Dynactin serves as an adaptor that allows the dynein motor to bind cargoes, but how dynactin associates with its diverse complement of subcellular binding partners remains mysterious. We show that the “pointed-end complex” of dynactin is a bipartite structural domain that stabilizes dynactin and supports its binding to different subcellular structures.
Abstract
Dynactin is an essential part of the cytoplasmic dynein motor that enhances motor processivity and serves as an adaptor that allows dynein to bind cargoes. Much is known about dynactin's interaction with dynein and microtubules, but how it associates with its diverse complement of subcellular binding partners remains mysterious. It has been suggested that cargo specification involves a group of subunits referred to as the “pointed-end complex.” We used chemical cross-linking, RNA interference, and protein overexpression to characterize interactions within the pointed-end complex and explore how it contributes to dynactin's interactions with endomembranes. The Arp11 subunit, which caps one end of dynactin's Arp1 filament, and p62, which binds Arp11 and Arp1, are necessary for dynactin stability. These subunits also allow dynactin to bind the nuclear envelope prior to mitosis. p27 and p25, by contrast, are peripheral components that can be removed without any obvious impact on dynactin integrity. Dynactin lacking these subunits shows reduced membrane binding. Depletion of p27 and p25 results in impaired early and recycling endosome movement, but late endosome movement is unaffected, and mitotic spindles appear normal. We conclude that the pointed-end complex is a bipartite structural domain that stabilizes dynactin and supports its binding to different subcellular structures.
INTRODUCTION
Microtubule-based transport is of key importance for the localization and motility of endomembranes and is critical for the events of cell division. Plus end–directed movement is powered by motors of the kinesin family, which contains 14 distinct subtypes (Miki et al., 2001). A single motor, cytoplasmic dynein, is responsible for the vast majority of minus end–directed (i.e., centripetal) transport. Dynein function in vivo depends upon dynactin, a protein complex first identified as an essential cofactor for dynein-based vesicle motility in vitro (Schroer and Sheetz, 1991). A large body of work has demonstrated that dynactin is required for dynein function at all stages of the cell cycle (Schroer, 2004; Schroer and Cheong, 2012). How dynein activity is regulated and how cargoes are selected and discriminated remains unclear, but dynactin is believed to be critical for both processes.
Dynactin is an ≈1-MDa assembly of 11 distinct components organized into three major structural domains. A short Arp1 filament (Arp1/actin) is the core of the dynactin structure (Schafer et al., 1994). This is capped by the actin-binding protein, CapZ α/β, at one end, and a unique assemblage of four subunits (Arp11, p62, p27, and p25; the “pointed-end complex”) at the other (Schafer et al., 1994; Eckley et al., 1999). The “sidearm/shoulder” (p150Glued, dynamitin, and p24) (Eckley et al., 1999), which associates with CapZ, allows dynein binding via a direct interaction between p150Glued and the dynein IC (DIC; Karki and Holzbaur, 1995; Vaughan and Vallee, 1995; King et al., 2003). p150Glued also binds microtubules, which enhances the processivity of individual dynein molecules and may contribute to motor function in other ways (Waterman-Storer et al., 1995; King and Schroer, 2000; Ross et al., 2006).
In addition to binding dynein and influencing motor activity, dynactin allows dynein to bind its cargoes. The molecular details of dynactin–cargo interactions are only beginning to be defined, but published work suggests that Arp1 may interact with membranes directly (Holleran et al., 1996, 2001; Muresan et al., 2001). The function of the pointed-end complex subunits is less clear. This structure binds neither dynein nor microtubules, and its p62, p27, and p25 subunits are not present in yeasts, indicating that they are dispensable for dynactin's most basic functions (Schroer, 2004). One possibility is that they help target dynein/dynactin to internal membranes. Evidence for this has been obtained in the filamentous fungi, Aspergillus nidulans, in which the p25 orthologue has been implicated in endosome movement (Zhang et al., 2011), and Neurospora crassa, in which p25, p62, and Arp11 null mutants exhibit reduced organelle transport and abnormal vacuole distribution (Lee et al., 2001).
In the present study, we characterize subunit–subunit interactions within the pointed-end complex and demonstrate that this dynactin domain mediates selective binding to nuclei and a subset of endocytic membranes. Interactions among Arp11, p62, p27, and p25 were mapped by chemical cross-linking. Subunit functions were explored using RNA interference (RNAi) to deplete Cos7 cells of the four subunits and, for comparison, p150Glued. Arp11 and p62 were found to be essential for preservation of dynactin structure, whereas p150Glued, p27, and p25 were not. p27/p25 and p150Glued function in membrane localization and mitosis was explored using immunofluorescence and live-cell imaging. Because depletion of Arp11 or p62 led to loss of the entire dynactin molecule, we evaluated the function of just these subunits in cargo binding using overexpression. Together these approaches revealed that the pointed-end complex contains two distinct binding activities, comprising the subunit pairs p62/Arp11 and p27/p25. These two entities allow dynactin to bind, and target dynein to, different cellular components in interphase and at the onset of mitosis, allowing dynein to power distinct types of motility at different stages of the cell cycle.
RESULTS
Intersubunit interactions within the pointed-end complex
Dynactin's pointed-end complex was initially described as a tetramer of subunits that resisted dynactin disassembly with the chaotropic salt, potassium iodide (Eckley et al., 1999). In some experiments, we noted that the complex dissociated into two heterodimers, p62/Arp11 and p27/p25. To learn more about protein–protein interactions within the pointed-end complex, we subjected intact dynactin to covalent cross-linking using the cysteine cross-linker bismaleimidohexane (BMH; Figure 1). Immunoblotting revealed several bands that reacted with antibodies to two or more components, indicative of cross-linked protomers (Figure 1B). In addition to a cross-linked species that contained all four components (band a), cross-links were seen among Arp11, p62, and p25 (band b); Arp11 and p62 (band c); Arp11, p25, and p27 (band d); Arp11 and p25 (band e); and p25 and p27 (band f). These results indicate Arp11 contacts both p62 and p25 directly and suggest p27 is located peripherally (see cartoon in Figure 1C).
FIGURE 1:
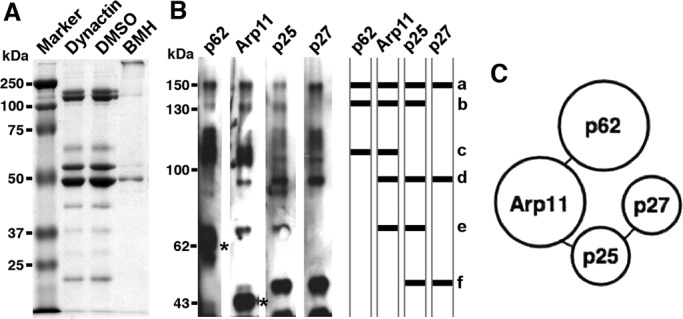
Protein–protein interactions in the pointed-end complex. Purified bovine dynactin was subjected to BMH cross-linking, and the reaction products were evaluated by (A) SDS–PAGE (10% gel, Coomassie Blue stain) or (B) immunoblotting for p62, Arp11, p25, and p27. The asterisks mark the positions of un–cross-linked p62 and Arp11. a, p62/Arp11/p25/p27; b, p62/Arp11/p25; c, p62/Arp11; d, Arp11/p25/p27; e, Arp11/p25; and f, p25/p27. The cross-linking results are summarized in the simplified cartoon in (C), which shows proposed subunit interactions.
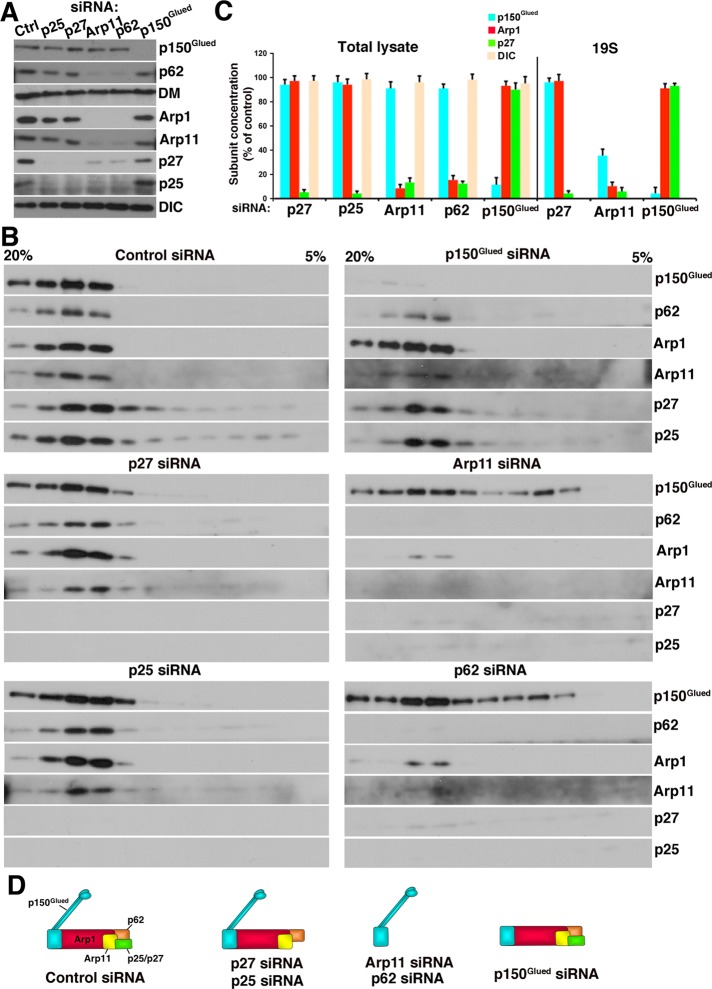
Effects of dynactin subunit depletion on dynactin integrity
To learn more about the functions of pointed-end complex components, we performed a systematic RNAi analysis. Cos7 cells were transfected with p62, Arp11, p27, or p25 small interfering RNAs (siRNAs) and cultured for 72 h to allow subunit depletion. p150Glued knockdown was performed for comparison. Cell lysates were analyzed for the abundance of the target subunits, plus dynamitin and Arp1 (Figure 2A). All siRNA treatments resulted in >90% depletion of the intended subunit. Although the present analysis is focused on Cos7 cells, we obtained similar results in HeLa cells. Of all the proteins we examined, only p150Glued could be depleted without impact on other dynactin subunits. Velocity sedimentation revealed that an ≈19S complex containing other dynactin subunits persisted (Figure 2, B and C). Depletion of any of the four pointed-end complex components, by contrast, resulted in loss of multiple dynactin polypeptides. Depletion of either p27 or p25 yielded loss of both subunits, but dynactin structure otherwise remained intact. Expression of siRNA-resistant p27 resulted in recovery of both p27 and p25 (Supplemental Figure S1). The finding that p27 and p25 are interdependent for stability is consistent with the observation that they are binding partners (Figure 1; see also Eckley et al., 1999).
FIGURE 2:
Effect of dynactin subunit depletion on dynactin integrity and composition. Detergent lysates of Cos7 cells transfected with control or dynactin subunit siRNAs were (A) immunoblotted directly or (B) subjected to velocity sedimentation into a 5–20% sucrose gradient and then immunoblotted for the dynactin subunits indicated or a control, DIC. DM, dynamitin. (C) Quantitation of subunit levels in the lysate and 19S fractions, respectively. The cartoons in (D) illustrate how depletion of different subunits affects dynactin structure.
Depletion of either Arp11 or p62 had a much more dramatic effect on dynactin structure. In addition to the target, multiple dynactin components, including Arp1, p27, and p25, were markedly diminished. Velocity sedimentation of Arp11-depleted cell lysates revealed that < 10% of dynactin (i.e., Arp1 at ≈19S) remained (Figure 2, B and C). Much of the p150Glued present in the lysate no longer sedimented at ≈19S, indicating it was not dynactin-associated. Depletion of p62 had an effect similar to Arp11 depletion on dynactin stability. The pool of ≈19S p150Glued that persisted in Arp11- or p62-depleted cells might be the result of abnormal association of p150Glued with other proteins, such as dynein, as reported in Arp11-depleted A. nidulans (Zhang et al., 2008).
Effects of dynactin subunit depletion on dynein-dependent behaviors
Dynein and dynactin contribute to a huge number of motile phenomena, including endomembrane movement, nuclear envelope breakdown, and mitotic spindle assembly (Karki and Holzbaur, 1999; Kardon and Vale, 2009). Initially, we planned to use RNAi to explore the role played by individual pointed-end complex subunits in these events. Because both Arp11 and p62 were found to be necessary for dynactin stability, it was not possible to examine their functions selectively. We refocused our efforts on evaluating the contribution to dynein-based events of the dynactin molecule as a whole (using Arp11 knockdown) and compared this with the effect of loss of the p27/p25 heterodimer (using p27 knockdown) or p150Glued (using p150Glued knockdown). Because the effects of p62 or p25 depletion were essentially the same as Arp11 or p27 depletion, respectively, we limited most of our remaining RNAi work to Arp11 and p27. To explore the functions of p62 and Arp11 in specific events, we used protein overexpression.
Dynactin is essential for mitosis (Echeverri et al., 1996; Gaglio et al., 1996; Howell et al., 2000; Yang et al., 2007, 2008; Rome et al., 2010), so we first examined mitotic spindle formation (Figure 3). As expected for cells lacking dynactin, profound spindle defects were seen following Arp11 siRNA treatment, with many cells exhibiting multipolar spindles. A similar result was obtained for p62 depletion. Depletion of p150Glued also yielded severe spindle assembly defects, consistent with the fact that this subunit is required for dynein binding. By contrast, cells depleted of p27 and p25 showed normal spindle morphology. This suggests that the p27/p25 dimer is not required for the dynactin-dependent events of mitotic spindle assembly and spindle pole focusing. Mitotic spindle structure also appeared normal in cells overexpressing either p62 or Arp11 (unpublished data).
FIGURE 3:
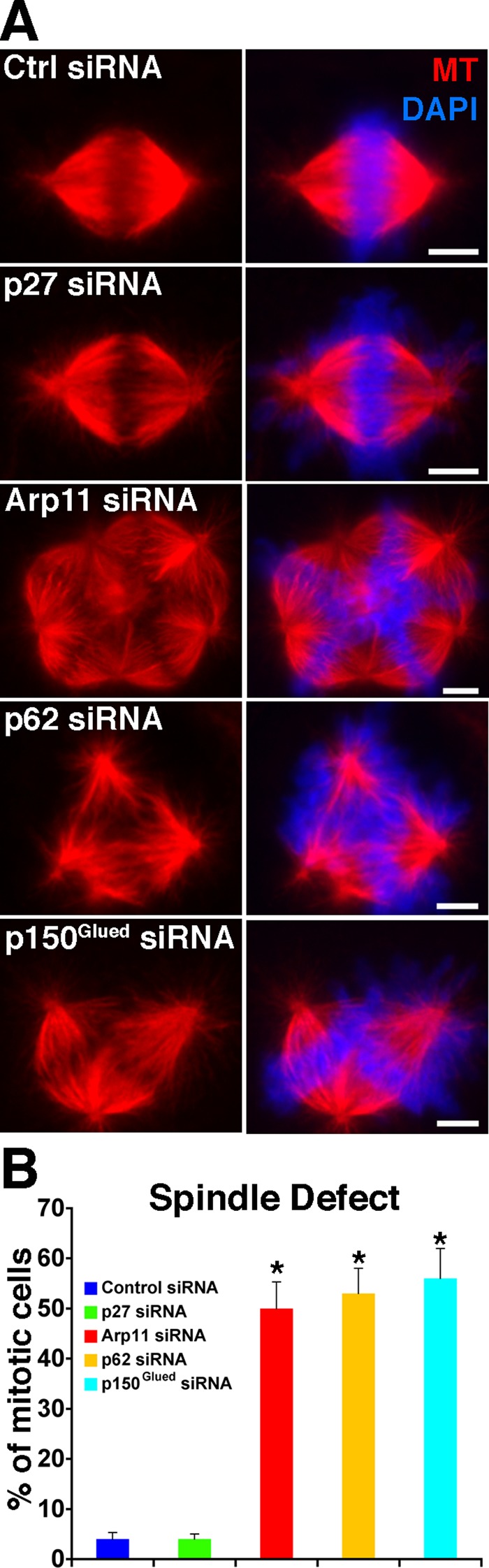
Effect of dynactin subunit depletion on mitotic spindle formation. (A) siRNA-treated Cos7 cells were stained for microtubules (red) and chromatin (DAPI, blue), and mitotic figures (shown) were identified by inspection. Scale bar: 5 μm. (B) Quantitative analysis of spindle disorganization (mean ± SD of three independent experiments; n > 300 mitotic cells/condition). Asterisks indicate values that are significantly different from controls (p < 0.05).
Conditions that impair dynein or dynactin structure or function commonly lead to redistribution of endomembrane compartments from the cell center to the periphery, coupled with a cessation of movement (Burkhardt et al., 1997; Valetti et al., 1999; LaMonte et al., 2002; King et al., 2003; Wassmer et al., 2009). To evaluate how dynactin subunit loss affects membrane organization, we examined the steady-state localizations of the Golgi complex (giantin), late endosomes (LAMP1), and recycling endosomes (transferrin receptor [TfR]) via immunofluorescence. We were surprised to find the distributions of these markers to be unchanged (Figure S2). This suggests that the compartments have dynactin-independent mechanisms for recruiting dynein or the dynactin bound to them turns over very slowly, which would make it resistant to siRNA treatment.
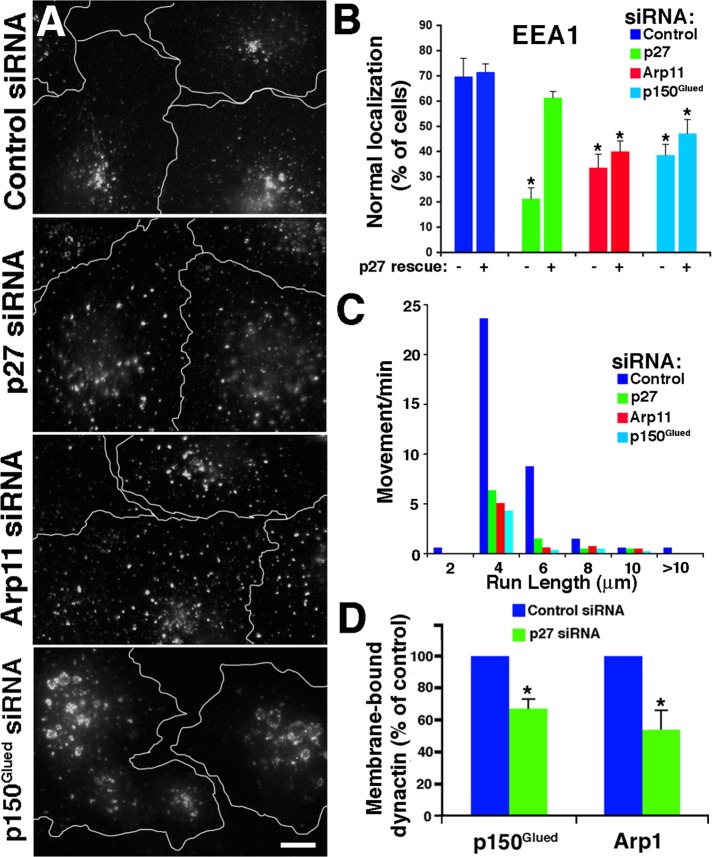
The p25 orthologue in the filamentous fungus, A. nidulans, contributes to the association of dynein with Rab5-positive (i.e., early) endosomes (Zhang et al., 2011). To explore whether p27/p25 participates in early endosome binding of dynactin in metazoan cells, we examined the steady-state localization and motile behavior of early endosome antigen 1 (EEA1)-positive endosomes (Figure 4). For comparison, we evaluated cells treated with Arp11 or p150Glued siRNAs. Cells lacking dynactin (Arp11 RNAi) showed a redistribution of EEA1 punctae from the cell center toward the periphery (Figure 4A). The EEA1-positive structures in cells depleted of p150Glued were also redistributed toward the periphery, and appeared swollen and exhibited patchy, irregular staining. Early endosome distribution in cells depleted of p27 and p25 was very similar to what was seen in cells depleted of Arp11, with EEA1 punctae distributed throughout the cell. Cotransfection with a siRNA-resistant p27 “rescue” plasmid returned EEA1 localization to normal (Figure 4B).
FIGURE 4:
Effect of dynactin subunit depletion on early endosome localization and motility. (A) siRNA-treated Cos7 cells were stained for early endosomes (EEA1). Scale bar: 5 μm. (B) Quantitative analysis of the effects of dynactin subunit depletion on the steady-state localization of EEA1-positive early endosomes. The left-hand bars (−) correspond to cells transfected with siRNAs alone; the right-hand bars (+) correspond to cells cotransfected with a cDNA encoding an siRNA-resistant form of p27. The values shown are the mean ± SD from three independent experiments (n > 300 cells per condition). (C) Motility of GFP-EEA1–labeled particles (from five cells, 7 min 20 s total per condition). (D) Quantitative analysis of membrane-associated dynactin subunits in cells depleted of p27/p25. Membranes were prepared by flotation (see Materials and Methods), and p150Glued and Arp1 levels were determined by quantitative immunoblotting (mean ± SD; three independent experiments). (B and D) *, Values significantly different from controls (p < 0.05).
To determine whether redistribution to the periphery reflected a suppression of dynactin-dependent movement, we imaged live cells expressing EEA1–green fluorescent protein (EEA1-GFP). In control cells, fluorescent punctae were seen to undergo long-range (>2 μm) translocations at velocities of 0.5 ± 0.3 μm/s (Figure 4C and Supplemental Movies S1–S4). Transfection with any dynactin subunit siRNA yielded a profound inhibition of motility, with fewer than 20% of the punctae showing normal movement.
Cells depleted of p27 and p25 contain normal amounts of dynactin (Figure 2), but their early endosome movement is profoundly impaired. This suggests that p27 and/or p25 are required for dynactin binding to early endosomes, a possibility we wished to explore further. Immunostaining does not allow detection of dynein or dynactin on membranes, so we used a biochemical approach. Cytoplasmic membranes isolated from cells treated with control or p27 siRNAs were floated into a sucrose step gradient and then harvested, and dynactin subunit levels were determined using quantitative immunoblotting. Although levels of both p150Glued and Arp1 were unchanged in p27/p25-depleted cell lysates (Figure 2B) and cytosols (unpublished data), both these subunits were significantly reduced in the membrane samples, as compared with controls (p150Glued, 67 ± 6% of control; Arp1, 54 ± 12% of control; Figure 4D).
p27 and p25 are required for motility of a subset of endosomes
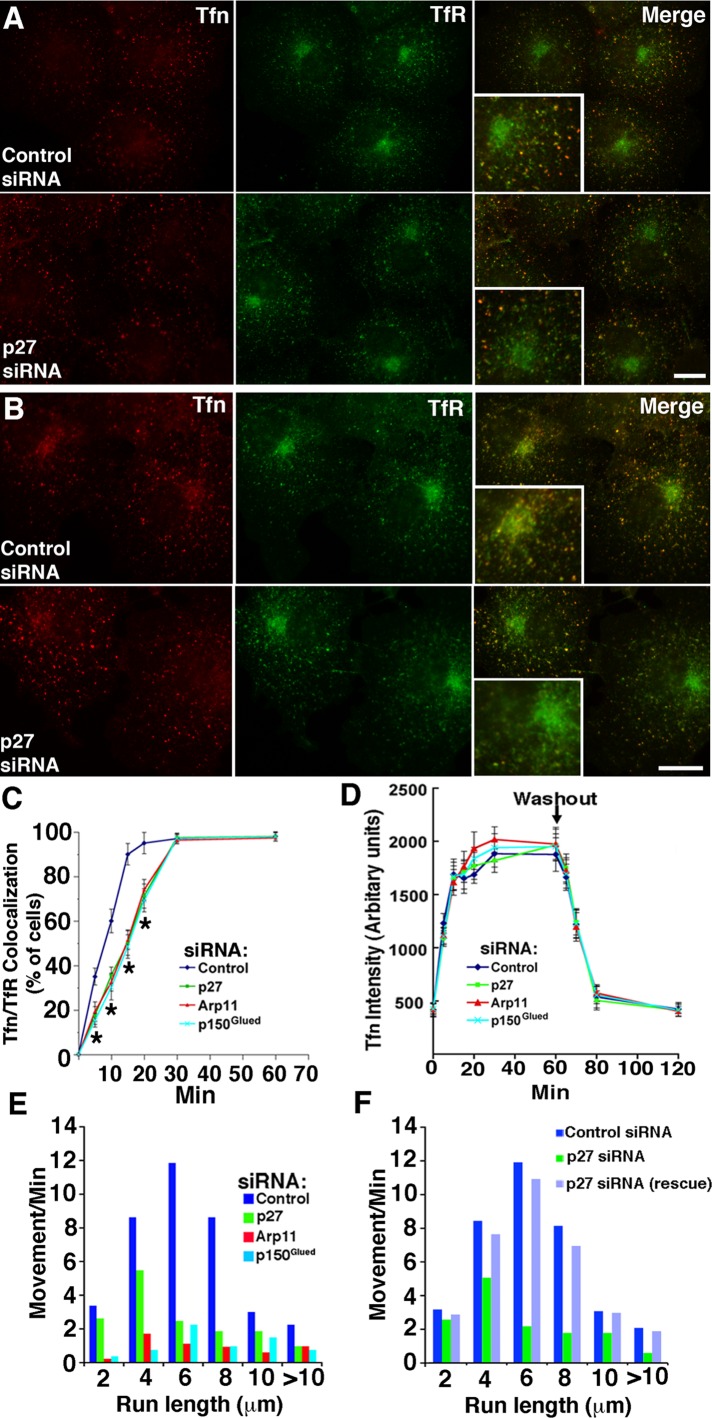
Our initial survey of membrane compartments (Figure S2) revealed no discernible effect of dynactin subunit depletion on the steady-state distribution of LAMP1-positive late endosomes or TfR-positive recycling endosomes. However, antibodies to LAMP1 and TfR label a variety of structures, many of which are known to be nonmotile. To assess the dynamic pool of late and recycling endosomes, we used externally added probes (Figures 5 and 6). Cells were pulse-labeled with fluorescently labeled α2-macroglobulin (α2M) or transferrin (Tfn), incubated for increasing chase times, and then fixed and stained with antibodies to follow trafficking of the probes through different endocytic compartments. In control cells, both Tfn and α2M first appeared in structures scattered throughout the cell (Figures 5A and 6A) that colocalized with the early endosome marker EEA1. Over time, Tfn accumulated in TfR-positive structures, and α2M accumulated in LAMP-1-positive structures. Both labeled compartments were concentrated near the cell center in agreement with previous reports that the structures carrying the probes undergo centripetal, dynein/dynactin-based movement (Yamashiro et al., 1989; Gruenberg and Maxfield, 1995). The redistribution of Tfn and α2M punctae to the cell center was impaired in cells treated with either Arp11 or p150Glued siRNAs (Figures 5C and 6, A and B). This verifies the importance of dynactin and its p150Glued subunit for motility of late and recycling endosomes (Valetti et al., 1999). Redistribution of Tfn particles, but not late endosomes carrying α2M, was delayed in cells depleted of p27/p25 (Figures 5, A and B, and 6, A and B). This suggests that the p27/p25 complex is required for binding of dynactin and recruitment of dynein to the vesicles that recycle Tfn to the cell surface, but not late endosomes.
FIGURE 5:
Tfn dynamics in cells depleted of dynactin subunits. (A and B) siRNA-treated Cos7 cells were incubated with Alexa Fluor 546-Tfn for (A) 5 min and (B) 15 min and then fixed and costained for TfR. Scale bar: 5 μm. The insets are 2×. (C) Cells (labeled with Tfn for increasing times) exhibiting colocalization of Tfn with perinuclear TfR were counted (mean ± SD of three independent experiments; n > 300 cells/condition). *, Values significantly different from controls (p < 0.05). (D) Kinetics of Tfn uptake and recycling. Cells were labeled with Tfn for increasing lengths of time (0–60 min). At the time points indicated, cells were fixed, and Tfn fluorescence (50 cells/condition) was quantified. (E) Analysis of particle movement in cells labeled with Alexa Fluor 555-Tfn for 2 min, then imaged in the interval between 10 and 15 min of chase. The data represent particle movements occurring in a total of 800 s of observation (80 s/cell, 10 cells/condition). (F) Cos7 cells cotransfected with control or p27 siRNA, plus pCAGIG-p27 as indicated (rescue). Data were collected as in (E).
FIGURE 6:
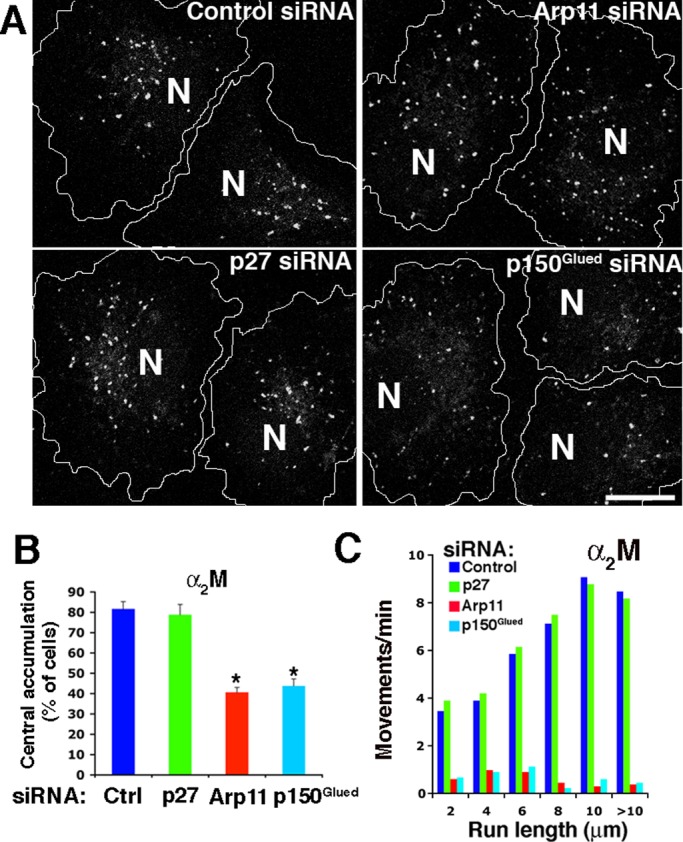
Late endosome dynamics in cells depleted of dynactin subunits. (A) siRNA-treated Cos7 cells were labeled for 30 min with Alexa Fluor 555-α2M, chased for 90 min, and then fixed. Cell borders are indicated. N, nucleus. Scale bar: 5 μm. (B) Quantitative analysis of α2M redistribution (mean ± SD from three independent experiments; n > 300 cells per condition). *, Values significantly different from controls (p < 0.05). (C) Analysis of late endosome movement in living cells labeled as in (A). The data represent particle movements occurring in a total of 800 s of observation (80 s/cell, 10 cells/condition).
To investigate this more deeply, we evaluated whether dynactin subunit depletion affected the kinetics of Tfn uptake and recycling. We observed no change in the Tfn cycle (Figure 5D), as expected based on a previous study in which dynamitin overexpression was found to have no effect (Valetti et al., 1999). We then examined the movement of Tfn punctae. Cells were pulse-labeled with fluorescent Tfn for 2 min and then chased for 10–15 min to capture motile events in the interval before the probe recycled completely out of the cell. As expected, given the impaired trafficking of Tfn to perinuclear TfR-positive structures (Figure 5C), the movement of Tfn punctae was markedly reduced in cells depleted of dynactin subunits (Figure 5E and Movies S5–S8). Expression of siRNA-resistant p27 in p27/p25-depleted cells fully restored Tfn particle movement (Figure 5F). These results verify that dynactin is required for movement of Tfn-loaded endosomes and strongly suggest that p27 and/or p25 are required for proper association of dynactin and dynein with these structures.
Redistribution of late endosomes loaded with α2M to the cell center was impaired in cells treated with either Arp11 or p150Glued siRNAs but not in cells depleted of p27/p25 (Figure 6B). This suggests that p27 and p25 are not necessary for late endosome motility. To test this directly, we used live-cell imaging. Late endosomes loaded with α2M for 30 min, which was followed by a 90-min chase, showed profoundly reduced long-range motility in cells treated with either Arp11 or p150Glued siRNAs (Figure 6C and Movies S9–S12). Similar results were obtained when endosomes were labeled using LAMP1-GFP (unpublished data). By contrast, long-range late endosome movement appeared completely normal in cells depleted of p27/p25. This suggests that p27 and p25 are not required for dynactin binding to late endosomes and subsequent dynein recruitment. The finding that p27 and p25 are required for long-range translocation of recycling endosomes, but not late endosomes, provides the first demonstration that dynactin can discriminate among vesicular cargoes.
p62 and Arp11 contribute to cell cycle–dependent nuclear envelope targeting of dynactin and dynein
As mentioned earlier, membrane-associated dynactin and dynein cannot be detected reliably via immunostaining. Instead, what is typically observed is staining of microtubule plus ends and centrosomes (Clark and Meyer, 1992; Quintyne et al., 1999; Quintyne and Schroer, 2002; Valetti et al., 1999; Vaughan et al., 1999). Dynactin localization and function at these sites and on membranes can be perturbed by overexpression of some dynactin subunits, but neither p62 nor Arp11 appears to have a strong effect (Quintyne et al., 1999; Quintyne and Schroer, 2002; Eckley and Schroer, 2003). Dynein also localizes to the nuclear envelope in a subpopulation of cells that correspond to those in late G2, just prior to mitotic prophase (Salina et al., 2002; Hebbar et al., 2008; Splinter et al., 2010; Bolhy et al., 2011). This has been reported to involve the dynein-binding protein Bicaudal D2 (BICD2; Splinter et al., 2010), but the role of dynactin has not been explored, nor is it known what subunit(s) might participate.
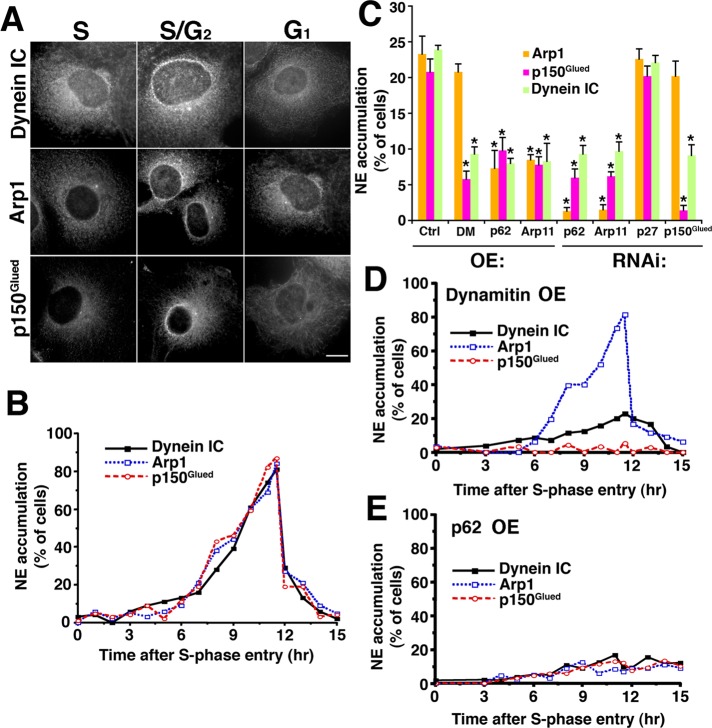
We first sought to determine whether dynactin associates with the nuclear envelope in a cell cycle–dependent manner. Cos7 cells were synchronized at the G1/S boundary, using a double thymidine block, and were then allowed to pass into S phase, through mitosis, and into G1 of the next cell cycle. Cells fixed at different time points were then immunostained and scored for a perinuclear ring of dynein or dynactin (Figure 7, A and B). Neither dynein nor dynactin accumulated on the nuclear envelope until several hours after S-phase entry. From this point on, nuclear envelope staining for both dynein and dynactin was observed in an increasing number of cells until nuclear envelope breakdown occurred (11–11.5 h after S-phase entry; Quintyne and Schroer, 2002). When the nuclear envelope reformed in early G1 of the next cell cycle, the typical subcellular distributions of both dynein and dynactin were reestablished. Parallel analysis of asynchronous cell populations revealed that ∼20–25% of cells exhibited nuclear envelope staining for dynein and dynactin (Figure 7C, “Ctrl”). We conclude that this represents the proportion in late S/G2.
FIGURE 7:
Effect of dynactin subunit RNAi and overexpression on dynein and dynactin binding to the nuclear envelope. (A and B) Cell cycle–dependent recruitment of dynactin and dynein to the nuclear envelope. (A) Cos7 cells were synchronized using a double thymidine block, released, and then fixed and labeled with antibodies to DIC or dynactin subunits (Arp1, p150Glued) at different times after release (S = 0 h; S/G2 = 10 h; G1 = 17 h). Scale bars: 5 μm. (B) Percentage of cells showing nuclear accumulation of dynein, Arp1, or p150Glued (n = 200 per time point). (C) Unsynchronized Cos7 cells were transfected with siRNAs or cDNAs encoding GFP-tagged versions of the subunits indicated, fixed, stained for dynein or dynactin, and then scored for nuclear envelope accumulation. *, Values significantly different from controls (p < 0.05). (D and E) Cos7 cells were synchronized at the G1/S boundary by double thymidine block, microinjected with GFP-tagged dynamitin (D) or p62 (E) expression plasmids, and released from the block for increasing times; the appearance of dynein or dynactin subunits on the nuclear envelope was evaluated as in (A).
To evaluate the roles played by individual dynactin subunits in nuclear envelope binding, we used a combination of RNAi and protein overexpression. Because p150Glued is the dynactin subunit that binds dynein, we expected that p150Glued depletion would abrogate dynein binding to the nuclear envelope. Cells treated with p150Glued siRNAs showed normal levels of Arp1 on the nuclear envelope, but dynein was significantly reduced (Figures 7C and S3B), as predicted. This indicates that dynactin that lacks p150Glued can still bind the nuclear envelope via the Arp1 filament. Cells overexpressing dynamitin, which dissociates p150Glued from the Arp1 filament (Echeverri et al., 1996; Melkonian et al., 2007), also exhibited normal Arp1 binding to nuclei, but p150Glued and dynein were diminished (Figure 7C). To verify that recruitment of the Arp1 filament to the nuclear envelope was cell cycle–dependent, we synchronized cells and then overexpressed dynamitin acutely via nuclear cDNA microinjection. Arp1 accumulated at the nuclear envelope over the same time course as in control cells, but p150Glued and dynein accumulation was dramatically reduced (Figure 7D). Together these findings indicate that the binding of dynein to nuclei is strongly enhanced by cell cycle–dependent binding of dynactin and utilizes p150Glued.
Dynactin binding to nuclei appears to be mediated by one or more components of the Arp1 filament. p62 overexpression significantly reduces dynein recruitment to the nuclear envelope (Salina et al., 2002), suggesting that the pointed-end complex plays a role. As expected, cells treated with siRNAs to either p62 or Arp11 showed minimal recruitment of Arp1 to the nuclear envelope, and p150Glued and dynein were significantly reduced as well (Figures 7C and S3).
To learn more about the roles played by p62 and Arp11 in nuclear envelope binding of dynactin and dynein, we overexpressed the two subunits in asynchronous cells and evaluated the effects (Figure 7C). About half of cells expressing p62 and a quarter of cells expressing Arp11 exhibited nuclear accumulation of the overexpressed protein (Figure S3A). Cells exhibiting this staining pattern contained duplicated centrosomes that stained for dynein, indicating they were primarily in late S/G2 (Quintyne and Schroer, 2002). This indicates that p62 and Arp11 by themselves have the capacity to bind the nuclear envelope in a cell cycle–dependent manner. Nuclear envelope accumulation is also seen in cells depleted of dynactin via Arp11 or p62 siRNA treatment, indicating that incorporation into dynactin is not required (Figure S4). Very few cells (<10%) overexpressing p62 or Arp11 exhibited nuclear envelope staining for dynactin or dynein (Figure 7C). This suggests that free p62 and Arp11 bind the nuclear envelope, which interferes with dynactin binding and subsequent dynein recruitment. To verify that nuclear envelope binding was cell cycle–dependent, we examined synchronized cells in which p62 had been acutely expressed by nuclear cDNA microinjection. Neither dynactin nor dynein accumulated significantly on nuclei at any cell cycle stage (Figure 7E), indicating that free p62 can indeed interfere with dynein/dynactin recruitment to nuclei in late G2.
To determine whether other components of the entire pointed-end complex were required for nuclear envelope binding, we evaluated the effect of p27/p25 depletion. The cells were found to exhibit normal staining for both dynactin and dynein (Figures 7C and S3), indicating that the p27/p25 component of the pointed-end complex is not required. Taken together, these experiments demonstrate that dynactin binds the nuclear envelope in a cell cycle–dependent manner and that the pointed-end complex subunits p62 and Arp11 play important roles in this process. Dynactin binding to the nuclear envelope involves different subunits from those that target dynactin to early and recycling endosomes, indicating that the pointed-end complex is a pleiotropic targeting module that supports multiple, distinct binding activities.
DISCUSSION
In this study, we explored the function and organization of dynactin's pointed-end complex. This part of the dynactin molecule contains three subunits, p62, p27, and p25, not present in yeasts and thus presumed not to be required for dynactin's most fundamental role as a dynein activator. The fourth pointed-end complex subunit, Arp11 (Arp10p in yeast), binds Arp1 directly and serves to cap the end of the filament (Eckley and Schroer, 2003; Clark and Rose, 2006). p62 has also been reported to bind Arp1 (Garces et al., 1999; Karki et al., 2000). p27 and p25 have been proposed to contribute to, and perhaps specify, the association of dynactin with dynein cargoes (Schroer, 2004). We find that the pointed-end complex comprises separable entities that contribute in distinct ways to dynactin integrity and function. Arp11 and p62 are required for dynactin stability, and thus all dynactin-dependent behaviors. They also allow dynactin to bind the nuclear envelope just prior to mitosis, which contributes to dynein recruitment and efficient nuclear envelope breakdown. The p27/p25 heterodimer “fine-tunes” dynein/dynactin activity. It can be removed without affecting dynactin stability and most dynactin-dependent, dynein-based motile phenomena. However, the binding of dynactin to a subset of endocytic membranes, and movement of these same membranes, is impaired. Although p27 and p25 may not be required for assembly of mitotic spindles, they are necessary for full recruitment of the important cell cycle kinase Plk1 to kinetochores in mitotic cells. Plk1 binding to p27 is required for proper execution of the spindle assembly checkpoint (Yeh et al., unpublished data).
Our protein cross-linking results provide new insights about the intermolecular interactions that occur among pointed-end complex subunits and allow us to speculate on how they associate with other dynactin subunits. The findings that p62 contacts Arp11 and p27 contacts p25 are consistent with the results of siRNA and overexpression experiments in which loss or overabundance of either binding partner phenocopies the other. Both Arp11 and p62 have been reported to interact with Arp1 (Garces et al., 1999; Karki et al., 2000; Eckley and Schroer, 2003). If these proteins favor Arp1 stability, this would explain why loss of either leads to loss of the entire Arp1 minifilament. p27 and p25 are not essential for dynactin integrity, but instead appear to depend on association with dynactin for their own stability. Our cross-linking data suggest that the p27/p25 heterodimer is bound to the rest of the dynactin molecule via an interaction between p25 and Arp11. It is not uncommon for components of protein complexes to show interdependent stability. Depletion of cytoplasmic dynein heavy chain results in loss of other dynein subunits (Palmer et al., 2009), and the Arp2/3 complex (Steffen et al., 2006), retromer (Seaman, 2004), and septins (Estey et al., 2010) show a similar behavior.
A well-known consequence of dynactin or dynein inhibition is the fragmentation and/or mislocalization of endomembrane compartments (Burkhardt et al., 1997; Valetti et al., 1999; King et al., 2003). We were surprised to find that many compartments whose steady-state localizations are widely assumed to depend on dynactin (e.g., the Golgi complex, late endosomes, the TfR-positive recycling compartment) were insensitive to the loss of p150Glued or the entire dynactin molecule. This may reflect the fact that a considerable subset of the structures that comprise these organelles is not motile, as observed when they are tagged with GFP reporters (Wassmer et al., 2009). This observation is also consistent with the notion that some compartments, such as the Golgi complex, have dynactin-independent mechanisms for dynein binding (Yadav et al., 2012). Additional motile and/or tethering activities may also contribute to steady-state morphology. The dynactin associated with these organelles may also be exceptionally stable; it is known that motor complexes that have been assembled onto membranes do not readily exchange with the cytosolic pool (Niclas et al., 1996), which itself can have a half-life of a day or more (Brown et al., 2005). If the membrane-associated pool is even more long-lived, it would be largely insensitive to RNAi.
The full range of mechanisms by which dynactin and dynein bind membranes is still being determined, and it is clear that binding modes vary among organelles. Associations between Arp1 and Golgi-associated spectrin have been reported to contribute to Golgi targeting (Holleran et al., 1996, 2001; Muresan et al., 2001); other spectrin isoforms may allow dynactin to bind elsewhere. Dynactin–Golgi binding has also been suggested to involve Rab6 (Short et al., 2002), and dynein can bind the Golgi complex directly via golgin-160 (Yadav et al., 2012). p150Glued, which has been identified as a binding partner for peripherally associated membrane proteins in a number of studies, is thought to associate with Rab7-RILP on late endosomes (Jordens et al., 2001; Johansson et al., 2007; Rocha et al., 2009), Jip4/Arf6 on recycling endosomes (Montagnac et al., 2009), the sorting nexin SNX6 (Wassmer et al., 2009), and huntingtin and its associated protein HAP1 (Engelender et al., 1997; Caviston et al., 2007). These proteins all bind p150Glued in the same vicinity, suggesting a generic and possibly mutually exclusive mode of interaction.
The pointed-end complex subunits p27 and/or p25 provide an additional binding mode that may involve interaction with integral components of the membrane. Although these proteins are not found in yeasts, the dependence of organelle movement and distribution on p25 in the filamentous fungi A. nidulans and N. crassa (Lee et al., 2001; Zhang et al., 2011) suggests that membrane binding is a highly conserved function. It is not clear from our work whether p27, p25, or both are most important, because depletion of either results in loss of the other. A common feature of all p25 sequences is an abundance of hydrophobic residues, suggesting that this protein might interact with membranes directly.
The present study examines the dynein-targeting mechanism that brings the nascent spindle toward the nucleus to facilitate nuclear envelope breakdown. Although we have pinpointed the dynactin subunits p62 and Arp11 as the components that allow cell cycle–dependent binding, the nuclear envelope binding partner(s) remains unknown. Dynactin perturbation or depletion does not completely prevent dynein binding to the nuclear envelope, indicating that dynein can be recruited in multiple ways. Cell cycle–dependent recruitment of the dynein/dynactin complex to the nucleus has been reported to involve binding between the nuclear pore complex component, RanBP2/Nup358, and BICD2, a scaffolding protein that stabilizes the dynein/dynactin complex via interactions with subunits other than p62 and Arp11 (Splinter et al., 2010). The dynein pathway components NudE/Nde1 and Lis1 also contribute to dynein and dynactin recruitment to the nuclear envelope in prophase (Hebbar et al., 2008), once again via interactions with the nuclear pore complex (Bolhy et al., 2011). In all cases, the molecular basis of cell cycle–dependent binding remains undefined, although it is presumed to involve cell cycle phosphorylation of dynactin and/or its binding partner. Although both p62 and Arp11 have been reported to be phosphorylated (Hoffert et al., 2006; Rigbolt et al., 2011), this is not at Cdk1 consensus sites and neither protein appears in published mitotic phosphoproteomes. Regardless, phosphorylation may contribute to the regulation of these proteins’ binding activities. More analysis will be needed to clarify how dynactin mediates the dynein-based motile activities that underlie membrane organization and dynamics.
MATERIALS AND METHODS
Antibodies
Antibodies used in this study were as follows: p150Glued: mAb P41920 (BD Biosciences, Franklin Lakes, NJ); pAb UP502 (a gift from E. Holzbaur, University of Pennsylvania); Arp1: mAb 45A, rabbit antibody to recombinant Arp1 (a gift from J. Lees-Miller, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY); p62: mAb 62B (Schafer et al., 1994; and ProteintechGroup, Chicago, IL); Arp11: an affinity-purified rabbit antibody (provided by M. Way, Cancer Research UK, London, UK; and Proteintech Group); dynamitin: clone 25 (BD Biosciences); dynein IC: mAb 74.1 (Millipore, Billerica, MA); p27: mAb 27A was generated in a standard hybridoma screen in which animals were immunized using pointed-end complex purified from bovine brain dynactin as in Eckley et al. (1999); p25: pAb JH3350 was raised by immunizing rabbits with gel band–purified mouse p25 expressed in Escherichia coli and Proteintech Group; tubulin: α-tubulin mAb DM1A (Sigma-Aldrich, St. Louis, MO); antibody YL1/2 (Serotec, Oxford, UK); 650952 rabbit anti-tubulin (ICN Pharmaceuticals, Costa Mesa, CA); affinity-purified rabbit antibody against peptide KVEGEGEEEGEEY (a gift from E. Karsenti, European Molecular Biology Laboratory, Heidelberg, Germany); mAb against giantin (a gift from A. Linstedt, Carnegie Mellon University, Pittsburgh, PA); GFP: rabbit pAb (Invitrogen, Carlsbad, CA); rabbit anti-phosphohistone 3 (Ser-10; Sigma-Aldrich); calnexin antibody H-70 (Santa Cruz Biotechnology, Santa Cruz, CA); EEA1: clone 14 (BD Biosciences). The transferrin receptor mAb (clone H68.4) was from Invitrogen. LAMP-1 (clone H4A3) was from the Developmental Studies Hybridoma Bank (Iowa City, IA). Alexa Fluor 488–, Alexa Fluor 568–, and Alexa Fluor 633–conjugated secondary antibodies were purchased from Invitrogen, and alkaline phosphatase–conjugated secondary antibodies were purchased from Tropix (Carlsbad, CA).
Chemical cross-linking
Purified bovine brain dynactin (Bingham et al., 1998) was cross-linked using BMH (Pierce, Rockford, IL), as previously described (Mullins et al., 1997). Stock solutions of BMH (15 mM = 10X) were prepared in dry dimethyl sulfoxide (DMSO) immediately before use. Dynactin (10 μg) was diluted to 0.3 mg/ml in PMEE buffer (35 mM PIPES, 5 mM MgSO4, 1 mM ethylene glycol tetraacetic acid [EGTA], 0.5 mM EDTA, pH 7.2), and an aliquot was gently mixed with an appropriate volume of the cross-linker to yield a final volume of 50 μl or more. All cross-linking reactions were carried out for 1 h at room temperature and quenched with an equal volume of 1 M Tris-HCl (pH 8.0) on ice for 15 min before being mixed with SDS–PAGE loading buffer and separated on an acrylamide gel. Cross-linked species were identified by immunoblotting with antibodies to pointed-end complex subunits. An untreated control sample was incubated with dry DMSO, quenched, and analyzed in parallel with the treated samples.
Cell culture, transfection, microinjection, and synchronization
Cos7 and HeLa cells were grown in DMEM plus 1% glutamine, penicillin, streptomycin, and 10% fetal bovine serum. For live-cell imaging, cells were grown in HEPES-buffered DMEM in glass bottom dishes (MatTek, Ashland, MA). For overexpression, cDNA transfections and microinjections were performed as previously described (Quintyne et al., 1999; Quintyne and Schroer, 2002). Cell synchronization was performed as in (Quintyne and Schroer, 2002).
siRNAs and cDNAs
Negative control siRNA or siRNAs against dynactin subunits were synthesized and purchased from Ambion (Austin, TX) or Thermo Scientific (Waltham, MA). The following primers were used: negative control (5′-UAAGGCUAUGAAGAGAUACdTdT-3′), p150Glued (5′-GGUAUCUGACACGCUCCUAdTdT-3′ or 5′-GCAAGCGAGCAGAUCUAUGdTdT-3’), p27 (5′-CCACCUAAAGA AGACUAUGdTdG-3′), p25 (5′-GCAAAGGUGUUGCAUUCUUdTdT-3′), Arp11 (5′-GCGCGUACUUGCUUUGUAAdTdT-3′), and p62 (5′-GCUGGUGCAUCCAUCAGUAdTdT-3′).
cDNAs encoding GFP-p62, GFP-Arp11, and GFP-dynamitin were as previously described (GFP-p62: Salina et al., 2002; GFP-Arp11: Eckley and Schroer, 2003; GFP-dynamitin: Quintyne et al., 1999; Valetti et al., 1999). EEA1-GFP and LAMP1-GFP were kindly provided by S. Corvera (University of Massachusetts) and E. Dell'Angelica (University of California, Los Angeles), respectively.
For RNAi, Cos-7 cells were electroporated with a mixture of 1.33 nM siRNA and 5 μg plasmid DNA encoding a fluorescence protein reporter to allow transfected cells to be identified. Some experiments used pmRFP-N1 (red fluorescent protein; Clontech, Mountain View, CA); others used pCAGIG (GFP; Addgene, Cambridge, MA; Matsuda and Cepko, 2004). pCAGIG allows expression to be driven by the chicken actin promoter and provides coexpression of any protein of interest plus enhanced GFP via an internal ribosomal entry site. This vector was also used to generate the p27 rescue vector, pCAGIG–p27, as follows: the full-length open reading frame of mouse p27, which lacks the target sequence of the human p27 siRNA, was amplified from an expressed sequence tag clone (AA073653) and subcloned into EcoRI- and XhoI-digested pCAGIG.
For electroporation, 107 Cos-7 cells were resuspended in 0.5 ml OPTI-MEM and electroporated at 240 V using a BTX Electro Cell Manipulator 600 (Holliston, MA). Cells were plated on dishes with gelatin-coated coverslips and grown for 72 h before being processed for fluorescence or biochemical assays. For membrane isolation and dynactin subunit quantification, cells were electroporated with siRNAs alone.
Cell fractionation
For analysis of total dynactin subunit content, transfected cells on two 10-cm dishes were harvested and solubilized with lysis buffer (20 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 0.2% NP-40) containing protease inhibitors. Cell lysates were prepared by centrifugation at 44,700 rpm (189,435 × g [average]) in a Beckman SW55 Ti rotor for 30 min. For evaluation of dynactin integrity, supernatant proteins were sedimented into 5–20% sucrose gradients at 30,500 rpm (88,195 × g [average]) for 16 h, and gradient fractions were analyzed by immunoblotting.
For membrane isolation, cells were homogenized in TMEE buffer (50 mM Tris, pH 7.4, 5 mM MgSO4, 1 mM EGTA, 0.5 mM EDTA plus protease inhibitors) using a ball-bearing homogenizer. Nuclei were pelleted at 1600 rpm for 10 min at 4°C (Dynac 420101, 4-place, 50-ml swinging-bucket rotor; Franklin Lakes, NJ). Membranes were pelleted at 44,700 rpm for 30 min at 4°C (SW55Ti rotor). The pellet was suspended in 2 M sucrose in TMEE buffer at a ratio of 1:4 (vol/vol) and overlayered stepwise with 1.2 M and 0.2 M sucrose in TMEE. The gradient was centrifuged at 44,700 rpm for 60 min at 4°C (SW55Ti rotor), and the floated membranes were collected from the interface of the 1.2 M/0.2 M sucrose layer. Membranes were diluted to 700 μl in TMEE and repelleted at 44,700 rpm for 30 min at 4°C, and the final pellet was dissolved in 0.5% CHAPS in TMEE for 30 min on ice.
Quantitative immunoblotting
Quantitative immunoblotting was carried out using horseradish peroxidase–conjugated anti-mouse or anti-rabbit secondary antibody and ECL Plus (GE Healthcare, Waukesha, WI). Samples (15 μg protein) were serially diluted 1:2 and then subjected to SDS–PAGE and immunoblotting. Blots were scanned using a GE Typhoon 9410 in fluorescence mode using the following settings: laser: 457 nm; emission filter: 520BP 40; PMT: 650; sensitivity: normal; pixel size: 100 μm. Image files (TIFF) were exported into Image J (National Institutes of Health) for quantification. Levels of cytosolic and membrane-associated dynactin subunits were normalized to the levels of tubulin and calnexin, respectively, in the same samples run in parallel. Levels of 19S dynactin subunits were normalized to the level of DIC.
Immunofluorescence microscopy
Cells were fixed using −20°C methanol or 4% paraformaldehyde at room temperature in phosphate-buffered saline with 2 mM MgCl2 and 0.2 mM CaCl2, then permeabilized with 0.0375% saponin, and blocked with 0.5% bovine serum albumin. Images of immunostained cells were acquired on a Zeiss Axiovert 200 microscope equipped with a Plan-Apochromat 100×/1.4 numerical aperture (NA) objective and a Cooke SensiCam camera (ASI, Romulus, MI) binned 1 × 1. Saturation was avoided by selecting an exposure time on the basis of a histogram of the fluorescence intensity in each pixel. For quantitative analysis of fluorescence intensities, images from all samples were collected under identical illumination and exposure conditions. The images were exported and analyzed using Image J.
Endocytic probe uptake
Human α2M (Calbiochem, San Diego, CA) was conjugated with Alexa Fluor 555 using an Invitrogen protein-labeling kit according to the manufacturer's instructions. Tfn conjugated with Alexa Fluor 546 or Alexa Fluor 555 (only used for cells transfected with pCAGIG) was obtained from Invitrogen. Before Tfn uptake, cells were serum-starved (>1 h at 37°C). Analysis of Alexa Fluor 546-Tfn (10 μg/ml) uptake and recycling kinetics in cells cotransfected with siRNAs and pCAGIG was performed as described (Valetti et al., 1999). For quantification of the total fluorescence intensity of Tfn, the region of interest was defined by the cell margin.
Live-cell imaging was performed 72 h after cells were cotransfected with siRNA and the reporter pCAGIG (lacking the mouse p27 insert). Only cells expressing GFP were evaluated for particle motility. For visualization of recycling endosome motility, serum-starved cells were pulse-fed 10 μg/ml Alexa Fluor 555-Tfn for 2 min at 37°C; Tfn particle motility was then recorded in the interval between 10 and 15 min of chase. For visualization of late endosome motility, cells were fed 60–100 μg/ml Alexa Fluor 555-α2M for 30 min and imaged during an interval 90–120 min later.
Quantitative analysis of particle movements
Movies were acquired on a Zeiss LSM 510 VIS confocal microscope equipped with a Plan-Apochromat 63×/1.4 NA oil objective, Argon and He-Ne lasers (excitation at 488 and 543 nm, respectively), and a PMT detector. Laser-scanning microscopy image stacks of each time series were imported into Image J. Displacement was determined in two dimensions within the plane of focus of the objective. Kymographs measuring the displacement over time of moving particles were generated using the Multiple Kymograph plug-in (provided by J. Rietdorf and A. Seitz, EMBL; www.embl.de/eamnet/html/body_kymograph.html). Velocity, which is directly proportional to the slope of the kymograph trace, was determined by calculating the arc tangent from linear regions of the kymograph, which represents directed motion (Gibbs et al., 2004). All motile particles from 10 independent siRNA-transfected cells were analyzed.
Supplementary Material
Acknowledgments
We thank Silvia Corvera, Esteban Dell'Angelica, Erika Holzbaur, Eric Karsenti, Jim Lees-Miller, Adam Linstedt, and Michael Way for cDNAs and antibody reagents. We are grateful to Irun Bhan and Frances Cheong for their corroborative work on p62 overexpression, to Steven Gill for his initial work on p62, to Formosa Chen for purification of p25 antibody, and to members of the Schroer laboratory for their advice and comments on the manuscript. This work was supported by a National Institutes of Health grant (RO1 GM 44589) to T.A.S.
Abbreviations used:
- α2M
α2-macroglobulin
- BICD2
Bicaudal D2
- BMH
bismaleimidohexane
- DIC
dynein IC
- DMSO
dimethyl sulfoxide
- EEA1
early endosome antigen 1
- EGTA
ethylene glycol tetraacetic acid
- GFP
green fluorescent protein
- NA
numerical aperture
- RNAi
RNA interference
- siRNA
small interfering RNA
- Tfn
transferrin
- TfR
transferrin receptor
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-07-0496) on August 23, 2012.
Present addresses: *Harriet L. Wilkes Honors College of Florida Atlantic University, Jupiter, FL 33458;
†Image Informatics and Computational Biology Unit, Laboratory of Genetics, National Institute on Aging, National Institutes of Health, Baltimore, MD 21224.
REFERENCES
- Bingham JB, King SJ, Schroer TA. Purification of dynactin and dynein from brain tissue. Methods Enzymol. 1998;298:171–184. doi: 10.1016/s0076-6879(98)98017-x. [DOI] [PubMed] [Google Scholar]
- Bolhy S, Bouhlel I, Dultz E, Nayak T, Zuccolo M, Gatti X, Vallee R, Ellenberg J, Doye V. A Nup133-dependent NPC-anchored network tethers centrosomes to the nuclear envelope in prophase. J Cell Biol. 2011;192:855–871. doi: 10.1083/jcb.201007118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CL, Maier KC, Stauber T, Ginkel LM, Wordeman L, Vernos I, Schroer TA. Kinesin-2 is a motor for late endosomes and lysosomes. Traffic. 2005;6:1114–1124. doi: 10.1111/j.1600-0854.2005.00347.x. [DOI] [PubMed] [Google Scholar]
- Burkhardt JK, Echeverri CJ, Nilsson T, Vallee RB. Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J Cell Biol. 1997;139:469–484. doi: 10.1083/jcb.139.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviston JP, Ross JL, Antony SM, Tokito M, Holzbaur EL. Huntingtin facilitates dynein/dynactin-mediated vesicle transport. Proc Natl Acad Sci USA. 2007;104:10045–10050. doi: 10.1073/pnas.0610628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SW, Meyer DI. Centractin is an actin homologue associated with the centrosome. Nature. 1992;359:246–250. doi: 10.1038/359246a0. [DOI] [PubMed] [Google Scholar]
- Clark SW, Rose MD. Arp10p is a pointed-end-associated component of yeast dynactin. Mol Biol Cell. 2006;17:738–748. doi: 10.1091/mbc.E05-05-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri CJ, Paschal BM, Vaughan KT, Vallee RB. Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J Cell Biol. 1996;132:617–633. doi: 10.1083/jcb.132.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckley DM, Gill SR, Melkonian KA, Bingham JB, Goodson HV, Heuser JE, Schroer TA. Analysis of dynactin subcomplexes reveals a novel actin-related protein associated with the Arp1 minifilament pointed end. J Cell Biol. 1999;147:307–320. doi: 10.1083/jcb.147.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckley DM, Schroer TA. Interactions between the evolutionarily conserved, actin-related protein, Arp11, actin, and Arp1. Mol Biol Cell. 2003;14:2645–2654. doi: 10.1091/mbc.E03-01-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelender S, Sharp AH, Colomer V, Tokito MK, Lanahan A, Worley P, Holzbaur EL, Ross CA. Huntingtin-associated protein 1 (HAP1) interacts with the p150Glued subunit of dynactin. Hum Mol Genet. 1997;6:2205–2212. doi: 10.1093/hmg/6.13.2205. [DOI] [PubMed] [Google Scholar]
- Estey MP, Di Ciano-Oliveira C, Froese CD, Bejide MT, Trimble WS. Distinct roles of septins in cytokinesis: SEPT9 mediates midbody abscission. J Cell Biol. 2010;191:741–749. doi: 10.1083/jcb.201006031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio T, Saredi A, Bingham JB, Hasbani MJ, Gill SR, Schroer TA, Compton DA. Opposing motor activities are required for the organization of the mammalian mitotic spindle pole. J Cell Biol. 1996;135:399–414. doi: 10.1083/jcb.135.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garces JA, Clark IB, Meyer DI, Vallee RB. Interaction of the p62 subunit of dynactin with Arp1 and the cortical actin cytoskeleton. Curr Biol. 1999;9:1497–1500. doi: 10.1016/s0960-9822(00)80122-0. [DOI] [PubMed] [Google Scholar]
- Gibbs D, Azarian SM, Lillo C, Kitamoto J, Klomp AE, Steel KP, Libby RT, Williams DS. Role of myosin VIIa and Rab27a in the motility and localization of RPE melanosomes. J Cell Sci. 2004;117:6473–6483. doi: 10.1242/jcs.01580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenberg J, Maxfield FR. Membrane transport in the endocytic pathway. Curr Opin Cell Biol. 1995;7:552–563. doi: 10.1016/0955-0674(95)80013-1. [DOI] [PubMed] [Google Scholar]
- Hebbar S, Mesngon MT, Guillotte AM, Desai B, Ayala R, Smith DS. Lis1 and Ndel1 influence the timing of nuclear envelope breakdown in neural stem cells. J Cell Biol. 2008;182:1063–1071. doi: 10.1083/jcb.200803071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffert JD, Pisitkun T, Wang G, Shen RF, Knepper MA. Quantitative phosphoproteomics of vasopressin-sensitive renal cells: regulation of aquaporin-2 phosphorylation at two sites. Proc Natl Acad Sci USA. 2006;103:7159–7164. doi: 10.1073/pnas.0600895103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holleran EA, Ligon LA, Tokito M, Stankewich MC, Morrow JS, Holzbaur EL. βIII spectrin binds to the Arp1 subunit of dynactin. J Biol Chem. 2001;276:36598–36605. doi: 10.1074/jbc.M104838200. [DOI] [PubMed] [Google Scholar]
- Holleran EA, Tokito MK, Karki S, Holzbaur ELF. Centractin (ARP1) associates with spectrin revealing a potential mechanism to link dynactin to intracellular organelles. J Cell Biol. 1996;135:1815–1829. doi: 10.1083/jcb.135.6.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BJ, Hoffman DB, Fang G, Murray AW, Salmon ED. Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J Cell Biol. 2000;150:1233–1250. doi: 10.1083/jcb.150.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M, Rocha N, Zwart W, Jordens I, Janssen L, Kuijl C, Olkkonen VM, Neefjes J. Activation of endosomal dynein motors by stepwise assembly of Rab7-RILP-p150Glued, ORP1L, and the receptor βIII spectrin. J Cell Biol. 2007;176:459–471. doi: 10.1083/jcb.200606077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordens I, Fernandez-Borja M, Marsman M, Dusseljee S, Janssen L, Calafat J, Janssen H, Wubbolts R, Neefjes J. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr Biol. 2001;11:1680–1685. doi: 10.1016/s0960-9822(01)00531-0. [DOI] [PubMed] [Google Scholar]
- Kardon JR, Vale RD. Regulators of the cytoplasmic dynein motor. Nat Rev Mol Cell Biol. 2009;10:854–865. doi: 10.1038/nrm2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki S, Holzbaur EL. Affinity chromatography demonstrates a direct binding between cytoplasmic dynein and the dynactin complex. J Biol Chem. 1995;270:28806–28811. doi: 10.1074/jbc.270.48.28806. [DOI] [PubMed] [Google Scholar]
- Karki S, Holzbaur ELF. Cytoplasmic dynein and dynactin in cell division and intracellular transport. Curr Opin Cell Biol. 1999;11:45–53. doi: 10.1016/s0955-0674(99)80006-4. [DOI] [PubMed] [Google Scholar]
- Karki S, Tokito MK, Holzbaur EL. A dynactin subunit with a highly conserved cysteine-rich motif interacts directly with Arp1. J Biol Chem. 2000;275:4834–4839. doi: 10.1074/jbc.275.7.4834. [DOI] [PubMed] [Google Scholar]
- King SJ, Brown CL, Maier KC, Quintyne NJ, Schroer TA. Analysis of the dynein-dynactin interaction in vitro and in vivo. Mol Biol Cell. 2003;14:5089–5097. doi: 10.1091/mbc.E03-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SJ, Schroer TA. Dynactin increases the processivity of the cytoplasmic dynein motor. Nat Cell Biol. 2000;2:20–24. doi: 10.1038/71338. [DOI] [PubMed] [Google Scholar]
- LaMonte BH, Wallace KE, Holloway BA, Shelly SS, Ascano J, Tokito M, Van Winkle T, Howland DS, Holzbaur EL. Disruption of dynein/dynactin inhibits axonal transport in motor neurons causing late-onset progressive degeneration. Neuron. 2002;34:715–727. doi: 10.1016/s0896-6273(02)00696-7. [DOI] [PubMed] [Google Scholar]
- Lee IH, Kumar S, Plamann M. Null mutants of the Neurospora actin-related protein 1 pointed-end complex show distinct phenotypes. Mol Biol Cell. 2001;12:2195–2206. doi: 10.1091/mbc.12.7.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Cepko CL. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci USA. 2004;101:16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkonian KA, Maier KC, Godfrey JE, Rodgers M, Schroer TA. Mechanism of dynamitin-mediated disruption of dynactin. J Biol Chem. 2007;282:19355–19364. doi: 10.1074/jbc.M700003200. [DOI] [PubMed] [Google Scholar]
- Miki H, Setou M, Kaneshiro K, Hirokawa N. All kinesin superfamily protein, KIF, genes in mouse and human. Proc Natl Acad Sci USA. 2001;98:7004–7011. doi: 10.1073/pnas.111145398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnac G, Sibarita JB, Loubery S, Daviet L, Romao M, Raposo G, Chavrier P. ARF6 Interacts with JIP4 to control a motor switch mechanism regulating endosome traffic in cytokinesis. Curr Biol. 2009;19:184–195. doi: 10.1016/j.cub.2008.12.043. [DOI] [PubMed] [Google Scholar]
- Mullins RD, Stafford WF, Pollard TD. Structure, subunit topology, and actin-binding activity of the Arp2/3 complex from Acanthamoeba. J Cell Biol. 1997;136:331–343. doi: 10.1083/jcb.136.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muresan V, Stankewich MC, Steffen W, Morrow JS, Holzbaur EL, Schnapp BJ. Dynactin-dependent, dynein-driven vesicle transport in the absence of membrane proteins: a role for spectrin and acidic phospholipids. Mol Cell. 2001;7:173–183. doi: 10.1016/s1097-2765(01)00165-4. [DOI] [PubMed] [Google Scholar]
- Niclas J, Allan VJ, Vale RD. Cell cycle regulation of dynein association with membranes modulates microtubule-based organelle transport. J Cell Biol. 1996;133:585–593. doi: 10.1083/jcb.133.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer KJ, Hughes H, Stephens DJ. Specificity of cytoplasmic dynein subunits in discrete membrane-trafficking steps. Mol Biol Cell. 2009;20:2885–2899. doi: 10.1091/mbc.E08-12-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintyne NJ, Gill SR, Eckley DM, Crego CL, Compton DA, Schroer TA. Dynactin is required for microtubule anchoring at centrosomes. J Cell Biol. 1999;18:321–334. doi: 10.1083/jcb.147.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintyne NJ, Schroer TA. Distinct cell cycle-dependent roles for dynactin and dynein at centrosomes. J Cell Biol. 2002;159:245–254. doi: 10.1083/jcb.200203089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigbolt KT, Prokhorova TA, Akimov V, Henningsen J, Johansen PT, Kratchmarova I, Kassem M, Mann M, Olsen JV, Blagoev B. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci Signal. 2011;4:rs3. doi: 10.1126/scisignal.2001570. [DOI] [PubMed] [Google Scholar]
- Rocha N, Kuijl C, van der Kant R, Janssen L, Houben D, Janssen H, Zwart W, Neefjes J. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150Glued and late endosome positioning. J Cell Biol. 2009;185:1209–1225. doi: 10.1083/jcb.200811005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rome P, Montembault E, Franck N, Pascal A, Glover DM, Giet R. Aurora A contributes to p150glued phosphorylation and function during mitosis. J Cell Biol. 2010;189:651–659. doi: 10.1083/jcb.201001144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JL, Wallace K, Shuman H, Goldman YE, Holzbaur EL. Processive bidirectional motion of dynein-dynactin complexes in vitro. Nat Cell Biol. 2006;8:562–570. doi: 10.1038/ncb1421. [DOI] [PubMed] [Google Scholar]
- Salina D, Bodoor K, Eckley DM, Schroer TA, Rattner JB, Burke B. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell. 2002;108:97–107. doi: 10.1016/s0092-8674(01)00628-6. [DOI] [PubMed] [Google Scholar]
- Schafer DA, Gill SR, Cooper JA, Heuser JE, Schroer TA. Ultrastructural analysis of the dynactin complex: an actin-related protein is a component of a filament that resembles F-actin. J Cell Biol. 1994;126:403–412. doi: 10.1083/jcb.126.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer T, Cheong FK-Y. Role of dynactin in dynein-mediated motility. In: King SM, editor. Dyneins: Structure, Biology and Disease. Oxford: Academic Press; 2012. pp. 505–522. [Google Scholar]
- Schroer TA. Dynactin. Annu Rev Cell Dev Biol. 2004;20:759–779. doi: 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- Schroer TA, Sheetz MP. Two activators of microtubule-based vesicle transport. J Cell Biol. 1991;115:1309–1318. doi: 10.1083/jcb.115.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MN. Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J Cell Biol. 2004;165:111–122. doi: 10.1083/jcb.200312034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short B, Preisinger C, Schaletzky J, Kopajtich R, Barr FA. The Rab6 GTPase regulates recruitment of the dynactin complex to Golgi membranes. Curr Biol. 2002;12:1792–1795. doi: 10.1016/s0960-9822(02)01221-6. [DOI] [PubMed] [Google Scholar]
- Splinter D, et al. Bicaudal D2, dynein, and kinesin-1 associate with nuclear pore complexes and regulate centrosome and nuclear positioning during mitotic entry. PLoS Biol. 2010;8:e1000350. doi: 10.1371/journal.pbio.1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen A, Faix J, Resch GP, Linkner J, Wehland J, Small JV, Rottner K, Stradal TE. Filopodia formation in the absence of functional WAVE- and Arp2/3-complexes. Mol Biol Cell. 2006;17:2581–2591. doi: 10.1091/mbc.E05-11-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valetti C, Wetzel DM, Schrader M, Hasbani MJ, Gill SR, Kreis TE, Schroer TA. Role of dynactin in endocytic traffic: effects of dynamitin overexpression and colocalization with CLIP-170. Mol Biol Cell. 1999;10:4107–4120. doi: 10.1091/mbc.10.12.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan KT, Tynan SH, Faulkner NE, Echeverri CJ, Vallee RB. Colocalization of cytoplasmic dynein with dynactin and CLIP-170 at microtubule distal ends. J Cell Sci. 1999;112:1437–1447. doi: 10.1242/jcs.112.10.1437. [DOI] [PubMed] [Google Scholar]
- Vaughan KT, Vallee RB. Cytoplasmic dynein binds dynactin through a direct interaction between the intermediate chains and p150Glued. J Cell Biol. 1995;131:1507–1516. doi: 10.1083/jcb.131.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassmer T, et al. The retromer coat complex coordinates endosomal sorting and dynein-mediated transport, with carrier recognition by the trans-Golgi network. Dev Cell. 2009;17:110–122. doi: 10.1016/j.devcel.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer CM, Karki S, Holzbaur EL. The p150Glued component of the dynactin complex binds to both microtubules and the actin-related protein centractin (Arp-1) Proc Natl Acad Sci USA. 1995;92:1634–1638. doi: 10.1073/pnas.92.5.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro DJ, Borden LA, Maxfield FR. Kinetics of α2-macroglobulin endocytosis and degradation in mutant and wild-type Chinese hamster ovary cells. J Cell Physiol. 1989;139:377–382. doi: 10.1002/jcp.1041390221. [DOI] [PubMed] [Google Scholar]
- Yadav S, Puthenveedu MA, Linstedt AD. Golgin160 recruits the dynein motor to position the Golgi apparatus. Dev Cell. 2012;23:153–165. doi: 10.1016/j.devcel.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Cameron LA, Maddox PS, Salmon ED, Danuser G. Regional variation of microtubule flux reveals microtubule organization in the metaphase meiotic spindle. J Cell Biol. 2008;182:631–639. doi: 10.1083/jcb.200801105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Tulu US, Wadsworth P, Rieder CL. Kinetochore dynein is required for chromosome motion and congression independent of the spindle checkpoint. Curr Biol. 2007;17:973–980. doi: 10.1016/j.cub.2007.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wang L, Zhuang L, Huo L, Musa S, Li S, Xiang X. Arp11 affects dynein-dynactin interaction and is essential for dynein function in Aspergillus nidulans. Traffic. 2008;9:1073–1087. doi: 10.1111/j.1600-0854.2008.00748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Yao X, Fischer L, Abenza JF, Penalva MA, Xiang X. The p25 subunit of the dynactin complex is required for dynein-early endosome interaction. J Cell Biol. 2011;193:1245–1255. doi: 10.1083/jcb.201011022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.