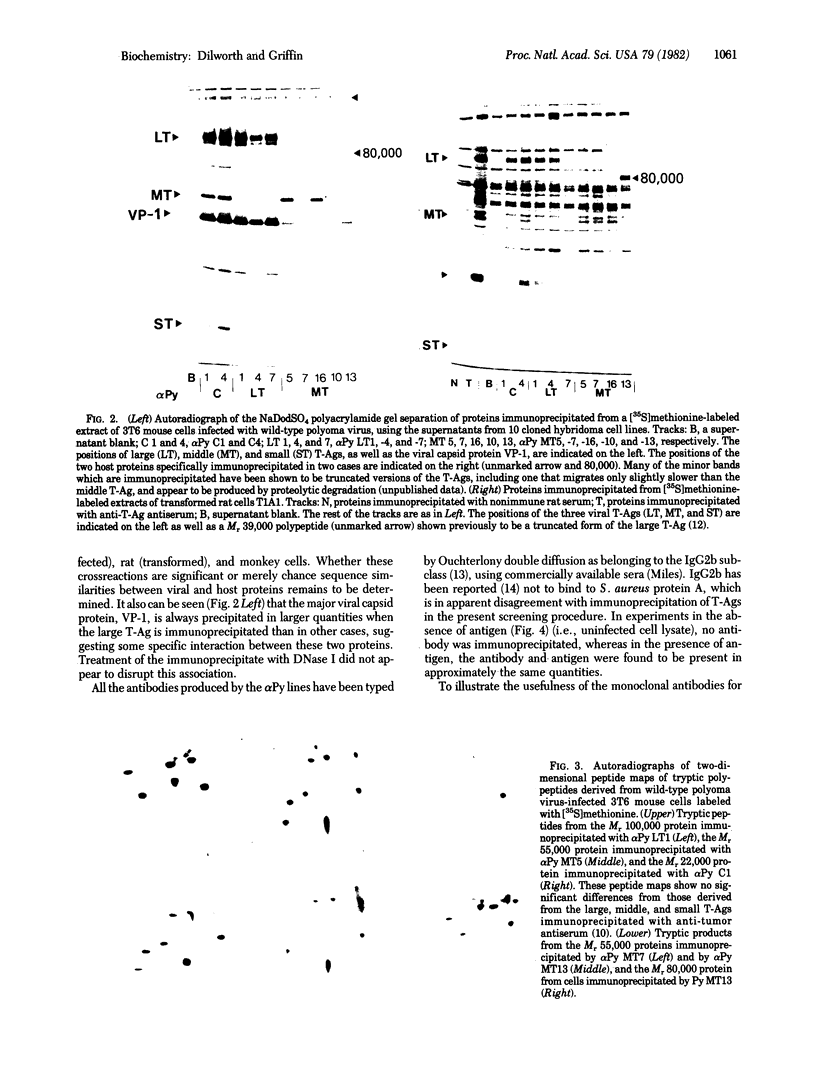
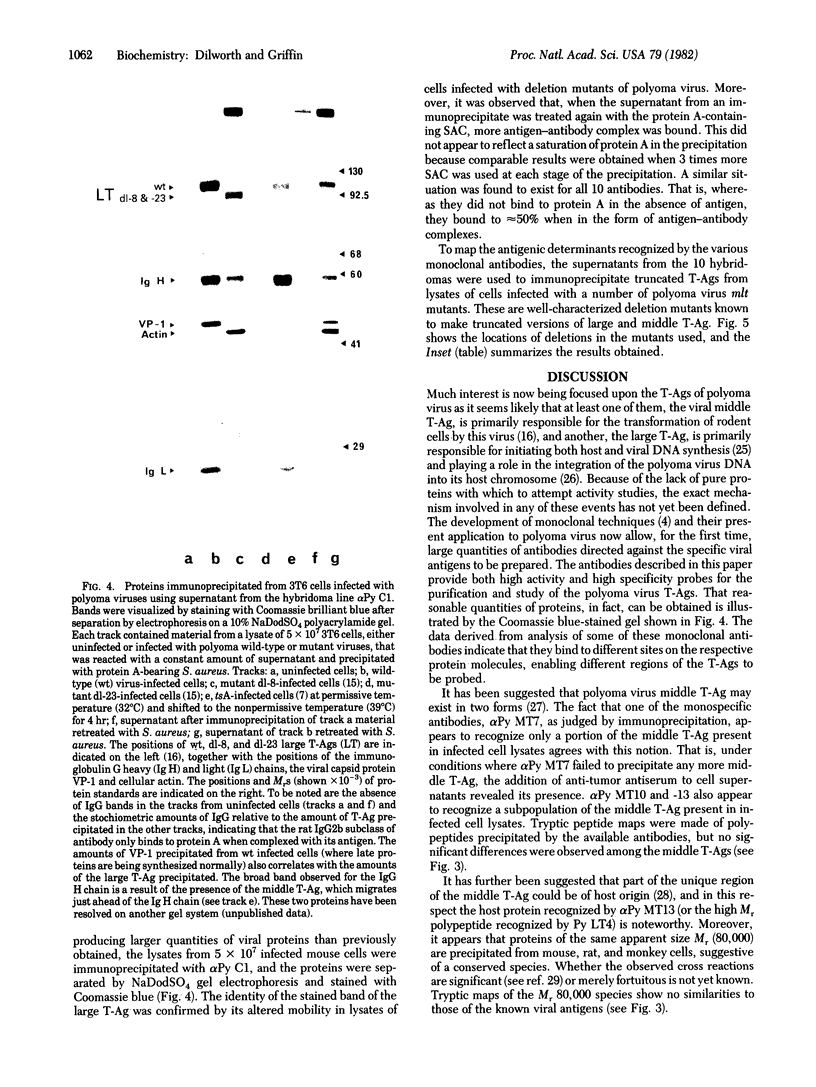
Abstract
A method is described for the production of hybridoma cell lines secreting antibodies directed against the polyoma virus tumor antigens (T-Ags). Ten cloned lines have been established by this procedure and designated alpha Py Cl and C4, with activities against all three T-Ags; alpha Py LT1, -4 and -7, with activities against large T-Ag only; and alpha Py MT5, -7, -10, -13, and -16, with activities against middle T-Ag only. Some of the antibodies appear to react also with specific cellular protein. Data from studies on alpha Py MT7 point to the possible existence of more than one antigenic form of the viral middle T-Ag and, similarly, from studies on alpha Py LT7, to more than one form of large T-Ag. The approximate positions on the polyoma virus genome of the sequences encoding antigenic determinants have been mapped by using defined polyoma virus deletion mutants. The availability of these monoclonal antibodies allows the isolation of large amounts of viral T-Ags.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bazin H., Beckers A., Querinjean P. Three classes and four (sub)classes of rat immunoglobulins: IgM, IgA, IgE and IgG1, IgG2a, IgG2b, IgG2c. Eur J Immunol. 1974 Jan;4(1):44–48. doi: 10.1002/eji.1830040112. [DOI] [PubMed] [Google Scholar]
- Della Valle G., Fenton R. G., Basilico C. Polyoma large T antigen regulates the integration of viral DNA sequences into the genome of transformed cells. Cell. 1981 Feb;23(2):347–355. doi: 10.1016/0092-8674(81)90130-6. [DOI] [PubMed] [Google Scholar]
- FRIED M. ISOLATION OF TEMPERATURE-SENSITIVE MUTANTS OF POLYOMA VIRUS. Virology. 1965 Apr;25:669–671. doi: 10.1016/0042-6822(65)90098-x. [DOI] [PubMed] [Google Scholar]
- Francke B., Eckhart W. Polyoma gene function required for viral DNA synthesis. Virology. 1973 Sep;55(1):127–135. doi: 10.1016/s0042-6822(73)81014-1. [DOI] [PubMed] [Google Scholar]
- Friedmann T., Esty A., LaPorte P., Deininger P. The nucleotide sequence and genome organization of the polyoma early region: extensive nucleotide and amino acid homology with SV40. Cell. 1979 Jul;17(3):715–724. doi: 10.1016/0092-8674(79)90278-2. [DOI] [PubMed] [Google Scholar]
- Gaudray P., Clertant P., Cuzin F. ATP phosphohydrolase (ATPase) activity of a polyoma virus T antigen. Eur J Biochem. 1980 Aug;109(2):553–560. doi: 10.1111/j.1432-1033.1980.tb04827.x. [DOI] [PubMed] [Google Scholar]
- Griffin B. E., Fried M., Cowie A. Polyoma DNA: a physical map. Proc Natl Acad Sci U S A. 1974 May;71(5):2077–2081. doi: 10.1073/pnas.71.5.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin B. E., Maddock C. New classes of viable deletion mutants in the early region of polyoma virus. J Virol. 1979 Sep;31(3):645–656. doi: 10.1128/jvi.31.3.645-656.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Brocklehurst J. R., Dulbecco R. Virus-specific proteins in the plasma membrane of cells lytically infected or transformed by pol-oma virus. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4666–4670. doi: 10.1073/pnas.74.10.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y. Polyoma virus-specific 55K protein isolated from plasma membrane of productively infected cells is virus-coded and important for cell transformation. Virology. 1979 Oct 15;98(1):261–266. doi: 10.1016/0042-6822(79)90545-2. [DOI] [PubMed] [Google Scholar]
- Ito Y., Spurr N., Griffin B. E. Middle T antigen as primary inducer of full expression of the phenotype of transformation by polyoma virus. J Virol. 1980 Jul;35(1):219–232. doi: 10.1128/jvi.35.1.219-232.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Spurr N. Polyoma virus T antigens expressed in transformed cells: significance of middle T antigen in transformation. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):149–157. doi: 10.1101/sqb.1980.044.01.017. [DOI] [PubMed] [Google Scholar]
- Kamen R., Favaloro J., Parker J., Treisman R., Lania L., Fried M., Mellor A. Comparison of polyoma virus transcription in productively infected mouse cells and transformed rodent cell lines. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):63–75. doi: 10.1101/sqb.1980.044.01.009. [DOI] [PubMed] [Google Scholar]
- Kennett R. H. Cell fusion. Methods Enzymol. 1979;58:345–359. doi: 10.1016/s0076-6879(79)58149-x. [DOI] [PubMed] [Google Scholar]
- Kessler S. W. Rapid isolation of antigens from cells with a staphylococcal protein A-antibody adsorbent: parameters of the interaction of antibody-antigen complexes with protein A. J Immunol. 1975 Dec;115(6):1617–1624. [PubMed] [Google Scholar]
- Köhler G., Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975 Aug 7;256(5517):495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lane D. P., Hoeffler W. K. SV40 large T shares an antigenic determinant with a cellular protein of molecular weight 68,000. Nature. 1980 Nov 13;288(5787):167–170. doi: 10.1038/288167a0. [DOI] [PubMed] [Google Scholar]
- Magnusson G., Berg P. Construction and analysis of viable deletion mutants of polyoma virus. J Virol. 1979 Nov;32(2):523–529. doi: 10.1128/jvi.32.2.523-529.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson G., Nilsson M. G., Dilworth S. M., Smolar N. Characterization of polyoma mutants with altered middle and large T-antigens. J Virol. 1981 Sep;39(3):673–683. doi: 10.1128/jvi.39.3.673-683.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medgyesi G. A., Füst G., Gergely J., Bazin H. Classes and subclasses of rat immunoglobulins: interaction with the complement system and with staphylococcal protein A. Immunochemistry. 1978 Feb;15(2):125–129. doi: 10.1016/0161-5890(78)90052-4. [DOI] [PubMed] [Google Scholar]
- Schaffhausen B., Benjamin T. L. Comparison of phosphorylation of two polyoma virus middle T antigens in vivo and in vitro. J Virol. 1981 Oct;40(1):184–196. doi: 10.1128/jvi.40.1.184-196.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolar N., Griffin B. E. DNA sequences of polyoma virus early deletion mutants. J Virol. 1981 Jun;38(3):958–967. doi: 10.1128/jvi.38.3.958-967.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeda E., Arrand J. R., Smolar N., Griffin B. E. Sequence from early region of polyoma virus DNA containing viral replication origin and encoding small, middle and (part of) large T antigens. Cell. 1979 Jun;17(2):357–370. doi: 10.1016/0092-8674(79)90162-4. [DOI] [PubMed] [Google Scholar]
- Soeda E., Arrand J. R., Smolar N., Walsh J. E., Griffin B. E. Coding potential and regulatory signals of the polyoma virus genome. Nature. 1980 Jan 31;283(5746):445–453. doi: 10.1038/283445a0. [DOI] [PubMed] [Google Scholar]
- Soeda E., Maruyama T., Arrand J. R., Griffin B. E. Host-dependent evolution of three papova viruses. Nature. 1980 May 15;285(5761):165–167. doi: 10.1038/285165a0. [DOI] [PubMed] [Google Scholar]
- Türler H. The tumor antigens and the early functions of polyoma virus. Mol Cell Biochem. 1980 Sep 15;32(2):63–93. doi: 10.1007/BF00227801. [DOI] [PubMed] [Google Scholar]
- Walter G., Hutchinson M. A., Hunter T., Eckhart W. Antibodies specific for the polyoma virus middle-size tumor antigen. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4882–4886. doi: 10.1073/pnas.78.8.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]