Abstract
Two major transcriptional regulators of Caenorhabditis elegans bodywall muscle (BWM) differentiation, hlh-1 and unc-120, are expressed in muscle where they are known to bind and regulate several well-studied muscle-specific genes. Simultaneously mutating both factors profoundly inhibits formation of contractile BWM. These observations were consistent with a simple network model in which the muscle regulatory factors drive tissue-specific transcription by binding selectively near muscle-specific targets to activate them. We tested this model by measuring the number, identity, and tissue-specificity of functional regulatory targets for each factor. Some joint regulatory targets (218) are BWM-specific and enriched for nearby HLH-1 binding. However, contrary to the simple model, the majority of genes regulated by one or both muscle factors are also expressed significantly in non-BWM tissues. We also mapped global factor occupancy by HLH-1, and created a genetic interaction map that identifies hlh-1 collaborating transcription factors. HLH-1 binding did not predict proximate regulatory action overall, despite enrichment for binding among BWM-specific positive regulatory targets of hlh-1. We conclude that these tissue-specific factors contribute much more broadly to the transcriptional output of muscle tissue than previously thought, offering a partial explanation for widespread HLH-1 occupancy. We also identify a novel regulatory connection between the BWM-specific hlh-1 network and the hlh-8/twist nonstriated muscle network. Finally, our results suggest a molecular basis for synthetic lethality in which hlh-1 and unc-120 mutant phenotypes are mutually buffered by joint additive regulation of essential target genes, with additional buffering suggested via newly identified hlh-1 interacting factors.
Gene networks that govern cell-type-specificity typically center around a few core transcription factors that interact directly, both physically and genetically, with “terminal differentiation” regulatory target genes (Davidson 2007). For the muscle gene network, these core factors are evolutionarily conserved in vertebrates and invertebrates, consisting of bHLH factors of the MyoD family and members of the MADS family (Fukushige et al. 2006). Decades of detailed genetic and molecular studies of selected “model” muscle genes showed that core factors interact physically and functionally with their transcriptional enhancers and promoters. This led to a parsimonious working model in which core factor occupancy specified all muscle-restricted transcription and thus defined the terminal differentiation state. It is now possible to test this and to probe more deeply how the core regulators act individually, additively, and/or synergistically on their targets. In principle, it is straightforward to build and compare a global physical map of factor occupancy determined by ChIP-seq (Johnson et al. 2007) with a corresponding perturbation map of factor function whose global output is measured by mRNA-seq (Mortazavi et al. 2008). Many differentiation systems now have good genomic maps of one kind but not the other due to various technical and biological limitations, but Caenorhabditis elegans bodywall muscle (BWM) is especially amenable to both kinds of mapping. In particular, its core BWM transcription factors, hlh-1 and unc-120, comprise a synthetic embryonic lethal pair. This permits each factor to be eliminated individually and the regulatory impact measured in muscle tissue, thus avoiding the problems of other systems in which mutation of a single factor eliminates the tissue entirely. This comparison can also address questions emerging from related systems, including mammalian myogenesis (Cao et al. 2010), in which factor occupancy maps are revealing much more pervasive physical occupancy across the genome than was initially expected.
Nematodes have three distinct muscle regulatory networks that establish and maintain the differentiated states for their respective tissues: bodywall muscle, nonstriated muscles (NSM), and pharyngeal muscle (PhM). Each core network has a dedicated transcription factor (hlh-1 in BWM, hlh-8 in NSM, ceh-22 in PhM) (Fig. 1A; Chen et al. 1992, 1994; Williams and Waterston 1994; Fukushige et al. 2006; Lei et al. 2009). These dedicated factors are joined by semidedicated factors expressed in multiple muscle types and muscle-associated cells (muscle-associated GLR cells, coelomocytes, and the contractile somatic gonad) but not in other tissues (unc-120 in both NSM and BWM) (Baugh et al. 2005a; Fukushige et al. 2006), and they are joined by more general factors that act in both nonmuscle and muscle tissues.
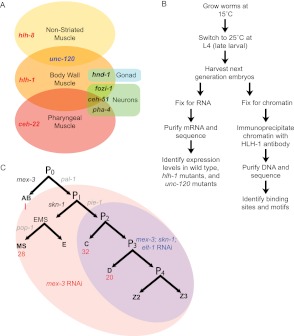
Figure 1.
Experimental flow and muscle differentiation network. (A) The three types of nematode muscle and their associated transcription factors. (B) The experiments performed across three RNAi conditions (mex-3; skn-1; elt-1 RNAi, mex-3 RNAi, and no RNAi) in N2, hlh-1(cc561), and unc-120(st364). (C) The embryonic development lineages with BWM cell number in normal embryos (red) are highlighted to show which lineages are enhanced under the RNAi conditions (mex-3 RNAi and mex-3; skn-1; elt-1 RNAi).
BWM is functionally analogous to the skeletal muscle of vertebrates and insects (Albertson and Thomson 1976; Chen et al. 1994; Fukushige et al. 2006), being the most prominent muscle in the animal by cell number and mass (81 embryonic and 14 post-embryonic BWM cells) (Sulston and Horvitz 1977; Sulston et al. 1983). Five transcription factors are known to regulate BWM: hlh-1, unc-120, hnd-1, ceh-51, and fozi-1 (Fig. 1A; Harfe et al. 1998a; Mathies et al. 2003; Fukushige et al. 2006; Amin et al. 2007; Broitman-Maduro et al. 2009). Ectopic expression of some can convert early blastomeres to muscle, based on myosin reporter assays, and hlh-1 is the most efficient, with all five up-regulating endogenous hlh-1 and unc-120 (Fukushige and Krause 2005; Fukushige et al. 2006; Broitman-Maduro et al. 2009). unc-120 is the most critical hlh-1 collaborator, based on its synthetic lethality with hlh-1 (Baugh et al. 2005b) and its expression throughout BWM (Baugh et al. 2005a; Fukushige et al. 2006). In contrast, the other factors are confined to developmentally early times of specification or very early differentiation and are restricted to subsets of BWM or are not specific to muscle (Baugh et al. 2003; Amin et al. 2007; Yanai et al. 2008; Broitman-Maduro et al. 2009). For these reasons, we consider hlh-1 and unc-120 to be the core regulators for the differentiated BWM network.
Detailed knowledge of connectivity between hlh-1 or unc-120 and their downstream targets comes from studies of specific target genes, myo-3, unc-54, and pat-3, where binding of the factor was observed at a specific (mutable and essential) cis-regulatory module (CRM) (Francis and Waterston 1985; Fukushige et al. 2006; Lei et al. 2009). These targets and their CRMs serve as internal standards for genomic assays in this work. Whether specific binding and action of HLH-1 and/or UNC-120 proteins regulates the hundreds of additional BWM genes has been untested, and the extent of individual versus shared connectivity is unknown. Recent studies that mapped HLH-1 protein occupancy across the entire genome (Gerstein et al. 2010; Lei et al. 2010) showed widespread binding with proximity to both muscle-specific and nonmuscle genes, but those studies did not directly address the relationship between binding and genome-wide regulatory dependence on the transcription factors.
Nonstriated muscles comprise a minor fraction of C. elegans (four embryonic, 16 post-embryonic muscles, and 10 contractile somatic gonad sheath cells) (Sulston and Horvitz 1977; Sulston et al. 1983). NSM uses hlh-8 as its major transcriptional regulator (Harfe et al. 1998b; Corsi et al. 2000; Liu and Fire 2000) along with unc-120 (Fukushige et al. 2006) and, in a subset of the NSM, mls-1 (Kostas and Fire 2002; Reece-Hoyes et al. 2007). Ectopic hlh-8 produces NSM phenotypes in other cell types (Harfe et al. 1998b; Wang et al. 2006; Zhao et al. 2007). hlh-8 and hlh-1 are transiently coexpressed in the post-embryonic M cell, whose progeny ultimately include 14 BWM cells expressing only HLH-1, 16 NSM cells expressing only hlh-8, and two nonmuscle coelomocytes (Sulston and Horvitz 1977). The molecular and regulatory relationship of BWM and NSM networks is a second focus of this work, based on our findings of crosstalk between them.
Here, we construct a C. elegans BWM genomic resource consisting of RNA-seq transcriptomes of the wild-type, hlh-1 mutant, and unc-120 mutant BWM plus ChIP-seq HLH-1 factor occupancy. Cell type classifications (BWM, general, or non-BWM) are assigned to regulatory target genes by comparing transcriptome measurements from BWM-enriched embryos and normal embryos. We then dissect unique and shared regulatory contributions from each factor by comparing transcriptomes of wild-type embryos with those of hlh-1 mutant and unc-120 mutant embryos. This reveals the regulatory influence of each factor on muscle-specific versus broadly expressed genes. We also provide a genetic resource of previously unknown hlh-1 interacting factors identified via a synthetic RNAi screen. Finally, we measure the number, location, and DNA sequence motif composition of in vivo HLH-1 bound regions to evaluate how biochemical factor binding is related to regulatory impact (Fig. 1B).
Prior genetic studies showed that no single factor in the core BWM network is essential for muscle differentiation (Baugh et al. 2005b; Fukushige et al. 2006; Broitman-Maduro et al. 2009), suggesting there is partial “redundancy” between factors, although no specific molecular explanation was suggested. Among genes affected by hlh-1 mutation in our study, one coherent set of transcription factors includes hlh-8/twist, which is known to positively regulate NSM differentiation. We discuss how this finding, plus other properties of the hlh-1/unc-120 network, contributes to the tolerance of worm BWM myogenesis to hlh-1 and unc-120 mutation.
Results
Increasing muscle by respecification reduces nonmuscle background
Our overall study design for genomic measurements is shown in Figure 1B. We increased BWM in embryos by knocking down known specification genes for non-BWM lineages that act prior to hlh-1 and unc-120 (Fig. 1C). Only one-sixth of C. elegans normally becomes bodywall muscle (Sulston et al. 1983). This presents signal-to-noise problems for ChIP and transcriptome experiments by diluting signal and obscuring any signal's cell type source. Increasing the proportion of BWM can ameliorate these problems, but prior methods (Bowerman et al. 1992; Draper et al. 1996; Page et al. 1997; Baugh et al. 2005a) had specific disadvantages for our purposes (Methods). We increased muscle content without directly augmenting the muscle network itself by respecifying nonmuscle fates to a muscle fate. RNAi knockdown of mex-3 can double muscle (Draper et al. 1996), while joint knockdown of mex-3, skn-1, and elt-1 is expected to convert over 80% of cells to BWM. mex-3 acts three cell divisions before HLH-1 expression (Draper et al. 1996; Hunter and Kenyon 1996), skn-1 acts two or three cell divisions beforehand (Bowerman et al. 1992; Blackwell et al. 1994), and elt-1 acts around the time hlh-1 will be activated (Spieth et al. 1991; Page et al. 1997; Michaux et al. 2001) but still permits hlh-1 expression.
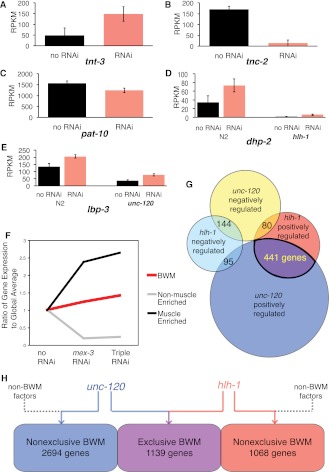
We assayed three conditions: no RNAi (empty vector); mex-3 RNAi only; and elt-1, mex-3, and skn-1 triple RNAi. Since knocking down multiple genes via RNAi can significantly reduce the efficiency of each individual knockdown (Gonczy et al. 2000; Gouda et al. 2010), we concatenated RNAi coding sequences to produce a single transcript. As expected, muscle-specific transcripts such as tnt-2 and tnt-3 were enriched by the RNAi strategy across two biological replicates (Fig. 2A). Known nonmuscle genes, such as tnc-2, were reduced with RNAi (Fig. 2B). Genes broadly expressed in both BWM and non-BWM, such as pat-10, were not significantly affected by RNAi (Fig. 2C). Muscle from the triple RNAi sample, unlike mex-3 alone, should be dominated by the C and D lineages, at the expense of the MS lineage. The mex-3 RNAi condition, which doubles the BWM contribution compared with wild type, is included to retain MS-derived muscle for observation (Figs. 1C, 2F).
Figure 2.
Mutation and RNAi-based muscle enrichment impact gene expression levels. (A) Muscle troponin T tnt-3 exemplifies genes enriched in muscle-enhanced embryos (red). Muscle-normal expression (black) is nonzero since these animals retain significant muscle. (B) Non-BWM troponin C tnc-2 exemplifies genes depleted in muscle-enriched embryos. BWM-enriched animals (red) have reduced non-BWM tissue, so little expression is seen. (C) Troponin pat-10, which is expressed in both BWM and non-BWM, exhibits a negligible net change. (D) dhp-2 (dihydropyrimidinase) and (E) lbp-3 (lipid-binding) are both muscle-enriched. dhp-2 is affected by hlh-1 loss of function, and lbp-3 is affected by unc-120 loss-of-function, suggesting positive regulation by the BWM transcription factors. (F) Group averages of the ratiometric change for the muscle-enriched genes (those significantly up-regulated in muscle-enriched animals), nonmuscle-enriched (those significantly down-regulated in muscle-enriched animals) genes, and annotated BWM genes (which significantly overlap with NSM and PhM genes and are, therefore, more broadly expressed) are plotted against the RNAi feeding conditions. (G) Overlap of hlh-1 and unc-120 regulated genes, both positively and negatively regulated, including the 441 genes positively regulated by both unc-120 and hlh-1. The number of genes in the other intersects are listed. (H) BWM transcription factors regulate more broadly expressed RNAs than BWM-specific ones. A total of 1139 genes whose RNA levels are significantly regulated by hlh-1 and/or unc-120 are expressed exclusively in BWM; hlh-1 and unc-120 significantly regulated 1068 and 2694 broadly expressed genes. (RPKM) Reads per kilobase of gene structure model per million reads.
RNA-seq reveals hlh-1 and unc-120 regulatory targets, many of which are shared
We quantified transcriptomes from total polyA+ RNA using RNA-seq (Mortazavi et al. 2008). The ∼400-min developmental time point (twofold through threefold stage at 25°C in wild-type animals) was used to ensure that BWM cells had already been specified, thus capturing embryos during differentiation. Due to concerns over variations in timing across mutants and RNAi conditions, we verified that both hnd-1 and ceh-51 had shut down in all animals (RPKM < 2), indicating that our samples across replicates and conditions represent middle to late differentiation. To identify regulatory targets of hlh-1 and unc-120, we compared wild type and temperature-sensitive hlh-1(cc561) and unc-120(st364) mutants cultured at the nonpermissive temperature, both with and without RNAi feeding. When the hlh-1(cc561) parent generations are elevated to the nonpermissive temperature prior to egg fertilization at the L4 stage, no detectable HLH-1 remains in the resulting embryos, and there is no maternal or zygotic effect (Chen et al. 1994), as confirmed here by immunoprecipitation (Methods).
We defined hlh-1 and unc-120 regulatory targets as genes whose expression differs significantly (average expression ± one standard deviation) between wild-type embryos and the mutant hlh-1(cc561) or unc-120(st364) under RNAi-treated conditions (mex-3 and mex-3; elt-1; skn-1 triple RNAi) (Fig. 1B). Among 13,216 genes expressed above background, 1445 are hlh-1 regulatory targets by these relatively conservative criteria, and we expect them to include both direct and indirect mechanisms (Table 1). Of these, 837 decreased significantly (one standard deviation) in the mutant, consistent with a simple positive mechanism of action for HLH-1 (Lei et al. 2009; see Discussion). Conversely, 608 genes were up-regulated by loss of HLH-1. These are explained most simply by indirect negative mechanisms, although the mammalian hlh-1 ortholog, MyoD, can act as a direct negative factor at a few target genes (Berkes et al. 2004; Dilworth et al. 2004; Penn et al. 2004). Mutation of unc-120 significantly reduced RNA levels from 2718 genes in muscle-enhanced embryos (Table 1), while 956 genes had significantly higher transcript levels.
Table 1.
Impact of hlh-1 mutation on expression levels in BWM-enriched worms
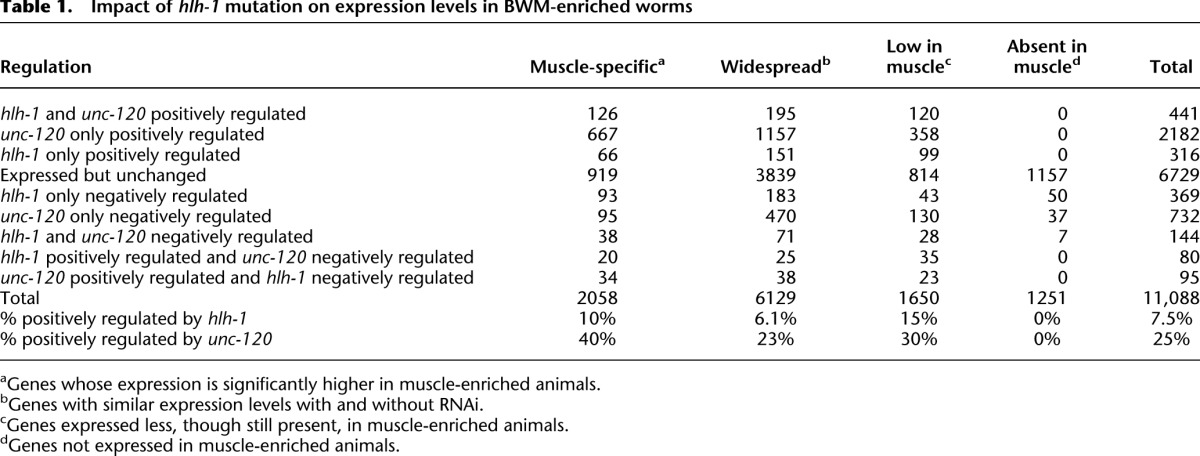
Among 592 transcription factor genes detectably expressed in our samples, 22 were up-regulated and 12 down-regulated in hlh-1 mutant embryos, while 13 were up-regulated and 103 down-regulated in unc-120 mutants (Supplemental Table S1). tbp-1 and nhr-63, regulated by hlh-1, also interact with hlh-1 in an RNAi synthetic lethal screen (see below), making them especially strong candidate members of a more complete BWM transcription network (Discussion). hlh-1 has a proportionally more negative effect than unc-120 on transcription factors, which may reflect their differing roles in BWM and NSM. A specific and unexpected example of hlh-1 negative regulation was hlh-8. Because hlh-8 is the major positive transcriptional regulator of NSM (Corsi et al. 2000, 2002; Liu and Fire 2000), this observation suggests a previously unknown regulatory connection between the BWM and NSM networks. In the unc-120 mutants, the transcript level of unc-120 itself was up-regulated, suggesting that there is a negative autoregulatory feedback loop. unc-120 was not, however, significantly affected by hlh-1 mutation. This result differs from a prior report of unc-120 regulation by hlh-1 (Yanai et al. 2008), although that study was performed at earlier developmental time-points during specification and used a different significance criteria and measurement technology (RT-PCR).
Seven hundred and sixty genes were significantly regulated by both unc-120 and hlh-1 (P < 0.001 for joint regulation), thus offering an explanation for these genes' continued though diminished expression when either factor is mutated. Four hundred and forty-one of these genes were positively regulated by both factors and 144 were jointly negatively regulated (Table 1; Fig. 2G). An additional 175 genes were divergently regulated between the two factors. The negative and mixed groups include genes best explained by differential regulation between BWM and NSM regulatory networks (Discussion).
hlh-1 and unc-120 regulate many BWM and non-BWM muscle genes
We next asked how hlh-1 and unc-120 regulatory target status is parsed among genes expressed preferentially in BWM, genes expressed in both BWM and other tissues, and genes expressed exclusively elsewhere. The last group serves as a measure of noncell autonomous effects and background. Bodywall muscle-enriched genes were defined by comparing RNA-seq of N2 control (no RNAi) embryos with RNAi-treated muscle-enriched animals. Overall, 2058 genes had expression levels significantly higher (one standard deviation) in BWM-enriched worms than in control, including many classical markers of BWM (Supplemental Table S2). From a previously described set of muscle structural genes (Fox et al. 2008), our set included 20 of their 38 genes, with the discordant 18 being explained by their known expression in pharynx or other tissues. Among 2901 genes expressed preferentially in nonmuscle tissues (Table 1), 1251 appeared entirely restricted to nonmuscle. The true number of genes with this “nonmuscle” pattern is almost certainly higher because our assay is not sensitive to genes expressed at low levels in only a few cells, and there are many such genes in C. elegans.
As expected, some classic BWM differentiation genes were among the most strongly down-regulated by hlh-1 loss (Fig. 2D) or unc-120 loss (Fig. 2E). RNA levels for these genes decreased significantly (>one standard deviation), but none lost all detectable RNA (see Discussion). At the other extreme, a different subgroup of BWM genes, including tni-1 and major actins and myosins, were unaffected by either hlh-1 or unc-120 mutation (Supplemental Fig. S1; Supplemental Table S2). Both unc-120 and hlh-1 contribute widely to BWM and non-BWM exclusive genes (Fig. 2H). Overall, known BWM genes displayed a broad range of quantitative responses to hlh-1 and unc-120 mutation, in both fractional and absolute change. This suggests different regulatory strength contributions from them and, implicitly, from additional transcription factors interacting with subsets of target genes (Supplemental Fig. S1). Of 2058 genes preferentially expressed in muscle, 10% (212) depended significantly on hlh-1 for expression and 40% (827) depended significantly on unc-120 (Table 1). Of all hlh-1 positive regulatory targets, 212 are muscle-preferred, 371 widespread, and 254 depleted but present in BWM. Likewise, unc-120 positive regulatory targets are distributed with 827 being muscle-preferred, 1390 widely expressed, and 501 being depleted but present in BWM.
hlh-1, but not unc-120, negatively regulates some NSM genes
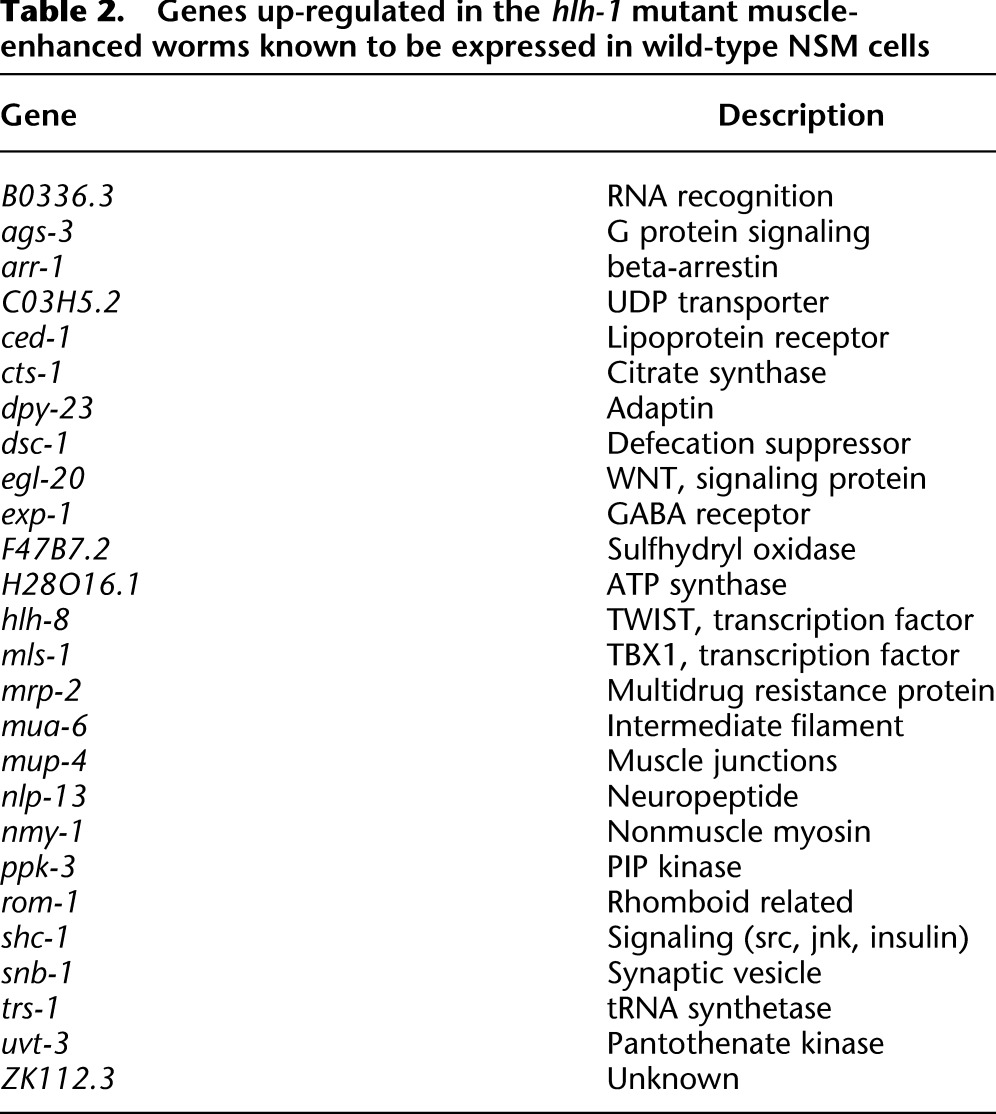
An unexpected result was that NSM annotated genes were prominent among the group of 307 genes up-regulated specifically in the BWM-enriched hlh-1 mutant animals and not in wild-type animals (Table 2). Prominent in this group was hlh-8, the central regulator in the NSM differentiation network (Corsi et al. 2000), and mls-1, another transcription factor in the NSM network (Kostas and Fire 2002). These findings were unexpected since triple-RNAi treatment in wild-type embryos abolished, or at least reduced, the entire NSM, including enteric muscles and the M cell lineage. This suggests that a subnetwork of BWM genes behaves differently than the rest of the tissue and that genes in this group are candidate regulatory targets of hlh-8 and/or mls-1. Overall, 9.5% (195 genes) of the BWM preferred expression group were annotated in WormBase as also expressed in normal NSM, and, of these, 26 were up-regulated along with hlh-8 and mls-1. BWM/NSM shared genes showed significant overlap with hlh-1 positively regulated target genes (hypergeometric P < 0.001). Ninety-five percent of NSM genes, including hlh-8 and mls-1, were not detectably elevated in unc-120 mutants, meaning it is likely that repression of NSM circuitry is specific to hlh-1. Though unaffected in BWM-enhanced wild-type embryos, hlh-8 was positively regulated by unc-120 in muscle-normal animals, presumably by acting in the NSM (which is absent in the BWM-enhanced condition).
Table 2.
Genes up-regulated in the hlh-1 mutant muscle-enhanced worms known to be expressed in wild-type NSM cells
Synthetic PAT screen for coregulators of hlh-1 and mediators of hlh-8/hlh-1 crosstalk
As shown above, unc-120 partly explains the robustness of worm myogenesis to hlh-1 mutation, but other factors might perform a similar function for additional hlh-1 targets. To find other regulators that collaborate with hlh-1, we performed a feeding RNAi synthetic paralysis-at-twofold (Pat; WBPhenotype:0000053) phenotype analysis in the hlh-1(cc561) mutant background, using a library of 512 genes that encode known and suspected transcription factors. In nematodes, elongation of the embryo depends on muscle contractions (Williams and Waterston 1994). The Pat phenotype, therefore, serves as a readily scored surrogate for major BWM failure. As expected, unc-120 scored strongly in this assay. Other strong interactors included ceh-20, grh-1, tbp-1, lin-26, pos-1, oma-2, nhr-4, nhr-46, nhr-63, nhr-116, hmg-1.2, hnd-1, and ceh-51 (Supplemental Table S5). The Pat phenotype suggests that each of these contributes to expression of one or more genes needed for differentiation of muscle in the absence of hlh-1.
A majority of these hlh-1 genetic interacting factors are themselves regulated by hlh-1 and/or unc-120. TATA-binding protein (tbp-1) is part of the transcription initiation complex and is positively regulated by both hlh-1 and unc-120, suggesting elevated demand for it by some muscle differentiation genes. nhr-63 is negatively regulated by hlh-1, while nhr-116 is negatively regulated by unc-120. lin-26, pos-1, oma-2, nhr-4, and nhr-46 are positively regulated by unc-120, suggesting feed-forward loops that are familiar structures in developmental circuits. nhr-63, grh-1, and ceh-20 are normally expressed in NSM, so they are candidates for genes that could interact with both hlh-1 and the hlh-8/mls-1 circuitry.
Genes positively regulated by hlh-1 and unc-120 are enriched for HLH-1 occupancy, which is widespread
To map sites of HLH-1 occupancy in vivo, we performed chromatin immunoprecipitation from RNAi fed wild-type embryos (both mex-3 and triple RNAi) with an anti-HLH-1 antibody, followed by DNA sequencing (ChIP-seq) (Johnson et al. 2007; Zhong et al. 2010). The most prominent signals were consistent in all conditions (Supplemental Fig. S2), but BWM-enrichment was important for detecting the majority of HLH-1 ChIP signals. We evaluated ChIP-seq signal intensities and locations relative to background (Pepke et al. 2009) to produce a high confidence set of 1047 peaks appearing in both RNAi conditions and a more inclusive union set of 9415 peaks appearing in either RNAi condition (mex-3 RNAi yielded 7021 peaks and triple RNAi yielded 3441 peaks; examples in Fig. 3A,B). The peak yield was similar to that of Lei et al. (2010), who recovered 20,143 peaks in their ChIP-seq experiment that they narrowed to 4016 high-confidence peaks (Lei et al. 2010). Their use of a different antibody likely accounts for much of the difference in peak identification, though the overlap was statistically significant (P < 0.001). For their ChIP-chip analysis, they used the same antibody but a different enrichment and detection technique (Lei et al. 2010), leading to a similar sample size as our high-confidence set of peaks and a statistically significant overlap (P < 0.001). The muscle enrichment and regulatory dependence of genes near peaks from both broad and stringent sets were comparable, but the stringent group was more strongly enriched. Thus, >50% and 20% of our hlh-1 positively regulated gene list was captured in the broad and stringent HLH-1-bound sets, respectively.
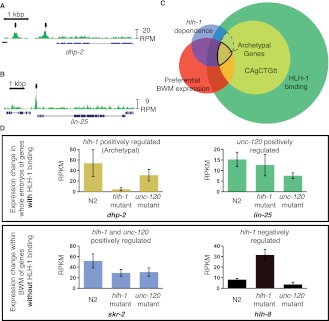
Figure 3.
HLH-1 ChIP-seq binding is associated with, but not predictive of, regulation. HLH-1 binds to the genes (A) dhp-2 and (B) lin-25 (arrows). (C) Venn diagram shows four criteria for rating interactions of BWM preferred expression (red circle, 2175 genes), hlh-1 regulation of expression (blue circle, 757 genes), HLH-1 ChIP-seq binding (green circle, 9519 genes), and the presence of a local HLH-1 binding motif (yellow circle, 3469 genes). The intersect of 78 genes is highlighted as the “Archetypal Muscle List.” (D) Gene expression (RPKM levels from RNA-seq) levels illustrate different regulatory dependency patterns for hlh-1 and unc-120, with (upper) or without (lower) detectable nearby HLH-1 occupancy. dhp-2 (gold from panel C) represents archetypal muscle genes, positively regulated by hlh-1 with significant HLH-1 occupancy. lin-25 (green, from C) is positively regulated by unc-120 but not hlh-1, even though it has HLH-1 occupancy. skr-2 (blue, from C) is positively regulated by both hlh-1 and unc-120. hlh-8 (black) represents a class up-regulated only in hlh-1 mutant BWM, suggesting indirect negative regulation.
Eighty-nine percent of the stringent set of HLH-1-occupied regions were within 5 kb 5′-ward of an annotated gene start (including regions that also fell within an upstream gene), with 36% of those concentrated in the proximal 500 bp. Sixteen percent of regions were in introns, 6.6% in exons, and only 1.2% in 3′ UTRs (Supplemental Table S3). The 5-kb 5′-ward, 500-bp proximal, and exon sequences were enriched genome-wide for HLH-1 ChIP peaks (P < 0.01), while other regions were depleted or not enriched (peaks per kb).
Because hlh-1 is a highly cell-type-specific activating transcription factor, an initial expectation was that most BWM-specific genes would have one or more adjacent HLH-1 ChIP regions. For specific CRMs and promoters previously shown to drive BWM expression (Okkema et al. 1993; Krause et al. 1994), this was true, with ChIP signals at expected locations (Supplemental Fig. S2). Genome-wide, 59.7% of the 941 annotated BWM genes had HLH-1 occupancy (broad set) within the gene body or 5 kb upstream (Supplemental Table S4), while 54% of our BWM-enriched expression gene set did. Sixty-seven percent of genes near a stringent HLH-1 peak (i.e., within 5 kb of the start site) are expressed at a significant level (RPKM > 3) in BWM (P < 0.001). However, the vast majority (80% in the stringent set and 87% in the broad set) were not muscle-preferred in their expression pattern (Fig. 3C; Discussion). Rather, the majority are expressed widely in muscle and nonmuscle tissue.
Genes whose expression depended positively on hlh-1 (Table 1; dhp-2 in Fig. 3D) were significantly, but not strikingly, enriched for HLH-1 occupancy within 5 kb upstream or in the gene body, compared with other genes in the genome (57% vs. 49%, P < 0.001) (Supplemental Table S4), while negatively regulated targets were not enriched (48% vs. 49%; hlh-8 in Fig. 3D). The overlap was on par with that of Lei et al. (2010), with our high-confidence peaks and their ChIP-chip analysis both yielding 5% of the occupied genes depending on hlh-1, while their ChIP-seq analysis yielded 9% of occupied genes depending on hlh-1 to our 10% for our broader data set. Genes depending on unc-120 (Table 1) were also more likely to have HLH-1 occupancy than the rest of the genome (53% vs. 49%, P < 0.001) (lin-25 in Fig. 3D). Genes jointly up-regulated by both hlh-1 and unc-120 function were similarly enriched for HLH-1 occupancy (54% vs. 49%, P < 0.002), though it is not required (skr-2 in Fig. 3D).
hlh-8, mls-1, and grl-26 were among 608 genes under negative regulation by hlh-1 but not by direct binding, according to the ChIP data (Fig. 3D). hlh-8, in particular, had no HLH-1 binding in its gene body nor within 20 kb upstream of the TSS or 10 kb downstream.
ChIP-regions contain the canonical HLH-1 binding motif and novel associated motifs
HLH-1-occupied regions were used to derive overrepresented sequence motifs. We expected to identify motifs responsible for direct HLH-1 binding, together with possibly collaborating motifs, since the latter are often present in functional cis-regulatory modules (Davidson 2007). Two substantially different motif discovery algorithms found similar motifs (Methods). The primary motif was AACAGCTG (Fig. 4A, first motif), which is an E-box family motif (CANNTG is the known general motif for the bHLH family). The core hexamer matches previous HLH-1 motif determinations from yeast one-hybrid assays (Grove et al. 2009), in vivo ChIP-chip and ChIP-seq (Lei et al. 2010), and mammalian MyoD (Cao et al. 2010), while the adjacent AA produces a more specific site which is analogous, but not identical, to the most highly preferred myogenin binding octa-E-box in mouse (CAGCTGRR) (Cao et al. 2010; A Kirilusha and B Wold, in prep.). Six other motifs (Fig. 4A; Supplemental Fig. S3) plus GA- and CT-simple repeat-rich regions (Guhathakurta et al. 2002; GuhaThakurta et al. 2004) were found. Expanding the search radius from 50 bp to 100 bp found a second E-box: CAACTG (web logo not shown), reported previously as a secondary site for HLH-1 binding (Grove et al. 2009; Lei et al. 2009).
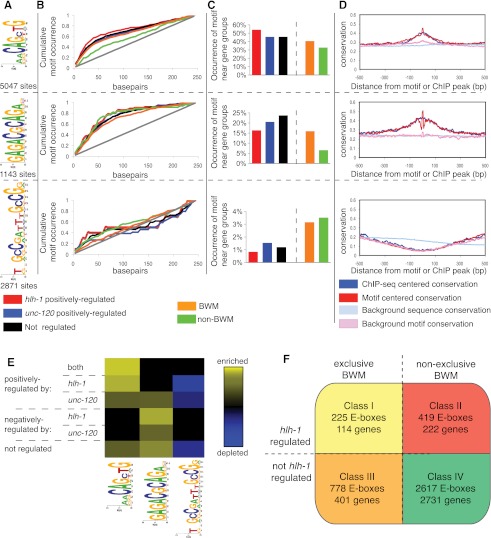
Figure 4.
HLH-1-associated motifs correlate with directional expression control. (A) The web logo position-specific frequency matrix (PSFM) diagrams for three representative motifs and the accompanying number of sites identified near HLH-1 occupancy (250-bp radius). (B) The relative locations of three motifs compared to their experimentally identified binding sites (analyzed per Ozdemir et al. 2011). The AACAGCTG motif is centered on the called ChIP-seq peak (50% within ±25 bp of the peak for hlh-1 positively regulated or muscle-enriched genes). The GAGACGCAGA motif (second panel) is less central (within 75 bp). The GCCGatttGCCG motif (third panel) shows no significant centrality. The gray line represents a uniform distribution. (C) The occurrence of each motif within ±250 bp of the HLH-1 occupancy peak near genes (ChIP regions within 5 kb of a gene TSS) belonging to expression groups is shown. The E-box shows the greatest enrichment for genes characterized as hlh-1 positively regulated (first panel). The GAGACGCAGA motif is more closely associated with unc-120 positively regulated genes (second panel), whereas the GCCGatttGCCG motif is enriched near genes absent in BWM (third panel). (D) The conservation across sequenced nematodes (elegans, briggsae, remanei, and brenneri) of ChIP-seq identified regions with the three motifs is shown. Conservation around the in vivo binding (blue) and around the motif (red) is shown compared to background (light blue and pink) (Ozdemir et al. 2011), with higher values representing a higher level of conservation. The E-box and GAGACGCAGA motifs, along with their surrounding sequences, are strongly conserved, while the GCCGatttGCCG motif is not at all conserved. (E) Heat maps show the level of motif enrichment (yellow) or depletion (blue) for the CAgCTGtt, GAGACGCAGA, and GCCGatttGCCG motifs near broadly expressed genes that are similarly regulated (y-axis). The E-box is enriched near genes positively regulated by hlh-1 and unc-120. The GAGACGCAGA motif is enriched near genes negatively regulated by hlh-1 and positively regulated by unc-120. The GCCGatttGCCG motif is depleted near genes positively regulated by either factor. (F) There are four classes of E-boxes observed: Class I contains muscle E-boxes that are bound by HLH-1, and it is predicted that mutation of these sites will lead to changes in expression, as the nearby genes are both specific to BWM and regulated (positively or negatively, in contrast to the Archetypal Genes, which are exclusively positively regulated) by hlh-1; Class II contains E-boxes that are similarly functional but are near genes not exclusively expressed in BWM; Class III contains E-boxes that are not required for expression but likely make contributions to nearby genes that are expressed exclusively in BWM; and Class IV contains seemingly nonfunctional E-boxes that are not required for expression or associated with BWM expression.
Analyzed across all HLH-1 ChIP regions with a 250-bp radius, the two E-boxes and the GAGACGCAGA motif (Fig. 4A, second motif), for which there is no known factor, were strongly centered near ChIP-seq summits, with the most statistically significant central concentration being in muscle-specific and unc-120 positively regulated genes (P < 0.05; Fig. 4E; Supplemental Fig. S4). The centered position argues that a motif is partly or solely responsible for the observed ChIP signal. Other motifs were more evenly distributed in the tested regions (Fig. 4B; Supplemental Fig. S3), consistent with accessory or independent roles. As expected, the HLH-1 octabox motif was most highly overrepresented among genes expressed preferentially in muscle (Fig. 4E).
HLH-1 ChIP-seq peaks near specific functional subsets of genes were analyzed for motif discovery, position, and frequency. Gene groups tested were those (1) strongly positively dependent on hlh-1 for expression, (2) strongly negatively regulated by hlh-1, (3) absent in bodywall muscle, (4) dependent on unc-120 for expression, (5) less stringently dependent on hlh-1 for expression, and (6) dependent on both hlh-1 and unc-120. Motifs identified above were rediscovered within some subsets, in addition to two novel candidates: AAAANNNNNAAA and GCCGATTTGCCG (Fig. 4A, third motif; Supplemental Fig. S3, sixth motif). The GCCGATTTGCCG motif was specifically associated with genes that do not positively depend on hlh-1 and with genes that do depend on unc-120. In fact, this motif was selectively depleted from the positively regulated HLH-1 gene set (P < 0.01).
HLH-1-bound regions are preferentially conserved
If HLH-1-occupied E-box motif instances located near hlh-1 regulated genes are functionally significant, we expect them to be preferentially conserved in evolution. Moreover, we expect functional HLH-1 binding sites to be embedded in larger domains of conservation that typify cis-regulatory modules. This was the case around our set of HLH-1-occupied sites, with preferential conservation among sequenced nematodes of ±200 bp (Fig. 4D). This conservation was not restricted to sites near HLH-1 regulated genes; rather, the larger set of HLH-1 ChIP regions located near genes that were not regulated by hlh-1 or were not BWM-specific displayed similar preferential conservation (Supplemental Fig. S3B). This suggests that HLH-1 ChIP signals overall identify functionally important sequences, but that these need not be adjacent to muscle-specific or HLH-1-dependent genes. Among the new candidate motifs, three others show preferential conservation, while GCCGATTTGCCG did not (Supplemental Fig. S3).
Discussion
Through analysis of the BWM differentiation network, we uncovered a significant overlap in transcription factor function, helping to explain the redundancy of the core factors, and a surprising lack of muscle factor target specificity to BWM. This work expanded loss-of-function analysis for BWM regulation to cover the entire transcriptome, finding that 21% (4359) of C. elegans genes are significantly affected by mutation of either hlh-1 or unc-120. It was found that 3.7% (760) were affected by both regulators (Table 1), and these were highly enriched for BWM annotation. Their pattern of regulatory dependence helps to explain how and why hlh-1 and unc-120 act as a synthetic lethal pair in the embryo. However, an equally strong result was that 71% of jointly regulated genes and 79% of HLH-1 or 69% of UNC-120 single targets are not tissue-specific—rather, they are substantially regulated by the muscle-specific factors in BWM and, presumably, by other unknown regulators in nonmuscle tissues. By integrating our mapping of regulatory connectivity with in vivo physical HLH-1 occupancy, we were able to define a set of “archetypal” direct transcriptional targets for hlh-1 (Fig. 3C and below); identify biologically pertinent indirect regulatory relationships, including the major NSM-specific regulator, hlh-8 (Fig. 5A,B,C; below); and define a set of HLH-1 occupancy sites located near broadly expressed genes. Since a significant number of broadly expressed HLH-1-occupied loci were functionally affected by hlh-1 mutation (220 genes), we conclude that hlh-1 either originated as a highly muscle-specific factor that has been drafted over time to help regulate widely expressed target genes in the specific context of muscle, or that hlh-1 was originally a more general factor whose role was narrowed to muscle tissue early in animal evolution. New hlh-1 collaborating factors expand the BWM differentiation regulatory network and network orthology based on an RNAi screen for muscle failure.
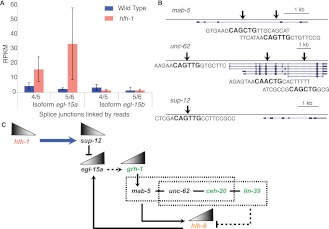
Figure 5.
hlh-1 negatively regulates the NSM transcription factor hlh-8. (A) Differential splicing of different isoforms of egl-15 (a and b) is hlh-1-regulated in BWM. Splicing isoform b, specific to BWM, is unaffected by the mutation, while isoform a, specific to NSM, is up-regulated specifically in the mutant muscle. (B) HLH-1 binding sites local to the genes mab-5, unc-62, and sup-12 are shown, with arrows representing the ChIP-seq peaks. Each gene contains an E-box motif characteristic of hlh-1 occupancy. sup-12 is the only one whose expression depends on hlh-1, but the other genes may be regulated in part by hlh-1. (C) sup-12 depends on HLH-1 binding for expression (blue arrow). The mRNA-binding protein SUP-12 inhibits the splicing variant EGL-15a (Kuroyanagi et al. 2007). In the absence of hlh-1, grh-1 is up-regulated and may be controlled (dashed line) by egl-15a (Zhong and Sternberg 2006) to regulate mab-5 (Venkatesan et al. 2003). In turn, MAB-5 competes with LIN-39 to interact with CEH-20 and UNC-62 in some cells to effect target expression or repression (Liu et al. 2006; Jiang et al. 2009; Potts et al. 2009). They may act on hlh-8, which is known in some cells to depend on ceh-20 and mab-5 (Kenyon 1986; Liu et al. 2006; Jiang et al. 2009). Therefore, in the absence of hlh-1, sup-12 decreases—leading to an increase in egl-15a and grh-1. GRH-1 and EGL-15a work to activate the MAB-5/UNC-62/CEH-20 Hox/Pbx complex to up-regulate the normally repressed hlh-8. This pathway is supported by the appearance of grh-1, ceh-20, and lin-39 (shown in green) in the synthetic PAT screen as being integral for muscle formation.
unc-120/hlh-1 compensation is based on overlapping roles in regulatory target control
In nematodes, myogenesis is robust to mutation of either unc-120 or hlh-1 (Baugh and Hunter 2006; Fukushige et al. 2006); by contrast, the hlh-1 bHLH ortholog, myogenin, is absolutely required for mammalian differentiation, and, in Drosophila, the unc-120 MADS family ortholog, Mef2 (also known as D-MEF2), is absolutely required (Black and Olson 1998). Nevertheless, the common theme is that bodywall or skeletal muscle differentiation in all three phyla uses both bHLH and MADS regulators. Our results help to explain the C. elegans network's unique behavior in three ways.
First, 760 genes are jointly regulated (Table 1), and the regulatory contributions from the two factors are roughly additive rather than highly synergistic. Significant residual expression (>30%) was observed in each mutant strain for the vast majority of shared positively regulated targets (89%), and only one gene (tag-10) lost more than 90% of its expression (Supplemental Fig. S1C). Though all these numbers are sensitive to thresholds, the qualitative results remained unchanged even when the stringency was significantly increased. The troponin gene family, several of whose members have been studied individually, nicely illustrates the varied regulatory specificity across multiple muscle types as well as in nonmuscle tissue (Supplemental Table S6). unc-120's large pool of positive targets may be partly explained by a broader activating role in both BWM and NSM, rather than just one muscle type, meaning that genes expressed in both may be regulated primarily by unc-120. hlh-1's smaller pool of regulatory targets and proportionally larger role in repression may reflect its narrow and specific function in BWM alone. This appears to include the capacity to silence the NSM network and potentially other nonmuscle gene networks. We cannot say yet how joint regulation is encoded in the cis-regulatory DNA of most target genes, but the specific list of HLH-1-occupied candidate CRMs belonging to jointly regulated worm genes we produce (below) provides the field with hundreds of specific starting points for direct tests via transgenic assays.
Second, newly identified hlh-1 interacting DNA binding factors (CEH-20, NHR-63, GRH-1, HMG-1.2, and LIN-39) are strong candidates to explain how important muscle genes, especially those with no unc-120 response, can continue to be expressed without hlh-1. Whether they act on many genes or only on small specific subsets will now be testable by performing experiments like those done above for hlh-1 and unc-120. Though the DNA binding motifs for these new hlh-1 interactors are unknown to us, our motif discovery analysis of hlh-1-bound regions produced candidates for combinatoric regulation. By several criteria, the most impressive of these is GAGACGCAGA, which is present in 1143 of 9447 HLH-1-bound regions, is most highly enriched in BWM-expressed genes, is preferentially conserved, and is centrally concentrated near HLH-1 ChIP-seq regions. The motif is enriched near genes that are positively regulated by unc-120 and negatively regulated by hlh-1. Together, these facts argue that it binds a significant collaborating factor.
Third, hlh-8 and mls-1 were among the genes strongly up-regulated in hlh-1 mutants, suggesting that a small and specific subgroup of NSM muscle genes contribute to what is otherwise BWM myogenesis if hlh-1 is gone. Among hlh-1 interacting factors identified in the RNAi screen, hmg-1.2 is also expressed normally in NSM and could function as part of this intersecting circuit. Target genes that are divergently regulated by unc-120 and hlh-1 might be explained as additional genes normally necessary in NSM (a domain of unc-120 regulation) but not in BWM.
Crosstalk between NSM and BWM regulators without wholesale tissue conversion
The finding that hlh-8 and mls-1 are strongly up-regulated by hlh-1 mutation raises several questions. At the tissue level, does up-regulation of hlh-8 and mls-1 produce a wholesale transformation of BWM into NSM? It appears not, since many BWM-specific genes were readily detected in hlh-1 mutant RNA. Of 104 genes annotated as expressed in wild-type BWM but not in NSM, all continued to be expressed significantly in hlh-1 mutant embryos, with or without RNAi feeding. At the network level, are muscle genes normally expressed in both NSM and BWM similarly expressed in hlh-1 mutants, as would be expected if they are primarily positively regulated by unc-120, hlh-8, and/or mls-1? Indeed, the vast majority (552 of 596) of genes annotated for both BWM and NSM expression in wild-type worms were similarly expressed in hlh-1 mutants. Only four BWM/NSM shared genes were reduced by more than two standard deviations from their wild-type level in the mutant. However, hlh-8 up-regulation is not sufficient to explain muscle differentiation in the absence of hlh-1, because it is not synthetic muscle-lethal with hlh-1.
At the level of circuit structure and molecular mechanism, why and how is hlh-8 switched on in the absence of hlh-1, and what does this imply about their relationship in normal muscle development? Negative regulation of hlh-8 by hlh-1 is likely to be by an indirect mechanism, partly because hlh-1 is known as a positive regulator of its direct targets. In addition, we detected no HLH-1 occupancy near hlh-8 or mls-1 via ChIP-seq, even at the most relaxed peak calling stringency. Our RNA data show that known positive regulators of hlh-8, such as unc-62, ceh-20, and mab-5, are all present in both wild-type and hlh-1 mutant animals, making activation of hlh-8 highly plausible when negative regulation mediated indirectly by hlh-1 is relieved (Harfe et al. 1998a).
Drawing on our data and additional studies, we propose a specific model for regulation of hlh-8 by hlh-1 (Fig. 5). From the synthetic PAT phenotype analysis, ceh-20, lin-39, and grh-1 were identified as strong genetic hlh-1 interactors. By independent criteria, each of these is also a candidate to help activate hlh-8. grh-1 positively regulates mab-5 (Venkatesan et al. 2003), and mab-5/unc-62/ceh-20 positively regulates hlh-8 in NSM (Liu and Fire 2000). Furthermore, we found that egl-15a and grh-1 are up-regulated in our hlh-1 mutants (Fig. 5A), and there is additional genetic precedent in Drosophila for interaction between mab-5 and egl-15 (Zhong and Sternberg 2006). EGL-15/FGFR is necessary for proper sex myoblast (NSM) migration (Stern and Horvitz 1991). The primary splicing variant in NSM, EGL-15a, is down-regulated by SUP-12, which destroys EGL-15a but not EGL-15b, the primary splicing variant in BWM (Kuroyanagi et al. 2007). We found that the EGL-15a RNA splice isoform is up-regulated in hlh-1 mutants. sup-12 expression depends on HLH-1 activity, and sup-12 has a high confidence HLH-1 occupancy domain (Fig. 5B). We therefore suggest that in normal BWM, HLH-1 drives SUP-12 to down-regulate EGL-15a and grh-1, while in hlh-1 mutant muscle, SUP-12 is not expressed, and EGL-15a and grh-1 increase, thus activating mab-5/unc-62/ceh-20 and leading to up-regulation of hlh-8 and some of its NSM target genes (Fig. 5C). This should especially favor target genes normally expressed in both NSM and BWM, since collaborating factors from BWM are present.
There is also evidence for reciprocal repression of BWM by NSM, since hlh-8 mutants have an unstable and sometimes higher number of BWM cells and their sex-specific muscles disappear (Corsi et al. 2000, 2002). Up-regulation of hlh-8 in BWM in the absence of hlh-1 is reminiscent of the connection reported in the post-embryonic M lineage (Harfe et al. 1998a), and regulation of normal M lineage development, which generates both NSM and BWM, might account for the crosstalk we see in BWM upon hlh-1 mutation.
Defining “archetypal” hlh-1 target genes and their candidate CRMs
We distilled a set of 78 genes and associated candidate cis-acting regulatory modules that meet four criteria for being “archetypal” regulatory and molecular hlh-1 targets: (1) They are expressed preferentially in BWM; (2) they display significantly reduced RNA levels in hlh-1 mutants; (3) they have HLH-1 occupancy at one or more sites in our ChIP-seq data; and (4) the HLH-1 occupancy region contains one or more instances of the extended myogenic “octa E-box” (AACAGCTG) (Supplemental Table S7; Fig. 3C). An additional 154 genes satisfy criteria 2, 3, and 4 but are also strongly expressed in tissues other than BWM. These genes apparently depend on HLH-1 in the context of muscle and on other factors elsewhere.
Membership in our list of candidate hlh-1 BWM CRMs did not use DNA sequence conservation as an initial criterion, since recently evolved active instances may exist and will be pertinent to the network. This allowed us to ask if the candidate canonical muscle CRM group is preferentially conserved, and it was. Preferential conservation encompassed a region of ±150 bp relative to the HLH-1 ChIP peak (Fig. 4D), showing that this group of candidate CRMs has been under pressure to function. The muscle octa-Ebox is even more highly conserved than the surrounding domain, suggesting that it drives binding of functional consequence.
The archetypal muscle HLH-1 targets were defined without using unc-120 data, yet they are highly enriched in unc-120 regulatory targets. Thus, 56% of the hlh-1 archetypal loci are positively regulated by unc-120, while only 18% of all genes are regulated by unc-120. Similarly, 52% of genes satisfying criteria 2, 3, and 4 were positive regulatory targets of unc-120. Finally, of the genes in the archetypal group that are also unc-120-regulated, 20% contain an 85% match instance to GAGACGCAGA within their HLH-1-occupied region.
The archetypal HLH-1 target genes and candidate CRMs have been defined by intentionally stringent multiple-measurement intersection to help learn the defining and shared characteristics of BWM regulation. It is, therefore, an underestimate of the BWM group and highlights the important role hlh-1 plays in regulating genes not specific to the BWM group.
HLH-1 occupancy versus hlh-1 regulatory impact
Expression of hlh-1 is specific to the bodywall muscle system, the phenotype of hlh-1 null mutants is myogenic, and hlh-1 orthologs across metazoan phyla regulate muscle development and differentiation. This relative simplicity made it possible to address some questions about factor occupancy that have been difficult in systems with larger genomes and more complex organization. First, factor occupancy alone, as measured by HLH-1 ChIP-seq, is a permissive condition for regulating gene expression, but it is not powerfully predictive of regulatory activity. In isolation, HLH-1 occupancy had low specificity for hlh-1-dependent RNA expression at nearby genes or TSS. HLH-1 occupancy in an independent study (Lei et al. 2010) had almost identical specificity (9% in Lei et al. [2010] versus 10% in this analysis). The Lei et al. (2010) study also concluded that many sites were upstream of nonmuscle genes. Their conclusion that binding was not predictive of enhancer activity mirrors our conclusion that it is not predictive of regulatory activity (Lei et al. 2010). Similarly, the majority of mouse MyoD occupancy sites are located closest to nonmuscle-specific genes (Cao et al. 2010). Substantial technical issues surrounding assay sensitivity, combined with consequences from assigning binding regions to genes by an overly simple proximity algorithm, are probably responsible for some lack of predictive power. Nevertheless, the data are most consistent with a majority of detected HLH-1 occupancy binding events having no regulatory effect or an effect too small to measure. Where hlh-1 regulation is detected, the majority of it is associated with genes that are not specific for BWM alone.
From HLH-1 ChIP-seq regions, we refined the HLH-1 binding motif preferentially affiliated with HLH-1-regulated loci (AACAGCTG) and showed it is the dominant, centrally located driver for HLH-1 genome occupancy. The distribution of additional novel motifs from HLH-1-occupied regions, among the functionally distinct target gene groups, provided insights into BWM/NSM cis-regulatory logic. The GAGACGCAGA has a high rate of co-occurrence with the octa-Ebox and presence in HLH-1 ChIP regions. It is a plausible candidate to bind UNC-120 or an intimate UNC-120 collaborator, as the motif is preferentially concentrated near genes positively regulated by unc-120, significantly enriched in genes annotated for NSM and BWM (Fig. 4E; Supplemental Fig. S4), and present in more than half of archetypal BWM candidate CRMs. Several other discovered motifs also colocalize with HLH-1 binding, but more weakly, and these are reasonable candidates to collaborate with HLH-1 in other target gene subgroups (Supplemental Fig. S3). For example, CGnnGCGAGACCC is enriched near genes positively regulated by hlh-1 but negatively regulated by unc-120. This pattern is expected for BWM genes that must be turned off in NSM, a group with which it has significant overlap. In contrast, GCCGatttGCCG is selectively depleted near genes positively regulated by either hlh-1 or unc-120, suggesting that this motif mediates a function that is orthogonal to muscle differentiation (Fig. 4E). None of the newly discovered motifs closely resemble known transcription factor binding sites, although some bear resemblance to previously reported muscle-associated motifs (Guhathakurta et al. 2002; GuhaThakurta et al. 2004), and they are all candidates to bind hlh-1 interacting factors from the RNAi screen.
Methods
Additional details may be found in the Supplemental Material.
General methods and strains
We obtained C. elegans strains N2, PD4605 (hlh-1(cc561)), and RW364 (unc-120(st364)) from the Caenorhabditis Genetics Center (CGC) and cultured them using standard methods (Brenner 1974). To increase the proportion of muscle, we chose to knock down early specification genes (mex-3, elt-1, and skn-1) to permit muscle specification (Fig. 1C; Supplemental Material).
RNAi feeding
Bacteria (Ahringer Lab RNAi Library) were used for control (HT115) and mex-3 RNAi feeding, in addition to RNAi knockdown for the synthetic PAT screen. The elt-1, mex-3 and skn-1 inserts were fused in a single vector for triple RNAi feeding (Gouda et al. 2010). Knocking down multiple RNA transcripts can suffer from poor efficiency (Gonczy et al. 2000), but our concatenation technique maintained high penetrance. Effectiveness was measured in aliquots from each biological replicate, with 100% of animals being affected by RNAi (100% embryonic-lethal, with 0% making it to the comma stage morphologically). Costaining with phalloidin and DAPI revealed significant enrichment of myosin in N2 worms, though not every cell was converted to muscle. Synchronized worms were grown on seeded NGM special plates with IPTG and carboxy-penicillin at 15°C until the L4 stage, and then at 25°C until gravid adults began egg-laying. Gravid adults were bleached, and the eggs were shaken at 25°C in S-complete media (five embryos/μL) for 400 min to ensure muscle differentiation but avoid tissue necrosis (Fig. 1B).
ChIP-seq
Immunoprecipitation with an existing anti-HLH-1 polyclonal antibody (Lei et al. 2009) was performed in N2 and hlh-1(cc561) animals with a modified protocol (Weinmann and Farnham 2002). While not a null, the mutation effectively destroys HLH-1 function (Lei et al. 2009), probably through non-sense mediated decay (Harfe et al. 1998a), and no signal was seen above background. Embryos were freeze-cracked in 2% formaldehyde on dry ice five times, fixed for 30 min, and then quenched for 5 min. The embryos were washed, lysed, and sonicated (Misonex, output 3.5) with a microtip for 15 30-sec pulses with 1-min intervals. Ten percent of the sample was set aside as control. The antibody was added to the chromatin prep and allowed to mix for 16 h at 4°C. Four sequential aliquots of 200 μL of magnetic beads (Invitrogen Dynabeads M-280 Sheep anti-Rabbit IgG) were then added for 4 h to extract the antibody. Beads were washed, the complexes eluted, and the DNA organically purified and quantified (Invitrogen Qubit Fluorometer). For ChIP sequencing, the average number of reads was 17 ± 2 million reads, the average number of unique reads was 12 ± 1 million reads, and quality control failed on an average of 0.5 ± 0.1 million reads (Supplemental Table S8).
RNA-seq
For RNA-based sequencing, embryos were flash-frozen in TRIzol (Sigma) and freeze-cracked on dry ice five times. The embryos were passed through a 21 G needle and a 25 G needle (10× each) to shear the eggshell. The RNA was precipitated, treated with Turbo DNase (Ambion), and dT-purified (Invitrogen Dynabeads Oligo-dT). External quantification standards were spiked in to the mRNA, which was then fragmented to an average length of 200 nt by heating at 94°C in the presence of Mg++ for 90 sec. The fragmented mRNA was then random-primed with hexamers for reverse transcription first strand synthesis, followed by nick translation second strand synthesis using a double-stranded cDNA synthesis kit (Invitrogen). Comparisons were made between two biological replicates that were independently sequenced for each condition, except for untreated hlh-1 mutants, for which only a single pooled sample was sequenced. For RNA sequencing, the average number of reads was 26 ± 9 million reads, the average number of unique reads was 20 ± 7 million reads, and the average number of repeat reads was 7 ± 4 million reads (Supplemental Table S8). Quality control failed on an average of 0.5 ± 0.3 million reads per sequenced lane.
Library making and sequencing
The standard single amplification Illumina library-making protocol was used, including end repair, adaptor ligation, gel purification, and PCR amplification. Flowcell generation and sequencer running followed the Illumina protocol. All sequencing data is publically available. All genomic data are DNA-sequence-based and publicly available through GEO (GSE28561, GSE28562, GSE28563).
RNAi feeding for synthetic lethal screening
Bacteria from the OpenBioSystems RNAi library and the Ahringer RNAi library were used for RNAi feeding of L4 animals for 36 h at 25°C. Adults were then transferred to fresh plates for egg-laying for 4 h at 25°C. Adults were removed, and embryos were allowed to develop for 18–24 h prior to scoring. Embryos were scored for developmental progression using a dissecting microscope. The stage of developmental arrest in embryonic lethal worms was noted as during the twofold stage (Pat) or otherwise.
Data analysis
WormBase release WS190 was used for all analysis. Read mapping and read processing were performed with Bowtie and ERANGE (Pepke et al. 2009). Python was used to perform calculations described in the text. Genes associated with stress response (e.g., heat shock genes) were monitored for signs of damage or stress. We looked for enriched motifs near the ChIP-identified binding sites using MEME on sequences within various radii of the binding site. A greedy algorithm-based motif finder reproducibly identified the major nonrepeat motifs found with MEME. Enrichments were determined by χ2 and hypergeometric statistical analysis.
Data access
All sequencing data have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) for “HLH-1 binding in muscle-enriched embryos and RNA expression in muscle-enriched embryos across different mutations” (accession number GSE28563, including GSM707199–GSM707213), “Genome-wide maps of HLH-1 binding in muscle-enriched embryos” (accession number GSE28561, including GSM707199–GSM707202), and “Genome-wide RNA expression in muscle-enriched embryos across different mutations” (accession number GSE28562, including GSM707203–GSM707213).
Acknowledgments
Some nematode strains used in this work were provided by the the Caenorhabditis Genetics Center, funded by the NIH National Center for Research Resources (NCRR). We thank M. Krause for the anti-HLH-1 antibody, K. Fisher for her assistance in generating density graphs, S. Pepke for her assistance in generating conservation graphs, A. Kirilusha for his motif counter, H. Amrhein for assistance in managing genomic databases, M. Anaya and J. Vielmetter for purification of the antibody, I. Antoshechkin and L. Schaeffer for library making and sequencing, J. DeModena for assistance in obtaining reagents and helping grow worms, and Wormbase. This work was supported by NIGMS (GM084389 to P.W.S. and B.J.W.), the HHMI, with which P.W.S. is an investigator, the National Human Genome Research Institute (NHGRI), the Beckman Institute at Caltech, and the NCRR.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.133306.111.
References
- Albertson DG, Thomson JN 1976. The pharynx of Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci 275: 299–325 [DOI] [PubMed] [Google Scholar]
- Amin NM, Hu K, Pruyne D, Terzic D, Bretscher A, Liu J 2007. A Zn-finger/FH2-domain containing protein, FOZI-1, acts redundantly with CeMyoD to specify striated body wall muscle fates in the Caenorhabditis elegans postembryonic mesoderm. Development 134: 19–29 [DOI] [PubMed] [Google Scholar]
- Baugh LR, Hunter CP 2006. MyoD, modularity, and myogenesis: Conservation of regulators and redundancy in C. elegans. Genes Dev 20: 3342–3346 [DOI] [PubMed] [Google Scholar]
- Baugh LR, Hill AA, Slonim DK, Brown EL, Hunter CP 2003. Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development 130: 889–900 [DOI] [PubMed] [Google Scholar]
- Baugh LR, Hill AA, Claggett JM, Hill-Harfe K, Wen JC, Slonim DK, Brown EL, Hunter CP 2005a. The homeodomain protein PAL-1 specifies a lineage-specific regulatory network in the C. elegans embryo. Development 132: 1843–1854 [DOI] [PubMed] [Google Scholar]
- Baugh LR, Wen JC, Hill AA, Slonim DK, Brown EL, Hunter CP 2005b. Synthetic lethal analysis of Caenorhabditis elegans posterior embryonic patterning genes identifies conserved genetic interactions. Genome Biol 6: R45 doi: 10.1186/gb-2005-6-5-r45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkes CA, Bergstrom DA, Penn BH, Seaver KJ, Knoepfler PS, Tapscott SJ 2004. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol Cell 14: 465–477 [DOI] [PubMed] [Google Scholar]
- Black BL, Olson EN 1998. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol 14: 167–196 [DOI] [PubMed] [Google Scholar]
- Blackwell TK, Bowerman B, Priess JR, Weintraub H 1994. Formation of a monomeric DNA binding domain by Skn-1 bZIP and homeodomain elements. Science 266: 621–628 [DOI] [PubMed] [Google Scholar]
- Bowerman B, Eaton BA, Priess JR 1992. skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell 68: 1061–1075 [DOI] [PubMed] [Google Scholar]
- Brenner S 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broitman-Maduro G, Owraghi M, Hung WW, Kuntz S, Sternberg PW, Maduro MF 2009. The NK-2 class homeodomain factor CEH-51 and the T-box factor TBX-35 have overlapping function in C. elegans mesoderm development. Development 136: 2735–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Yao Z, Sarkar D, Lawrence M, Sanchez GJ, Parker MH, MacQuarrie KL, Davison J, Morgan MT, Ruzzo WL, et al. 2010. Genome-wide MyoD binding in skeletal muscle cells: A potential for broad cellular reprogramming. Dev Cell 18: 662–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Krause M, Draper B, Weintraub H, Fire A 1992. Body-wall muscle formation in Caenorhabditis elegans embryos that lack the MyoD homolog hlh-1. Science 256: 240–243 [DOI] [PubMed] [Google Scholar]
- Chen L, Krause M, Sepanski M, Fire A 1994. The Caenorhabditis elegans MYOD homologue HLH-1 is essential for proper muscle function and complete morphogenesis. Development 120: 1631–1641 [DOI] [PubMed] [Google Scholar]
- Corsi AK, Kostas SA, Fire A, Krause M 2000. Caenorhabditis elegans twist plays an essential role in non-striated muscle development. Development 127: 2041–2051 [DOI] [PubMed] [Google Scholar]
- Corsi AK, Brodigan TM, Jorgensen EM, Krause M 2002. Characterization of a dominant negative C. elegans Twist mutant protein with implications for human Saethre-Chotzen syndrome. Development 129: 2761–2772 [DOI] [PubMed] [Google Scholar]
- Davidson EH 2007. The regulatory genome: Gene regulatory networks in development and evolution Academic Press, San Diego, CA. [Google Scholar]
- Dilworth FJ, Seaver KJ, Fishburn AL, Htet SL, Tapscott SJ 2004. In vitro transcription system delineates the distinct roles of the coactivators pCAF and p300 during MyoD/E47-dependent transactivation. Proc Natl Acad Sci 101: 11593–11598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper BW, Mello CC, Bowerman B, Hardin J, Priess JR 1996. MEX-3 is a KH domain protein that regulates blastomere identity in early C. elegans embryos. Cell 87: 205–216 [DOI] [PubMed] [Google Scholar]
- Fox JE, Burow ME, McLachlan JA, Miller CA 3rd 2008. Detecting ligands and dissecting nuclear receptor-signaling pathways using recombinant strains of the yeast Saccharomyces cerevisiae. Nat Protoc 3: 637–645 [DOI] [PubMed] [Google Scholar]
- Francis GR, Waterston RH 1985. Muscle organization in Caenorhabditis elegans: Localization of proteins implicated in thin filament attachment and I-band organization. J Cell Biol 101: 1532–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushige T, Krause M 2005. The myogenic potency of HLH-1 reveals wide-spread developmental plasticity in early C. elegans embryos. Development 132: 1795–1805 [DOI] [PubMed] [Google Scholar]
- Fukushige T, Brodigan TM, Schriefer LA, Waterston RH, Krause M 2006. Defining the transcriptional redundancy of early bodywall muscle development in C. elegans: Evidence for a unified theory of animal muscle development. Genes Dev 20: 3395–3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. 2010. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330: 1775–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczy P, Echeverri C, Oegema K, Coulson A, Jones SJ, Copley RR, Duperon J, Oegema J, Brehm M, Cassin E, et al. 2000. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature 408: 331–336 [DOI] [PubMed] [Google Scholar]
- Gouda K, Matsunaga Y, Iwasaki T, Kawano T 2010. An altered method of feeding RNAi that knocks down multiple genes simultaneously in the nematode Caenorhabditis elegans. Biosci Biotechnol Biochem 74: 2361–2365 [DOI] [PubMed] [Google Scholar]
- Grove CA, De Masi F, Barrasa MI, Newburger DE, Alkema MJ, Bulyk ML, Walhout AJ 2009. A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell 138: 314–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guhathakurta D, Schriefer LA, Hresko MC, Waterston RH, Stormo GD 2002. Identifying muscle regulatory elements and genes in the nematode Caenorhabditis elegans. Pac Symp Biocomput 7: 425–436 [DOI] [PubMed] [Google Scholar]
- GuhaThakurta D, Schriefer LA, Waterston RH, Stormo GD 2004. Novel transcription regulatory elements in Caenorhabditis elegans muscle genes. Genome Res 14: 2457–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe BD, Branda CS, Krause M, Stern MJ, Fire A 1998a. MyoD and the specification of muscle and non-muscle fates during postembryonic development of the C. elegans mesoderm. Development 125: 2479–2488 [DOI] [PubMed] [Google Scholar]
- Harfe BD, Vaz Gomes A, Kenyon C, Liu J, Krause M, Fire A 1998b. Analysis of a Caenorhabditis elegans Twist homolog identifies conserved and divergent aspects of mesodermal patterning. Genes Dev 12: 2623–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter CP, Kenyon C 1996. Spatial and temporal controls target pal-1 blastomere-specification activity to a single blastomere lineage in C. elegans embryos. Cell 87: 217–226 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Shi H, Liu J 2009. Two Hox cofactors, the Meis/Hth homolog UNC-62 and the Pbx/Exd homolog CEH-20, function together during C. elegans postembryonic mesodermal development. Dev Biol 334: 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B 2007. Genome-wide mapping of in vivo protein-DNA interactions. Science 316: 1497–1502 [DOI] [PubMed] [Google Scholar]
- Kenyon C 1986. A gene involved in the development of the posterior body region of C. elegans. Cell 46: 477–487 [DOI] [PubMed] [Google Scholar]
- Kostas SA, Fire A 2002. The T-box factor MLS-1 acts as a molecular switch during specification of nonstriated muscle in C. elegans. Genes Dev 16: 257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, Harrison SW, Xu SQ, Chen L, Fire A 1994. Elements regulating cell- and stage-specific expression of the C. elegans MyoD family homolog hlh-1. Dev Biol 166: 133–148 [DOI] [PubMed] [Google Scholar]
- Kuroyanagi H, Ohno G, Mitani S, Hagiwara M 2007. The Fox-1 family and SUP-12 coordinately regulate tissue-specific alternative splicing in vivo. Mol Cell Biol 27: 8612–8621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei H, Liu J, Fukushige T, Fire A, Krause M 2009. Caudal-like PAL-1 directly activates the bodywall muscle module regulator hlh-1 in C. elegans to initiate the embryonic muscle gene regulatory network. Development 136: 1241–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei H, Fukushige T, Niu W, Sarov M, Reinke V, Krause M 2010. A widespread distribution of genomic CeMyoD binding sites revealed and cross validated by ChIP-Chip and ChIP-Seq techniques. PLoS ONE 5: e15898 doi: 10.1371/journal.pone.0015898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Fire A 2000. Overlapping roles of two Hox genes and the exd ortholog ceh-20 in diversification of the C. elegans postembryonic mesoderm. Development 127: 5179–5190 [DOI] [PubMed] [Google Scholar]
- Liu H, Strauss TJ, Potts MB, Cameron S 2006. Direct regulation of egl-1 and of programmed cell death by the Hox protein MAB-5 and by CEH-20, a C. elegans homolog of Pbx1. Development 133: 641–650 [DOI] [PubMed] [Google Scholar]
- Mathies LD, Henderson ST, Kimble J 2003. The C. elegans Hand gene controls embryogenesis and early gonadogenesis. Development 130: 2881–2892 [DOI] [PubMed] [Google Scholar]
- Michaux G, Legouis R, Labouesse M 2001. Epithelial biology: Lessons from Caenorhabditis elegans. Gene 277: 83–100 [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Okkema PG, Harrison SW, Plunger V, Aryana A, Fire A 1993. Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics 135: 385–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdemir A, Fisher-Aylor KI, Pepke S, Samanta M, Dunipace L, McCue K, Zeng L, Ogawa N, Wold BJ, Stathopoulos A 2011. High resolution mapping of Twist to DNA in Drosophila embryos: Efficient functional analysis and evolutionary conservation. Genome Res 21: 566–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page BD, Zhang W, Steward K, Blumenthal T, Priess JR 1997. ELT-1, a GATA-like transcription factor, is required for epidermal cell fates in Caenorhabditis elegans embryos. Genes Dev 11: 1651–1661 [DOI] [PubMed] [Google Scholar]
- Penn BH, Bergstrom DA, Dilworth FJ, Bengal E, Tapscott SJ 2004. A MyoD-generated feed-forward circuit temporally patterns gene expression during skeletal muscle differentiation. Genes Dev 18: 2348–2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepke S, Wold B, Mortazavi A 2009. Computation for ChIP-seq and RNA-seq studies. Nat Methods 6: S22–S32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts MB, Wang DP, Cameron S 2009. Trithorax, Hox, and TALE-class homeodomain proteins ensure cell survival through repression of the BH3-only gene egl-1. Dev Biol 329: 374–385 [DOI] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Shingles J, Dupuy D, Grove CA, Walhout AJ, Vidal M, Hope IA 2007. Insight into transcription factor gene duplication from Caenorhabditis elegans Promoterome-driven expression patterns. BMC Genomics 8: 27 doi: 10.1186/1471-2164-8-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spieth J, Shim YH, Lea K, Conrad R, Blumenthal T 1991. elt-1, an embryonically expressed Caenorhabditis elegans gene homologous to the GATA transcription factor family. Mol Cell Biol 11: 4651–4659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern MJ, Horvitz HR 1991. A normally attractive cell interaction is repulsive in two C. elegans mesodermal cell migration mutants. Development 113: 797–803 [DOI] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol 56: 110–156 [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol 100: 64–119 [DOI] [PubMed] [Google Scholar]
- Venkatesan K, McManus HR, Mello CC, Smith TF, Hansen U 2003. Functional conservation between members of an ancient duplicated transcription factor family, LSF/Grainyhead. Nucleic Acids Res 31: 4304–4316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Zhao J, Corsi AK 2006. Identification of novel target genes of CeTwist and CeE/DA. Dev Biol 293: 486–498 [DOI] [PubMed] [Google Scholar]
- Weinmann AS, Farnham PJ 2002. Identification of unknown target genes of human transcription factors using chromatin immunoprecipitation. Methods 26: 37–47 [DOI] [PubMed] [Google Scholar]
- Williams BD, Waterston RH 1994. Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J Cell Biol 124: 475–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai I, Baugh LR, Smith JJ, Roehrig C, Shen-Orr SS, Claggett JM, Hill AA, Slonim DK, Hunter CP 2008. Pairing of competitive and topologically distinct regulatory modules enhances patterned gene expression. Mol Syst Biol 4: 163 doi: 10.1038/msb.2008.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Wang P, Corsi AK 2007. The C. elegans Twist target gene, arg-1, is regulated by distinct E box promoter elements. Mech Dev 124: 377–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W, Sternberg PW 2006. Genome-wide prediction of C. elegans genetic interactions. Science 311: 1481–1484 [DOI] [PubMed] [Google Scholar]
- Zhong M, Niu W, Lu ZJ, Sarov M, Murray JI, Janette J, Raha D, Sheaffer KL, Lam HY, Preston E, et al. 2010. Genome-wide identification of binding sites defines distinct functions for Caenorhabditis elegans PHA-4/FOXA in development and environmental response. PLoS Genet 6: e1000848 doi: 10.1371/journal.pgen.1000848 [DOI] [PMC free article] [PubMed] [Google Scholar]