like repressor CatR regulating the major catalase gene in Streptomyces coelicolor. J. Biol. Chem. 275: 38254–38260 [DOI] [PubMed] [Google Scholar]
- 37. Hahn J-S, Oh S-Y, Roe J-H. 2002. Role of OxyR as a peroxide-sensing positive regulator in Streptomyces coelicolor A3(2). J. Bacteriol. 184: 5214–5222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Haridas V, et al. 2005. Avicinylation (thioesterification): a protein modification that can regulate the response to oxidative and nitrosative stress. Proc. Natl. Acad. Sci. U. S. A. 102: 10088–10093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Harrison A, et al. 2007. The OxyR regulon in nontypeable Haemophilus influenzae. J. Bacteriol. 189: 1004–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hausladen A, Privalle CT, Keng T, De Angelo J, Stamler JS. 1996. Nitrosative stress: activation of the transcription factor OxyR. Cell 86: 719–729 [DOI] [PubMed] [Google Scholar]
- 41. Hayashi K, Ohsawa T, Kobayashi K, Ogasawara N, Ogura M. 2005. The H2O2 stress-responsive regulator PerR positively regulates srfA expression in Bacillus subtilis. J. Bacteriol. 187: 6659–6667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. He X, et al. 2010. Oral-derived bacterial flora defends its domain by recognizing and killing intruders—a molecular analysis using Escherichia coli as a model intestinal bacterium. Microb. Ecol. 60: 655–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Helmann JD. 2011. Bacillithiol, a new player in bacterial redox homeostasis. Antioxid. Redox Signal. 15: 123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Helmann JD. 2002. OxyR: a molecular code for redox sensing. Sci. STKE 2002(157): pe46 doi:10.1126/stke.20002.157.pe46 [DOI] [PubMed] [Google Scholar]
- 45. Heo Y-J, et al. 2010. The major catalase gene (katA) of Pseudomonas aeruginosa PA14 is under both positive and negative control of the global transactivator OxyR in response to hydrogen peroxide. J. Bacteriol. 192: 381–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Herbig AF, Helmann JD. 2001. Roles of metal ions and hydrogen peroxide in modulating the interaction of the Bacillus subtilis PerR peroxide regulon repressor with operator DNA. Mol. Microbiol. 41: 849–859 [DOI] [PubMed] [Google Scholar]
- 47. Hong M, Fuangthong M, Helmann JD, Brennan RG. 2005. Structure of an OhrR-ohrA operator complex reveals the DNA binding mechanism of the MarR family. Mol. Cell 20: 131–141 [DOI] [PubMed] [Google Scholar]
- 48. Horsburgh MJ, Clements MO, Crossley H, Ingham E, Foster SJ. 2001. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect. Immun. 69: 3744–3754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ieva R, et al. 2008. OxyR tightly regulates catalase expression in Neisseria meningitidis through both repression and activation mechanisms. Mol. Microbiol. 70: 1152–1165 [DOI] [PubMed] [Google Scholar]
- 50. Imlay JA. 2008. Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 77: 755–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Imlay JA. 2003. Pathways of oxidative damage. Annu. Rev. Microbiol. 57: 395–418 [DOI] [PubMed] [Google Scholar]
- 52. Jacquamet L, et al. 2009. Structural characterization of the active form of PerR: insights into the metal-induced activation of PerR and Fur proteins for DNA binding. Mol. Microbiol. 73: 20–31 [DOI] [PubMed] [Google Scholar]
- 53. Jonssona I-M, Lindholma C, Luongb TT, Leeb CY, Tarkowskia A. 2008. mgrA regulates staphylococcal virulence important for induction and progression of septic arthritis and sepsis. Microbes Infect. 10: 1229–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kaito C, Morishita D, Matsumoto Y, Kurokawa K, Sekimizu K. 2006. Novel DNA binding protein SarZ contributes to virulence in Staphylococcus aureus. Mol. Microbiol. 62: 1601–1617 [DOI] [PubMed] [Google Scholar]
- 55. Keyer K, Imlay JA. 1996. Superoxide accelerates DNA damage by elevating free-iron levels. Proc. Natl. Acad. Sci. U. S. A. 93: 13635–13640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kim SO, et al. 2002. OxyR: a molecular code for redox-related signaling. Cell 109: 383–396 [DOI] [PubMed] [Google Scholar]
- 57. Klomsiri C, Panmanee W, Dharmsthiti S, Vattanaviboon P, Mongkolsuk S. 2005. Novel roles of ohrR-ohr in Xanthomonas sensing, metabolism, and physiological adaptive response to lipid hydroperoxide. J. Bacteriol. 187: 3277–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. 2007. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130: 797–810 [DOI] [PubMed] [Google Scholar]
- 59. Kona J, Brinck T. 2006. A combined molecular dynamics simulation and quantum chemical study on the mechanism for activation of the OxyR transcription factor by hydrogen peroxide. Org. Biomol. Chem. 4: 3468–3478 [DOI] [PubMed] [Google Scholar]
- 60. Kullik I, Stevens J, Toledano MB, Storz G. 1995. Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for DNA binding and multimerization. J. Bacteriol. 177: 1285–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kullik I, Toledano MB, Tartaglia LA, Storz G. 1995. Mutational analysis of the redox-sensitive transcriptional regulator OxyR: regions important for oxidation and transcriptional activation. J. Bacteriol. 177: 1275–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lamb C, Dixon RA. 1997. The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48: 251–275 [DOI] [PubMed] [Google Scholar]
- 63. Lan L, Murray TS, Kazmierczak BI, He C. 2010. Pseudomonas aeruginosa OspR is an oxidative stress sensing regulator that affects pigment production, antibiotic resistance and dissemination during infection. Mol. Microbiol. 75: 76–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lau GW, Britigan BE, Hassett DJ. 2005. Pseudomonas aeruginosa OxyR is required for full virulence in rodent and insect models of infection and for resistance to human neutrophils. Infect. Immun. 73: 2550–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lee C, et al. 2004. Redox regulation of OxyR requires specific disulfide bond formation involving a rapid kinetic reaction path. Nat. Struct. Mol. Biol. 11: 1179–1185 [DOI] [PubMed] [Google Scholar]
- 66. Lee J-W, Helmann JD. 2006. Biochemical characterization of the structural Zn2+ site in the Bacillus subtilis peroxide sensor PerR. J. Biol. Chem. 281: 23567–23578 [DOI] [PubMed] [Google Scholar]
- 67. Lee J-W, Helmann JD. 2006. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature 440: 363–367 [DOI] [PubMed] [Google Scholar]
- 68. Lee J-W, Soonsanga S, Helmann JD. 2007. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc. Natl. Acad. Sci. U. S. A. 104: 8743–8748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Li H, Singh AK, McIntyre LM, Sherman LA. 2004. Differential gene expression in response to hydrogen peroxide and the putative PerR regulon of Synechocystis sp. strain PCC 6803. J. Bacteriol. 186: 3331–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Liochev SI, Fridovich I. 1992. Fumarase C, the stable fumarase of Escherichia coli, is controlled by the soxRS regulon. Proc. Natl. Acad. Sci. U. S. A. 89: 5892–5896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Liu Y, Bauer SC, Imlay JA. 2011. The YaaA protein of the Escherichia coli OxyR regulon lessens hydrogen peroxide toxicity by diminishing the amount of intracellular unincorporated iron. J. Bacteriol. 193: 2186–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Luong TT, Dunman PM, Murphy E, Projan SJ, Lee CY. 2006. Transcription profiling of the mgrA regulon in Staphylococcus aureus. J. Bacteriol. 188: 1899–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Luong TT, Newell SW, Lee CY. 2003. mgr, a novel global regulator in Staphylococcus aureus. J. Bacteriol. 185: 3703–3710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Melstrom KAJ, et al. 2007. Cytotoxicity of Pseudomonas secreted exotoxins requires OxyR expression. J. Surg. Res. 143: 50–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mongkolsuk S, Helmann JD. 2002. Regulation of inducible peroxide stress responses. Mol. Microbiol. 45: 9–15 [DOI] [PubMed] [Google Scholar]
- 76. Mongkolsuk S, et al. 2002. The repressor for an organic peroxide-inducible operon is uniquely regulated at multiple levels. Mol. Microbiol. 44: 793–802 [DOI] [PubMed] [Google Scholar]
- 77. Moore CM, Nakano MM, Wang T, Ye RW, Helmann JD. 2004. Response of Bacillus subtilis to nitric oxide and the nitrosating agent sodium nitroprusside. J. Bacteriol. 186: 4655–4664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Morikawa K, et al. 2006. Bacterial nucleoid dynamics: oxidative stress response in Staphylococcus aureus. Genes Cells 11: 409–423 [DOI] [PubMed] [Google Scholar]
- 79. Mukhopadhyay A, et al. 2007. Cell-wide responses to low-oxygen exposure in Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 189: 5996–6010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Mukhopadhyay P, Zheng M, Bedzyk LA, LaRossa RA, Storz G. 2004. Prominent roles of the NorR and Fur regulators in the Escherichia coli transcriptional response to reactive nitrogen species. Proc. Natl. Acad. Sci. U. S. A. 101: 745–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Newberry KJ, Fuangthong M, Panmanee W, Mongkolsuk S, Brennan RG. 2007. Structural mechanism of organic hydroperoxide induction of the transcription regulator OhrR. Mol. Cell 28: 652–664 [DOI] [PubMed] [Google Scholar]
- 82. Ochsner UA, Vasil ML, Eyad A, Kislay P, Hassett DJ. 2000. Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF. J. Bacteriol. 182: 4533–4544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Oh S-Y, Shin J-H, Roe J-H. 2007. Dual role of OhrR as a repressor and an activator in response to organic hydroperoxides in Streptomyces coelicolor. J. Bacteriol. 189: 6284–6292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Paget MS, Buttner MJ. 2003. Thiol-based regulatory switches. Annu. Rev. Genet. 37: 91–121 [DOI] [PubMed] [Google Scholar]
- 85. Panmanee W, et al. 2002. OhrR, a transcription repressor that senses and responds to changes in organic peroxide levels in Xanthomonas campestris pv. phaseoli. Mol. Microbiol. 45: 1647–1654 [DOI] [PubMed] [Google Scholar]
- 86. Panmanee W, Vattanaviboon P, Poole LB, Mongkolsuk S. 2006. Novel organic hydroperoxide-sensing and responding mechanisms for OhrR, a major bacterial sensor and regulator of organic hydroperoxide stress. J. Bacteriol. 188: 1389–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Pomposiello PJ, Demple B. 2002. Global adjustment of microbial physiology during free radical stress. Adv. Microb. Physiol. 46: 319–341 [DOI] [PubMed] [Google Scholar]
- 88. Poole LB. 2005. Bacterial defenses against oxidants: mechanistic features of cysteine-based peroxidases and their flavoprotein reductases. Arch. Biochem. Biophys. 433: 240–254 [DOI] [PubMed] [Google Scholar]
- 89. Poor CB, Chen PR, Duguid EM, Rice PA, He C. 2009. Crystal structures of the reduced, sulfenic acid, and mixed disulfide forms of SarZ, a redox active global regulator in Staphylococcus aureus. J. Biol. Chem. 284: 23517–23524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ramsey MM, Whiteley M. 2009. Polymicrobial interactions stimulate resistance to host innate immunity through metabolite perception. Proc. Natl. Acad. Sci. U. S. A. 106: 1578–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Rea R, Hill C, Gahan CGM. 2005. Listeria monocytogenes PerR mutants display a small-colony phenotype, increased sensitivity to hydrogen peroxide, and significantly reduced murine virulence. Appl. Environ. Microbiol. 71: 8314–8322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Robinson JM. 2009. Phagocytic leukocytes and reactive oxygen species. Histochem. Cell Biol. 131: 465–469 [DOI] [PubMed] [Google Scholar]
- 93. Schell MA. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47: 597–626 [DOI] [PubMed] [Google Scholar]
- 94. Seaver LC, Imlay JA. 2001. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J. Bacteriol. 183: 7173–7181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Seaver LC, Imlay JA. 2001. Hydrogen peroxide fluxes and compartmentalization inside growing Escherichia coli. J. Bacteriol. 183: 7182–7189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Seib KL, et al. 2007. Characterization of the OxyR regulon of Neisseria gonorrhoeae. Mol. Microbiol. 63: 54–68 [DOI] [PubMed] [Google Scholar]
- 97. Semchyshyn H, Bagnyukova T, Storey K, Storey V. 2005. Hydrogen peroxide increases the activities of soxRS regulon enzymes and the levels of oxidized proteins and lipids in Escherichia coli. Cell Biol. Int. 29: 898–902 [DOI] [PubMed] [Google Scholar]
- 98. Seth D, Hausladen A, Wang Y-J, Stamler JS. 2012. Endogenous protein S-nitrosylation in E. coli: regulation by OxyR. Science 336: 470–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Shanks RM, et al. 2007. A Serratia marcescens OxyR homolog mediates surface attachment and biofilm formation. J. Bacteriol. 189: 7262–7272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Sobota JM, Imlay JA. 2011. Iron enzyme ribulose-5-phosphate 3-epimerase in Escherichia coli is rapidly damaged by hydrogen peroxide but can be protected by manganese. Proc. Natl. Acad. Sci. U. S. A. 108: 5402–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Soonsanga S, Lee J-W, Helmann JD. 2008. Conversion of Bacillus subtilis OhrR from a 1-Cys to a 2-Cys peroxide sensor. J. Bacteriol. 190: 5738–5745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Soonsanga S, Lee J-W, Helmann JD. 2008. Oxidant-dependent switching between reversible and sacrificial oxidation pathways for Bacillus subtilis OhrR. Mol. Microbiol. 68: 678–986 [DOI] [PubMed] [Google Scholar]
- 103. Storz G, Tartaglia LA, Ames BM. 1990. Transcriptional regulator of oxidative stress inducible genes: direct activation by oxidation. Science 248: 189–194 [DOI] [PubMed] [Google Scholar]
- 104. Sund CJ, et al. 2008. The Bacteroides fragilis transcriptome response to oxygen and H2O2: the role of OxyR and its effect on survival and virulence. Mol. Microbiol. 67: 129–142 [DOI] [PubMed] [Google Scholar]
- 105. Tao K. 1999. In vivo oxidation-reduction kinetics of OxyR, the transcriptional activator for an oxidative stress-inducible regulon in Escherichia coli. FEBS Lett. 457: 90–92 [DOI] [PubMed] [Google Scholar]
- 106. Tao K, Zou C, Fujita N, Ishihama A. 1995. Mapping of the OxyR protein contact site in the C-terminal region of RNA polymerase α subunit. J. Bacteriol. 177: 6740–6744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Tartaglia LA, Gimeno CJ, Storz G, Ames BN. 1992. Multidegenerate DNA recognition by the OxyR transcriptional regulator. J. Biol. Chem. 267: 2038–2045 [PubMed] [Google Scholar]
- 108. Toledano MB, et al. 1994. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell 78: 897–909 [DOI] [PubMed] [Google Scholar]
- 109. Traoré DAK, et al. 2006. Crystal structure of the apo-PerR-Zn protein from Bacillus subtilis. Mol. Microbiol. 61: 1211–1219 [DOI] [PubMed] [Google Scholar]
- 110. Traoré DAK, et al. 2009. Structural and functional characterization of 2-oxo-histidine in oxidized PerR protein. Nat. Chem. Biol. 5: 53–59 [DOI] [PubMed] [Google Scholar]
- 111. Tseng H-J, McEwan AG, Apicella MA, Jennings MP. 2003. OxyR acts as a repressor of catalase expression in Neisseria gonorrhoeae. Infect. Immun. 71: 550–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. van Vliet AHM, Baillon M-LA, Penn CW, Ketley JM. 1999. Campylobacter jejuni contains two fur homologs: characterization of iron-responsive regulation of peroxide stress defense genes by the PerR repressor. J. Bacteriol. 181: 6371–6376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Wallecha A, Correnti J, Munster V, van der Woude M. 2003. Phase variation of Ag43 is independent of the oxidation state of OxyR. J. Bacteriol. 185: 2203–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Wallecha A, Munster V, Correnti J, Chan T, van der Woude M. 2002. Dam- and OxyR-dependent phase variation of agn43: essential elements and evidence for a new role of DNA methylation. J. Bacteriol. 184: 3338–3347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Wang X, et al. 2006. Mutational analysis to define an activating region on the redox-sensitive transcriptional regulator OxyR. J. Bacteriol. 188: 8335–8342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Wei Q, et al. 2012. Global regulation of gene expression by OxyR in an important human opportunistic pathogen. Nucleic Acids Res. 40: 4320–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Wen Y-T, et al. 2011. Differential secretomics of Streptococcus pyogenes reveals a novel peroxide regulator (PerR)-regulated extracellular virulence factor mitogen factor 3 (MF3). Mol. Cell. Proteomics 10: M110.007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Wu H-J, et al. 2006. PerR controls Mn-dependent resistance to oxidative stress in Neisseria gonorrhoeae. Mol. Microbiol. 60: 401–416 [DOI] [PubMed] [Google Scholar]
- 119. Wu J, Lin X, Xie H, Burne R. 2008. OxyR is involved in coordinate regulation of expression of fimA and sod genes in Porphyromonas gingivalis. FEMS Microbiol. Lett. 282: 188–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Yin L, et al. 2010. DRA0336, another OxyR homolog, involved in the antioxidation mechanisms in Deinococcus radiodurans. J. Microbiol. 48: 473–479 [DOI] [PubMed] [Google Scholar]
- 121. Zeller T, et al. 2007. Regulation of hydrogen peroxide-dependent gene expression in Rhodobacter sphaeroides: regulatory functions of OxyR. J. Bacteriol. 189: 3784–3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Zhang A, et al. 1998. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J. 17: 6061–6068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Zheng M, Aslund F, Storz G. 1998. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279: 1718–1721 [DOI] [PubMed] [Google Scholar]
- 124. Zheng M, Doan B, Schneider TD, Storz G. 1999. OxyR and SoxRS regulation of fur. J. Bacteriol. 181: 4639–4643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Zheng M, et al. 2001. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J. Bacteriol. 183: 4562–4570 [DOI] [PMC free article] [PubMed] [Google Scholar]