SUMMARY
Sirtuins are a group of NAD+-dependent enzymes that post-translationally modify histones and other proteins. Among seven mammalian sirtuins, SIRT1 has been the most extensively studied and has been demonstrated to play a critical role in all major metabolic organs and tissues. SIRT1 regulates glucose and lipid homeostasis in the liver, modulates insulin secretion in pancreatic islets, controls insulin sensitivity and glucose uptake in skeletal muscle, increases adiponectin expression in white adipose tissue and controls food intake and energy expenditure in the brain. Recently, SIRT3 has been demonstrated to modulate insulin sensitivity in skeletal muscle and systemic metabolism, and Sirt3-null mice manifest characteristics of metabolic syndrome on a high-fat diet. Thus, it is reasonable to believe that enhancing the activities of SIRT1 and SIRT3 may be beneficial for Type 2 diabetes. Although it is controversial, the SIRT1 activator SRT1720 has been reported to be effective in improving glucose metabolism and insulin sensitivity in animal models. More research needs to be conducted so that we can better understand the physiological functions and molecular mechanisms of sirtuins in order to therapeutically target these enzymes for diabetes treatment.
Sirtuins are evolutionarily conserved
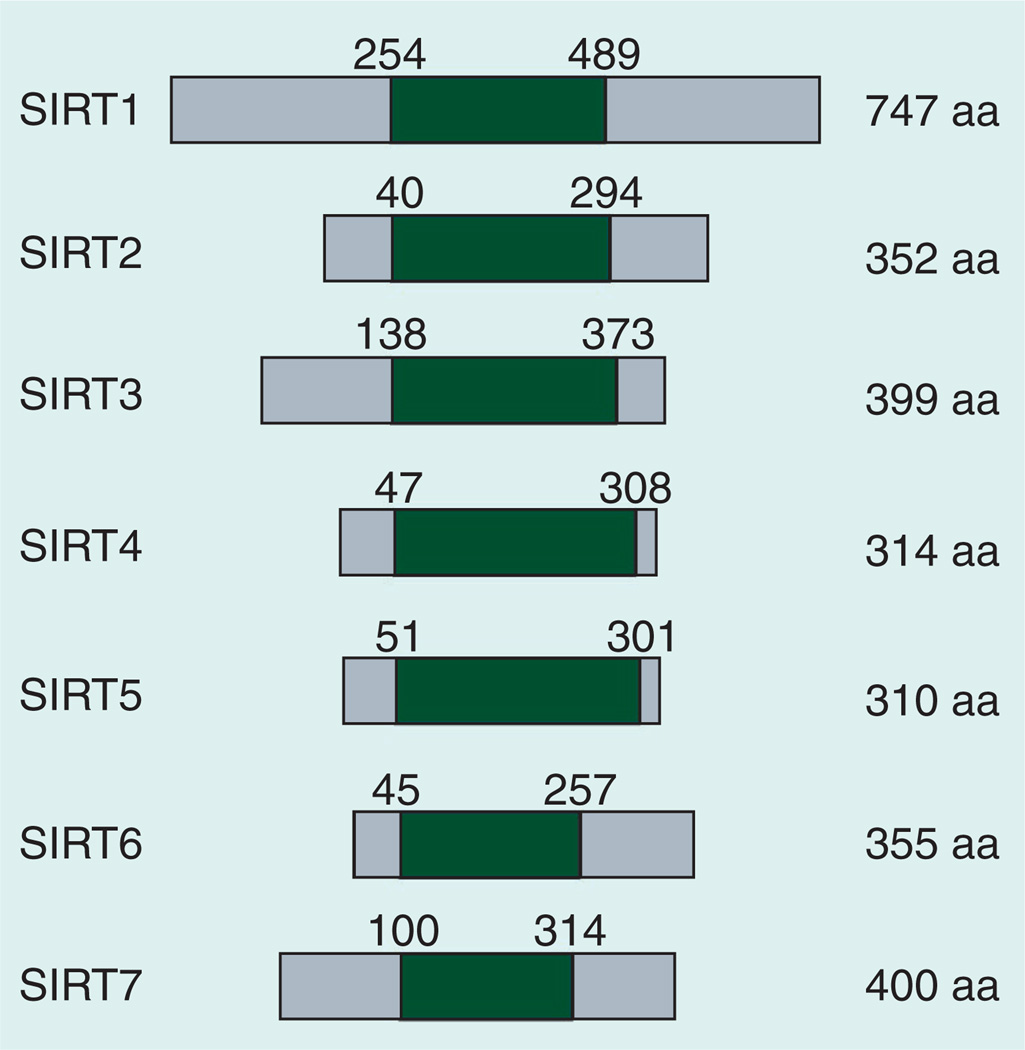
Sirtuins are a family of proteins that are homologous to yeast Sir2. Sirtuins are evolutionarily conserved as they exist in a wide range of organisms, from bacteria to mammals. In humans, there are seven sirtuins (SIRT1–SIRT7). These sirtuins have different subcellular localizations: SIRT1 (also found in the cytoplasm) and SIRT6 are found in the nucleus, SIRT2 is primarily found in the cytoplasm (but is also found in nucleus), SIRT3, SIRT4 and SIRT5 are present in mitochondria, and SIRT7 is found in the nucleolus [1–3]. All sirtuins share well-conserved domains for NAD+ and peptide binding, which are also called sirtuin domains (Figure 1) [4]. At least four distinct enzymatic activities have been characterized in sirtuin family members: deacetylation (the transfer of an acetyl group from the substrate to ADP-ribose moiety of NAD+ and the generation of O-acetyl-ADP-ribose and nicotinamide [NAM]), ADP-ribosylation (the transfer of ADP-ribose moiety of NAD+ to the substrate), desuccinylation (the transfer of a succinyl group from the substrate to the ADP-ribose moiety of NAD+) and demalonylation (the transfer of a malonyl group to the ADP-ribose moiety of NAD+) [5–12]. Since NAD+ is required for the enzymatic activities of sirtuins, it has been suggested that they may be important metabolic sensors [13]. Although it is not fully established how sirtuins respond to NAD+ flux in cells, changes in the NAD+/NADH ratio or the reaction product NAM have been demonstrated to be critical [14–19]. The NAD+ levels, at least in mouse livers, oscillate in a circadian manner that peaks both late in the day and also late in the night [20,21]. However, SIRT1 activity does not seem to follow exactly the same pattern because either SIRT1 protein levels or activities only peak at night in mice [22,23]. This apparent inconsistency between NAD+ levels and SIRT1 activity has not been resolved yet.
Figure 1. Human sirtuin proteins and their domain structure.
The conserved sirtuin domain is shaded. The protein sequences and annotations are based on the NCBI Gene data set.
To date, the deacetylation function of sirtuins has been best characterized among all the sirtuin enzymatic activities. It is well documented that acetylation/deacetylation play pleiotropic roles in all biological systems, including gene transcription, protein stability, signal transduction and metabolism [24–27]. Protein acetylation status and sirtuin activities are dynamic and change in response to different metabolic conditions [24–27]. Mammalian sirtuins, except SIRT4, have characterized deacetylation activities on a variety of protein substrates described in Table 1.
Table 1.
The identified substrates of sirtuins.
| Sirtuin | Known substrates | Process involved | Ref. |
|---|---|---|---|
| SIRT1 | ACSS1: K661 | Conversion of acetate to acetyl-CoA | [169] |
| AR: K630 | Androgen receptor signaling | [170] | |
| BMAL1: K537 | Circadian rhythms | [23] | |
| β-catenin | Suppression of tumorigenesis | [171] | |
| Cortactin | Cell migration | [172] | |
| CREB: K136 | Glucose and lipid metabolism | [81] | |
| CRTC2: K628 | Gluconeogenesis | [87] | |
| DNMT1: K1349, 1415 | DNA methylation | [173] | |
| eNOS: K496, K506 | Endothelium vasodilation | [174] | |
| FOXO1: K242, K245, K262, K274, K294, K559 | Transcription, autophagy, among others | [175–177] | |
| FOXO3: K242, K245, K259, K271, K290, K569 | Transcription | [177,178] | |
| FOXO4: K186, K189, K215, K237, K407 | Transcription | [177] | |
| Histone H1: K26 | Heterochromatin formation | [179] | |
| H2A.x | DNA damage response | [180] | |
| H2A.z: K15 | Cardiac hypertrophy | [181] | |
| Histone H3: K9 | Chromatin remodeling | [179] | |
| Histone H3: K56 | Chromatin remodeling | [182] | |
| Histone H4: K16 | Chromatin remodeling | [179] | |
| HIV Tat: K50 | HIV transcription | [183] | |
| HNF4α | Transcription | [184] | |
| Ku70: K539, K542 | DNA repair | [148] | |
| LKB1: K48 | Cell proliferation and metabolism | [130] | |
| LXRα: K432 | Lipid metabolism | [97] | |
| c-Myc: K323 | Cell proliferation | [185] | |
| MyoD: K99, K102, K104 | Muscle differentiation | [186] | |
| p53: K317, K370, K382 | Cell survival and stress response | [187,188] | |
| p300: K102, K1024 | Protein acetylation | [189] | |
| PARP1 | Cell survival | [190] | |
| PCAF | DNA damage response | [186] | |
| PER2 | Circadian rhythms | [22] | |
| PGC-1α: K77, K144, K183, K253, K270, K277, K320, K412, K441, K450, K757, K778 | Mitochondrial biogenesis and metabolism | [84] | |
| PGC-1β | Glucose and lipid metabolism | [191] | |
| NF-κBRelA/p65: K310 | Transcription | [192] | |
| Rb: K873, 874 | Cell cycle control | [193] | |
| Smad7: K64, K70 | Apoptosis | [194] | |
| SREBP-1c: K289, K309 | Lipid metabolism | [95] | |
| SUV39H1: K266 | Heterochromatin formation | [195] | |
| TAF(I)68 | rDNA transcription | [196] | |
| Zyxin | Cytoskeletal dynamics | [197] | |
| SIRT2 | CDC20 | Mitosis | [31] |
| CDH1 | Mitosis | [31] | |
| FOXO1: K262, K265, K274 | Adipogenesis and autophagy | [33,35] | |
| FOXO3: K242, K259, K290, K569 | Oxidative stress and ubiquitination | [30,116] | |
| Histone H3: K56 | DNA damage response | [198] | |
| Histone H4: K16 | Mitosis | [3] | |
| NF-κBRelA/p65: K310 | Transcription | [34] | |
| p53 | Cell survival and stress response | [199] | |
| p300: K418, K423, K1542, K1546, K1549, K1699, K1704, K1707 | Chromatin remodeling | [200] | |
| Par3: K831, K848, K881, K1327 | Myelination | [201] | |
| PEPCK: K70, K71, K594 | Gluconeogenesis | [32] | |
| PRLR | Prolactin receptor dimerization | [202] | |
| α-tubulin: K40 | Mitosis | [36] | |
| SIRT3 | ACSS2: K635 | Conversion of acetate to acetyl-CoA | [169,203] |
| ALDH2: K377 | Alcohol metabolism | [38] | |
| CYPD: K166 | Mitochondrial PTP control | [204] | |
| GDH | Glutamate oxidation and insulin secretion | [56] | |
| HMGCS2: K310, K447, K473 | Ketogenesis | [50] | |
| IDH2 | Anti-oxidative stress | [48] | |
| LCAD: K42 | Fatty acid oxidation | [53] | |
| MRPL10: K124, K162, K196 | Mitochondrial protein synthesis | [47] | |
| NDUF9A | Mitochondrial ETC | [54] | |
| OTC: K88 | Urea cycle | [43] | |
| SDH | Mitochondrial ETC | [205] | |
| SOD2: K53, K68, K89, K122 | Anti-oxidative stress | [51] | |
| SIRT4 | GDH | Glutamate oxidation and insulin secretion | [8,9] |
| SIRT5 | CPS1 | Urea cycle | [59] |
| CS | Citric acid cycle | [11] | |
| GDH | Amino acid-induced insulin secretion | [11] | |
| GOT2 | Amino acid metabolism | [11] | |
| HMGCS2 | Ketogenesis | [11] | |
| MDH | Citric acid cycle | [11] | |
| TST | Cyanide detoxification | [11] | |
| SIRT6 | CtIP: K432, K526, K604 | DNA repair | [66] |
| H3K9 | Chromatin remodeling | [69] | |
| H3K56 | Chromatin remodeling | [67,206] | |
| PARP1: K521 (mono-ADP-ribosylation) | DNA repair | [60] | |
| SIRT7 | p53 | Cell survival | [72] |
ETC: Electron transport chain; PTP: Permeability transition pore; Rb: Retinoblastoma.
As a founding member, SIRT1 can deacetylate histone (H1K26, H3K9 and H4K16) and non-histone substrates (e.g., FOXO1/3, PGC-1α, p53, NF-κB, CREB, TORC2, LXR, MyoD, MEF2, SREBP-1 and BMAL1; see Table 1). Thus, SIRT1 has pleiotropic cellular functions including cell proliferation, survival, anti-inflammation, antistress and metabolism [28,29]. SIRT2 deacetylates α-tubulin, PEPCK, FOXO3 and others for the regulation of mitosis, chromatin remodeling, gene transcription, autophagy and metabolism [3,30–37]. SIRT3 is a predominant mitochondrial sirtuin that deacetylates numerous proteins or enzymes, such as ACCS2, ALDH2, LCAD and SOD2, which are involved in diverse metabolic functions and antioxidative stress [38–56]. SIRT4 can inhibit glutamate dehydrogenase (GDH) activity and amino acid-stimulated insulin secretion in pancreatic β cells through NAD+-dependent ADP-ribosylation [8,9,57]. SIRT5 can deacetylate and desuccinylate CPS1 to promote ammonia detoxification through the urea cycle [11,58,59]. SIRT6 is involved in glucose homeostasis, chromatin remodeling and DNA repair through deacetylation of histone 3, lysines 9 and 56, CtIP and PARP1, respectively [60–70]. With regard to SIRT7, only p53 has been reported to be a substrate in the regulation of cell survival and cardiac function [71–74]. Since the scope of this review focuses on diabetes, here I mainly summarize the metabolic functions of sirtuins in Table 2 and discuss them as follows.
Table 2.
Sirtuin metabolic functions.
| Gene | Functions | Ref. |
|---|---|---|
| SIRT1 | Pancreatic β cells | |
| ▪ Increases insulin gene transcription | [75–79] | |
| ▪ Decreases UCP2 gene transcription | [75,76,78,207] | |
| ▪ Increases glucose-stimulated insulin secretion | [75–79] | |
| ▪ Protects against cytokine toxicity via NF-κβ downregulation | [79] | |
| ▪ Protects against oxidative stress via upregulation of NeuroD and MafA | [175] | |
| Liver | ||
| ▪ Regulates gluconeogenesis | [32,80,82–88] | |
| ▪ Inhibits glycolysis | [84,89,90] | |
| ▪ Inhibits lipogenesis | [91,95,96] | |
| ▪ Increases fatty acid oxidation | [92] | |
| ▪ Inhibits cholesterol biosynthesis | [96–98] | |
| ▪ Improves hepatic insulin sensitivity | [82] | |
| ▪ Inhibits oxidative stress, inflammation and ER stress | [82,92,94,208] | |
| ▪ Regulates circadian rhythms | [22,23] | |
| ▪ Modulates NAD biosynthesis | [20,21] | |
| Brain | ||
| ▪ Mediobasal hypothalamic SIRT1 inhibits hepatic glucose production | [117] | |
| ▪ Central SIRT1 positively regulates food intake and energy expenditure | [118–124] | |
| ▪ Regulates physical activities | [123] | |
| Skeletal muscle | ||
| ▪ Increases insulin sensitivity | [99,103] | |
| ▪ Increases mitochondrial biogenesis | [100–102] | |
| ▪ Increases fatty acid oxidation | [100,104] | |
| Adipose tissue | ||
| ▪ Increases adiponectin biosynthesis and secretion | [109–111] | |
| ▪ Increases lipolysis | [112,113] | |
| ▪ Inhibits adipogenesis | [112] | |
| SIRT2 | Liver | |
| ▪ Increases hepatic gluconeogenesis via deacetylation of PEPCK | [32] | |
| Adipose tissue | ||
| ▪ Decreases oxidative stress via deacetylation of FOXO3 | [116] | |
| ▪ Inhibits adipocyte differentiation via deacetylation of FOXO1 | [35,115] | |
| SIRT3 | Liver | |
| ▪ Regulates energy homeostasis via control of ETC complexes I and II | [54] | |
| ▪ Reduces oxidative stress via deacetylation of SOD2 | [51] | |
| ▪ Increases fatty acid oxidation via deacetylation of LCAD and others | [39,43,50,53 | |
| ▪ Increases ketone body production via deacetylation of HMGCS2 | [50] | |
| ▪ Promotes the urea cycle via deacetylation of OTC | [43] | |
| Skeletal muscle | ||
| ▪ Induced by fasting, caloric restriction and exercise | [105–107] | |
| ▪ Decreased by high-fat diet | [40,41,107] | |
| ▪ Inhibits mitochondrial protein synthesis via deacetylation of MRPL10 | [47] | |
| ▪ Increases mitochondrial oxidation | [39,40] | |
| ▪ Decreases ROS and insulin resistance | [40,108] | |
| ▪ Increases mitochondrial biogenesis | [108] | |
| Brain | ||
| ▪ Protects against age-related hearing loss by reducing oxidative damage | [48] | |
| SIRT4 | Pancreatic β cells | |
| ▪ Decreases insulin secretion via inhibition of GDH | [8,9] | |
| SIRT5 | Liver | |
| ▪ Regulates the urea cycle via deacetylation of CPS1 | [58,59] | |
| SIRT6 | Liver | |
| ▪ Inhibits glycolysis and lipogenesis via deacetylation of H3K9 | [64] | |
| ▪ Increases fatty acid oxidation via deacetylation of H3K9 | [64] | |
| Brain | ||
| ▪ Regulates somatic growth and adiposity via deacetylation of H3K9 and H3K56 in the brain | [63] | |
| Skeletal muscle | ||
| ▪ Inhibits basal- and insulin-stimulated glucose uptake | [61,62] | |
| Adipose tissue | ||
| ▪ Decreases triglyceride synthesis via downregulation of DGAT1 | [65] | |
| ▪ Inhibits glucose uptake | [61] | |
| SIRT7 | Regulates Pol I transcription and p53 function, particularly protects against stress, apoptosis and inflammation in the heart | [72,74] |
ETC: Electron transport chain; GDH: Glutamate dehydrogenase; ROS: Reactive oxygen species.
Sirtuin functions in pancreatic β cells
SIRT1 and SIRT4 have been reported to play roles in pancreatic β cells. SIRT1 can increase insulin gene transcription and glucose-stimulated insulin secretion (GSIS). This has been confirmed in Sirt1-knockout and β-cell-specific overexpression transgenic mice (BESTO) [75–78]. GSIS was blunted in the knockout mice and elevated in the BESTO mice. One potential mechanism may be the repression of the UCP2 gene expression by SIRT1. Moreover, SIRT1 also protects β cells against inflammation-induced apoptosis [79]. By contrast, SIRT4 inhibits amino acid-induced insulin secretion through the ADP-ribosylation of GDH [8,9].
Sirtuin functions in the liver
Glucose metabolism
Several sirtuins have been found to play important roles in the liver. SIRT1 has been reported to regulate hepatic gluconeogenesis. However, reports are still conflicting with regard to either activation or inhibition of this process. Motta et al. reported that SIRT1 might inhibit gluconeogenesis through suppression of PEPCK gene expression, which is one of the key enzymes in the gluconeogenic process [80]. Qiang et al. suggest that SIRT1 suppresses hepatic gluconeogenesis via direct deacetylation and inhibition of CREB, a critical transcription factor for PEPCK [81]. Recently, Wang et al. have found that SIRT1 also suppresses the hepatic expression of PEPCK and glucose 6-phosphatase genes via upregulation of Rictor, a unique component of the mTORC2 complex, and subsequent inhibition of FOXO1 activity [82]. Several other reports suggest that SIRT1 might upregulate gluconeogenic genes through the activation of FOXO1 and PGC-1α via direct deacetylation [83–86]. A study has indicated a time-dependent switch from early CRTC2 to late FOXO1 activation; however, SIRT1 plays dual roles by not only inhibiting CRTC2 but also promoting FOXO1 activity [87]. SIRT1 can also activate gluconeogenesis via inhibition of STAT3 [88]. Moreover, there is also a feedback mechanism in which the SIRT1/FOXO1 pathway induces the expression of SHP and subsequently, SHP inhibits FOXO1 transcriptional activity [83]. Apparently the regulation of hepatic gluconeogenesis by SIRT1 is quite complex and dynamic under different conditions [81–88]. It requires more systemic study to pinpoint how SIRT1 is involved in gluconeogenesis in a time course. SIRT2 also regulates hepatic gluconeogenesis through direct deacetylation of PEPCK [32]. Additionally, SIRT1 and SIRT6 regulate glycolysis through distinct mechanisms. SIRT1 may repress the expression of glucokinase and pyruvate kinase genes through deacetylation of PGC-1α and FOXO1 [84,89,90], whereas SIRT6 suppresses these two genes via deacetylation of H3K9 [64].
Lipid metabolism
In addition to the regulation of glucose metabolism, SIRT1 also modulates hepatic lipid homeostasis. This function has been demonstrated by Sirt1 knockout and transgenic mouse models. The liver-specific Sirt1 knockout mice develop hepatic steatosis on a high-fat diet [91,92], whereas the overexpression of Sirt1 specifically in the liver or globally, protects mice from developing fatty liver disease [93,94]. Several potential mechanisms may contribute to the beneficial effects of SIRT1 in triglyceride metabolism. First, SIRT1 increases hepatic fatty acid oxidation, possibly through the activation of PGC-1α/PPARα [92]. Second, SIRT1 improves cellular functions by reducing oxidative and endoplasmic reticulum (ER) stress [94]. Third, SIRT1 can inhibit lipogenesis through deacetylation-induced SREBP-1c degradation and downregulation of ChREBP gene expression [91,95,96]. However, constitutive systemic overexpression of Sirt1 in mice also causes elevated hepatic and circulating levels of triglycerides and the inhibition of CREB by Sirt1-mediated deacetylation may play a critical in role in this dysregulation [81]. SIRT1 has also been reported to control cholesterol homeostasis. Knockdown or knockout of Sirt1 in mouse livers leads to an elevated level of total hepatic cholesterol. This may be attributed to the dysregulation of SREBP-2 and LXR protein stability and activities [96–98]. SIRT3 also plays an important role in lipid metabolism through deacetylation of some key enzymes including long chain acyl-CoA dehydrogenase for fatty acid oxidation and 3-hydroxy-3-methylglutaryl-CoA synthase 2 for ketogenesis [39,43,50,53]. SIRT6 can improve lipid homeostasis through inhibition of lipogenesis and activation of fatty acid oxidation by modulating the expression of numerous genes such as FASN, ACC1, ELOVL6, SCD1, CPT1 and ACOX1 [64]. By contrast, SIRT4 has been shown to inhibit fatty acid oxidation in the liver and skeletal muscle [57].
Sirtuin functions in skeletal muscle
Sirtuins also play critical roles in skeletal muscle physiology. SIRT1 has been demonstrated to increase insulin sensitivity and insulin-stimulated glucose uptake in skeletal muscle or myotubes by downregulating PTP1B [99]. Mitochondrial quality and quantity are critical for skeletal muscle function. Significantly, SIRT1 can enhance mitochondrial biogenesis, and thus increases muscle function, partly through the activation of PGC-1α [100–102]. SIRT1 also mediates calorie restriction (CR)-enhanced insulin sensitivity via the inhibition of STAT3-induced p55α/p50α of PI3K [103]. SIRT1 also boosts fatty acid oxidation in the skeletal muscle [100,104]. As a mitochondrial sirtuin, SIRT3 can be induced by fasting, caloric restriction and exercise [105–107]. SIRT3 activation can lead to increased mitochondrial biogenesis and function and can decrease oxidative stress [40,105–108]. By contrast, SIRT6 inhibits basal- and insulin-stimulated glucose uptake in the skeletal muscle by the inhibition of Akt phosphorylation and the suppression of recruitment of the glucose transporters, GLUT1 and GLUT4, to plasma membrane [62].
Sirtuin functions in adipose tissue
Adipose tissue has been increasingly appreciated to play a critical role in energy homeostasis. At least three sirtuins (SIRT1, 2 and 6) have been implicated in adipose functions. SIRT1 can increase the expression of adiponectin, an important adipokine for energy metabolism, in part through deacetylation of FOXO1 [109–111]. SIRT1 also inhibits adipogenesis and activates lipolysis through repression of PPARγ, a key adipose regulator [112]. Through deacetylation and activation of FOXO1, SIRT1 induces the transcription of adipose triglyceride lipase and promotes lipolysis in adipose tissue [113]. In 3T3-L1 adipocytes, SIRT1 knockdown decreases insulin signaling and insulin-stimulated glucose uptake [114]. SIRT2 can also inhibit adipocyte differentiation, partly through FOXO1 deacetylation [35,115]. In addition, SIRT2 activates FOXO3 in adipocytes to reduce oxidative stress [116]. SIRT6 has been reported to downregulate the expression of the DGAT1 gene and decrease triglyceride biosynthesis [65].
Sirtuin functions in the brain
The brain plays a key role in the integration of systemic energy homeostasis. Interestingly, mediobasal hypothalamic SIRT1 has been implicated in the suppression of hepatic glucose production by resveratrol [117]. Moreover, CNS SIRT1 positively regulates food intake and energy expenditure [118–124]. Inhibition of central Sirt1 blocks ghrelin-induced food intake in rodents partly through regulation of p53 and melanocortin 4 receptor pathways [118,119,124]. Hypothalamic SIRT1 also mediates dietary restriction-induced neural adaption by increasing physical activity and body temperature [123]. Through chromatin remodeling, particularly deacetylation of H3K9 and H3K56, neural SIRT6 regulates normal somatic growth and adiposity and protects against obesity [63].
An integrative view of sirtuins in metabolism
As described above, sirtuins play distinct roles in different tissues, which are consistent with the tissue-specific functions. In general, each sirtuin has unique functions in a given tissue. For example, SIRT1 promotes mitochondrial biogenesis in skeletal muscle and SIRT5 detoxifies ammonia in the liver [58,59,101,125]. Our current understanding is that sirtuins also functionally cooperate to achieve systemic homeostasis. For example, SIRT1 and SIRT3 both control fatty acid oxidation; however, SIRT1 does so at the gene transcription level mostly through PGC-1α, whereas SIRT3 directly controls the activity of metabolic enzymes involved in the fatty acid oxidation (FAO) process [126]. Another example is that although both SIRT1 and SIRT2 regulate hepatic gluconeogenesis, again, SIRT1 acts via the control of PEPCK and G6pc gene expression (up or down) while SIRT2 directly deacetylates PEPCK and increases its stability [32,80,82–88]. Regarding SIRT4, only a few studies have been carried out but they mostly reveal an inhibitory effect on metabolism – inhibition of amino acid-stimulated insulin secretion in pancreatic β cells as well as inhibition of fatty acid oxidation in the liver and skeletal muscle [8,9,57].
Targeting sirtuins for diabetes therapeutics
As summarized above, most, if not all, sirtuins have been implicated in metabolism and energy homeostasis. Therefore, they may become useful therapeutic targets for diabetes and the metabolic syndrome. Although there are conflicting reports in the literature, resveratrol has been directly or indirectly linked to SIRT1 for metabolic functions, possibly involving AMPK and other pathways [14,93,127–135]. Indeed, resveratrol and SIRT1 share numerous similar effects. For example, resveratrol has been shown to improve insulin sensitivity in obese animals and humans, increase mitochondrial biogenesis and function and protect against inflammation (Table 3) [101,136–140]. A recent report has shown that 30-day resveratrol supplementation increases activated AMPK and protein levels of SIRT1 and PGC-1α in skeletal muscle of obese subjects, decreases triglycerides in the liver and improves the homeostatic model assessment index and inflammatory markers [141]. Resveratrol can also improve dyslipidemia, ketoacidosis and inflammation in Type 1 diabetic rats [136,142]. Resveratrol is also found to increase GSIS in pancreatic β cells [77]. In most cases, the processes that resveratrol is involved in are also partially related to SIRT1 functions. However, it should be pointed out that it is still controversial as to whether resveratrol directly acts on SIRT1, since AMPK deficiency blunts resveratrol effects in mice [128]. Additionally, some reports suggest that SIRT1 overexpression may also cause increased lipogenesis [81,143,144]. More mechanistic studies are needed to clarify how and when SIRT1 contributes to beneficial or adverse effects on lipid metabolism.
Table 3.
Sirtuin activators and metabolic effects.
| Compound name | Metabolic effects | Ref. |
|---|---|---|
| Resveratrol | Improves insulin sensitivity | [99,117,140,141,209] |
| Improves hyperglycemia | [140,141] | |
| Increases mitochondrial biogenesis | [101,137] | |
| Increases glucose uptake | [210–214] | |
| Increases lipid transport and β-oxidation in skeletal muscle | [137,141] | |
| Regulates hepatic gluconeogenesis | [117,140] | |
| Improves ketoacidosis and muscle protein degradation in T1DM | [136] | |
| Increases insulin secretion in pancreatic β cells | [77,215] | |
| Improves hepatic steatosis and hepatocyte ballooning | [208,216,217] | |
| Improves dyslipidemia | [93,218–220] | |
| Inhibits adipogenesis | [112,114,221,222] | |
| Protects against inflammation | [139,141,223,224] | |
| Protects against oxidative stress | [138,209,223,225] | |
| SRT1720 | Improves glucose homeostasis | [104,155] |
| Increases insulin sensitivity | [104,155] | |
| Increases mitochondrial function | [104,155] | |
| Reduces lipogenic gene expression | [96,155,226] | |
| NMN | Protects against inflammation in pancreatic islets | [227] |
| Increases glucose-stimulated insulin secretion | [160,162,207] | |
| Improves glucose tolerance in obesity | [162] | |
| Improves glucose and lipid homeostasis in age-induced diabetes | [162,207] | |
T1DM: Type 1 diabetes mellitus.
CR has been demonstrated to be effective for extending lifespan and improving age-related abnormalities [145–147]. Although it is still controversial whether sirtuins may increase lifespan, the current literature tends to suggest that there is some connection between sirtuin functions and CR effects. First, it has been reported that SIRT1 gene expression is induced in fasting and CR-treated mice, rats and human subjects [84,148–150]. Second, transgenic and knockout mouse models indicate that SIRT1 may mediate some of the CR effects. For instance, Sirt1 knockout mice are irresponsive to CR in activity changes [151]. Mice carrying the Sirt1 transgene in the β-actin locus also manifest numerous characteristics of CR, including increased physical activity, decreased blood cholesterol, insulin and glucose as well as delayed reproduction [152]. Third, the metabolic functions of SIRT1, particularly in glucose, lipid and mitochondrial biogenesis, implicate its role in CR [84,96,97,112]. However, whether SIRT1 mediates CR-induced longevity is debatable because some yeast strains are still long-lived in the absence of Sir2 (yeast homolog of SIRT1) and overexpression or activation of SIRT1 in mice does not increase lifespan on a regular diet [133,152–155]. Additionally, SIRT1 is likely to function differently in a tissue-specific manner. Chen et al. have reported that Sirt1 activities increase in skeletal muscle and white adipose tissue but decrease in the liver on a CR regimen [156]. More intriguingly, different regions of the brain respond differently to CR in the expression of Sirt1 gene in mice – an increase in the cortex and hippocampus and a decrease in the midbrain and cerebellum [157]. In addition to SIRT1, other sirtuins may be also implicated in CR. For instance, SIRT3 has been shown to mediate some of the anti-oxidative stress effects by CR in the liver and in the brain, including cochlear neurons [48,51].
In pursuit of the development of small-molecule activators of sirtuins, several compounds including SRT1720 have been reported to specifically activate SIRT1 and manifest pharmacological efficacy on Type 2 diabetes in rodents [155]. However, whether those small molecules directly bind to SIRT1 is still debatable [158,159]. Another approach involving an increase in cellular NAD+ levels has been also tested in mouse models. NAD+ biosynthesis comprises of at least four pathways according to substrates: tryptophan (de novo synthesis), nicotinic acid, nicotinamide and nicotinamide riboside. It has been reported that the rate-limiting enzyme for the salvage biosynthesis of NAD+ – nicotinamide phosphoribosyltransferase (NAMPT) – plays a critical role in NAD+ homeostasis and insulin secretion in mice [15,160,161]. As one of the key intermediates of NAD+ biosynthesis, nicotinamide mononucleotide (NMN; a product of the NAMPT enzyme) has been shown to have protective effects against obesity- and aging-induced diabetes [162]. Daily injections of 500 mg/kg body weight of NMN for 5–7 days improves glucose intolerance and insulin resistance in high-fat-induced diabetic mice. Similar treatment in old diabetic mice also demonstrated an improvement in glucose and lipid homeostasis [162].
Since no drug targeting sirtuins has yet been approved for clinical use and only limited animal studies have been performed, the advantages and disadvantages of sirtuin activators compared to current diabetes drugs are not clear. Some reports suggest that SIRT1 might mediate part of the metformin effects in the suppression of hepatic gluconeogenesis, the activation of fatty acid oxidation in skeletal muscle and protection against hyperglycemia-induced retina damage [14,163,164]. It is possible that AMPK activation by metformin may also stimulate SIRT1 activity [14]. Interestingly, it is hypothalamic SIRT1 but not AMPK that mediates resveratrol-suppressed hepatic glucose production [117]. Moreover, SIRT1 might also activate AMPK according to several lines of evidence [93,127,130,164–167]. Since sirtuins can be modulated by increasing cellular NAD+ levels, nutriceutical approaches by providing NAD+ biosynthesis substrates or intermediate metabolites, such as NMN, may be also useful in the prevention or treatment of Type 2 diabetes [160,162,168].
Conclusion & future perspective
Sirtuins have pleiotropic functions in metabolism including glucose and lipid metabolism, energy expenditure and insulin secretion. Certainly, we still have many unanswered questions with respect to sirtuin biology and their therapeutic potentials. For example, what determines sirtuin activities? Why do Sirt2/3/4/5 deficient mice have mild phenotype? How do we design specific assays for each sirtuin? How can we target sirtuins for specific diseases such as diabetes without complications? Through future investigation, we should be able to address those questions and choose the best available approach for drug development.
Practice Points.
-
■
Exercise and diet should always be considered when managing Type 2 diabetes.
-
■
Sirtuins may be partly attributed to the beneficial effects of calorie restriction and exercise. Although the underlying mechanisms are not quite clear, SIRT1 and SIRT3 have been demonstrated to mediate some of the salutary effects of calorie restriction and exercise in animal models, including an increase in mitochondrial function and protection against oxidative stress.
-
■
Resveratrol may act partly through SIRT1 to improve glucose and lipid metabolism in diabetics.
-
■
Strategies to boost NAD biosynthesis may be beneficial for metabolic homeostasis.
Acknowledgments
XC Dong’s research was supported by the National Institute of Diabetes And Digestive And Kidney Diseases (R00DK077505 and R01DK091592).
Footnotes
Financial & competing interests disclosure
The author has no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell. 2005;16(10):4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J. Biol. Chem. 2007;282(9):6823–6832. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- 3.Vaquero A, Scher MB, Lee DH, et al. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006;20(10):1256–1261. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sauve AA, Wolberger C, Schramm VL, Boeke JD. The biochemistry of sirtuins. Annu. Rev. Biochem. 2006;75:435–465. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- 5.Tanny JC, Dowd GJ, Huang J, Hilz H, Moazed D. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell. 1999;99(7):735–745. doi: 10.1016/s0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- 6. Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403(6771):795–800. doi: 10.1038/35001622.. ▪▪ This seminal work demonstrated that yeast Sir2 and mouse Sir2 homologs are NAD+-dependent histone deacetylases.
- 7. Smith JS, Brachmann CB, Celic I, et al. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl Acad. Sci. USA. 2000;97(12):6658–6663. doi: 10.1073/pnas.97.12.6658.. ▪▪ Demonstrates that sirtuin 1 homologs from Archaea, eubacteria, yeast and human can deacetylate histones in an NAD+-dependent manner and are regulated by NPT1, an enzyme involved in NAD+ biosynthesis.
- 8. Haigis MC, Mostoslavsky R, Haigis KM, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126(5):941–954. doi: 10.1016/j.cell.2006.06.057.. ▪ Demonstrates that SIRT4 regulates insulin secretion through ADP-ribosylation of glutamate dehydrogenase.
- 9.Ahuja N, Schwer B, Carobbio S, et al. Regulation of insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase. J. Biol. Chem. 2007;282(46):33583–33592. doi: 10.1074/jbc.M705488200. [DOI] [PubMed] [Google Scholar]
- 10.Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J. Biol. Chem. 2005;280(22):21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- 11. Du J, Zhou Y, Su X, et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334(6057):806–809. doi: 10.1126/science.1207861.. ▪ Reports that SIRT5 has lysine demalonylation and desuccinylation enzymatic activities.
- 12.Peng C, Lu Z, Xie Z, et al. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol. Cell Proteomics. 2011;10(12):M111.012658. doi: 10.1074/mcp.M111.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haigis MC, Guarente LP. Mammalian sirtuins – emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20(21):2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 14. Canto C, Gerhart-Hines Z, Feige JN, et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458(7241):1056–1060. doi: 10.1038/nature07813.. ▪ Reports that AMPK can regulate SIRT1 activity, suggesting that SIRT1 activation by resveratrol might be mediated through AMPK.
- 15.Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J. Biol. Chem. 2004;279(49):50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- 16.Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature. 2003;423(6936):181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast Sir2 and human SIRT1. J. Biol. Chem. 2002;277(47):45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- 18.Lin SJ, Ford E, Haigis M, Liszt G, Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 2004;18(1):12–16. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin SJ, Kaeberlein M, Andalis AA, et al. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418(6895):344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- 20.Ramsey KM, Yoshino J, Brace CS, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324(5927):651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324(5927):654–657. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asher G, Gatfield D, Stratmann M, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134(2):317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 23.Nakahata Y, Kaluzova M, Grimaldi B, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134(2):329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choudhary C, Kumar C, Gnad F, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 25.Wang Q, Zhang Y, Yang C, et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327(5968):1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao S, Xu W, Jiang W, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327(5968):1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imai S, Guarente L. Ten years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases. Trends Pharmacol. Sci. 2010;31(5):212–220. doi: 10.1016/j.tips.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guarente L, Franklin H. Epstein lecture: sirtuins, aging, and medicine. N. Engl. J. Med. 2011;364(23):2235–2244. doi: 10.1056/NEJMra1100831. [DOI] [PubMed] [Google Scholar]
- 29.Yu J, Auwerx J. Protein deacetylation by SIRT1: an emerging key post-translational modification in metabolic regulation. Pharmacol. Res. 2010;62(1):35–41. doi: 10.1016/j.phrs.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang F, Chan CH, Chen K, Guan X, Lin HK, Tong Q. Deacetylation of FOXO3 by SIRT1 or SIRT2 leads to Skp2-mediated FOXO3 ubiquitination and degradation. Oncogene. 2011;31(12):1546–1557. doi: 10.1038/onc.2011.347. [DOI] [PubMed] [Google Scholar]
- 31.Kim HS, Vassilopoulos A, Wang RH, et al. SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell. 2011;20(4):487–499. doi: 10.1016/j.ccr.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang W, Wang S, Xiao M, et al. Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol. Cell. 2011;43(1):33–44. doi: 10.1016/j.molcel.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Y, Yang J, Liao W, et al. Cytosolic FOXO1 is essential for the induction of autophagy and tumour suppressor activity. Nat. Cell. Biol. 2010;12(7):665–675. doi: 10.1038/ncb2069. [DOI] [PubMed] [Google Scholar]
- 34.Rothgiesser KM, Erener S, Waibel S, Luscher B, Hottiger MO. SIRT2 regulates NF-κB dependent gene expression through deacetylation of p65 Lys310. J. Cell. Sci. 2010;123(Pt 24):4251–4258. doi: 10.1242/jcs.073783. [DOI] [PubMed] [Google Scholar]
- 35.Jing E, Gesta S, Kahn CR. SIRT2 regulates adipocyte differentiation through FOXO1 acetylation/deacetylation. Cell. Metab. 2007;6(2):105–114. doi: 10.1016/j.cmet.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human SIR2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell. 2003;11(2):437–444. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 37.Dryden SC, Nahhas FA, Nowak JE, Goustin AS, Tainsky MA. Role for human SIRT2 NAD-dependent deacetylase activity in control of mitotic exit in the cell cycle. Mol. Cell. Biol. 2003;23(9):3173–3185. doi: 10.1128/MCB.23.9.3173-3185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu Z, Bourdi M, Li JH, et al. SIRT3-dependent deacetylation exacerbates acetaminophen hepatotoxicity. EMBO Rep. 2011;12(8):840–846. doi: 10.1038/embor.2011.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kendrick AA, Choudhury M, Rahman SM, et al. Fatty liver is associated with reduced SIRT3 activity and mitochondrial protein hyperacetylation. Biochem. J. 2011;433(3):505–514. doi: 10.1042/BJ20100791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jing E, Emanuelli B, Hirschey MD, et al. Sirtuin-3 (Sirt3) regulates skeletal muscle metabolism and insulin signaling via altered mitochondrial oxidation and reactive oxygen species production. Proc. Natl Acad. Sci. USA. 2011;108(35):14608–14613. doi: 10.1073/pnas.1111308108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hirschey MD, Shimazu T, Jing E, et al. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol. Cell. 2011;44(2):177–190. doi: 10.1016/j.molcel.2011.07.019.. ▪ Suggests that SIRT3 may play a critical role in the development of metabolic syndrome.
- 42.Hirschey MD, Shimazu T, Capra JA, Pollard KS, Verdin E. SIRT1 and SIRT3 deacetylate homologous substrates: AceCS1,2 and HMGCS1,2. Aging (Albany NY) 2011;3(6):635–642. doi: 10.18632/aging.100339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hallows WC, Yu W, Smith BC, et al. SIRT3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol. Cell. 2011;41(2):139–149. doi: 10.1016/j.molcel.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Finley LW, Haas W, Desquiret-Dumas V, et al. Succinate dehydrogenase is a direct target of sirtuin 3 deacetylase activity. PLoS ONE. 2011;6(8):e23295. doi: 10.1371/journal.pone.0023295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Finley LW, Carracedo A, Lee J, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1α destabilization. Cancer Cell. 2011;19(3):416–428. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bell EL, Emerling BM, Ricoult SJ, Guarente L. SIRT3 suppresses hypoxia inducible factor 1α and tumor growth by inhibiting mitochondrial ROS production. Oncogene. 2011;30(26):2986–2996. doi: 10.1038/onc.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Y, Cimen H, Han MJ, et al. NAD+-dependent deacetylase SIRT3 regulates mitochondrial protein synthesis by deacetylation of the ribosomal protein MRPL10. J. Biol. Chem. 2010;285(10):7417–7429. doi: 10.1074/jbc.M109.053421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Someya S, Yu W, Hallows WC, et al. SIRT3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010;143(5):802–812. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shulga N, Wilson-Smith R, Pastorino JG. Sirtuin-3 deacetylation of cyclophilin D induces dissociation of hexokinase II from the mitochondria. J. Cell. Sci. 2010;123(Pt 6):894–902. doi: 10.1242/jcs.061846. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Shimazu T, Hirschey MD, Hua L, et al. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl coA synthase 2 and regulates ketone body production. Cell. Metab. 2010;12(6):654–661. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell. Metab. 2010;12(6):662–667. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 52.Kim HS, Patel K, Muldoon-Jacobs K, et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 2010;17(1):41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hirschey MD, Shimazu T, Goetzman E, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464(7285):121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ahn BH, Kim HS, Song S, et al. A role for the mitochondrial deacetylase SIRT3 in regulating energy homeostasis. Proc. Natl Acad. Sci. USA. 2008;105(38):14447–14452. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang H, Yang T, Baur JA, et al. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130(6):1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lombard DB, Alt FW, Cheng HL, et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol. Cell. Biol. 2007;27(24):8807–8814. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nasrin N, Wu X, Fortier E, et al. SIRT4 regulates fatty acid oxidation and mitochondrial gene expression in liver and muscle cells. J. Biol. Chem. 2010;285(42):31995–32002. doi: 10.1074/jbc.M110.124164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ogura M, Nakamura Y, Tanaka D, et al. Overexpression of SIRT5 confirms its involvement in deacetylation and activation of carbamoyl phosphate synthetase 1. Biochem. Biophys. Res. Commun. 2010;393(1):73–78. doi: 10.1016/j.bbrc.2010.01.081. [DOI] [PubMed] [Google Scholar]
- 59.Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137(3):560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mao Z, Hine C, Tian X, et al. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332(6036):1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhong L, D’Urso A, Toiber D, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1α. Cell. 2010;140(2):280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiao C, Kim HS, Lahusen T, et al. SIRT6 deficiency results in severe hypoglycemia by enhancing both basal and insulin-stimulated glucose uptake in mice. J. Biol. Chem. 2010;285(47):36776–36784. doi: 10.1074/jbc.M110.168039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schwer B, Schumacher B, Lombard DB, et al. Neural sirtuin 6 (Sirt6) ablation attenuates somatic growth and causes obesity. Proc. Natl Acad. Sci. USA. 2010;107(50):21790–21794. doi: 10.1073/pnas.1016306107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim HS, Xiao C, Wang RH, et al. Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell. Metab. 2010;12(3):224–236. doi: 10.1016/j.cmet.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kanfi Y, Peshti V, Gil R, et al. SIRT6 protects against pathological damage caused by diet-induced obesity. Aging Cell. 2010;9(2):162–173. doi: 10.1111/j.1474-9726.2009.00544.x. [DOI] [PubMed] [Google Scholar]
- 66.Kaidi A, Weinert BT, Choudhary C, Jackson SP. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science. 2010;329(5997):1348–1353. doi: 10.1126/science.1192049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Yang B, Zwaans BM, Eckersdorff M, Lombard DB. The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle. 2009;8(16):2662–2663. doi: 10.4161/cc.8.16.9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kawahara TL, Michishita E, Adler AS, et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-κB-dependent gene expression and organismal life span. Cell. 2009;136(1):62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Michishita E, McCord RA, Berber E, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452(7186):492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mostoslavsky R, Chua KF, Lombard DB, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124(2):315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 71.Grob A, Roussel P, Wright JE, McStay B, Hernandez-Verdun D, Sirri V. Involvement of SIRT7 in resumption of rDNA transcription at the exit from mitosis. J. Cell. Sci. 2009;122(Pt 4):489–498. doi: 10.1242/jcs.042382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vakhrusheva O, Smolka C, Gajawada P, et al. SIRT7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ. Res. 2008;102(6):703–710. doi: 10.1161/CIRCRESAHA.107.164558. [DOI] [PubMed] [Google Scholar]
- 73.Vakhrusheva O, Braeuer D, Liu Z, Braun T, Bober E. SIRT7-dependent inhibition of cell growth and proliferation might be instrumental to mediate tissue integrity during aging. J. Physiol. Pharmacol. 2008;59(Suppl. 9):201–212. [PubMed] [Google Scholar]
- 74.Ford E, Voit R, Liszt G, Magin C, Grummt I, Guarente L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 2006;20(9):1075–1080. doi: 10.1101/gad.1399706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bordone L, Motta MC, Picard F, et al. Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol. 2006;4(2):e31. doi: 10.1371/journal.pbio.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moynihan KA, Grimm AA, Plueger MM, et al. Increased dosage of mammalian Sir2 in pancreatic beta cells enhances glucose-stimulated insulin secretion in mice. Cell. Metab. 2005;2(2):105–117. doi: 10.1016/j.cmet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 77.Vetterli L, Brun T, Giovannoni L, Bosco D, Maechler P. Resveratrol potentiates glucose-stimulated insulin secretion in INS-1E beta-cells and human islets through a SIRT1-dependent mechanism. J. Biol. Chem. 2011;286(8):6049–6060. doi: 10.1074/jbc.M110.176842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu J, Zhang F, Yan M, et al. WldS enhances insulin transcription and secretion via a SIRT1-dependent pathway and improves glucose homeostasis. Diabetes. 2011;60(12):3197–3207. doi: 10.2337/db11-0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee JH, Song MY, Song EK, et al. Overexpression of SIRT1 protects pancreatic beta-cells against cytokine toxicity by suppressing the nuclear factor-κB signaling pathway. Diabetes. 2009;58(2):344–351. doi: 10.2337/db07-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Motta MC, Divecha N, Lemieux M, et al. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116(4):551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 81.Qiang L, Lin HV, Kim-Muller JY, Welch CL, Gu W, Accili D. Proatherogenic abnormalities of lipid metabolism in SirT1 transgenic mice are mediated through Creb deacetylation. Cell. Metab. 2011;14(6):758–767. doi: 10.1016/j.cmet.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang RH, Kim HS, Xiao C, Xu X, Gavrilova O, Deng CX. Hepatic Sirt1 deficiency in mice impairs mTorc2/Akt signaling and results in hyperglycemia, oxidative damage, and insulin resistance. J. Clin. Invest. 2011;121(11):4477–4490. doi: 10.1172/JCI46243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wei D, Tao R, Zhang Y, White MF, Dong XC. Feedback regulation of hepatic gluconeogenesis through modulation of SHP/Nr0b2 gene expression by Sirt1 and FoxO1. Am. J. Physiol. Endocrinol. Metab. 2011;300(2):E312–E320. doi: 10.1152/ajpendo.00524.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature. 2005;434(7029):113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 85.Erion DM, Yonemitsu S, Nie Y, et al. SirT1 knockdown in liver decreases basal hepatic glucose production and increases hepatic insulin responsiveness in diabetic rats. Proc. Natl Acad. Sci. USA. 2009;106(27):11288–11293. doi: 10.1073/pnas.0812931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Frescas D, Valenti L, Accili D. Nuclear trapping of the forkhead transcription factor FoxO1 via Sirt-dependent deacetylation promotes expression of glucogenetic genes. J. Biol. Chem. 2005;280(21):20589–20595. doi: 10.1074/jbc.M412357200. [DOI] [PubMed] [Google Scholar]
- 87.Liu Y, Dentin R, Chen D, et al. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008;456(7219):269–273. doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nie Y, Erion DM, Yuan Z, et al. STAT3 inhibition of gluconeogenesis is downregulated by SirT1. Nat. Cell. Biol. 2009;11(4):492–500. doi: 10.1038/ncb1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hirota K, Sakamaki J, Ishida J, et al. A combination of HNF-4 and Foxo1 is required for reciprocal transcriptional regulation of glucokinase and glucose-6-phosphatase genes in response to fasting and feeding. J. Biol. Chem. 2008;283(47):32432–32441. doi: 10.1074/jbc.M806179200. [DOI] [PubMed] [Google Scholar]
- 90.Zhang W, Patil S, Chauhan B, et al. FoxO1 regulates multiple metabolic pathways in the liver: effects on gluconeogenic, glycolytic, and lipogenic gene expression. J. Biol. Chem. 2006;281(15):10105–10117. doi: 10.1074/jbc.M600272200. [DOI] [PubMed] [Google Scholar]
- 91.Wang RH, Li C, Deng CX. Liver steatosis and increased ChREBP expression in mice carrying a liver specific SIRT1 null mutation under a normal feeding condition. Int. J. Biol. Sci. 2010;6(7):682–690. doi: 10.7150/ijbs.6.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009;9(4):327–338. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hou X, Xu S, Maitland-Toolan KA, et al. SIRT1 regulates hepatocyte lipid metabolism through activating AMP-activated protein kinase. J. Biol. Chem. 2008;283(29):20015–20026. doi: 10.1074/jbc.M802187200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschop MH. Sirt1 protects against high-fat diet-induced metabolic damage. Proc. Natl Acad. Sci. USA. 2008;105(28):9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ponugoti B, Kim DH, Xiao Z, et al. SIRT1 deacetylates and inhibits SREBP-1C activity in regulation of hepatic lipid metabolism. J. Biol. Chem. 2010;285(44):33959–33970. doi: 10.1074/jbc.M110.122978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Walker AK, Yang F, Jiang K, et al. Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev. 2010;24(13):1403–1417. doi: 10.1101/gad.1901210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li X, Zhang S, Blander G, Tse JG, Krieger M, Guarente L. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol. Cell. 2007;28(1):91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 98.Rodgers JT, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc. Natl Acad. Sci. USA. 2007;104(31):12861–12866. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sun C, Zhang F, Ge X, et al. SIRT1 improves insulin sensitivity under insulin-resistant conditions by repressing PTP1B. Cell. Metab. 2007;6(4):307–319. doi: 10.1016/j.cmet.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 100.Gerhart-Hines Z, Rodgers JT, Bare O, et al. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1α. EMBO J. 2007;26(7):1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lagouge M, Argmann C, Gerhart-Hines Z, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1α. Cell. 2006;127(6):1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 102.Li L, Pan R, Li R, et al. Mitochondrial biogenesis and peroxisome proliferator-activated receptor-γ coactivator-1α (PGC-1α) deacetylation by physical activity: intact adipocytokine signaling is required. Diabetes. 2011;60(1):157–167. doi: 10.2337/db10-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schenk S, McCurdy CE, Philp A, et al. Sirt1 enhances skeletal muscle insulin sensitivity in mice during caloric restriction. J. Clin. Invest. 2011;121(11):4281–4288. doi: 10.1172/JCI58554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Feige JN, Lagouge M, Canto C, et al. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell. Metab. 2008;8(5):347–358. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 105.Gurd BJ, Holloway GP, Yoshida Y, Bonen A. In mammalian muscle, SIRT3 is present in mitochondria and not in the nucleus; and SIRT3 is upregulated by chronic muscle contraction in an adenosine monophosphate-activated protein kinase-independent manner. Metabolism. 2011 doi: 10.1016/j.metabol.2011.09.016. (Epub ahead of print). [DOI] [PubMed] [Google Scholar]
- 106.Hokari F, Kawasaki E, Sakai A, Koshinaka K, Sakuma K, Kawanaka K. Muscle contractile activity regulates Sirt3 protein expression in rat skeletal muscles. J. Appl. Physiol. 2010;109(2):332–340. doi: 10.1152/japplphysiol.00335.2009. [DOI] [PubMed] [Google Scholar]
- 107.Palacios OM, Carmona JJ, Michan S, et al. Diet and exercise signals regulate SIRT3 and activate AMPK and PGC-1α in skeletal muscle. Aging (Albany NY) 2009;1(9):771–783. doi: 10.18632/aging.100075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kong X, Wang R, Xue Y, et al. Sirtuin 3, a new target of PGC-1α, plays an important role in the suppression of ROS and mitochondrial biogenesis. PLoS ONE. 2010;5(7):e11707. doi: 10.1371/journal.pone.0011707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Banks AS, Kon N, Knight C, et al. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell. Metab. 2008;8(4):333–341. doi: 10.1016/j.cmet.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Qiang L, Wang H, Farmer SR. Adiponectin secretion is regulated by SIRT1 and the endoplasmic reticulum oxidoreductase Ero1-l alpha. Mol. Cell. Biol. 2007;27(13):4698–4707. doi: 10.1128/MCB.02279-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Qiao L, Shao J. SIRT1 regulates adiponectin gene expression through Foxo1-C/enhancer-binding protein alpha transcriptional complex. J. Biol. Chem. 2006;281(52):39915–39924. doi: 10.1074/jbc.M607215200. [DOI] [PubMed] [Google Scholar]
- 112.Picard F, Kurtev M, Chung N, et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-γ. Nature. 2004;429(6993):771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chakrabarti P, English T, Karki S, et al. SIRT1 controls lipolysis in adipocytes via FOXO1-mediated expression of ATGL. J. Lipid Res. 2011;52(9):1693–1701. doi: 10.1194/jlr.M014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fischer-Posovszky P, Kukulus V, Tews D, et al. Resveratrol regulates human adipocyte number and function in a Sirt1-dependent manner. Am. J. Clin. Nutr. 2010;92(1):5–15. doi: 10.3945/ajcn.2009.28435. [DOI] [PubMed] [Google Scholar]
- 115.Wang F, Tong Q. SIRT2 suppresses adipocyte differentiation by deacetylating FOXO1 and enhancing FOXO1’s repressive interaction with PPARγ. Mol. Biol. Cell. 2009;20(3):801–808. doi: 10.1091/mbc.E08-06-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang F, Nguyen M, Qin FX, Tong Q. SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging Cell. 2007;6(4):505–514. doi: 10.1111/j.1474-9726.2007.00304.x. [DOI] [PubMed] [Google Scholar]
- 117.Knight CM, Gutierrez-Juarez R, Lam TK, et al. Mediobasal hypothalamic SIRT1 is essential for resveratrol’s effects on insulin action in rats. Diabetes. 2011;60(11):2691–2700. doi: 10.2337/db10-0987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cakir I, Perello M, Lansari O, Messier NJ, Vaslet CA, Nillni EA. Hypothalamic Sirt1 regulates food intake in a rodent model system. PLoS ONE. 2009;4(12):e8322. doi: 10.1371/journal.pone.0008322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dietrich MO, Antunes C, Geliang G, et al. Agrp neurons mediate Sirt1’s action on the melanocortin system and energy balance: roles for Sirt1 in neuronal firing and synaptic plasticity. J. Neurosci. 2010;30(35):11815–11825. doi: 10.1523/JNEUROSCI.2234-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ramadori G, Fujikawa T, Anderson J, et al. SIRT1 deacetylase in SF1 neurons protects against metabolic imbalance. Cell Metab. 2011;14(3):301–312. doi: 10.1016/j.cmet.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ramadori G, Fujikawa T, Fukuda M, et al. SIRT1 deacetylase in POMC neurons is required for homeostatic defenses against diet-induced obesity. Cell Metab. 2010;12(1):78–87. doi: 10.1016/j.cmet.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sasaki T, Kim HJ, Kobayashi M, et al. Induction of hypothalamic Sirt1 leads to cessation of feeding via agouti-related peptide. Endocrinology. 2010;151(6):2556–2566. doi: 10.1210/en.2009-1319. [DOI] [PubMed] [Google Scholar]
- 123.Satoh A, Brace CS, Ben-Josef G, et al. SIRT1 promotes the central adaptive response to diet restriction through activation of the dorsomedial and lateral nuclei of the hypothalamus. J. Neurosci. 2010;30(30):10220–10232. doi: 10.1523/JNEUROSCI.1385-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Velasquez DA, Martinez G, Romero A, et al. The central Sirtuin 1/p53 pathway is essential for the orexigenic action of ghrelin. Diabetes. 2011;60(4):1177–1185. doi: 10.2337/db10-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gurd BJ, Yoshida Y, McFarlan JT, et al. Nuclear SIRT1 activity, but not protein content, regulates mitochondrial biogenesis in rat and human skeletal muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011;301(1):R67–R75. doi: 10.1152/ajpregu.00417.2010. [DOI] [PubMed] [Google Scholar]
- 126.Lomb DJ, Laurent G, Haigis MC. Sirtuins regulate key aspects of lipid metabolism. Biochim. Biophys. Acta. 2010;1804(8):1652–1657. doi: 10.1016/j.bbapap.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 127.Zu Y, Liu L, Lee MY, et al. SIRT1 promotes proliferation and prevents senescence through targeting LKB1 in primary porcine aortic endothelial cells. Circ. Res. 2010;106(8):1384–1393. doi: 10.1161/CIRCRESAHA.109.215483. [DOI] [PubMed] [Google Scholar]
- 128. Um JH, Park SJ, Kang H, et al. AMP-activated protein kinase-deficient mice are resistant to the metabolic effects of resveratrol. Diabetes. 2010;59(3):554–563. doi: 10.2337/db09-0482.. ▪ Suggests that resveratrol may activate SIRT1 through AMPK-mediated pathway.
- 129.Ruderman NB, Xu XJ, Nelson L, et al. AMPK and SIRT1: a long-standing partnership? Am. J. Physiol. Endocrinol. Metab. 2010;298(4):E751–E760. doi: 10.1152/ajpendo.00745.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lan F, Cacicedo JM, Ruderman N, Ido Y. SIRT1 modulation of the acetylation status, cytosolic localization, and activity of LKB1. Possible role in AMP-activated protein kinase activation. J. Biol. Chem. 2008;283(41):27628–27635. doi: 10.1074/jbc.M805711200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fulco M, Cen Y, Zhao P, et al. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev. Cell. 2008;14(5):661–673. doi: 10.1016/j.devcel.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Canto C, Jiang LQ, Deshmukh AS, et al. Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell. Metab. 2010;11(3):213–219. doi: 10.1016/j.cmet.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Burnett C, Valentini S, Cabreiro F, et al. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature. 2011;477(7365):482–485. doi: 10.1038/nature10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Howitz KT, Bitterman KJ, Cohen HY, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425(6954):191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 135.Gerhart-Hines Z, Dominy JE, Jr, Blattler SM, et al. The cAMP/PKA pathway rapidly activates SIRT1 to promote fatty acid oxidation independently of changes in NAD+ Mol. Cell. 2011;44(6):851–863. doi: 10.1016/j.molcel.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chen KH, Cheng ML, Jing YH, Chiu DT, Shiao MS, Chen JK. Resveratrol ameliorates metabolic disorders and muscle wasting in streptozotocin-induced diabetic rats. Am. J. Physiol. Endocrinol. Metab. 2011;301(5):E853–E863. doi: 10.1152/ajpendo.00048.2011. [DOI] [PubMed] [Google Scholar]
- 137.Chen LL, Zhang HH, Zheng J, et al. Resveratrol attenuates high-fat diet-induced insulin resistance by influencing skeletal muscle lipid transport and subsarcolemmal mitochondrial beta-oxidation. Metabolism. 2011;60(11):1598–1609. doi: 10.1016/j.metabol.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 138.Kao CL, Chen LK, Chang YL, et al. Resveratrol protects human endothelium from H(2)O(2)-induced oxidative stress and senescence via SirT1 activation. J. Atheroscler. Thromb. 2010;17(9):970–979. doi: 10.5551/jat.4333. [DOI] [PubMed] [Google Scholar]
- 139.Olholm J, Paulsen SK, Cullberg KB, Richelsen B, Pedersen SB. Anti-inflammatory effect of resveratrol on adipokine expression and secretion in human adipose tissue explants. Int. J. Obes. (Lond.) 2010;34(10):1546–1553. doi: 10.1038/ijo.2010.98. [DOI] [PubMed] [Google Scholar]
- 140.Ramadori G, Gautron L, Fujikawa T, Vianna CR, Elmquist JK, Coppari R. Central administration of resveratrol improves diet-induced diabetes. Endocrinology. 2009;150(12):5326–5333. doi: 10.1210/en.2009-0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Timmers S, Konings E, Bilet L, et al. Calorie restriction-like effects of 30 days of resveratrol supplementation on energy metabolism and metabolic profile in obese humans. Cell. Metab. 2011;14(5):612–622. doi: 10.1016/j.cmet.2011.10.002.. ▪ Demonstrates that resveratrol has beneficial effects in obese humans such as improved insulin resistance, lower blood glucose, triglycerides and inflammation markers.
- 142.Su HC, Hung LM, Chen JK. Resveratrol, a red wine antioxidant, possesses an insulin-like effect in streptozotocin-induced diabetic rats. Am. J. Physiol. Endocrinol. Metab. 2006;290(6):E1339–E1346. doi: 10.1152/ajpendo.00487.2005. [DOI] [PubMed] [Google Scholar]
- 143.Caton PW, Nayuni NK, Khan NQ, Wood EG, Corder R. Fructose induces gluconeogenesis and lipogenesis through a SIRT1-dependent mechanism. J. Endocrinol. 2011;208(3):273–283. doi: 10.1530/JOE-10-0190. [DOI] [PubMed] [Google Scholar]
- 144.Wu D, Qiu Y, Gao X, Yuan XB, Zhai Q. Overexpression of SIRT1 in mouse forebrain impairs lipid/glucose metabolism and motor function. PLoS ONE. 2011;6(6):e21759. doi: 10.1371/journal.pone.0021759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bordone L, Guarente L. Calorie restriction, SIRT1 and metabolism: understanding longevity. Nat. Rev. Mol. Cell. Biol. 2005;6(4):298–305. doi: 10.1038/nrm1616. [DOI] [PubMed] [Google Scholar]
- 146.Fontana L, Partridge L, Longo VD. Extending healthy life span – from yeast to humans. Science. 2010;328(5976):321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Holloszy JO, Fontana L. Caloric restriction in humans. Exp. Gerontol. 2007;42(8):709–712. doi: 10.1016/j.exger.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cohen HY, Miller C, Bitterman KJ, et al. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305(5682):390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 149.Civitarese AE, Carling S, Heilbronn LK, et al. Calorie restriction increases muscle mitochondrial biogenesis in healthy humans. PLoS Med. 2007;4(3):e76. doi: 10.1371/journal.pmed.0040076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Nisoli E, Tonello C, Cardile A, et al. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. 2005;310(5746):314–317. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- 151.Chen D, Steele AD, Lindquist S, Guarente L. Increase in activity during calorie restriction requires Sirt1. Science. 2005;310(5754):1641. doi: 10.1126/science.1118357. [DOI] [PubMed] [Google Scholar]
- 152.Bordone L, Cohen D, Robinson A, et al. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell. 2007;6(6):759–767. doi: 10.1111/j.1474-9726.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- 153.Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2004;2(9):E296. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Delaney JR, Sutphin GL, Dulken B, et al. Sir2 deletion prevents lifespan extension in 32 long-lived mutants. Aging Cell. 2011;10(6):1089–1091. doi: 10.1111/j.1474-9726.2011.00742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Milne JC, Lambert PD, Schenk S, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of Type 2 diabetes. Nature. 2007;450(7170):712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chen D, Bruno J, Easlon E, et al. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev. 2008;22(13):1753–1757. doi: 10.1101/gad.1650608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chen D, Steele AD, Hutter G, et al. The role of calorie restriction and SIRT1 in prion-mediated neurodegeneration. Exp. Gerontol. 2008;43(12):1086–1093. doi: 10.1016/j.exger.2008.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Pacholec M, Bleasdale JE, Chrunyk B, et al. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J. Biol. Chem. 2010;285(11):8340–8351. doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Dai H, Kustigian L, Carney D, et al. SIRT1 activation by small molecules: kinetic and biophysical evidence for direct interaction of enzyme and activator. J. Biol. Chem. 2010;285(43):32695–32703. doi: 10.1074/jbc.M110.133892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Revollo JR, Korner A, Mills KF, et al. Nampt/PBEF/Visfatin regulates insulin secretion in beta cells as a systemic NAD biosynthetic enzyme. Cell. Metab. 2007;6(5):363–375. doi: 10.1016/j.cmet.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tao R, Wei D, Gao H, Liu Y, DePinho RA, Dong XC. Hepatic FoxOs regulate lipid metabolism via modulation of expression of the nicotinamide phosphoribosyltransferase gene. J. Biol. Chem. 2011;286(16):14681–14690. doi: 10.1074/jbc.M110.201061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Yoshino J, Mills KF, Yoon MJ, Imai S. Nicotinamide mononucleotide, a key NAD+ intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell. Metab. 2011;14(4):528–536. doi: 10.1016/j.cmet.2011.08.014.. ▪ Suggests that an increase in NAD+ biosynthetic substrates may improve metabolic fitness in diabetics.
- 163.Caton PW, Nayuni NK, Kieswich J, Khan NQ, Yaqoob MM, Corder R. Metformin suppresses hepatic gluconeogenesis through induction of SIRT1 and GCN5. J. Endocrinol. 2010;205(1):97–106. doi: 10.1677/JOE-09-0345. [DOI] [PubMed] [Google Scholar]
- 164.Zheng Z, Chen H, Li J, et al. Sirtuin 1-mediated cellular metabolic memory of high glucose via the LKB1/AMPK/ROS pathway and therapeutic effects of metformin. Diabetes. 2012;61(1):217–228. doi: 10.2337/db11-0416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhuo L, Fu B, Bai X, et al. NAD blocks high glucose induced mesangial hypertrophy via activation of the sirtuins-AMPK-mTOR pathway. Cell Physiol. Biochem. 2011;27(6):681–690. doi: 10.1159/000330077. [DOI] [PubMed] [Google Scholar]
- 166.Wang P, Xu TY, Guan YF, et al. Nicotinamide phosphoribosyltransferase protects against ischemic stroke through SIRT1-dependent adenosine monophosphate-activated kinase pathway. Ann. Neurol. 2011;69(2):360–374. doi: 10.1002/ana.22236. [DOI] [PubMed] [Google Scholar]
- 167.Shen Z, Liang X, Rogers CQ, Rideout D, You M. Involvement of adiponectin-SIRT1-AMPK signaling in the protective action of rosiglitazone against alcoholic fatty liver in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2010;298(3):G364–G374. doi: 10.1152/ajpgi.00456.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Caton PW, Kieswich J, Yaqoob MM, Holness MJ, Sugden MC. Nicotinamide mononucleotide protects against pro-inflammatory cytokine-mediated impairment of mouse islet function. Diabetologia. 2011;54(12):3083–3092. doi: 10.1007/s00125-011-2288-0. [DOI] [PubMed] [Google Scholar]
- 169.Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc. Natl Acad. Sci. USA. 2006;103(27):10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Fu M, Liu M, Sauve AA, et al. Hormonal control of androgen receptor function through SIRT1. Mol. Cell. Biol. 2006;26(21):8122–8135. doi: 10.1128/MCB.00289-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Firestein R, Blander G, Michan S, et al. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PLoS ONE. 2008;3(4):e2020. doi: 10.1371/journal.pone.0002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zhang Y, Zhang M, Dong H, et al. Deacetylation of cortactin by SIRT1 promotes cell migration. Oncogene. 2009;28(3):445–460. doi: 10.1038/onc.2008.388. [DOI] [PubMed] [Google Scholar]
- 173.Peng L, Yuan Z, Ling H, et al. SIRT1 deacetylates the DNA methyltransferase 1 (DNMT1) protein and alters its activities. Mol. Cell. Biol. 2011;31(23):4720–4734. doi: 10.1128/MCB.06147-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Mattagajasingh I, Kim CS, Naqvi A, et al. SIRT1 promotes endothelium-dependent vascular relaxation by activating endothelial nitric oxide synthase. Proc. Natl Acad. Sci. USA. 2007;104(37):14855–14860. doi: 10.1073/pnas.0704329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kitamura YI, Kitamura T, Kruse JP, et al. FoxO1 protects against pancreatic beta cell failure through NeuroD and MafA induction. Cell. Metab. 2005;2(3):153–163. doi: 10.1016/j.cmet.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 176.Daitoku H, Hatta M, Matsuzaki H, et al. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc. Natl Acad. Sci. USA. 2004;101(27):10042–10047. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Calnan DR, Brunet A. The FoxO code. Oncogene. 2008;27(16):2276–2288. doi: 10.1038/onc.2008.21. [DOI] [PubMed] [Google Scholar]
- 178.Brunet A, Sweeney LB, Sturgill JF, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303(5666):2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 179.Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol. Cell. 2004;16(1):93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 180.Yamagata K, Kitabayashi I. Sirt1 physically interacts with Tip60 and negatively regulates Tip60-mediated acetylation of H2AX. Biochem. Biophys. Res. Commun. 2009;390(4):1355–1360. doi: 10.1016/j.bbrc.2009.10.156. [DOI] [PubMed] [Google Scholar]
- 181.Chen IY, Lypowy J, Pain J, et al. Histone H2A.z is essential for cardiac myocyte hypertrophy but opposed by silent information regulator 2α. J. Biol. Chem. 2006;281(28):19369–19377. doi: 10.1074/jbc.M601443200. [DOI] [PubMed] [Google Scholar]
- 182.Yuan J, Pu M, Zhang Z, Lou Z. Histone H3-K56 acetylation is important for genomic stability in mammals. Cell Cycle. 2009;8(11):1747–1753. doi: 10.4161/cc.8.11.8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Pagans S, Pedal A, North BJ, et al. SIRT1 regulates HIV transcription via Tat deacetylation. PLoS Biol. 2005;3(2):e41. doi: 10.1371/journal.pbio.0030041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Yang J, Kong X, Martins-Santos ME, et al. Activation of SIRT1 by resveratrol represses transcription of the gene for the cytosolic form of phosphoenolpyruvate carboxykinase (GTP) by deacetylating hepatic nuclear factor 4α. J. Biol. Chem. 2009;284(40):27042–27053. doi: 10.1074/jbc.M109.047340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Yuan J, Minter-Dykhouse K, Lou Z. A c-Myc-SIRT1 feedback loop regulates cell growth and transformation. J. Cell. Biol. 2009;185(2):203–211. doi: 10.1083/jcb.200809167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Fulco M, Schiltz RL, Iezzi S, et al. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol. Cell. 2003;12(1):51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- 187.Luo J, Nikolaev AY, Imai S, et al. Negative control of p53 by Sir2α promotes cell survival under stress. Cell. 2001;107(2):137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 188.Vaziri H, Dessain SK, Ng Eaton E, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107(2):149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 189.Bouras T, Fu M, Sauve AA, et al. SIRT1 deacetylation and repression of p300 involves lysine residues 1020/1024 within the cell cycle regulatory domain 1. J. Biol. Chem. 2005;280(11):10264–10276. doi: 10.1074/jbc.M408748200. [DOI] [PubMed] [Google Scholar]
- 190.Rajamohan SB, Pillai VB, Gupta M, et al. SIRT1 promotes cell survival under stress by deacetylation-dependent deactivation of poly(ADP-ribose) polymerase 1. Mol. Cell. Biol. 2009;29(15):4116–4129. doi: 10.1128/MCB.00121-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kelly TJ, Lerin C, Haas W, Gygi SP, Puigserver P. GCN5-mediated transcriptional control of the metabolic coactivator PGC-1beta through lysine acetylation. J. Biol. Chem. 2009;284(30):19945–19952. doi: 10.1074/jbc.M109.015164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Yeung F, Hoberg JE, Ramsey CS, et al. Modulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23(12):2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Wong S, Weber JD. Deacetylation of the retinoblastoma tumour suppressor protein by SIRT1. Biochem. J. 2007;407(3):451–460. doi: 10.1042/BJ20070151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Kume S, Haneda M, Kanasaki K, et al. SIRT1 inhibits transforming growth factor beta-induced apoptosis in glomerular mesangial cells via Smad7 deacetylation. J. Biol. Chem. 2007;282(1):151–158. doi: 10.1074/jbc.M605904200. [DOI] [PubMed] [Google Scholar]
- 195.Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450(7168):440–444. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- 196.Muth V, Nadaud S, Grummt I, Voit R. Acetylation of TAF(I)68, a subunit of TIF-IB/SL1, activates RNA polymerase I transcription. EMBO J. 2001;20(6):1353–1362. doi: 10.1093/emboj/20.6.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Fujita Y, Yamaguchi A, Hata K, Endo M, Yamaguchi N, Yamashita T. Zyxin is a novel interacting partner for SIRT1. BMC Cell. Biol. 2009;10:6. doi: 10.1186/1471-2121-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459(7243):113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Jin YH, Kim YJ, Kim DW, et al. Sirt2 interacts with 14-13-3 beta/γ and down-regulates the activity of p53. Biochem. Biophys. Res. Commun. 2008;368(3):690–695. doi: 10.1016/j.bbrc.2008.01.114. [DOI] [PubMed] [Google Scholar]
- 200.Black JC, Mosley A, Kitada T, Washburn M, Carey M. The SIRT2 deacetylase regulates autoacetylation of p300. Mol. Cell. 2008;32(3):449–455. doi: 10.1016/j.molcel.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Beirowski B, Gustin J, Armour SM, et al. Sir-two-homolog 2 (Sirt2) modulates peripheral myelination through polarity protein Par-3/atypical protein kinase C (aPKC) signaling. Proc. Natl Acad. Sci. USA. 2011;108(43):E952–E961. doi: 10.1073/pnas.1104969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Ma L, Gao JS, Guan Y, et al. Acetylation modulates prolactin receptor dimerization. Proc. Natl Acad. Sci. USA. 2010;107(45):19314–19319. doi: 10.1073/pnas.1010253107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc. Natl Acad. Sci. USA. 2006;103(27):10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Hafner AV, Dai J, Gomes AP, et al. Regulation of the mPTP by SIRT3-mediated deacetylation of CypD at lysine 166 suppresses age-related cardiac hypertrophy. Aging (Albany NY) 2010;2(12):914–923. doi: 10.18632/aging.100252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Cimen H, Han MJ, Yang Y, Tong Q, Koc H, Koc EC. Regulation of succinate dehydrogenase activity by SIRT3 in mammalian mitochondria. Biochemistry. 2010;49(2):304–311. doi: 10.1021/bi901627u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Michishita E, McCord RA, Boxer LD, et al. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle. 2009;8(16):2664–2666. doi: 10.4161/cc.8.16.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Ramsey KM, Mills KF, Satoh A, Imai S. Age-associated loss of Sirt1-mediated enhancement of glucose-stimulated insulin secretion in beta cell-specific Sirt1-overexpressing (BESTO) mice. Aging Cell. 2008;7(1):78–88. doi: 10.1111/j.1474-9726.2007.00355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Li Y, Xu S, Giles A, et al. Hepatic overexpression of SIRT1 in mice attenuates endoplasmic reticulum stress and insulin resistance in the liver. FASEB J. 2011;25(5):1664–1679. doi: 10.1096/fj.10-173492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Brasnyo P, Molnar GA, Mohas M, et al. Resveratrol improves insulin sensitivity, reduces oxidative stress and activates the Akt pathway in Type 2 diabetic patients. Br. J. Nutr. 2011;106(3):383–389. doi: 10.1017/S0007114511000316. [DOI] [PubMed] [Google Scholar]
- 210.Penumathsa SV, Thirunavukkarasu M, Zhan L, et al. Resveratrol enhances GLUT-4 translocation to the caveolar lipid raft fractions through AMPK/Akt/eNOS signalling pathway in diabetic myocardium. J. Cell Mol. Med. 2008;12(6A):2350–2361. doi: 10.1111/j.1582-4934.2008.00251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Park CE, Kim MJ, Lee JH, et al. Resveratrol stimulates glucose transport in C2C12 myotubes by activating AMP-activated protein kinase. Exp. Mol. Med. 2007;39(2):222–229. doi: 10.1038/emm.2007.25. [DOI] [PubMed] [Google Scholar]
- 212.Deng JY, Hsieh PS, Huang JP, Lu LS, Hung LM. Activation of estrogen receptor is crucial for resveratrol-stimulating muscular glucose uptake via both insulin-dependent and -independent pathways. Diabetes. 2008;57(7):1814–1823. doi: 10.2337/db07-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Breen DM, Sanli T, Giacca A, Tsiani E. Stimulation of muscle cell glucose uptake by resveratrol through sirtuins and AMPK. Biochem. Biophys. Res. Commun. 2008;374(1):117–122. doi: 10.1016/j.bbrc.2008.06.104. [DOI] [PubMed] [Google Scholar]
- 214.Minakawa M, Kawano A, Miura Y, Yagasaki K. Hypoglycemic effect of resveratrol in Type 2 diabetic model db/db mice and its actions in cultured L6 myotubes and RIN-5F pancreatic beta-cells. J. Clin. Biochem. Nutr. 2011;48(3):237–244. doi: 10.3164/jcbn.10-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Chen WP, Chi TC, Chuang LM, Su MJ. Resveratrol enhances insulin secretion by blocking K(ATP) and K(V) channels of beta cells. Eur. J. Pharmacol. 2007;568(1–3):269–277. doi: 10.1016/j.ejphar.2007.04.062. [DOI] [PubMed] [Google Scholar]
- 216.Bujanda L, Hijona E, Larzabal M, et al. Resveratrol inhibits nonalcoholic fatty liver disease in rats. BMC Gastroenterol. 2008;8:40. doi: 10.1186/1471-230X-8-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Tauriainen E, Luostarinen M, Martonen E, et al. Distinct effects of calorie restriction and resveratrol on diet-induced obesity and fatty liver formation. J. Nutr. Metab. 2011;2011:525094. doi: 10.1155/2011/525094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Zang M, Xu S, Maitland-Toolan KA, et al. Polyphenols stimulate AMP-activated protein kinase, lower lipids, and inhibit accelerated atherosclerosis in diabetic LDL receptor-deficient mice. Diabetes. 2006;55(8):2180–2191. doi: 10.2337/db05-1188. [DOI] [PubMed] [Google Scholar]
- 219.Rivera L, Moron R, Zarzuelo A, Galisteo M. Long-term resveratrol administration reduces metabolic disturbances and lowers blood pressure in obese Zucker rats. Biochem. Pharmacol. 2009;77(6):1053–1063. doi: 10.1016/j.bcp.2008.11.027. [DOI] [PubMed] [Google Scholar]
- 220.Ahn J, Cho I, Kim S, Kwon D, Ha T. Dietary resveratrol alters lipid metabolism-related gene expression of mice on an atherogenic diet. J. Hepatol. 2008;49(6):1019–1028. doi: 10.1016/j.jhep.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 221.Chen S, Li Z, Li W, Shan Z, Zhu W. Resveratrol inhibits cell differentiation in 3T3-L1 adipocytes via activation of AMPK. Can. J. Physiol. Pharmacol. 2011;89(11):793–799. doi: 10.1139/y11-077. [DOI] [PubMed] [Google Scholar]
- 222.Rayalam S, Yang JY, Ambati S, Della-Fera MA, Baile CA. Resveratrol induces apoptosis and inhibits adipogenesis in 3T3-L1 adipocytes. Phytother. Res. 2008;22(10):1367–1371. doi: 10.1002/ptr.2503. [DOI] [PubMed] [Google Scholar]
- 223.Ghanim H, Sia CL, Korzeniewski K, et al. A resveratrol and polyphenol preparation suppresses oxidative and inflammatory stress response to a high-fat, high-carbohydrate meal. J. Clin. Endocrinol. Metab. 2011;96(5):1409–1414. doi: 10.1210/jc.2010-1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Lee SM, Yang H, Tartar DM, et al. Prevention and treatment of diabetes with resveratrol in a non-obese mouse model of Type 1 diabetes. Diabetologia. 2011;54(5):1136–1146. doi: 10.1007/s00125-011-2064-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Kitada M, Kume S, Imaizumi N, Koya D. Resveratrol improves oxidative stress and protects against diabetic nephropathy through normalization of Mn-SOD dysfunction in AMPK/SIRT1-independent pathway. Diabetes. 2011;60(2):634–643. doi: 10.2337/db10-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Yamazaki Y, Usui I, Kanatani Y, et al. Treatment with SRT1720, a SIRT1 activator, ameliorates fatty liver with reduced expression of lipogenic enzymes in MSG mice. Am. J. Physiol. Endocrinol. Metab. 2009;297(5):E1179–E1186. doi: 10.1152/ajpendo.90997.2008. [DOI] [PubMed] [Google Scholar]
- 227.Caton PW, Kieswich J, Yaqoob MM, Holness MJ, Sugden MC. Nicotinamide mononucleotide protects against pro-inflammatory cytokine-mediated impairment of mouse islet function. Diabetologia. 2011;54(12):3083–3092. doi: 10.1007/s00125-011-2288-0. [DOI] [PubMed] [Google Scholar]