Abstract
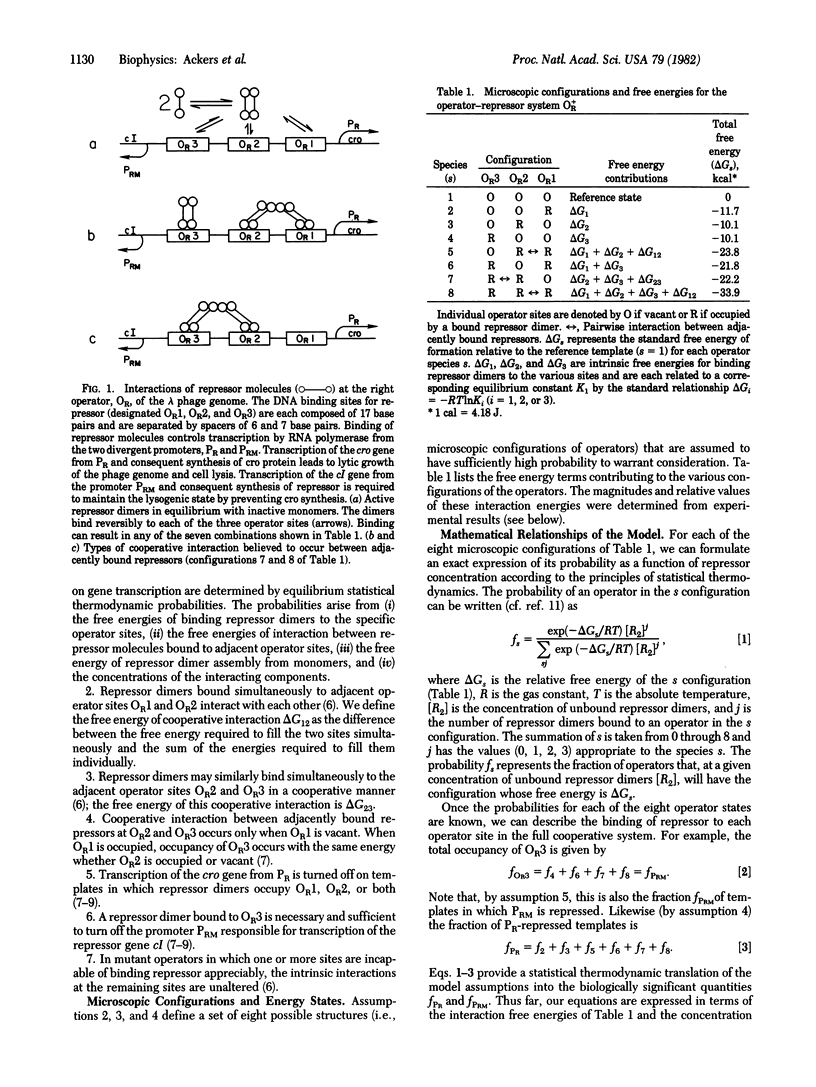
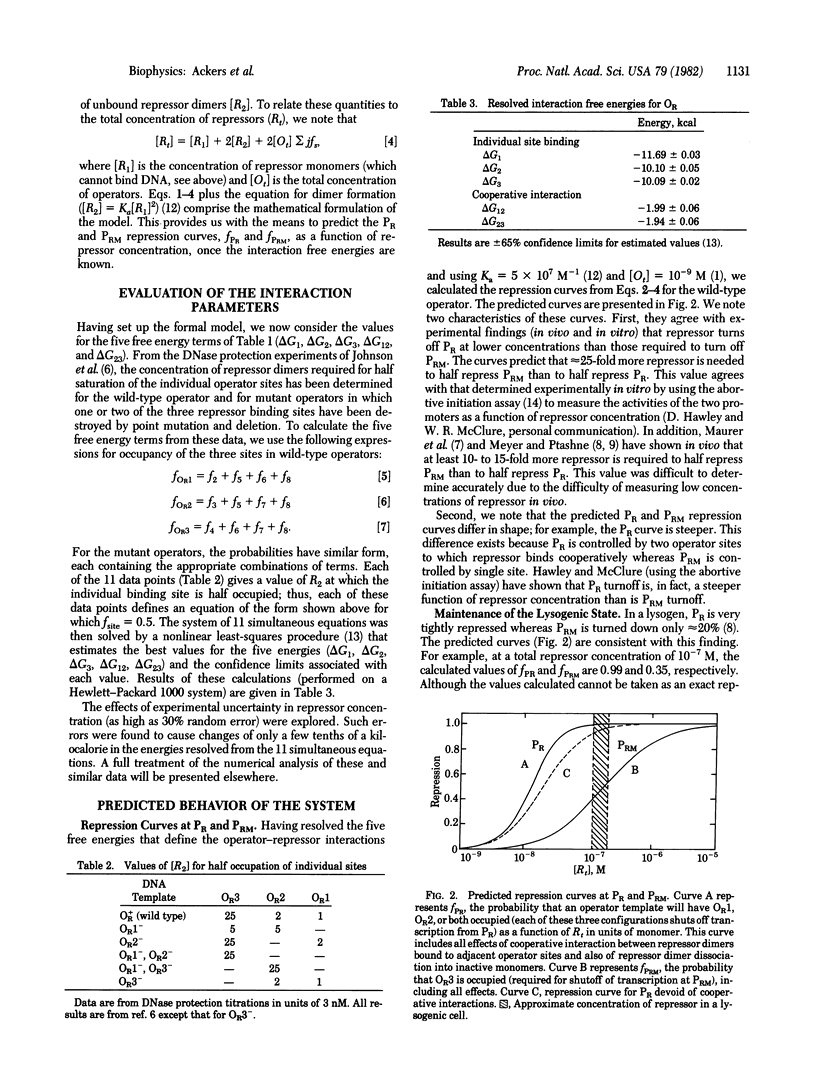
A statistical thermodynamic model has been developed to account for the cooperative interactions of the bacteriophage lambda repressor with the lambda right operator. The model incorporates a general theory for quantitatively interpreting cooperative site-specific equilibrium binding data. Values for all interaction parameters of the model have been evaluated at 37 degrees C, 0.2 M KCl, from results of DNase protection experiments in vitro [A. D. Johnson, B. J. Meyer, & M. Ptashne, Proc. Natl. Acad. Sci. USA (1979) 76, 5061-5065]. With these values, the model predicts repression curves at the divergent promoters PR and PRM that control transcription of genes coding for the regulatory proteins cro and repressor, respectively. At physiological repressor concentrations, repression at PR is predicted to be nearly complete whereas PRM is predicted to remain highly active. The results demonstrate the importance of cooperative interactions between repressor dimers bound to the adjacent operator sites OR1 and OR2 in maintaining a stable lysogenic state and in allowing efficient switchover to the lytic state during induction.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bailone A., Levine A., Devoret R. Inactivation of prophage lambda repressor in vivo. J Mol Biol. 1979 Jul 5;131(3):553–572. doi: 10.1016/0022-2836(79)90007-x. [DOI] [PubMed] [Google Scholar]
- Brent R., Ptashne M. The lexA gene product represses its own promoter. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1932–1936. doi: 10.1073/pnas.77.4.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunaway M., Manly S. P., Matthews K. S. Model for lactose repressor protein and its interaction with ligands. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7181–7185. doi: 10.1073/pnas.77.12.7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein I. R. Cooperative and non-cooperative binding of large ligands to a finite one-dimensional lattice. A model for ligand-oligonucleotide interactions. Biophys Chem. 1978 Sep;8(4):327–339. doi: 10.1016/0301-4622(78)80015-5. [DOI] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. D., Meyer B. J., Ptashne M. Interactions between DNA-bound repressors govern regulation by the lambda phage repressor. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5061–5065. doi: 10.1073/pnas.76.10.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. D., Poteete A. R., Lauer G., Sauer R. T., Ackers G. K., Ptashne M. lambda Repressor and cro--components of an efficient molecular switch. Nature. 1981 Nov 19;294(5838):217–223. doi: 10.1038/294217a0. [DOI] [PubMed] [Google Scholar]
- Johnson M. L., Halvorson H. R., Ackers G. K. Oxygenation-linked subunit interactions in human hemoglobin: analysis of linkage functions for constituent energy terms. Biochemistry. 1976 Nov 30;15(24):5363–5371. doi: 10.1021/bi00669a024. [DOI] [PubMed] [Google Scholar]
- Kao-Huang Y., Revzin A., Butler A. P., O'Conner P., Noble D. W., von Hippel P. H. Nonspecific DNA binding of genome-regulating proteins as a biological control mechanism: measurement of DNA-bound Escherichia coli lac repressor in vivo. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4228–4232. doi: 10.1073/pnas.74.10.4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N. L., Gielow W. O., Wallace R. G. Mechanism of araC autoregulation and the domains of two overlapping promoters, Pc and PBAD, in the L-arabinose regulatory region of Escherichia coli. Proc Natl Acad Sci U S A. 1981 Feb;78(2):752–756. doi: 10.1073/pnas.78.2.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer R., Meyer B., Ptashne M. Gene regulation at the right operator (OR) bacteriophage lambda. I. OR3 and autogenous negative control by repressor. J Mol Biol. 1980 May 15;139(2):147–161. doi: 10.1016/0022-2836(80)90302-2. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure W. R. Rate-limiting steps in RNA chain initiation. Proc Natl Acad Sci U S A. 1980 Oct;77(10):5634–5638. doi: 10.1073/pnas.77.10.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D., von Hippel P. H. Theoretical aspects of DNA-protein interactions: co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. J Mol Biol. 1974 Jun 25;86(2):469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- Meyer B. J., Maurer R., Ptashne M. Gene regulation at the right operator (OR) of bacteriophage lambda. II. OR1, OR2, and OR3: their roles in mediating the effects of repressor and cro. J Mol Biol. 1980 May 15;139(2):163–194. doi: 10.1016/0022-2836(80)90303-4. [DOI] [PubMed] [Google Scholar]
- Meyer B. J., Ptashne M. Gene regulation at the right operator (OR) of bacteriophage lambda. III. lambda repressor directly activates gene transcription. J Mol Biol. 1980 May 15;139(2):195–205. doi: 10.1016/0022-2836(80)90304-6. [DOI] [PubMed] [Google Scholar]
- Ogden S., Haggerty D., Stoner C. M., Kolodrubetz D., Schleif R. The Escherichia coli L-arabinose operon: binding sites of the regulatory proteins and a mechanism of positive and negative regulation. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3346–3350. doi: 10.1073/pnas.77.6.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V., Chadwick P., Ptashne M. Active form of two coliphage repressors. Nature. 1970 Jul 4;227(5253):41–44. doi: 10.1038/227041a0. [DOI] [PubMed] [Google Scholar]
- Ptashne M., Jeffrey A., Johnson A. D., Maurer R., Meyer B. J., Pabo C. O., Roberts T. M., Sauer R. T. How the lambda repressor and cro work. Cell. 1980 Jan;19(1):1–11. doi: 10.1016/0092-8674(80)90383-9. [DOI] [PubMed] [Google Scholar]
- Reichardt L., Kaiser A. D. Control of lambda repressor synthesis. Proc Natl Acad Sci U S A. 1971 Sep;68(9):2185–2189. doi: 10.1073/pnas.68.9.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975 May 25;94(3):441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- Schwarz G. On the analysis of linear binding effects associated with curved Scatchard plots. Biophys Chem. 1976 Dec;6(1):65–76. doi: 10.1016/0301-4622(76)80062-2. [DOI] [PubMed] [Google Scholar]
- von Hippel P. H., Revzin A., Gross C. A., Wang A. C. Non-specific DNA binding of genome regulating proteins as a biological control mechanism: I. The lac operon: equilibrium aspects. Proc Natl Acad Sci U S A. 1974 Dec;71(12):4808–4812. doi: 10.1073/pnas.71.12.4808. [DOI] [PMC free article] [PubMed] [Google Scholar]


