Abstract
Mitochondrial protein tyrosine phosphorylation is an important mechanism for the modulation of mitochondrial functions. In the present study, we have identified novel substrates of c-Src in mitochondria and investigated their function in the regulation of oxidative phosphorylation. The Src family kinase inhibitor PP2 {amino-5-(4-chlorophenyl)-7-(t-butyl) pyrazolo [3,4d] pyrimidine} exhibits significant reduction of respiration. Similar results were obtained from cells expressing kinase-dead c-Src, which harbours a mitochondrial-targeting sequence. Phosphorylation-site analysis selects c-Src targets, including NDUFV2 (NADH dehydrogenase [ubiquinone] flavoprotein 2) at Tyr193 of respiratory complex I and SDHA (succinate dehydrogenase A) at Tyr215 of complex II. The phosphorylation of these sites by c-Src is supported by an in vivo assay using cells expressing their phosphorylation-defective mutants. Comparison of cells expressing wild-type proteins and their mutants reveals that NDUFV2 phosphorylation is required for NADH dehydrogenase activity, affecting respiration activity and cellular ATP content. SDHA phosphorylation shows no effect on enzyme activity, but perturbed electron transfer, which induces reactive oxygen species. Loss of viability is observed in T98G cells and the primary neurons expressing these mutants. These results suggest that mitochondrial c-Src regulates the oxidative phosphorylation system by phosphorylating respiratory components and that c-Src activity is essential for cell viability.
Keywords: cell death, energy metabolism, mitochondrion, Src, tyrosine kinase, reactive oxygen species (ROS)
Abbreviations: ANT, adenine nucleotide translocase; BN, Blue native; CA, constitutive-active; COX, cytochrome c oxidase; CSK, C-terminal Src kinase; DDM, n-dodecyl-β-D-maltoside; 2-DE, two-dimensional PAGE; HE, hydroethidine; HEK, human embryonic kidney; IPG, immobilized pH gradient; KD, kinase-dead; LDH, lactate dehydrogenase; mAb, monoclonal antibody; MAP2, microtubule-associated protein 2; MTS, mitochondria-targeting sequence; NBT, Nitro Blue Tetrazolium; NDUFB10, NADH dehydrogenase [ubiquinone] 1β subcomplex subunit 10; NDUFV2, NADH dehydrogenase [ubiquinone] flavoprotein 2; PI, propidium iodide; PMS, phenazine methosulfate; PP2, amino-5-(4-chlorophenyl)-7-(t-butyl) pyrazolo [3,4d] pyrimidine; ROS, reactive oxygen species; SDHA, succinate dehydrogenase A; SFK, Src family kinase; UCP, uncoupling protein; VLCAD, very long chain acyl-CoA dehydrogenase; WT, wild-type
INTRODUCTION
Mitochondrial serine/threonine phosphorylation has been investigated in terms of the mitochondrial compartments in which it occurs, and the results of the mitochondrial serine/threonine phosphoproteome have previously been reported [1–3]. Tyrosine phosphorylation has also been implicated in mitochondrial regulation, in which the energy production system of a cell is modulated by the modification of mitochondrial proteins [4]. Several non-receptor-type and receptor-type tyrosine kinases, including c-Src, Fyn, Lyn, Fgr, CSK (C-terminal Src kinase), Abl and EGFR (epidermal growth factor receptor), have been observed in mitochondria [5–8], and SFKs (Src family kinases) are major agents in mitochondrial tyrosine phosphorylation [9]. However, only a few mitochondrial-tyrosine phosphorylated proteins have been characterized to date. Previously, it was demonstrated that c-Src is required for the activity of the mitochondrial electron transport chain through the direct phosphorylation of subunit II of COX (cytochrome c oxidase), although the phosphorylation sites were not identified [6]. In addition, the phosphorylation of Tyr304 in the catalytic subunit I of COX in concert with activation of the cAMP-dependent pathway leads to the suppression of enzyme activity [10].
A number of mitochondrial proteins have been identified as tyrosine phosphorylated using different proteomic approaches, and the phosphorylation sites have been reported. These include cytochrome c [11], enzymes of the tricarboxylic acid cycle, such as malate dehydrogenase and succinate CoA-ligase [12], long chain acyl CoA synthetase 1, a voltage-dependent anion channel [13], glycerol-3-phosphate dehydrogenase, creatine kinase, the ATP synthase ϵ chain, ANT (adenine nucleotide translocase) 1 and ANT2 [14]. It has also been reported that Tyr543 and Tyr604 of SDHA (succinate dehydrogenase A) are phosphorylated by Fgr [15] and that Tyr194 of ANT1 is phosphorylated by c-Src and Lck [16]. However, the physiological roles of their phosphorylation are not fully understood, and further investigations are needed to elucidate the roles of tyrosine phosphorylation in the molecular functions of mitochondria.
In the present study, we have identified novel mitochondrial targets of c-Src kinase, NDUFV2 {NADH dehydrogenase [ubiquinone] flavoprotein 2}, which is phosphorylated at Tyr193, and SDHA, which is phosphorylated at Tyr215. We have further demonstrated that phosphorylation of these proteins is required for the regulation of the respiratory electron transfer complex I and complex II systems, as well as for efficient energy production and cell survival. These results suggest that c-Src activity is essential for mitochondrial functions and cell viability.
MATERIALS AND METHODS
Antibodies and chemicals
Mouse anti-FLAG M2 mAb (monoclonal antibody), anti-FLAG M2 affinity gel, mouse anti-MAP2 (microtubule-associated protein 2) mAb, NBT (Nitro Blue Tetrazolium), PMS (phenazine methosulfate), HE (hydroethidine) and PI (propidium iodide) were purchased from Sigma; rabbit polyclonal anti-NDUFV2 antibody was from Abcam; rabbit polyclonal anti-c-Src antibody and anti-SDHA antibody were from Cell Signaling Technology; mouse anti-β-tubulin mAb was from Santa Cruz Biotechnology; mouse anti-cytochrome c mAb, poly-D-lysine and laminin were from BD Biosciences; mouse anti-phosphotyrosine (4G-10) mAb was from Millipore; 3–12% Bis-Tris native gel, MitoTracker Red reagent, penicillin, streptomycin, Neurobasal® medium, B27 supplements and Versene were from Invitrogen; PP2 {amino-5-(4-chlorophenyl)-7-(t-butyl) pyrazolo [3,4d] pyrimidine} was from Calbiochem; Hoechst 33342 and DDM (n-dodecyl-β-D-maltoside) were from Dojindo; FuGENE-HD was from Roche; the CytoTox-ONE LDH (lactate dehydrogenase) assay kit was from Promega; and 9.25 MBq γ[32P]ATP was from PerkinElmer. All other chemicals and reagents were of the highest grade commercially available.
Cell cultures and transfection
Human T98G glioblastoma cells were cultivated in RPMI 1640 medium (Sigma) supplemented with 10% FBS (fetal bovine serum) in a humidified atmosphere of 5% CO2 and 95% air at 37°C. Transfection was carried out using FuGENE-HD according to the manufacturer's recommended protocol, and the cells were used for experiments 48 h or 72 h after transfection.
Primary neurons and nucleofection
This study was carried out in compliance with the Guideline for Animal Experimentation at Fukushima Medical University with an effort to minimize the number of animals used and their suffering. Primary neurons were prepared from 17-day-old embryonic ICR (Institute of Cancer Research) mouse neocortex as described previously [17]. In brief, embryonic neocortex was dissected and incubated with Versene at room temperature (25°C) for 12 min. Cells were then mechanically dissociated with a fire-narrowed Pasteur pipette in the culture medium. Isolated neurons were plated at a density of 6.3×104 cells/cm2 on wells coated with poly-D-lysine and laminin and maintained at 37°C and 5% CO2 in Neurobasal® medium supplemented with 2% B-27, 500 μM glutamine, 50 units/ml penicillin and 50 μg/ml streptomycin. Transfection was accomplished using a Mouse Neuron Nucleofector kit and device (Lonza) as described previously [18]. The transfection was evaluated on day 3 after transfection.
Expression plasmid construction and mutagenesis
Full-length cDNAs for human c-Src, NDUFV2 and SDHA were obtained by RT (reverse transcription)–PCR using total RNA from normal human lung fibroblast TIG7 cells, and their sequence was confirmed by BigDye sequencing (Applied Biosystems) as described previously [19]. Lys298 of c-Src was replaced with a methionine residue (KD-c-Src: kinase-dead type) and Tyr530 was replaced with a phenylalanine residue (CA-c-Src: constitutive-active type) using the PrimeSTAR mutagenesis kit (Takara) and confirmed [19]. Similarly, Tyr193 of NDUFV2 was replaced with phenylalanine (Y193F-NDUFV2) and Tyr215 of SDHA was replaced with phenylalanine (Y215F-SDHA). The cDNAs for KD-c-Src, CA-c-Src, NDUFV2, Y193F-NDUFV2, SDHA and Y215F-SDHA were subcloned into the mammalian expression plasmid pcDNA3 (Life Technologies) harbouring a FLAG tag at the C-terminus. For predominant expression in mitochondria, the MTS (mitochondria-targeting sequence; residues 1–40 of the amino acid sequence) of VLCAD (very long chain acyl-CoA dehydrogenase) was fused to the N-termini of C-FLAG-tagged c-Src. C-GFP (green fluorescent protein)-tagged plasmids, MTS-KD-c-Src, NDUFV2, Y193F-NDUFV2, SDHA and Y215F-SDHA, were produced by inserting the full-length cDNAs into pAcGFP1-N1 (Clontech).
Preparation of the mitochondria-enriched fraction
Mitochondria were isolated from cultured cells by differential centrifugation as described previously [19]. Cells were homogenized in a Potter glass homogenizer with H-Buffer (10 mM Tris/HCl, pH 7.4, and 250 mM sucrose) and centrifuged at 800 g for 1 min at 4°C. The supernatant was centrifuged at 6000 g for 5 min, and the resulting pellet, the crude mitochondrial fraction, was suspended in H-Buffer. The suspension was layered over a discontinuous sucrose gradient consisting of 1.1 M and 1.6 M sucrose in 10 mM Tris/HCl, pH 7.4, and centrifuged for 3 h at 37000 rev./min at 4°C (TLS-55 rotor, Optima™ TLX ultracentrifuge, Beckman). The interface was collected in 10 mM Tris/HCl, pH 7.4, and centrifuged at 6000 g for 5 min. The resulting pellets were suspended in 10 mM Tris/HCl, pH 7.4, and used for experiments after confirming the presence of the mitochondrial marker cytochrome c.
Western blotting and immunoprecipitation
Cell pellets or the mitochondria-enriched fraction were solubilized in lysis buffer (20 mM Tris/HCl, pH 7.5, 0.15 M NaCl, 5 mM EDTA, 1% Triton X-100, 10 mM NaF, 12 mM 2-glycerophosphate and 1 mM Na3VO4) containing aprotinin (10 μg/ml), leupeptin (10 μg/ml) and PMSF (1 mM). After incubating on ice for 15 min, the lysates were clarified by centrifugation at 12000 g for 15 min. After protein determination by a protein assay reagent (Bio-Rad Laboratories), the supernatants (20 μg) were subjected to SDS/PAGE (12.5% gel) and transferred to PVDF filter membranes (Millipore). The membranes were blocked with 5% (w/v) non-fat dried skimmed milk powder in TBS (Tris-buffered saline) containing 0.05% Tween 20 and incubated with primary antibodies. Blots were probed with goat anti-mouse antibody coupled to HRP (horseradish peroxidase) (Bio-Rad Laboratories), and the positive signals were visualized by ECL (enhanced chemiluminescence) (Perkin Elmer). For immunoprecipitation, the supernatants were incubated with anti-FLAG M2 affinity gel for 2 h and washed with washing buffer (20 mM Tris/HCl, pH 7.5, 0.15 M NaCl, 5 mM EDTA and 1 mM PMSF), and the precipitated proteins were blotted with an anti-phosphotyrosine antibody.
2-DE (two-dimensional PAGE)
The mitochondria enriched fraction was solubilized in lysis buffer [7 M urea, 2 M thiourea, 4% CHAPS, 1% IPG (immobilized pH gradient) buffer, 1 mM benzamidine, 25 μg/ml leupeptin, 20 μg/ml pepstatin A, 20 μg/ml aprotinin, 1 mM Na3VO4, 1 μM microcystin-LR and 20 mM dithiothreitol], and the lysate was clarified by centrifugation at 39000 rev./min for 30 min (TLS-55 rotor, Optima™ TLX ultracentrifuge). After protein determination by a Bio-Rad Laboratories protein assay reagent, the supernatants (225 μg) were processed for isoelectric focusing as described previously [19] using IPG gel strips (pI 4–7, pI 3–10, 18 cm, GE Healthcare) and SDS/PAGE (12.5% gels). The gels were removed from glass plates and subjected to Western blotting.
Indirect immunofluorescence
Cells growing on glass coverslips were incubated with MitoTracker Red (Life Technologies) for 15 min at 37°C, followed by fixation with 4% (w/v) paraformaldehyde for 15 min at room temperature. The cells were permeabilized with 0.1% Triton X-100 in PBS containing 3% BSA for 1 h at 4°C, and incubated with primary antibodies. Then, the cells were reacted with anti-rabbit IgG or anti-mouse IgG conjugated with Alexa Fluor® 488/546 (Life Technologies) for 2 h at 4°C, and observed under a confocal laser-scanning microscope system (FV-1000D, Olympus).
Oxygen consumption analysis
Mitochondrial oxygen consumption was determined using a Clark-type oxygen electrode (Strathkelvin Instruments) as described previously [20,21]. The mitochondria-enriched fraction was incubated in oxygen measurement buffer (225 mM mannitol, 75 mM sucrose, 10 mM KCl, 20 mM Tris/HCl 7.4, 0.1 mM EDTA, 3 mM potassium phosphate, 5 mM succinate and 5 mM glutamate) at 30°C. After recording state 4 of the respiration reaction, ADP was added to a final concentration of 200 μM to induce state 3.
In vitro kinase assay
c-Src proteins recovered on an anti-FLAG M2 affinity gel from T98G cells expressing C-FLAG-tagged CA-c-Src were incubated with or without synthetic peptides (200 μg/ml) in kinase buffer consisting of 20 mM Hepes/NaOH, pH 7.4, 10 mM MgCl2 50 μM ATP and 92.5 kBq of γ-[32P]ATP in a total volume of 50 μl at 30°C for 20 min as described previously [22]. The synthetic peptides used were INDNYYEDLTAK [(NDUFV2 WT (wild-type)], INDNYFEDLTAK (NDUFV2 Y193F mutant), RYDTSYFVE (SDHA WT) and RYDTSFFVE (SDHA Y215F mutant). After incubation, the phosphorylation products were collected on to a P81 filter (Whatman), washed thoroughly with 175 mM phosphoric acid and subjected to scintillation counting. Specific activity was calculated by subtracting the radioactivity in control samples.
BN (Blue native)-PAGE and in-gel staining for dehydrogenase activity
The mitochondria-enriched fraction was solubilized in BN-lysis buffer (50 mM NaCl, 50 mM imidazole/HCl, pH 7.0, 5 mM 6-aminohexanoic acid and 1% DDM), and the lysate was clarified by centrifugation at 39000 rev./min for 15 min (TLS-55 rotor, Optima™ TLX ultracentrifuge). The resultant supernatant (50 μg of protein) was supplemented with 10% glycerol and 0.5% Coomassie Blue G-250 dye and subjected to BN-PAGE using Bis-Tris native gels as described previously [23]. NADH dehydrogenase (complex I) and succinate dehydrogenase (complex II) activities were evaluated by in-gel staining using NBT. After electrophoresis, the gels were equilibrated in reaction buffer (2 mM Tris/HCl, pH 7.4, and 0.2 mM NADH for NADH dehydrogenase, or 5 mM Tris/HCl, pH 7.4, 20 mM succinate and 0.2 mM PMS for succinate dehydrogenase) without NBT for 10 min, and then incubated in fresh reaction buffer with NBT at room temperature for 10–20 min. The reactions were stopped by fixing the gels in 45% methanol and 10% acetic acid. The gels were destained overnight in the same solution to remove residual Coomassie Blue G-250 dye. Band intensities were quantified using Image J software.
Measurement of cellular ATP content
T98G cells were transfected with pcDNA3 harbouring WT and mutants of various cDNAs. After 24 h of transfection, the cells were plated in 96-well dishes at a density of 10000 cells/well and then cultured for 24 h. ATP content was measured using an ATP luminescence assay kit (TOYO INK) according to the manufacturer's instructions. Light emission was recorded using a Turner TD-20/20 luminometer (Turner Designs).
Measurement of ROS (reactive oxygen species)
ROS were detected using the fluorescence probe HE. Transfected T98G cells were loaded with HE (50 μM), and the fluorescence intensity of HE was observed over 30 min by time-lapse imaging using a confocal laser-scanning microscope system (FV-1000D, Olympus) with excitation at 488 nm and emission at 575 nm.
Determination of cell viability
Transfected T98G cells were treated with the membrane-permeable fluorescent dye Hoechst 33342 (10 μg/ml) and the impermeable dye PI (5 μg/ml), and the number of cells stained with Hoechst 33342 and PI were individually counted in five different fields chosen at random. The ratio of PI-positive cells to total cells stained with Hoechst 33342 was calculated as described previously [24]. The viability of primary neurons was assessed by measuring LDH in culture medium using a CytoTox-ONE LDH assay kit (Promega) according to the manufacturer's recommended protocol as described previously [25].
RESULTS
Involvement of c-Src in the mitochondrial respiratory system
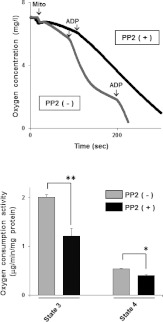
First, we confirmed the levels of c-Src in mitochondria prepared from human cell lines such as T98G, A172, HEK (human embryonic kidney)-293 and RPE (retinal pigment epithelium). The protein contents of c-Src were twice or more in the mitochondria-enriched fraction derived from all cells tested compared with whole-cell lysates (Supplementary Figure S1 at http://www.BiochemJ.org/bj/447/bj4470281add.htm). The blots were also probed for cytochrome c as a mitochondrial marker, and β-tubulin as a cytoplasmic marker. We detected cytochrome c, but not β-tubulin, in the mitochondria-enriched fraction, indicating pivotal roles of c-Src in mitochondria. In order to understand its function in the energy production system, we studied whether respiratory activity is affected by PP2, a selective inhibitor of SFKs. As shown in Figure 1, state 3 and state 4 oxygen consumptions were inhibited by 40% and 25% respectively in T98G cells treated with PP2 when the mitochondria-enriched fractions were incubated with glutamate and succinate to drive complex I- and complex II-dependent electron transport, and state 3 was induced by adding ADP. Similar results were obtained from T98G cells expressing MTS-KD-c-Src, a dominant-negative form harbouring the MTS of VLCAD (Figure 3 and Supplementary Figure S2A at http://www.BiochemJ.org/bj/447/bj4470281add.htm). These results suggest that mitochondrial c-Src kinases regulate energy production through direct phosphorylation of respiratory components.
Figure 1. Regulation of mitochondrial respiratory activity by c-Src.
The mitochondrial fraction was prepared from T98G cells treated with either 1 μM PP2 or vehicle for 1 h, and oxygen consumption was measured using glutamate and succinate as substrates. After recording state 4 of the respiration reaction, state 3 oxygen consumption was induced by the addition of ADP and the rates were compared with (+) and without (−) PP2. The oxygen consumption rate was normalized to the amount of mitochondrial protein. The experiments were repeated three times, and representative results are shown in the upper panel and quantitative data in the lower panel. Results are means±S.E.M. Statistical significance was evaluated by Student's t test (*P<0.05; **P<0.01).
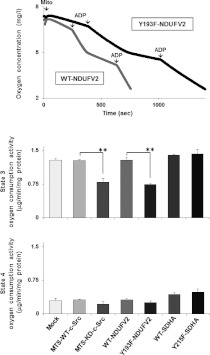
Figure 3. Effect of phosphorylation of NDUFV2 and SDHA on respiratory activity.
The mitochondria-enriched fraction was prepared from T98G cells expressing WT and the indicated mutant proteins, and state 3 and state 4 oxygen consumptions were measured using glutamate and succinate as substrates. The experiments were repeated three times, and representative results are shown in the top panel and quantitative data in the bottom two panels. Results are means±S.E.M. Statistical significance was evaluated by Student's t test (**P<0.01).
c-Src phosphorylates NDUFV2 at Tyr193 and SDHA at Tyr215 in vitro and in vivo
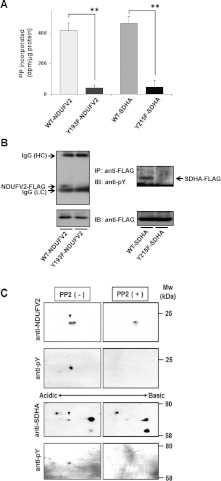
The molecular targets and phosphorylation sites of respiratory complex components by c-Src were analysed using NetPhosK 1.0 (http://www.cbs.dtu.dk/services/NetPhosK/). Tyr193 of NDUFV2, a flavoprotein subunit of complex I, and Tyr215 of SDHA, a flavoprotein subunit of complex II, were selected with the highest score for phosphorylation by c-Src (Supplementary Table S1 at http://www.BiochemJ.org/bj/447/bj4470281add.htm). The phosphorylation of NDUFV2 at Tyr193 and SDHA at Tyr215 by c-Src was examined by an in vitro kinase assay using purified preparations of a CA-c-Src and synthetic peptides, WT INDNYYEDLTAK or mutant INDNYFEDLTAK for NDUFV2 and WT RYDTSYFVE or mutant RYDTSFFVE for SDHA respectively. CA-c-Src phosphorylated the WT peptides efficiently, but not the mutant peptides (Figure 2A). The phosphorylation of NDUFV2 at Tyr193 and SDHA at Tyr215 were also detected in vivo. An anti-phosphotyrosine antibody clearly detected tyrosine-phosphorylated NDUFV2–FLAG and SDHA–FLAG in immunoprecipitates prepared from T98G cells expressing WT-NDUFV2–FLAG and WT-SDHA–FLAG, whereas the band was faint for cells expressing the phosphorylation-defective mutants Y193F-NDUFV2–FLAG and Y215F-SDHA–FLAG (Figure 2B). In addition, we performed 2-DE-Western blotting analysis using mitochondrial extracts prepared from T98G cells treated with PP2. Figure 2(C) shows major changes in immunoreactive protein spots corresponding to NDUFV2 and SDHA, which appeared in acidic and basic forms in control mitochondria, but mainly in the basic forms in mitochondria from cells treated with PP2. To examine whether the acidic spots represent the tyrosine-phosphorylated form of NDUFV2 and SDHA, the blots were also probed with an anti-phosphotyrosine antibody. We detected the tyrosine-phosphorylated form of NDUFV2 and SDHA in control mitochondria, but not in mitochondria from cells treated with PP2. These results confirm that c-Src phosphorylates NDUFV2 at Tyr193 and SDHA at Tyr215 in vitro and in vivo.
Figure 2. Phosphorylation of NDUFV2 at Tyr193 and SDHA at Tyr215 by c-Src.
(A) In vitro kinase assays were carried out using purified C-FLAG-tagged CA-c-Src and synthetic peptides with the sequence INDNYYEDLTAK (WT-NDUFV2) or INDNYFEDLTAK (Y193F-NDUFV2), and RYDTSYFVE (WT-SDHA) or RYDTSFFVE (Y215F-SDHA), containing the putative c-Src-targeting site of NDUFV2 and SDHA respectively. Specific activity was calculated by subtracting the radioactivity measured in the absence of substrate peptide. The experiment was repeated three times and representative results are shown as means±S.E.M. Statistical significance was evaluated by Student's t test (**P<0.01). (B) Immunoprecipitates obtained from T98G cells expressing C-FLAG-tagged WT-NDUFV2, Y193F-NDUFV2, WT-SDHA or Y215F-SDHA with anti-FLAG M2 affinity gel were separated by SDS/PAGE and blotted with an anti-phosphotyrosine antibody. Total mitochondrial proteins were analysed using an anti-FLAG antibody (bottom). IB, immunoblot; IP, immunoprecipitation. (C) Mitochondrial proteins prepared from T98G cells treated with either 10 μM PP2 (right-hand panels) or vehicle (left-hand panels) for 1 h were separated by 2-DE, and the blots were probed with anti-NDUFV2 (upper) or anti-SDHA (lower) antibody. The blots were re-probed with an anti-phosphotyrosine antibody. Spots for tyrosine-phosphorylated NDUFV2 and SDHA are indicated with arrowheads.
Phosphorylation of NDUFV2 is required for respiration and ATP production
To assess the effect of Tyr193 of NDUFV2 on respiratory activity, oxygen consumption rates of the mitochondria-enriched fraction prepared from T98G cells expressing the WT (WT-NDUFV2) or Y193F mutant (Y193F-NDUFV2) were examined. State 3 oxygen consumption was reduced by 43% in T98G cells expressing the Y193F mutant as compared with cells expressing the WT (Figure 3). This reduction was almost the same level when state 3 oxygen consumption was observed in the mitochondria-enriched fraction derived from T98G cells expressing MTS-KD-c-Src (40% reduction as compared with WT cells). On the other hand, state 4 oxygen consumption was not significantly affected by the expression of the mutants as compared with WT cells.
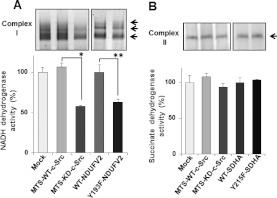
The effects of this phosphorylation on the molecular complex formation and dehydrogenase activity were examined using BN-PAGE. The bands corresponding to complex I were compared between the mitochondria-enriched fractions prepared from cells expressing WT-NDUFV2 and Y193F-NDUFV2. As shown in Figure 4(A), the NADH dehydrogenase activity of complex I was significantly affected (~38%) in cells expressing Y193F-NDUFV2 as compared with those expressing the WT. This indicates that the phosphorylation of NDUFV2 at Tyr193 is essential for the NADH dehydrogenase activity of complex I. A significant inhibition of the NADH dehydrogenase activity (~42%) was observed in the case of MTS-KD-c-Src. In parallel, cellular ATP contents were reduced by 25% and 20% in cells expressing MTS-KD-c-Src and Y193F-NDUFV2 respectively compared with their controls (Figure 5). These results suggest that mitochondrial c-Src modulates cellular ATP content by affecting the mitochondrial respiratory system.
Figure 4. Effect of phosphorylation of NDUFV2 and SDHA on dehydrogenase activity.
Mitochondrial extracts obtained from T98G cells expressing WT and the indicated mutant proteins were separated by BN-PAGE, and NADH dehydrogenase activity (A) and succinate dehydrogenase activity (B) were detected using NBT as an electron acceptor. The experiment was repeated three times; a representative gel image is shown in the upper panel, and quantitative data are shown in the lower panel. Results are means±S.E.M. Statistical significance was evaluated by Student's t test (*P<0.05, **P<0.01).
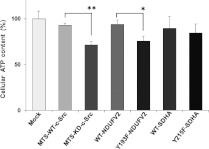
Figure 5. Effect of phosphorylation of NDUFV2 and SDHA on ATP production.
Cellular ATP contents in T98G cells expressing WT and the indicated mutant proteins were measured. The experiment was repeated three times, and representative results are shown as means±S.E.M. Statistical significance was evaluated by Student's t test (*P<0.05, **P<0.01).
The effects of SDHA at Tyr215 by mitochondrial c-Src on respiration activity were also examined using the mitochondria-enriched fraction prepared from T98G cells expressing the WT (WT-SDHA) or Y215F mutant (Y215F-SDHA). In contrast with the above results, state 3 oxygen consumption (Figure 3), enzyme activity (Figure 4B) and ATP production (Figure 5) were not significantly affected by this phosphorylation-defective mutant.
Perturbation of SDHA phosphorylation induces ROS generation
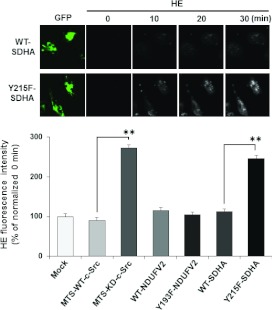
The above results indicate that the dysregulation of oxidative phosphorylation by reduced phosphorylation of NDUFV2 at Tyr193 and SDHA at Tyr215 might cause impaired energy metabolism and enhancement of ROS generation by electron leakage. ROS generation was monitored using the fluorescence probe HE. As shown in Figure 6, T98G cells expressing MTS-KD-c-Src produced a significant amount of ROS, whereas WT-transfected cells left only a trace. A significant enhancement in amounts of ROS was also observed in T98G cells expressing Y215F-SDHA, but not in cells expressing Y193F-NDUFV2, compared with WT cells (Figure 6). These results suggest that the phosphorylation of SDHA at Tyr215 is important for efficient electron transfer in complex II and the prevention of ROS generation.
Figure 6. Effect of phosphorylation of NDUFV2 and SDHA on ROS generation.
ROS production in T98G cells expressing WT and the indicated mutant proteins were measured. The cells were incubated for 30 min with the fluorescent dye HE for ROS detection, and the fluorescence intensity was monitored every 15 s. The experiments were repeated three times, and representative micrographs collected every 10 min are shown in the upper panel and the quantitative ratio of F30 to F0 (fluorescence intensities at 30 min and 0 min) in the lower panel. Results are means±S.E.M. Statistical significance was evaluated by Student's t test (**P<0.01).
Requirement of phosphorylation of NDUFV2 and SDHA for cell survival
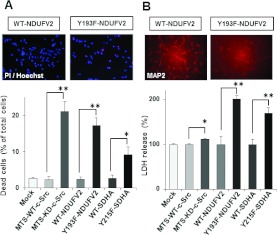
Since ATP depletion and excess ROS generation lead to cell death, we examined cell viability in T98G cells expressing MTS-KD-c-Src, Y193F-NDUFV2 or Y215F-SDHA using the fluorescent probes PI and Hoechst 33342. There was a significant increase in the number of dead cells expressing MTS-KD-c-Src, Y193F-NDUFV2 and Y215F-SDHA mutants compared with WT cells (Figure 7A). Similar results were obtained using primary neuron cultures expressing MTS-KD-c-Src, Y193F-NDUFV2 and Y215F-SDHA. Cell viability was assessed by expression of MAP2, a specific neuronal marker, and release of LDH into culture medium. The number of MAP2-positive cells was significantly affected in primary neurons by the expression of MTS-KD-c-Src, Y193F-NDUFV2 and Y215F-SDHA as compared with WT cells. A significant increase in LDH release was also observed in cells expressing MTS-KD-c-Src, Y193F-NDUFV2 and Y215F-SDHA (Figure 7B). These results suggest that phosphorylation of NDUFV2 at Tyr193 and SDHA at Tyr215 by mitochondrial c-Src play an essential role in cell survival.
Figure 7. Effect of phosphorylation of NDUFV2 and SDHA on cell viability.
(A) Cell viability in T98G cells expressing WT and the indicated mutant proteins was determined with PI/Hoechst staining. (B) Viability of primary cortical neurons was assessed by MAP2 staining and an LDH release assay. The experiments were repeated three times, and representative micrographs are shown in the upper panels and quantitative data in the lower panels. Results are means±S.E.M. Statistical significance was evaluated by Student's t test (*P<0.05, **P<0.01).
DISCUSSION
Although the expression levels of c-Src are relatively high in mitochondria, its function and molecular targets are not fully understood. A recent study suggested that NDUFB10 (NADH dehydrogenase [ubiquinone] 1β subcomplex subunit 10), a respiratory complex I component, is phosphorylated by c-Src kinase, and its phosphorylation is required for the preservation of respiratory complex I function [26]. In addition, other research groups have shown Tyr55 and Tyr142 of NDUFB10 to be phosphorylation sites by phosphoproteomic analysis [27,28]. However, the phosphorylation sites affecting complex I function have not been determined. In the present study, we identified NDUFV2 and SDHA as substrates for c-Src kinase in mitochondria, and succeeded in demonstrating that their phosphorylation by c-Src is essential for efficient electron transfer and oxidative phosphorylation in the respiratory system. NDUFV2 and SDHA are flavoproteins that are key components of the catalytic cores of respiratory complex I and II. Phosphorylation at Thr164 of NDUFV2 is suggested by phosphoproteomic analysis [29], although the kinase responsible has not been identified [30]. Tyr543 and Tyr604 in SDHA are also phosphorylated by Fgr tyrosine kinase, a member of the SFKs [15]. However, the physiological significance of the phosphorylation of these components remains unclear. In the present study, we demonstrate phosphorylation of Tyr193 of NDUFV2 and Tyr215 of SDHA by c-Src kinase in mitochondria. We used synthetic peptides, 2-DE-Western blotting and cells expressing mutants in which the c-Src target tyrosine residues were replaced with phenylalanine to confirm that c-Src in fact phosphorylates these sites on NDUFV2 and SDHA in mitochondria (Figure 2), and that these phosphorylations are closely associated with state 3 respiration (Figure 3) and ATP synthesis (Figure 5). On the basis of these results, we conclude that c-Src phosphorylates NUDFV2 at Tyr193 and SDHA at Tyr215, and that these phosphorylations are required for the electron transfer activities of respiratory complexes I and II.
It is of interest that the attenuation of c-Src-mediated phosphorylation of these components significantly reduced state 3 respiration (Figure 3) and enhanced ROS generation (Figure 6). Cells expressing the phosphorylation-defective Y193F mutant of NUDFV2 exhibited a significant loss of ATP content (Figure 5) and an increased rate of cell death (Figure 7), but no increase in ROS production. In contrast, cells expressing the Y215F mutant of SDHA showed significant ROS production (Figure 6) and an enhancement in cell death (Figure 7), whereas no changes in ATP content were observed. These results suggest different effects of c-Src-mediated phosphorylation on the molecular functions of NUDFV2 and SDHA. Since NDUFV2 locates in an NADH acceptor site of complex I and the Y193F mutant of NDUFV2 shows significantly reduced dehydrogenase activity (Figure 4), it is possible that phosphorylation by c-Src is involved in the reaction mechanism of the NADH dehydrogenase of complex I. In this case, electron transfer from NADH to FMN would be totally suppressed in the Y193F mutant, and ATP production would be affected, but ROS generation induced by the electron leakage would not occur in mutant cells. Alternatively, phosphorylation of SDHA at Tyr215 plays an important role in electron transfer from FADH2 to ubiquinone. A previous report on the crystal structure of porcine mitochondrial respiratory complex II has advanced our understanding of the regulatory mechanism of complex II [31], and conformational analysis reveals that Tyr215 locates at an FAD-binding domain between FAD in SDHA and an iron–sulfur cluster in SDHB. It has been shown that RNAi (RNA interference) at SDHB, an iron–sulfur protein, results in enhanced ROS generation [32], suggesting that electron transfer from FADH2 to the iron–sulfur cluster is a critical point for the generation of ROS in complex II. Thus it is conceivable that Tyr215 functions as a key residue in this electron transfer. Further studies on the molecular conformations of these sites are required to clarify the exact mechanisms by which their phosphorylation affects the electron transport chain.
It is known that dysregulation of the oxidative phosphorylation system causes impaired energy metabolism and, on occasion, an enhancement of ROS generation by electron leakage, suggesting the involvement of this system in cell death. For example, treating HEK-293 cells with the respiratory complex I inhibitor rotenone or complex II inhibitor TTFA (2-thenoyltrifluoroacetone) causes cell death and an enhancement of ROS generation [33]. Our previous study also demonstrated that VLCAD, a rate-limiting enzyme in the fatty acid β-oxidation pathway, is regulated by phosphorylation at Ser586, and that the suppression of this phosphorylation causes a perturbation in its enzyme activity, the induction of apoptotic cell death accompanied by enhanced ROS generation [19]. These results suggest the importance of post-translational modifications, including phosphorylation, of protein components for mitochondrial functions. More studies, such as those involving phosphoproteomic approaches, are needed to understand this issue. On the other hand, emerging evidence shows that impaired mitochondrial functions are involved in the pathogenesis of various disorders including neurodegenerative diseases, such as Alzheimer's disease, Parkinson's disease and Huntington's disease [34]. These patients exhibit reduced energy metabolism, loss of respiratory complex activity and increased oxidative stress in affected neurons and brain regions [35–37]. However, the cause of the mitochondrial alterations in the cells of these patients is not fully understood. In the present study, we demonstrated that phosphorylation of NDUFV2 and SDHA by c-Src is required for neuronal survival (Figure 7B). Therefore it may be necessary to study phosphorylation of respiratory chain components in these diseases.
The activities of protein kinases in mitochondria are important for the regulation of mitochondrial functions. A previous study revealed that SFKs are major players in mitochondrial tyrosine phosphorylation [9] and that a significant decrease in state 3 is observed when brain mitochondria are treated with the SFK inhibitor PP2 [7]. In the present study, we have also demonstrated that T98G cells treated with PP2, or those predominantly expressing a negative regulator of c-Src, MTS-KD-c-Src, exhibit a significant reduction in state 3 respiration and cellular ATP content (Figures 1, 3 and 5). On the basis of these results, we conclude that mitochondrial c-Src plays a critical role in the energy homoeostasis of cells by its involvement in the regulation of oxidative phosphorylation. On the other hand, a significant decrease in state 4 respiration was observed when T98G cells were treated with PP2, but not those expressing MTS-KD-c-Src (Figures 1 and 3). These results indicate the involvement of other Src family kinases in the regulatory mechanism of state 4 oxygen consumption, which represents respiration due to proton leakage across the mitochondrial inner membrane [38,39]. UCPs (uncoupling proteins) are inner membrane proteins specialized in inducible proton conductance. In recent studies, it was proposed that UCP1 and UCP3 are the tyrosine-phosphorylated proteins [40,41], and that UCP3 is activated by phosphorylation, leading to increased proton leakage. Thus it is possible that phosphorylation of UCPs by Src family kinases in the mitochondrial compartment may be involved in the molecular mechanism underlying the proton conductance.
The regulatory mechanism underlying mitochondrial c-Src activity remains elusive. Preliminary results demonstrate that the basal level of c-Src kinase activity is high, and that it seems to be constitutively active in mitochondria (results not shown). In addition, the level of a negative regulator of c-Src, such as CSK, expression and the amount of phosphorylation of the CSK target site in c-Src (Tyr530) are lower in the mitochondrial fraction than in plasma membrane fractions (results not shown). Proteins associated with c-Src in mitochondria differ completely from those associated with c-Src in the plasma membrane (results not shown). Thus it is conceivable that the relative activity of mitochondrial c-Src might be due to a low level of CSK and different activator molecules for c-Src activity in mitochondria.
In summary, we have identified new targets for c-Src in mitochondria, Tyr193 of NDUFV2 and Tyr215 of SDHA, which are both required for normal electron transfer in respiration complex I and complex II respectively and for oxidative phosphorylation. The attenuation of NDUFV2 phosphorylation induces cell death, probably through impaired NADH dehydrogenase activity and ATP production. On the other hand, a decrease in SDHA phosphorylation induces cell death through the production of ROS, but has no affect on enzyme activity or ATP levels. These results suggest that c-Src activity is important for the regulation of mitochondrial functions and cell viability. Further studies on the regulation of mitochondrial c-Src will provide additional insight into the energy production system and ROS generation.
Online data
AUTHOR CONTRIBUTION
Specifically, Masato Ogura and Yoshimi Homma designed the research. Masato Ogura and Junko Yamaki performed the experiments. Masato Ogura, Junko Yamaki and Miwako Homma analysed the data and interpreted the results. Masato Ogura and Yoshimi Homma wrote the paper. All of the authors participated in the preparation of the paper and approved the final version.
FUNDING
This work was supported by the Japan Society for the Promotion of Science (Grant-in-Aid for Scientific Research) [grant numbers 23790341 (to M.O.) and 60192324 (to Y.H.)] and Fukushima Medical University (grant for project research).
References
- 1.Pagliarini D. J., Dixon J. E. Mitochondrial modulation: reversible phosphorylation takes center stage? Trends Biochem. Sci. 2006;31:26–34. doi: 10.1016/j.tibs.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 2.Lee J., Xu Y., Chen Y., Sprung R., Kim S. C., Xie S., Zhao Y. Mitochondrial phosphoproteome revealed by an improved IMAC method and MS/MS/MS. Mol. Cell. Proteomics. 2007;6:669–676. doi: 10.1074/mcp.M600218-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reinders J., Wagner K., Zahedi R. P., Stojanovski D., Eyrich B., Van der Laan M., Rehling P., Sickmann A., Pfanner N., Meisinger C. Profiling phosphoproteins of yeast mitochondria reveals a role of phosphorylation in assembly of the ATP synthase. Mol. Cell. Proteomics. 2007;6:1896–1906. doi: 10.1074/mcp.M700098-MCP200. [DOI] [PubMed] [Google Scholar]
- 4.Salvi M., Brunati A. M., Toninello A. Tyrosine phosphorylation in mitochondria: a new frontier in mitochondrial signaling. Free Radical Biol. Med. 2005;38:1267–1277. doi: 10.1016/j.freeradbiomed.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Salvi M., Brunati A. M., Bordin L., La Rocca N., Clari G., Toninello A. Characterization and location of Src dependent tyrosine phosphorylation in rat brain mitochondria. Biochim. Biophys. Acta. 2002;1589:181–195. doi: 10.1016/s0167-4889(02)00174-x. [DOI] [PubMed] [Google Scholar]
- 6.Miyazaki T., Neff L., Tanaka S., Horne W. C., Baron R. Regulation of cytochrome c oxidase activity by c-Src in osteoclasts. J. Cell Biol. 2003;160:709–718. doi: 10.1083/jcb.200209098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Augereau O., Claverol S., Boudes N., Basurko M. J., Bonneu M., Rossignol R., Mazat J. P., Letellier T., Dachary-Prigent J. Identification of tyrosine-phosphorylated proteins of the mitochondrial oxidative phosphorylation machinery. Cell. Mol. Life Sci. 2005;62:1478–1488. doi: 10.1007/s00018-005-5005-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Livigni A., Scorziello A., Agnese S., Adornetto A., Carlucci A., Garbi C., Castaldo L., Annunziato L., Avvedimento E. V., Feliciello A. Mitochondrial AKAP121 links cAMP and src signaling to oxidative metabolism. Mol. Biol. Cell. 2006;17:263–271. doi: 10.1091/mbc.E05-09-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tibaldi E., Brunati A. M., Massimino M. L., Stringaro A., Colone M., Agostinelli E., Arancia G., Toninello A. Src-tyrosine kinases are major agents in mitochondrial tyrosine phosphorylation. J. Cell. Biochem. 2008;104:840–849. doi: 10.1002/jcb.21670. [DOI] [PubMed] [Google Scholar]
- 10.Lee L., Salomon A. R., Ficarro S., Mathes I., Lottspeich F., Grossman L. I., Huttemann M. cAMP-dependent tyrosine phosphorylation of subunit I inhibits cytochrome c oxidase activity. J. Biol. Chem. 2005;280:6094–6100. doi: 10.1074/jbc.M411335200. [DOI] [PubMed] [Google Scholar]
- 11.Lee L., Salomon A. R., Yu K., Doan J. W., Grossman L. I., Huttemann M. New prospects for an old enzyme: mammalian cytochrome c is tyrosine-phosphorylated in vivo. Biochemistry. 2006;45:9121–9128. doi: 10.1021/bi060585v. [DOI] [PubMed] [Google Scholar]
- 12.Rush J., Moritz A., Lee K. A., Guo A., Goss V. L., Spek E. J., Zhang H., Zha X. M., Olakiewicz R. D., Comb J. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 13.Distler A. M., Kerner J., Hoppel C. L. Post-translational modifications of rat liver mitochondrial outer membrane proteins identified by mass spectrometry. Biochim. Biophys. Acta. 2007;1774:628–636. doi: 10.1016/j.bbapap.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewandrowski U., Sickmann A., Cesaro L., Brunati A. M., Toninello A., Salvi M. Identification of new tyrosine phosphorylated proteins in rat brain mitochondria. FEBS Lett. 2008;582:1104–1110. doi: 10.1016/j.febslet.2008.02.077. [DOI] [PubMed] [Google Scholar]
- 15.Salvi M., Morrice N. A., Brunati A. M., Toninello A. Identification of the flavoprotein of succinate dehydrogenase and aconitase as in vitro mitochondrial substrates of Fgr tyrosine kinase. FEBS Lett. 2007;581:5579–5585. doi: 10.1016/j.febslet.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 16.Feng J., Lucchinetti E., Enkavi G., Wang Y., Gehrig P., Roschitzki B., Schaub M. C., Tajkhorshid E., Zaugg K., Zaugg M. Tyrosine phosphorylation by Src within the cavity of the adenine nucleotide translocase 1 regulates ADP/ATP exchange in mitochondria. Am. J. Physiol. Cell. Physiol. 2010;298:C740–C748. doi: 10.1152/ajpcell.00310.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogura M., Taniura H., Nakamichi N., Yoneda Y. Upregulation of the glutamine transporter through transactivation mediated by cAMP/protein kinase A signals toward exacerbation of vulnerability to oxidative stress in rat neocortical astrocytes. J. Cell. Physiol. 2007;212:375–385. doi: 10.1002/jcp.21031. [DOI] [PubMed] [Google Scholar]
- 18.Zeitelhofer M., Vessey J. P., Xie Y., Tübing F., Thomas S., Kiebler M., Dahm R. High-efficiency transfection of mammalian neurons via nucleofection. Nat. Protoc. 2007;2:1692–1704. doi: 10.1038/nprot.2007.226. [DOI] [PubMed] [Google Scholar]
- 19.Kabuyama Y., Suzuki T., Nakazawa N., Yamaki J., Homma M. K., Homma Y. Dysregulation of very long chain acyl-CoA dehydrogenase coupled with lipid peroxidation. Am. J. Physiol.: Cell. Physiol. 2010;298:C107–C113. doi: 10.1152/ajpcell.00231.2009. [DOI] [PubMed] [Google Scholar]
- 20.Gottlieb E., Armour S. M., Thompson C. B. Mitochondrial respiratory control is lost during growth factor deprivation. Proc. Natl. Acad. Sci. U.S.A. 2002;99:12801–12806. doi: 10.1073/pnas.202477599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wegrzyn J., Potla R., Chwae Y. J., Sepuri N. B., Zhang Q., Koeck T., Derecka M., Szczepanek K., Szelag M., Gornicka A., et al. Function of mitochondrial Stat3 in cellular respiration. Science. 2009;323:793–797. doi: 10.1126/science.1164551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Homma M. K., Wada I., Suzuki T., Yamaki J., Krebs E. G., Homma Y. CK2 phosphorylation of eukaryotic translation initiation factor 5 potentiates cell cycle progression. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15688–15693. doi: 10.1073/pnas.0506791102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wittig I., Karas M., Schägger H. High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol. Cell. Proteomics. 2007;6:1215–1225. doi: 10.1074/mcp.M700076-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Ogura M., Takarada T., Nakamichi N., Kawagoe H., Sako A., Nakazato R., Yoneda Y. Exacerbated vulnerability to oxidative stress in astrocytic C6 glioma cells with stable overexpression of the glutamine transporter slc38a1. Neurochem. Int. 2011;58:504–511. doi: 10.1016/j.neuint.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 25.Khanna S., Roy S., Park H. A., Sen C. K. Regulation of c-Src activity in glutamate-induced neurodegeneration. J. Biol. Chem. 2007;282:23482–23490. doi: 10.1074/jbc.M611269200. [DOI] [PubMed] [Google Scholar]
- 26.Hebert-Chatelain E., Jose C., Gutierrez Cortes N., Dupuy J. W., Rocher C., Dachary-Prigent J., Letellier T. Preservation of NADH ubiquinone-oxidoreductase activity by Src kinase-mediated phosphorylation of NDUFB10. Biochim. Biophys. Acta. 2012;1817:718–725. doi: 10.1016/j.bbabio.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 27.Moritz A., Li Y., Guo A., Villén J., Wang Y., MacNeill J., Kornhauser J., Sprott K., Zhou J., Possemato A., et al. Akt-RSK-S6 kinase signaling networks activated by oncogenic receptor tyrosine kinases. Sci. Signaling. 2010;3:ra64. doi: 10.1126/scisignal.2000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cesaro L., Salvi M. Mitochondrial tyrosine phosphoproteome: new insights from an up-to-date analysis. Biofactors. 2010;36:437–450. doi: 10.1002/biof.123. [DOI] [PubMed] [Google Scholar]
- 29.Hopper R. K., Carroll S., Aponte A. M., Johnson D. T., French S., Shen R. F., Witzmann F. A., Harris R. A., Balaban R. S. Mitochondrial matrix phosphoproteome: effect of extra mitochondrial calcium. Biochemistry. 2006;45:2524–2536. doi: 10.1021/bi052475e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olsen J. V., Vermeulen M., Santamaria A., Kumar C., Miller M. L., Jensen L. J., Gnad F., Cox J., Jensen T. S., Nigg E. A., et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signaling. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 31.Sun F., Huo X., Zhai Y., Wang A., Xu J., Su D., Bartlam M., Rao Z. Crystal structure of mitochondrial respiratory membrane protein complex II. Cell. 2005;121:1043–1057. doi: 10.1016/j.cell.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 32.Guzy R. D., Sharma B., Bell E., Chandel N. S., Schumacker P. T. Loss of the SdhB, but not the SdhA, subunit of complex II triggers reactive oxygen species-dependent hypoxia-inducible factor activation and tumorigenesis. Mol. Cell. Biol. 2008;28:718–731. doi: 10.1128/MCB.01338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y., McMillan-Ward E., Kong J., Israels S. J., Gibson S. B. Mitochondrial electron-transport-chain inhibitors of complexes I and II induce autophagic cell death mediated by reactive oxygen species. J. Cell. Sci. 2007;120:4155–4166. doi: 10.1242/jcs.011163. [DOI] [PubMed] [Google Scholar]
- 34.Mattson M. P., Gleichmann M., Cheng A. Mitochondria in neuroplasticity and neurological disorders. Neuron. 2008;60:748–766. doi: 10.1016/j.neuron.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mosconi L., De Santi S., Li J., Tsui W. H., Li Y., Boppana M., Laska E., Rusinek H., de Leon M. J. Hippocampal hypometabolism predicts cognitive decline from normal aging. Neurobiol. Aging. 2008;29:676–692. doi: 10.1016/j.neurobiolaging.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schapira A. H. Mitochondria in the aetiology and pathogenesis of Parkinson's disease. Lancet Neurol. 2008;7:97–109. doi: 10.1016/S1474-4422(07)70327-7. [DOI] [PubMed] [Google Scholar]
- 37.Walker F. O. Huntington's disease. Lancet. 2007;369:218–228. doi: 10.1016/S0140-6736(07)60111-1. [DOI] [PubMed] [Google Scholar]
- 38.Brand M. D., Nicholls D. G. Assessing mitochondrial dysfunction in cells. Biochem. J. 2011;435:297–312. doi: 10.1042/BJ20110162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Divakaruni A. S., Brand M. D. The regulation and physiology of mitochondrial proton leak. Physiology (Bethesda) 2011;26:192–205. doi: 10.1152/physiol.00046.2010. [DOI] [PubMed] [Google Scholar]
- 40.Rikova K., Guo A., Zeng Q., Possemato A., Yu J., Haack H., Nardone J., Lee K., Reeves C., Li Y., et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 41.Kelly O. M., McNamara Y. M., Manzke L. H., Meegan M. J., Porter R. K. The preservation of in vivo phosphorylated and activated uncoupling protein 3 (UCP3) in isolated skeletal muscle mitochondria following administration of 3,4-methylenedioxymethamphetamine (MDMA aka ecstasy) to rats/mice. Mitochondrion. 2012;12:110–119. doi: 10.1016/j.mito.2011.03.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.