Abstract
In plants, fatty acids (FAs) and FA-derived complex lipids are major carbon and energy reserves in seeds. They are essential components of cellular membranes and cellular signal or hormone molecules. Although TRANSPARENT TESTA2 (TT2) is well studied for its function in regulating proanthocyanidin biosynthesis in the seed coat, little attention has been given to its role in affecting seed FA accumulation and tolerance to environmental stresses. We demonstrate that the tt2 mutation remarkably increased the seed FA content, decreased seed weight, and altered the FA composition. The increase in FA content in the tt2 seeds was due to the relative decrease of seed coat proportion as well as the more efficient FA synthesis in the tt2 embryo. Microarray analysis revealed that tt2 mutation up-regulated a group of genes critical to FA biosynthesis and embryonic development. The mutation also altered the gene expressions that respond to stress. The microarray analysis discovered that the increase in FA accumulation of the tt2 seeds were accompanied by the significant up-regulation of FUSCA3, a transcriptional factor for embryonic development and FATTY ACID ELONGASE1, which catalyzes the elongation of FA chains. Moreover, lower seed protein accumulation during seed maturation also contributed to the increased seed FA accumulation in tt2 mutants. This study advances the understanding of the TT2 gene in seed FA accumulation and abiotic stresses during seed germination and seedling establishment.
In Arabidopsis (Arabidopsis thaliana), mature seeds consist of three major components, namely, the embryo, endosperm, and seed coat. Fatty acids (FAs) and FA-derived complex lipids are among the major energy reserves predominantly stored in the seed embryo, which develops into the vegetative plant. FAs and FA-derived lipids facilitate successful seed germination and seedling establishment (Li et al., 2006). Seed development can be divided into two main steps, embryonic morphogenesis (EM) and maturation (Baud and Lepiniec, 2009). EM is initiated by double fertilization within anatropous bitegmic ovules. Through a series of programmed cell divisions, the embryo acquires the basic architecture of the plant (Mayer and Jürgens, 1998; Jenik et al., 2007). At 6 d after pollination (DAP), the embryo of Arabidopsis becomes torpedo shaped, and EM is accomplished. The maturation phase from 7 to 20 DAP is characterized by the accumulation of storage compounds, such as seed FA and proteins, and the acquisition of dormancy and desiccation tolerance (Goldberg et al., 1994; Baud et al., 2002; Fait et al., 2006). Finally, during the last few days of seed maturation, storage compound synthesis ends and the embryo becomes metabolically quiescent. The accumulation of seed FA sharply increases between 7 and 17 DAP with a maximum level of 18 DAP. A slight decrease in seed lipid content has been observed in several oilseed species, including Arabidopsis, at the very end of the maturation process (Turnham and Northcote, 1983; Eastmond and Rawsthorne, 2000; Baud et al., 2002). During the embryonic maturation phase of Arabidopsis, the endosperm is almost degraded and reduced to one cell layer surrounding the embryo (Baud and Lepiniec, 2009). As a tissue system, the endosperm serves as nutrition for the developing embryo and/or germinating seedling (Stone et al., 2008). Around 10% of the FAs present in whole seeds are accumulated in the endosperm (Penfield et al., 2004). In contrast, the seed coat that surrounds the embryo and the endosperm is derived from ovular tissue and is therefore strictly of maternal origin. The mature seed coat of Arabidopsis is composed of four cell layers (from outside to inside the seed): An epidermis and a palisade layer composes the outer integument, whereas a crushed layer and an endothelial layer form the inner integument (Debeaujon et al., 2000). The seed coat plays an important role in embryonic nutrition during seed development and in protecting the embryo against detrimental agents from the environment (Mohamedyasseen et al., 1994; Weber et al., 1996; Debeaujon et al., 2000).
Seed coats are consisted of some important compounds like flavonoids, which are unique to higher plants as secondary metabolites involved in physiological functions, such as dormancy or viability. Flavonoids also serve as beneficial micronutrients in human and animal diets. Three major classes of flavonoids, namely, anthocyanins (red to purple pigments), flavonols (colorless to pale yellow pigments), and proanthocyanidins (colorless pigments that turn brown), also known as condensed tannins, are present in Arabidopsis. They give the red, purple, and brown pigmentation to flowers, fruits, and seeds (Koes et al., 1994; Shirley, 1996; Mol et al., 1998; Nesi et al., 2001; Lepiniec et al., 2006). Proanthocyanidins specifically accumulate in the innermost cell layer of the seed coat, called the endothelium (Devic et al., 1999; Nesi et al., 2001). The defect in flavonoid pigmentation results in mutants such as the transparent testa (tt) and the tt glabra (Shirley et al., 1995), different from another kind of seed coat mutations with abnormal testa structure, such as the glabra2 mutant (Rerie et al., 1994). Coordinated growth and the development of the seed coat, endosperm, and embryo is essential for the reserve accumulation of FAs and proanthocyanidins in mature seeds. Although the mechanisms of FA and proanthocyanidins biosynthesis have been extensively studied, little attention has been given to their relationship in determining the final seed storage compounds.
To understand this relationship, the seed FA content and composition of tt2 mutant plants were investigated. TT2 features an R2R3 MYB domain protein located mostly in the nucleus, which is consistent with its regulatory function. It is a major limiting regulator in the proanthocyanidin regulatory network and predominantly expressed during early seed development. The loss of TT2 function specifically affects seed pigmentation and dramatically reduces the expression of several structural genes involved in tannin metabolism. Gain-of-function experiments have demonstrated that TT2 induces ectopic BAN expression, which is regarded as the first enzyme committed to proanthocyanidin biosynthesis (Nesi et al., 2001). Despite knowledge regarding the function of TT2 in proanthocyanidin biosynthesis, the role of TT2 in affecting seed FA storage has been poorly understood. In this research, previously uncharacterized functions of TT2, i.e. the negative effect on seed FA accumulation and the positive effect on stress tolerance during seed germination and seedling establishment were identified.
RESULTS
Increase in FA Content and Alteration of FA Composition in Mature tt2 Seeds
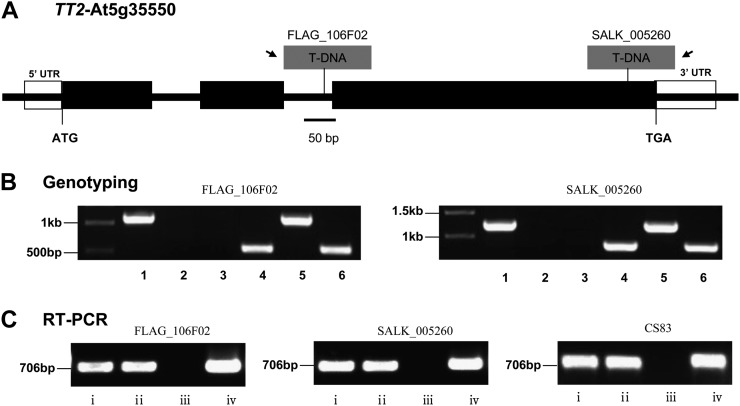
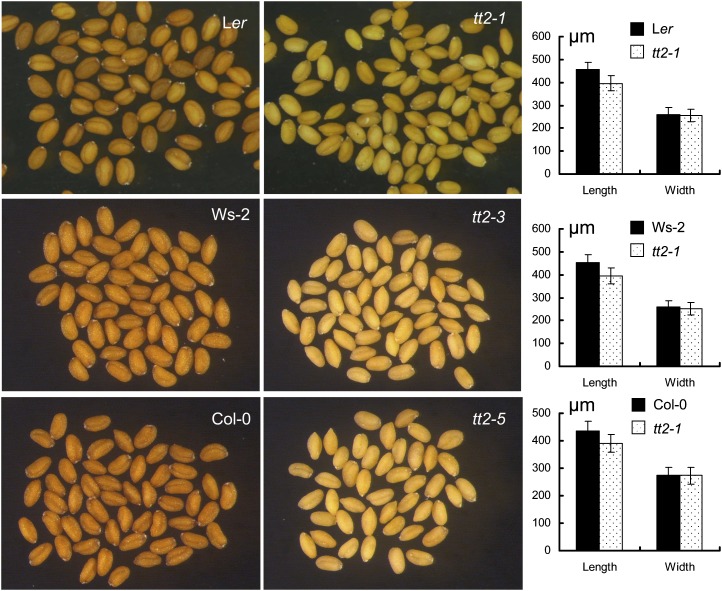
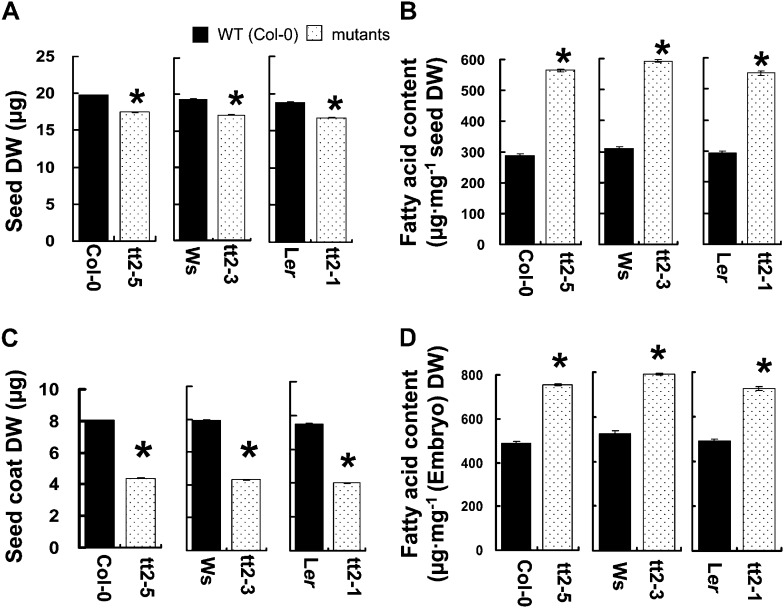
Two mutant lines of the TT2 gene, CS83 (tt2-1; Nesi et al., 2001) and SALK_005260 (tt2-5), a newly nomenclatured allele in this article, were provided by the Arabidopsis Biological Resources Center in the United States. Another mutant line, FLAG_106F02 (tt2-3; Nesi et al., 2001), was obtained from the National Institute for Agricultural Research, France. Figure 1A shows the structure of the TT2 gene and the respective insertion sites of the two allelic mutants. To investigate the effects of the tt2 mutations on seed FA accumulation, the mutant lines (tt2-3 and tt2-5) were backcrossed with their respective wild-type parents in case other mutations were present in the mutant lines. Genotyping by PCR (Fig. 1B) indicated the presence of the homozygous tt2-3 and tt2-5 mutants, which completely lack the transcription of TT2 gene, as detected by reverse transcriptase (RT)-PCR (Fig. 1C). On the other hand, the mutant tt2-1 was created by x-ray irradiation and was difficult to genotype by PCR. Thus, the yellow seed coat trait (Fig. 2) was used as an indicator for genotyping and the mutant plants were further confirmed by RT-PCR for null TT2 transcripts (Fig. 1C). The tt2 mutant seeds are smaller in size than their respective wild-type controls (Fig. 2). In terms of dry weight (DW), the mutant seeds upon maturation were approximately 12% lighter than their corresponding wild-type seeds (Fig. 3A). In contrast, the mutant embryos were approximately 11% to 13% heavier than those of the respective wild types (Fig. 3, A and C).
Figure 1.
Identification of tt2 mutant lines using the primers given in Supplemental Tables S1 and S2. A, Molecular characterization of the tt2 mutation. Structure of the TT2 gene showing the position of the T-DNA insertions in FLAG_106F02 (tt2-3) and SALK_005260 (tt2-5) mutants. Black boxes represent exons while open boxes stand for untranslated regions (UTRs). The arrows indicate the left border on T-DNA. B, PCR-based genotyping of the SALK mutants. 1, 3, and 5 stand for bands of LP and RP primers for genotyping wild types and homozygous and heterozygous SALK mutants, respectively; 2, 4, and 6 stand for bands of RP and BP primers for genotyping wild types and homozygous and heterozygous SALK mutants, respectively; LP and RP refer to the TT2 gene-specific primers and BP refer to T-DNA right-border primer given in Supplemental Table S1. C, RT-PCR was used to measure the TT2 gene expression in the SALK mutants. i and ii represent bands of EF1αA4 for homozygous SALK mutants and the wild type, respectively; iii and iv represent bands of primers specific for the TT2 gene for homozygous SALK mutants and the wild type, respectively.
Figure 2.
Microscopic observation of tt2 mature seeds. Quantitative comparisons of the length and width of a seed between tt2 mutant lines and their respective wild-type controls are shown in a graph neighboring each pair of the wild type and mutant.
Figure 3.
Comparison between the tt2 mutant lines and their respective wild types. A, Seed DW. B, FA content. C, Seed coat DW (including endosperm). D, FA of embryo. Data presented are mean values of three independent experiments (three biological repeats) ± se. The asterisks (*) indicate significant difference (P ≤ 0.05) compared with the wild type.
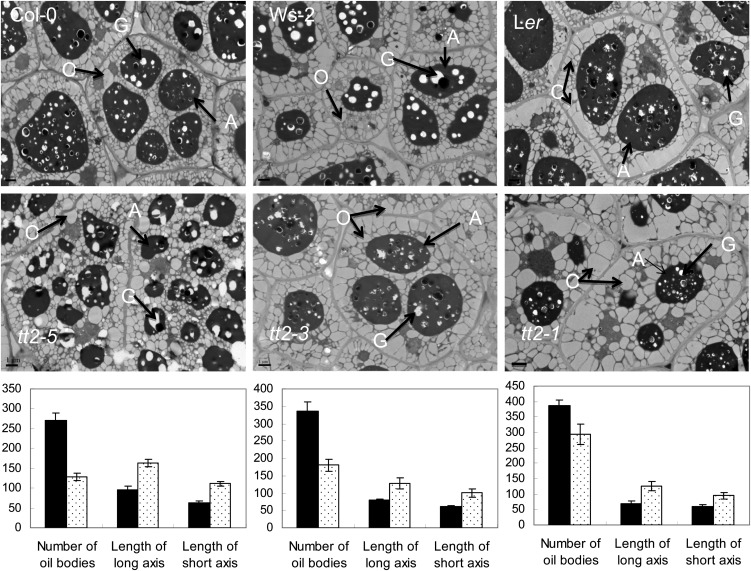
The seed FA content and FA composition in mature tt2 seeds were analyzed using gas chromatography. As shown in Table I and Figure 3B, the tt2 seeds demonstrated 86% to 95% higher FA content than the wild-type seeds. The degree of increase depended on the allelic variations at the TT2 locus (Fig. 3B). Regardless of the mutated allele, the increase in FA content of the mutant seeds was attributed to the increased proportion of the embryo and the increase in FAs in the embryos (Fig. 3, C and D). Moreover, the increase in FA content was accompanied by the alteration of FA composition. In the mature mutant seeds, the proportions of 16-C and 18-C FAs, with an exception of 18:1, decreased significantly, whereas the proportions of 20-C, 22-C, and 24-C FAs (very-long-chain FAs [VLCFAs]; C ≥ 20) increased significantly (Table I). Subcellular structure of the tt2 mutant lines (tt2-3, tt2-5, and tt2-1) was presented in comparison with that of their respective wild-type controls (Fig. 4). The average number of oil bodies that were contained in a cell as well as the average lengths of long and short axis of an oil body were also shown. It is obvious that the tt2 cells had much greater number and larger sizes of oil bodies contained.
Table I. Comparison of FA composition and total FA content at mature seed stage between tt2 mutant lines and their respective wild types.
Values are expressed in μg mg−1 seed DW (in roman) and mol/% (in boldface). Data reported are mean values of five independent experiments (five biological repeats) ± se. Arrows indicate the significant differences (P ≤ 0.05).
| 16:0 | 18:0 | 18:1 | 18:2 | 18:3 | 20:0 | 20:1 | 22:0 | 22:1 | 24:0 | Sum | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Col-0 | 27.78 ± 0.80 | 8.04 ± 0.27 | 26.13 ± 0.52 | 107.51 ± 2.44 | 77.62 ± 1.26 | 3.85 ± 0.09 | 34.87 ± 0.41 | 0.71 ± 0.05 | 2.96 ± 0.32 | 0.45 ± 0.05 | 289.92 ± 5.66 |
| tt2-5 | 48.92 ± 0.92 | 14.69 ± 0.46 | 60.97 ± 0.76 | 201.79 ± 3.17 | 135.24 ± 2.18 | 9.68 ± 0.11 | 83.97 ± 0.68 | 1.54 ± 0.23 | 7.31 ± 0.11 | 1.13 ± 0.05 | 565.24 ± 4.29 |
| Ws-2 | 28.09 ± 1.12 | 9.96 ± 0.61 | 26.59 ± 0.98 | 113.26 ± 2.27 | 82.50 ± 1.85 | 5.13 ± 0.12 | 38.39 ± 1.08 | 0.78 ± 0.07 | 3.59 ± 0.15 | 0.76 ± 0.07 | 309.05 ± 6.99 |
| tt2-3 | 50.24 ± 0.64 | 17.21 ± 0.11 | 59.41 ± 0.55 | 206.21 ± 1.15 | 142.30 ± 2.66 | 12.56 ± 0.13 | 91.24 ± 1.42 | 1.92 ± 0.12 | 9.91 ± 0.31 | 1.89 ± 0.11 | 592.89 ± 4.19 |
| Ler | 26.81 ± 0.39 | 9.79 ± 0.39 | 27.22 ± 1.29 | 108.91 ± 1.32 | 77.91± 1.18 | 4.67 ± 0.11 | 36.13 ± 0.85 | 0.75 ± 0.15 | 3.01 ± 0.28 | 0.55 ± 0.05 | 295.75 ± 5.27 |
| tt2-1 | 46.06 ± 1.09 | 16.44 ± 0.58 | 63.85 ± 1.42 | 180.26 ± 2.22 | 138.96 ± 1.47 | 10.88 ± 0.17 | 84.87 ± 1.32 | 1.77 ± 0.05 | 6.98 ± 0.12 | 1.29 ± 0.05 | 551.36 ± 6.63 |
| Col-0 | 9.58 ± 0.79 | 2.77 ± 0.31 | 9.01 ± 0.33 | 37.08 ± 3.27 | 26.77 ± 3.68 | 1.33 ± 0.13 | 12.03 ± 0.56 | 0.24 ± 0.07 | 1.02 ± 0.31 | 0.16 ± 0.03 | |
| tt2-5 | 8.65 ± 0.86↓ | 2.60 ± 0.47↓ | 10.79 ± 0.39↑ | 35.70 ± 4.12↓ | 23.93 ± 4.15↓ | 1.71 ± 0.15↑ | 14.86 ± 0.75↑ | 0.27 ± 0.03↑ | 1.29 ± 0.23↑ | 0.20 ± 0.05↑ | |
| Ws-2 | 9.09 ± 1.23 | 3.22 ± 0.53 | 8.60 ± 0.52 | 36.65 ± 3.15 | 26.69 ± 3.95 | 1.66 ± 0.17 | 12.42 ± 1.23 | 0.25 ± 0.06 | 1.16 ± 0.18 | 0.25 ± 0.07 | |
| tt2-3 | 8.47 ± 0.86↓ | 2.9 ± 0.15↓ | 10.02 ± 0.31↑ | 34.78 ± 2.07↓ | 24.00 ± 4.62↓ | 2.12 ± 0.21↑ | 15.39 ± 1.56↑ | 0.32 ± 0.08↑ | 1.67 ± 0.19↑ | 0.32 ± 0.09↑ | |
| Ler | 9.07 ± 0.42 | 3.31 ± 0.38 | 9.20 ± 0.67 | 36.83 ± 2.98 | 26.34 ± 3.17 | 1.58 ± 0.15 | 12.22 ± 0.98 | 0.25 ± 0.05 | 1.02 ± 0.26 | 0.19 ± 0.03 | |
| tt2-1 | 8.35 ± 1.13↓ | 2.98 ± 0.51↓ | 11.58 ± 0.73↑ | 32.69 ± 3.01↓ | 25.20 ± 3.02↓ | 1.97 ± 0.31↑ | 15.39 ± 1.63↑ | 0.32 ± 0.07↑ | 1.27 ± 0.35↑ | 0.23 ± 0.06↑ |
Figure 4.
Comparison of the number and size of oil bodies in mature seed cells between the wild type and tt2 mutant lines in pairs. Arrows in TEM images point to oil bodies (O), aleurone grains (A), and globoids (G). Bar = 1 μm. The quantitative data are shown in a graph under each pair of mutant and wild type. The number of oil bodies are mean value of 120 cells from six seeds. The length of long axis and short axis were calculated based on all oil bodies that were contained in 120 cells. The y axis of the graphs stands for numbers as well as 1% μm.
These results demonstrate that the tt2 mutation led to defective proanthocyanidin biosynthesis in the seed coat, significant increases in the embryo weight and FA content, and alternation of the FA composition, and changes in oil body number and size within a mature seed cell.
Differentially Expressed Genes between the tt2-5 and the Wild Type in Developing Seeds at 6 and 16 DAP
We elaborately selected two critical developing stages for comparison of expressional profiles between a tt2 mutant (tt2-5) and its wild-type control (Columbia-0 [Col-0]). At 6 DAP, EM is completed and seed FA accumulation starts (Baud and Lepiniec, 2009), whereas, at 16 DAP, large differences in seed FA content and FA composition between the mutant and the wild type were identified. As shown in Table II, the total FA content in the tt2-5 seeds was 420.99 μg/mg, significantly higher than 252.45 μg/mg in the wild-type seeds. The FA content of developing seeds at 16 DAP accounted for more than 75% of the final FA content in mature seeds in both genotypes (Tables I and II). The accumulation of seed FAs increased sharply during the late embryonic maturation stages from 16 to 18 DAP and reached the final seed FA content at the end of EM maturation, indicating an active involvement of the genes that contribute to seed FA accumulation. Therefore, molecular analysis of developing seeds at 6 and 16 DAP could provide clues to figure out the downstream targets of TT2 involved in FA biosynthesis and understand the molecular mechanism underlying TT2-regulated seed FA accumulation.
Table II. Comparison of FA composition and total FA content at 16 DAP between tt2-5 and Col-0.
Data reported are mean values of five independent experiments (five biological repeats) ± se. Values are expressed as μg mg−1 DW (in roman) and mol/% (in boldface).
| Genotypes | 16:0 | 18:0 | 18:1 | 18:2 | 18:3 | 20:0 | 20:1 | 22:0 | 22:1 | 24:0 | Sum |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Col-0 | 24.34 ± 0.59 | 6.66 ± 0.24 | 21.29 ± 0.83 | 92.05 ± 0.74 | 68.09 ± 1.51 | 3.34 ± 0.12 | 33.68 ± 0.36 | 0.28 ± 0.02 | 2.38 ± 0.12 | 0.35 ± 0.01 | 252.46 ± 3.26 |
| tt2-5 | 36.37 ± 0.38 | 10.74 ± 0.21 | 48.40 ± 1.02 | 145.65 ± 1.48 | 104.29 ± 0.72 | 6.30 ± 0.20 | 63.34 ± 1.05 | 0.64 ± 0.03 | 4.63 ± 0.23 | 0.62 ± 0.01 | 420.98 ± 3.05 |
| Col-0 | 9.64 ± 0.35 | 2.64 ± 0.23 | 8.43 ± 0.53 | 36.46 ± 0.52 | 26.97 ± 0.65 | 1.32 ± 0.09 | 13.34 ± 0.27 | 0.11 ± 0.01 | 0.94 ± 0.09 | 0.14 ± 0.01 | |
| tt2-5 | 8.64 ± 0.29 | 2.55 ± 0.19 | 11.50 ± 0.63 | 34.60 ± 1.09 | 24.77 ± 0.39 | 1.50 ± 0.13 | 15.05 ± 0.91 | 0.15 ± 0.02 | 1.10 ± 0.16 | 0.15 ± 0.01 |
Microarray analysis revealed that 503 differentially expressed genes (DEGs) were identified, among which 447 were up-regulated (Supplemental Table S3) and 56 were down-regulated in the tt2 mutant (Supplemental Table S4) at the seed developmental stage 6 DAP. Functional analysis of these 503 DEGs discovered that 51 (11.4%) and 59 (13.2%) of the up-regulated genes are related to carbohydrate metabolism and general protein synthesis, respectively. However, only 13 (2.9%) of the up-regulated genes are involved in lipid metabolism and embryogenesis. Many of the DEGs are related to stress/defense response (Table II; Supplemental Tables S3 and S4). Therefore, the mutation may promote seed starch and protein synthesis and alters the responses to environmental stresses during EM.
At 16 DAP, 746 DEGs were identified. Of these, 289 were up-regulated (Supplemental Table S5) and 457 were down-regulated (Supplemental Table S6) in the developing mutant seeds. Functional analysis unveiled that the expression of genes involved in lipid metabolism and embryogenesis was significantly altered in the tt2-5 seeds. Relative to the 6 DAP stage, a greater number of DEGs were involved in lipid metabolism and embryogenesis at 16 DAP. More than 17% of the down-regulated genes, particularly the ribosomal protein genes, were involved in general protein synthesis (Supplemental Table S6). Up to 48 down-regulated genes (10.5%) and 24 up-regulated genes (8.4%) were related to carbohydrate metabolism. A greater number of DEGs involved in carbohydrate metabolism were identified in the tt2-5 seeds at 16 DAP than at 6 DAP. The largest class of DEGs at 6 DAP and at 16 DAP between the mutant and the wild type was related to stress/defense response (Tables III and IV; Supplemental Tables S3–S6). Therefore, the tt2-5 mutation may decrease the rate of protein and starch syntheses during embryonic maturation, and gave rise to a different response to environmental stresses.
Table III. Functional classification of DEGs in developing seeds of tt2-5 plants at 6 DAP.
Functional classification of the DEGs was performed using the biological process category of Arabidopsis Gene Ontology (http://www.geneontology.com). Percentage (far-right column) refers to the ratio of genes of each functional category relative to total up-regulated or down-regulated DEGs identified in the microarray experiment. The DEGs with log2 ratios greater than 1.00 or less than −1.00 (only GO Slim IDs with P values equal to or less than 0.5) were listed.
| Category | Up-Regulated DEGs |
Down-regulated DEGs |
||||||
|---|---|---|---|---|---|---|---|---|
| ≥2 | 1–2 | Total | Percentage | ≤−2 | −2 to −1 | Total | Percentage | |
| Log2 ratio | ||||||||
| Metabolism | ||||||||
| Flavonoid metabolism | 0 | 1 | 1 | 0.2 | 3 | 0 | 3 | 5.4 |
| Lipid metabolism | 1 | 6 | 7 | 1.6 | 0 | 2 | 2 | 3.6 |
| Carbohydrate metabolism | 15 | 36 | 51 | 11.4 | 1 | 5 | 6 | 10.7 |
| Nucleic acid | 0 | 5 | 5 | 1.1 | 0 | 1 | 1 | 1.8 |
| Amino acid and protein | 7 | 52 | 59 | 13.2 | 2 | 4 | 6 | 10.7 |
| Growth and development | ||||||||
| Leaf and root development | 0 | 1 | 1 | 0.2 | ||||
| Embryo and seed development | 1 | 5 | 6 | 1.3 | 1 | 0 | 1 | 1.8 |
| Flower development | 2 | 1 | 3 | 0.7 | ||||
| Cell growth | 2 | 14 | 16 | 3.6 | 1 | 1 | 2 | 3.6 |
| Cytoskeleton structure | 1 | 0 | 1 | 1.8 | ||||
| Hormone | 4 | 25 | 29 | 6.5 | 2 | 1 | 3 | 5.4 |
| Stress/defense response | 27 | 123 | 150 | 33.6 | 5 | 13 | 18 | 32.1 |
| Cell regulation | ||||||||
| Transcriptional regulation | 11 | 37 | 48 | 10.7 | 1 | 1 | 2 | 3.6 |
| Signaling transduction | 5 | 19 | 24 | 5.4 | 1 | 7 | 8 | 14.3 |
| Electron transport | 0 | 6 | 6 | 1.3 | ||||
| Transport facilitation | 4 | 14 | 18 | 4.0 | ||||
| Other | 5 | 18 | 23 | 5.1 | 0 | 3 | 3 | 5.4 |
Table IV. Functional classification of DEGs in developing seeds of tt2-5 plants at 16 DAP.
Functional classification of the DEGs was performed using the biological process category of Arabidopsis Gene Ontology (http://www.geneontology.com). Percentage (far-right column) refers to the ratio of genes of each functional category relative to total up-regulated or down-regulated DEGs identified in the microarray experiment. The DEGs with log2 ratios greater than 1.00 or less than −1.00 (only GO Slim IDs with P values equal to or less than 0.5) were listed.
| Category | Up-regulated DEGs |
Down-regulated DEGs |
||||||
|---|---|---|---|---|---|---|---|---|
| ≥2 | 1–2 | Total | Percentage | ≤−2 | −2 to −1 | Total | Percentage | |
| Log2 ratio | ||||||||
| Metabolism | ||||||||
| Flavonoid metabolism | 0 | 4 | 4 | 1.3 | ||||
| Lipid metabolism | 2 | 21 | 23 | 8.0 | 1 | 6 | 7 | 1.5 |
| Carbohydrate metabolism | 0 | 24 | 24 | 8.4 | 9 | 39 | 48 | 10.5 |
| Nucleic acid | 0 | 7 | 7 | 2.4 | 4 | 28 | 32 | 7.0 |
| Amino acid and protein | 1 | 30 | 31 | 10.7 | 6 | 75 | 81 | 17.7 |
| Growth and development | ||||||||
| Leaf and root development | 0 | 1 | 1 | 0.3 | 0 | 9 | 9 | 2.0 |
| Embryo and seed development | 2 | 4 | 6 | 2.1 | 0 | 23 | 23 | 5.0 |
| Flower development | 0 | 6 | 6 | 2.1 | 1 | 11 | 12 | 2.6 |
| Cell growth | 1 | 19 | 20 | 6.9 | 2 | 18 | 20 | 4.4 |
| Cytoskeleton structure | 1 | 9 | 10 | 3.5 | 2 | 18 | 20 | 4.4 |
| Hormone | 0 | 11 | 11 | 3.8 | 4 | 13 | 17 | 3.7 |
| Stress/defense response | 5 | 66 | 71 | 24.6 | 13 | 65 | 78 | 17.1 |
| Cell regulation | ||||||||
| Transcriptional regulation | 0 | 2 | 2 | 0.7 | 2 | 40 | 42 | 9.2 |
| Signaling transduction | 0 | 15 | 15 | 5.2 | 1 | 27 | 28 | 6.1 |
| Transport facilitation | 0 | 39 | 39 | 13.5 | 1 | 12 | 13 | 2.8 |
| Other | 0 | 19 | 19 | 6.6 | 2 | 25 | 27 | 5.9 |
According to Beisson et al. (2003) and Baud and Lepiniec (2009), 33 genes encode key enzymes or enzyme subunits that catalyze FA synthesis (FAS) in the plastids and endoplasmic reticulum during seed development. Most of these genes were slightly up-regulated in the tt2-5 mutant at 6 and 16 DAP, with log2 ratios lower than 1.00 (Supplemental Tables S7 and S8). Notably, FATTY ACID ELONGASE1 (FAE1; or KCS1; At4g34520), which encodes condensing enzymes for VLCFA synthesis (James et al., 1995; Millar and Kunst, 1997) was significantly up-regulated in the tt2-5 developing seeds at 16 DAP (Supplemental Tables S3 and S5). Only 2.9% of all up-regulated genes (log2 ratios ≥ 1.00, P values ≤ 0.5) in the tt2-5 seeds are related to lipid metabolism and embryonic development at 6 DAP (Table II). In contrast, the proportion of the up-regulated genes involved in lipid metabolism and embryonic development in the tt2 seeds increased to 10.1% at 16 DAP (log2 ratios ≥ 1.00, P values ≤ 0.5; Table IV).
FA biosynthesis is regulated by several key transcription factors (TFs), including WRINKLED1 (WRI1; At3g54320), LEAFY COTYLEDON1 (LEC1; At1g21970), LEC2 (At1g28300), ABSCISIC ACID INSENSITIVE3 (ABI3; At3g24650), and FUSCA3 (FUS3; At3g26790). No significant differences in TF expression between the tt2-5 mutant and the wild type (Col-0) at 6 and 16 DAP were observed, except for FUS3 expression, which was significantly increased at 16 DAP in tt2-5 relative to the wild type (Col-0; Supplemental Tables S3–S8).
Confirmation of the Up-Regulated Genes for FA Synthesis and Embryonic Development at Different Developmental Stages in the tt2 Seeds
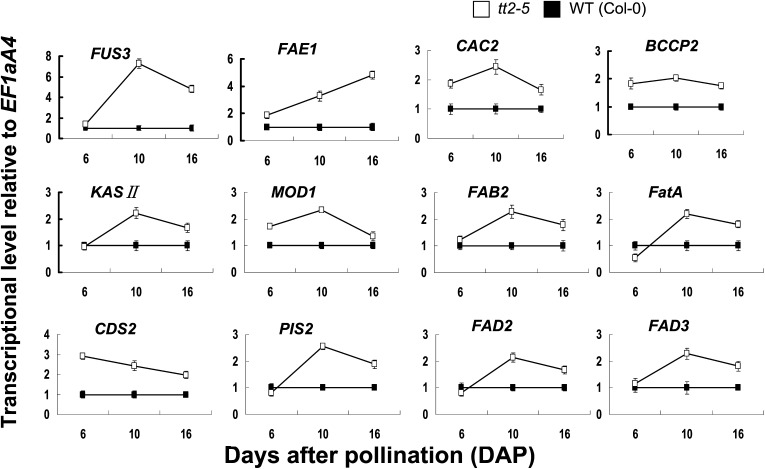
To confirm the up-regulation of genes for FA biosynthesis and embryonic development in the tt2-5 mutant, quantitative (q)RT-PCR was performed. As shown in Figure 5, the relative FUS3 expression remained unchanged at 6 DAP, then increased dramatically from 6 to 16 DAP, and reached its peak at 10 DAP in the tt2-5 mutant compared with the wild type. The expression of the genes that encode the critical FA biosynthetic enzymes, such as BCCP2 (At5g15530), CAC2 (At5g35360), KASII (At1g74960), MOD1 (At2g05990), FAB2 (At2g43710), FatA (At3g25110), CDS2 (At4g22340), PIS2 (At4g38570), FAD2 (At3g12120), FAD3 (At2g29980), and FAE1 (At4g34520), which catalyze various steps in the FA biosynthetic pathway, was also investigated. As detailed in Figure 5, except for CDS2 and FAE1, the expression of the 11 genes in the tt2-5 mutant were up-regulated slightly at 16 DAP and peaked at the 10 DAP stage by about 2-fold that of the wild type (Col-0). The relative CDS2 expression peaked at 6 DAP and decreased at 10 and 16 DAP. However, it was always higher than that of the wild type. The relative FAE1 expression in the tt2-5 mutant was less than 2-fold that of the wild type at 6 DAP, but it increased dramatically from 6 to 16 DAP and peaked at 16 DAP.
Figure 5.
Time-course analysis of the up-regulated genes for FAS and embryo development at different developing stages in tt2 seeds. RNA samples were extracted from developing seeds at three different developmental stages, including 6, 10, and 16 DAP, and gene expression level was estimated by qRT-PCR using EF1aA4 as the internal standard. Values are the means of two replicates carried out on cDNA dilutions obtained from two independent RNA extractions.
Taken together, the tt2-5 mutation promotes the expression of FUS3, a TF for embryonic development, and the genes encoding critical enzymes catalyzing FA biosynthesis.
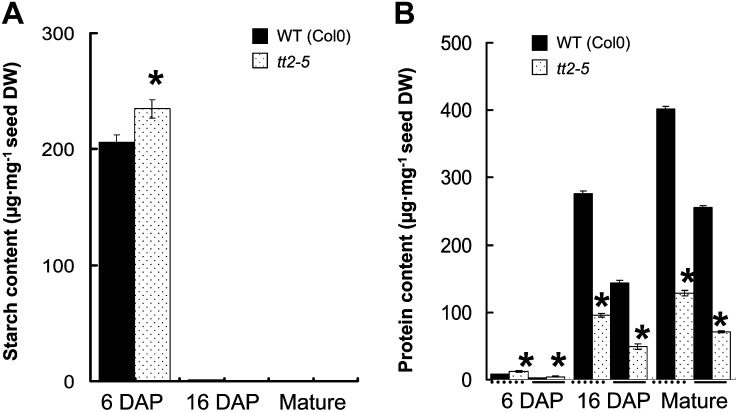
Dynamic Comparison of Seed Starch and Protein Accumulation between tt2-5 and the Wild Type (Col-0)
Indicated by the microarray analysis, the loss of TT2 function alters the expression of genes related to carbohydrate and protein biosynthesis (Tables II and III; Supplemental Tables S4–S7). The tt2-5 and Col-0 were, therefore, compared at 6 DAP, 16 DAP, and the mature stage for the starch and protein contents. As shown in Figure 6A, the starch content of the wild type (Col-0; 205.86 μg/mg DW) and tt2-5 (234.75 μg/mg DW) exhibited the highest levels in the developing seeds at 6 DAP. The mutant seeds contained significantly higher starch than the wild-type seeds. The starch content decreased markedly later during EM in both genotypes. In the seeds at 16 DAP and mature stage, only traces of starch could be detected in both the wild type and the mutant. Figure 6B shows the increase of seed total protein and storage protein contents from 6 DAP to the mature stage in the wild type and tt2-5 mutant. The seed protein contents in the tt2-5 seed were significantly higher than that of the wild type at 6 DAP (Fig. 6B), which is consistent with the finding that the up-regulated genes outnumbered the down-regulated genes for carbohydrate and protein metabolism in the tt2-5 mutant (Tables III and IV; Supplemental Tables S3 and S4). The expression of the genes involved in protein metabolism significantly changed at 16 DAP and the number of up-regulated genes was much less than those of down-regulated genes (Tables III and IV; Supplemental Tables S5 and S6). This may give rise to the lower content of seed total and storage proteins in the tt2-5 seeds at 16 DAP as well as at mature stage. This was helpful to the FA accumulation in tt2-5 mutant line on the other hand.
Figure 6.
Evaluation of starch and protein content in developing Arabidopsis seeds between Col-0 and tt2-5. A, Starch. B, Protein contents were analyzed at three different developmental stages, including 6, 16 DAP, and mature stage, on the DW basis. The dash underlines of B indicate total protein content, and the solid underlines show seed storage protein content. Data presented are mean values of three independent experiments (biological repeats) ± se. The asterisks (*) indicate significant difference (P ≤ 0.05) compared with the wild type.
Alteration of Responses to Stress in the tt2 Mutant Lines
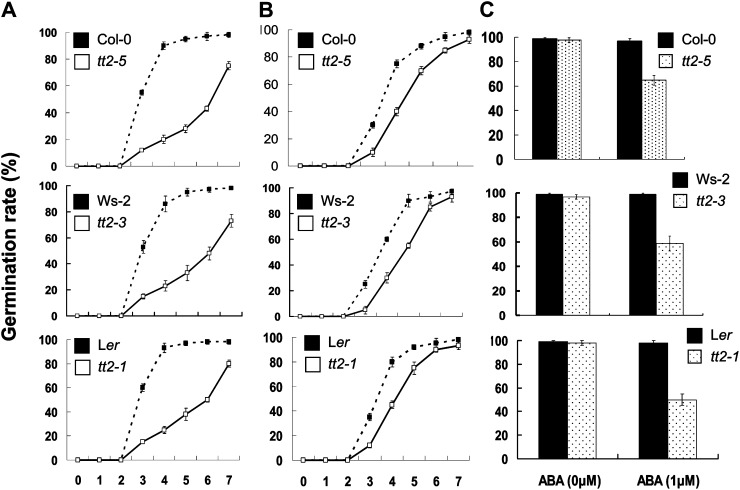
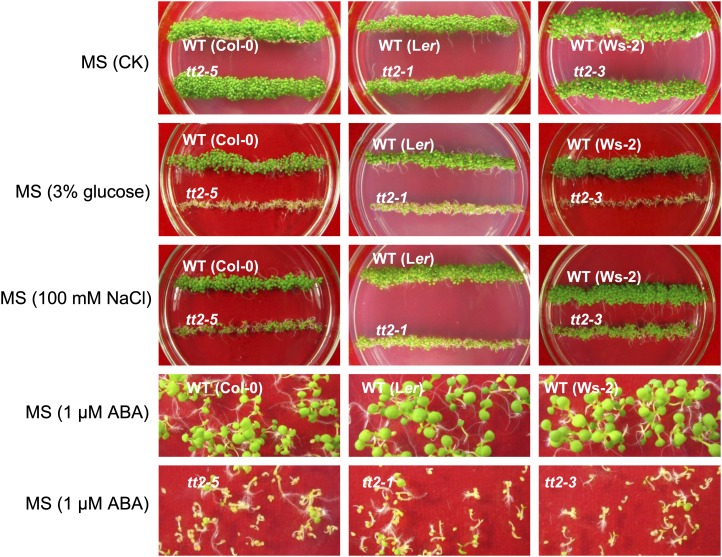
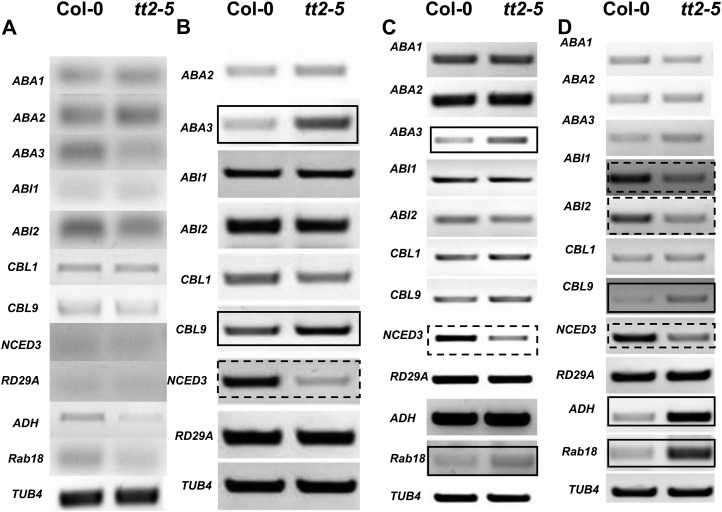
To determine the effect of the tt2 mutation on stress responses, seed germination and seedling establishment were investigated in medium containing 3% (w/v) Glc, 100 mm NaCl, or 1 μm abscisic acid (ABA). Seed germination rate and seedling growth on the medium without any stress treatment were similar between the tt2 mutant lines and their respective wild-type controls. In the medium containing 3% (w/v) Glc, the rate of seed germination and vigor of the tt2 mutant lines were significantly lower than that of their respective wild-type controls. Seven days after sowing, only about three-fourth of the tt2 mutant seeds germinated. In contrast, more than 97% of the wild-type seeds germinated (Fig. 7). All wild-type plants reached the two-leaf stage 10 d after sowing (DAS) unlike the tt2 plants that failed to develop normal cotyledons and green leaves in the medium (Fig. 8). Moreover, a comparison of the expression of genes related to osmotic stress and ABA between the wild type (Col-0) and the tt2-5 plants through semiquantitative RT-PCR unveiled that Glc stress distinctly represses NCED3 and induces the expression of ABA3 and CBL9 in the tt2-5 mutant plants. In contrast, the expression of ABA2, ABI1, ABI2, CBL1, and RD29A in the tt2-5 seedlings was similar to that in the wild-type plants (Fig. 9B). Under the 100 mm NaCl treatment, the seed germination vigor of the tt2-5 plants 7 DAS was lower than that of the wild type (Fig. 7). The cotyledons and green leaves developed poorly in the tt2 mutant lines, which indicates that the loss of TT2 function induces sensitivity to salinity stress (Fig. 8). Semiquantitative RT-PCR analysis shows that NCED3 is significantly down-regulated, whereas ABA3 and Rab18 were up-regulated in the tt2-5 seedlings 10 DAS (Fig. 9C).
Figure 7.
Comparison of seed germination rate under stressed environments between tt2 mutants and their respective wild-type controls. Seed germination was scored daily after the radical tips had fully emerged from the seed coats. Data are the means ± se from three independent experiments evaluating 100 seeds. A, Time courses of seed germination on 1× MS agar medium containing 3% (w/v) Glc. B, Time courses of seed germination on 1× MS agar medium containing 100 mm NaCl. C, Seed germination rate of 7 DAS on 1× MS agar medium containing 1 μm ABA.
Figure 8.
Comparison of seedling establishment under stressed environments between tt2 mutant lines and their respective wild-type controls. The pictures were taken 10 DAS, showing the situations on the control medium (without stress), 1× MS agar medium containing 3% (w/v) Glc, 1× MS agar medium containing 100 mm NaCl, and 1× MS agar medium containing 1 μm ABA.
Figure 9.
Comparison of osmotic-stress- and ABA-related gene expressions between the wild type (Col-0) and tt2-5 under stressed environments. Total RNA was isolated from the whole seedling 10 DAS. RT-PCR was performed with osmotic-stress- and ABA-related gene primers. The cDNA samples were normalized by RT-PCR using a tubulin probe as an internal control. A, 1× MS agar medium without stress. B, 1× MS agar medium containing 3% (w/v) Glc. C, 1× MS agar medium containing 100 mm NaCl. D, 1× MS agar medium containing 1 μm ABA. Boxes of solid lines indicate an up-regulation of a gene in the tt2-5 mutant; boxes of dashed lines show a down-regulation of a gene in the tt2-5 mutant.
The tt2-5 mutants were also sensitive to exogenous ABA (1 μm) during seed germination and seedling establishment. Compared with the wild type, the tt mutants exhibited low seed germination rates (Fig. 7) and badly developed seedlings in the Murashige and Skoog (MS) medium containing 1 μm ABA (Fig. 8). The expression of ABI1, ABI2, and NCED3 was distinctly repressed. However, the expression of CBL9, ADH, and Rab18 was obviously induced in the tt2-5 mutant plants (Fig. 9D). The experiments with the tt2-3 and tt2-1 mutant lines obtained similar results (data not shown), which indicate that TT2 may directly or indirectly confers tolerance to Glc, salinity, and ABA stresses during seed germination and seedling establishment.
DISCUSSION
Factors that control the overall amount of FA stored in the seeds are still largely unknown. In this article, the novel function of the TT2 in affecting seed FA accumulation and abiotic stress tolerance was demonstrated. TT2, which encodes a R2R3 MYB domain protein, is a key regulator of proanthocyanidin biosynthesis in the Arabidopsis seed coat (Nesi et al., 2001). We found that the tt2 mutation resulted in considerably increased seed FA content (86%–95% depending on mutated allele; Table I; Fig. 3B), decreased seed size (Fig. 2), and seed weight (Fig. 3A), and altered FA composition (Table I). The increase in FA content in the tt2 seeds accompanied by the relative decrease of seed coat proportion (Fig. 3C), more sufficient FAS, and reduced protein accumulation in the tt2 embryo (Figs. 3D and 6). On the other hand, the tt2 mutant lines demonstrated poor tolerance to stressful environments (Fig. 7–9). Microarray analysis unveiled that TT2 regulates genes critical to lipid metabolism, embryonic development, and stress/defense responses (Tables III and IV; Supplemental Tables S3–S8).
Loss of TT2 Function Results in More Sufficient FA Synthesis
In Arabidopsis, TT2, together with TT GLABRA1 and TT8, synergistically control the expression of biosynthetic genes in the proanthocyanidin biosynthetic pathway, including TT12, DFR, LDOX, and BAN (Pelletier et al., 1997; Nesi et al., 2000, 2001; Debeaujon et al., 2003; Lepiniec et al., 2006). Consistent with previous studies, the tt2 mutation significantly down-regulated the expression of DFR, LDOX, and BAN in the developing seeds at 6 DAP (Supplemental Table S5) and slightly down-regulated TT12 (data not shown). The loss of TT2 gene function up-regulated the TT10 and the RNS1 in the developing seeds at the 16 DAP stage (Supplemental Table S6). The reduction of proanthocyanidins in the seed coat caused the tt2 seeds to appear yellow (Debeaujon et al., 2000) and have smaller sizes and reduced weights (Figs. 2 and 3A). Their embryos were, however, much heavier than those of the wild type (Fig. 3, A and C).
FAs, a class of prominent metabolites in seeds, are predominantly stored in embryo and play essential roles during the growth and development of all living organisms. Although the biochemistry of FA biosynthesis and the regulatory mechanism of FA metabolism have been well studied in Arabidopsis, little attention has been given to the regulation of TT2 in the metabolism of seed FA and other seed metabolites. Malonyl-CoA formed from acetyl-CoA catalyzed by ACCase is the precursor for seed FA biosynthesis (Lepiniec et al., 2006; Baud and Lepiniec, 2009). ACCase is the key switch that controls FA flux, which must be highly dynamic in responding to variable signals and kept at a low threshold to allow tight control. Therefore, ACCase acts as a sensor or a gating system to monitor the overall flux of FA biosynthesis (Mu et al., 2008). As ACCase subunits, BCCP2 and CAC2 were up-regulated during tt2-5 seed EM (Fig. 5). More active ACCase should lead to increased FA accumulation in the tt2 seeds. Moreover, total protein and storage protein contents in the developing seeds at the 16 DAP and the mature seeds of tt2-5 were much lower than that of the wild type (Col-0; Fig. 6), which may be helpful for seed FA accumulation in the mutant. Therefore, TT2 could be a reasonable choice for genetic engineering toward higher seed oil content in oil crops, such as Brassica spp. oilseeds.
Expression of the genes critical for FAS is coordinately regulated (Ruuska et al., 2002; Baud et al., 2003). Loss of function of the TT2 gene up-regulated many FA biosynthetic genes in the plastid and endoplasmic reticulum. Of these enzymes, the BCCP2 and CAC2, subunits of ACCase, catalyze the initial synthesis step, the formation of malonyl-CoA from acetyl-CoA. A key reaction is to transfer acetyl-CoA and malonyl-ACP to 3-ketoacyl-ACP, which is catalyzed by 3-ketoacyl-ACP synthase. The enzyme MOD1 functions as an enoyl-ACP reductase, whereas FAB2 (SSI2) encodes a stearoyl-ACP desaturase as the ssi2 mutants increase 18:0 and reduce 18:1 FA. In the endoplasmic reticulum, CDP-diacylglycerol is formed from 1,2-diacylglycerol-3-phosphate catalyzed by CDS2 and transferred to phosphatidylinositol by PIS2. FAD2 is essential for polyunsaturated lipid synthesis (Okuley et al., 1994), and FAD3 is responsible for the synthesis of 18:3 FA from phospholipids, which uses cytochrome b5 as an electron donor (Shah et al., 1997). FAE1 is essential for the synthesis of VLCFAs in seeds, extending the chain length of FAs from C18 to C20 and C22. In this study, most FA synthetic genes were only moderately up-regulated, including the genes for ACCase. In contrast, FAE1 was up-regulated remarkably (Fig. 5; Supplemental Table S6). As a consequence of this increased expression, the biosynthesis of VLCFAs, such as C20:0, C20:1, and C22:1 was much more efficient than other FAs, such as C16:0, C18:0, C18:2, and C18:3 in the tt2 seeds at 16 DAP (Table I; Supplemental Table S3). The synthesis of C18:1 in the tt2 seeds was more sufficient than that in the wild-type seeds (Table I). This could be caused by the remarkably increased expression of KASII and FAB2, which are essential in transferring 16:0-ACP into 18:1-ACP in plastids (Fig. 5).
FA biosynthesis is regulated at multiple levels, of which transcriptional control is important for regulating the FA biosynthesis pathway (Ohlrogge and Jaworski, 1997; Millar et al., 2000). The key TFs regulating FA biosynthesis may include WRI1, LEC1, LEC2, FUS3, and ABI3. WRI1 encodes a transcriptional regulator of the AP2/EREB family that targets enzymes in late glycolysis and in the plastidial FA biosynthetic network (Focks and Benning, 1998; Cernac and Benning, 2004). LEC1 encodes the HAP3 subunit of the CCAAT binding factor and plays essential roles in FA biosynthesis (Lee et al., 2003; Mu et al., 2008). LEC2, FUS3, and ABI3 encode B3 domain TFs of functional importance for the regulation of seed maturation (Luerssen et al., 1998; Stone et al., 2001; To et al., 2006). These five TFs have unique and overlapping functions in regulating embryonic development and FA biosynthetic pathway in Arabidopsis (To et al., 2006; Wang et al., 2007; Santos-Mendoza et al., 2008; Baud and Lepiniec, 2009). In this study, the tt2 mutation did not cause significant changes in the expression levels of WRI1, LEC1, LEC2, and ABI3 (Supplemental Tables S8 and S9). However, the FUS3 expression level increased remarkably (Fig. 5; Supplemental Table S6). FUS3 is expressed in seeds mostly during seed filling, and promotes oil deposition by synchronizing genes in the FA biosynthetic pathway to increase the transcript levels of CAC2, KASI, FAB2, FAD3, BCCP2, ACP1, and ACP5, etc. (Wang et al., 2007; Yamamoto et al., 2010). On the other hand, it also plays an active role in positively regulating the expression of photosynthetic genes during early seed development (Yamamoto et al., 2010). Therefore, it was possible that the increased expression of FUS3 in the tt2 mutant could have resulted in more photosynthates, which might be further partitioned into seed FA deposits.
According to Debeaujon et al. (2003), TT2 expression is limited to the testa of early developing seeds. However, FA is mainly synthesized in embryos. It is not likely that TT2 directly regulates the FAS in embryos. Possibly, the deficiency of flavonoids, the precursors of proanthocyanidins, in tt2 mutant causes more sufficient FA biosynthesis in the embryo, because flavonoids, permeable from testa to embryo, are potential inhibitors of FA biosynthesis. They suppress the expression of FabG and FabI, that encode important reductases involved in chain elongation (Zhang and Rock, 2004). The tt2 mutation, therefore, indirectly promoted the FA accumulation due to the removal of the flavonoid repression. In nature, there are two basic types of FAS architectures. The prototypical FAS type I is found in mammals and consists of a single gene that produces a polypeptide, which contains all of the reaction centers required to produce a FA (Smith et al., 2003). In contrast, the dissociated FAS type II system, in which each single step on the FAS pathway is catalyzed with a unique protein encoded by a separate gene, is found in bacteria, plants, and parasites (White et al., 2005).
Loss of TT2 Function Leads to Reduced Tolerance to Environmental Stress during Seedling Establishment
FAS is a primary metabolic pathway essential for the functioning of plant cells. FAs or FA derivatives act as signal molecules or hormones, carbon and energy storage, and surface layer protecting the plant from environmental stress (Ohlrogge and Jaworski, 1997; Hong et al., 2008; Mu et al., 2008). The seed coat plays an important role in the protection against harmful microorganisms and unfavorable environmental conditions (Mohamedyasseen et al., 1994; Debeaujon et al., 2000). This study shows that the tt2 mutant line were much more sensitive to abiotic stresses caused by high concentrations of Glc, NaCl, or ABA in the growth medium (Fig. 7–9). The transcription levels of genes related to osmotic stress and ABA were different between the tt2 and the wild type under stressful environments. The NCED3 expression in the tt2 significantly decreased under a stressful environment. NCED3 encodes 9-cis-epoxycarotenoid dioxygenase, which functions as a key enzyme in ABA biosynthesis (Fig. 7–9). The microarray analysis revealed that the largest class of DEGs between the tt2 mutant and the wild type was related to stress/defense response (Tables II and III; Supplemental Tables S4–S7).
It is also possible that the deficiency of flavonoids gave rise to the alternation of the expression pattern of stress-responsive gene during young seedling establishment. On the other hand, the stress-sensitive phenotypes of tt2 mutant lines are because of increased permeability of stressful elements through the tt2 seed coats.
This work advances the understanding of TT2 functions in seed FA accumulation and abiotic stresses during seed germination and seedling establishment. However, the mechanisms by which TT2 regulate FA accumulation in seeds and stress responses during young seedling establishment are not completely clear.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) accessions Col-0, Landsberg erecta (Ler), and Wassilewskija-2 (Ws-2) were used as the wild-type controls according to the background of the mutant. The seeds of these lines were prechilled at 4°C for 2 to 4 d and then surface sterilized with 6% (w/v) NaClO solution for 6 to 8 min. Subsequently, the seeds were washed five to seven times with distilled, deionized water and then sown onto solid MS stock medium. After about 2 weeks, the healthy seedlings were transferred into autoclaved soil in a flowerpot 7 cm in diameter. The plants were fertilized three times at two true leaf stages, the rosette leaf stage and the bolting stage. The fertilizer (Shanghai Wintong Chemicals Company) contained 20% total nitrogen (5.97% nitrate nitrogen, 3.92% ammonia nitrogen, and 10.11% urea nitrogen), 20% water-soluble phosphorus (P2O5), 20% water-soluble potassium (K2O), 0.07% water-soluble magnesium, 0.009% boron, and 0.02% copper, 0.1% iron, 0.05% manganese, 0.002% molybdenum, 0.02% zinc, and 0.0005% cobalt (all chelated). The plants were grown under long daytime conditions (16 h) in an air-conditioned growth room, at 24°C and 18°C day and night temperatures, respectively. The light intensity for day was 100 μmol m−2 s−1, as measured on the base of a plant.
Analysis of FAs, Seed Starch, and Protein
The extraction and analysis of FAs were carried out as follows (Poirier et al., 1999; Mu et al., 2008). About 10 mg of intact seeds were heated at 80°C in methanol solution containing 1 m HCl for 2 h. The FA methyl esters were extracted with 2 mL hexane and 2 mL of 0.9% (w/v) NaCl, and the organic phase was used for analysis by gas chromatography, using methyl heptadecanoate as an internal standard. The machine (Shimadzu, GC-2014) was equipped with a flame ionization detector and a 30 m (length) × 0.25 mm (i.d.) × 0.5 μm (liquid membrane thickness) column (Supelco wax-10, Supelco, Cat. no. 24079). The initial column temperature was maintained at 160°C for 1 min, then increased by 4°C min−1 to 240°C, and held for 16 min at the final temperature. After the run, the peaks corresponding to each FA species were identified by their characteristic retention times. Concentrations of each sample were normalized against the internal control.
To analyze the FA content and FA composition in the embryo, the seed envelopes that are composed of the integuments plus the endosperm layer were left on the MS medium when the seeds germinated, and then were carefully collected. Up to 100 seed envelopes for each of the three replicates were collected, dried at the similar condition as the mature seeds, and then weighed until the weight was unchanged. The weight of the seed envelopes was subtracted from that of the whole seed, leaving the weight of the embryo. The FA content of the seed embryo was calculated using the FA content of the whole seed divided by the net weight of the seed embryo.
The seed starch content was determined as follows (Mccleary et al., 1994; Baud et al., 2002). Up to 20 seeds were ground twice in 2 × 500 μL of 80% (v/v) ethanol at 4°C. After centrifugation (15,000g, 10 min, 4°C), the resulting pellet was dried at 50°C for 30 min and then homogenized in 150 μL of 50 mm MOPS buffer (pH 7.0) containing 15.4 units of heat-stable α-amylase. The suspension was incubated at 100°C for 6 min. Then, 200 μL of 0.2 m sodium acetate buffer (pH 4.8) containing 26.1 units of amyloglucosidase were added, and the suspension was incubated at 50°C for 30 min with regular shaking. After centrifugation, the Glc in the supernatant liquid was used for starch quantification.
The quantification of seed storage and total protein was adapted from Baud et al. (2002). Twenty seeds were homogenized in 200 μL of extraction buffer containing 50 mm HEPES, 5 mm MgCl2, 5 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, 1 mm EDTA, and 10% (v/v) ethylene glycol (pH 7.5) in the presence of Polyclar. After centrifugation (17,530g, 10 min, 4°C), the supernate was used for storage protein measurements according to Bradford (1976). The remaining proteins insoluble in the extraction buffer were recovered by treatment with 1 m sodium hydroxide and measured as described above. In both assays, gammaglobulin (Bio-Rad) was used for calibration.
Morphologic Observations of Mature Seeds
The seeds for observation were selected from the siliques harvested from the basal part of a major inflorescence in this study. The mature seeds of the wild type and the tt2 mutant lines were randomly selected and were photographed using an OLYMPUS SZ 61 stereomicroscope. Six seeds of each genotype were randomly selected for sectioning. Perpendicular transections were produced and the sections with the largest oval-shaped surface area were selected randomly for the oil body observation. The oil bodies were observed under transmission electron microscopy (TEM). A number of 20 cells from one seed were observed for the oil bodies contained. The average number of oil bodies within a cell was calculated based on six seeds. TEM observation was performed as follows. The main steps, double fixation, dehydration, infiltration and embedding, and ultrathin sectioning are described in detail as follows. Double fixation: The Arabidopsis seeds were first fixed with 2.5% glutaraldehyde in phosphate buffer (0.1 m, pH 7.0) for about 16 h and then washed three times in the phosphate buffer (0.1 m, pH 7.0) for 25 min. The seeds were then postfixed with 1% OsO4 in phosphate buffer (0.1 m, pH 7.0) for 2 h and washed three times in phosphate buffer (0.1 m, pH 7.0) for 25 min, respectively. Dehydration: The specimen was dehydrated using a graded series of ethanol (50%, 70%, 80%, 90%, 95%, and 100%) for about 20 min, respectively, and then transferred to absolute acetone for 20 min. Infiltration: The specimen was placed in a 1:1 mixture of absolute acetone and the final Spurr resin mixture for 2 h at room temperature and then transferred to a 1:3 mixture of absolute acetone and the final Spurr resin mixture for 5 h and to final Spurr resin mixture overnight. Embedding and ultrathin sectioning: The specimen was placed in Eppendorf tubes containing embedding medium and heated at 70°C for about 9 h. The specimen sections were stained with uranyl acetate and alkaline lead citrate for 15 min, respectively, and observed under a TEM (JEM-1230).
Microarray Hybridization and Data Analysis
The flowers of the wild type (Col-0) and tt2-5 were tagged with different-colored threads to indicate the DAP. Only the seeds from the siliques on the primary shoots were utilized in this study. Three independent biological replicates were conducted for the wild type (Col-0) and tt2-5 in the microarray experiment. Probe labeling and chip hybridization were carried out through the Affymetrix custom service (CapitalBio) following the standard protocol (http://www.affymetrix.com/support/technical/manual/expr-ession_manual.affx). Normalization was performed according to the standard Affymetrix protocol to allow the comparison of the samples for each set of experiments. The Excel add in for significance analysis of microarrays was used to identify DEGs between the control and the T-DNA insertion mutant line tt2-5. Functional classification of the DEGs was performed using the biological process category of Arabidopsis Gene Ontology (http://www.geneontology.com). The percentage (far-right column) refers to the ratio of genes relative to the total up-regulated or down-regulated DEGs in each functional category. For the microarray experiment, three independent biological replicates were conducted, and the DEGs with log2 ratios ≥ 1.00 or ≤ −1.00 (only GO Slim IDs with P values ≤ 0.5) were analyzed.
Analysis of Gene Expression by RT-PCR and qRT-PCR
The RNA samples were isolated using the Invisorb spin plant RNA mini kit (Invitek; microarray experiments) following the manufacturer’s instructions. The RNA samples were treated with RNase-free DNAse I (New England Biolabs) to remove any trace genomic DNA. For semiquantitative RT-PCR and qRT-PCR templates, first-strand complementary DNA (cDNA) were synthesized in a 20-μL reaction solution containing approximately 0.5 and 3 μg total RNA, respectively, using PrimerScript RT (Takara) and oligo (dT) 12-18 as a primer. EF1aA4 and UBIQUITIN4 (AT5G44340) were used as internal controls in the RT-PCR and qRT-PCR. All primer pairs used in the RT-PCR and qRT-PCR analyses are listed in Supplemental Table S1. The real-time amplification reactions were performed using the SYBR Green kit (Takara) and iCycler iQ thermocycler (Bio-Rad) according to the manuals from the manufacturers.
Analysis of the Seed Germination Rate and Seedling Establishment of tt2-5 Mutant Lines under Stressed Conditions
The seeds used for the germination assays were harvested at nearly the same time from plants grown under the same growth conditions as previously described, allowed to mature for 5 weeks at room temperature, and stored in a dry cabinet (approximately 15% moisture level). The Arabidopsis seeds were surface sterilized and then placed on MS medium containing 3% (w/v) Glc, 100 mm NaCl, or 1 μm ABA as previously described. The seed germination frequency (defined as radicle emergence) was scored daily (Cernac et al., 2006). Arabidopsis plants at 10 DAS were photographed and harvested for RNA analysis.
Statistical Analysis
A completely randomized block design with at least three biological replicates was conducted for each experiment in this study. Data were classified with Win-Excel and analyzed via an ANOVA using the SPSS statistical package (version 8.0). Comparisons between the treatment means were made via a Tukey’s test at a probability level of P ≤ 0.05.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Primers used for RT-PCR and qRT-PCR analyses.
Supplemental Table S2. Primers used for the genotyping of the tt2 mutants.
Supplemental Table S3. A list of up-regulated genes in the developing seeds of SALK_005260 plants at 6 DAP.
Supplemental Table S4. A list of down-regulated genes in the developing seeds of SALK_005260 plants at 6 DAP.
Supplemental Table S5. A list of up-regulated genes in the developing seeds of SALK_005260 plants at 16 DAP.
Supplemental Table S6. A list of down-regulated genes in the developing seeds of SALK_005260 plants at 16 DAP.
Supplemental Table S7. A list of TFs and genes encoding key enzymes for FA biosynthesis at 6 DAP.
Supplemental Table S8. A list of TFs and genes encoding key enzymes for FA biosynthesis at 16 DAP.
Glossary
- FAs
fatty acids
- EM
embryonic morphogenesis
- DAP
d after pollination
- RT
reverse transcriptase
- DW
dry weight
- VLCFAs
very-long-chain fatty acids
- Col-0
Columbia-0
- DEGs
differentially expressed genes
- TFs
transcription factors
- q
quantitative
- qRT
quantitative reverse transcriptase
- ABA
abscisic acid
- DAS
d after sowing
- FAS
fatty acid synthesis
- Ler
Landsberg erecta
- Ws-2
Wassilewskija-2
- TEM
transmission electron microscopy
- MS
Murashige and Skoog
- cDNA
complementary DNA
References
- Baud S, Boutin JP, Miquel M, Lepiniec L, Rochat C. (2002) An integrated overview of seed development in Arabidopsis thaliana ecotype WS. Plant Physiol Biochem 40: 151–160 [Google Scholar]
- Baud S, Guyon V, Kronenberger J, Wuillème S, Miquel M, Caboche M, Lepiniec L, Rochat C. (2003) Multifunctional acetyl-CoA carboxylase 1 is essential for very long chain fatty acid elongation and embryo development in Arabidopsis. Plant J 33: 75–86 [DOI] [PubMed] [Google Scholar]
- Baud S, Lepiniec L. (2009) Regulation of de novo fatty acid synthesis in maturing oilseeds of Arabidopsis. Plant Physiol Biochem 47: 448–455 [DOI] [PubMed] [Google Scholar]
- Beisson F, Koo AJK, Ruuska S, Schwender J, Pollard M, Thelen JJ, Paddock T, Salas JJ, Savage L, Milcamps A, et al. (2003) Arabidopsis genes involved in acyl lipid metabolism: a 2003 census of the candidates, a study of the distribution of expressed sequence tags in organs, and a web-based database. Plant Physiol 132: 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Cernac A, Andre C, Hoffmann-Benning S, Benning C. (2006) WRI1 is required for seed germination and seedling establishment. Plant Physiol 141: 745–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernac A, Benning C. (2004) WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J 40: 575–585 [DOI] [PubMed] [Google Scholar]
- Debeaujon I, Léon-Kloosterziel KM, Koornneef M. (2000) Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol 122: 403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Nesi N, Perez P, Devic M, Grandjean O, Caboche M, Lepiniec L. (2003) Proanthocyanidin-accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development. Plant Cell 15: 2514–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devic M, Guilleminot J, Debeaujon I, Bechtold N, Bensaude E, Koornneef M, Pelletier G, Delseny M. (1999) The BANYULS gene encodes a DFR-like protein and is a marker of early seed coat development. Plant J 19: 387–398 [DOI] [PubMed] [Google Scholar]
- Eastmond PJ, Rawsthorne S. (2000) Coordinate changes in carbon partitioning and plastidial metabolism during the development of oilseed rape embryos. Plant Physiol 122: 767–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fait A, Angelovici R, Less H, Ohad I, Urbanczyk-Wochniak E, Fernie AR, Galili G. (2006) Arabidopsis seed development and germination is associated with temporally distinct metabolic switches. Plant Physiol 142: 839–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focks N, Benning C. (1998) wrinkled1: a novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol 118: 91–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg RB, de Paiva G, Yadegari R. (1994) Plant embryogenesis: zygote to seed. Science 266: 605–614 [DOI] [PubMed] [Google Scholar]
- Hong JK, Choi HW, Hwang IS, Kim DS, Kim NH, Choi S, Kim YJ, Hwang BK. (2008) Function of a novel GDSL-type pepper lipase gene, CaGLIP1, in disease susceptibility and abiotic stress tolerance. Planta 227: 539–558 [DOI] [PubMed] [Google Scholar]
- James DW, Jr, Lim E, Keller J, Plooy I, Ralston E, Dooner HK. (1995) Directed tagging of the Arabidopsis FATTY ACID ELONGATION1 (FAE1) gene with the maize transposon activator. Plant Cell 7: 309–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenik PD, Gillmor CS, Lukowitz W. (2007) Embryonic patterning in Arabidopsis thaliana. Annu Rev Cell Dev Biol 23: 207–236 [DOI] [PubMed] [Google Scholar]
- Koes RE, Quattrocchio F, Mol JNM. (1994) The flavonoid biosynthetic-pathway in plants—function and evolution. Bioessays 16: 123–132 [Google Scholar]
- Lee HS, Fischer RL, Goldberg RB, Harada JJ. (2003) Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proc Natl Acad Sci USA 100: 2152–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepiniec L, Debeaujon I, Routaboul JM, Baudry A, Pourcel L, Nesi N, Caboche M. (2006) Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol 57: 405–430 [DOI] [PubMed] [Google Scholar]
- Li YH, Beisson F, Pollard M, Ohlrogge J. (2006) Oil content of Arabidopsis seeds: the influence of seed anatomy, light and plant-to-plant variation. Phytochemistry 67: 904–915 [DOI] [PubMed] [Google Scholar]
- Luerssen H, Kirik V, Herrmann P, Miséra S. (1998) FUSCA3 encodes a protein with a conserved VP1/AB13-like B3 domain which is of functional importance for the regulation of seed maturation in Arabidopsis thaliana. Plant J 15: 755–764 [DOI] [PubMed] [Google Scholar]
- Mayer U, Jürgens G. (1998) Pattern formation in plant embryogenesis: a reassessment. Semin Cell Dev Biol 9: 187–193 [DOI] [PubMed] [Google Scholar]
- Mccleary BV, Solah V, Gibson TS. (1994) Quantitative measurement of total starch in cereal flours and products. J Cereal Sci 20: 51–58 [Google Scholar]
- Millar AA, Kunst L. (1997) Very-long-chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. Plant J 12: 121–131 [DOI] [PubMed] [Google Scholar]
- Millar AA, Smith MA, Kunst L. (2000) All fatty acids are not equal: discrimination in plant membrane lipids. Trends Plant Sci 5: 95–101 [DOI] [PubMed] [Google Scholar]
- Mohamedyasseen Y, Barringer SA, Splittstoesser WE, Costanza S. (1994) The role of seed coats in seed viability. Bot Rev 60: 426–439 [Google Scholar]
- Mol J, Grotewold E, Koes R. (1998) How genes paint flowers and seeds. Trends Plant Sci 3: 212–217 [Google Scholar]
- Mu J, Tan H, Zheng Q, Fu F, Liang Y, Zhang J, Yang X, Wang T, Chong K, Wang XJ, et al. (2008) LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in Arabidopsis. Plant Physiol 148: 1042–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L. (2000) The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12: 1863–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L. (2001) The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13: 2099–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge JB, Jaworski JG. (1997) Regulation of fatty acid synthesis. Annu Rev Plant Physiol Plant Mol Biol 48: 109–136 [DOI] [PubMed] [Google Scholar]
- Okuley J, Lightner J, Feldmann K, Yadav N, Lark E, Browse J. (1994) Arabidopsis FAD2 gene encodes the enzyme that is essential for polyunsaturated lipid synthesis. Plant Cell 6: 147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier MK, Murrell JR, Shirley BW. (1997) Characterization of flavonol synthase and leucoanthocyanidin dioxygenase genes in Arabidopsis: further evidence for differential regulation of “early” and “late” genes. Plant Physiol 113: 1437–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Rylott EL, Gilday AD, Graham S, Larson TR, Graham IA. (2004) Reserve mobilization in the Arabidopsis endosperm fuels hypocotyl elongation in the dark, is independent of abscisic acid, and requires PHOSPHOENOLPYRUVATE CARBOXYKINASE1. Plant Cell 16: 2705–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier Y, Ventre G, Caldelari D. (1999) Increased flow of fatty acids toward β-oxidation in developing seeds of Arabidopsis deficient in diacylglycerol acyltransferase activity or synthesizing medium-chain-length fatty acids. Plant Physiol 121: 1359–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rerie WG, Feldmann KA, Marks MD. (1994) The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev 8: 1388–1399 [DOI] [PubMed] [Google Scholar]
- Ruuska SA, Girke T, Benning C, Ohlrogge JB. (2002) Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14: 1191–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Mendoza M, Dubreucq B, Baud S, Parcy F, Caboche M, Lepiniec L. (2008) Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J 54: 608–620 [DOI] [PubMed] [Google Scholar]
- Shah S, Xin ZG, Browse J. (1997) Overexpression of the FAD3 desaturase gene in a mutant of Arabidopsis. Plant Physiol 114: 1533–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley BW. (1996) Flavonoid biosynthesis: ‘new’ functions for an ‘old’ pathway. Trends Plant Sci 1: 377–382 [Google Scholar]
- Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM. (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J 8: 659–671 [DOI] [PubMed] [Google Scholar]
- Smith S, Witkowski A, Joshi AK. (2003) Structural and functional organization of the animal fatty acid synthase. Prog Lipid Res 42: 289–317 [DOI] [PubMed] [Google Scholar]
- Stone SL, Braybrook SA, Paula SL, Kwong LW, Meuser J, Pelletier J, Hsieh TF, Fischer RL, Goldberg RB, Harada JJ. (2008) Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: implications for somatic embryogenesis. Proc Natl Acad Sci USA 105: 3151–3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ. (2001) LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA 98: 11806–11811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To A, Valon C, Savino G, Guilleminot J, Devic M, Giraudat J, Parcy F. (2006) A network of local and redundant gene regulation governs Arabidopsis seed maturation. Plant Cell 18: 1642–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnham E, Northcote DH. (1983) Changes in the activity of acetyl-CoA carboxylase during rape-seed formation. Biochem J 212: 223–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HY, Guo JH, Lambert KN, Lin Y. (2007) Developmental control of Arabidopsis seed oil biosynthesis. Planta 226: 773–783 [DOI] [PubMed] [Google Scholar]
- Weber H, Borisjuk L, Wobus U. (1996) Controlling seed development and seed size in Vicia faba: a role for seed coat-associated invertases and carbohydrate state. Plant J 10: 823–834 [Google Scholar]
- White SW, Zheng J, Zhang YM, Rock (2005) The structural biology of type II fatty acid biosynthesis. Annu Rev Biochem 74: 791–831 [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Kagaya Y, Usui H, Hobo T, Takeda S, Hattori T. (2010) Diverse roles and mechanisms of gene regulation by the Arabidopsis seed maturation master regulator FUS3 revealed by microarray analysis. Plant Cell Physiol 51: 2031–2046 [DOI] [PubMed] [Google Scholar]
- Zhang YM, Rock CO. (2004) Evaluation of epigallocatechin gallate and related plant polyphenols as inhibitors of the FabG and FabI reductases of bacterial type II fatty-acid synthase. J Biol Chem 279: 30994–31001 [DOI] [PubMed] [Google Scholar]