Background: Replicative DNA polymerases δ and ϵ are believed to synthesize lagging and leading strands, respectively.
Results: Human DNA polymerases α/δ and ϵ segregate during S phase and DNA polymerase ϵ alone remains bound to lamins.
Conclusion: DNA polymerases δ and ϵ act independently in late S phase
Significance: Human cell DNA replication may mechanistically differ from prokaryotic replication.
Keywords: Chromatin, Chromatin Immunoprecipitation (ChiP), DNA Polymerase, DNA Replication, DNA-Protein Interaction
Abstract
DNA polymerases (Pol) α, δ, and ϵ replicate the bulk of chromosomal DNA in eukaryotic cells, Pol ϵ being the main leading strand and Pol δ the lagging strand DNA polymerase. By applying chromatin immunoprecipitation (ChIP) and quantitative PCR we found that at G1/S arrest, all three DNA polymerases were enriched with DNA containing the early firing lamin B2 origin of replication and, 2 h after release from the block, with DNA containing the origin at the upstream promoter region of the MCM4 gene. Pol α, δ, and ϵ were released from these origins upon firing. All three DNA polymerases, Mcm3 and Cdc45, but not Orc2, still formed complexes in late S phase. Reciprocal ChIP of the three DNA polymerases revealed that at G1/S arrest and early in S phase, Pol α, δ, and ϵ were associated with the same nucleoprotein complexes, whereas in late S phase Pol ϵ and Pol α/δ were largely associated with distinct complexes. At G1/S arrest, the replicative DNA polymerases were associated with lamins, but in late S phase only Pol ϵ, not Pol α/δ, remained associated with lamins. Consistently, Pol ϵ, but not Pol δ, was found in nuclear matrix fraction throughout the cell cycle. Therefore, Pol ϵ and Pol α/δ seem to pursue their functions at least in part independently in late S phase, either by physical uncoupling of lagging strand maturation from the fork progression, or by recruitment of Pol δ, but not Pol ϵ, to post-replicative processes such as translesion synthesis or post-replicative repair.
Introduction
Three DNA polymerases (Pol)4 α, δ, and ϵ replicate the bulk of the eukaryotic genome (for reviews, see Refs. 1–3). Pol α is unique among DNA polymerases by having an intrinsic primase. It is therefore able to start DNA synthesis de novo (reviewed in Refs. 4 and 5). The primase acts as a DNA-dependent RNA polymerase synthesizing an RNA primer of about 10 bases long, which is then extended by the DNA polymerase activity of Pol α complex to about 30 bases. For duplication of simian virus 40 (SV40) DNA, a classic model system for eukaryotic DNA replication, replication factor C is specifically bound to these primers and expels Pol α. Replication factor C then loads the ring-shaped proliferating cell nuclear antigen (PCNA) to form a sliding clamp around the double-stranded DNA at the primer end, and recruits Pol δ, which synthesizes both leading strand DNA and Okazaki fragments of the lagging strand, the latter being then processed to a continuous strand (for review, see Ref. 6). Besides Pol α and δ, a third large DNA polymerase, Pol ϵ, was found to be essential for yeast Sacharomyces cerevisiae (7), and it was found to be involved in synthesis of chromosomal DNA in human cells (8–10). It is also required for efficient DNA synthesis in Xenopus egg extracts (11). It has been recently found that S. cerevisiae Pol δ and ϵ harboring mutations that confer specific mutation patterns to the enzymes, sign their mutational signatures to lagging and leading strand, respectively (2, 12, 13). Based on this evidence and on former work (for review, see Ref. 14) it is safe to conclude that Pol δ is a main player in synthesis of lagging strand DNA, whereas Pol ϵ is predominantly involved in the synthesis of the leading strand DNA.
However, there is also evidence according to which the division of labor between Pol δ and ϵ may be more complex than a simple splitting between lagging and leading strands, respectively. The deletion of the domain containing polymerase and proofreading exonuclease motifs from S. cerevisiae causes growth and replication defects but the deletion is not lethal (15, 16), indicating that in this case, like in SV40 DNA replication, Pol δ is able to synthesize both strands. Furthermore, when the proofreading activity of Pol δ is mutationally inactivated, the mutation rate is significantly higher than in cells having analogous mutation in Pol ϵ (17, 18). Amino acid substitutions in the polymerase domain of Pol δ also seem to generate a higher increase in the mutation rates and cause more severe growth defects than analogous amino acid substitutions in Pol ϵ (19). Further evidence conflicting with the current model comes from studies of human cells. We previously found that (i) a neutralizing antibody against Pol ϵ inhibits DNA synthesis in permeabilized nuclei more efficiently in the early S phase than in the late S phase, whereas the contrary is true for antibodies against Pol δ, and that (ii) trapping of Pol ϵ to nascent DNA remained nearly constant throughout the S phase, whereas Pol δ was three to four times more intensely cross-linked to nascent DNA in late compared with early S phase, and that (iii) the chromatin-bound fraction of Pol δ, unlike Pol ϵ, increased in the late S phase (20). These results suggest that the contribution of Pol δ to DNA synthesis increases toward the late S phase, whereas that of Pol ϵ either decreases or remains constant. In contrast, Fuss and Linn (21) proposed that Pol ϵ acts in the replication of heterochromatin during late S phase based on the observation that in immunofluorescense microscopy, the enzyme is neighboring PCNA foci and sites of DNA synthesis in early S phase but co-localizes with these sites in late S phase. Our previous study also suggested that ultrastructural localization of the Pol δ and ϵ were essentially distinct although minor colocalization was also detected (20). Depletion of the activity of Pol δ or ϵ in higher eukaryotes causes distinct defects for genome duplication (10, 22), arguing for different contributions to DNA replication. All these observations were not expected if Pol ϵ and δ are part of the same replication fork complex, acting on the leading and the lagging strands, respectively, in a similar manner as Pol III in Escherichia coli cells (for review, see Ref. 23). They could still be explained if (i) Pol δ is involved in delayed maturation of accumulating Okazaki fragments or both strands independent of Pol ϵ, (ii) Pol δ, but not Pol ϵ is increasingly involved in post-replicative processes such as DNA translesion synthesis or post-replicative DNA recombination, or if other than in yeast, (iii) the share of labor of Pol δ and ϵ has changed from yeast to human, i.e. human Pol ϵ acts more at early origins of replication and Pol δ more at late origins of replication on both strands. To address these questions we applied chromatin immunoprecipitation (ChIP) techniques and quantitative PCR to study association and release of the three replicative DNA polymerases, Pol α, δ, and ϵ, with DNA from two origins of replication, the lamin B2 (LB2) gene origin and the upstream promoter region (UPR) of MCM4 gene, firing at 0 and 2 h, respectively, after S phase entry, and to study co-existence of the three replicative DNA polymerases, origin recognition complex (ORC) subunit Orc2, a component of the Mcm-helicase complex Mcm3, and cell division control protein Cdc45 in cross-linked nucleoprotein complexes during S phase. We also studied the presence of lamins A/C in these complexes. The results reveal that Pol α, δ, and ϵ are loaded to and released from both origins of replication during S phase. In G1/S arrested cells Pol α, δ, and ϵ are present in highly purified nucleoprotein complexes containing 200–1000-base pair long DNA fragments and they are associated with lamins. In late S phase Pol α and δ are segregated from Pol ϵ and lamins A/C, whereas Pol ϵ remains associated with lamins. Based on this study and other studies that have been previously published we propose a model according to which Pol ϵ bound to nuclear matrix synthesizes the leading strand and Pol δ synthesizes the lagging strand, but the latter partly trails behind to process still immature lagging strand DNA, and possibly also leading strand DNA, or fulfills post-replicative tasks such as DNA translesion synthesis after Pol ϵ has essentially completed its job.
EXPERIMENTAL PROCEDURES
Antibodies
Primary antibodies used are listed in supplemental Table S1. Rabbit polyclonal K31 antibody against human Pol δ catalytic subunit was raised against a polypeptide corresponding to amino acids 108–276, and rabbit polyclonal K32 and K33 antibodies against human Pol δ catalytic subunit were raised against peptides corresponding to amino acids 297–542. The antigens were expressed as GST fusion proteins and purified by preparative SDS-PAGE after thrombin cleavage of the GST as described (9). Secondary antibodies for Western blotting were either alkaline phosphatase-conjugated or horseradish peroxidase-conjugated (Jackson ImmunoResearch or Chemicon).
Cell Cultivation, Synchronization, and Cell Cycle Analysis
HeLa CCL2 monolayer cells (American Type Culture Collection ATCC, Manassas, VA) were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and antibiotics (Invitrogen) at 37 °C in a 5% carbon dioxide atmosphere. Cells were synchronized by arresting them at G1/S phase with a double thymidine block (28) and then either used as such (0 h) or released from the block to proceed in S phase by replacing the medium containing excess thymidine with medium without addition of thymidine for the indicated times. Human T98G glioblastoma cells (ATCC CRL 1690) were cultured in Eagle's minimum essential medium (Sigma) supplemented with Earle's salts, 10% fetal bovine serum, l-glutamine, nonessential amino acids, and antibiotics at 37 °C in a 5% carbon dioxide atmosphere. T98G cells were starved in medium containing 0.5% serum for 6 days followed by addition of conditioned, complete medium. The quality of the synchrony was analyzed by flow cytometry of propidium iodine-stained cells (29) using a CyFlow Space flow cytometer (Partec GmbH, Münster, Germany) or a Cell Lab Quanta SC flow cytometer (Beckman Coulter).
Chromatin Immunoprecipitation
In vivo cross-linking and subsequent chromatin immunoprecipitations (ChIPs) were performed from double thymidine synchronized, and exponentially growing monolayer HeLa cells as described (30). The steps are essentially (i) in vivo cross-linking treatment of cells with formaldehyde, (ii) homogenization of cells and collection of nuclear material, (iii) isolation of nucleoproteins by CsCl gradient centrifugation, (iv) sonication and digestion of the nucleoproteins with micrococcal nuclease optimized to result in DNA fragments of 200–1000 base pairs (supplemental Fig. S1A), (v) immunoprecipitation with a selected protein antibody, and (vi) division of the immunoprecipitate for Western blotting and DNA extraction. CsCl gradient centrifugation for the isolation of nucleoprotein complexes was performed as follows. Nuclear material was obtained from the swollen cells disrupted by a Dounce homogenization and washed as described (30) and suspended in low-salt NSB buffer (0.1 m NaCl, 10 mm Tris-Cl, 0.1% Nonidet P-40, 1 mm EDTA, pH 8.0). The material was then loaded onto a step gradient made up of cesium chloride solutions of 1.7, 1.5, and 1.3 mg/ml in the gradient buffer (0.5% Sarcosyl, 1 mm EDTA, 20 mm Tris-Cl, pH 8.0). Ultracentrifugation was performed at 37,000 rpm for 24 h at 18 °C with Sorvall ultracentrifuge Ultra Pro 80 using TH-641 swinging bucket rotor. Compact nucleoprotein pellet from the middle of the gradient (supplemental Fig. S1B) was then dialyzed and treated further as described (30).
For Western blotting, proteins were eluted from cross-linked nucleoproteins and for DNA extraction, proteins were hydrolyzed by proteinase K and DNA was purified by phenol-chloroform extraction and ethanol precipitation. Immunoprecipitation reactions contained 5 μg of K19 and 10 μg of K27 for Pol ϵ, 2.5 μg each of K30, K31, K32, and K33 for Pol δ, 5 μg of p140 for Pol α, 5 μg of N18 for lamin A/C, or 5 μg of rabbit IgG as negative control. For colorimetric immunodetection of Pol ϵ, δ, and α, lamin A/C, Mcm3, Cdc45, and Orc2 a mixture of G1A, H3B, and E24C antibodies, a mixture of K30, K31, K32, and K33 antibodies, p140 antibody, lamin A/C (N18) antibody, Mcm3 (N20) antibody, Cdc45 (3G10) antibody, and Orc2 (3B7) antibody were used, respectively. Semi-quantitative analysis of the Western blots was performed with the Analysis Toolbox of the ImageQuant TL program suite (GE Healthcare). Co-immunoprecipitation efficiencies were calculated relative to the signal of the same protein in the input.
Quantitative PCR Analysis
Quantitative PCR (qPCR) reactions were run with the Opticon Monitor program in Chromo 4 Peltier Thermal Cycler (MJ Research). The LB2 gene origin of replication (31) and the origin located in the UPR of the MCM4 gene (32) were utilized in this study, and PCR analysis was performed from the areas shown (Figs. 1 and 2). The primers used are presented in supplemental Table S2. The quantity of specific DNA in the immunoprecipitate is given as a relative ratio of DNA precipitated over input DNA.
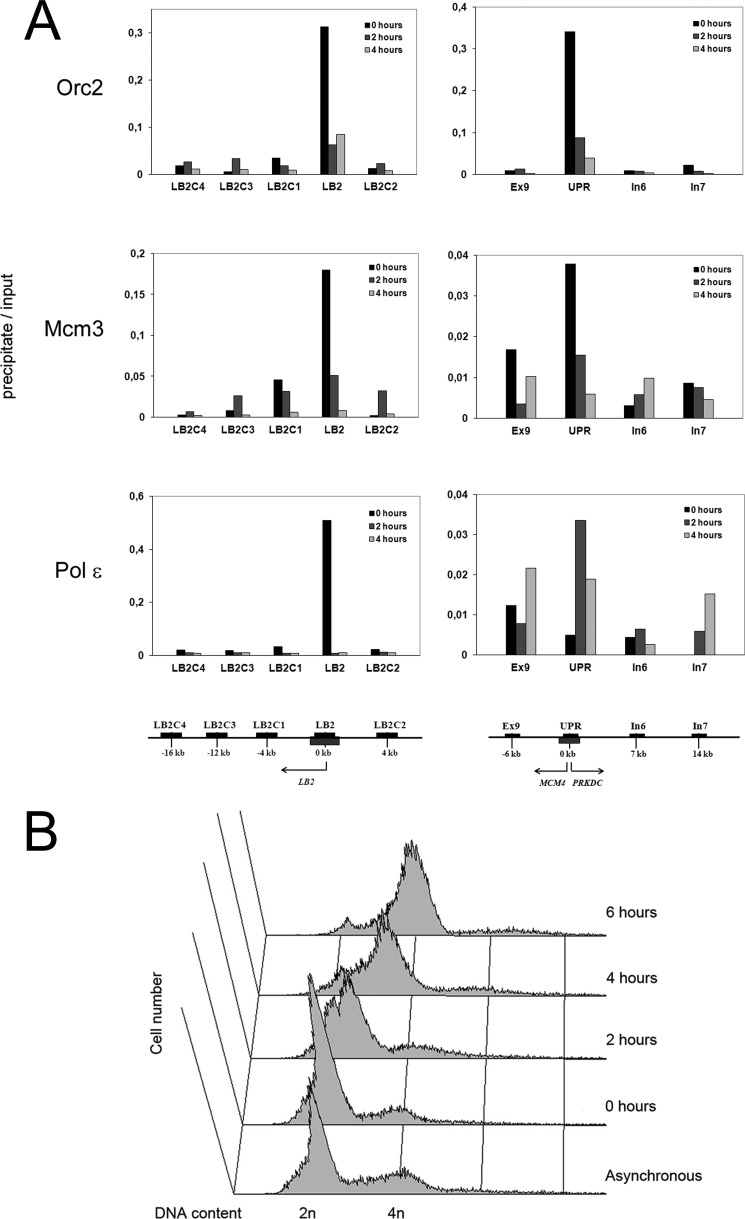
FIGURE 1.
Time course of association of replication proteins with two human origins of DNA replication. A, association with replication origins at the LB2 gene (left panels) and at the UPR of the MCM4 gene (right panels) with nucleoprotein complexes isolated by immunoprecipitation of S phase Orc2, Mcm3, and Pol ϵ. HeLa cells were synchronized by double thymidine block to early S phase (0 h) and released to proceed in S phase (2 and 4 h). Nucleoproteins derived from CsCl centrifugation were sonicated and digested with micrococcal nuclease as described under “Experimental Procedures.” Soluble nucleoproproteins, 1 mg/500 μl, were taken for immunoprecipitation with the cognate antibodies, the protein was digested with proteinase K and DNA isolated. From 5 to 10 ng of DNA were obtained from the immunoprecipitate, 1/20 was used for quantitative PCR (precipitate). DNA from soluble nucleoproteins representing genomic DNA was isolated and treated in the similar manner, except that the immunoprecipitation step was omitted, and analyzed by quantitative PCR (input). The values are mean values from two distinct experiments. The regions analyzed by qPCR are indicated in the panels and their locations are shown below the panels, for LB2 origin and UPR origin, see the left and right panels, respectively. The LB2 gene is on the left site of the replication origin as indicated by an arrow. The MCM4 gene is on the left and PRKDC gene on right site of the replication origin as presented by arrows. The MCM4 gene encodes the minichromosome maintenance protein 4 and the PRKDC gene the catalytic subunit of the DNA-dependent protein kinase. The origin regions are shown as gray boxes below the axis. Black bars above the axis represent regions that were amplified by qPCR. B, cell cycle progression of G1/S-arrested cells released to progress into S phase as verified by FACS of propidium iodine-treated cells.
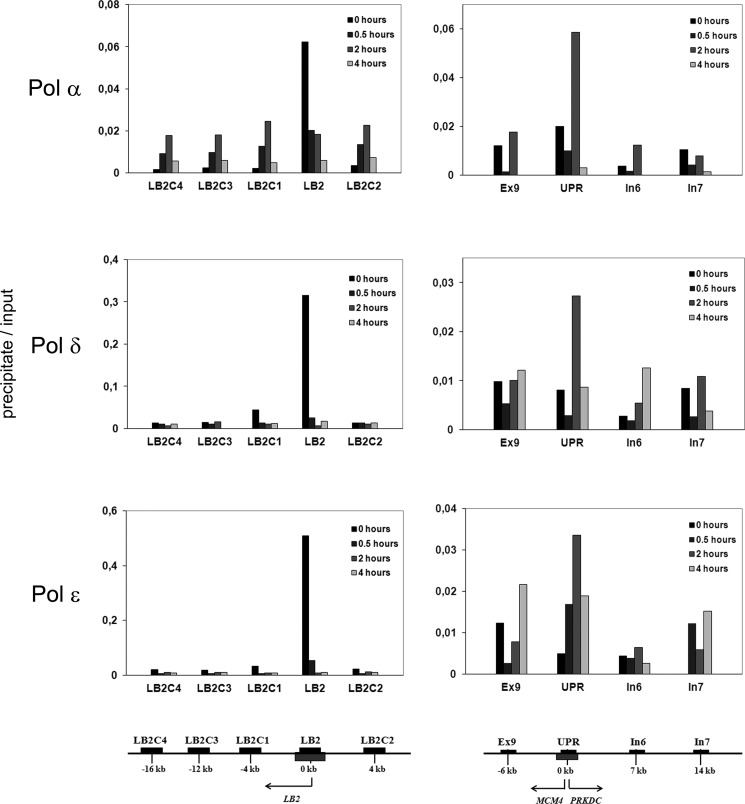
FIGURE 2.
Time course of the association of origin of replication DNA within the LB2 gene (left panels) and the UPR of the MCM4 gene (right panels) with nucleoprotein complexes isolated by immunoprecipition of S phase Pol α, δ, and ϵ. The regions analyzed by qPCR are indicated in the panels and their locations are shown below the panels, the LB2 origin in the left panel and UPR origin in the right panel. The origins are shown as gray boxes. Black bars represent regions that were amplified by quantitative PCR. The genomic organization and the location of the PCR products are identical to Fig. 1. The cells were synchronized by double thymidine block to early S phase (0 h) and released to proceed in S phase (0.5, 2, and 4 h). Nucleoproteins derived from CsCl centrifugation were sonicated and digested with micrococcal nuclease as described under “Experimental Procedures.” Soluble nucleoproproteins, 1 mg/500 μl, were taken for immunoprecipitation with the cognate antibodies, the protein was digested with proteinase K, and DNA isolated. 5 to 10 ng of DNA was obtained from the immunoprecipitate, 1/20 was used for quantitative PCR (precipitate). DNA from soluble nucleoproteins representing genomic DNA was isolated and treated in the similar manner, except that the immunoprecipitation step was omitted, and analyzed by quantitative PCR (input). The values are mean values from two distinct experiments.
Cell Fractionation Studies
100-mm plates of T98G cells were cooled to 4 °C, washed twice with cold TBS (150 mm NaCl, 20 mm Tris-Cl, pH 7.5) and twice with hypotonic KM buffer (10 mm NaCl, 1 mm MgCl2, 2 mm DTT, and 10 mm MOPS-NaOH, pH 7.0). Cells were then lysed by hypotonic extraction with 1 ml of KM buffer containing 0.5% Nonidet P-40 for 30 min with occasional gentle agitation. The resulting supernatant represented the detergent-soluble fraction (S). The cell remnants were washed twice with KAc buffer (5 mm potassium acetate, 0.5 mm MgCl2, 2 mm DTT, and 30 mm HEPES-KOH, pH 7.4) followed by incubation with 1 ml of DNase I solution (150 mm NaCl, 1.5 mm CaCl2, 6 mm MgCl2, complete EDTA-free protease inhibitors, 50 μg/ml of RNase A, 50 units/ml of DNase I, and 40 mm Tris-Cl, pH 8.0) at room temperature for one-half hour. The supernatant containing proteins released by the DNase I digest represented the chromatin-bound fraction (B) and was collected. The remaining cell matrix was washed twice with DNase solution (without DNase I and RNase A) and solubilized by addition of 1 ml of lysis buffer (100 mm NaCl, 0.5 mm MgCl2, 0.5 mm DTT, 5 mm KCl, 0.5% SDS, and 20 mm HEPES-KOH, pH 7.7). The insoluble matrix fraction (M) was collected using a cell scraper. Total cell extract (T) was prepared by washing a parallel 100-mm plate twice with cold TBS followed by lysis with lysis buffer and cell scraper. All samples were rapidly frozen in liquid nitrogen and stored at −70 °C. For Western detection, samples corresponding to an equal number of cells of each fraction were separated by SDS-PAGE, transferred onto a PVDF membrane and detected using chemiluminescence reagents (Pierce and Bio-Rad). Antibodies PC10, 3G10, PDG-1E8, and a combination of G1A, H3B, and E24C were used for detection of PCNA, Cdc45, Pol δ and ϵ, respectively.
RESULTS
Association of Pol ϵ Defines Replication Timing of the Lamin B2 Gene Origin and the MCM4 Gene UPR Origin
In the yeast S. cerevisiae, Pol ϵ has been found to be the main leading strand replicase, whereas Pol δ replicates the bulk of the lagging strand. Considering the central importance of DNA replication for cell function, one could expect a similar share of labor also at the human replication fork. On the other hand, studies of replication dynamics of human DNA polymerases have left the possibility that Pol ϵ may be more strongly involved in either early S phase DNA synthesis (20), or in replication of heterochromatin in late S phase (21). As an attempt to address this issue, we first studied enrichment of the DNA from two replication origins, the LB2 gene origin (31) and the UPR origin at the MCM4 gene (30) in nucleoprotein complexes derived by the ChIP method (see “Experimental Procedures”) with Orc2, Mcm3, and Pol ϵ antibodies. In HeLa cells arrested in early S phase by double thymidine block, LB2 DNA was strongly associated with immunoprecipitates of Orc2, Mcm3, and Pol ϵ (Fig. 1A), the enrichment being 10–20-fold compared with proximal regions. At 2 and 4 h after release from the thymidine arrest enrichment of the LB2 DNA in Mcm3 immunoprecipitates disappeared, whereas Orc2 immunoprecipitates still showed about 3–4-fold enrichment even at 4 h after release. In contrast, enrichment of the LB2 DNA in Pol ϵ immunoprecipitates was very low already at 0.5 h and practically disappeared at 2 h after release. Enrichment of DNA of the UPR of the MCM4 gene in immunoprecipitates was not as pronounced as enrichment of DNA of the LB2 region. As seen for LB2, MCM4 UPR enrichment was also highest in immunoprecipitates of Orc2 and Mcm3 from G1/S-arrested cells at 0 h showing about 20-fold MCM4 UPR enrichment for Orc2 immunoprecipitations and 3–7-fold for Mcm3 immunoprecipitations (Fig. 1A). These results confirmed previous studies that found that Orc2 (30) and Mcm3 (32) bind to the MCM4 UPR origin already in G1 phase and remain bound with the origin DNA in early S phase. In contrast, no enrichment of MCM4 UPR was detectable at 0 h in Pol ϵ immunoprecipitates. Instead, enrichment of MCM4 UPR origin DNA in Pol ϵ immunoprecipitates peaked at 2 h after the release from the arrest and its qPCR detection showed a 4–6-fold higher level when compared with proximal regions of MCM4 UPR origin, although its enrichment in Orc2 and Mcm3 immunoprecipitates had already decreased down to 4- and 2-fold, respectively. Therefore, loading of Pol ϵ to and subsequent release from this origin of replication takes place significantly later than at the LB2 origin. These findings suggest that the MCM4 UPR origin fires about 2 h later than the LB2 origin. The latter was already previously shown to be an early firing origin in HeLa cells (33). As enrichment of the MCM4 UPR origin DNA in Orc2 and Pol ϵ immunoprecipitate persists up to 4 h after the release from the thymidine arrest (Fig. 1A) (30), this origin fires in mid-S phase, and probably with less efficiency than the LB2 origin.
DNA Polymerases α, δ, and ϵ Are All Loaded to and Released from Both LB2 Origin and MCM4 Gene UPR Origin
We then analyzed the origin DNA of nucleoprotein complexes purified from precipitates of each replicative DNA polymerase. All three Pol α, δ, and ϵ associated strongly with the LB2 origin in G1/S-arrested cells at 0 h (Fig. 2). Enrichment of nascent DNA was about 20-fold for each enzyme complex. The replicative DNA polymerases were rapidly released from LB2 origin after cells proceeded in S phase because enrichment of LB2 origin DNA dropped rapidly to only 2–3-fold as early as 0.5 h after release from the double thymidine block. Obviously the three DNA polymerases moved away from the origin along with proceeding replication forks. The firing of the origin and progression of the forks at G1/S-arrested cells immediately after release of the block obviously takes place with a relatively high synchrony, as the flanking regions did not show any enrichment after 0.5 h. It seems that all three DNA polymerases are involved in the early DNA synthesis from this early origin of replication. For the later firing origin in the UPR of the MCM4 gene, enrichment of the origin fragment reached a maximum at 2 h with a 3–6-fold increase of that of the control DNA, and this applied equally to immunoprecipitates of all three DNA polymerases (Fig. 2). In contrast, no enrichment of the MCM4 UPR origin DNA was found at 0- or 4-h time points. Enrichment of MCM4 UPR origin DNA at 2 h in precipitates of all three replicative DNA polymerases provides further evidence that this origin fires later than the LB2 origin. These results suggest that the three replicative DNA polymerases are all involved in early synthesis of DNA from both the LB2 origin and the MCM4 UPR origin that fires about 2 h later than the LB2 origin. Altogether, loading and release of all three replicative DNA polymerases at these two origins of replication suggests that they are all involved in the initiation and progression of DNA replication from both origins. It is therefore not likely that the share of labor at the fork differs in the later firing MCM4 UPR origin from the early firing LB2 origin. Therefore, these results do not provide an explanation for the more active role of Pol δ in overall replicative DNA synthesis in late S phase when compared with Pol ϵ (20).
In G1/S-arrested Cells All Three Replicative DNA Polymerases Are Present in the Same Nucleoprotein Complexes, but in Late S Phase Pol ϵ Behaves Distinct from Pol α/δ
If Pol δ and ϵ act at all replication forks as proposed by studies on yeast cells (13, 14) and confirmed here for selected origins in human cells, it could be expected that the two replicases act as a complex responsible for simultaneous synthesis of both strands like Pol III in E. coli cells (for review, see Ref. 22). We therefore studied the presence of all three replicative DNA polymerases, the ORC subunit Orc2, and the Mcm helicase component Mcm3 in highly purified nucleoprotein complexes isolated by ChIP at different stages of S phase. We included also Cdc45, because this protein forms on one hand an integral part of the Cdc45-MCM-GINS (CMG) complex considered the active form of the replicative DNA helicase (10, 34, 35). On the other hand, Cdc45 also appears to mediate the contact between the MCM proteins and the replicative DNA polymerases (see Ref. 27, reviewed in Ref. 36). Orc2, Mcm3, and Cdc45 were present in nucleoprotein complexes in cells arrested in G1/S and separately precipitated individually with antibodies against the three replicative DNA polymerases (Fig. 3A), although the Orc2 signal was very weak in Pol δ precipitates. ChIP performed late in S phase at 6 h showed that Mcm3 and Cdc45 were still present in all three DNA polymerase precipitates, whereas Orc2 was absent. These results are consistent with earlier studies revealing that origin recognition complex proteins are not present at progressing replication forks (37, 38), whereas Mcm3 and Cdc45 travel along with the forks (37). At 6 h, firing of origins is rare compared with progressive forks and therefore, the contacts between ORC complex proteins and elongation proteins have obviously been lost. In contrast, Mcm3 and Cdc45 are expected to be present in cross-linked nucleoprotein complexes purified by immunoprecipitation with antibodies against replicative DNA polymerases if these DNA polymerases were present at progressing forks. As this is the case (Fig. 3A), the results suggest that all three replicative DNA polymerases act at forks, or close to forks still at the 6-h time point. Although it should be noted that the length of the DNA at cross-linked nucleoprotein complexes is 200–1000 bp, and therefore the presence of the three replicative DNA polymerases in these complexes may not indicate a direct interaction between them.
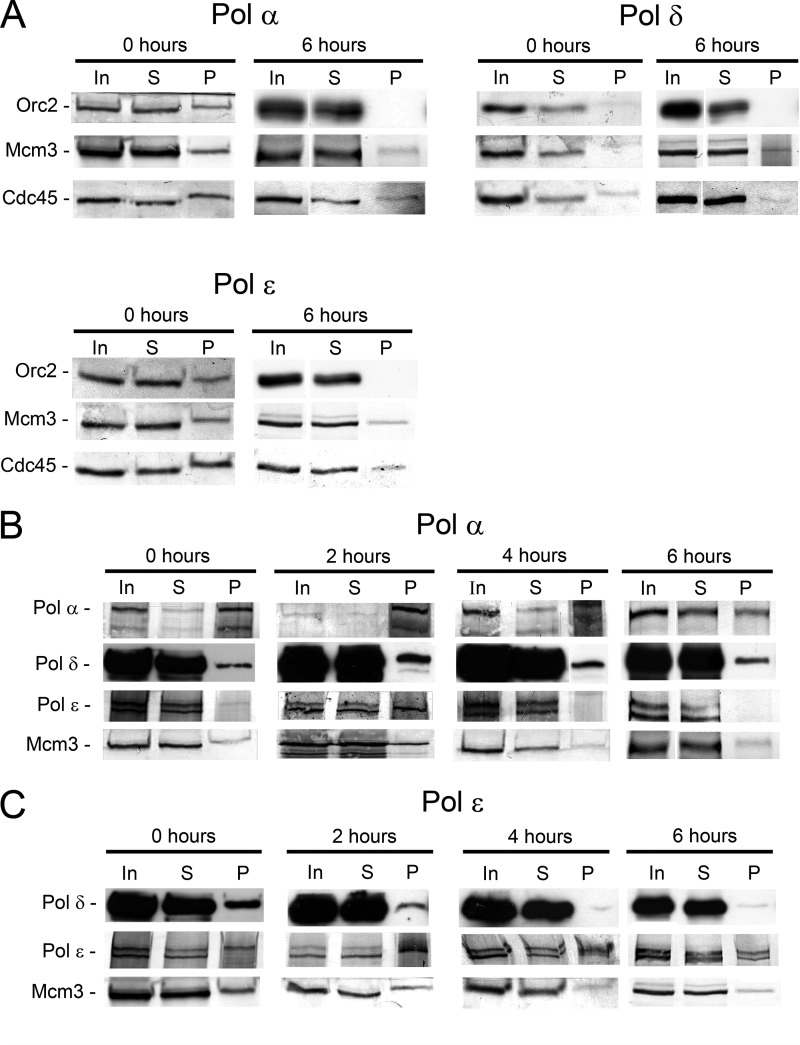
FIGURE 3.
A, association of Orc2, Mcm3, and Cdc45 with S phase nucleoprotein complexes isolated by precipitation with antibodies against replicative DNA polymerase. B, association of Pol δ and Pol ϵ with early S phase nucleoprotein complexes immunoprecipitated with Pol α antibodies. C, association of Pol δ with S phase nucleoprotein complexes immunoprecipitated with Pol ϵ antibodies. The cells were synchronized by double thymidine block to G1/S (0 h) and released to proceed in S phase. The antibody used for immunoprecipitation is indicated above each panel and the antibody used for Western blotting on the left site of the panels. Nucleoproteins were isolated and treated as described under “Experimental Procedures” for chromatin immunoprecipitation. Nucleoproteins derived from CsCl centrifugation were sonicated and digested with micrococcal nuclease as described under “Experimental Procedures.” Soluble nucleoproproteins, 1 mg/500 μl, were taken for immunoprecipitation with the cognate antibodies. 10 μl of soluble nucleoprotein complex (In), 10 μl of supernatant after precipitation (S), and the entire precipitate (P) were loaded onto gel.
If all three DNA polymerases are associated with stable replication complexes comparable to the Pol III holoenzyme in E. coli, nucleoprotein complexes should have a comparable protein composition, including replicative DNA polymerases and other replication factors, independent of the polymerase antibody that was used for ChIP. To address this we studied the presence of Pol δ and ϵ in nucleoprotein complexes precipitated with Pol α antibodies. Pol δ and Pol ϵ were both present in these immunoprecipitates from G1/S-arrested cells at 0 h (Fig. 3B) as can be expected on their obvious presence at early firing replication origins at this time (see above). Pol δ was present at all later time points studied 2, 4, and 6 h in Pol α immunoprecipitates, but Pol ϵ was no longer detectable at 4 and 6 h, indicating, that in late S phase Pol α and Pol δ were still present in the same nucleoprotein complexes, suggesting that they still acted at the same forks, but Pol ϵ was essentially no longer present in these complexes. Consistently, when nucleoprotein fragments were immunoprecipitated with Pol ϵ antibodies, Pol δ was present in seemingly large quantities at 0 h and in smaller quantities still at 2 h, but despite the very high sensitivity of the antibodies used here for detection of Pol δ, only a weak band is detectable at 4 and 6 h (Fig. 3C). The corresponding experiment with Pol δ was not informative, because the signal for Pol α and ϵ in Pol δ-chromatin precipitates was very weak throughout S phase (supplemental Fig. S3). The extensive purification procedure for the isolation of the nucleoprotein includes a CsCl density gradient centrifugation that efficiently separates the nucleoproteins from free protein complexes and free nucleic acids. Therefore, the ChIPs analyzed here represent the chromatin-associated fractions of the DNA polymerases, and not soluble pools of nonproductive enzymes. Taken together these results suggest that the three replicative DNA polymerases are all loaded to the origin of replication and co-exist in nucleoprotein complexes purified with DNA polymerase antibodies, but already at 4 h in S phase, Pol α and Pol δ are processing DNA in complexes that are essentially free of Pol ϵ. Altogether, these results suggest that in late S phase Pol ϵ and Pol α/δ act essentially independently, although the three DNA polymerases are all still in nucleoprotein complexes containing Mcm3 and Cdc45 proteins.
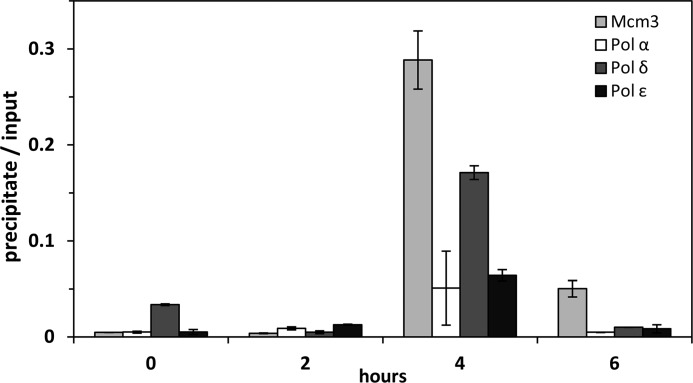
Because we are not aware of any well characterized origins of DNA replication fired late in S phase, we decided to analyze the association of the replicative polymerases and Mcm3 with β-satellites, a repetitive element that has been proposed to represent heterochromatin and to be replicated exclusively late in S phase (39). By selecting a repetitive element, we sought to be able to detect progressing replication forks. Performing quantitative real time PCR with a primer pair specific for the 68-bp β-satellite, we observed a significant enrichment of these repeats in ChIPs of Mcm3 as well as Pol α, δ, and ϵ at 4 h in S phase (Fig. 4). This result not only supports the view that β-satellites are mainly replicated late in S phase, but also suggests that the three Pol all participate in the replication of these sequences.
FIGURE 4.
Time course of the association of β-satellite DNA with nucleoprotein complexes isolated by immunoprecipition of S phase Mcm3, Pol α, δ, and ϵ. The association was studied by qPCR with primers specific for the 24-bp β-satellite repeat as indicated under “Experimental Procedures.” This repeat has previously been suggested to be replicated in late S phase in human cells (39). The cells were synchronized by double thymidine block to early S phase (0 h) and released to proceed in S phase (2, 4, and 6 h).
DNA Polymerase ϵ, but Not DNA Polymerase δ Is Associated with Nuclear Matrix throughout the Cell Cycle
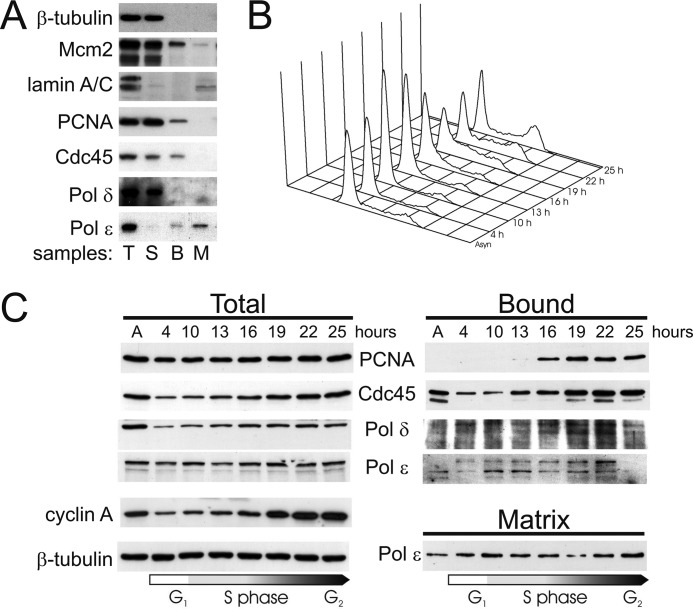
We studied nuclear association of Pol δ and ϵ by fractionation of human T98G glioblastoma cells to further analyze the distinct contexts in which Pol δ and ϵ may act. T98G cells respond to serum deprivation and can therefore be synchronized without interference with the checkpoint response. Hypotonic extraction in the presence of Nonidet P-40 was performed to release soluble proteins including the nucleoplasmic fraction. Nuclear remnants were then treated with DNase I to release chromatin-bound proteins and the remaining insoluble material was classified as “matrix” fraction representing largely the nuclear matrix. Total cell extract was prepared from parallel plates and all extracts were analyzed by Western blotting using marker antibodies (supplemental Table S1, Fig. 5A). β-Tubulin was completely found in the soluble fraction indicating that bound and matrix fractions were largely free of soluble proteins. Mcm2 appeared both in soluble and DNA-bound fractions as expected (40). Lamins A/C could be detected almost exclusively in the matrix fraction, whereas the soluble and chromatin-bound fractions were largely free of lamins A/C, demonstrating that these fractions were substantially free of contaminating nuclear matrix. In asynchronous cells PCNA, Cdc45, and Pol δ were predominantly in the soluble fraction, a minor part of each being bound to chromatin, and none to the matrix (Fig. 5A). Pol ϵ behaved in a different manner. The majority of the enzyme was in the matrix fraction, a small amount was released by DNase I, and only very little was found to be soluble.
FIGURE 5.
Chromatin and matrix association of replicative DNA polymerases during S phase. T98G cells synchronized by serum stimulation were subjected to subnuclear fractionation and relevant proteins were analyzed by Western blotting. A, analysis of marker proteins and selected replication factors in fractionated, asynchronous T98G cells. T, total cell extract; S, soluble fraction; B, chromatin bound fraction; and M, matrix fraction. B, cell cycle progression of the cell synchronization of T98G cells was employed for fractionation. Propidium iodide-stained cells at the indicated times after serum stimulation were analyzed by flow cytometry. C, time course for the occurrence of Pol δ, Pol ϵ, and Cdc45 in fractions from serum-stimulated synchronous T98G cells. Times after the addition of serum are presented on the top of the panel.
To analyze the nuclear dynamic of replication proteins during S phase, T98G cells were starved in G0 phase by serum deprivation and induced to grow by re-addition of serum. Samples from the indicated time points were analyzed by flow cytometry to verify synchronous re-entry into the cycle (Fig. 5B). PCNA and Pol δ became bound to chromatin after cells had entered S phase at 16 h (Fig. 5C, supplemental Fig. S2). Cdc45 remained bound to chromatin throughout the cell cycle, but there was an increase in binding toward late S phase. It appears that chromatin association of Cdc45 preceded that of PCNA and Pol δ, consistent with the role of the former in initiation of DNA replication (41). Pol ϵ behaved completely different from the other proteins analyzed. In contrast to these replication proteins that were bound to chromatin or were in the soluble fraction (Fig. 5C, supplemental Fig. S2), Pol ϵ was mainly in the insoluble matrix fraction resistant to detergent and DNase extraction, and its abundance in the bound fraction peaked in mid-S phase at 22 h (Fig. 5, B and C). Only a small portion of Pol ϵ was soluble or remained chromatin-bound throughout the cell cycle (supplemental Fig. S2B). Notably, the amount of a form of Pol ϵ with reduced electrophoretic mobility increased in the chromatin fraction, whereas the form with normal electrophoretic mobility decreased at the same time (Fig. 5C, supplemental Fig. 2B). The observed difference in the association with nuclear fractions and its time course are consistent with our previous study on replicative polymerases during S phase (20) and with the results presented above. In particular, it is noteworthy that Pol ϵ, unlike Pol δ remains essentially bound to nuclear matrix throughout the cell cycle (Fig. 5C).
The Three Replicative DNA Polymerases Associate with Lamins A/C at G1/S Arrest, but Only DNA Polymerase ϵ Remains Bound to Lamins in Late S Phase
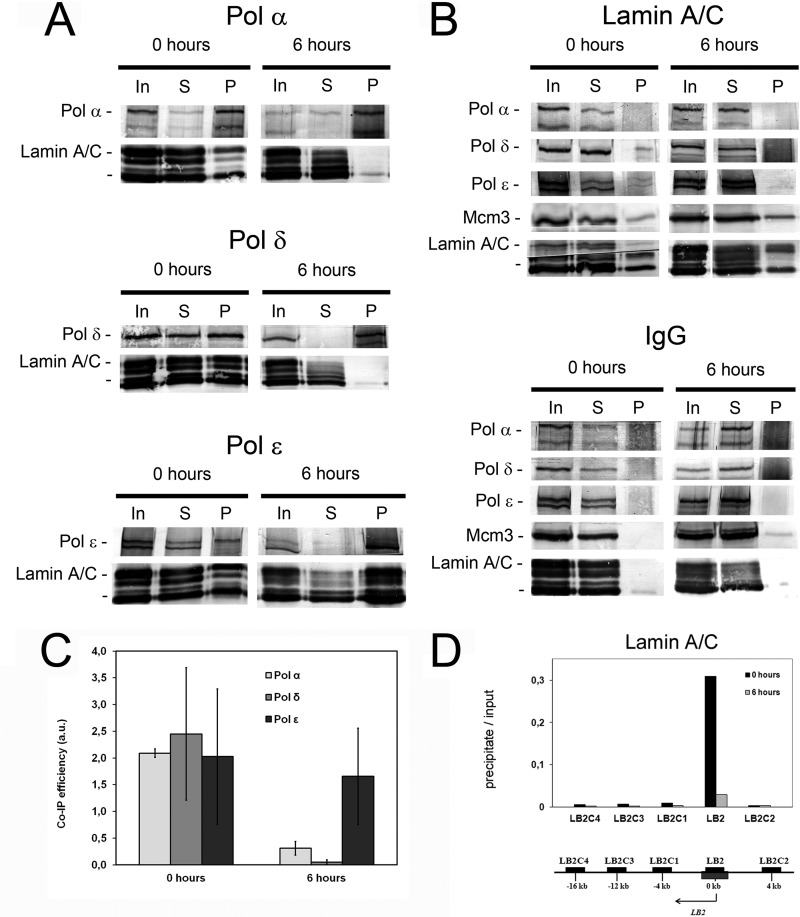
Association of Pol ϵ with nuclear matrix prompted us to study whether lamins would be components of replicative nucleoprotein complexes purified by ChIP. We found that in nucleoprotein complexes purified by immunoprecipitation with DNA polymerase antibodies from G1/S-arrested cells, lamins A/C were abundant in precipitates of all three replicative polymerases (Fig. 6, A and C). This is not surprising taking into account that Pol ϵ is continuously bound to nuclear matrix, and all three replicative polymerases seem to be bound to replication initiation sites in these conditions (above). When nucleoprotein complexes were immunoprecipitated with antibodies against lamin A/C that are structural proteins and hence very much more abundant than replicative polymerases, signals of Pol ϵ and δ, and Mcm3, but not of Pol α, were still obtained (Fig. 6B).
FIGURE 6.
Association of DNA polymerases with lamins A/C in S phase. HeLa cells were synchronized by double thymidine block to early S phase (0 h) and released to proceed in S phase (6 h). A, co-precipitation of lamin A/C in chromatin immunoprecipitates of Pol α, δ, and ϵ. B, co-precipitation of replicative DNA polymerases in lamin A/C chromatin immunoprecipitates. The antibody used for immunoprecipitation is indicated on top and the antibody used for Western blotting on left side of the panels. Nucleoproteins for chromatin immunoprecipitation were isolated and treated as described under “Experimental Procedures.” Nucleoproteins derived from CsCl centrifugation were sonicated and digested with micrococcal nuclease as described under “Experimental Procedures.” Soluble nucleoproproteins, 1 mg/500 μl, were taken for immunoprecipitation with the cognate antibodies. 10 μl of soluble nucleoprotein complex (In), 10 μl of supernatant after precipitation (S), and the entire precipitate (P) were loaded onto the gel. C, quantification of the lamin A/C in chromatin immunoprecipitates with antibodies against Pol α, δ, and ϵ, respectively. The lamin A/C signal in the precipitate of the cognate DNA polymerase was quantified relative to the signal in the input on the same blot. The values represent the mean ± S.D. of two to four independent experiments. D, association of LB2 origin DNA with lamins A/C at 0 and 6 h after release from double thymidine arrest in nucleoprotein complexes. Nucleoproteins derived from CsCl centrifugation were sonicated and digested with micrococcal nuclease as described under “Experimental Procedures.” Soluble nucleoproproteins, 1 mg/500 μl, were taken for immunoprecipitation with the cognate antibodies, the protein was digested with proteinase K, and DNA isolated. 5 to 10 ng of DNA were obtained from the immunoprecipitate, 1/20 was used for quantitative PCR (precipitate). DNA from soluble nucleoproteins representing genomic DNA was isolated and treated in the similar manner, except that the immunoprecipitation step was omitted, and analyzed by quantitative PCR (input). The values are mean values from two distinct experiments.
At 6 h a strong signal from lamins A/C was obtained in the Pol ϵ ChIP, but essentially no signal in immunoprecipitates of Pol α and Pol δ (Fig. 6, A and C). Consistently, reciprocal ChIPs with lamin A/C antibodies resulted in signals for Pol ϵ and Mcm3, but not for Pol α and Pol δ (Fig. 6B). These results indicate that binding of Pol ϵ to nuclear matrix (Fig. 5) is most likely mediated by lamins. Control precipitates utilizing a nonspecific IgG fraction analyzed on the same membrane did not yield a discernible signal for any of the proteins analyzed, thereby confirming the specificity of the ChIPs (Fig. 6B).
Analysis of the DNA of the nucleoprotein complexes isolated from G1/S-arrested cells by lamin A/C antibodies revealed about 20-fold enrichment of LB2 origin DNA when compared with proximal regions, but at 6 h this enrichment had essentially disappeared (Fig. 6D). Obviously, since associated with DNA elongating replication complexes, lamins A/C were also released from the origin of replication after firing, and, as suggested above, retained its association with Pol ϵ but not with Pol α/δ. Thus, whereas in early S phase, nucleoprotein complexes containing all three replicative polymerases, Mcm3, Cdc45, Orc2, and lamin A/C were detectedable, in late S phase, Pol α/δ and Pol ϵ represented distinguishable complexes, the latter marked by the presence of lamin A/C (Figs. 3 and 6).
DISCUSSION
Applying the ChIP technique, we found that in HeLa cells all three replicative DNA polymerases, Pol α, δ, and ϵ, are bound to and released from both early firing LB2 origin and MCM4 UPR origin that was found to fire 2 h later in mid-S phase (Figs. 1 and 2). Nucleoprotein complexes purified by immunoprecipitation with Pol α, δ, or ϵ antibodies from S phase cells were highly enriched with DNA fragments representing the two origins at the time preceding the firing, followed by a rapid decrease after firing. Therefore, both replicases are loaded to the two origins and disappear together with Mcm3, when replication forks have moved away from the origin. This is consistent with studies in yeast suggesting that Pol ϵ acts mainly at leading strand and Pol δ mainly at lagging strand synthesis (for review, see Ref. 3).
All three DNA polymerases were associated with the same nucleoprotein complexes in G1/S-arrested cells, as can be expected if they all bind to origins of replication and are associated with each other or are even physically coupled at the replication fork at this time. In late S phase the three polymerases are all still likely to be at replication forks, as shown by the presence of Mcm3 and Cdc45 proteins in nucleoprotein complexes purified by immunoprecipitation with cognate Pol antibodies (Fig. 3A). However, in late S phase Pol δ and α seem to be essentially devoid of Pol ϵ, and vice versa, suggesting that Pol α/δ and Pol ϵ are now mainly associated with distinct complexes. This suggests that Pol δ and ϵ are physically uncoupled in late S phase. This is consistent with the fact that Pol ϵ alone remains associated with nuclear matrix, obviously through lamin A/C, whereas Pol α and δ are only associated with lamins in early S phase, most likely as components of replication complexes.
How can the lack of association between Pol α/δ and Pol ϵ in late S phase be explained? First, the share of labor could be different in late compared with early S phase, such that late S phase forks have a different DNA polymerase composition. The co-localization of Pol ϵ with sites of DNA synthesis in late, but not early S phase (21), would be consistent with an augmented role of Pol ϵ in late S phase replication. Nevertheless, profiling of replication errors generated by an asymmetric mutator variant of Pol δ in the budding yeast indicate that Pol δ synthesizes the lagging-strand throughout the genome (13), and argues strongly against such a model. We found in a previous study that in late S phase, Pol δ is associated more strongly with newly synthesized DNA and chromatin than Pol ϵ, and that replication in isolated nuclei is more strongly inhibited in late S than in early S phase by a monoclonal antibody inhibiting Pol δ (20). Therefore, alternatively, this can be explained in a manner proposed by Pavlov and Shcherbakova (14). In their model Pol ϵ in principle replicates the leading strand, but is switched to Pol δ at pause sites or sites of translesion synthesis. In this way, Pol ϵ would synthesize most of the leading strand, but Pol δ would synthesize the parts of the leading strand after replication fork arrest and the restart of DNA synthesis. The fact that the Pol domain of Pol ϵ is not essential for viability of yeast (15) and that Pol ϵ is not needed for SV40 DNA replication (6, 8, 9) are two examples of the ability of Pol δ to substitute for Pol ϵ at the leading strand. Alternatively, it is possible that replicative Pol α, δ, and ϵ in late S phase are increasingly involved in tasks that are no longer closely associated with the replication fork. It has been reported that a considerable amount of DNA translesion synthesis may be performed after and independent of DNA replication (42). There may also be a differential requirement of DNA polymerases, e.g. for the termination of DNA replication, or for the rescue of stalled and reversed forks by recombinational processes.
Finally, Pol δ and ϵ may be physically separated in late S phase but are still synthesizing DNA at the lagging and leading strand of the same forks. This view is supported by the fact that all three replicative polymerases are associated with Mcm3 in the ChIP of highly purified nucleoprotein fractions (Fig. 3). Furthermore, β-satellite repeats become enriched in ChIP of Mcm3 as well as Pol α, δ, and ϵ 4 h after release from the thymidine block, consistent with the β-satellites being replicated late in S phase (39) under participation of all three replicative polymerases. We consider the possibility that synthesis of the lagging strand is distributive, i.e. a new molecule of Pol α and Pol δ is recruited from a nucleoplasmic pool, and that the lagging strand replication machinery may be released from the fork during initiation of the following lagging strand. In this way, maturation of several successive Okazaki fragments may be ongoing simultaneously, and Pol δ molecules may remain associated with chromatin after completion and joining of Okazaki fragments. Such a model would explain both the increasing association of Pol δ with chromatin (Fig. 5B) as well as the increased involvement in DNA synthesis as S phase progresses (20). In contrast, the leading strand would be synthesized largely progressively by the same Pol ϵ molecule attached to the nuclear scaffold. There is no evidence on heterodimer formation by Pol δ and ϵ (see e.g. Ref. 43) that would represent a human counterpart of the E. coli Pol III holoenzyme dimer or trimer that is stably associated with the core replication proteins (44, 45) and carries out simultaneous synthesis of both strands. Pol α, δ, and ϵ are all accumulated at replication roadblocks (46), but they did not form a stable complex with a replication progression complex (47). And although coupling of Pol α and ϵ with the Cdc45-Mcm2–7-GINS (CMG) helicase complex has been described (48, 49), replicases are easily uncoupled from DNA unwinding after inhibition of DNA synthesis (50) or after DNA damage (51). Therefore, transient association and dissociation of the replicative DNA polymerases with the CMG complex is likely to allow their flexible utilization for the duplication of complex human genome.
It is possible that a combination of all three, different involvement of the Pol δ and ϵ in DNA replication-associated processes such as DNA translesion synthesis, increasing involvement of the replicative DNA polymerases in late S phase in tasks that are no longer closely associated with replication forks, and repeated recruitment of new Pol δ molecules from a nucleoplasmic pool for the lagging strand synthesis, account for the dynamics of Pols α, δ, and ϵ in their mutual association described here.
Supplementary Material
Acknowledgment
We are indebted to Leena Pääkkönen for excellent technical assistance.
This work was supported by Academy of Finland Grants 106986 (to H. P.) and 123082 (to J. E. S.).

This article contains supplemental Tables S1 and S2 and Figs. S1–S3.
- Pol
- polymerase(s)
- PCNA
- proliferating cell nuclear antigen
- ORC
- origin recognition complex
- qPCR
- quantitative PCR
- UPR
- upstream promoter region.
REFERENCES
- 1. Stillman B. (2008) DNA polymerases at the replication fork in eukaryotes. Mol. Cell 30, 259–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nick McElhinny S. A., Gordenin D. A., Stith C. M., Burgers P. M., Kunkel T. A. (2008) Division of labor at the eukaryotic replication fork. Mol. Cell 30, 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Johansson E., Macneill S. A. (2010) The eukaryotic replicative DNA polymerases take shape. Trends Biochem. Sci. 35, 339–347 [DOI] [PubMed] [Google Scholar]
- 4. Nasheuer H. P., Grosse F. (1988) DNA polymerase α-primase from calf thymus. Determination of the polypeptide responsible for primase activity. J. Biol. Chem. 263, 8981–8988 [PubMed] [Google Scholar]
- 5. Nasheuer H. P., Smith R., Bauerschmidt C., Grosse F., Weisshart K. (2002) Initiation of eukaryotic DNA replication. Regulation and mechanisms. Prog. Nucleic Acids Res. Mol. Biol. 72, 41–94 [DOI] [PubMed] [Google Scholar]
- 6. Waga S., Stillman B. (1998) The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67, 721–751 [DOI] [PubMed] [Google Scholar]
- 7. Morrison A., Araki H., Clark A. B., Hamatake R. K., Sugino A. (1990) A third essential DNA polymerase in S. cerevisiae. Cell 62, 1143–1151 [DOI] [PubMed] [Google Scholar]
- 8. Zlotkin T., Kaufmann G., Jiang Y., Lee M. Y., Uitto L., Syväoja J., Dornreiter I., Fanning E., Nethanel T. (1996) DNA polymerase ϵ may be dispensable for SV40, but not cellular, DNA replication. EMBO J. 15, 2298–2305 [PMC free article] [PubMed] [Google Scholar]
- 9. Pospiech H., Kursula I., Abdel-Aziz W., Malkas L., Uitto L., Kastelli M., Vihinen-Ranta M., Eskelinen S., Syväoja J. E. (1999) A neutralizing antibody against human DNA polymerase ϵ inhibits cellular but not SV40 DNA replication. Nucleic Acids Res. 27, 3799–3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bermudez V. P., Farina A., Raghavan V., Tappin I., Hurwitz J. (2011) Studies on human DNA polymerase ϵ and GINS complex and their role in DNA replication. J. Biol. Chem. 286, 28963–28977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shikata K., Sasa-Masuda T., Okuno Y., Waga S., Sugino A. (2006) The DNA polymerase activity of Pol ϵ holoenzyme is required for rapid and efficient chromosomal DNA replication in Xenopus egg extracts. BMC Biochem. 7, 21–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pursell Z. F., Isoz I., Lundström E. B., Johansson E., Kunkel T. A. (2007) Yeast DNA polymerase ϵ participates in leading-strand DNA replication. Science 317, 127–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Larrea A. A., Lujan S. A., Nick McElhinny S. A., Mieczkowski P. A., Resnick M. A., Gordenin D. A., Kunkel T. A. (2010) Genome-wide model for the normal eukaryotic DNA replication fork. Proc. Natl. Acad. Sci. U.S.A. 107, 17674–17679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pavlov Y. I., Shcherbakova P. V. (2010) DNA polymerases at the eukaryotic fork. 20 Years Later. Mutat. Res. 685, 45–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kesti T., Flick K., Keränen S., Syväoja J. E., Wittenberg C. (1999) DNA polymerase ϵ catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol. Cell 3, 679–685 [DOI] [PubMed] [Google Scholar]
- 16. Dua R., Levy D. L., Campbell J. L. (1999) Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiae Pol ϵ and its unexpected ability to support growth in the absence of the DNA polymerase domain. J. Biol. Chem. 274, 22283–22288 [DOI] [PubMed] [Google Scholar]
- 17. Morrison A., Johnson A. L., Johnston L. H., Sugino A. (1993) Pathway correcting DNA replication errors in Saccharomyces cerevisiae. EMBO J. 12, 1467–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Morrison A., Sugino A. (1994) The 3′–>5′ exonucleases of both DNA polymerases delta and epsilon participate in correcting errors of DNA replication in Saccharomyces cerevisiae. Mol. Gen. Genet. 242, 289–296 [DOI] [PubMed] [Google Scholar]
- 19. Pavlov Y. I., Shcherbakova P. V., Kunkel T. A. (2001) In vivo consequences of putative active site mutations in yeast DNA polymerases α, ϵ, δ, and ζ. Genetics 159, 47–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rytkönen A. K., Vaara M., Nethanel T., Kaufmann G., Sormunen R., Läärä E., Nasheuer H. P., Rahmeh A., Lee M. Y., Syväoja J. E., Pospiech H. (2006) Distinctive activities of DNA polymerases during human DNA replication. FEBS J. 273, 2984–3001 [DOI] [PubMed] [Google Scholar]
- 21. Fuss J., Linn S. (2002) Human DNA polymerase ϵ colocalizes with proliferating cell nuclear antigen and DNA replication late, but not early, in S phase. J. Biol. Chem. 277, 8658–8666 [DOI] [PubMed] [Google Scholar]
- 22. Fukui T., Yamauchi K., Muroya T., Akiyama M., Maki H., Sugino A., Waga S. (2004) Distinct roles of DNA polymerases δ and ϵ at the replication fork in Xenopus egg extracts. Genes to Cells 9, 179–191 [DOI] [PubMed] [Google Scholar]
- 23. McHenry C. S. (2011) DNA replicases from a bacterial perspective. Annu. Rev. Biochem. 80, 403–436 [DOI] [PubMed] [Google Scholar]
- 24. Stadlbauer F., Brueckner A., Rehfuess C., Eckerskorn C., Lottspeich F., Förster V., Tseng B. Y., Nasheuer H. P. (1994) DNA replication in vitro by recombinant DNA polymerase α-primase. Eur. J. Biochem. 222, 781–793 [DOI] [PubMed] [Google Scholar]
- 25. Uitto L., Halleen J., Hentunen T., Höyhtyä M., Syväoja J. E. (1995) Structural relationship between DNA polymerases ϵ and ϵ* and their occurrence in eukaryotic cells. Nucleic Acids Res. 23, 244–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mäkiniemi M., Hillukkala T., Tuusa J., Reini K., Vaara M., Huang D., Pospiech H., Majuri I., Westerling T., Mäkelä T. P., Syväoja J. E. (2001) BRCT domain-containing protein TopBP1 functions in DNA replication and damage response. J. Biol. Chem. 276, 30399–30406 [DOI] [PubMed] [Google Scholar]
- 27. Bauerschmidt C., Pollok S., Kremmer E., Nasheuer H. P., Grosse F. (2007) Interactions of human Cdc45 with the Mcm2–7 complex, the GINS complex, and DNA polymerases δ and ϵ during S phase. Genes Cells 12, 745–758 [DOI] [PubMed] [Google Scholar]
- 28. Johnson R. T., Downes C. S., Meyn R. E. (1993) in The Cell Cycle: A Practical Approach, pp. 1–24, IRL Press, Oxford, United Kingdom [Google Scholar]
- 29. Vindeløv L. L., Christensen I. J., Nissen N. I. (1983) Standardization of high-resolution flow cytometric DNA analysis by the simultaneous use of chicken and trout red blood cells as internal reference standards. Cytometry 3, 328–331 [DOI] [PubMed] [Google Scholar]
- 30. Ladenburger E. M., Keller C., Knippers R. (2002) Identification of a binding region for human origin recognition complex proteins 1 and 2 that coincides with an origin of DNA replication. Mol. Cell Biol. 22, 1036–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Abdurashidova G., Deganuto M., Klima R., Riva S., Biamonti G., Giacca M., Falaschi A. (2000) Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science 287, 2023–2026 [DOI] [PubMed] [Google Scholar]
- 32. Schaarschmidt D., Ladenburger E. M., Keller C., Knippers R. (2002) Human Mcm proteins at a replication origin during the G1 to S phase transition. Nucleic Acids Res. 30, 4176–4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Giacca M., Zentilin L., Norio P., Diviacco S., Dimitrova D., Contreas G., Biamonti G., Perini G., Weighardt F., Riva S. (1994) Fine mapping of a replication origin of human DNA. Proc. Natl. Acad. Sci. U.S.A. 91, 7119–7123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ilves I., Petojevic T., Pesavento J. J., Botchan M. R. (2010) Activation of the MCM2–7 helicase by association with Cdc45 and GINS proteins. Mol. Cell 37, 247–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Costa A., Ilves I., Tamberg N., Petojevic T., Nogales E., Botchan M. R., Berger J. M. (2011) The structural basis for MCM2–7 helicase activation by GINS and Cdc45. Nat. Struct. Mol. Biol. 18, 471–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pospiech H., Grosse F., Pisani F. M. (2010) The initiation step of eukaryotic DNA replication. Subcell. Biochem. 50, 79–104 [DOI] [PubMed] [Google Scholar]
- 37. Aparicio O. M., Weinstein D. M., Bell S. P. (1997) Components and dynamics of DNA replication complexes in S. cerevisiae. Redistribution of MCM proteins and Cdc45p during S phase. Cell 91, 59–69 [DOI] [PubMed] [Google Scholar]
- 38. Xu W., Aparicio J. G., Aparicio O. M., Tavaré S. (2006) Genome-wide mapping of ORC and Mcm2p binding sites on tiling arrays and identification of essential ARS consensus sequences in S. cerevisiae. BMC Genomics 7, 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ten Hagen K. G., Cohen S. N. (1993) Timing of replication of β satellite repeats of human chromosomes. Nucleic Acids Res. 21, 2139–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Todorov I. T., Attaran A., Kearsey S. E. (1995) BM28, a human member of the MCM2–3-5 family, is displaced from chromatin during DNA replication. J. Cell Biol. 129, 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bell S. P., Dutta A. (2002) DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71, 333–374 [DOI] [PubMed] [Google Scholar]
- 42. Karras G. I., Jentsch S. (2010) The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell 141, 255–267 [DOI] [PubMed] [Google Scholar]
- 43. Rytkönen A. K., Hillukkala T., Vaara M., Sokka M., Jokela M., Sormunen R., Nasheuer H. P., Nethanel T., Kaufmann G., Pospiech H., Syväoja J. E. (2006) DNA polymerase ϵ associates with the elongating form of RNA polymerase II and nascent transcripts. FEBS J. 273, 5535–5549 [DOI] [PubMed] [Google Scholar]
- 44. McInerney P., Johnson A., Katz F., O'Donnell M. (2007) Characterization of a triple DNA polymerase replisome. Mol. Cell 27, 527–538 [DOI] [PubMed] [Google Scholar]
- 45. Georgescu R. E., Yao N. Y., O'Donnell M. (2010) Single-molecule analysis of the Escherichia coli replisome and use of clamps to bypass replication barriers. FEBS Lett. 584, 2596–2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pacek M., Tutter A. V., Kubota Y., Takisawa H., Walter J. C. (2006) Localization of MCM2–7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol. Cell 21, 581–587 [DOI] [PubMed] [Google Scholar]
- 47. Gambus A., Jones R. C., Sanchez-Diaz A., Kanemaki M., van Deursen F., Edmondson R. D., Labib K. (2006) GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 8, 358–366 [DOI] [PubMed] [Google Scholar]
- 48. Lou H., Komata M., Katou Y., Guan Z., Reis C. C., Budd M., Shirahige K., Campbell J. L. (2008) Mrc1 and DNA polymerase ϵ function together in linking DNA replication and the S phase checkpoint. Mol. Cell 32, 106–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gambus A., van Deursen F., Polychronopoulos D., Foltman M., Jones R. C., Edmondson R. D., Calzada A., Labib K. (2009) A key role for Ctf4 in coupling the MCM2–7 helicase to DNA polymerase α within the eukaryotic replisome. EMBO J. 28, 2992–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pacek M., Walter J. C. (2004) A requirement for MCM7 and Cdc45 in chromosome unwinding during eukaryotic DNA replication. EMBO J. 23, 3667–3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Byun T. S., Pacek M., Yee M. C., Walter J. C., Cimprich K. A. (2005) Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 19, 1040–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.