Abstract
One of the greatest thrills a biomedical researcher may experience is seeing the product of many years of dedicated effort finally make its way to the patient. As a team, we have worked for the past eight years to discover a drug that could treat a devastating childhood neuromuscular disease, spinal muscular atrophy (SMA). Here, we describe the journey that has led to a promising drug based on the biology underlying the disease.
SMA is caused by the homozygous loss of the survival of motor neuron 1, telomeric (SMN1) gene; either by deletion or rarely by mutation. In its most severe form, it is the leading genetic cause of infant mortality, with children rarely living beyond two years of age. There are no approved therapies for SMA, with medical care focused mainly on supportive and palliative measures. It is this dire need that has motivated the authors to work collaboratively together to identify a potential therapy for these children. As we describe here, this has been a wonderful journey over the past 8 years that was built off of a strong scientific foundation in basic RNA biology, neuroscience and antisense technology.
SMA background
Spinal muscular atrophy (SMA) is an autosomal recessive neuromuscular disease characterized by degeneration of the motor neurons in the anterior horn of the spinal cord, resulting in atrophy of the voluntary muscles of the limbs and trunk (Lefebvre et al., 1995; Crawford and Pardo, 1996). It is the most common genetic cause of infant mortality, and a major cause of childhood morbidity due to weakness. SMA is caused by deletions or loss-of-function mutations in the SMN1 gene on chromosome 5q13 (Lefebvre et al., 1995); how loss of the SMN protein causes disease is not well understood. The SMN1 premRNA undergoes alternative splicing, with greater than 90% of the mature transcripts derived from the SMN1 gene containing exon 7, which makes a full-length protein product. Humans have a paralogue gene called SMN2, also on chromosome 5, which differs from SMN1 by 11 nucleotides but has an identical coding sequence. One of the nucleotide changes between SMN1 and SMN2 genes is a C-to-T transition within exon 7, and although it is a synonymous change, it weakens the 3′ splice site, resulting in skipping of exon 7. Because of the less efficient splicing of exon 7, 80–90% of the transcripts derived from the SMN2 gene skip exon 7, which codes for a protein product that is rapidly degraded. The limited amount of full-length protein made from the SMN2 gene does not fully compensate for loss of the SMN1 gene. We reasoned that antisense oligonucleotides (ASOs) would be the most direct approach for increasing SMN2 exon 7 inclusion and restoring functional levels of the SMN protein.
Antisense oligonucleotides: Versatile tools to target RNA
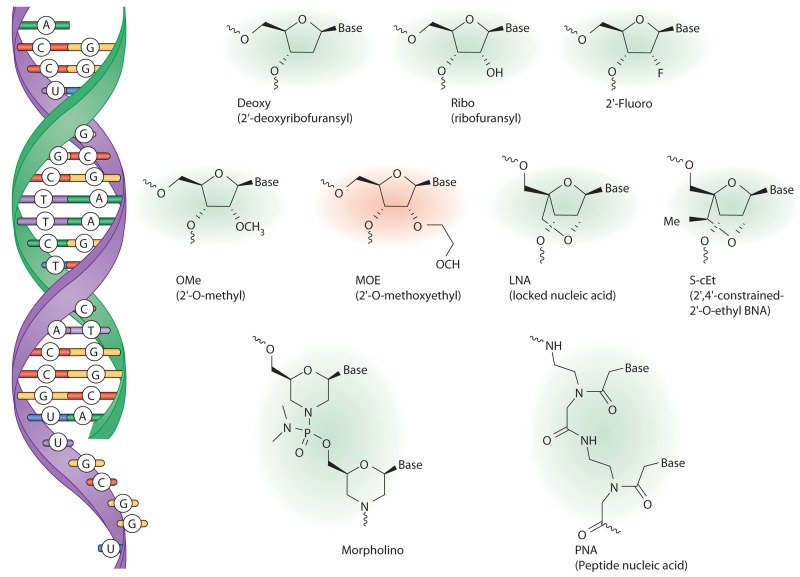
ASOs bind to RNA through Watson-Crick base paring (Fig. 1). Once bound to the target RNA, there are multiple mechanisms by which antisense-based drugs alter its function, including promoting its degradation, interfering with pre-mRNA processing, blocking access to the RNA of specific proteins such as RNA-binding proteins and ribosome subunits, and disrupting the secondary and tertiary structure of the RNA (Bennett and Swayze, 2010; Kole et al., 2012). The mechanism by which an ASO elicits these effects is dependent upon the class of RNA, where on the RNA the ASO binds, and the chemical composition of the ASO.
Figure 1.
Nucleotide analogues used in antisense oligonucleotide drugs. Antisense oligonucleotides (green) bind to the target RNA (purple) by Watson-Crick base pairing (left). Chemical structures of various nucleotides or nucleotide analogues commonly used in antisense drugs are shown. The antisense oligonucleotide developed by our group to improve SMN2 splicing has the 2′-MOE modification (red).
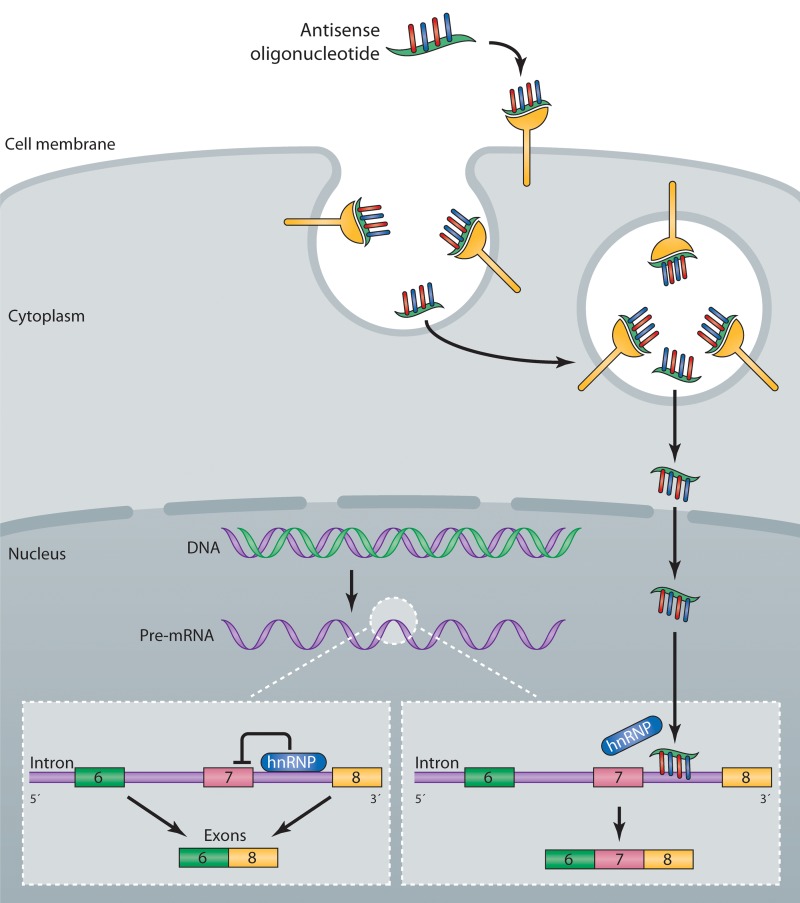
Various chemical modifications of individual nucleotide subunits of the oligonucleotide can enhance the pharmaceutical properties of antisense-based drugs (Fig. 1; Bennett and Swayze, 2010). One of the better characterized chemical modifications, the 2′-O-methoxyethyl (2′-MOE) modification, has been used in 25 different antisense drugs in clinical development, including the SMA antisense drug currently in clinical trials (ISIS-SMNRx). ASOs containing 2′-MOE and other ribose modifications (Fig. 1) typically have phosphorothioate modifications (substitution of sulfur for one of the nonbridging oxygen atoms) to provide additional stability against nuclease degradation, increase protein binding and enhance cellular uptake. The phosphorothioate-modified oligonucleotides are readily taken up by cells in tissues and bind to target RNA, producing the desired antisense effects (Bennett and Swayze, 2010). The mechanism(s) by which ASOs enter cells include at least two endocytic uptake mechanisms (Fig. 2; Geary et al., 2009; Koller et al., 2011), but overall remains poorly understood.
Figure 2.
Mechanism of action of an antisense drug that modulates SMN2 splicing. Single-stranded antisense oligonucleotides (ASO) are taken up into cells by an endocytic process via interaction with proteins expressed on the surface of cells (Koller et al., 2011). The ASOs escape the endosome and enter the nucleus, where they bind to the SMN2 pre-mRNA. Binding of the ASO to the RNA displaces an hnRNP protein that normally represses splicing of exon 7, resulting in the production of a mature mRNA that includes exon 7, which is translated into the full-length SMN protein (Rigo et al., 2012).
A candidate drug for SMA has to be pharmacologically active in motor neurons and other cell types in the central nervous system (CNS). ASOs do not cross an intact blood–brain barrier, but there are several approved methods and devices available for delivery of drugs into the cerebrospinal fluid including direct injection into the intrathecal space (Penn, 2003; Erdine and De Andrés, 2006). We have found that ASOs injected in this manner distribute broadly into CNS tissues with highest concentrations found in neurons, microglial cells, and astrocytes in spinal cord and cortical regions of the brain (Butler et al., 2005; Smith et al., 2006; Passini et al., 2011; Kordasiewicz et al., 2012).
Identification of ASOs that target SMN
The molecular mechanism for SMN1 and SMN2 exon 7 splicing had been previously characterized by A.R. Krainer and others (Hua et al., 2008; Lorson et al., 2010; Singh and Singh, 2011). In addition to the core splicing signals that flank exon 7—such as the 5′ and 3′ splice sites, polypyrimidine tract, and branch point sequence—positive and negative cis-regulatory sequences within exon 7 and in the flanking introns fine tune splicing. For the SMN1 and SMN2 pre-mRNAs, exonic enhancers bind splicing activators such as the SR protein SRSF1 and the SR-like protein Tra2-β1 within exon 7 (Hofmann et al., 2000; Cartegni and Krainer, 2002). It is the SRSF1 binding site that is weakened by the C-to-T substitution at nucleotide 6 in SMN2 exon 7, resulting in the predominant skipping of this exon. In addition to splicing enhancer sequences, splicing silencer sequences have been identified, including one in SMN2 exon 7 that has been reported to be strengthened as a result of the C-to-T transition.
An early approach first used by A.R. Krainer to enhance SMN2 exon 7 inclusion was to recruit factors that activate splicing to the pre-mRNA through the use of bifunctional ASOs. The ESSENCE (exon-specific splicing enhancement by small chimeric effectors) includes a peptide nucleic acid (Fig. 1) as the antisense moiety to hybridize to exon 7 of the SMN2 pre-mRNA, covalently linked to a small peptide that mimics the RS activation domain of SR proteins, effectively making a synthetic SR protein (Cartegni and Krainer, 2003). This publication caught the attention of C.F. Bennett and caused him to contact A.R. Krainer for a potential collaboration to further extend his findings.
To further optimize the ESSENCE strategy, we explored additional binding sites on the SMN2 pre-mRNA for targeting the antisense moiety. We screened 2′-MOE–modified ASOs without the RS peptide and identified several ASOs that were effective at promoting exon 7 inclusion in patient fibroblasts, suggesting that appending the RS peptide was not essential for activity (Hua et al., 2007). Eliminating the protein recruitment appendage (peptide or nucleic acid) from the ASO greatly simplified the molecule and was an important step toward improving its pharmacokinetic and toxicological properties and importantly also reducing the complexity of manufacturing. These studies were extended to identify additional sites in the SMN2 pre-mRNA sensitive to ASOs (Hua et al., 2008). In particular, a site in intron 7 adjacent to the 5′ splice site, termed ISS-N1, was identified by us and others, and targeting this site with ASOs resulted in almost complete SMN2 exon 7 inclusion (Singh et al., 2006, 2009; Hua et al., 2008). The binding site comprises a bipartite hnRNP A1-dependent intronic splicing silencer, which represses exon 7 inclusion (Fig. 2; Hua et al., 2008). Active ASOs targeting this region were found to compete with hnRNP A1 and A2 for binding to the RNA, thereby preventing the repressors from binding to the transcript (Fig. 2; Rigo et al., 2012). A variety of chemically modified ASOs, all targeting ISS-N1, promoted SMN2 exon 7 inclusion (Fig. 1). Surprisingly, we found that ASOs with the same sequence but different chemical modifications could produce opposite effects on exon 7 inclusion (Rigo et al., 2012), demonstrating that ASOs can be used to displace RNA-binding proteins from specific transcripts, but in some cases they can be used to recruit specific RNA binding proteins to RNA transcripts in a highly controlled manner. By choosing not to ignore an unexpected result, we discovered an important nuance of how RNA–oligonucleotide duplexes are recognized by RNA binding proteins that can be exploited by antisense drugs to modulate gene expression.
Pharmacology of antisense drugs that target SMN
Having identified several antisense drug candidates in cell culture, we next moved into mouse models. Because of the short lifespan of mice engineered to mimic the most severe form of SMA, proof-of-concept experiments were performed in mouse models of SMA in which the mice were either heterozygous for the mouse Smn1 gene or had the mouse Smn1 gene deleted and expressed four copies of a human SMN2 transgene (Hsieh-Li et al., 2000). The homozygous Smn1-deleted mice have a normal lifespan and the most notable pathology is necrosis of the tail and ear pinnae, which occurs within 4 wk of birth. After systemic administration, the phosphorothioate and 2′-MOE–modified ASOs targeting the ISS-N1 site increased SMN2 exon 7 inclusion in a dose- and time-dependent manner in liver, kidney, and skeletal muscle, but not CNS tissues (Hua et al., 2008), consistent with the biodistribution of the modified ASO (Geary et al., 2001, 2003). Administration of the ASO into the lateral ventricles of mice resulted in a dose-dependent increase in exon 7 containing transcripts in motor neurons and other cells in the CNS (Hua et al., 2010). Administration of the ASO in utero significantly delayed tail and ear necrosis in the mice (Hua et al., 2010). These studies demonstrated that treatment of mice expressing the human SMN2 transgene with a 2′-MOE–modified ASO can increase exon 7 inclusion from 20% to greater than 90% of the transcripts derived from the transgene in peripheral and CNS tissues. The ASO was well tolerated at doses that promoted almost complete exon 7 inclusion (Hua et al., 2010).
These results were extended to models of severe SMA in which newborn mice were administered a single dose of 2′-MOE ASO at d 0 or d 1 after birth (Hua et al., 2011; Passini et al., 2011). In both severe mouse models the ASO delayed the loss of motor neurons, preserved neuromuscular junctions, improved muscle physiology, and increased survival.
Recent publications have indicated that in addition to motor neuron pathology a number of pathological changes in peripheral tissues of the severe SMA mouse models may contribute to their early demise (Vitte et al., 2004; Bevan et al., 2010; Heier et al., 2010; Shababi et al., 2010). We were curious if systemic treatment with an ASO could enhance survival in a mouse model of severe SMA. Therefore, we treated mice on d 1 and 3 after birth with increasing doses of the 2′-MOE–modified ASO by subcutaneous or intraperitoneal injection (Hua et al., 2011). Surprisingly, treating systemically markedly improved survival of the mice, with some mice living greater than 400 d after birth, versus 10 d in the control mice (Hua et al., 2011). Systemic administration of the ASO in newborn mice did result in a small increase in SMN2 exon 7 inclusion in CNS tissue, consistent with newborn mice having an immature blood–brain barrier. Mice dosed subcutaneously with the ASO on postnatal d 1 and 3 demonstrated marked improvements in motor neuron function and morphology of the neuromuscular junction and muscle tissue, demonstrating that systemic treatment of neonatal mice did improve motor neuron health. In addition, improvements in cardiac pathology were also noted. Interestingly, we observed that circulating levels of IGF1 were reduced in mice with severe SMA, which was reversed by systemic treatment with the ASO. A hallmark of the severe SMA mice is that they have slow growth rates compared with mSmn1 heterozygous littermates. Considering that IGF1 deficiency leads to dwarfism, loss of IGF1 may contribute to the smaller size of the severe SMA mice compared with mSmn1 heterozygous mice. The observation that severe SMA mice are deficient in IGF1 was recently confirmed by another research group (Murdocca et al., 2012). In aggregate, these studies suggest that marked improvement of survival in the severe SMA mouse model may require SMN protein replacement in peripheral tissues, in addition to the CNS, which may explain in part the robust effects observed with systemic versus central gene replacement therapies (Foust et al., 2010; Passini et al., 2010; Valori et al., 2010). It remains to be determined whether these findings are only relevant to the severe SMA mouse model or if they translate to patients. Although the cardiac changes and peripheral vascular perfusion abnormalities reported in mice have occasionally been reported in severely affected SMA patients (El-Matary et al., 2004; Araujo et al., 2009), they do not appear to be a common feature of the disease. More work is required to determine whether these findings in mice will translate to patients with SMA, perhaps depending on the severity of disease.
Based upon these preclinical studies in mice and nonhuman primates, we have advanced the above-mentioned antisense drug into clinical development. A challenge for the program is that no large animal species expresses the SMN2 gene, making it difficult to demonstrate a positive pharmacological effect in preclinical studies. To help determine dose ranges to be used in the clinical trials, we carefully measured the ASO tissue concentrations in the hSMN2 transgenic mice and determined the degree of SMN2 exon 7 inclusion (the pharmacodynamic effect) at each tissue concentration. Based upon these studies, we determined that tissue concentrations ranging from 1 to 5 µg/g tissue after bolus injection was sufficient to achieve 50–90% exon 7 inclusion. Studies in nonhuman primates demonstrated that these concentrations could be achieved after a single intrathecal bolus injection, and tissue concentrations were maintained for several months after dosing (unpublished data). The preclinical studies that support filing an Investigation New Drug (IND) application with the U.S. FDA for the SMN2 splicing ASO (ISIS SMNRx) have been completed, and a phase 1 trial of the drug has been initiated (ClinicalTrials.gov Identifier NCT01494701).
Conclusion
This project serves as an example of how scientists in both academia and industry can successfully collaborate to identify a drug candidate for a rare disease. The collaboration played to the strengths of the parties involved with A.R. Krainer, Y. Hua, and F. Rigo providing expertise in RNA splicing and cell biology of the SMN protein; C.F. Bennett and F. Rigo providing expertise in antisense technology and drug development; and all parties contributing basic understanding of disease mechanisms. We would be remiss if we did not acknowledge the support we received from nonprofit patient advocacy groups, the SMA Foundation, Families of SMA, and Muscular Dystrophy Association. These groups not only provided financial support for the early and risky stages of the project, but also provided scientific and clinical advice for the project. Furthermore, they facilitated the presentation of the data to scientific and clinical peers at various venues, allowing for important constructive critique of the project. The data generated from this collaborative SMA project provided us with the confidence needed to advance the drug into clinical development and potentially benefit patients with a severe disease who, at present, have no other therapeutic options. Our sincere hope is that our efforts ultimately benefit these patients.
Acknowledgments
We would like to acknowledge The Muscular Dystrophy Association for funding studies in A.R. Krainer’s laboratory and Isis Pharmaceutical. In addition, A.R. Krainer was supported by grants from The SMA Foundation and National Institutes of Health grant R37 GM42699-22. Illustrations were provided by Neil Smith, www.neilsmithillustration.co.uk.
Footnotes
Abbreviations used in this paper:
- ASO
- antisense oligonucleotide
- 2′-MOE
- 2′-O-methoxyethyl
- CNS
- central nervous system
- SMA
- spinal muscular atrophy
- SMN
- Survival motor neuron
References
- Araujo Ade.Q., Araujo M., Swoboda K.J. 2009. Vascular perfusion abnormalities in infants with spinal muscular atrophy. J. Pediatr. 155:292–294 10.1016/j.jpeds.2009.01.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett C.F., Swayze E.E. 2010. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 50:259–293 10.1146/annurev.pharmtox.010909.105654 [DOI] [PubMed] [Google Scholar]
- Bevan A.K., Hutchinson K.R., Foust K.D., Braun L., McGovern V.L., Schmelzer L., Ward J.G., Petruska J.C., Lucchesi P.A., Burghes A.H., Kaspar B.K. 2010. Early heart failure in the SMNDelta7 model of spinal muscular atrophy and correction by postnatal scAAV9-SMN delivery. Hum. Mol. Genet. 19:3895–3905 10.1093/hmg/ddq300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler M., Hayes C.S., Chappell A., Murray S.F., Yaksh T.L., Hua X.-Y. 2005. Spinal distribution and metabolism of 2′-O-(2-methoxyethyl)-modified oligonucleotides after intrathecal administration in rats. Neuroscience. 131:705–715 10.1016/j.neuroscience.2004.11.038 [DOI] [PubMed] [Google Scholar]
- Cartegni L., Krainer A.R. 2002. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat. Genet. 30:377–384 10.1038/ng854 [DOI] [PubMed] [Google Scholar]
- Cartegni L., Krainer A.R. 2003. Correction of disease-associated exon skipping by synthetic exon-specific activators. Nat. Struct. Biol. 10:120–125 10.1038/nsb887 [DOI] [PubMed] [Google Scholar]
- Crawford T.O., Pardo C.A. 1996. The neurobiology of childhood spinal muscular atrophy. Neurobiol. Dis. 3:97–110 10.1006/nbdi.1996.0010 [DOI] [PubMed] [Google Scholar]
- El-Matary W., Kotagiri S., Cameron D., Peart I. 2004. Spinal muscle atrophy type 1 (Werdnig-Hoffman disease) with complex cardiac malformation. Eur. J. Pediatr. 163:331–332 10.1007/s00431-004-1437-6 [DOI] [PubMed] [Google Scholar]
- Erdine S., De Andrés J. 2006. Drug delivery systems. Pain Pract. 6:51–57 10.1111/j.1533-2500.2006.00059.x [DOI] [PubMed] [Google Scholar]
- Foust K.D., Wang X., McGovern V.L., Braun L., Bevan A.K., Haidet A.M., Le T.T., Morales P.R., Rich M.M., Burghes A.H., Kaspar B.K. 2010. Rescue of the spinal muscular atrophy phenotype in a mouse model by early postnatal delivery of SMN. Nat. Biotechnol. 28:271–274 10.1038/nbt.1610 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Geary R.S., Watanabe T.A., Truong L., Freier S., Lesnik E.A., Sioufi N.B., Sasmor H., Manoharan M., Levin A.A. 2001. Pharmacokinetic properties of 2′-O-(2-methoxyethyl)-modified oligonucleotide analogs in rats. J. Pharmacol. Exp. Ther. 296:890–897 [PubMed] [Google Scholar]
- Geary R.S., Yu R.Z., Watanabe T., Henry S.P., Hardee G.E., Chappell A., Matson J.E., Sasmor H., Cummins L., Levin A.A. 2003. Pharmacokinetics of a tumor necrosis factor-alpha phosphorothioate 2′-O-(2-methoxyethyl) modified antisense oligonucleotide: comparison across species. Drug Metab. Dispos. 31:1419–1428 10.1124/dmd.31.11.1419 [DOI] [PubMed] [Google Scholar]
- Geary R.S., Wancewicz E.V., Matson J.E., Pearce M., Siwkowski A., Swayze E., Bennett F. 2009. Effect of dose and plasma concentration on liver uptake and pharmacologic activity of a 2′-methoxyethyl modified chimeric antisense oligonucleotide targeting PTEN. Biochem. Pharmacol. 78:284–291 10.1016/j.bcp.2009.04.013 [DOI] [PubMed] [Google Scholar]
- Heier C.R., Satta R., Lutz C., DiDonato C.J. 2010. Arrhythmia and cardiac defects are a feature of spinal muscular atrophy model mice. Hum. Mol. Genet. 19:3906–3918 10.1093/hmg/ddq330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann Y., Lorson C.L., Stamm S., Androphy E.J., Wirth B. 2000. Htra2-beta 1 stimulates an exonic splicing enhancer and can restore full-length SMN expression to survival motor neuron 2 (SMN2). Proc. Natl. Acad. Sci. USA. 97:9618–9623 10.1073/pnas.160181697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh-Li H.M., Chang J.G., Jong Y.J., Wu M.H., Wang N.M., Tsai C.H., Li H. 2000. A mouse model for spinal muscular atrophy. Nat. Genet. 24:66–70 10.1038/71709 [DOI] [PubMed] [Google Scholar]
- Hua Y., Vickers T.A., Baker B.F., Bennett C.F., Krainer A.R. 2007. Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol. 5:e73 10.1371/journal.pbio.0050073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y., Vickers T.A., Okunola H.L., Bennett C.F., Krainer A.R. 2008. Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am. J. Hum. Genet. 82:834–848 10.1016/j.ajhg.2008.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y., Sahashi K., Hung G., Rigo F., Passini M.A., Bennett C.F., Krainer A.R. 2010. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 24:1634–1644 10.1101/gad.1941310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y., Sahashi K., Rigo F., Hung G., Horev G., Bennett C.F., Krainer A.R. 2011. Peripheral SMN restoration is essential for long-term rescue of a severe SMA mouse model. Nature. 478:123–126 10.1038/nature10485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kole R., Krainer A.R., Altman S. 2012. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat. Rev. Drug Discov. 11:125–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller E., Vincent T.M., Chappell A., De S., Manoharan M., Bennett C.F. 2011. Mechanisms of single-stranded phosphorothioate modified antisense oligonucleotide accumulation in hepatocytes. Nucleic Acids Res. 39:4795–4807 10.1093/nar/gkr089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordasiewicz H.B., Stanek L.M., Wancewicz E.V., Mazur C., McAlonis M.M., Pytel K.A., Artates J.W., Weiss A., Cheng S.H., Shihabuddin L.S., et al. 2012. Sustained therapeutic reversal of Huntington’s disease by transient repression of huntingtin synthesis. Neuron. 74:1031–1044 10.1016/j.neuron.2012.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre S., Bürglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., et al. 1995. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 80:155–165 10.1016/0092-8674(95)90460-3 [DOI] [PubMed] [Google Scholar]
- Lorson C.L., Rindt H., Shababi M. 2010. Spinal muscular atrophy: mechanisms and therapeutic strategies. Hum. Mol. Genet. 19(R1):R111–R118 10.1093/hmg/ddq147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdocca M., Malgieri A., Luchetti A., Saieva L., Dobrowolny G., De Leonibus E., Filareto A., Quitadamo M.C., Novelli G., Musarò A., Sangiuolo F. 2012. IPLEX administration improves motor neuron survival and ameliorates motor functions in a severe mouse model of SMA. Mol. Med. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passini M.A., Bu J., Roskelley E.M., Richards A.M., Sardi S.P., O’Riordan C.R., Klinger K.W., Shihabuddin L.S., Cheng S.H. 2010. CNS-targeted gene therapy improves survival and motor function in a mouse model of spinal muscular atrophy. J. Clin. Invest. 120:1253–1264 10.1172/JCI41615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passini M.A., Bu J., Richards A.M., Kinnecom C., Sardi S.P., Stanek L.M., Hua Y., Rigo F., Matson J.E., Hung G., et al. 2011. Antisense oligonucelotides delivered to the mouse CNS ameliorate symptoms of severe spinal muscular atrophy. Sci. Transl. Med. 3:72r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn R.D. 2003. Intrathecal medication delivery. Neurosurg. Clin. N. Am. 14:381–387 10.1016/S1042-3680(03)00016-0 [DOI] [PubMed] [Google Scholar]
- Rigo F., Hua Y., Chun S.J., Prakash T.P., Krainer A.R., Bennett C.F. 2012. Modulation of gene expression by antisense oligonucleotide directed recruitment of specific proteins to RNA. Nat. Chem. Biol. 8:556–561 10.1038/nchembio.939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shababi M., Habibi J., Yang H.T., Vale S.M., Sewell W.A., Lorson C.L. 2010. Cardiac defects contribute to the pathology of spinal muscular atrophy models. Hum. Mol. Genet. 19:4059–4071 10.1093/hmg/ddq329 [DOI] [PubMed] [Google Scholar]
- Singh N.N., Singh R.N. 2011. Alternative splicing in spinal muscular atrophy underscores the role of an intron definition model. RNA Biol. 8:600–606 10.4161/rna.8.4.16224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N.K., Singh N.N., Androphy E.J., Singh R.N. 2006. Splicing of a critical exon of human Survival Motor Neuron is regulated by a unique silencer element located in the last intron. Mol. Cell. Biol. 26:1333–1346 10.1128/MCB.26.4.1333-1346.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N.N., Shishimorova M., Cao L.C., Gangwani L., Singh R.N. 2009. A short antisense oligonucleotide masking a unique intronic motif prevents skipping of a critical exon in spinal muscular atrophy. RNA Biol. 6:341–350 10.4161/rna.6.3.8723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R.A., Miller T.M., Yamanaka K., Monia B.P., Condon T.P., Hung G., Lobsiger C.S., Ward C.M., McAlonis-Downes M., Wei H., et al. 2006. Antisense oligonucleotide therapy for neurodegenerative disease. J. Clin. Invest. 116:2290–2296 10.1172/JCI25424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valori C.F., Ning K., Wyles M., Mead R.J., Grierson A.J., Shaw P.J., Azzouz M. 2010. Systemic delivery of scAAV9 expressing SMN prolongs survival in a model of spinal muscular atrophy. Sci. Transl. Med. 2:35ra42. [DOI] [PubMed] [Google Scholar]
- Vitte J.M., Davoult B., Roblot N., Mayer M., Joshi V., Courageot S., Tronche F., Vadrot J., Moreau M.H., Kemeny F., Melki J. 2004. Deletion of murine Smn exon 7 directed to liver leads to severe defect of liver development associated with iron overload. Am. J. Pathol. 165:1731–1741 10.1016/S0002-9440(10)63428-1 [DOI] [PMC free article] [PubMed] [Google Scholar]