Abstract
Background
Up to 7% of patients with severe-to-profound deafness do not benefit from cochlear implantation. Given the high surgical implantation and clinical management cost of cochlear implantation (> $1 million lifetime cost), prospective identification of the worst performers would reduce unnecessary procedures and healthcare costs. Because cochlear implants bypass the membranous labyrinth but rely on the spiral ganglion for functionality, we hypothesize that cochlear implant (CI) performance is dictated in part by the anatomic location of the cochlear pathology that underlies the hearing loss. As a corollary, we hypothesize that because genetic testing can identify sites of cochlear pathology, it may be useful in predicting CI performance.
Methods
29 adult CI recipients with idiopathic adult-onset severe-to-profound hearing loss were studied. DNA samples were subjected to solution-based sequence capture and massively parallel sequencing using the OtoSCOPE® platform. The cohort was divided into three CI performance groups (good, intermediate, poor) and genetic causes of deafness were correlated with audiometric data to determine whether there was a gene-specific impact on CI performance.
Results
The genetic cause of deafness was determined in 3/29 (10%) individuals. The two poor performers segregated mutations in TMPRSS3, a gene expressed in the spiral ganglion, while the good performer segregated mutations in LOXHD1, a gene expressed in the membranous labyrinth. Comprehensive literature review identified other good performers with mutations in membranous labyrinth-expressed genes; poor performance was associated with spiral ganglion-expressed genes.
Conclusions
Our data support the underlying hypothesis that mutations in genes preferentially expressed in the spiral ganglion portend poor CI performance while mutations in genes expressed in the membranous labyrinth portend good CI performance. Although the low mutation rate in known deafness genes in this cohort likely relates to the ascertainment characteristics (postlingual hearing loss in adult CI recipients), these data suggest that genetic testing should be implemented as part of the CI evaluation to test this association prospectively.
Keywords: Cochlear implant performance, spiral ganglion, hearing loss, genetic testing, massively parallel sequencing
1. Introduction
While hearing aids are effective for mild-to-moderate degrees of hearing loss, when the loss is severe-to-profound, cochlear implantation is the better habilitation option for improved language and speech production.(Tomblin et al., 1999) Cochlear implantation is a common procedure (over 41,000 adults and 25,000 children receive an implant annually in the US) with a large associated healthcare cost ($24,475 surgical cost and over $1,000,000 lifetime cost per patient).(2010; Cheng et al., 2000; Mohr et al., 2000) Despite the upfront surgical cost, in most cases cochlear implantation is a cost-effective treatment that improves quality of life and results in a net savings to society.(Cheng et al., 2000; Colletti et al., 2011; Turchetti et al., 2011) However, among cochlear implant (CI) recipients, 3–7% do not benefit from implantation.(Archbold, Nikolopoulos & Lloyd-Richmond, 2009; Raine et al., 2008)
Currently, there is no method to identify poor CI performers prior to surgery, as many pre-surgical phenotypic traits of poor performers are indistinguishable from those of excellent performers: all have severe-profound hearing loss, similar audiometric patterns, and favorable temporal bone imaging findings. Given that a CI ‘bypasses’ cochlear hair cells and directly stimulates spiral ganglion neurons, we hypothesized that CI performance might be affected by the site of the deafness-causing cochlear pathology and that genetic deafness would adversely impact CI performance if the mutant protein caused primary spiral ganglion pathology as opposed to membranous labyrinth/hair cell damage. To test this hypothesis, we completed comprehensive genetic analysis in a cohort of adult CI users. In persons in whom we could identify a genetic cause of hearing loss, we sought to correlate the expression pattern of the relevant gene with CI performance.
2. Material and Methods
2.1 Study participants
Eligibility criteria for this study included: severe-to-profound, postlingual hearing loss (pure tone average ≥85dB hearing threshold), at least three years of audiologic follow-up, and cochlear implantation by one of three surgeons at the University of Iowa from 1986–2005. All patients were Caucasian and had normal inner ear and auditory nerve anatomy as assessed by temporal bone imaging. Exclusion criteria included: prelingual or perilingual deafness, non-Caucasian race, acquired hearing loss including prenatal infection, TORCH (toxoplasmosis, rubella, CMV, HSV) infection, aminoglycoside exposure, otoacoustic trauma, meningitis, syndromic presentation (congenital abnormalities in addition to hearing loss), low birth weight, inner ear abnormalities, or hyperbilirubinemia. All patients were enrolled in accordance with the University of Iowa Institutional Review Board guidelines and gave written informed consent prior to their involvement.
2.2 Targeted capture and massively parallel sequencing
DNA was extracted from peripheral whole blood obtained by venipuncture using standard methods.(Grimberg et al., 1989) Targeted sequence capture and massively parallel sequencing of all genes implicated in non-syndromic hearing loss were performed using the OtoSCOPE® genomic enrichment platform as described previously.(Shearer et al., 2010) In brief, patient genomic DNA was sheared and Illumina sequencing adaptors were ligated after blunt end-repair and A-tailing. The prepared DNA library and complementary biotin-labeled RNA baits, designed to target all exonic regions associated with non-syndromic deafness, were then hybridized. After capture of targeted regions using streptavidin-coated beads, non-targeted DNA was washed away, and targeted DNA was amplified to generate an enriched library for sequencing on the Illumina HiSeq2000.
2.3 Sequence analysis
Read mapping was performed using BWA and variants were called using the GATK Unified Genotyper.(Li & Durbin, 2010; McKenna et al., 2010) Variant analysis was completed using our custom variant calling pipeline. We used 40X depth of coverage as a minimum threshold for calling variants. Variations were annotated using population-scale data from publicly available databases (1000 Genomes project and Exome Variant Server) and filtered for variants that were exonic and non-synonymous, splice site, or indels and rare (< 2% allele frequency). Any reported deafness-causing mutations were identified using our Deafness Variation Database (http://deafnessvariationdatabase.org). All potentially clinically relevant variants were confirmed with Sanger sequencing.
2.4 Cochlear implant performance
The speech perception tests used to measure CI performance were chosen based on the recommendations of the Committee on Hearing and Equilibrium of the American Academy of Otolaryngology-Head and Neck Surgery.(Luxford & Ad Hoc Subcommittee of the Committee on Hearing and Equilibrium of the American Academy of Otolaryngology-Head and Neck Surgery, 2001) Patients were followed for at least three years post-implantation with the Consonant-Nucleus-Consonant (CNC) word test and the Hearing in Noise Test (HINT), and the mean score from multiple testing sessions beginning one year after implantation was calculated for each respective test.(Nilsson, Soli & Sullivan, 1994; PETERSON & LEHISTE, 1962) Two lists of 50 CNC words were presented at 60 dBA in quiet from a front-facing loudspeaker (0°) at 1m distance. Four lists of 12 HIN T sentences were administered in noise at +10 S/N ratio using 11-talker babble at 0°-azimuth. CNC scoring was based on percent-correct performance at both the word and the phoneme levels and the HINT sentences were scored by dividing the total number of key words correctly identified by the total number of key words possible. Individual as well as combined scores for these two tests were obtained and used to stratify patients by performance.
2.5 Statistical analyses
Patients were subdivided into three groups based on CI performance. Good performers were defined as those who performed one standard deviation above the mean on the combined performance measure (average of HINT and CNC scores), and poor performers were defined as those performing one standard deviation below the mean on the combined performance measure. Intermediate performers performed within one standard deviation of the mean. Differences between performance groups were evaluated by one-way ANOVA and confirmed with post hoc Bonferroni adjustments for the following variables: age of initial onset of hearing loss, age of onset of severe-profound hearing loss, duration of hearing loss progression, age at implantation, length of severe-profound hearing loss prior to implantation, and residual hearing (pure tone average prior to implantation). Differences in age and sex between groups were evaluated with the chi-squared test. Due to limited sample size comparisons of individual genes based on expression pattern were descriptive only.
2.6 Literature Review
A comprehensive literature review was completed such that all identifiable studies examining genotype-phenotype interactions in cochlear implant patients were tabulated. A pubmed search was completed with keywords “cochlear implant performance” and “genetics”. In addition, all individual genes included on the OtoSCOPE platform were individually searched in pubmed using the gene name and “cochlear implant”.
3. Results
3.1 CI Performance
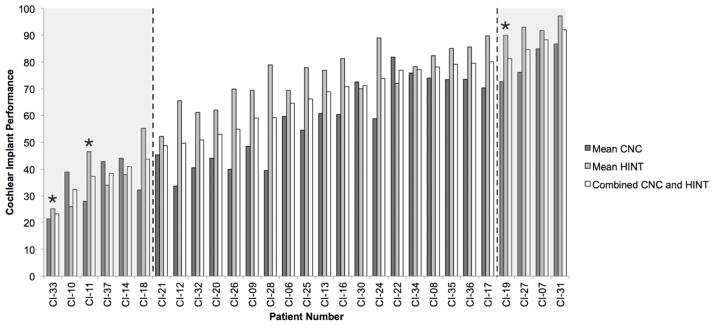
Twenty-nine of the 30 adult CI patients enrolled in the study met the inclusion criteria. The CNC and HINT were highly correlated (r = 0.78, p < 0.01) and the combined score effectively stratified groups (Figure 1). Sex and device type were not statistically different between performance groups. One-way ANOVA analysis revealed no differences between performance groups for the following variables: age of initial onset of hearing loss, age of onset of severe-profound hearing loss, duration of hearing loss progression, age at implantation, length of severe-profound hearing loss prior to implantation, and residual hearing (pure tone average prior to implantation).
Figure 1.
Cochlear implant performance by patient. HINT, CNC, and combined scores are shown. Data is sorted by combined score. *indicates individuals for which the genetic cause of hearing loss was determined in this study. Poor, intermediate, and good performance groups are separated by dashed lines.
3.2 Sequencing Results
An average of 65 million sequence reads were generated per sample, with an average of 85% of reads mapping to the reference sequence (human genome version hg19) and 52% overlapping targeted regions. The average depth-of-coverage per targeted base was 3,486 reads (standard deviation (SD) 1,921) with 97% (SD 0.5%) of targeted bases having 40X coverage, our variant calling threshold.
3.3 Mutation Detection and Clinical History
A genetic cause for the presenting hearing loss was identified in 3 of 29 patients (10%) (Figure 1; Table 1). Two patients, CI-11 and CI-33, were found to carry pathogenic mutations in TMPRSS3 (DFNB8/10). In the third patient, we identified pathogenic mutations in the gene LOXHD1 (DFNB77).(Edvardson et al., 2011; Grillet et al., 2009)
Table 1.
Pathogenic mutations identified in this study. Higher GERP scores (−11.6 to 5.82) represent increased conservation.
| Individual | Gene | Variant | SIFT | PolyPhen | PhyloP | Mutation Taster | MAF EVP | GERP Conservation | CI Performance (Combined) | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| CI-11 | TMPRSS3 | p.Ala138Glu | D | B | D | D | 0.10% | 3.3 | 37 | (Weegerink et al., 2011b) |
| p.Arg216Cys | D | D | D | D | 0.01% | 4.95 | (Elbracht et al., 2007) | |||
| CI-19 | LOXHD1 | p.Gly398Glu | D | - | - | - | 0.50% | - | 81.3 | This study |
| p.Arg383X | Non-sense | Non-sense | Non-sense | Non-sense | 0.10% | - | This study | |||
| CI-33 | TMPRSS3 | p.Ala138Glu | D | B | D | D | 0.10% | 3.3 | 23 | (Weegerink et al., 2011b) |
| p.Ala306Thr | D | D | D | D | Not Present | 4.37 | (Weegerink et al., 2011b) |
Abbreviations: B = Benign, D = Damaging. MAF = minor allele frequency, ESP = exome sequencing project (http://evs.gs.washington.edu/EVS/)
CI-11 is a 58-year-old male with a history of postlingual progressive hearing loss that began at age five and progressed to severe-to-profound sensorineural hearing loss (SNHL) by age 45. The family history was remarkable for hearing loss in 3 of 11 siblings and two nephews. The patient underwent left-sided cochlear implantation at age 47 with an Advanced Bionics CII device. At implantation his residual hearing (pure tone average implanted side) was 93 dB HL. This patient was a poor CI performer as assessed by his HINT (47), CNC (28), and combined performance (37) scores (Figure 1). OtoSCOPE® testing identified two previously reported deafness-causing mutations in TMPRSS3: p.Ala138Glu and p.Arg216Cys.(Elbracht et al., 2007; Hutchin et al., 2005) (Table 1).
CI-33 is a 58-year-old female with a history of postlingual progressive hearing loss that began at the age of six and progressed to severe-to-profound SNHL by the age of 17. The patient had a family history of hearing loss including her two brothers (3 of 8 siblings affected). The patient underwent left-sided cochlear implantation at age 32 with a Ineraid device. Prior to implantation her residual hearing (pure tone average implanted side) was 98 dB HL. This patient subsequently underwent explantation and reimplantation with an Advanced Bionics CII device due to hardware infection (pedestal). This patient was a poor CI performer as assessed by her HINT (25), CNC (21), and combined performance (23) scores (Figure 1). OtoSCOPE® testing identified two previously reported deafness-causing mutations in TMPRSS3: p.Ala138Glu and p.Ala306Thr (Table 1).
Individual CI-19 is a 77-year-old female with a history of adult onset, progressive hearing loss that began at the age of 40 and progressed to severe-to-profound hearing loss at age 67. The patient has a positive family history of hearing loss in one maternal aunt, however, her parents, siblings, and children have normal hearing. She underwent right cochlear implantation at age 68 after four years of severe-to-profound hearing loss with an Advanced Bionics Clarion HiFocus II-CII device. Prior to implantation her residual hearing (pure tone average implanted side) was 97 dB HL. Despite two years of severe-to-profound deafness prior to implantation, this patient was a good CI performer (HINT (90), CNC (73), combined (81.3)) (Figure 1). Genetic analysis revealed compound heterozygosity for two novel variants in LOXHD1, p.Gly398Glu and p.Arg383X. Neither of these variants had been reported, however the missense p.Gly398Glu variant is predicted to be damaging by the only in silico prediction available for this variant, and both variants are either not present or very rare in large population-scale genomic databases (Table 1).
3.4 Analysis of Previously Reported Cochlear Implantation Performance
We performed a systematic literature review and identified 70 studies examining the effect of genetic mutation on CI performance (Table 2).
Table 2.
Summary of previous studies examining the effect genetic mutation on cochlear implant performance. Genes are divided by their location of primary expression. Data from the present study is included under the corresponding gene. Mitochondrial genes are not listed.
| Gene | Protein | Function | Phenotype | Studies (Individuals) | CI Performance | Reference |
|---|---|---|---|---|---|---|
| MEMBRANOUS LABYRINTH EXPRESSED GENES
| ||||||
| GJB2 | gap junction protein, beta 2 | ion homeostasis | SNHL | 25 (307) | Good | * |
| LOXHD1 | lipoxygenase homology domain-containing 1 | stereociliary protein | SNHL | 3 (11) | Good | (Edvardson et al., 2011; Grillet et al., 2009) |
| OTOF | otoferlin | synaptic transmission (exocytosis at auditory ribbon synapse) | SNHL (auditory neuropathy) | 4 (12) | Good | (Loundon et al., 2005; Rodriguez-Ballesteros et al., 2003; Rouillon et al., 2006; Wu et al., 2011b) |
| CDH23 | cadherin-related 23 | structural (cell adhesion) | USH1 | 2 (3) | Good | (Liu et al., 2008; Pennings et al., 2006) |
| MYO7A | myosin VIIA | motor protein (hair bundle) | USH1 | 1 (6) | Good | (Pennings et al., 2006) |
| KCNQ1 | KCNQ1 downstream neighbor protein | ion homeostasis | Jervell and Lange-Nielsen syndrome | 2 (8) | Good | (Chorbachi et al., 2002; Siem et al., 2008) |
| TMC1 | transmembrane channel-like 1 | unknown | Progressive SNHL | 1 (2) | Good | (Makishima et al., 2004) |
| COCH | coagulation factor C homolog, cochlin | structural (extracellular matrix) | Progressive SNHL | 1 (11) | Good | (Vermeire et al., 2006) |
| SLC26A4 | solute carrier family 26, member 4 | ion homeostasis | EVA and SNHL | 5 (49) | Good | (de Wolf et al., 2010; Lai et al., 2012; Vescan et al., 2002; Wu et al., 2008; Wu et al., 2011a) |
| MYH9 | myosin, heavy chain 9, non-muscle | motor protein (hair cell) | Progressive SNHL | 2(6) | Variable | (Hildebrand et al., 2006; Lalwani et al., 1997) |
| POU3F4 | pou-domain class 3 transcription factor 4 | transcription factor | SNHL, attention disorders, motor delay | 2 (7) | Variable | (Lee et al., 2009; Stankovic et al., 2010) |
|
| ||||||
| SPIRAL GANGLION GENES
| ||||||
| TMPRSS3 | transmembrane protease, serine 3 | unknown | Progressive SNHL | 3 (15) | Variable | (Elbracht et al., 2007; Weegerink et al., 2011b) |
| CHD7 | chromodomain-helicase-DNA-binding protein 7 | transcription factor | CHARGE | 1 (8) | Poor | (Lanson et al., 2007; Song et al., 2011) |
| DDP1/TIMM8a | mitochondrial import inner membrane translocase subunit Tim8 A | mitochondrial protein transport | Deafness-dystonia-optic neuronopathy | 1 (1) | Poor | (Brookes et al., 2008) |
GJB2 references include: (Angeli et al., 2011; Arndt et al., 2010; Bauer et al., 2003; Chora et al., 2010; Choung et al., 2008; Connell et al., 2007; Cullen et al., 2004; Dahl et al., 2003; Dalamon et al., 2009; Daneshi et al., 2011; Fukushima et al., 2002; Gerard et al., 2010; Green et al., 2002; Karamert et al., 2011; Lustig et al., 2004; Matsushiro et al., 2002; Reinert, Honegger & Gurtler, 2010; Sinnathuray et al., 2004a; Sinnathuray et al., 2004b; Taitelbaum-Swead et al., 2006; Weegerink et al., 2011a; Wrobel et al., 2008; Wu et al., 2008; Wu et al., 2011a; Yoshikawa et al., 2011.
3.4.1 Membranous Labyrinth Expressed Genes
Genotype-phenotype correlations have been reported for 11 genes, which are primarily expressed in the membranous labyrinth (Table 2). The majority of studies have investigated the impact of GJB2 (25 studies, 307 individuals), SLC26A4 (5 studies, 49 individuals), and OTOF (4 studies, 12 individuals) mutations on CI performance. Mutations in these three genes are associated with good performance. The remaining genes (LOXHD1, MYH9, CDH23, MYO7A, KCNQ1, TMC1, COCH, POU3F4) have not been as well studied. These eight genes have also been associated with good cochlear implant performance with the exception of MYH9 and POU3F4, which demonstrate variable performance (Table 2).
3.4.2 Mitochondrial
Mutations in mitochondrial genes cause both syndromic and non-syndromic forms of deafness and also increase susceptibility to aminoglycoside-associated hearing loss. In the cochlea, both the stria vascularis and hair cells have extremely high ATP demands and are mitochondrial rich. Several studies of patients with non-syndromic, syndromic and aminoglycoside-associated mitochondrial hearing loss have demonstrated good CI performance.(Counter et al., 2001; Tono et al., 2001; Ulubil, Furze & Angeli, 2006)
3.4.3 Spiral Ganglion Expressed Genes
There have been very few investigations into genotype-phenotype correlations in genes primarily expressed in the spiral ganglion. TMPRSS3 is a transmembrane serine protease that may be involved in cleavage of neurotrophins in spiral ganglion neurons.(Guipponi et al., 2002; Weegerink et al., 2011b) The two persons we identified were compound heterozygotes for reported TMPRSS3 mutations and poor CI performers. Our results conflict with two reports of TMPRSS3-related deafness, which demonstrate good performance. In one report, describing four affected siblings, no performance data are provided although the paper reports that bilateral cochlear implantation was performed “with good results”.(Elbracht et al., 2007) The second report provides more details and notes improved CNC scores after implantation, with nine persons showing better performance than the control group.(Weegerink et al., 2011b) Neither study reports the age at implantation or duration of deafness prior to implantation.
Two other spiral ganglion expressed genes, CHD7 and DDP1/TIMM8a, result in syndromic forms of deafness and have been associated with poor CI performance. Mutation of CHD7 results in CHARGE syndrome (coloboma, heart defects, choanal atresia, retarded growth and development, genital hypoplasia, ear abnormalities). One study of persons with confirmed CHD7-related hearing loss noted variable CI performance (5 good, 3 poor), while another study of individuals with CHARGE syndrome with unknown genotype demonstrated limited benefit.(Lanson et al., 2007; Song et al., 2011) Mutation of DDP1/TIMM8a results in deafness-dystonia-optic neuronopathy syndrome (Mohr-Tranebjærg syndrome, DFN-1), which is associated with SNHL and neurologic manifestations (visual impairment, dystonia, cognitive defects). Histopathology in such individuals demonstrates near complete absence of spiral ganglion neurons and as expected these patients perform poorly with CIs.(Brookes et al., 2008; Nadol & Merchant, 2001) Genotype-phenotype correlations for CHD7 and DDP1/TIMM8a are confounded by cognitive impairment.
3.4.4 Auditory Neuropathy
Auditory neuropathy is a heterogeneous group of disorders with a characteristic clinical phenotype: functioning outer hair cells (normal OAE) and aberrant neurotransmission (abnormal or absent ABR). The location of the underlying lesion (hair cell, ribbon synapse, spiral ganglion neuron, or auditory nerve) in auditory neuropathy is much debated and likely differs based on the genetic defect. For a detailed review of auditory neuropathy and genetics the reader is directed to Santarelli’s review.(Santarelli, 2010) A recent systematic review of the literature regarding CI performance in children with auditory neuropathy has demonstrated that implantation is generally beneficial.(Roush et al., 2011) In general, similar outcomes have been demonstrated in adult CI recipients with auditory neuropathy, however there are reports of more variable performance.(Mason et al., 2003; Teagle et al., 2010) In aggregate, these reports are consistent with the observation that the most common genetic cause of auditory neuropathy is mutation of OTOF.(Loundon et al., 2005; Rodriguez-Ballesteros et al., 2003; Rouillon et al., 2006; Wu et al., 2011b) Its translated protein, otoferlin, is required for synaptic vesicle fusion in inner hair cells.
4. Discussion
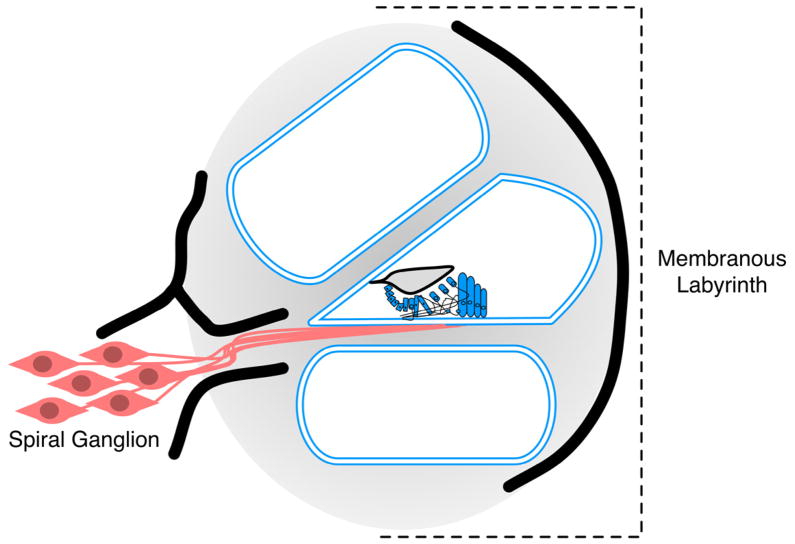
The advent of massively parallel sequencing technology has revolutionized the diagnosis of genetic hearing loss by making it possible to screen all known deafness genes simultaneously.(Brownstein et al., 2011; Shearer et al., 2010; Tang et al., 2012) This breakthrough may impact cochlear implantation by making it possible to diagnose genetic causes of hearing loss preoperatively. We hypothesized that the expression pattern of the involved gene would impact CI performance. We expected that mutations in spiral ganglion expressed genes would portend poor performance, whereas mutations in membranous labyrinth expressed genes, which are bypassed by a CI would result in good performance (Figure 2). Although, we only identified causative mutations in three individuals, those with mutations in TMPRSS3, which is expressed in the spiral ganglion, did perform worse than the individual with mutations in LOXHD1, which is expressed in the stereocilia. It is notable that there were no differences in residual hearing or length of deafness prior to implantation between good and poor performance groups. As the cause of deafness was uncovered in very few patients in this cohort, we examined other published genotype-phenotype correlations in order to draw conclusions on the impact of expression pattern of the involved gene on CI performance.
Figure 2.
Expression pattern of hearing loss genes. Genes with established genotype-phenotype correlations are included. Genes expressed in the membranous labyrinth (labeled blue) include: GJB2, SLC26A4, OTOF, LOXHD1, KCNQ1, CDH23, MYO7A, POU3F4, MYH9, TMC1, and COCH. Genes expressed in the spiral ganglion (labeled red) include: TMPRSS3, CHD7, and DDP1/TIMM8a. (Adapted from the Hereditary Hearing Loss Homepage, hereditaryhearingloss.org, with permission from Van Camp G and Smith RJH)
4.1 Comprehensive Literature Review
Although much has been published on correlations of CI outcome and genetics (over 70 publications) few robust conclusions and little prognostic information can be drawn (Table 2). Previous study has been limited to retrospective case series with small sample sizes, variable and poorly categorized performance measures, poor categorization of known confounding variables such as residual hearing loss and duration of deafness prior to implantation, few accounts of poor CI performance, and few gene-to-gene comparisons. We recommend that standardized performance measures be used in future investigations of genotype-phenotype correlations in the CI population. Specifically, in our experience the HINT and CNC performance measures best stratify adult CI performance because these tests challenge even the best performers. Each measure offers different challenges to the listener: the difficulty of the HINT greatly increases as the amount of background noise is increased, while the CNC test challenges the listener by presenting single words without the contextual cues of a sentence. This testing battery is recommended by the Committee on Hearing and Equilibrium of the American Academy of Otolaryngology-Head and Neck Surgery.(Luxford & Ad Hoc Subcommittee of the Committee on Hearing and Equilibrium of the American Academy of Otolaryngology-Head and Neck Surgery, 2001)
In general, the data reported here support our hypothesis that CI recipients with mutations in membranous labyrinth expressed genes will be good CI performers while those with mutations in spiral ganglion expressed genes will be poor performers with more variable outcomes (Figure 2). However, there are several discordances. We identified reports of two membranous labyrinth expressed genes associated with variable performance - MYH9 and POU3F4. Robust MYH9 expression has been demonstrated in inner hair cells, outer hair cells, the spiral limbus, and spiral ligament with only low expression in the spiral ganglion.(Mhatre et al., 2006) A study of five members of an Australian family with CIs and MYH9-associated hearing loss demonstrated excellent performance on the CNC word test, however there is also a report of an individual with MYH9-related hearing loss who demonstrated poor CI performance.(Hildebrand et al., 2006; Lalwani et al., 1997) Based on our hypothesis, we would expect the latter individual to perform well but believe his poor performance may be attributable to the duration of deafness prior to implantation, an established cause of poor CI performance.(Clopton, Spelman & Miller, 1980; Nadol, Young & Glynn, 1989; Rubinstein et al., 1999) Importantly, the five Australian individuals received their implants after a short duration of profound deafness. In the case of POU3F4-related deafness, the comorbid cognitive impairment and cochlear dysplasia likely contribute to the variable CI performance thereby confounding genotype-phenotype correlations.
With respect to genes expressed in the spiral ganglion neurons, our results differ from two reports, which examined genotype-phenotype correlations in individuals with TMPRSS3-related deafness. TMPRSS3 encodes a transmembrane serine protease, which is expressed in the spiral ganglion.(Guipponi et al., 2002) We predicted such individuals would be poor CI performers, which our results corroborated. The two contradictory reports include 10 patients, 9 of whom were likely less than 30 years old at the time of the report.(Elbracht et al., 2007; Weegerink et al., 2011b) This contrasts greatly from the patients in our cohort who were in their sixth decade at the time of testing. In addition, neither report notes the age at implantation and in one, the measures used to evaluate performance are not provided making comparisons difficult.(Elbracht et al., 2007). Given that TMPRSS3-associated hearing loss is progressive, one might predict that CI performance in the patients detailed in these two reports will decline with time, which is consistent with histological studies of a mouse model of TMPRSS3-related deafness (a truncating mutation of Tmprss3, Y260X) demonstrating progressive spiral ganglion neuron degeneration.(Fasquelle et al., 2011) Two other spiral ganglion expressed genes, CHD7 and DDP1/TIMM8a, were associated with poor performance, however these results are confounded by comorbid cognitive impairment.
Another interesting group of CI recipients are persons with auditory neuropathy. Although auditory neuropathy is a heterogeneous group of disorders, 55–87% of cases are due to mutations in OTOF. The translated protein, otoferlin, is expressed in the inner and outer hair cells and is believed to be involved in synaptic vesicle exocytosis at the afferent hair cell ribbon synapse.(Rodriguez-Ballesteros et al., 2008; Yasunaga et al., 1999) As such, persons with OTOF auditory neuropathy initially display a classic phenotype (normal otoacoustic emission (OAE) and absent auditory brainstem response (ABR) until the hearing loss progress and outer hair cells also degenerate) and would be expected to be good CI performers. However other forms of auditory neuropathy are associated with variable CI performance.(Teagle et al., 2010) It is possible that this variability correlates with variability in the site of the underlying defect. Thus, while we would expect good CI performance with lesions affecting the hair cell or hair cell synaptic ribbon, we would expect poor CI performance if the spiral ganglion neurons and/or auditory nerve degenerate.
Given these preliminary results we believe that genetic testing will eventually become a standard part of the preoperative cochlear implant evaluation. As additional studies are completed genotype-phenotype correlations will become more robust and offer valuable prognostic information. In addition, the establishment of the genetic diagnosis of hearing loss in individual patients will allow for more patient-centered care. A potential example of this is patients whose deafness results from mutations in spiral ganglion-expressed genes. As has been demonstrated for TMPRSS3, hearing loss is progressive, perhaps due to continued loss of spiral ganglion neurons. If progressive hearing loss in this population is secondary to spiral ganglion degeneration, it is possible that following such patients with ABR may prove a useful metric for the ‘amount’ of remaining neuronal substrate. We would predict that progressive loss of ABR thresholds would coincide with decreasing cochlear implant performance.
There are several limitations associated with this study, most notably the small number of persons with an identifiable genetic cause of hearing loss. This outcome reflects the certain inclusion of many cases of acquired or complex causes of hearing loss in the adult population we studied. A secondary limitation is the OtoSCOPE® platform itself, which has a ‘solve’ rate of just over 50% for all comers with presumed non-syndromic hearing loss (unpublished data). This solve rate reflects the vast heterogeneity of non-syndromic hearing loss and implies that many deafness-causing genes await discovery. Nevertheless, the availability of comprehensive genetic testing for deafness now makes it possible to complete rigorous studies to correlate genotype with phenotype in CI patients. The data we have presented support the completion of this type of study. Establishing genotype-phenotype correlations may make it possible to tailor CI programming strategies to a patient’s specific needs as determined by their genetic cause of hearing loss, provide genetic-based CI performance metrics, and reduce the likelihood of ineffective cochlear implantation thereby decreasing healthcare costs.
5. Conclusions
Our data support the underlying hypothesis that mutations in genes preferentially expressed in the spiral ganglion portend poor CI performance while mutations in genes expressed in the membranous labyrinth portend good CI performance. Although the low mutation rate in known deafness genes in this cohort likely relates to the ascertainment characteristics (postlingual hearing loss in adult CI recipients), these data suggest that genetic testing should be implemented as part of the CI evaluation to test this association prospectively.
Highlights.
We hypothesize the site of the genetic defect impacts cochlear implant outcome
We apply comprehensive genetic testing and literature review to test our hypothesis
We demonstrate mutations affecting the spiral ganglion portend poor outcome
Mutations affecting membranous labyrinth expressed genes portend good outcome
Genetic testing should become a standard part of cochlear implant evaluation
Acknowledgments
We would like to acknowledge Richard Gibbs and Donna Muzny for their help with sequencing.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- National Institute on Deafness and Other Communication Disorders Quick Facts. 2010;2011(1/11):1. [Google Scholar]
- Angeli SI, Suarez H, Lopez A, Balkany TJ, Liu XZ. Influence of DFNB1 status on expressive language in deaf children with cochlear implants. Otol Neurotol. 2011;32(9):1437–1443. doi: 10.1097/MAO.0b013e31823387f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archbold SM, Nikolopoulos TP, Lloyd-Richmond H. Long-term use of cochlear implant systems in paediatric recipients and factors contributing to non-use. Cochlear Implants Int. 2009;10(1):25–40. doi: 10.1179/cim.2009.10.1.25. [DOI] [PubMed] [Google Scholar]
- Arndt S, Aschendorff A, Schild C, Beck R, Maier W, Laszig R, Birkenhager R. A novel dominant and a de novo mutation in the GJB2 gene (connexin-26) cause keratitis-ichthyosis-deafness syndrome: implication for cochlear implantation. Otol Neurotol. 2010;31(2):210–215. doi: 10.1097/MAO.0b013e3181cc09cd. [DOI] [PubMed] [Google Scholar]
- Bauer PW, Geers AE, Brenner C, Moog JS, Smith RJ. The effect of GJB2 allele variants on performance after cochlear implantation. Laryngoscope. 2003;113(12):2135–2140. doi: 10.1097/00005537-200312000-00015. [DOI] [PubMed] [Google Scholar]
- Brookes JT, Kanis AB, Tan LY, Tranebjaerg L, Vore A, Smith RJ. Cochlear implantation in deafness-dystonia-optic neuronopathy (DDON) syndrome. Int J Pediatr Otorhinolaryngol. 2008;72(1):121–126. doi: 10.1016/j.ijporl.2007.08.019. [DOI] [PubMed] [Google Scholar]
- Brownstein Z, Friedman LM, Shahin H, Oron-Karni V, Kol N, Rayyan AA, Parzefall T, Lev D, Shalev S, Frydman M, Davidov B, Shohat M, Rahile M, Lieberman S, Levy-Lahad E, Lee MK, Shomron N, King MC, Walsh T, Kanaan M, Avraham KB. Targeted genomic capture and massively parallel sequencing to identify genes for hereditary hearing loss in middle eastern families. Genome Biol. 2011;12(9):R89. doi: 10.1186/gb-2011-12-9-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AK, Rubin HR, Powe NR, Mellon NK, Francis HW, Niparko JK. Cost-utility analysis of the cochlear implant in children. JAMA. 2000;284(7):850–856. doi: 10.1001/jama.284.7.850. [DOI] [PubMed] [Google Scholar]
- Chora JR, Matos TD, Martins JH, Alves MC, Andrade SM, Silva LF, Ribeiro CA, Antunes MC, Fialho MG, Caria MH. DFNB1-associated deafness in Portuguese cochlear implant users: prevalence and impact on oral outcome. Int J Pediatr Otorhinolaryngol. 2010;74(10):1135–1139. doi: 10.1016/j.ijporl.2010.06.014. [DOI] [PubMed] [Google Scholar]
- Chorbachi R, Graham JM, Ford J, Raine CH. Cochlear implantation in Jervell and Lange-Nielsen syndrome. Int J Pediatr Otorhinolaryngol. 2002;66(3):213–221. doi: 10.1016/s0165-5876(02)00181-7. [DOI] [PubMed] [Google Scholar]
- Choung YH, Shin YR, Kim HJ, Kim YC, Ahn JH, Choi SJ, Jeong SY, Park K. Cochlear implantation and connexin expression in the child with keratitis-ichthyosis-deafness syndrome. Int J Pediatr Otorhinolaryngol. 2008;72(6):911–915. doi: 10.1016/j.ijporl.2008.02.023. [DOI] [PubMed] [Google Scholar]
- Clopton BM, Spelman FA, Miller JM. Estimates of essential neural elements for stimulation through a cochlear prosthesis. Ann Otol Rhinol Laryngol Suppl. 1980;89(2 Pt 2):5–7. doi: 10.1177/00034894800890s202. [DOI] [PubMed] [Google Scholar]
- Colletti L, Mandala M, Shannon RV, Colletti V. Estimated net saving to society from cochlear implantation in infants: a preliminary analysis. Laryngoscope. 2011;121(11):2455–2460. doi: 10.1002/lary.22131. [DOI] [PubMed] [Google Scholar]
- Connell SS, Angeli SI, Suarez H, Hodges AV, Balkany TJ, Liu XZ. Performance after cochlear implantation in DFNB1 patients. Otolaryngol Head Neck Surg. 2007;137(4):596–602. doi: 10.1016/j.otohns.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Counter PR, Hilton MP, Webster D, Wardell T, Taylor RW, Besley G, Turnbull DM, Robinson PJ. Cochlear implantation of a patient with a previously undescribed mitochondrial DNA defect. J Laryngol Otol. 2001;115(9):730–732. doi: 10.1258/0022215011908766. [DOI] [PubMed] [Google Scholar]
- Cullen RD, Buchman CA, Brown CJ, Copeland BJ, Zdanski C, Pillsbury HC, 3rd, Shores CG. Cochlear implantation for children with GJB2-related deafness. Laryngoscope. 2004;114(8):1415–1419. doi: 10.1097/00005537-200408000-00019. [DOI] [PubMed] [Google Scholar]
- Dahl HH, Wake M, Sarant J, Poulakis Z, Siemering K, Blamey P. Language and speech perception outcomes in hearing-impaired children with and without connexin 26 mutations. Audiol Neurootol. 2003;8(5):263–268. doi: 10.1159/000071998. [DOI] [PubMed] [Google Scholar]
- Dalamon V, Lotersztein V, Lipovsek M, Beheran A, Mondino ME, Diamante F, Pallares N, Diamante V, Elgoyhen AB. Performance of speech perception after cochlear implantation in DFNB1 patients. Acta Otolaryngol. 2009;129(4):395–398. doi: 10.1080/00016480802566295. [DOI] [PubMed] [Google Scholar]
- Daneshi A, Hassanzadeh S, Emamdjomeh H, Mohammadi SH, Arzhangi S, Farhadi M, Najmabadi H. Prevalence of GJB2-associated deafness and outcomes of cochlear implantation in Iran. J Laryngol Otol. 2011;125(5):455–459. doi: 10.1017/S0022215110002999. [DOI] [PubMed] [Google Scholar]
- de Wolf MJ, Honings J, Joosten FB, Hoefsloot L, Mylanus EA, Cremers CW. Two siblings with progressive, fluctuating hearing loss after head trauma, treated with cochlear implantation. J Laryngol Otol. 2010;124(1):86–89. doi: 10.1017/S0022215109990296. [DOI] [PubMed] [Google Scholar]
- Edvardson S, Jalas C, Shaag A, Zenvirt S, Landau C, Lerer I, Elpeleg O. A deleterious mutation in the LOXHD1 gene causes autosomal recessive hearing loss in Ashkenazi Jews. Am J Med Genet A. 2011;155A(5):1170–1172. doi: 10.1002/ajmg.a.33972. [DOI] [PubMed] [Google Scholar]
- Elbracht M, Senderek J, Eggermann T, Thurmer C, Park J, Westhofen M, Zerres K. Autosomal recessive postlingual hearing loss (DFNB8): compound heterozygosity for two novel TMPRSS3 mutations in German siblings. J Med Genet. 2007;44(6):e81. doi: 10.1136/jmg.2007.049122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasquelle L, Scott HS, Lenoir M, Wang J, Rebillard G, Gaboyard S, Venteo S, Francois F, Mausset-Bonnefont AL, Antonarakis SE, Neidhart E, Chabbert C, Puel JL, Guipponi M, Delprat B. Tmprss3, a transmembrane serine protease deficient in human DFNB8/10 deafness, is critical for cochlear hair cell survival at the onset of hearing. J Biol Chem. 2011;286(19):17383–17397. doi: 10.1074/jbc.M110.190652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima K, Sugata K, Kasai N, Fukuda S, Nagayasu R, Toida N, Kimura N, Takishita T, Gunduz M, Nishizaki K. Better speech performance in cochlear implant patients with GJB2-related deafness. Int J Pediatr Otorhinolaryngol. 2002;62(2):151–157. doi: 10.1016/s0165-5876(01)00619-x. [DOI] [PubMed] [Google Scholar]
- Gerard JM, Deggouj N, Hupin C, Buisson AL, Monteyne V, Lavis C, Dahan K, Gersdorff M. Evolution of communication abilities after cochlear implantation in prelingually deaf children. Int J Pediatr Otorhinolaryngol. 2010;74(6):642–648. doi: 10.1016/j.ijporl.2010.03.010. [DOI] [PubMed] [Google Scholar]
- Green GE, Scott DA, McDonald JM, Teagle HF, Tomblin BJ, Spencer LJ, Woodworth GG, Knutson JF, Gantz BJ, Sheffield VC, Smith RJ. Performance of cochlear implant recipients with GJB2-related deafness. Am J Med Genet. 2002;109(3):167–170. doi: 10.1002/ajmg.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillet N, Schwander M, Hildebrand MS, Sczaniecka A, Kolatkar A, Velasco J, Webster JA, Kahrizi K, Najmabadi H, Kimberling WJ, Stephan D, Bahlo M, Wiltshire T, Tarantino LM, Kuhn P, Smith RJ, Muller U. Mutations in LOXHD1, an evolutionarily conserved stereociliary protein, disrupt hair cell function in mice and cause progressive hearing loss in humans. Am J Hum Genet. 2009;85(3):328–337. doi: 10.1016/j.ajhg.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimberg J, Nawoschik S, Belluscio L, McKee R, Turck A, Eisenberg A. A simple and efficient non-organic procedure for the isolation of genomic DNA from blood. Nucleic Acids Res. 1989;17(20):8390. doi: 10.1093/nar/17.20.8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guipponi M, Vuagniaux G, Wattenhofer M, Shibuya K, Vazquez M, Dougherty L, Scamuffa N, Guida E, Okui M, Rossier C, Hancock M, Buchet K, Reymond A, Hummler E, Marzella PL, Kudoh J, Shimizu N, Scott HS, Antonarakis SE, Rossier BC. The transmembrane serine protease (TMPRSS3) mutated in deafness DFNB8/10 activates the epithelial sodium channel (ENaC) in vitro. Hum Mol Genet. 2002;11(23):2829–2836. doi: 10.1093/hmg/11.23.2829. [DOI] [PubMed] [Google Scholar]
- Hildebrand MS, de Silva MG, Gardner RJ, Rose E, de Graaf CA, Bahlo M, Dahl HH. Cochlear implants for DFNA17 deafness. Laryngoscope. 2006;116(12):2211–2215. doi: 10.1097/01.mlg.0000242089.72880.f8. [DOI] [PubMed] [Google Scholar]
- Hutchin T, Coy NN, Conlon H, Telford E, Bromelow K, Blaydon D, Taylor G, Coghill E, Brown S, Trembath R, Liu XZ, Bitner-Glindzicz M, Mueller R. Assessment of the genetic causes of recessive childhood non-syndromic deafness in the UK - implications for genetic testing. Clin Genet. 2005;68(6):506–512. doi: 10.1111/j.1399-0004.2005.00539.x. [DOI] [PubMed] [Google Scholar]
- Karamert R, Bayazit YA, Altinyay S, Yilmaz A, Menevse A, Gokdogan O, Gokdogan C, Ant A. Association of GJB2 gene mutation with cochlear implant performance in genetic non-syndromic hearing loss. Int J Pediatr Otorhinolaryngol. 2011;75(12):1572–1575. doi: 10.1016/j.ijporl.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Lai R, Hu P, Zhu F, Zhu G, Vivero R, Peng A, Wu W, Xiao Z, Liu X, Xie D. Genetic diagnosis and cochlear implantation for patients with nonsyndromic hearing loss and enlarged vestibular aqueduct. J Laryngol Otol. 2012;126(4):349–355. doi: 10.1017/S002221511100346X. [DOI] [PubMed] [Google Scholar]
- Lalwani AK, Linthicum FH, Wilcox ER, Moore JK, Walters FC, San Agustin TB, Mislinski J, Miller MR, Sinninger Y, Attaie A, Luxford WM. A five-generation family with late-onset progressive hereditary hearing impairment due to cochleosaccular degeneration. Audiol Neurootol. 1997;2(3):139–154. doi: 10.1159/000259237. [DOI] [PubMed] [Google Scholar]
- Lanson BG, Green JE, Roland JT, Lalwani AK, Jr, Waltzman SB. Cochlear implantation in Children with CHARGE syndrome: therapeutic decisions and outcomes. Laryngoscope. 2007;117(7):1260–1266. doi: 10.1097/MLG.0b013e31806009c9. [DOI] [PubMed] [Google Scholar]
- Lee HK, Lee SH, Lee KY, Lim EJ, Choi SY, Park RK, Kim UK. Novel POU3F4 mutations and clinical features of DFN3 patients with cochlear implants. Clin Genet. 2009;75(6):572–575. doi: 10.1111/j.1399-0004.2009.01181.x. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XZ, Angeli SI, Rajput K, Yan D, Hodges AV, Eshraghi A, Telischi FF, Balkany TJ. Cochlear implantation in individuals with Usher type 1 syndrome. Int J Pediatr Otorhinolaryngol. 2008;72(6):841–847. doi: 10.1016/j.ijporl.2008.02.013. [DOI] [PubMed] [Google Scholar]
- Loundon N, Marcolla A, Roux I, Rouillon I, Denoyelle F, Feldmann D, Marlin S, Garabedian EN. Auditory neuropathy or endocochlear hearing loss? Otol. Neurotol. 2005;26(4):748–754. doi: 10.1097/01.mao.0000169044.63970.4a. [DOI] [PubMed] [Google Scholar]
- Lustig LR, Lin D, Venick H, Larky J, Yeagle J, Chinnici J, Polite C, Mhatre AN, Niparko JK, Lalwani AK. GJB2 gene mutations in cochlear implant recipients: prevalence and impact on outcome. Arch Otolaryngol Head Neck Surg. 2004;130(5):541–546. doi: 10.1001/archotol.130.5.541. [DOI] [PubMed] [Google Scholar]
- Luxford WM Ad Hoc Subcommittee of the Committee on Hearing, Equilibrium of the American Academy of Otolaryngology-Head, Neck Surgery. Minimum speech test battery for postlingually deafened adult cochlear implant patients. Otolaryngol Head Neck Surg. 2001;124(2):125–126. doi: 10.1067/mhn.2001.113035. [DOI] [PubMed] [Google Scholar]
- Makishima T, Kurima K, Brewer CC, Griffith AJ. Early onset and rapid progression of dominant nonsyndromic DFNA36 hearing loss. Otol Neurotol. 2004;25(5):714–719. doi: 10.1097/00129492-200409000-00011. [DOI] [PubMed] [Google Scholar]
- Mason JC, De Michele A, Stevens C, Ruth RA, Hashisaki GT. Cochlear implantation in patients with auditory neuropathy of varied etiologies. Laryngoscope. 2003;113(1):45–49. doi: 10.1097/00005537-200301000-00009. [DOI] [PubMed] [Google Scholar]
- Matsushiro N, Doi K, Fuse Y, Nagai K, Yamamoto K, Iwaki T, Kawashima T, Sawada A, Hibino H, Kubo T. Successful cochlear implantation in prelingual profound deafness resulting from the common 233delC mutation of the GJB2 gene in the Japanese. Laryngoscope. 2002;112(2):255–261. doi: 10.1097/00005537-200202000-00011. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhatre AN, Li Y, Atkin G, Maghnouj A, Lalwani AK. Expression of Myh9 in the mammalian cochlea: localization within the stereocilia. J Neurosci Res. 2006;84(4):809–818. doi: 10.1002/jnr.20993. [DOI] [PubMed] [Google Scholar]
- Mohr PE, Feldman JJ, Dunbar JL, McConkey-Robbins A, Niparko JK, Rittenhouse RK, Skinner MW. The societal costs of severe to profound hearing loss in the United States. Int J Technol Assess Health Care. 2000;16(4):1120–1135. doi: 10.1017/s0266462300103162. [DOI] [PubMed] [Google Scholar]
- Nadol JB, Merchant SN., Jr Histopathology and molecular genetics of hearing loss in the human. Int J Pediatr Otorhinolaryngol. 2001;61(1):1–15. doi: 10.1016/s0165-5876(01)00546-8. [DOI] [PubMed] [Google Scholar]
- Nadol JB, Young YS, Jr, Glynn RJ. Survival of spiral ganglion cells in profound sensorineural hearing loss: implications for cochlear implantation. Ann Otol Rhinol Laryngol. 1989;98(6):411–416. doi: 10.1177/000348948909800602. [DOI] [PubMed] [Google Scholar]
- Nilsson M, Soli SD, Sullivan JA. Development of the Hearing in Noise Test for the measurement of speech reception thresholds in quiet and in noise. J Acoust Soc Am. 1994;95(2):1085–1099. doi: 10.1121/1.408469. [DOI] [PubMed] [Google Scholar]
- Pennings RJ, Damen GW, Snik AF, Hoefsloot L, Cremers CW, Mylanus EA. Audiologic performance and benefit of cochlear implantation in Usher syndrome type I. Laryngoscope. 2006;116(5):717–722. doi: 10.1097/01.mlg.0000205167.08415.9e. [DOI] [PubMed] [Google Scholar]
- PETERSON GE, LEHISTE I. Revised CNC lists for auditory tests. J Speech Hear Disord. 1962;27:62–70. doi: 10.1044/jshd.2701.62. [DOI] [PubMed] [Google Scholar]
- Raine CH, Summerfield Q, Strachan DR, Martin JM, Totten C. The cost and analysis of nonuse of cochlear implants. Otol Neurotol. 2008;29(2):221–224. doi: 10.1097/mao.0b013e31815c25a1. [DOI] [PubMed] [Google Scholar]
- Reinert J, Honegger F, Gurtler N. High homogeneity in auditory outcome of pediatric CI-patients with mutations in Gap-Junction-Protein Beta2. Int J Pediatr Otorhinolaryngol. 2010;74(7):791–795. doi: 10.1016/j.ijporl.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Ballesteros M, del Castillo FJ, Martin Y, Moreno-Pelayo MA, Morera C, Prieto F, Marco J, Morant A, Gallo-Teran J, Morales-Angulo C, Navas C, Trinidad G, Tapia MC, Moreno F, del Castillo I. Auditory neuropathy in patients carrying mutations in the otoferlin gene (OTOF) Hum Mutat. 2003;22(6):451–456. doi: 10.1002/humu.10274. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Ballesteros M, Reynoso R, Olarte M, Villamar M, Morera C, Santarelli R, Arslan E, Meda C, Curet C, Volter C, Sainz-Quevedo M, Castorina P, Ambrosetti U, Berrettini S, Frei K, Tedin S, Smith J, Cruz Tapia M, Cavalle L, Gelvez N, Primignani P, Gomez-Rosas E, Martin M, Moreno-Pelayo MA, Tamayo M, Moreno-Barral J, Moreno F, del Castillo I. A multicenter study on the prevalence and spectrum of mutations in the otoferlin gene (OTOF) in subjects with nonsyndromic hearing impairment and auditory neuropathy. Hum Mutat. 2008;29(6):823–831. doi: 10.1002/humu.20708. [DOI] [PubMed] [Google Scholar]
- Rouillon I, Marcolla A, Roux I, Marlin S, Feldmann D, Couderc R, Jonard L, Petit C, Denoyelle F, Garabedian EN, Loundon N. Results of cochlear implantation in two children with mutations in the OTOF gene. Int J Pediatr Otorhinolaryngol. 2006;70(4):689–696. doi: 10.1016/j.ijporl.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Roush P, Frymark T, Venediktov R, Wang B. Audiologic management of auditory neuropathy spectrum disorder in children: a systematic review of the literature. Am J Audiol. 2011;20(2):159–170. doi: 10.1044/1059-0889(2011/10-0032). [DOI] [PubMed] [Google Scholar]
- Rubinstein JT, Parkinson WS, Tyler RS, Gantz BJ. Residual speech recognition and cochlear implant performance: effects of implantation criteria. Am J Otol. 1999;20(4):445–452. [PubMed] [Google Scholar]
- Santarelli R. Information from cochlear potentials and genetic mutations helps localize the lesion site in auditory neuropathy. Genome Med. 2010;2(12):91. doi: 10.1186/gm212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer AE, DeLuca AP, Hildebrand MS, Taylor KR, Gurrola J, 2nd, Scherer S, Scheetz TE, Smith RJ. Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proc Natl Acad Sci U S A. 2010;107(49):21104–21109. doi: 10.1073/pnas.1012989107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siem G, Fruh A, Leren TP, Heimdal K, Teig E, Harris S. Jervell and Lange-Nielsen syndrome in Norwegian children: aspects around cochlear implantation, hearing, and balance. Ear Hear. 2008;29(2):261–269. doi: 10.1097/aud.0b013e3181645393. [DOI] [PubMed] [Google Scholar]
- Sinnathuray AR, Toner JG, Clarke-Lyttle J, Geddis A, Patterson CC, Hughes AE. Connexin 26 (GJB2) gene-related deafness and speech intelligibility after cochlear implantation. Otol Neurotol. 2004a;25(6):935–942. doi: 10.1097/00129492-200411000-00013. [DOI] [PubMed] [Google Scholar]
- Sinnathuray AR, Toner JG, Geddis A, Clarke-Lyttle J, Patterson CC, Hughes AE. Auditory perception and speech discrimination after cochlear implantation in patients with connexin 26 (GJB2) gene-related deafness. Otol Neurotol. 2004b;25(6):930–934. doi: 10.1097/00129492-200411000-00012. [DOI] [PubMed] [Google Scholar]
- Song MH, Cho HJ, Lee HK, Kwon TJ, Lee WS, Oh S, Bok J, Choi JY, Kim UK. CHD7 mutational analysis and clinical considerations for auditory rehabilitation in deaf patients with CHARGE syndrome. PLoS One. 2011;6(9):e24511. doi: 10.1371/journal.pone.0024511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankovic KM, Hennessey AM, Herrmann B, Mankarious LA. Cochlear implantation in children with congenital X-linked deafness due to novel mutations in POU3F4 gene. Ann Otol Rhinol Laryngol. 2010;119(12):815–822. doi: 10.1177/000348941011901205. [DOI] [PubMed] [Google Scholar]
- Taitelbaum-Swead R, Brownstein Z, Muchnik C, Kishon-Rabin L, Kronenberg J, Megirov L, Frydman M, Hildesheimer M, Avraham KB. Connexin-associated deafness and speech perception outcome of cochlear implantation. Arch Otolaryngol Head Neck Surg. 2006;132(5):495–500. doi: 10.1001/archotol.132.5.495. [DOI] [PubMed] [Google Scholar]
- Tang W, Qian D, Ahmad S, Mattox D, Todd NW, Han H, Huang S, Li Y, Wang Y, Li H, Lin X. A Low-Cost Exon Capture Method Suitable for Large-Scale Screening of Genetic Deafness by the Massively-Parallel Sequencing Approach. Genet Test Mol Biomarkers. 2012 doi: 10.1089/gtmb.2011.0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teagle HF, Roush PA, Woodard JS, Hatch DR, Zdanski CJ, Buss E, Buchman CA. Cochlear implantation in children with auditory neuropathy spectrum disorder. Ear Hear. 2010;31(3):325–335. doi: 10.1097/AUD.0b013e3181ce693b. [DOI] [PubMed] [Google Scholar]
- Tomblin JB, Spencer L, Flock S, Tyler R, Gantz B. A comparison of language achievement in children with cochlear implants and children using hearing aids. J Speech Lang Hear Res. 1999;42(2):497–509. doi: 10.1044/jslhr.4202.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tono T, Kiyomizu K, Matsuda K, Komune S, Usami S, Abe S, Shinkawa H. Different clinical characteristics of aminoglycoside-induced profound deafness with and without the 1555 A-->G mitochondrial mutation. ORL J Otorhinolaryngol Relat Spec. 2001;63(1):25–30. doi: 10.1159/000055702. [DOI] [PubMed] [Google Scholar]
- Turchetti G, Bellelli S, Palla I, Berrettini S. Systematic review of the scientific literature on the economic evaluation of cochlear implants in adult patients. Acta Otorhinolaryngol Ital. 2011;31(5):319–327. [PMC free article] [PubMed] [Google Scholar]
- Ulubil SA, Furze AD, Angeli SI. Cochlear implantation in a patient with profound hearing loss with the A1555G mitochondrial DNA mutation and no history of aminoglycoside exposure. J Laryngol Otol. 2006;120(3):230–232. doi: 10.1017/S002221510500318X. [DOI] [PubMed] [Google Scholar]
- Vermeire K, Brokx JP, Wuyts FL, Cochet E, Hofkens A, De Bodt M, Van de Heyning PH. Good speech recognition and quality-of-life scores after cochlear implantation in patients with DFNA9. Otol Neurotol. 2006;27(1):44–49. doi: 10.1097/01.mao.0000187240.33712.01. [DOI] [PubMed] [Google Scholar]
- Vescan A, Parnes LS, Cucci RA, Smith RJ, MacNeill C. Cochlear implantation and Pendred’s syndrome mutation in monozygotic twins with large vestibular aqueduct syndrome. J Otolaryngol. 2002;31(1):54–57. doi: 10.2310/7070.2002.19332. [DOI] [PubMed] [Google Scholar]
- Weegerink NJ, Pennings RJ, Huygen PL, Hoefsloot LH, Cremers CW, Kunst HP. Phenotypes of two Dutch DFNA3 families with mutations in GJB2. Ann Otol Rhinol Laryngol. 2011a;120(3):191–197. doi: 10.1177/000348941112000308. [DOI] [PubMed] [Google Scholar]
- Weegerink NJ, Schraders M, Oostrik J, Huygen PL, Strom TM, Granneman S, Pennings RJ, Venselaar H, Hoefsloot LH, Elting M, Cremers CW, Admiraal RJ, Kremer H, Kunst HP. Genotype-phenotype correlation in DFNB8/10 families with TMPRSS3 mutations. J Assoc Res Otolaryngol. 2011b;12(6):753–766. doi: 10.1007/s10162-011-0282-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel M, Magierska-Krzyszton M, Szyfter K, Mietkiewska D, Szyfter W, Rydzanicz M, Szyfter K, Karlik M. Comparison of rehabilitation results in deaf patients with and without genetically related hearing loss. Cochlear Implants Int. 2008;9(3):132–142. doi: 10.1179/cim.2008.9.3.132. [DOI] [PubMed] [Google Scholar]
- Wu CC, Lee YC, Chen PJ, Hsu CJ. Predominance of genetic diagnosis and imaging results as predictors in determining the speech perception performance outcome after cochlear implantation in children. Arch Pediatr Adolesc Med. 2008;162(3):269–276. doi: 10.1001/archpediatrics.2007.59. [DOI] [PubMed] [Google Scholar]
- Wu CC, Liu TC, Wang SH, Hsu CJ, Wu CM. Genetic characteristics in children with cochlear implants and the corresponding auditory performance. Laryngoscope. 2011a;121(6):1287–1293. doi: 10.1002/lary.21751. [DOI] [PubMed] [Google Scholar]
- Wu CC, Liu TC, Wang SH, Hsu CJ, Wu CM. Genetic characteristics in children with cochlear implants and the corresponding auditory performance. Laryngoscope. 2011b;121(6):1287–1293. doi: 10.1002/lary.21751. [DOI] [PubMed] [Google Scholar]
- Yasunaga S, Grati M, Cohen-Salmon M, El-Amraoui A, Mustapha M, Salem N, El-Zir E, Loiselet J, Petit C. A mutation in OTOF, encoding otoferlin, a FER-1-like protein, causes DFNB9, a nonsyndromic form of deafness. Nat Genet. 1999;21(4):363–369. doi: 10.1038/7693. [DOI] [PubMed] [Google Scholar]
- Yoshikawa S, Kawano A, Hayashi C, Nishiyama N, Kawaguchi S, Furuse H, Ikeda K, Suzuki M, Nakagawa M. The clinical features of patients with the homozygous 235delC and the compound-heterozygous Y136X/G45E of the GJB2 mutations (Connexin 26) in cochlear implant recipients. Auris Nasus Larynx. 2011;38(4):444–449. doi: 10.1016/j.anl.2010.11.012. [DOI] [PubMed] [Google Scholar]