Plants and pathogens evolve in response to each other. This co-evolutionary arms race is fueled by genetic variation underlying the recognition of pathogen proteins by the host and the defeat of host defenses by the pathogen. Together with new mutations, genetic diversity in populations of both the host and pathogen represent a pool of possible variants to maintain adaptation via natural selection.
Drastic changes in genetic diversity in crop species have occurred as a consequence of domestication. Whether changes in the genetic composition of these host populations also have affected genetic diversity in pathogen species is, so far, poorly understood. Advances in comparative genomics and population genomic approaches open new avenues to study adaptive processes in plant pathogens and to infer the impact of agro-ecosystems on the evolution of pathogen populations. Here we summarize new insights gained from comparative genome studies and population genomics in host-pathogen systems.
1. What Are the Evolutionary Consequences of Domestication in Crop Species?
In the process of domestication, crop species have undergone striking changes in both morphology and physiology. At the genetic level, these phenotypic changes have been brought by strong directional selection on a few genes [1]. Genome-wide analyses of domesticated crop species and their wild relatives have provided new insights into the evolutionary consequences of domestication. While only a modest, yet growing, number of genes associated with domestication have been identified [2]–[4], genome-wide “footprints” of domestication are well documented in several crop species [5]–[8]. Most notably the level of genetic variation has been dramatically reduced in many domesticated species relative to their wild ancestors [1]. This is best explained by intense selection on a small subset of genotypes exhibiting desirable phenotypes. Such strong directional selection also entails a number of important evolutionary side effects. First, genetic diversity can be swept away in genomic regions flanking strongly selected domestication genes [9]. Second, strong selection on one gene can reduce the efficacy of selection at neighboring loci, which, in return, may lead to an accumulation of slightly deleterious mutations. This process was first documented as an accumulation of non-synonymous mutations in Asian rice [5] and has since been termed the “cost” of domestication (see, e.g., [1], [10]). The loss of variation and the cost of domestication in genomes of crop species may compromise the level of natural defenses against pathogens and render them more susceptible than their wild relatives.
2. How Does the Agro-Ecosystem Affect Evolution of Plant Pathogens?
Agro-ecosystems brought by domestication have favored the emergence and specialization of new pathogen species by providing a new ecological niche. High densities of host individuals, genetic homogeneity of host populations, and recently the facilitated long distance dispersal of propagules by anthropogenic activities are factors that are greatly conducive for the propagation of pathogens in agro-ecosystems [11]. However, successful propagation in the field also poses challenges for pathogens: the new agricultural environment imposes strong directional selection on the pathogen genome, notably on genes controlling the defeat of crop resistance genes and resistance towards pesticides. Speciation in an agricultural environment can also have drastic implications for the population biology of pathogens as exemplified by the potato late blight pathogen Phythophthora infestans [12], the rice blast pathogen Magnaporthe oryzae [13], and the yellow rust pathogen Puccinia striiformis [14]. The emergence of these three species was associated with a strong reduction in sexual recombination and the spread of only few specialized clonal lineages. Notably, genetic diversity among “domesticated” asexual pathogen lineages was significantly reduced compared to their wild relatives. However, even when sexual recombination is maintained in the pathogen, the transition to an agricultural host can entail founder events where substantial genetic variation is lost. Comparisons of levels of genetic diversity in pathogens occurring on domesticated versus non-cultivated hosts have confirmed this hypothesis [15], [16]. Substantial loss of genetic diversity is often associated with speciation and specialization to a crop host, yet several studies report rapid emergence and adaptation of plant pathogens to crop species [17]. This states the paradox that despite relatively modest levels of genetic variation, pathogen populations can readily adapt to agro-ecosystems and rapidly respond to new introduced resistance genes, fungicides, and other disease control agents.
3. How Have Plant Pathogens Emerged in Agro-Ecosystems?
Comparative genomics and population genomic analyses have provided new insights into genome evolution, speciation, and the origin of pathogenicity traits. A main conclusion from these studies is that fungal pathogens can exhibit very high levels of genome plasticity and plasticity itself may be instrumental in enabling the emergence of new virulence traits [18], [19].
In the plant pathogen Verticillium dahliae a population genomics analysis was used to identify a determinant of race specificity [20]. Genome re-sequencing of four “race 1” isolates and six “race 2” isolates of V. dahliae led to the identification of a 50 kb fragment only present in race 1 isolates. Subsequent RNAseq data revealed only a single highly expressed ORF in this region. Experimental analyses confirmed the determining role of this gene as a virulence factor in race 1 strains of the pathogen. V. dahliae is a predominantly asexual pathogen and the acquisition of this virulence factor, possibly through a horizontal gene transfer event, underlines the importance of genome plasticity in adaptive evolution of fungal pathogens.
In addition to horizontal gene transfer, new pathogenicity traits may emerge from the crossing of genetically rather distant individuals. Population genomic analysis of the grass pathogen Zymoseptoria pseudotritici revealed a particular evolutionary history for this species [21]. The genome of Z. pseudotritici harbors a mosaic of long fragments without any polymorphisms interspersed with fragments exhibiting two distinct haplotypes. This pattern is consistent with a recent interspecific mating of just two genotypes resulting in a successful new species formation via hybrid speciation. The underlying molecular traits responsible for pathogenicity in the hybrid still remain to be identified. However, the broad distribution of Z. pseudotritici at its center of origin demonstrates how new genomic combinations mediated through interspecific hybridization can lead to the successful emergence of new pathogens.
4. How Do Fungi Adapt and Specialize to their Environment?
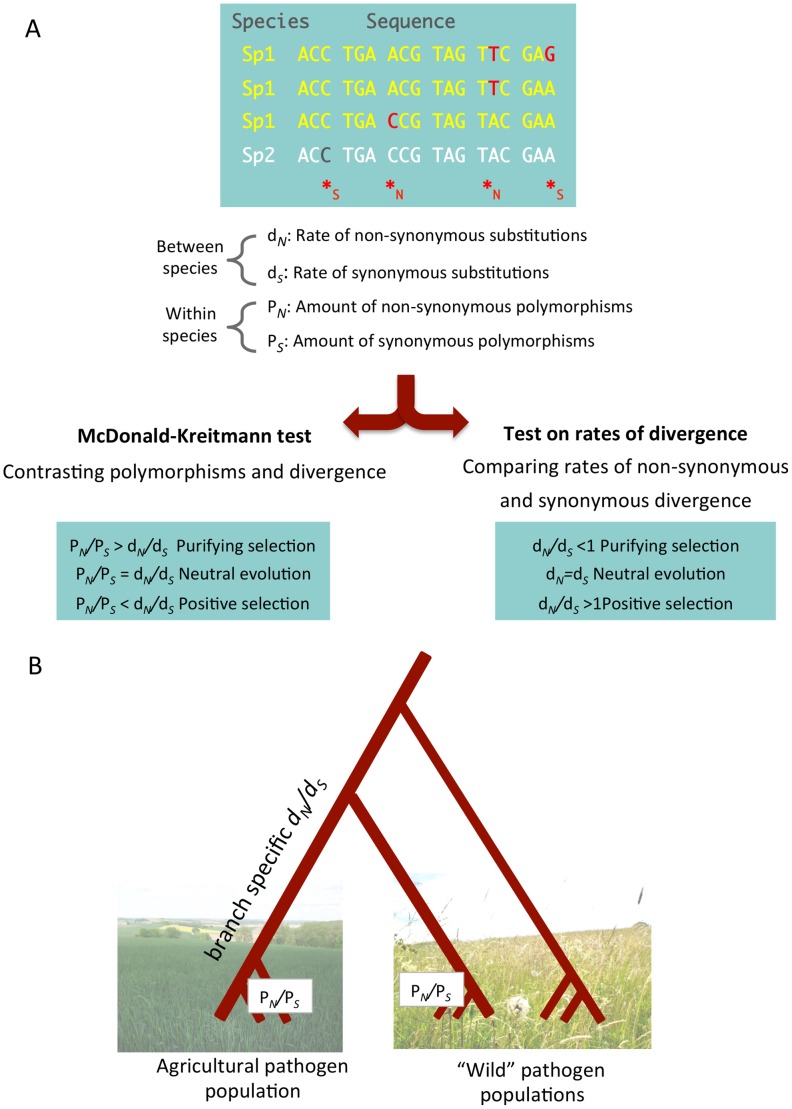
Population genomics surveys allow the inference of genome-wide distributions of polymorphisms within a species and allow the detection of synonymous and non-synonymous polymorphisms in coding sequences [22]. Some important parameters can be inferred from genome-wide distributions of polymorphisms. Traces of selective sweeps can be visualized as local drops in nucleotide diversity in regions surrounding a swept locus. Such genomic patterns may lead to the identification of strongly selected alleles in a population [23]. Moreover, measures of population differentiation such as the parameter Fst can reveal loci contributing to divergent adaptive evolution among sub-populations. Deviations from a genome-wide average Fst value can reflect recent divergent selection of alleles responsible for local adaptation. The relative abundance of non-synonymous and synonymous polymorphisms (PN and PS) furthermore measures the direct effect of natural selection removing slightly deleterious non-synonymous variants in coding sequences (purifying selection). Most genes are expected to evolve under purifying selection (i.e., PN/PS<1), however a local increase in PN/PS may reveal those few genes where new non-synonymous variants are favored by natural selection (positive selection). If genomic data from one or more related species are available, rates of non-synonymous (d N) and synonymous (d S) substitutions can be assessed to contrast within and between species patterns of variation (Figure 1). Such tests allow the detection of ancient selection in homologous genes of related species.
Figure 1. A population genomic approach allows the identification of synonymous and non-synonymous polymorphisms and substitutions.
Based on these parameters, amounts of adaptive evolution and the strength of purifying selection can be quantified. (A) A multi-species (between species) and multi-genotype (within species) alignment. Nucleotide positions can be categorized as either synonymous (*S) or non-synonymous (*N). Comparison of sequences from distinct species allows the detection of sites that have undergone substitutions, while the comparison of individuals of the same species allows the detection of non-synonymous (PN) and synonymous (PS) polymorphisms. While PN and PS reflect present time nucleotide variation in a species, the two rates of divergence dN and dS inform on types of selection during the past divergence of the species. Contrasting the rates of polymorphism and divergence (using a McDonald-Kreitmann-based test [24]) provides a finer grained picture of ongoing levels of purifying selection. (B) During the divergence of a new pathogen species in an agricultural environment, non-synonymous and synonymous substitutions have accumulated in the genome. A branch-specific model [28] can infer rates of non-synonymous and synonymous substitutions as dN and dS. The dN/dS ratio provides insight into the amount of fixed substitutions since the species divergence. An increased dN/dS ratio reflects either a relaxation of purifying selection (accumulation of slightly deleterious mutations) or the fixation of adaptive mutations. To assess the strength of purifying selection we can compare ratios of non-synonymous to synonymous polymorphisms (PN/PS). A comparison of PN/PS ratios between populations can illustrate differences in evolutionary rates under different environmental conditions. Pictures are courtesy of Julien Dutheil.
A number of other approaches have been developed to explore genome data and to characterize patterns of natural selection. These include methods to quantify adaptive evolution and purifying selection as well as coalescence models to infer genome evolution, recombination patterns, and effective population sizes [24], [25].
Ongoing adaptation and footprints of natural selection were reported in an elegant population genomics study of the non-pathogenic model species Neurospora crassa. Here the authors looked for regions exhibiting distinctly high levels of population differentiation [26] among 48 N. crassa isolates representing two geographically isolated populations evolving in different climatic environments. The highest Fst values between subpopulations of N. crassa were found in genomic islands encoding genes involved in temperature response and circadian function suggesting adaptation to different temperature and photoperiod ranges.
A population genomics approach was also used to study patterns of adaptive evolution in the wheat pathogen Mycosphaerella graminicola and to assess whether the specialization to an agricultural environment entailed a “domestication cost” [27]. To do so, two closely related species occurring only on hosts in natural grasslands were compared to M. graminicola. Patterns of non-synonymous and synonymous polymorphisms (PN and PS) were analyzed jointly with rates of branch-specific estimates of substitutions (dN and dS) to assess the effect of natural selection on coding sequences. A significant finding was that the transition to an agro-ecosystem did not entail an evolutionary “cost” in M. graminicola. Evolution in M. graminicola is instead characterized by the efficient fixation of new beneficial mutations enabling the pathogen to adapt to its new host and environment as reflected by currents ratios of PN/PS in the three species and branch-specific rates of d N and d S. Sexual recombination in the pathogen has likely also played a significant role in the purging of deleterious mutations.
The above-mentioned studies exemplify the broad potential of population genomic analyses in evolutionary studies of plant pathogens including the identification of strongly selected genes, estimates of evolutionary potential, and inferences of past demographic histories and current levels of adaptive evolution.
5. What's Next: Population Genomics as a Tool in the Development of Improved Disease Management Strategies
Population genomics studies will boost our understanding of adaptive evolution of plant pathogens in agro-ecosystems. The first studies on fungal pathogens have so far shown that fungal pathogens readily adapt to the agricultural environment and have revealed strong footprints of natural selection on their genome-wide diversity. Several studies have documented unexpectedly high levels of genome plasticity in fungal pathogens, allowing the acquisition of new genetic material and the presence of supernumerary chromosomes. We still need to understand the drivers of genome plasticity and their importance for adaptation. An equally important endeavor will be to understand further how the evolutionary potential of pathogen populations is maintained in spite of strong directional selection within homogenous host environments.
In light of the increasing need to control plant pathogens, we consider the close integration of evolutionary genomics with experimental studies will be essential to describe and predict the emergence, establishment, and adaptation of plant pathogens in agro-ecosystems. A combination of evolutionary analyses of genome-wide patterns of genetic diversity in crops and pathogens combined with targeted experiments, including functional studies and experimental evolution on pathogens, will be important to guide the much-needed design of novel and sustainable strategies to slow down the emergence and spread of pathogens [10], [27].
Funding Statement
The authors received no specific funding for this study.
References
- 1. Glemin S, Bataillon T (2009) A comparative view of the evolution of grasses under domestication. New Phytol 183: 273–290. [DOI] [PubMed] [Google Scholar]
- 2. Wright SI, Bi IV, Schroeder SG, Yamasaki M, Doebley JF, et al. (2005) The effects of artificial selection on the maize genome. Science 308: 1310–1314. [DOI] [PubMed] [Google Scholar]
- 3. Wang E, Wang J, Zhu X, Hao W, Wang L, et al. (2008) Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat Genet 40: 1370–1374. [DOI] [PubMed] [Google Scholar]
- 4. Komatsuda T, Pourkheirandish M, He C, Azhaguvel P, Kanamori H, et al. (2007) Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc Nat Acad Sci U S A 104: 1424–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Caicedo AL, Williamson SH, Hernandez RD, Boyko A, Fledel-Alon A, et al. (2007) Genome-wide patterns of nucleotide polymorphism in domesticated rice. PLoS Genet 3: e163 doi:10.1371/journal.pgen.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Haudry A, Cenci A, Ravel C, Bataillon T, Brunel D, et al. (2007) Grinding up wheat: a massive loss of nucleotide diversity since domestication. Mol Biol Evol 24: 1506–1517. [DOI] [PubMed] [Google Scholar]
- 7. Lu J, Tang T, Tang H, Huang J, Shi S, et al. (2006) The accumulation of deleterious mutations in rice genomes: a hypothesis on the cost of domestication. Trends in Genetics 22: 126–131. [DOI] [PubMed] [Google Scholar]
- 8. Hufford MB, Xu X, van Heerwaarden J, Pyhajarvi T, Chia J-M, et al. (2012) Comparative population genomics of maize domestication and improvement. Nat Genet 44: 808–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gillespie JH (2000) Genetic drift in an infinite population: the pseudohitchhiking model. Genetics 155: 909–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Morrell PL, Buckler ES, Ross-Ibarra J (2012) Crop genomics: advances and applications. Nat Rev Genet 13: 85–96. [DOI] [PubMed] [Google Scholar]
- 11. Stukenbrock EH, McDonald BA (2008) The origins of plant pathogens in agro-ecosystems. Annu Rev Phytopathol 46: 75–100. [DOI] [PubMed] [Google Scholar]
- 12. Goodwin SB, Cohen BA, Fry WE (1994) Panglobal distribution of a single clonal lineage of the Irish potato famine fungus. Proc Nat Acad Sci U S A 91: 11591–11595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Couch BC, Fudal I, Lebrun M-H, Tharreau D, Valent B, et al. (2005) Origins of host-specific populations of the blast pathogen Magnaporthe oryzae in crop domestication with subsequent expansion of pandemic clones on rice and weeds of rice. Genetics 170: 613–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bahri B, Leconte M, Ouffroukh A, De Vallavieille-Pope C, Enjalbert J (2009) Geographic limits of a clonal population of wheat yellow rust in the Mediterranean region. Mol Ecol 18: 4165–4179. [DOI] [PubMed] [Google Scholar]
- 15. Zaffarano PL, McDonald BA, Linde CC (2008) Rapis speciation following recent host shifts in the plant pathogenic fungus Rhyncosporium. Evolution 62: 1418–1436. [DOI] [PubMed] [Google Scholar]
- 16. Gladieux P, Zhang X-G, RÓLdan-Ruiz I, Caffier V, Leroy T, et al. (2010) Evolution of the population structure of Venturia inaequalis, the apple scab fungus, associated with the domestication of its host. Mol Ecol 19: 658–674. [DOI] [PubMed] [Google Scholar]
- 17. Fisher MC, Henk DA, Briggs CJ, Brownstein JS, Madoff LC, et al. (2012) Emerging fungal threats to animal, plant and ecosystem health. Nature 484: 186–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ma L-J, van der Does HC, Borkovich KA, Coleman JJ, Daboussi M-J, et al. (2010) Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464: 367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Coleman JJ, Rounsley SD, Rodriguez-Carres M, Kuo A, Wasmann CC, et al. (2009) The genome of Nectria haematococca: contribution of supernumerary chromosomes to gene expansion. PLoS Genet 5: e1000618 doi:10.1371/journal.pgen.1000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. de Jonge R, Peter van Esse H, Maruthachalam K, Bolton MD, Santhanam P, et al. (2012) Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc Nat Aca Sci U S A 109: 5110–5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stukenbrock EH, Christiansen FB, Hansen TT, Dutheil JY, Schierup MH (2012) Fusion of two divergent fungal individuals led to the recent emergence of a unique widespread pathogen species. Proc Nat Aca Sci U S A 109: 10954–10959 doi:10.1073/pnas.1201403109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Luikart G, England PR, Tallmon D, Jordan S, Taberlet P (2003) The power and promise of population genomics: from genotyping to genome typing. Nat Rev Genet 4: 981–994. [DOI] [PubMed] [Google Scholar]
- 23. Nygaard S, Braunstein A, Malsen G, Van Dongen S, Gardner PP, et al. (2010) Long- and short-term selective forces on malaria parasite genomes. PLoS Genet 6: e1001099 doi:10.1371/journal.pgen.1001099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Welch JJ (2006) Estimating the genomewide rate of adaptive protein evolution in Drosophila. Genetics 173: 821–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dutheil JY, Ganapathy G, Hobolth A, Mailund T, Uyenoyama MK, et al. (2009) Ancestral population genomics: the coalescent hidden Markov model approach. Genetics 183: 259–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ellison CE, Hall C, Kowbel D, Welch J, Brem RB, et al. (2011) Population genomics and local adaptation in wild isolates of a model microbial eukaryote. Proc Nat Aca Sci U S A 108: 2831–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stukenbrock EH, Bataillon T, Dutheil JY, Hansen TT, Li R, et al. (2011) The making of a new pathogen: insights from comparative population genomics of the domesticated wheat pathogen Mycosphaerella graminicola and its wild sister species. Genome Res 21: 2157–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang Z, Nielsen R (2002) Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol 19: 908–917. [DOI] [PubMed] [Google Scholar]