The insertion of transgenes into the plastid genome (plastome) has proved to be an effective alternative to nuclear transformation for producing stable transformants expressing heterologous proteins. Recently, methodology, characteristics, and possible fields of application of plastome transformation have been extensively reviewed (Lössl and Waheed, 2011; Scotti et al., 2012). From these reports, plastid transformation turns out to be a very promising tool for the production of recombinant proteins in plants, yet some limitations must still be overcome. Although the plastome of many species has been successfully transformed, an efficient and reproducible transformation procedure easily adoptable by any plant laboratory has been established only in tobacco (Nicotiana tabacum) and to a minor extent in two other solanaceous species (potato [Solanum tuberosum] and tomato [Solanum lycopersicum]) as well as in a few other species (Maliga and Bock, 2011). Moreover, up to now, the steady-state level of a recombinant protein coded by a gene inserted into the plastome has failed to be predictable. In spite of a huge number of reports on successful foreign protein production in plastids, above all proteins with a pharmaceutical interest, in many cases the expression level of other recombinant proteins in the transplastomic plants appears to be very low or even undetectable (Birch-Machin et al., 2004; Bellucci et al., 2005; Wirth et al., 2006). We believe that this is mainly due to our limited knowledge of the mechanisms in plastids influencing the maintenance of protein homeostasis, or proteostasis. This term refers to a complex network of biological pathways capable of maintaining the dynamic equilibrium of the protein pool, thus allowing the cell to get used to environmental changes (Balch et al., 2008). This concept can also be extended to the subcellular level, especially for those organelles with an endosymbiotic origin like the plastids and mitochondria (Waller, 2012). These organelles retain a whole series of mechanisms for the preservation of their protein balance, including specific proteases, transcriptional and translational control, as well as molecular chaperones and enzymes useful in protein folding. Therefore, it is important to develop basic studies in factors regulating protein synthesis, stability, folding, targeting, and accumulation in plastids, including the protein quality control that contributes to the functional integrity of proteins. Our intention here is not to provide a comprehensive review of the mechanisms that regulate plastid proteostasis but to discuss some recent insights into this field that might bring beneficial applications in plastid biotechnology for transgene expression and foreign protein accumulation.
TRANSCRIPTIONAL CONTROL IS AN IMPORTANT STEP BUT NOT THE MAIN REGULATOR OF PLASTID GENE EXPRESSION
In spite of the prokaryotic origin and the similarity to its ancestors of the gene expression machinery, the presence of introns, RNA editing, and processing of mRNA polycistronic precursors make the chloroplast a unique organelle that combines eukaryotic and prokaryotic features (Stern et al., 2010). This uniqueness can be noticed in at least two cases: (1) the dual genetic origin of the plastid proteome, made up of proteins encoded either by the plastome or the nucleus; (2) the presence of unique ribosomal proteins, named plastid-specific ribosomal proteins, together with ribosomal proteins of prokaryotic origin (Tiller et al., 2012). As an endosymbiont, chloroplast has almost totally replaced the control of gene expression at the transcriptional level, switching to a predominantly posttranscriptional control (Marìn-Navarro et al., 2007). However, some transcriptional regulation may also take place, because plastid transcription is performed by three RNA polymerases, two monomeric, nucleus-encoded, phage-type enzymes (dubbed RPOTp and RPOTmp) and one multimeric, plastid-encoded, eubacteria-type enzyme (dubbed PEP), which need to interact with up to six nucleus-encoded transcription initiation factors in order to perform their activity (Liere and Börner, 2007; Lerbs-Mache, 2011). Hence, transcription is considered an important step for transgene expression, and it can be controlled by using appropriate promoters in the plastid transformation vectors. Most of the chloroplast transformation vectors utilize the strong promoter of the plastid ribosomal RNA operon (Prrn), but also other promoters like the plastidial psbA and clpP or the bacterial trc are sometimes used. The technology of plastid transformation, from the engineering of plastid transgenes in Escherichia coli cloning vectors to the obtainment of stable transplastomic plants, is clearly described in a recent review (Maliga and Bock, 2011), but here we intend to underline the importance of some specific motifs present in the chloroplast transformation vectors for transgene expression.
In addition to the promoters, the transgene expression cassettes commonly employed for plastid vector construction harbor other regulatory elements like 5′-untranslated regions (UTRs) and 3′-UTRs. These regions are both involved in the regulation of RNA stability and the translation efficiency of plastid transcripts (Zou et al., 2003; Pfalz et al., 2009). The exact modality for the correct positioning of the chloroplast ribosome on the translation initiation codon of mRNAs is still beyond our understanding, but most of the plastid mRNAs possess a Shine-Dalgarno (SD) sequence in the 5′-UTR. In prokaryotes, the SD sequence (GGAGG) is located between four and 12 nucleotides upstream from the start codon and, being complementary to the 3′ terminus of the 16S ribosomal RNA, mediates the binding of the ribosome to an mRNA. In order to find out the mechanism of SD sequence recognition, Drechsel and Bock (2011) analyzed translation initiation from mRNAs containing multiple SD sequences, demonstrating that the plastid ribosomes displayed a bias toward preferentially utilizing the 5′-most SD sequence. The facts that the location of plastidial SD elements is much more variable than in E. coli and that for some plastid genes SD-like sequences have turned out to be less important for translation initiation lead to the possibility that other mechanisms for translation initiation may exist (Marìn-Navarro et al., 2007; Scharff et al., 2011). Indeed, detailed analyses of the plastid 5′-UTRs have detected the presence of cis-elements that facilitate the interaction with nucleus-encoded RNA-binding proteins (Merhige et al., 2005). These associations between protein factors and the respective RNA sequences influence the translation initiation process. There is a little concern about the use of homologous regulatory elements in chloroplast transformation vectors due to the risk of potential recombination with the endogenous ones (Rogalski et al., 2006). Therefore, for example, to transform the tobacco plastome, Elghabi et al. (2011) utilized promoter and 3′-UTR sequences from Chlamydomonas reinhardtii in order to avoid unwanted recombination with the resident copies in the plastome. Conversely, in other laboratories, the use of homologous regulatory sequences is preferred, as shown in the case of the psbA 5′-UTR. It has been demonstrated that, despite the possible accumulation of foreign proteins using a heterologous psbA 5′-UTR, it occurs with a reduced efficiency in comparison with the results achieved when the homologous regulatory elements are used (Ruhlman et al., 2010).
The reason for these results is probably the preferential association to stromal RNA-binding proteins with the endogenous psbA 5′-UTR, whereas the heterologous one is an ineffective competitor. On the basis of the results obtained and after a taxonomic analysis of the region 200 bases upstream of the translation start codon, the authors suggested the use of species-specific regulatory elements for significant accumulation of foreign protein in transplastomic plants. The 5′-UTR of gene 10 of bacteriophage T7 (T7g10) has been successfully used in plastids achieving high protein expression (Kuroda and Maliga 2001) as well as intact or truncated 5′-UTR from highly expressed genes like rbcL, psbA, and atbP (Herz et al., 2005; Valkov et al., 2011). The inverted repeat sequence, typically present in the 3′-UTR of many plastid genes, can potentially form a stem-loop structure, thus becoming important in the stabilization and the accumulation of the transcript coded by the upstream sequence. This concept has been recently highlighted in transplastomic tobacco plants expressing several gfp gene constructs with the same 5′ regulatory sequence but different 3′-UTRs (Tangphatsornruang et al., 2011). Generally, 3′ regulatory elements derived from plastid or bacterial genes are used in plastid transformation vectors. The E. coli rrnB 3′ region seems to be the most stabilizing element both in tobacco and potato transplastomic plants, although a great mRNA stability and abundance does not always correspond to high heterologous protein expression (Tangphatsornruang et al., 2011; Valkov et al., 2011). Therefore, a list of the best promoters, 5′-UTRs, and 3′-UTR combinations utilized in different experiments to achieve the highest transcript accumulation and stability can surely be made (Maliga, 2002; Herz et al., 2005; Maliga and Bock, 2011; Tangphatsornruang et al., 2011; Valkov et al., 2011), but it is quite clear that the amount of transgene mRNA, even with good stability, does not always correspond to an elevated foreign protein synthesis and accumulation inside plastids (Eberhard et al., 2002). This is because the translational control is much more prevalent than transcriptional regulation for transgene expression (see below), and this in many cases smoothes the differences eventually present in mRNA abundance. Do the sequences downstream of the translation initiation codon affect transcript stability or translation? The answer in both cases is affirmative. Experimental results obtained by fusing N-terminal segments of highly expressed proteins in plastids to the transgene coding region indicate that it is possible to stabilize the cyanovirin-N mRNA by sequence insertions of 60 nucleotides or greater between the T7g10 5′-UTR and the cyanovirin-N coding region (Elghabi et al., 2011). With a similar experimental approach, it is also possible to demonstrate that 14 N-terminal amino acids encoded in the rbcL plastid gene are important determinants of translation efficiency (Kuroda and Maliga, 2001). These results encourage many scientists to empirically optimize the sequence of the region located immediately downstream of the start codon (Gray et al., 2011). If processing of polycistronic transgenic transcripts into monocistronic mRNAs is needed to increase the efficiency of translation, a small intercistronic expression element can be used to separate foreign genes of the same operon (Zhou et al., 2007).
Another aspect concerning heterologous protein expression inside plastids is that, so far, almost all plastid transformation vectors have been designed to obtain constitutive expression of the transgene. However, expression of a transgene product may result in a mutant phenotype or growth retardation due to the toxicity of the metabolite expressed or due to its interaction with photosynthesis or the plastid endomembrane system (Lössl et al., 2003; Hennig et al., 2007). A massive expression of a highly stable protein can also lead to the exhaustion of chloroplast protein synthesis capacity, resulting in delayed growth (Oey et al., 2009). To avoid deleterious effects caused by constitutive transgene expression, different strategies have been adopted, such as the one based on a nucleus-encoded, chloroplast-targeted, ethanol-inducible T7 RNA polymerase that controls plastid transgene transcription from a T7 promoter system (Lössl et al., 2005) or that depends on the control of transgene expression using the bacterial lac repressor (Mühlbauer and Koop, 2005). However, the use of these systems is strongly limited by the requested transformation of two separate cell compartments (nucleus and plastid) or by the missing of a tight regulation of gene expression in the uninduced status. Recently, the use of engineering riboswitches (Verhounig et al., 2010) has been proposed as a tool to control chloroplast transgene expression without the use of additional nuclear or plastid transgenes. The use of a synthetic theophylline riboswitch displays great regulatory properties, because the undetectable basal GFP expression in the transplastomic tobacco plants becomes rapidly detectable after application of the theophylline metabolite.
TRANSLATIONAL CONTROL AND PROTEIN CONFORMATION
Proteins coded by genes located in the plastome are synthesized on plastid 70S ribosomes, and the expression of these genes is mainly regulated during translation of the corresponding mRNAs (Zerges, 2004) and at the posttranslational level as well. Indeed, after synthesis, these proteins must rapidly fold into stable three-dimensional structures that often contemplate posttranslational modifications to perform their biological mission, avoiding aberrant folding and aggregation with the help of molecular chaperones (Hartl et al., 2011). The combined action of molecular chaperones and proteases maintains the quality control over protein structure and function (Wickner et al., 1999). Protein quality control components of plastids are very similar to bacteria. Chloroplasts contain members of five major chaperone families (Hsp100, Hsp90, Hsp70, Hsp60 or type 1 chaperonins, and small heat shock proteins) and several cochaperones, for example, J-domain proteins, GrpE, and Cpn10/20 (Schroda, 2004; Willmund et al., 2008). Chloroplasts also contain various proteases of bacterial ancestry, which are involved in numerous aspects of the biogenesis and the maintenance of these organelles. These proteases include the multigene families of stromal Ser Clp proteases and their AAA+ chaperones, thylakoid-bound FtsH metalloproteases, Lon, Deg, and several other proteases characterized in recent years (Adam et al., 2006).
Expression of the psbA gene, coding for the D1 protein, one of the core proteins of PSII, is an example of the complex regulatory strategies utilized in the chloroplasts of higher plants. D1 is damaged by light-induced oxidation and is degraded and resynthesized during the PSII repair cycle. In plant cells, a constant pool of psbA mRNA associated with the polysomes does exist, but the translation of D1 can be completed only when a PSII complex without D1 is available to accept the integration of the new D1 protein (Mulo et al., 2012). This observation leads to the conclusion that translation elongation is the prevalent mechanism for the regulation of psbA gene expression. The existence of a quality control on D1 is evident during the PSII repair cycle, which requires posttranslational phosphorylation of the D1 protein to regulate its degradation. Light induces phosphorylation and damage of the D1 protein, and the damaged PSII complexes migrate from the grana stacks to the stroma lamellae, where the D1 protein is dephosphorylated and degraded by the FtsH and DegP proteases (Tikkanen et al., 2011). Replacement of damaged D1 protein also requires the assistance of molecular chaperones that are integral thylakoid membrane proteins (LPA1 and PAM68) or a lumenal thylakoid protein (CYP38; Mulo et al., 2012).
Protein Sequences Impacting Protein Stability
Studies on transplastomic plants have confirmed that the expression of genes integrated into the plastome is mainly regulated after the translation of the mRNAs into the corresponding protein, whose stability is the key aspect that determines foreign protein accumulation (Bellucci et al., 2005; Zhou et al., 2008; Oey et al., 2009). Small proteins are empirically known to be more unstable (Ortigosa et al., 2010), but people working on plastid biotechnology are very interested in finding general determinants of protein stability in the plastids. For example, which are the signals on the protein structure that regulate the turnover of the photodamaged PSII proteins or the proteins of the oxygen-evolving complexes? Two recent reports suggest that in plastids the major stability determinants reside in the N terminus of the protein (Apel et al., 2010; Adam et al., 2011). This may help to explain why unstable foreign proteins are stabilized in plastids after the addition of N-terminal peptide fusions (Ye et al., 2001; Lenzi et al., 2008).
It is generally accepted that the identity of the N-terminal amino acid residue of a protein is related to its in vivo half-life, a principle known as the N-end rule (Varshavsky, 1996). N-terminal amino acids are classified as stabilizing or destabilizing residues, and in the eukaryotic cytosol, proteins carrying a destabilizing N-terminal residue are degraded by the ubiquitin-proteasome system. Primary destabilizing residues are recognized directly by the E3 ubiquitin ligases; conversely, secondary or tertiary destabilizing residues must be enzymatically modified (Cys also requires a previous nonenzymatic oxidation) before the corresponding protein can be targeted by E3 ubiquitin ligases to the proteasome. The N-end rule pathway is evolutionarily conserved, being present in all organisms examined (Graciet et al., 2010), including bacteria, even if the prokaryotic version of the rule differs in the hierarchical order of the destabilizing amino acid residues (Mogk et al., 2007) and in the associated proteolytic machinery represented by the Clp protease (Román-Hernández et al., 2009). Apel and coworkers (2010), by fusing N- and C-terminal sequences from five plastid proteins to GFP, showed that the N terminus harbors important information affecting the half-life of the protein, whereas the C terminus seems to have scarce influence on protein half-life. They also tested in one of the N-terminal-fused GFP constructs all 20 amino acids in the position after the initiator Met, suggesting for plastids the existence of an N-end rule-like pathway that is not fully homologous to the bacterial one. However, the observed differences in protein accumulation conferred by the five different N-terminal sequences cannot be explained only by the identity of the penultimate amino acid, so additional major determinants of plastid protein stability must reside in the N-terminal region of the protein. In an attempt to increase, in transplastomic tobacco plants, the accumulation and stability of the rotavirus VP6 protein, even Inka Borchers et al. (2012) indicated the vital importance of the N-terminal part for protein stability. The additional amino acids (up to five) at the 5′ end of the VP6 coding region increased the expression of the foreign gene in the transplastomic plants and stabilized the protein in aging leaves. However, the N and C termini are not the only ones to influence protein stability, because in other systems, for example in E. coli, the three-dimensional structure of internal protein domains turns out to be a key determinant of protein stability (Zoldák et al., 2009).
Protein Modifications Influencing Protein Stability
The first cotranslational modification applied to almost all plastid-encoded proteins is (as in bacteria and mitochondria) the N-terminal deformylation by the enzyme peptide deformylase. After the removal of the N-formyl group, if the second residue is small and uncharged (Cys or Ala, for example), the protein can then undergo N-terminal methionine excision (NME), catalyzed by Met aminopeptidase, which involves roughly 60% to 70% of the proteins in a given proteome (Meinnel et al., 2006, and refs. therein). Inhibition of NME induced by peptide deformylase inactivation indicates that NME is involved in the regulation of protein turnover in the chloroplast (Giglione et al., 2003). In particular, retention of the N-Met in D1 and D2 proteins, which usually undergo NME, can result in acceleration of their degradation (Giglione et al., 2003). D1 and D2 proteins are primary targets of photodamage, so a lot of work has been done on the proteases involved in their degradation, especially for D1 (Chi et al., 2012). In Arabidopsis (Arabidopsis thaliana), NME seems to regulate the accumulation of D1 and D2 proteins, in concerted action with chloroplast proteases (Adam et al., 2011). The authors showed that correctly NME-processed N termini of D1 and D2 are indispensable for both FtsH-mediated quality control of the proteins and repair-related degradation of D1 and D2. Moreover, inhibition of NME induces the degradation of the D1 and D2 unprocessed forms by proteases other than FtsH. Therefore, FtsH proteases should recognize the N terminus of both neosynthesized or damaged D1 and D2 proteins, and the open question is how FtsH complexes can recognize the N-terminal tail of these proteins (Komenda et al., 2007).
According to the N-end rule in bacteria and chloroplasts, proteins with an N-terminal Met are very stable. On the contrary, the retained N-terminal Met in D1 and D2 proteins, due to inhibition of NME, may act as a signal for degradation. Therefore, considering that the stability of other chloroplast proteins remains unaffected by the retention of their N-Met, N-Met might be necessary but not sufficient to reduce the half-life of a protein; thus, other protein modifications on or close to the N-Met are required to accelerate the protein degradation process (Meinnel et al., 2006). Indeed, other polypeptide modifications likely participate in regulating the stability of a protein. More than 300 different types of protein cotranslational or posttranslational modifications (PTMs) have been described (Zhao and Jensen, 2009), among which the N-terminal modifications can be grouped as follows: (1) removal of one or more residues such as NME or propeptide cleavage; (2) N-blocking of the intact or modified proteins such as N-α-terminal acetylation (NTA) or N-myristoylation; or (3) ligation events extending the length of the polypeptide (Bienvenut et al., 2011). These modifications can influence protein localization, activity, or stability. With regard to stability, some PTMs can produce destabilizing N-terminal residues in proteins through, for example, cleavage by endopeptidases in the cytosol, or removal of transit peptides in the stroma, or NME. Indeed, removal of the N-terminal Met in the cytosol from the initiating motif MCGGAII and the consequent exposition of the penultimate Cys seem to target the transcription factor RAP2.12 for degradation by the N-end rule pathway (Sasidharan and Mustroph, 2011). Moreover, in yeast, NTA and NME are part of the N-end rule pathway, and NTA targets proteins for degradation (Hwang et al., 2010).
Proteomics can be a very useful tool to achieve a comprehensive understanding of plastid protein stability. A recent review has provided un update of the current status of plastid proteome research, highlighting the importance of creating a high-quality plastid proteome atlas that should include, thanks to the improvements in mass spectrometry instrumentation, information on protein abundance, protein-protein interactions, and PTMs (van Wijk and Baginsky, 2011). Such an atlas can provide useful information for functional studies and systems biology, and if enriched with data on the protein half-life, it can also help to clarify the relationship between protein stability and PTMs in plastids. For instance, combining two-dimensional gel electrophoresis profiling with mass spectrometry and pulse-chase analyses, it has been shown in C. reinhardtii that the most unstable stromal proteins (half-life of less than 1 h) rarely possess an α-acetylated N terminus, in contrast to the most abundant proteins (Bienvenut et al., 2011). This suggests that, in contrast to what happens in the yeast cytosol, NTA could contribute to protein stabilization in the stromal proteome. In Arabidopsis, large-scale analysis of PTMs has recently been carried out using chloroplast preparations (Zybailov et al., 2008) and suspension cells as well as whole seedling lysate (Bienvenut et al., 2012). These studies have identified N-terminal modifications of a large sets of nucleus- and plastome-encoded plastid proteins, obtaining information on the protein N termini, the processing site for transit peptides of imported plastid proteins, and the frequencies of many PTMs. NTA, in particular, which is considered an essential cotranslational modification occurring mainly in the cytosol of eukaryotes, appears to be a widespread protein PTM also in the chloroplast, because a high proportion of nucleus-encoded chloroplast proteins undergo NTA of the mature protein after removal of the transit peptides (Bienvenut et al., 2012). The fact that for nucleus-encoded proteins the acetylated residues have a prevalence of Ala, Val, Ser, and Thr, whereas for chloroplast-encoded proteins the cotranslational acetylated N termini are Thr, Arg, or Pro, likely suggests the involvement of two different N-α-acetyl transferases (Zybailov et al., 2008).
TARGETING
To ensure proper folding and accumulation of a recombinant protein in transformant cells, it is necessary to target this protein to an appropriate cell compartment (Fig. 1). The plant secretory pathway and the accumulation of foreign protein in the endoplasmic reticulum (ER) is often the chosen localization. This is because a lot of information on protein quality control is available on the ER (Vitale and Ceriotti, 2004), and this knowledge greatly helps to delineate strategies to retain and accumulate heterologous proteins of interest in this subcellular compartment, for example by means of retention signals such as the KDEL/HDEL motif (Ma et al., 1995) or fusing the protein of interest with a protein domain capable of inducing ER-protein body formation (de Virgilio et al., 2008). Before the end of the 20th century, plastids were rarely the final compartment of localization of foreign proteins. Transgenes, inserted into the nucleus, contain at the 5′ end a sequence coding for a transit peptide that permits the transport of the polypeptide from the cytosol, where the transgene mRNA is translated, to the stroma through the main pathway to plastids, the TIC/TOC system present in the inner and outer envelopes (Jarvis, 2008). Recently, the advantages offered by chloroplast-based expression of heterologous genes, above all those related to the achievable high level of protein expression and transgene confinement, have convinced many researchers to use the plastome transformation technology for the accumulation of biomolecules in the chloroplasts of C. reinhardtii and plants (Lössl and Waheed, 2011; Michelet et al., 2011). Foreign proteins expressed in the chloroplast can be accumulated in the soluble fraction, composed of stroma and thylakoid lumen, or in the membranous fraction, formed by envelope and thylakoid membranes. The localization of these proteins within the chloroplast allows insertion in different environments with a more or less intense proteolytic activity. Moreover, each subcompartment owns its proteome, which contains a specific box of enzymes and other molecules involved in protein folding (Ferro et al., 2010; Agrawal et al., 2011). Targeting to the outer envelope membrane seems to be possible only for nucleus-encoded genes utilizing the TOC system or a “spontaneous” insertion mechanism.
Figure 1.
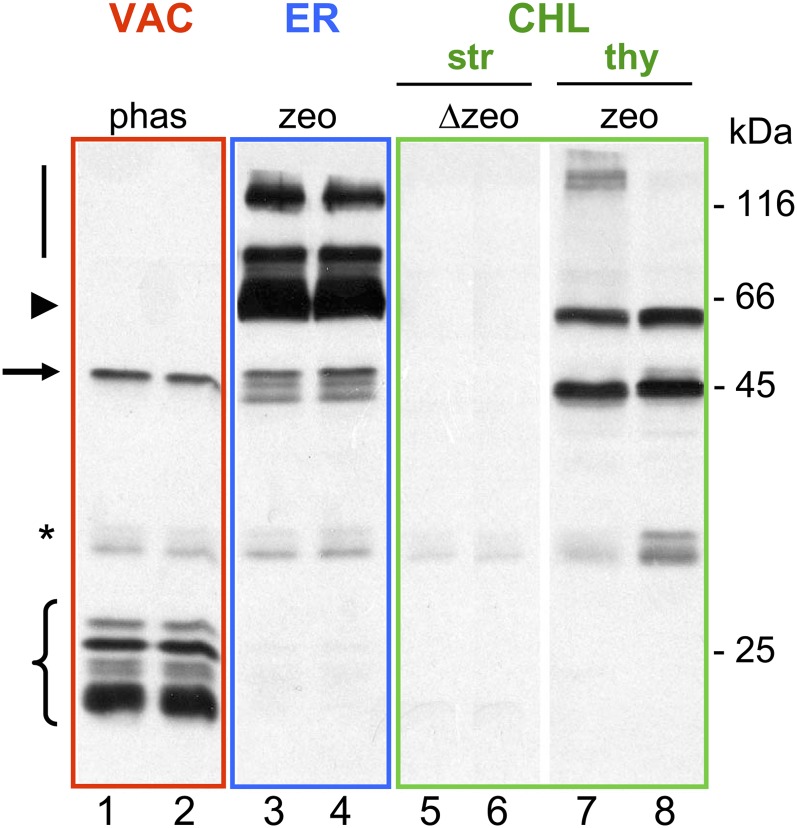
Expression of the bean (Phaseolus vulgaris) storage protein phaseolin and its mutated forms is very different according to the compartment of localization inside transgenic tobacco leaf cells. Proteins extracted from transformed tobacco plants were analyzed by western-blot analysis using anti-phaseolin antiserum. Lanes 1 and 2 show phaseolin (arrow) expressed from nucleus-transformed plants secreted to the vacuole (VAC), where it is degraded to 20- to 25-kD proteolytic fragments (brace). Phaseolin can be accumulated in the ER of transgenic nuclear plants, either adding to the polypeptide a KDEL retention signal (data not shown) or expressing phaseolin fused to a γ-zein N-terminal domain (the name of the fusion protein is zeolin; lanes 3 and 4). The accumulation of phaseolin in the ER was much higher than that in the vacuole, both as phaseolin-KDEL (data not shown) and as zeolin (arrowhead), which forms large aggregates (vertical bar) due to the formation of interchain disulfide bonds (Mainieri et al., 2004). In the chloroplast (CHL) of transplastomic plants expressing the phaseolin fusion protein zeolin, the recombinant protein devoid of its signal peptide (Δzeo) was barely detectable in the stroma (str; lanes 5 and 6); conversely, when targeted to the thylakoid membrane (thy; lanes 7 and 8), it is clearly detectable (De Marchis et al., 2011). Molecular mass in kD (numbers at right) is indicated. The asterisk indicates contaminant peptides.
Which are the protein-targeting pathways within chloroplasts? Endogenous chloroplast proteins, either nucleus encoded or plastome encoded, share the same internal routes. Once in the stroma, a protein can fold and perform its biological role or can be delivered to other compartments if it harbors specific sorting signals. The most studied sorting mechanisms inside the chloroplast concern the targeting of proteins to the thylakoids, even if other pathways have been identified to target proteins from the stroma to the inner envelope membrane and the space between the two envelope membranes (Jarvis, 2008; Skalitzky et al., 2011). At least four different targeting mechanisms, homologous to bacterial transport systems, have been discovered for the transport of proteins into and across the thylakoid membrane. Two of these insert membrane proteins: one employs a signal recognition particle (SRP)-dependent pathway, and the other one (called spontaneous) appears to require none of the known targeting factors (Ouyang et al., 2011). The other two pathways translocate lumenal proteins across the thylakoid membrane. One needs ATP hydrolysis and transports proteins in an unfolded conformation by a Sec-dependent machinery. The other one is the Tat pathway that accompanies folded proteins into the thylakoid lumen using a proton gradient force (Albiniak et al., 2012). The transgenic chloroplast system has been largely used to express recombinant proteins in the stroma, but few reports have analyzed other alternative intraplastidial localizations like the inner envelope membrane (Singh et al., 2008) or the thylakoid lumen (Tissot et al., 2008). The correct folding of a protein to reach its tertiary or quaternary structure often needs PTMs, and one of the positive characteristics of chloroplasts is that many PTMs have been described in this organelle, giving the possibility to accumulate a wide repertoire of functional heterologous proteins. Some of the chloroplast PTMs have been cited here in the previous sections (phosphorylation, NME, NTA, N-terminal myristoylation, and S-palmitoylation), but others have also been described, such as lipidation, oxidation, glutathionylation, and S-nitrosylation (Glenz et al., 2006, Armbruster et al., 2011). However, we need more detailed information on the occurrence of PTMs in different plastidial locations in order to decide where the recombinant protein of interest has to be addressed according to its specific folding requirements.
Localization of Recombinant Protein Containing Covalent Disulfide Bonds in the Chloroplast
For this purpose, an interesting case of study is the formation of covalent disulfide bonds required by many proteins for proper folding and/or their catalytic activity as enzymes. Moreover, disulfide linkage is also important for the immunogenicity of recombinant proteins to be used as vaccine antigens. Experimentally, it has been demonstrated that recombinant disulfide bond-containing proteins can be successfully expressed in transgenic chloroplasts, both in the stroma and in the lumen of thylakoids, but it is still not clear which of these two compartments is more suitable to allocate this kind of protein. A monomeric protein with intrachain disulfide bonds (Staub et al., 2000) or a multimeric protein with interchain disulfide bonds (Daniell et al., 2001) is biologically active when expressed in the stroma of plant chloroplasts. Furthermore, a soluble and functional monoclonal antibody forming interchain disulfide-bridged dimers can be expressed in the chloroplast of the alga C. reinhardtii (Mayfield et al., 2003). However, other recombinant proteins containing disulfide bonds form in the chloroplast dimers and higher order oligomers that are often accumulated, likely in the stroma, as insoluble aggregates (Fernández-San Millán et al., 2003; Arlen et al., 2007; Ruhlman et al., 2007; Boyhan and Daniell, 2011; Lee et al., 2011). Also, the human TGF-β3, which is a homodimer linked by disulfide bonds, when expressed in the chloroplast accumulates in unfolded insoluble aggregates, so it is necessary to refold the heterologous protein to show its biological activity (Gisby et al., 2011). Even if insoluble aggregates can be easily purified, denaturation and refolding of the recombinant protein significantly increase the cost of the production process, without ensuring the attainment of the native protein conformation (Yasukawa et al., 1995). In an attempt to increase the solubility of plastid-expressed human serum albumin (HSA) and thus avoid the formation of insoluble inclusion bodies, Sanz-Barrio et al. (2011) have overexpressed tobacco thioredoxins together with HSA. Thioredoxins enhance the solubility of recombinant proteins that cannot form disulfide bridges in microbial expression systems, but this strategy has failed to prevent the formation of HSA protein bodies within chloroplasts. One of the possible explanations for the formation of chloroplast inclusion bodies in the stroma is that the high translation rate and the high stability of the recombinant proteins alter the organelle protein homeostasis, causing the formation of aberrant protein-protein interactions (such as incorrect disulfide bonds) that cause accumulation in insoluble aggregates (Villar-Piqué et al., 2010; Gisby et al., 2011).
Recently, it has been proposed that the thylakoid lumen could be a more adequate environment than the stroma for the folding and activity of proteins that require the oxidation of Cys residues. When the enzyme alkaline phosphatase, a homodimer with two intramolecular disulfide bonds for each subunit, necessary for stability and catalytic activity of the molecule, was expressed both in the stroma and in the thylakoid lumen, the results indicated that sorting to the thylakoid lumen leads to larger amounts and more active enzyme (Bally et al., 2008). In addition, the Cys residues of another recombinant protein, capable of forming interchain disulfide bonds, can be oxidized if addressed to the thylakoid membranes (De Marchis et al., 2011). Conversely, the same proteins when localized into the stroma are unable to form this type of covalent linkage. A similar conclusion has been drawn for a nanobody with intramolecular disulfide bonds expressed both in the stroma and in the thylakoid lumen (Lentz et al., 2012). This is not surprising, as the thylakoid lumen, as well as the intermembrane space of mitochondria, is topologically equivalent to the periplasmic space of bacteria, and active thiol oxidation pathways are present in these compartments in order to promote protein folding and regulate protein function (Herrmann et al., 2009). Redox reactions are used for metabolic regulation in both the stroma and the thylakoid lumen (Dietz and Pfannschmidt, 2011), but in light conditions, the stroma should be a reducing environment with proteins maintained in a reduced state by thioredoxins that counteract the oxidative activity of molecular oxygen, whereas luminal enzymes are oxidatively activated (Buchanan and Luan, 2005). Moreover, the evidence for functional oxidoreductases such as the ER-resident protein disulfide isomerases (PDIs), which catalyze the formation, reduction, and isomerization of disulfide bonds, has been reported in C. reinhardtii but not in the stroma of plant chloroplasts (Trebitsh et al., 2001). Even though several chloroplast PDIs are described in proteomic studies, up to now only for one Arabidopsis PDI (At3g54960) has a chloroplast location been experimentally demonstrated, but neither its intraplastidial localization nor its biochemical function has been shown (Armbruster et al., 2009). Conversely, functional oxidoreductases have been identified in the Arabidopsis thylakoid membranes like LTO1, which has a thioredoxin-like domain exposed to the thylakoid lumen capable of catalyzing disulfide bond formation in the PsbO protein, a luminal subunit of PSII requiring for its stability the formation of a single intramolecular disulfide (Karamoko et al., 2011). Recently, other reports described two zinc finger proteins showing disulfide isomerase activity located in the thylakoid membrane and involved in the maintenance of PSII (Hall et al., 2010) or thylakoid biogenesis (Tanz et al., 2012). This information together demonstrates that a catalyzed thiol-disulfide process operates in the luminal side of the thylakoid membrane, with a thio-oxidizing pathway promoting the formation of disulfide bonds and a thio-reducing pathway to transfer reducing equivalents (Motohashi and Hisabori, 2010).
Foreign protein localization in the chloroplast seems to be of vital importance for getting properly folded and functional polypeptides; therefore, a thorough understanding of the mechanisms governing the various transport systems within the chloroplast, as well as the identification of a complete chloroplast proteome (enzymes, chaperones, complex subunits, etc.), are essential in order to obtain the best results. Chaperones have many functions in chloroplast biogenesis; for example, protein trafficking inside chloroplasts is assisted by chaperone systems that perform an essential role in facilitating protein translocation (Su and Li, 2010) and in protecting membrane proteins from unwanted aggregation (Jaru-Ampornpan et al., 2010). The targeting factor of the chloroplast SRP (cpSRP43) is a molecular chaperone that reverses the aggregation of its substrate proteins (Jaru-Ampornpan et al., 2010). This means that, en route to their destinations, membrane proteins translocated by the SRP pathway are protected from misfolding. Recently, it has been shown that a ER plant signal peptide is capable of carrying a fusion protein called zeolin, whose gene is inserted into the plastoma, to the thylakoid membranes, where the recombinant protein accumulates as trimers. In the stroma, the folding of the same protein without the signal peptide is hampered as the protein accumulates at low amounts in a monomeric form (De Marchis et al., 2011). Thus, we can speculate that whatever is the sorting mechanism that directs zeolin to the thylakoids, it probably also has a stabilizing effect on the recombinant protein (Fig. 2).
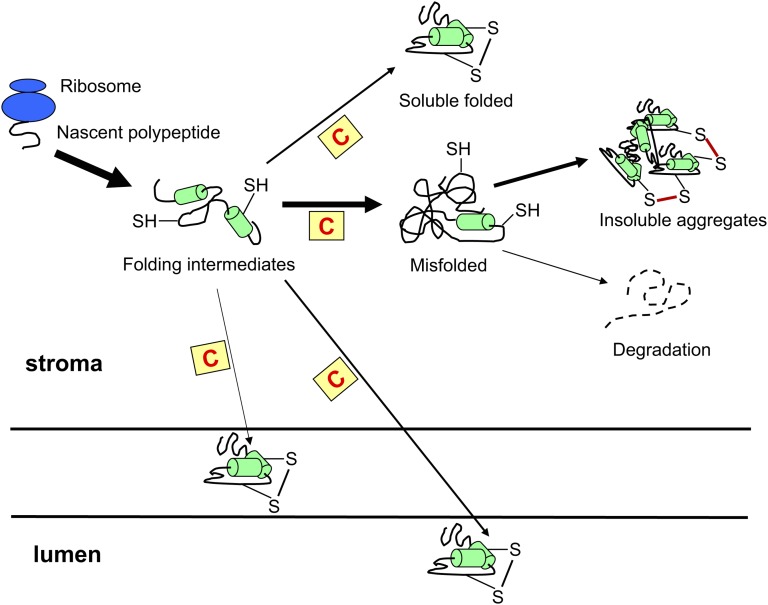
Figure 2.
Model of the possible site of accumulation in the chloroplast of recombinant proteins containing covalent disulfide bonds. Recombinant disulfide-containing proteins (represented for simplicity by a monomeric protein with an intrachain disulfide bridge) when expressed in transplastomic plants have been in most cases accumulated in the stroma. In this compartment, only a few studies reported the obtainment of soluble properly folded polypeptides, whereas the other recombinant proteins described were probably misfolded, often originating in insoluble aggregates with incorrect disulfide bonds (depicted by red lines). Alternatively, the misfolded proteins are targeted for degradation by stromal proteases, resulting in a low level of protein expression. Recently, recombinant proteins requiring the oxidation of Cys residues were also addressed by targeting signals to the thylakoid membrane or lumen, where they correctly folded. The thickness of the arrows is roughly proportional to the number of publications describing the expression of a recombinant protein in a chosen chloroplast subcompartment (for more details, see text). The yellow squares with a red C inside indicate chaperone pathways that assist in protein folding and trafficking.
CONCLUSION
Nowadays, different technologies can be adopted to transform a plant, and plastid biotechnology is one of the most promising ways. The future perspectives of plastid genetic engineering have been clearly described by Clarke et al. (2011), and here we try to emphasize that a big effort has to be made in understanding the factors that regulate plastid proteostasis (Fig. 3). More exhaustive studies on chloroplasts are necessary to elucidate the relationship between protein stability and the characteristics of its N-terminal part. Moreover, the fast-growing amount of data coming from proteomic studies can be utilized to create and/or to improve prediction tools for N-terminal modifications and their relationship with protein half-life, as illustrated for the online prediction tool TermiNator (http://www.isv.cnrs-gif.fr/terminator2/index.html), which predicts NME, NTA, N-terminal myristoylation, and S-palmitoylation of either prokaryotic or eukaryotic proteins originating from organellar or nuclear genomes. The role of chaperones in different plastid compartments also has to be clearly elucidated in order to understand how these molecules assist protein folding in plastids and how this information can be used in plastid genetic engineering. For example, several chaperones have been coexpressed together with recombinant proteins in the chloroplast to improve the accumulation of the latter. The results are encouraging only for the bacterial putative chaperonin open reading frame2 (De Cosa et al., 2001) but not for the ER chaperone binding protein and the chloroplast tobacco thioredoxins f and m (Bellucci et al., 2007; Sanz-Barrio et al., 2011). Another aspect to be taken into consideration is that the members of the plastid family are classified according to their metabolic specialization inside the cell and that more than 10 different variants can be identified like chloroplasts, chromoplasts, amyloplasts, etc. Proteostasis and gene expression in nongreen plastids are not well described, but some results show that these systems are quite distinct from those of chloroplasts (Kahlau and Bock, 2008; Valkov et al., 2009; Zhang et al., 2012). A major challenge for the future will be the identification of substrates for the chloroplast proteases in order to better understand the determinants of protein stability. Indeed, it remains unknown which protease(s) degrade the most plastid proteins. Searching for possible substrates is the objective of Stanne et al. (2009) to identify potential substrates for Clp protease in higher plants.
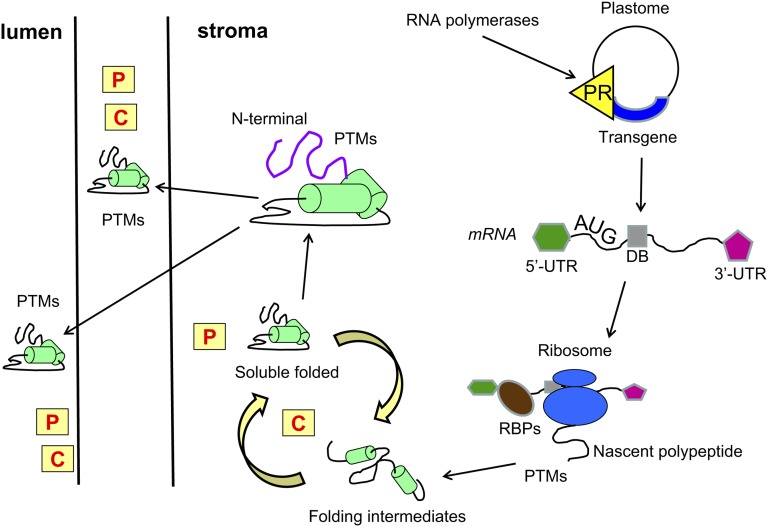
Figure 3.
Mechanisms and main players involved in the maintenance of plastid proteostasis, which affects foreign protein accumulation. Transcriptional and translational control, proteases, molecular chaperones, protein folding, and targeting are graphically illustrated. For discussion, see text. The yellow squares with a red C inside indicate chaperone pathways; the yellow squares with a red P inside indicate proteases. The yellow triangle with black PR inside indicates the transgene promoter. 5′-UTRs and 3′-UTRs are indicated, as well as the AUG translation initiation codon and the sequences downstream of it that affect transcript stability and/or translation (downstream box region [DB]). RBPs, RNA-binding proteins.
In conclusion, a cocktail of basic and applied studies on plastid proteostasis is needed in the coming years to predictably express foreign proteins from the plastid genome.
Acknowledgments
We thank Prof. Ralph Bock for critical reading of the manuscript. We sincerely apologize to all colleagues whose work could not be cited because of space constraints.
Glossary
- UTR
untranslated region
- SD
Shine-Dalgarno
- NME
N-terminal methionine excision
- PTM
posttranslational modification
- NTA
N-α-terminal acetylation
- ER
endoplasmic reticulum
- SRP
signal recognition particle
- HSA
human serum albumin
- PDI
protein disulfide isomerase
References
- Adam Z, Frottin F, Espagne C, Meinnel T, Giglione C. (2011) Interplay between N-terminal methionine excision and FtsH protease is essential for normal chloroplast development and function in Arabidopsis. Plant Cell 23: 3745–3760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam Z, Rudella A, van Wijk KJ. (2006) Recent advances in the study of Clp, FtsH and other proteases located in chloroplasts. Curr Opin Plant Biol 9: 234–240 [DOI] [PubMed] [Google Scholar]
- Agrawal GK, Bourguignon J, Rolland N, Ephritikhine G, Ferro M, Jaquinod M, Alexiou KG, Chardot T, Chakraborty N, Jolivet P, et al. (2011) Plant organelle proteomics: collaborating for optimal cell function. Mass Spectrom Rev 30: 772–853 [DOI] [PubMed] [Google Scholar]
- Albiniak AM, Baglieri J, Robinson C. (2012) Targeting of lumenal proteins across the thylakoid membrane. J Exp Bot 63: 1689–1698 [DOI] [PubMed] [Google Scholar]
- Apel W, Schulze WX, Bock R. (2010) Identification of protein stability determinants in chloroplasts. Plant J 63: 636–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlen PA, Falconer R, Cherukumilli S, Cole A, Cole AM, Oishi KK, Daniell H. (2007) Field production and functional evaluation of chloroplast-derived interferon-alpha2b. Plant Biotechnol J 5: 511–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster U, Hertle A, Makarenko E, Zühlke J, Pribil M, Dietzmann A, Schliebner I, Aseeva E, Fenino E, Scharfenberg M, et al. (2009) Chloroplast proteins without cleavable transit peptides: rare exceptions or a major constituent of the chloroplast proteome? Mol Plant 2: 1325–1335 [DOI] [PubMed] [Google Scholar]
- Armbruster U, Pesaresi P, Pribil M, Hertle A, Leister D. (2011) Update on chloroplast research: new tools, new topics, and new trends. Mol Plant 4: 1–16 [DOI] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW. (2008) Adapting proteostasis for disease intervention. Science 319: 916–919 [DOI] [PubMed] [Google Scholar]
- Bally J, Paget E, Droux M, Job C, Job D, Dubald M. (2008) Both the stroma and thylakoid lumen of tobacco chloroplasts are competent for the formation of disulphide bonds in recombinant proteins. Plant Biotechnol J 6: 46–61 [DOI] [PubMed] [Google Scholar]
- Bellucci M, De Marchis F, Mannucci R, Bock R, Arcioni S. (2005) Cytoplasm and chloroplasts are not suitable subcellular locations for β-zein accumulation in transgenic plants. J Exp Bot 56: 1205–1212 [DOI] [PubMed] [Google Scholar]
- Bellucci M, De Marchis F, Nicoletti I, Arcioni S. (2007) Zeolin is a recombinant storage protein with different solubility and stability properties according to its localization in the endoplasmic reticulum or in the chloroplast. J Biotechnol 131: 97–105 [DOI] [PubMed] [Google Scholar]
- Bienvenut WV, Espagne C, Martinez A, Majeran W, Valot B, Zivy M, Vallon O, Adam Z, Meinnel T, Giglione C. (2011) Dynamics of post-translational modifications and protein stability in the stroma of Chlamydomonas reinhardtii chloroplasts. Proteomics 11: 1734–1750 [DOI] [PubMed] [Google Scholar]
- Bienvenut WV, Sumpton D, Martinez A, Lilla S, Espagne C, Meinnel T, Giglione C. (2012) Comparative large-scale characterisation of plant versus mammal proteins reveals similar and idiosyncratic N-α-acetylation features. Mol Cell Proteomics 11: M111.015131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch-Machin I, Newell CA, Hibberd JM, Gray JC. (2004) Accumulation of rotavirus VP6 protein in chloroplasts of transplastomic tobacco is limited by protein stability. Plant Biotechnol J 2: 261–270 [DOI] [PubMed] [Google Scholar]
- Boyhan D, Daniell H. (2011) Low-cost production of proinsulin in tobacco and lettuce chloroplasts for injectable or oral delivery of functional insulin and C-peptide. Plant Biotechnol J 9: 585–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan BB, Luan S. (2005) Redox regulation in the chloroplast thylakoid lumen: a new frontier in photosynthesis research. J Exp Bot 56: 1439–1447 [DOI] [PubMed] [Google Scholar]
- Chi W, Sun X, Zhang L. (2012) The roles of chloroplast proteases in the biogenesis and maintenance of photosystem II. Biochim Biophys Acta 1817: 239–246 [DOI] [PubMed] [Google Scholar]
- Clarke JL, Daniell H, Nugent JM. (2011) Chloroplast biotechnology, genomics and evolution: current status, challenges and future directions. Plant Mol Biol 76: 207–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Lee SB, Panchal T, Wiebe PO. (2001) Expression of the native cholera toxin B subunit gene and assembly as functional oligomers in transgenic tobacco chloroplasts. J Mol Biol 311: 1001–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cosa B, Moar W, Lee SB, Miller M, Daniell H. (2001) Overexpression of the Bt cry2Aa2 operon in chloroplasts leads to formation of insecticidal crystals. Nat Biotechnol 19: 71–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marchis F, Pompa A, Mannucci R, Morosinotto T, Bellucci M. (2011) A plant secretory signal peptide targets plastome-encoded recombinant proteins to the thylakoid membrane. Plant Mol Biol 76: 427–441 [DOI] [PubMed] [Google Scholar]
- de Virgilio M, De Marchis F, Bellucci M, Mainieri D, Rossi M, Benvenuto E, Arcioni S, Vitale A. (2008) The human immunodeficiency virus antigen Nef forms protein bodies in leaves of transgenic tobacco when fused to zeolin. J Exp Bot 59: 2815–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz KJ, Pfannschmidt T. (2011) Novel regulators in photosynthetic redox control of plant metabolism and gene expression. Plant Physiol 155: 1477–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsel O, Bock R. (2011) Selection of Shine-Dalgarno sequences in plastids. Nucleic Acids Res 39: 1427–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard S, Drapier D, Wollman FA. (2002) Searching limiting steps in the expression of chloroplast-encoded proteins: relations between gene copy number, transcription, transcript abundance and translation rate in the chloroplast of Chlamydomonas reinhardtii. Plant J 31: 149–160 [DOI] [PubMed] [Google Scholar]
- Elghabi Z, Karcher D, Zhou F, Ruf S, Bock R. (2011) Optimization of the expression of the HIV fusion inhibitor cyanovirin-N from the tobacco plastid genome. Plant Biotechnol J 9: 599–608 [DOI] [PubMed] [Google Scholar]
- Fernández-San Millán A, Mingo-Castel A, Miller M, Daniell H. (2003) A chloroplast transgenic approach to hyper-express and purify human serum albumin, a protein highly susceptible to proteolytic degradation. Plant Biotechnol J 1: 71–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro M, Brugière S, Salvi D, Seigneurin-Berny D, Court M, Moyet L, Ramus C, Miras S, Mellal M, Le Gall S, et al. (2010) AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol Cell Proteomics 9: 1063–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglione C, Vallon O, Meinnel T. (2003) Control of protein life-span by N-terminal methionine excision. EMBO J 22: 13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisby MF, Mellors P, Madesis P, Ellin M, Laverty H, O’Kane S, Ferguson MW, Day A. (2011) A synthetic gene increases TGFβ3 accumulation by 75-fold in tobacco chloroplasts enabling rapid purification and folding into a biologically active molecule. Plant Biotechnol J 9: 618–628 [DOI] [PubMed] [Google Scholar]
- Glenz K, Bouchon B, Stehle T, Wallich R, Simon MM, Warzecha H. (2006) Production of a recombinant bacterial lipoprotein in higher plant chloroplasts. Nat Biotechnol 24: 76–77 [DOI] [PubMed] [Google Scholar]
- Graciet E, Mesiti F, Wellmer F. (2010) Structure and evolutionary conservation of the plant N-end rule pathway. Plant J 61: 741–751 [DOI] [PubMed] [Google Scholar]
- Gray BN, Yang H, Ahner BA, Hanson MR. (2011) An efficient downstream box fusion allows high-level accumulation of active bacterial beta-glucosidase in tobacco chloroplasts. Plant Mol Biol 76: 345–355 [DOI] [PubMed] [Google Scholar]
- Hall M, Mata-Cabana A, Akerlund HE, Florencio FJ, Schröder WP, Lindahl M, Kieselbach T. (2010) Thioredoxin targets of the plant chloroplast lumen and their implications for plastid function. Proteomics 10: 987–1001 [DOI] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. (2011) Molecular chaperones in protein folding and proteostasis. Nature 475: 324–332 [DOI] [PubMed] [Google Scholar]
- Hennig A, Bonfig K, Roitsch T, Warzecha H. (2007) Expression of the recombinant bacterial outer surface protein A in tobacco chloroplasts leads to thylakoid localization and loss of photosynthesis. FEBS J 274: 5749–5758 [DOI] [PubMed] [Google Scholar]
- Herrmann JM, Kauff F, Neuhaus HE. (2009) Thiol oxidation in bacteria, mitochondria and chloroplasts: common principles but three unrelated machineries? Biochim Biophys Acta 1793: 71–77 [DOI] [PubMed] [Google Scholar]
- Herz S, Füssl M, Steiger S, Koop HU. (2005) Development of novel types of plastid transformation vectors and evaluation of factors controlling expression. Transgenic Res 14: 969–982 [DOI] [PubMed] [Google Scholar]
- Hwang CS, Shemorry A, Varshavsky A. (2010) N-terminal acetylation of cellular proteins creates specific degradation signals. Science 327: 973–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inka Borchers AM, Gonzalez-Rabade N, Gray JC. (2012) Increased accumulation and stability of rotavirus VP6 protein in tobacco chloroplasts following changes to the 5′ untranslated region and the 5′ end of the coding region. Plant Biotechnol J 10: 422–434 [DOI] [PubMed] [Google Scholar]
- Jaru-Ampornpan P, Shen K, Lam VQ, Ali M, Doniach S, Jia TZ, Shan SO. (2010) ATP-independent reversal of a membrane protein aggregate by a chloroplast SRP subunit. Nat Struct Mol Biol 17: 696–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P. (2008) Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol 179: 257–285 [DOI] [PubMed] [Google Scholar]
- Kahlau S, Bock R. (2008) Plastid transcriptomics and translatomics of tomato fruit development and chloroplast-to-chromoplast differentiation: chromoplast gene expression largely serves the production of a single protein. Plant Cell 20: 856–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamoko M, Cline S, Redding K, Ruiz N, Hamel PP. (2011) Lumen Thiol Oxidoreductase1, a disulfide bond-forming catalyst, is required for the assembly of photosystem II in Arabidopsis. Plant Cell 23: 4462–4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komenda J, Tichy M, Prásil O, Knoppová J, Kuviková S, de Vries R, Nixon PJ. (2007) The exposed N-terminal tail of the D1 subunit is required for rapid D1 degradation during photosystem II repair in Synechocystis sp PCC 6803. Plant Cell 19: 2839–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda H, Maliga P. (2001) Complementarity of the 16S rRNA penultimate stem with sequences downstream of the AUG destabilizes the plastid mRNAs. Nucleic Acids Res 29: 970–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SB, Li B, Jin S, Daniell H. (2011) Expression and characterization of antimicrobial peptides Retrocyclin-101 and Protegrin-1 in chloroplasts to control viral and bacterial infections. Plant Biotechnol J 9: 100–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentz EM, Garaicoechea L, Alfano EF, Parreño V, Wigdorovitz A, Bravo-Almonacid FF. (2012) Translational fusion and redirection to thylakoid lumen as strategies to improve the accumulation of a camelid antibody fragment in transplastomic tobacco. Planta 236: 703–714 [DOI] [PubMed] [Google Scholar]
- Lenzi P, Scotti N, Alagna F, Tornesello ML, Pompa A, Vitale A, De Stradis A, Monti L, Grillo S, Buonaguro FM, et al. (2008) Translational fusion of chloroplast-expressed human papillomavirus type 16 L1 capsid protein enhances antigen accumulation in transplastomic tobacco. Transgenic Res 17: 1091–1102 [DOI] [PubMed] [Google Scholar]
- Lerbs-Mache S. (2011) Function of plastid sigma factors in higher plants: regulation of gene expression or just preservation of constitutive transcription? Plant Mol Biol 76: 235–249 [DOI] [PubMed] [Google Scholar]
- Liere K, Börner T. (2007) Transcription and transcriptional regulation in plastids. In R Bock, ed, Topics in Current Genetics: Cell and Molecular Biology of Plastids, Vol 19. Springer-Verlag, Berlin, pp 121–174
- Lössl A, Bohmert K, Harloff H, Eibl C, Mühlbauer S, Koop HU. (2005) Inducible trans-activation of plastid transgenes: expression of the R. eutropha phb operon in transplastomic tobacco. Plant Cell Physiol 46: 1462–1471 [DOI] [PubMed] [Google Scholar]
- Lössl A, Eibl C, Harloff HJ, Jung C, Koop HU. (2003) Polyester synthesis in transplastomic tobacco (Nicotiana tabacum L.): significant contents of polyhydroxybutyrate are associated with growth reduction. Plant Cell Rep 21: 891–899 [DOI] [PubMed] [Google Scholar]
- Lössl AG, Waheed MT. (2011) Chloroplast-derived vaccines against human diseases: achievements, challenges and scopes. Plant Biotechnol J 9: 527–539 [DOI] [PubMed] [Google Scholar]
- Ma JK, Hiatt A, Hein M, Vine ND, Wang F, Stabila P, van Dolleweerd C, Mostov K, Lehner T. (1995) Generation and assembly of secretory antibodies in plants. Science 268: 716–719 [DOI] [PubMed] [Google Scholar]
- Mainieri D, Rossi M, Archinti M, Bellucci M, De Marchis F, Vavassori S, Pompa A, Arcioni S, Vitale A. (2004) Zeolin: a new recombinant storage protein constructed using maize γ-zein and bean phaseolin. Plant Physiol 136: 3447–3456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maliga P. (2002) Engineering the plastid genome of higher plants. Curr Opin Plant Biol 5: 164–172 [DOI] [PubMed] [Google Scholar]
- Maliga P, Bock R. (2011) Plastid biotechnology: food, fuel, and medicine for the 21st century. Plant Physiol 155: 1501–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marìn-Navarro J, Manuell AL, Wu J, P Mayfield S. (2007) Chloroplast translation regulation. Photosynth Res 94: 359–374 [DOI] [PubMed] [Google Scholar]
- Mayfield SP, Franklin SE, Lerner RA. (2003) Expression and assembly of a fully active antibody in algae. Proc Natl Acad Sci USA 100: 438–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinnel T, Serero A, Giglione C. (2006) Impact of the N-terminal amino acid on targeted protein degradation. Biol Chem 387: 839–851 [DOI] [PubMed] [Google Scholar]
- Merhige PM, Both-Kim D, Robida MD, Hollingsworth MJ. (2005) RNA-protein complexes that form in the spinach chloroplast atpI 5′ untranslated region can be divided into two subcomplexes, each comprised of unique cis-elements and trans-factors. Curr Genet 48: 256–264 [DOI] [PubMed] [Google Scholar]
- Michelet L, Lefebvre-Legendre L, Burr SE, Rochaix JD, Goldschmidt-Clermont M. (2011) Enhanced chloroplast transgene expression in a nuclear mutant of Chlamydomonas. Plant Biotechnol J 9: 565–574 [DOI] [PubMed] [Google Scholar]
- Mogk A, Schmidt R, Bukau B. (2007) The N-end rule pathway for regulated proteolysis: prokaryotic and eukaryotic strategies. Trends Cell Biol 17: 165–172 [DOI] [PubMed] [Google Scholar]
- Motohashi K, Hisabori T. (2010) CcdA is a thylakoid membrane protein required for the transfer of reducing equivalents from stroma to thylakoid lumen in the higher plant chloroplast. Antioxid Redox Signal 13: 1169–1176 [DOI] [PubMed] [Google Scholar]
- Mühlbauer SK, Koop HU. (2005) External control of transgene expression in tobacco plastids using the bacterial lac repressor. Plant J 43: 941–946 [DOI] [PubMed] [Google Scholar]
- Mulo P, Sakurai I, Aro EM. (2012) Strategies for psbA gene expression in cyanobacteria, green algae and higher plants: from transcription to PSII repair. Biochim Biophys Acta 1817: 247–257 [DOI] [PubMed] [Google Scholar]
- Oey M, Lohse M, Kreikemeyer B, Bock R. (2009) Exhaustion of the chloroplast protein synthesis capacity by massive expression of a highly stable protein antibiotic. Plant J 57: 436–445 [DOI] [PubMed] [Google Scholar]
- Ortigosa SM, Fernández-San Millán A, Veramendi J. (2010) Stable production of peptide antigens in transgenic tobacco chloroplasts by fusion to the p53 tetramerisation domain. Transgenic Res 19: 703–709 [DOI] [PubMed] [Google Scholar]
- Ouyang M, Li X, Ma J, Chi W, Xiao J, Zou M, Chen F, Lu C, Zhang L. (2011) LTD is a protein required for sorting light-harvesting chlorophyll-binding proteins to the chloroplast SRP pathway. Nat Commun 2: 277. [DOI] [PubMed] [Google Scholar]
- Pfalz J, Bayraktar OA, Prikryl J, Barkan A. (2009) Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J 28: 2042–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalski M, Ruf S, Bock R. (2006) Tobacco plastid ribosomal protein S18 is essential for cell survival. Nucleic Acids Res 34: 4537–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Román-Hernández G, Grant RA, Sauer RT, Baker TA. (2009) Molecular basis of substrate selection by the N-end rule adaptor protein ClpS. Proc Natl Acad Sci USA 106: 8888–8893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhlman T, Ahangari R, Devine A, Samsam M, Daniell H. (2007) Expression of cholera toxin B-proinsulin fusion protein in lettuce and tobacco chloroplasts: oral administration protects against development of insulitis in non-obese diabetic mice. Plant Biotechnol J 5: 495–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhlman T, Verma D, Samson N, Daniell H. (2010) The role of heterologous chloroplast sequence elements in transgene integration and expression. Plant Physiol 152: 2088–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Barrio R, Millán AF, Corral-Martínez P, Seguí-Simarro JM, Farran I. (2011) Tobacco plastidial thioredoxins as modulators of recombinant protein production in transgenic chloroplasts. Plant Biotechnol J 9: 639–650 [DOI] [PubMed] [Google Scholar]
- Sasidharan R, Mustroph A. (2011) Plant oxygen sensing is mediated by the N-end rule pathway: a milestone in plant anaerobiosis. Plant Cell 23: 4173–4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharff LB, Childs L, Walther D, Bock R. (2011) Local absence of secondary structure permits translation of mRNAs that lack ribosome-binding sites. PLoS Genet 7: e1002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda M. (2004) The Chlamydomonas genome reveals its secrets: chaperone genes and the potential roles of their gene products in the chloroplast. Photosynth Res 82: 221–240 [DOI] [PubMed] [Google Scholar]
- Scotti N, Rigano MM, Cardi T. (2012) Production of foreign proteins using plastid transformation. Biotechnol Adv 30: 387–397 [DOI] [PubMed] [Google Scholar]
- Singh ND, Li M, Lee SB, Schnell D, Daniell H. (2008) Arabidopsis Tic40 expression in tobacco chloroplasts results in massive proliferation of the inner envelope membrane and upregulation of associated proteins. Plant Cell 20: 3405–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalitzky CA, Martin JR, Harwood JH, Beirne JJ, Adamczyk BJ, Heck GR, Cline K, Fernandez DE. (2011) Plastids contain a second sec translocase system with essential functions. Plant Physiol 155: 354–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanne TM, Sjögren LL, Koussevitzky S, Clarke AK. (2009) Identification of new protein substrates for the chloroplast ATP-dependent Clp protease supports its constitutive role in Arabidopsis. Biochem J 417: 257–268 [DOI] [PubMed] [Google Scholar]
- Staub JM, Garcia B, Graves J, Hajdukiewicz PT, Hunter P, Nehra N, Paradkar V, Schlittler M, Carroll JA, Spatola L, et al. (2000) High-yield production of a human therapeutic protein in tobacco chloroplasts. Nat Biotechnol 18: 333–338 [DOI] [PubMed] [Google Scholar]
- Stern DB, Goldschmidt-Clermont M, Hanson MR. (2010) Chloroplast RNA metabolism. Annu Rev Plant Biol 61: 125–155 [DOI] [PubMed] [Google Scholar]
- Su PH, Li HM. (2010) Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. Plant Cell 22: 1516–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangphatsornruang S, Birch-Machin I, Newell CA, Gray JC. (2011) The effect of different 3′ untranslated regions on the accumulation and stability of transcripts of a gfp transgene in chloroplasts of transplastomic tobacco. Plant Mol Biol 76: 385–396 [DOI] [PubMed] [Google Scholar]
- Tanz SK, Kilian J, Johnsson C, Apel K, Small I, Harter K, Wanke D, Pogson B, Albrecht V. (2012) The SCO2 protein disulphide isomerase is required for thylakoid biogenesis and interacts with LHCB1 chlorophyll a/b binding proteins which affects chlorophyll biosynthesis in Arabidopsis seedlings. Plant J 69: 743–754 [DOI] [PubMed] [Google Scholar]
- Tikkanen M, Grieco M, Aro EM. (2011) Novel insights into plant light-harvesting complex II phosphorylation and ‘state transitions.’ Trends Plant Sci 16: 126–131 [DOI] [PubMed] [Google Scholar]
- Tiller N, Weingartner M, Thiele W, Maximova E, Schöttler MA, Bock R. (2012) The plastid-specific ribosomal proteins of Arabidopsis thaliana can be divided into non-essential proteins and genuine ribosomal proteins. Plant J 69: 302–316 [DOI] [PubMed] [Google Scholar]
- Tissot G, Canard H, Nadai M, Martone A, Botterman J, Dubald M. (2008) Translocation of aprotinin, a therapeutic protease inhibitor, into the thylakoid lumen of genetically engineered tobacco chloroplasts. Plant Biotechnol J 6: 309–320 [DOI] [PubMed] [Google Scholar]
- Trebitsh T, Meiri E, Ostersetzer O, Adam Z, Danon A. (2001) The protein disulfide isomerase-like RB60 is partitioned between stroma and thylakoids in Chlamydomonas reinhardtii chloroplasts. J Biol Chem 276: 4564–4569 [DOI] [PubMed] [Google Scholar]
- Valkov VT, Gargano D, Manna C, Formisano G, Dix PJ, Gray JC, Scotti N, Cardi T. (2011) High efficiency plastid transformation in potato and regulation of transgene expression in leaves and tubers by alternative 5′ and 3′ regulatory sequences. Transgenic Res 20: 137–151 [DOI] [PubMed] [Google Scholar]
- Valkov VT, Scotti N, Kahlau S, Maclean D, Grillo S, Gray JC, Bock R, Cardi T. (2009) Genome-wide analysis of plastid gene expression in potato leaf chloroplasts and tuber amyloplasts: transcriptional and posttranscriptional control. Plant Physiol 150: 2030–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijk KJ, Baginsky S. (2011) Plastid proteomics in higher plants: current state and future goals. Plant Physiol 155: 1578–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. (1996) The N-end rule: functions, mysteries, uses. Proc Natl Acad Sci USA 93: 12142–12149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhounig A, Karcher D, Bock R. (2010) Inducible gene expression from the plastid genome by a synthetic riboswitch. Proc Natl Acad Sci USA 107: 6204–6209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar-Piqué A, Sabaté R, Lopera O, Gibert J, Torne JM, Santos M, Ventura S. (2010) Amyloid-like protein inclusions in tobacco transgenic plants. PLoS ONE 5: e13625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale A, Ceriotti A. (2004) Protein quality control mechanisms and protein storage in the endoplasmic reticulum: a conflict of interests? Plant Physiol 136: 3420–3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller RF. (2012) Second genesis of a plastid organelle. Proc Natl Acad Sci USA 109: 5142–5143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner S, Maurizi MR, Gottesman S. (1999) Posttranslational quality control: folding, refolding, and degrading proteins. Science 286: 1888–1893 [DOI] [PubMed] [Google Scholar]
- Willmund F, Dorn KV, Schulz-Raffelt M, Schroda M. (2008) The chloroplast DnaJ homolog CDJ1 of Chlamydomonas reinhardtii is part of a multichaperone complex containing HSP70B, CGE1, and HSP90C. Plant Physiol 148: 2070–2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth S, Segretin ME, Mentaberry A, Bravo-Almonacid F. (2006) Accumulation of hEGF and hEGF-fusion proteins in chloroplast-transformed tobacco plants is higher in the dark than in the light. J Biotechnol 125: 159–172 [DOI] [PubMed] [Google Scholar]
- Yasukawa T, Kanei-Ishii C, Maekawa T, Fujimoto J, Yamamoto T, Ishii S. (1995) Increase of solubility of foreign proteins in Escherichia coli by coproduction of the bacterial thioredoxin. J Biol Chem 270: 25328–25331 [DOI] [PubMed] [Google Scholar]
- Ye GN, Hajdukiewicz PT, Broyles D, Rodriguez D, Xu CW, Nehra N, Staub JM. (2001) Plastid-expressed 5-enolpyruvylshikimate-3-phosphate synthase genes provide high level glyphosate tolerance in tobacco. Plant J 25: 261–270 [DOI] [PubMed] [Google Scholar]
- Zerges W. (2004) Regulation of translation in chloroplasts. In H Daniell, C Chase, eds, Molecular Biology of Plant Organelles: Chloroplast and Mitochondria. Springer, Dordrecht, The Netherlands, pp 443–490
- Zhang J, Ruf S, Hasse C, Childs L, Scharff LB, Bock R. (2012) Identification of cis-elements conferring high levels of gene expression in non-green plastids. Plant J (in press) [DOI] [PubMed] [Google Scholar]
- Zhao Y, Jensen ON. (2009) Modification-specific proteomics: strategies for characterization of post-translational modifications using enrichment techniques. Proteomics 9: 4632–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Badillo-Corona JA, Karcher D, Gonzalez-Rabade N, Piepenburg K, Borchers A-MI, Maloney AP, Kavanagh TA, Gray JC, Bock R. (2008) High-level expression of human immunodeficiency virus antigens from the tobacco and tomato plastid genomes. Plant Biotechnol J 6: 897–913 [DOI] [PubMed] [Google Scholar]
- Zhou F, Karcher D, Bock R. (2007) Identification of a plastid intercistronic expression element (IEE) facilitating the expression of stable translatable monocistronic mRNAs from operons. Plant J 52: 961–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoldák G, Carstensen L, Scholz C, Schmid FX. (2009) Consequences of domain insertion on the stability and folding mechanism of a protein. J Mol Biol 386: 1138–1152 [DOI] [PubMed] [Google Scholar]
- Zou Z, Eibl C, Koop HU. (2003) The stem-loop region of the tobacco psbA 5′UTR is an important determinant of mRNA stability and translation efficiency. Mol Genet Genomics 269: 340–349 [DOI] [PubMed] [Google Scholar]
- Zybailov B, Rutschow H, Friso G, Rudella A, Emanuelsson O, Sun Q, van Wijk KJ. (2008) Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS ONE 3: e1994 [DOI] [PMC free article] [PubMed] [Google Scholar]