Summary
The concept of reaction similarity has been well-studied in terms of the overall transformation associated with a reaction, but not in terms of mechanism. We present the first method to give a quantitative measure of the similarity of reactions based upon their explicit mechanisms. Two approaches are presented to measure the similarity between individual steps of mechanisms: a fingerprint-based approach which incorporates relevant information on each mechanistic step, and an approach based only on bond formation, cleavage and changes in order. The overall similarity for two reaction mechanisms is then calculated using the Needleman-Wunsch alignment algorithm. An analysis of MACiE, a database of enzyme mechanisms, using our measure of similarity identifies some examples of convergent evolution of chemical mechanism. In many cases mechanism similarity is not reflected by similarity according to the EC system of enzyme classification. In particular, little mechanistic information is conveyed by the class level of the EC.
Keywords: reaction similarity, MACiE, enzyme reaction, enzyme mechanism, enzyme classification
Introduction
The concept of similarity is of central importance to the study of enzymes, but its meaning depends on the context. Enzymes may be regarded as similar if they are evolutionarily related – this is the measure used by evolutionary trees such as those in the database of protein domain families, Pfam.1 Alternatively, we may regard as similar those enzymes that share structural elements – this is the basis of the CATH,2 SCOP3 and DALI4 classification systems for proteins. Other classification systems consider enzyme function, such as the Enzyme Commission (EC) system,5 the Reaction Classification (RC) system6 and the RLCP classification,7 which are based upon the overall reaction catalysed by the enzyme. In addition, the RLCP classification also considers the mechanism type and structural features of the enzyme.
There is a need for a similarity measure for enzymes based on reaction mechanism. A growing body of evidence indicates that reaction steps are conserved during the evolution of enzyme function. Both Babbitt and Gerlt8 and Todd et al.9 have found that in functionally diverse superfamilies, a common chemical strategy may be used in the context of different overall transformations. By comparing the reaction steps of 27 pairs of homologous enzymes of different function, Bartlett et al.10 identified examples of mechanisms that share steps at the start, middle or end of the reaction but that catalyse different overall transformations. Although the EC classification system has proved its worth as a system for cataloguing and comparing the overall reactions catalysed by enzymes, it does not take into account the increasing amount of mechanistic information that is becoming available, and has several known limitations.11,12 For example, the four types of β-lactamase share the same EC number, but have different reaction mechanisms, most notably the difference between Classes A, C and D, all of which use the active site serine as the nucleophile, and Class B, which uses one or two zinc ions to effect the reaction. In addition, enzymes with very similar mechanisms can have quite different classifications under the EC system. For example, there are two phosphatidylinositol-specific phospholipase Cs which belong to different EC classes (EC 3.1.4.11 and EC 4.6.1.13), but the only difference in their mechanism is that in one case the reaction proceeds through a cyclic intermediate to hydrolysis, whereas in the other the major product is the cyclic structure.
Other hierarchical systems have been developed that use mechanistic information. The Structure-Function Linkage Database13 (SFLD) of Babbitt et al. uses an initial classification of enzymes into superfamilies based on conserved partial reactions (currently, only six superfamilies are included). Subsequent divisions are superfamily specific, but are typically based on conserved structural features. Gariev and Varfolomeev’s hierarchical classification of hydrolase catalytic sites14 combines structural information with the catalytic roles of active site residues. Although such hierarchical arrangements are useful from the point of view of classification, and incorporate expert knowledge of particular families of enzymes, they do not use or provide an explicit way to calculate the similarity of two reaction mechanisms.
Here we describe the development and application of a similarity measure for enzyme reaction mechanisms. This provides the first quantitative measure of similarity between reactions based on their explicit mechanisms. Previous methods for calculating similarity between reactions have at most included mechanistic information indirectly by means of physicochemical descriptors of the reactants and/or products that represent inductive and resonance effects around the reactive centre.15,16 These descriptors have also been used to classify enzyme reactions according to the EC system with an accuracy of 94% at the class level of the EC.17 However, as mechanism is not explicitly included, reactions with identical overall transformations but different mechanisms cannot be differentiated. Additionally, these methods do not allow the detection of similarity for reaction mechanisms that are not identical but do share some mechanistic steps. For example, in terms of mechanistic steps it is clear that an aldol addition is a subset of an aldol condensation, but this would not be apparent by simply considering the reactants and products.
Our method of measuring the similarity of enzyme reaction mechanisms uses the explicit mechanism of a reaction, as determined by chemical, structural and biological studies taken from the primary literature. It calculates a value for similarity by combining a sequence alignment algorithm with methods to measure the similarity of two mechanistic steps. Using this similarity measure, it is possible to calculate meaningful values for how similar two reaction mechanisms are, and to discover similarity/dissimilarity between reactions whether they are far apart or close together in terms of the conventional EC system.
Results
Most similar reaction pairs
In order to test whether the similarity measures we propose can detect similarities between enzyme mechanisms, we looked at the pairwise similarities between enzyme mechanisms in the MACiE database, which contains non-homologous enzymes covering a broad range of EC space.18 5% and 1% significance levels for the similarity values between two reaction mechanisms a and b were calculated based on the distribution of similarity values for random sequences of steps of the same length as a and b. Each mechanism step in the random sequences was chosen with replacement from the set of all steps in the database, and the procedure was repeated 10,000 times.
The pairwise similarities of all of the reactions in MACiE version 1.1 were calculated using the fingerprint method (FP), as well as using the bond change method (BC). Figure 1 shows the distribution of pairwise similarities in the dataset. The 20 most similar pairs of reactions for each method are shown in Table 1, along with the level up to which their EC codes agree with each other. Twelve pairs of reactions are highly scored by both the FP and BC methods, and in particular, both methods agree on seven of the top ten most similar reactions. All of the similarity values are significant at the 5% level of significance, and all but four are significant at the 1% level.
Figure 1.
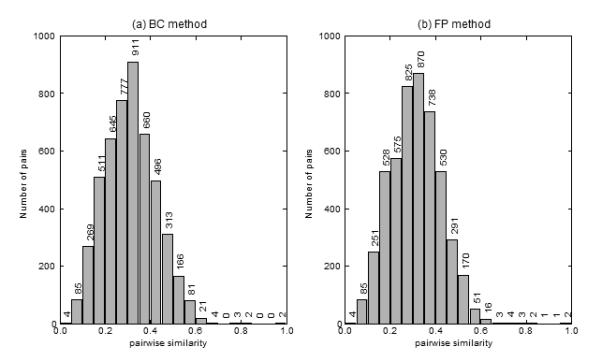
A histogram showing the distribution of pairwise similarities in the MACiE dataset for (a) the BC method and (b) the FP method. Bin sizes of 0.05 were used.
Table 1.
The most similar pairs of reactions in the dataset according to the Fingerprint (FP), and Bond Change (BC) methods. Reaction pairs highly scored by both FP and BC are shown in bold.
| BC | FP | |||||||
|---|---|---|---|---|---|---|---|---|
| Rank | Pair a | Score | Significance |
same
EC b |
Pair | Score | Significance |
same
EC |
| 1 | 27, 35 | 1.00 | 1% | 0 | 27, 35 | 1.00 | 1% | 0 |
| 2 | 26, 41 | 1.00 | 1% | 0 |
11,
40 |
1.00 | 1% | 0 |
| 3 | 11, 40 | 1.00 | 5% | 0 | 5, 94 | 0.92 | 1% | 1 |
| 4 | 17, 91 | 0.78 | 1% | 3 |
17,
91 |
0.89 | 1% | 3 |
| 5 | 5, 94 | 0.76 | 1% | 1 | 62, 63 | 0.82 | 1% | 3 |
| 6 | 32, 33 | 0.75 | 1% | 0 | 7, 21 | 0.81 | 1% | 3 |
| 7 | 92, 100 | 0.69 | 1% | 1 | 26, 41 | 0.76 | 1% | 0 |
| 8 | 2, 29 | 0.67 | 1% | 2 | 27, 48 |
0.75 | 1% | 0 |
| 9 | 62, 63 | 0.64 | 1% | 3 | 35, 48 | 0.75 | 1% | 1 |
| 10 | 7, 21 | 0.58 | 1% | 3 |
27,
86 |
0.74 | 1% | 0 |
| 11 | 39, 80 | 0.54 | 1% | 0 | 35, 86 | 0.74 | 1% | 2 |
| 12 | 53, 78 | 0.52 | 1% | 3 |
48,
86 |
0.74 | 1% | 1 |
| 13 | 11, 51 | 0.50 | 5% | 0 | 69, 83 | 0.72 | 1% | 0 |
| 14 | 19, 36 | 0.50 | 1% | 1 |
32,
33 |
0.68 | 1% | 0 |
| 15 | 24, 83 | 0.50 | 1% | 1 | 33, 99 | 0.67 | 1% | 0 |
| 16 | 27, 44 | 0.50 | 1% | 2 | 22, 96 |
0.65 | 1% | 0 |
| 17 | 27, 86 | 0.50 | 1% | 0 | 15, 50 | 0.64 | 5% | 0 |
| 18 | 35, 44 | 0.50 | 1% | 0 | 36, 47 |
0.64 | 1% | 1 |
| 19 | 35, 86 | 0.50 | 1% | 2 | 42, 83 | 0.64 | 5% | 2 |
| 20 | 40, 51 | 0.50 | 5% | 0 | 97, 98 |
0.64 | 1% | 2 |
| 21 | 48, 86 | 0.50 | 1% | 1 | ||||
| 22 | 69, 83 | 0.50 | 1% | 0 | ||||
The reaction numbered nn refers to MACiE entry M00nn.
This is the level up to which the two reactions share the same EC classification number.
Since the enzymes in MACiE are unique at the fourth level of the CATH code, except where the mechanism is significantly different, there should not be any duplicate mechanisms in MACiE. Preliminary use of the methods described here identified such a pair of duplicates, one of which was subsequently replaced (before the generation of the dataset used here). Any remaining mechanisms that are identical or highly similar according to our measures of mechanism similarity are considered to be examples of convergent evolution of chemical mechanism. Here we present a discussion of some of the most similar pairs of reaction mechanisms in our dataset according to our novel measures of similarity (Table 1). The reaction mechanisms themselves can be viewed at http://www-mitchell.ch.cam.ac.uk/macie/JMBPaper.
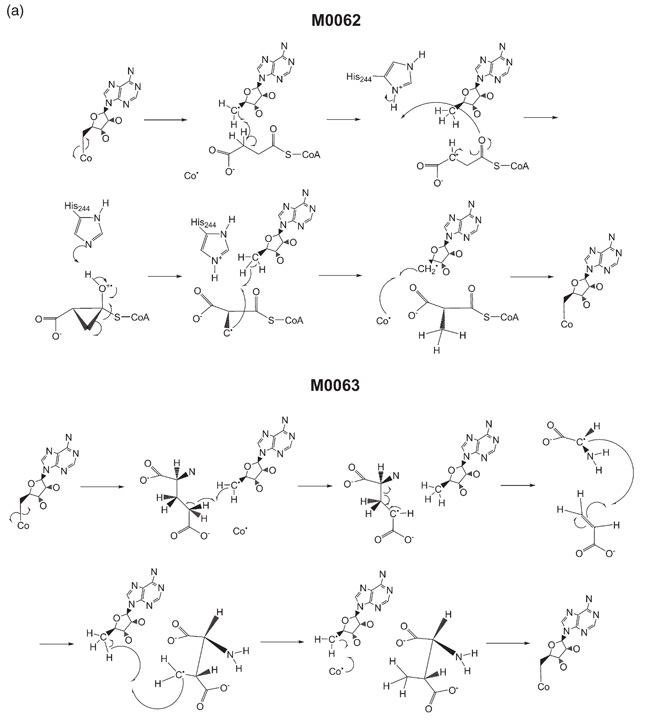
As a first example, we have taken two enzymes in the dataset whose mechanisms have been taken from the same literature source, and which we know to be very similar. M006219,20 and M006319 are two adenosylcobalamin-dependent isomerases, both classified as intramolecular transferases (EC 5.4), whose proposed mechanisms differ only in the order of steps 3 and 4 of their 6-step mechanisms (Figure 2(a)). These steps involve the 1,2-shift of a CH-COOH through a radical mechanism, but the two reactions differ in how this shift is accomplished. For M0062, the isomerisation occurs via an intramolecular bond formation (leading to a cyclic intermediate) followed by a bond cleavage, whereas for M0063 a bond cleavage occurs first followed by reattachment of the leaving group at a different position. Both the FP and BC methods are able to identify the similarities between the two mechanisms and rank them in the top ten most similar pairs in the dataset.
Figure 2(a).
The catalytic mechanisms of MACiE entries M0062, methylmalonyl-CoA mutase, and M0063, methylaspartate mutase.
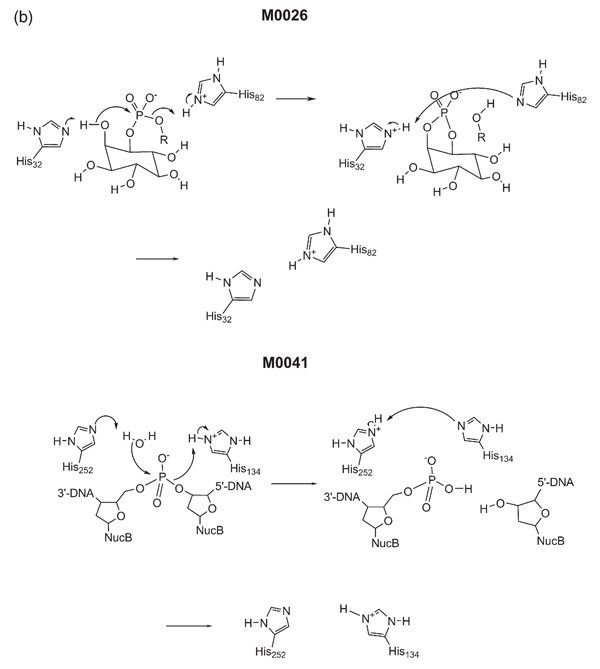
The measurement of mechanism similarity as defined by the BC or FP methods can be used to identify examples of mechanistic convergence. That is, cases where the same mechanisms are used by enzymes unrelated in sequence or structure. Reactions M002621,22,23 and M004124 are identical according to the BC method, and in the top ten according to the FP method. In terms of conventional similarity measures they are very different: they differ at the class level of the EC, have different catalytic domains, and M0041 catalyses the hydrolysis of DNA whereas M0026 involves the cleavage of diacylglycerol (DAG) from phosphatidylinositol diacylglycerol (Figure 2(b)). However, the general mechanisms of the reactions are very similar. A histidine residue abstracts a proton from a hydroxyl group which then attacks a phosphate ester, displacing RO−. This is protonated by another histidine residue, after which proton transfer occurs between the two histidine residues to return the enzyme to the native state. Since the attacking hydroxyl group is OH− for M0041 but OR− for M0026, the former is classified as a hydrolase (EC class 3) acting on an ester bond, whereas the latter is classified as a lyase (EC class 4) acting on P-O bonds.
Figure 2(b).
The catalytic mechanisms of MACiE entries M0026, phosphatidylinositol diacylglycerol-lyase, and M0041, deoxyribonuclease I.
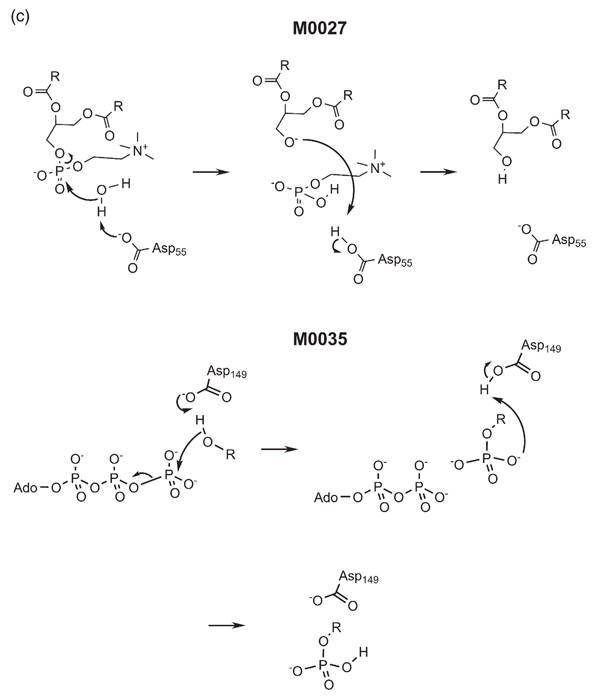
Both M002725 and M003526 also involve RO− attack on a phosphate ester (Figure 2(c)). The former hydrolyses a phosphatidylcholine, whereas the latter phosphorylates the b form of the phosphorylase enzyme. Again, although these may be regarded as quite different, the mechanisms are very similar. An aspartate residue abstracts a proton from a hydroxyl group which then attacks the phosphate ester, displacing DAG in the case of M0027 and ADP in M0035. Proton transfer occurs from the Asp to the leaving group to return the enzyme to the native state. Despite these similarities in mechanism, M0027 is classified as a hydrolase (EC class 3) acting on an ester bond, whereas M0035 is classified as a transferase (EC class 2) of phosphorus-containing groups.
Figure 2(c).
The catalytic mechanisms of MACiE entries M0027, phospholipase C, and M0035, phosphorylase kinase.
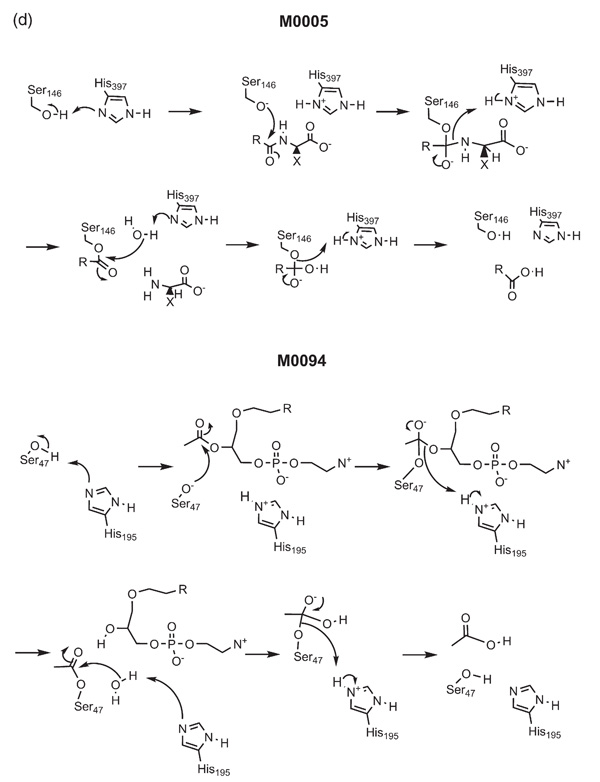
In the case of the previous mechanisms, similarity of mechanism might be inferred based on the overall reaction. This is less likely in the case of M000527 and M0094,28,29 where the former hydrolyses an amide bond but the latter hydrolyses an ester bond (Figure 2(d)). In both mechanisms, a histidine residue abstracts a proton from a serine which then attacks the carbonyl group of the amide or ester, and the amine or carboxylic acid is displaced. Next, a hydroxyl group attacks the carbonyl group of the enzyme-substrate complex, displacing the serine. Both enzymes are classified as hydrolases, acting either on a peptide bond (EC 3.4) or on an ester bond (EC 3.1). By using a measure of mechanism similarity, it is clear that the same mechanism is used to hydrolyse both functional groups. This similarity between enzymes may be due to the fact that the catalytic domains of both enzymes have similar structures (they have the same CATH code up to the topology level - 3.40.50.x), although they are not detectable homologues.
Figure 2(d).
The catalytic mechanisms of MACiE entries M0005, carboxypeptidase D, and M0094, 1-alkyl-2-acetylglycerophosphocholine esterase.
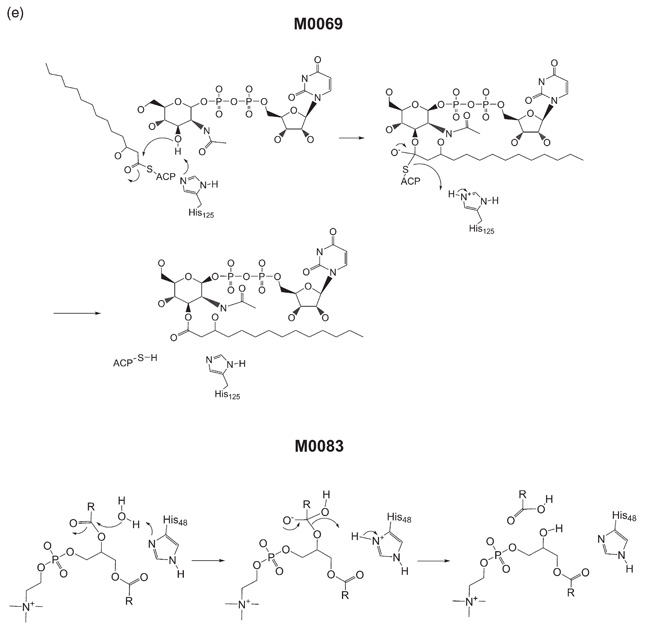
The apparent dissimilarity of the mechanisms of M006930 and M008331,32 (Figure 2(e)) according to the EC system highlights a deficiency of the hierarchical nature of that system. These enzymes differ at the class level of the EC. M0069 is a transesterification from an alcohol and a thioester to an ester and a thiol, and is classified as a transferase (EC 2). M0083 involves the hydrolysis of an ester and thus is classified as a hydrolase (EC 3). However, in terms of mechanism, the hydrolysis of an ester can be thought of as a special case of transesterification where the reacting alcohol is replaced by water, and the product ester is replaced by a carboxylic acid. As a review of the history of the EC nomenclature system points out, the division of enzymes into categories is somewhat artificial in that hydrolases in general could also be regarded as transferases.33 Although it may be consistent with how a biochemist thinks about reactions, this division can obscure the similarity between the mechanisms of related enzymes.
Figure 2(e).
The catalytic mechanisms of MACiE entries M0069, acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase, and M0083, phospholipase A2.
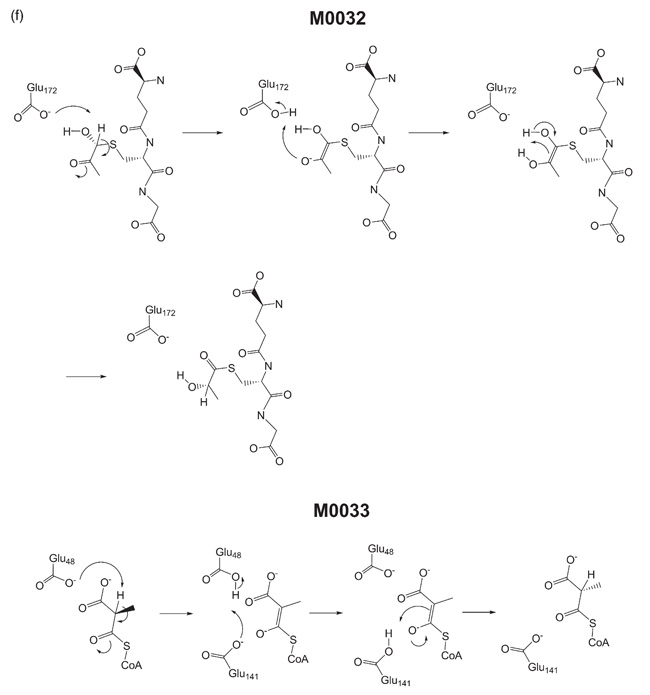
The direction in which a reaction occurs is not specified by the EC system; even if a reaction has only been observed in the reverse direction, it is written in the direction common to the subclass to which it belongs.33 M003234,35,36 (EC 4.4.1.5) and M003336,37 (EC 5.1.99.1) share the same CATH domain 3.10.180.10, and have similar mechanisms according to the BC and FP methods (Figure 2(f)), but their EC numbers differ at the class level. However, the classification of M0032 as a lyase acting on C-S bonds is probably misleading, as M0032 catalyses this reaction in the opposite direction (and entered as such in MACiE), and moreover, the enzyme does not catalyse the formation of the C-S bond – recent work has shown that the substrate of the enzyme is a thioacetal (formed spontaneously from methylglyoxal and glutathione).34 The overall reaction in M0032 is a 1,4-prototropic shift between a neighbouring alcohol and ketone, and so should probably be classed as an isomerase (EC class 5).
Figure 2(f).
The catalytic mechanisms of MACiE entries M0032, lactoylglutathione lyase, and M0033, methylmalonyl-CoA epimerase.
Searching using similarity methods
In addition to finding similarities between mechanisms that are already in the database, it is also possible to use similarity methods to search for enzyme reaction mechanisms that are similar to a query mechanism. This could be used by an experimentalist to find out whether there are known enzymes with similar mechanisms, as a step in the validation of a proposed mechanism. Using either the BC or FP method, it is quite easy to define a query. The alignment algorithm described in the Methods can then be applied to find similar reactions. In these searches, we are not looking for a complete match to the query reaction mechanism, but simply to find a reaction mechanism that contains the query. As a result, Equation 2 is not applied to the similarity value resulting from the alignment algorithm.
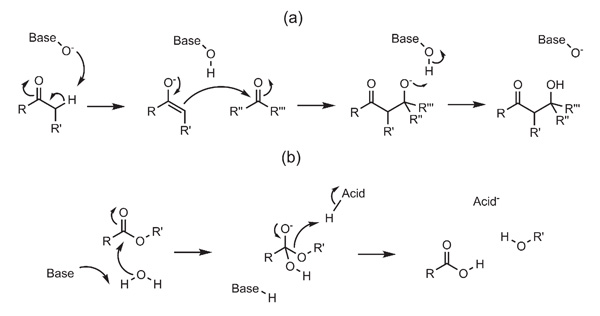
Figure 3(a) shows the mechanism of a base-catalysed aldol addition. Using the BC method to define a query, this can be described by the following sequence of bond changes:
Step 1, abstraction of α-H and formation of enolate: C=O → C-O, C-C → C=C, O-H formed, C-H cleaved
Step 2: enolate attack on carbonyl: C-O → C=O, C=C → C-C, C=O → C-O, C-C formed
Step 3: protonation of hydroxide: O-H formed, O-H cleaved
The MACiE database is annotated on a step-by-step basis. A mechanism step involving enolate attack on a carbonyl (Step 2 above) is annotated as an aldol addition. The top five matches in the database to a query consisting of Step 2 are the five mechanisms in the database that contain steps annotated as aldol additions. Two of these match exactly (M005338 and M007739,40; scores of 1.0), whereas the other three have a reduced score (M0052,41 M005942 and M007843; scores of 0.66) because the protonation step (Step 3 above) is included as part of the same step. Of the remaining five reactions in the top ten (M0089,44,45 0.5; M0081,46,47,48,49 0.43; M0031,50,51,52 0.43; M0070,53 0.33; M0051,54 0.33) two are of interest. M0070 is annotated as an “Assisted Keto-Enol Tautomerisation” and involves enolate attack on a base to form the ketone. M0031 is a possible false negative identified by this query; step 3 of M0031 involves enolate attack on the nitrogen analogue of a carbonyl, an imine. The score for this match is quite low compared to the true positives identified as it involves a C=N rather than a C=O, and in addition, the attacking group is an enol (rather than an enolate), which is deprotonated as part of the step.
Figure 3.
(a) A base-catalysed aldol addition. (b) A base-catalysed hydrolysis of an ester.
It is interesting to compare the results of the previous query with those of a query involving all of Steps 1 to 3 as described above. The top ten matches in the database include only three of the five annotated aldol additions in the dataset: M0052, M0053, and M0078 are found, but not M0059 or M0077. M0059 scores lower as the enolate is not formed in the typical way, but rather by cleavage of an OR bond, and protonation occurs as part of Step 2. In M0077, the enolate is formed as described in Step 1 above, but the abstracting base is RS− not RO−; in addition, instead of protonation of the hydroxide as Step 3, decomposition of the thiohemiketal intermediate occurs to give a ketone and a thiol. Six of the remaining top ten matches involve the formation of an enolate or enol (M0032, M0033, M0055,11,55 M0060,56,57 M006858,59 and M009560,61).
The base-catalysed hydrolysis of an ester (Figure 3(b)) can similarly be described in terms of bond changes, although there is some ambiguity as to the nature of the basic and acidic groups involved in proton abstraction and donation:
Step 1: C=O → C-O, base-H formed, C-O formed, O-H cleaved
Step 2: C-O → C=O, acid-H cleaved, C-O cleaved, O-H formed
If the base is a histidine and the acid is its protonated form, then Step 1 involves formation of an N-H bond, and Step 2 involves its cleavage. There are four exact matches in the database to this query: M0005, M0024,62,63 M0083 and M0094 (steps 4 and 5), all of which are valid hits. If instead we take the base as RO− and the acid as ROH, then there are two exact matches: M000264,65,66,67 and M0029,68 which again are valid. There is also an exact match, M0002, for the case where Step 1 involves formation of an N-H bond and Step 2 involves cleavage of an O-H bond, but this is not a valid hit as the two matching steps are not consecutive. A search was also made for a three step mechanism, where the proton abstraction occurs in a separate step to the attack on the carbonyl. Hits were found with both M0005 (steps 1, 2 and 5) and M0094 (steps 1, 2 and 3) in the case of the base being RNH, and the acid RN+, but only the latter involves consecutive steps.
Non-identical matches can also give an idea of how general a particular search query is, and whether it needs to be modified to pick up other relevant instances. The top ten matches to the initial query for the hydrolysis of an ester described above also include M0002 and M0029, which involve an oxygen-based acid and base rather than nitrogen-based ones. In addition, there are two matches to the hydrolysis of thioesters, and one to the hydrolysis of an amide ester.
Discussion
The two methods we have described in this paper for comparing mechanisms are based on different ideas for detection of similarity. Implicit in the FP method is the assumption that similar mechanisms will share features that are widely used to classify chemical reaction mechanisms. The BC method makes a weaker assumption; similar mechanisms will share a common sequence of bond changes. The examples of the use of these methods described in the Results section show that these methods are useful both for identifying meaningful similarities between reaction mechanisms, and for searching databases of reaction mechanisms with a query mechanism. Here we discuss differences between these methods, some of their shortcomings, as well as some possible ways to extend them to overcome these problems.
Although there is quite good agreement between the FP and BC methods regarding the pairs of reactions that are regarded as most similar in our dataset (Table 1), there are several examples of reactions that are highly ranked by one method but not the other. For example, the pair M0002 and M0029 is highly ranked by the BC method, but does not appear in the top twenty most similar pairs according to the FP method. These mechanisms both involve the hydrolysis of an amide bond, with a β-lactam as the substrate in M0002 and a linear amide in M0029. There are two main contributing factors to the difference in scores between the methods. Firstly, the C-N bond cleavage is intramolecular in M0002, but intermolecular in M0029, affecting the change in the number of species, the change in the number of cycles, as well as whether an enzyme substrate bond is cleaved. Secondly, the BC method is insensitive to differences in how protons are accounted for between the mechanisms (that is, although certain steps have the same bond formations/cleavages, the actual bonds involved are different). Although the FP measure of similarity has greater discriminating power compared to the BC method, it is also more prone to false positives. Of the pairs of reaction mechanisms shown in Table 1 that are not shared between the FP and BC methods, all but one pair (M0097, M0098) of those scored highly by the FP method may be regarded as false positives. That is, on inspection, the remaining pairs of mechanisms can not be regarded as similar. In contrast, none of the pairs of reactions in Table 1 identified as similar by the BC method are false positives.
While inspecting the results of the analysis of the MACiE database, a number of more sophisticated methods of measuring similarity occurred to us that would have resolved problems with particular false positives/negatives. The FP and BC methods discriminate between the identities of proton donors/acceptors where the reactive group is a hydroxyl (Glu, Asp), and those where the reactive group is an amine (His, Lys), due to the fact that an N-H bond is involved in one case but an O-H in the other. Since the exact identity of the proton donor/acceptor may not be important in assessing similarity, a better approach might be not to regard this as a normal bond change, but rather as a Donor-H bond change. For the FP method, rather than increment or decrement the charge on an oxygen or nitrogen, the charge on a Donor should be altered. As such bond changes are very common in MACiE, this could have a significant effect.
As an extension of the previous idea, we may also regard some bond changes as more similar than others. Both the FP and BC methods as we have described them do not consider the similarity of particular bond changes; that is, two bond changes are either identical or not. However, the formation of an N-H bond may be regarded as more similar to the formation of an O-H bond than to cleavage of a C-C bond. Both carbonyls and phosphate esters undergo similar chemistry which suggests that the decrease in bond order of a P=O bond might be regarded as similar to that of a C=O bond. At the other extreme, it may be useful to further discriminate between bond changes that involve different atom types. For example, the cleavage of a bond between a chlorine atom attached to an aromatic carbon atom would be regarded by an organic chemist as quite different to that involving a chlorine and a sp3-hybridised carbon. In this initial study, we have used simpler methods as we wished to avoid excessive parameterisation on a limited dataset.
Although the order of steps in a mechanism is, in principle, usually well-defined in terms of the lowest activation energy path between local minima on a free energy surface, there is some potential ambiguity in reported mechanisms. For example, it is sometimes not clear whether a particular proton transfer occurs before or after a particular bond formation or cleavage. Additionally, there are a few cases in our dataset where independent proton transfers are described as part of the same single step of a mechanism. Some descriptions of mechanisms may not adhere to common conventions; for example, it is usual to describe nucleophilic attack on a carbonyl group and subsequent loss of a leaving group as two sequential steps, but in some cases these are described as a single step. Such ambiguities will reduce the similarity values between two similar mechanisms unless the same conventions have been used. However, we note that in the querying examples discussed in the Results, the same reactions are identified despite minor modifications to the query, suggesting that the method is quite robust. It may be possible to allow for these ambiguities using a more sophisticated alignment procedure that allows the position of a particular proton transfer in a mechanism to be moved into a previous/subsequent step if it improves the match.
The two methods described in this paper should be regarded as starting points for the development of similarity measures for reaction mechanisms appropriate to particular studies, as there is no one solution appropriate for all studies. For example, a distance measure developed for enzyme reaction mechanisms would not be suitable for the reaction mechanisms found in typical organic chemistry reactions. In that case, an additional feature for the presence of a concerted reaction step might be appropriate, and radical reactions would be regarded as very dissimilar to heterolytic reactions.
Our measures have been developed with the MACiE database in mind, but for a study of the mechanisms of closely-related enzymes, a similarity measure focused on the specific features that differentiate those mechanisms might be more appropriate. An example of such a set of enzyme mechanisms is the enolase superfamily of the Structure-Function Linkage Database13 (SFLD). In this context, a superfamily is defined as a set of evolutionarily-related enzymes whose members retain a conserved aspect of function. The enolase superfamily conserves the ability to extract a proton alpha to a carboxylic acid to form an enolate intermediate. Based on the partial reactions given by the SFLD, the families contained in this superfamily are divided by the BC method into 4 sets: (a) galactonate dehydratase, L-fuconate dehydratase, glucarate dehydratase, osuccinylbenzoate synthase, enolase; (b) mandelate racemase, N-succinylamino acid racemase, muconate cycloisomerase, dipeptide epimerase; (c) methylaspartate ammonia-lyase; (d) chloromuconate cycloisomerase. Within each set, the mechanisms are identical. A comparison of sets (a), (b) and (c) gives pairwise similarities of 0.5, but set (d) is more distant (with similarities of 0.1, 0.3 and 0.2, respectively, with respect to sets (a), (b) and (c)). This division of the mechanisms into sets based on bond changes differs from the SFLD’s division of the enolase superfamily into subgroups based on conserved active site residues. For example, in the SFLD mandelate racemase is in a different subgroup from the other members of set (b). Since neither the BC method nor FP method incorporates any information regarding the identity of active site residues (beyond the identity of a donating or accepting atom), detailed comparisons of very similar enzymes may require the extension of these methods.
The EC system classifies enzymes based on the overall reaction catalysed, and does not consider mechanism. However, as shown by M0005 and M0094 in our dataset, enzymes with similar EC numbers often have similar mechanisms. In order to quantify the extent to which this occurs, we compared reaction mechanism similarity using the BC or FP method with similarity between overall transformations as classified by the EC. Table 2 shows that the greater the agreement between the EC numbers of two reactions, the more similar the mechanisms of the two reactions are in general. However, the first level of the EC (class) does not contain much information about the reaction mechanism, as the median rank for those reaction pairs sharing the same class is not much greater than the median rank for all reaction pairs. A large increase in the median rank occurs (in particular for the BC method) once the subclass is included. The subclass level includes generic information on the type of bond or group a particular enzyme acts on. The third level, the subsubclass, describes more specifically the type of bond or group the enzyme acts on, but according to Table 2 it does not in fact provide additional information regarding the mechanism.
Table 2.
The extent to which similarity according to the EC agrees with similarity in terms of mechanism.
| same ECa | Number of pairs | Median rank | |
|---|---|---|---|
| FP | BC | ||
| ≥0 (all) | 4950 | 2476 | 2476 |
| ≥1 | 1008 | 2384 | 2134 |
| ≥2 | 232 | 1747 | 1326 |
| ≥3 | 81 | 1670 | 1571 |
| 4 | 4 | 2595 | 1155 |
This is the level up to which the two reactions share the same EC classification.
Table 2 shows that out of 4950 pairs of reactions, there are four that share the same EC classification. The median rank of these reactions should not be regarded as significant both due to the small sample size and to the fact that these reactions have been chosen so that no two reactions share the same mechanism. Three of these pairs involve the β-lactamases M0002, M0015,69 and M0016,69 which share the EC classification 3.5.2.6. M0002 is a Class A β-lactamase which uses an activated serine to hydrolyse β-lactam rings. Its mechanism is different from those of the included Class B β-lactamases M0015 and M0016, both of which use zinc ions to catalyse the hydrolysis of the β-lactam by OH−. The mechanisms of M0016 and M0015 differ in that the former uses a single zinc ion to effect the reaction, whereas the latter uses two. The remaining pair of reactions that share the same EC number are M005411,70 and M0055, which are Type I and II 3-dehydroquinate dehydratases, respectively. These enzymes have no sequence similarity. Type I enzymes use a Schiff-base intermediate and catalyse the elimination of water with syn stereochemistry. Type II do not require metals or cofactors for activity, do not involve a covalent intermediate and catalyse the elimination reaction with trans stereochemistry.71
Conclusions
As more and more mechanistic data on enzymes become available, the ability to identify similar mechanisms in other enzymes is becoming more important. Such information may be used to identify mechanistically convergent or divergent enzymes, to study the link between structure and function, to perform literature searches, and to validate experimental results.
Here we have described two methods to measure the similarity of enzyme reaction mechanisms, a fingerprint (FP) method and a bond change (BC) method. When applied to the MACiE database of enzyme reactions, similarities between several reaction mechanisms are detected, often between enzymes with different EC classifications. The FP and BC methods place different emphasis on features of the mechanistic steps and result in slightly different measurements of similarity. In its current form the FP method is more susceptible to identifying false positives, although an appropriate weighting scheme may be able to overcome this problem.
Since the meaning of similarity depends on the context, we are continuing to search for better similarity methods for particular studies. The alignment method described here may be combined with any appropriate measure for the similarity of two reaction mechanism steps. Based on the methods described here we intend to provide a facility to query the MACiE database online, which will be a useful service for those elucidating the mechanisms of enzyme reactions.
Although the EC system plays a very important conceptual role in the classification of enzyme reactions, we hope that the ideas of enzyme mechanism similarity outlined here will encourage researchers to look beyond the hierarchical picture that the EC imposes and see that similarities and differences can exist between enzyme mechanisms whatever their classification.
Methods
Measurement of similarity of reaction steps
In our approach, the similarity of reactions depends upon the similarity of the reaction steps of the mechanisms. Any numerical measure of similarity of reaction steps is necessarily arbitrary and depends on the context. Here we describe two different methods for measuring the similarity of reaction steps, one of which takes into account a number of different features that are widely used in the classification of chemical reaction mechanisms (the FP method), and another which we regard as the simplest possible method that can yield a useful measure of similarity (the BC method).
In the fingerprint (FP) method, each reaction step in a reaction mechanism is represented by a list of 58 features which are used to measure the similarity between steps. The choice of features to include in the reaction step fingerprint affects how similar different reaction steps appear. Our approach was to include features that should allow reaction mechanisms of different Ingold classifications72 to be differentiated. In addition, we considered several features specific to enzyme reaction mechanisms, such as the presence of cofactors, and the formation or cleavage of enzyme substrate complexes. In general a reaction can be viewed as a change from the starting state (reactants) to the final state (products). Such a change in chemistry is characterised by the movement of electrons from their position in the reactants to their position in the products, with the following possible effects:
Bonds are changed in order. This is described by either bond formation (order change from 0 to 1), cleavage (order change from 1 to 0) or a bond order change (1 to 2, 2 to 1, and so on).
The formal charges on atoms increase or decrease.
Electron pairs are split up to form free radicals, or formed from two unpaired electrons.
In addition, there are other changes which can affect how a reaction might be classified:
A mechanistic step may result in a cyclic or a linear product. In the Ingold classification system this would be the difference between an intra- or intermolecular reaction. We include a feature for the change in the number of rings.
Ingold used the molecularity of the reaction in his classification, and it is therefore included in the FP method.
The full list of 58 features is as follows (all have integer values):
3 features: the number of reactants (the molecularity), the number of products minus the number of reactants, the number of cycles in the products minus the number of cycles in the reactants.
21 features: the total number of a particular bond type involved in a reaction. For example, the number of C-O bonds involved in the reaction.
4 features: Boolean values – is an enzyme substrate (ES) complex formed (1 feature) or cleaved (1 feature) or present in the reactants (1 feature)? Is a cofactor involved (1 feature)?
5 features: the total number of each type of bond order change: bond formation, bond cleavage, changes in order from 1 to 2, 2 to 1, 3 to 2.
16 features: for a particular atom type, the number that increase in charge as well as the number that decrease in charge.
4 features: Boolean values – are radicals involved (1 feature)? Does radical initiation (1 feature), propagation (1 feature), or termination (1 feature) occur?
5 features: Boolean values – does this step involve a radical based on N, C, O, S, or on another atom?
Once all of the reaction steps were encoded in this way, the Euclidean distances between reaction step fingerprints were calculated and stored in a pairwise distance matrix. Similarity values were derived from the distances by first normalising the distances to unity by dividing by the maximum distance value in the distance matrix for all the reactions in our dataset, and then subtracting the results from 1.
As an alternative measure of the similarity of a pair of reaction steps, we considered the simplest possible method that we thought could still yield useful information. In this bond change (BC) approach, we define similarity in terms of the bond changes that occur on going from the reactants to the products in each of the individual steps of the mechanism. Each reaction step is represented by a set of bond changes comprising the bond formations, bond cleavages, increases in bond order, and decreases in bond order associated with that step. Multiple bond changes of the same type are included multiple times. The similarity between two sets of bond changes is measured using the Tanimoto coefficient,73 T:
The Tanimoto coefficient has the characteristic that its value is in the range 0 to 1, and that in addition to scoring highly those reaction steps that share bond changes, it penalises bond changes that occur in one but not in the other.
Measurement of similarity of reaction mechanisms
Once pairwise similarity values have been calculated for the reaction steps of two reaction mechanisms, an overall reaction similarity can be calculated. First of all, corresponding reaction steps in the two reaction mechanisms must be matched up, or aligned, as well as possible. The problem of aligning two sets of reaction steps is analogous to that of aligning two DNA or protein sequences, a common and well-studied problem in bioinformatics.74,75 Very efficient methods have been developed to find the best alignment of two sequences.76,77 Some of the possible alignments include gaps. In protein sequences, our understanding of evolution suggests that the introduction of gaps due to insertions or deletions is less likely than the substitution of one amino acid by another. As a result, gaps are penalised and higher similarity values are found for sequences that align without gaps. In our approach for chemical reaction similarity, we will not penalise gaps. Not only is this the simplest approach, but it does not make any assumptions about how the similarity between a pair of reactions may have arisen. The Needleman-Wunsch algorithm for global alignment was used,76 using the reaction step pairwise similarity values instead of a substitution matrix.
The final step involves normalising the similarity value from the Needleman-Wunsch algorithm to take into account the maximum possible similarity that could have been calculated given the two sequences being compared. The equation used has the same form as that for the Tanimoto coefficient:
where Sij is the calculated similarity between two reaction mechanisms i and j, and NWij is the similarity score calculated from the alignment of reaction mechanisms i and j using the Needleman-Wunsch algorithm. Note that for the FP and BC methods NWii is equal to the number of reaction steps in reaction mechanism i.
Dataset
To test our proposed measures of reaction mechanism similarity, we used version 1.1 of the MACiE database18 of enzyme reaction mechanisms. MACiE 1.1 contains 100 reaction mechanisms (395 reaction steps) taken from the literature, covering 97 EC numbers and approximately one-third of EC subsubclasses for which mechanisms are available. Each reaction is well characterised, with a crystal structure deposited in the PDB, and a relatively well understood, or at least plausible, mechanism. The enzymes in MACiE are also unique at the fourth level of the CATH code (that is, they do not share a detectable common ancestor), except where the mechanism is significantly different.
The specific data used in this study is the version of MACiE current on 19 Sept 2006. A frozen copy of this dataset has been created, and is available as diagrams, annotation and Chemical Markup Language (CML) from http://www-mitchell.ch.cam.ac.uk/macie/JMBPaper.
Supplementary Material
Acknowledgements
We thank the EPSRC (G.L.H. and J.B.O.M.), the BBSRC (N.M.O.B. and J.B.O.M. - grant BB/C51320X/1), the Chilean Government’s Ministerio de Planificación y Cooperación (D.E.A) and Cambridge Overseas Trust (D.E.A.) for funding and Unilever for their financial support of the Centre for Molecular Science Informatics.
Footnotes
Supplementary Data Fingerprints and lists of bond changes for each of the 395 reactions steps, as well as matrices of FP and BC pairwise similarities for the 100 reaction mechanisms, are available as supplementary data.
References
- 1.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer ELL, Studholme DJ, Yeats C, Eddy SR. The Pfam protein families database. Nucl. Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Orengo CA, Michie AD, Jones S, Jones DT, Swindells MB, Thornton JM. CATH - a hierarchic classification of protein domain structures. Structure. 1997;5:1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- 3.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP - a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 4.Holm L, Sander C. Mapping the protein universe. Science. 1996;273:595–602. doi: 10.1126/science.273.5275.595. [DOI] [PubMed] [Google Scholar]
- 5.Nomenclature Committee of the International Union of Biochemistry and Molecular Biology. Webb EC. Enzyme Nomenclature: Recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology on the Nomenclature and Classification of Enzymes. 6th edn Academic Press; San Diego: 1992. [Google Scholar]
- 6.Kotera M, Okuno Y, Hattori M, Goto S, Kanehisa M. Computational assignment of the EC numbers for genomic-scale analysis of enzymatic reactions. J. Am. Chem. Soc. 2004;126:16487–16498. doi: 10.1021/ja0466457. [DOI] [PubMed] [Google Scholar]
- 7.Nagano N. EzCatDB: the Enzyme Catalytic-mechanism Database. Nucl. Acids Res. 2005;33:D407–D412. doi: 10.1093/nar/gki080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Babbitt PC, Gerlt JA. Understanding enzyme superfamilies. Chemistry as the fundamental determinant in the evolution of new catalytic activities. J. Biol. Chem. 1997;272:30591–30594. doi: 10.1074/jbc.272.49.30591. [DOI] [PubMed] [Google Scholar]
- 9.Todd AE, Orengo CA, Thornton JM. Evolution of function in protein superfamilies, from a structural perspective. J. Mol. Biol. 2001;307:1113–1143. doi: 10.1006/jmbi.2001.4513. [DOI] [PubMed] [Google Scholar]
- 10.Bartlett GJ, Borkakoti N, Thornton JM. Catalysing new reactions during evolution: economy of residues and mechanism. J. Mol. Biol. 2003;331:829–860. doi: 10.1016/s0022-2836(03)00734-4. [DOI] [PubMed] [Google Scholar]
- 11.Gourley DG, Shrive AK, Polikarpov I, Krell T, Coggins JR, Hawkins AR, Isaacs NW, Sawyer L. The two types of 3-dehydroquinase have distinct structures but catalyze the same overall reaction. Nat. Struct. Biol. 1999;6:521–525. doi: 10.1038/9287. [DOI] [PubMed] [Google Scholar]
- 12.Coutinho PM, Deleury E, Davies GJ, Henrissat B. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 2003;328:307–317. doi: 10.1016/s0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- 13.Pegg SC-H, Brown SD, Ojha S, Seffernick J, Meng EC, Morris JH, Chang PJ, Huang CC, Ferrin TE, Babbitt PC. Leveraging enzyme-structure function relationships for functional inference and experimental design: the Structure-Function Linkage Database. Biochemistry. 2006;45:2545–2555. doi: 10.1021/bi052101l. [DOI] [PubMed] [Google Scholar]
- 14.Gariev IA, Varfolomeev SD. Hierachical classification of hydrolases catalytic sites. Bioinformatics. 2006;22:2574–2576. doi: 10.1093/bioinformatics/btl413. [DOI] [PubMed] [Google Scholar]
- 15.Chen L, Gasteiger J. Knowledge discovery in reaction databases: landscaping organic reactions by a self-organizing neural network. J. Am. Chem. Soc. 1997;119:4033–4042. [Google Scholar]
- 16.Satoh H, Sacher O, Nakata T, Chen L, Gasteiger J, Funatsu K. Classification of organic reactions: similarity of reactions based on changes in the electronic features of oxygen atoms at the reaction sites. J. Chem. Inf. Comput. Sci. 1998;38:210–219. [Google Scholar]
- 17.Latino DARS, Aires-de-Sousa J. Genome-scale classification of metabolic reactions: a cheminformatics approach. Angew. Chem. Int. Ed. 2006;45:2066–2069. doi: 10.1002/anie.200503833. [DOI] [PubMed] [Google Scholar]
- 18.Holliday GL, Bartlett GJ, Almonacid DE, O’Boyle NM, Murray-Rust P, Thornton JM, Mitchell JBO. MACiE: a database of enzyme reaction mechanisms. Bioinformatics. 2005;21:4315–4316. doi: 10.1093/bioinformatics/bti693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marsh ENG, Drennan CL. Adenosylcobalamin-dependent isomerases: new insights into structure and mechanism. Curr. Opin. Chem. Biol. 2001;5:499–505. doi: 10.1016/s1367-5931(00)00238-6. [DOI] [PubMed] [Google Scholar]
- 20.Mancia F, Keep NH, Nakagawa A, Leadlay PF, McSweeney S, Rasmussen B, Böseke P, Diat O, Evans PR. How coenzyme B12 radicals are generated: the crystal structure of methylmalonyl-coenzyme A mutase at 2 A resolution. Structure. 1996;4:339–350. doi: 10.1016/s0969-2126(96)00037-8. [DOI] [PubMed] [Google Scholar]
- 21.Heinz DW, Essen LO, Williams RL. Structural and mechanistic comparison of prokaryotic and eukaryotic phosphoinositide-specific phospholipases C. J. Mol. Biol. 1998;275:635–650. doi: 10.1006/jmbi.1997.1490. [DOI] [PubMed] [Google Scholar]
- 22.Hondal RJ, Zhao Z, Kravchuk AV, Liao H, Riddle SR, Yue X, Bruzik KS, Tsai M-D. Mechanism of phosphatidylinositol-specific phospholipase C: a unified view of the mechanism of catalysis. Biochemistry. 1998;37:4568–4580. doi: 10.1021/bi972646i. [DOI] [PubMed] [Google Scholar]
- 23.Ryan M, Liu T, Dahlquist FW, Griffith OH. A catalytic diad involved in substrate-assisted catalysis: NMR study of hydrogen bonding and dynamics at the active site of phosphatidylinositol-specific phospholipase C. Biochemistry. 2001;40:9743–9750. doi: 10.1021/bi010958m. [DOI] [PubMed] [Google Scholar]
- 24.Jones SJ, Worrall AF, Connolly BA. Site-directed mutagenesis of the catalytic residues of bovine pancreatic deoxyribonuclease I. J. Mol. Biol. 1996;264:1154–1163. doi: 10.1006/jmbi.1996.0703. [DOI] [PubMed] [Google Scholar]
- 25.Martin SF, Hergenrother PF. Catalytic cycle of the phosphatidylcholine-preferring phospholipase C from Bacillus cereus. Solvent viscosity, deuterium isotope effects, and proton inventory studies. Biochemistry. 1999;38:4403–4408. doi: 10.1021/bi9821216. [DOI] [PubMed] [Google Scholar]
- 26.Skamnaki VT, Owen DJ, Noble ME, Lowe ED, Lowe G, Oikonomakos NG, Johnson LN. Catalytic mechanism of phosphorylase kinase probed by mutational studies. Biochemistry. 1999;38:14718–14730. doi: 10.1021/bi991454f. [DOI] [PubMed] [Google Scholar]
- 27.Bullock TL, Branchaud B, Remington SJ. Structure of the complex of L-benzylsuccinate with wheat serine carboxypeptidase II at 2.0-A resolution. Biochemistry. 1994;33:11127–11134. doi: 10.1021/bi00203a009. [DOI] [PubMed] [Google Scholar]
- 28.Ho YS, Sheffield PJ, Masuyama J, Arai H, Li J, Aoki J, Inoue K, Derewenda U, Derewenda ZS. Probing the substrate specificity of the intracellular brain platelet-activating factor acetylhydrolase. Protein Eng. 1999;12:693–700. doi: 10.1093/protein/12.8.693. [DOI] [PubMed] [Google Scholar]
- 29.Ho YS, Swenson L, Derewenda U, Serre L, Wei Y, Dauter Z, Hattori M, Adachi T, Aoki J, Arai H, Inoue K, Derewenda ZS. Brain acetylhydrolase that inactivates platelet-activating factor is a G-protein-like trimer. Nature. 1997;385:89–93. doi: 10.1038/385089a0. [DOI] [PubMed] [Google Scholar]
- 30.Wyckoff TJ, Raetz RC. The active site of Escherichia coli UDP-N-acetylglucosamine acyltransferase. Chemical modification and site-directed mutagenesis. J. Biol. Chem. 1999;274:27047–27055. doi: 10.1074/jbc.274.38.27047. [DOI] [PubMed] [Google Scholar]
- 31.Scott DL, Sigler PB. Structure and catalytic mechanism of secretory phospholipases A2. Adv. Protein Chem. 1994;45:53–88. doi: 10.1016/s0065-3233(08)60638-5. [DOI] [PubMed] [Google Scholar]
- 32.Segelke BW, Nguyen D, Chee R, Xuong NH, Dennis EA. Structures of two novel crystal forms of Naja naja naja phospholipase A2 lacking Ca2+ reveal trimeric packing. J. Mol. Biol. 1998;279:223–232. doi: 10.1006/jmbi.1998.1759. [DOI] [PubMed] [Google Scholar]
- 33.Tipton K, Boyce S. History of the enzyme nomenclature system. Bioinformatics. 2000;16:34–40. doi: 10.1093/bioinformatics/16.1.34. [DOI] [PubMed] [Google Scholar]
- 34.Cameron AD, Ridderström M, Olin B, Kavarana MJ, Creighton DJ, Mannervik B. Reaction mechanism of glyoxalase I explored by an X-ray crystallographic analysis of the human enzyme in complex with a transition state analogue. Biochemistry. 1999;38:13480–13490. doi: 10.1021/bi990696c. [DOI] [PubMed] [Google Scholar]
- 35.Himo F, Siegbahn PEM. Catalytic mechanism of glyoxalase I: a theoretical study. J. Am. Chem. Soc. 2001;123:10280–10289. doi: 10.1021/ja010715h. [DOI] [PubMed] [Google Scholar]
- 36.Armstrong RN. Mechanistic diversity in a metalloenzyme superfamily. Biochemistry. 2000;39:13625–13632. doi: 10.1021/bi001814v. [DOI] [PubMed] [Google Scholar]
- 37.McCarthy AA, Baker HM, Shewry SC, Patchett ML, Baker EN. Crystal structure of methylmalonyl-coenzyme A epimerase from P. shermanii: a novel enzymatic function on an ancient metal binding scaffold. Structure. 2001;9:637–646. doi: 10.1016/s0969-2126(01)00622-0. [DOI] [PubMed] [Google Scholar]
- 38.Howard BR, Endrizzi JA, Remington SJ. Crystal structure of Escherichia coli malate synthase G complexed with magnesium and glyoxylate at 2.0 A resolution: mechanistic implications. Biochemistry. 2000;39:3156–3168. doi: 10.1021/bi992519h. [DOI] [PubMed] [Google Scholar]
- 39.Mathieu M, Modis Y, Zeelen J. Ph., Engel CK, Abagyan RA, Ahlberg A, Rasmussen B, Lamzin VS, Kunau WH, Wierenga RK. The 1.8 A crystal structure of the dimeric peroxisomal 3-ketoacyl-CoA thiolase of Saccharomyces cerevisiae: implications for substrate binding and reaction mechanism. J. Mol. Biol. 1997;273:714–728. doi: 10.1006/jmbi.1997.1331. [DOI] [PubMed] [Google Scholar]
- 40.Modis Y, Wierenga RK. Crystallographic analysis of the reaction pathway of Zoogloea ramigera biosynthetic thiolase. J. Mol. Biol. 2000;297:1171–1182. doi: 10.1006/jmbi.2000.3638. [DOI] [PubMed] [Google Scholar]
- 41.Hall DR, Leonard GA, Reed CD, Watt CI, Berry A, Hunter WN. The crystal structure of Escherichia coli class II fructose-1, 6-bisphosphate aldolase in complex with phosphoglycolohydroxamate reveals details of mechanism and specificity. J. Mol. Biol. 1999;287:383–394. doi: 10.1006/jmbi.1999.2609. [DOI] [PubMed] [Google Scholar]
- 42.Carpenter EP, Hawkins AR, Frost JW, Browns KA. Structure of dehydroquinate synthase reveals an active site capable of multistep catalysis. Nature. 1998;394:299–302. doi: 10.1038/28431. [DOI] [PubMed] [Google Scholar]
- 43.Karpusas M, Branchaud B, Remington SJ. Proposed mechanism for the condensation reaction of citrate synthase: 1.9-A structure of the ternary complex with oxaloacetate and carboxymethyl coenzyme A. Biochemistry. 1990;29:2213–2219. [PubMed] [Google Scholar]
- 44.Lesburg CA, Zhai G, Cane DE, Christianson DW. Crystal structure of pentalenene synthase: mechanistic insights on terpenoid cyclization reactions in biology. Science. 1997;277:1820–1824. doi: 10.1126/science.277.5333.1820. [DOI] [PubMed] [Google Scholar]
- 45.Seemann M, Zhai G, Umezawa K, Cane D. Pentalenene synthase. Histidine-309 is not required for catalytic activity. J. Am. Chem. Soc. 1999;121:591–592. [Google Scholar]
- 46.Strater N, Schnappauf G, Braus G, Lipscomb WN. Mechanisms of catalysis and allosteric regulation of yeast chorismate mutase from crystal structures. Structure. 1997;5:1437–1452. doi: 10.1016/s0969-2126(97)00294-3. [DOI] [PubMed] [Google Scholar]
- 47.Ma J, Zheng X, Schnappauf G, Braus G, Karplus M, Lipscomb WN. Yeast chorismate mutase in the R state: simulations of the active site. Proc. Natl. Acad. Sci. USA. 1998;95:14640–14645. doi: 10.1073/pnas.95.25.14640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martí S, Andrés J, Moliner V, Silla E, Tuñón I, Bertrán J. Preorganization and reorganization as related factors in enzyme catalysis: the chorismate mutase case. Chem. Eur. J. 2003;9:984–991. doi: 10.1002/chem.200390121. [DOI] [PubMed] [Google Scholar]
- 49.Xue Y, Lipscomb WN. Location of the active site of allosteric chorismate mutase from Saccharomyces cerevisiae, and comments on the catalytic and regulatory mechanisms. Proc. Natl. Acad. Sci. USA. 1995;92:10595–10598. doi: 10.1073/pnas.92.23.10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Finer-Moore JS, Santi DV, Stroud RM. Lessons and conclusions from dissecting the mechanism of a bisubstrate enzyme: thymidylate synthase mutagenesis, function, and structure. Biochemistry. 2003;42:248–256. doi: 10.1021/bi020599a. [DOI] [PubMed] [Google Scholar]
- 51.Hyatt DC, Maley F, Montfort WR. Use of strain in a stereospecific catalytic mechanism: crystal structures of Escherichia coli thymidylate synthase bound to FdUMP and methylenetetrahydrofolate. Biochemistry. 1997;36:4585–4594. doi: 10.1021/bi962936j. [DOI] [PubMed] [Google Scholar]
- 52.Chiericatti G, Santi DV. Aspartate 221 of thymidylate synthase is involved in folate cofactor binding and in catalysis. Biochemistry. 1998;37:9038–9042. doi: 10.1021/bi9802770. [DOI] [PubMed] [Google Scholar]
- 53.Benning MM, Haller T, Gerlt JA, Holden HM. New reactions in the crotonase superfamily: structure of methylmalonyl CoA decarboxylase from Escherichia coli. Biochemistry. 2000;39:4630–4639. doi: 10.1021/bi9928896. [DOI] [PubMed] [Google Scholar]
- 54.Matte A, Tari LW, Goldie H, Delbaere LT. Structure and mechanism of phosphoenolpyruvate carboxykinase. J. Biol. Chem. 1997;272:8105–8108. doi: 10.1074/jbc.272.13.8105. [DOI] [PubMed] [Google Scholar]
- 55.Roszak AW, Robinson DA, Krell T, Hunter IS, Fredrickson M, Abell C, Coggins JR, Lapthorn AJ. The structure and mechanism of the type II dehydroquinase from Streptomyces coelicolor. Structure. 2002;10:493–503. doi: 10.1016/s0969-2126(02)00747-5. [DOI] [PubMed] [Google Scholar]
- 56.Oliva G, Fontes MRM, Garratt RC, Altamirano MM, Calcagno ML, Horjales E. Structure and catalytic mechanism of glucosamine 6-phosphate deaminase from Escherichia coli at 2.1 A resolution. Structure. 1995;3:1323–1332. doi: 10.1016/s0969-2126(01)00270-2. [DOI] [PubMed] [Google Scholar]
- 57.Montero-Moran GM, Lara-Gonzalez S, Alvarez-Anorve LI, Plumbridge JA, Calcagno ML. On the multiple functional roles of the active site histidine in catalysis and allosteric regulation of Escherichia coli glucosamine 6-phosphate deaminase. Biochemistry. 2001;40:10187–10196. doi: 10.1021/bi0105835. [DOI] [PubMed] [Google Scholar]
- 58.Tiffany KA, Roberts DL, Wang M, Paschke R, Mohsen AW, Vockley J, Kim JJ. Structure of human isovaleryl-CoA dehydrogenase at 2.6 A resolution: structural basis for substrate specificity. Biochemistry. 1997;36:8455–8464. doi: 10.1021/bi970422u. [DOI] [PubMed] [Google Scholar]
- 59.Thorpe C, Kim JJ. Structure and mechanism of action of the acyl-CoA dehydrogenases. FASEB J. 1995;9:718–725. doi: 10.1096/fasebj.9.9.7601336. [DOI] [PubMed] [Google Scholar]
- 60.Seemann JE, Schulz GE. Structure and mechanism of L-fucose isomerase from Escherichia coli. J. Mol. Biol. 1997;273:256–268. doi: 10.1006/jmbi.1997.1280. [DOI] [PubMed] [Google Scholar]
- 61.Collyer CA, Blow DM. Observations of reaction intermediates and the mechanism of aldose-ketose interconversion by D-xylose isomerase. Proc. Natl. Acad. Sci. USA. 1990;87:1362–1366. doi: 10.1073/pnas.87.4.1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang G, Liu RQ, Taylor KL, Xiang H, Price J, Dunaway-Mariano D. Identification of active site residues essential to 4-chlorobenzoyl-coenzyme A dehalogenase catalysis by chemical modification and site directed mutagenesis. Biochemistry. 1996;35:10879–10885. doi: 10.1021/bi9609533. [DOI] [PubMed] [Google Scholar]
- 63.Zheng Y-J, Bruice TC. On the dehalogenation mechanism of 4-chlorobenzoyl CoA by 4-chlorobenzoyl CoA dehalogenase: Insights from study based on the nonenzymatic reaction. J. Am. Chem. Soc. 1997;119:3868–3877. [Google Scholar]
- 64.Maveyraud L, Pratt RF, Samama JP. Crystal structure of an acylation transition-state analog of the TEM-1 beta-lactamase. Mechanistic implications for class A beta-lactamases. Biochemistry. 1998;37:2622–2628. doi: 10.1021/bi972501b. [DOI] [PubMed] [Google Scholar]
- 65.Pitarch J, Pascual-Ahuir J-L, Silla E, Tunon I. A quantum mechanics/molecular mechanics study of the acylation reaction of TEM1 beta-lactamase and penicillanate. J. Chem. Soc. Perkin Trans. 2000;2:761–767. [Google Scholar]
- 66.Castillo R, Silla E, Tunon I. Role of protein flexibility in enzymatic catalysis: Quantum mechanical-molecular mechanical study of the deacylation reaction in class A beta-lactamases. J. Am. Chem. Soc. 2002;124:1809–1816. doi: 10.1021/ja017156z. [DOI] [PubMed] [Google Scholar]
- 67.Hermann JC, Ridder L, Mulholland AJ, Holtje H-D. Identification of Glu166 as the general base in the acylation reaction of class A beta-lactamases through QM/MM modeling. J. Am. Chem. Soc. 2003;125:9590–9591. doi: 10.1021/ja034434g. [DOI] [PubMed] [Google Scholar]
- 68.Ortlund E, Lacount MW, Lewinski K, Lebioda L. Reactions of Pseudomonas 7A glutaminase-asparaginase with diazo analogues of glutamine and asparagine result in unexpected covalent inhibitions and suggests an unusual catalytic triad Thr-Tyr-Glu. Biochemistry. 2000;39:1199–1204. doi: 10.1021/bi991797d. [DOI] [PubMed] [Google Scholar]
- 69.Wang Z, Fast W, Valentine AM, Benkovic SJ. Metallo-beta-lactamase: structure and mechanism. Curr. Opin. Chem. Biol. 1999;3:614–622. doi: 10.1016/s1367-5931(99)00017-4. [DOI] [PubMed] [Google Scholar]
- 70.Leech AP, Boetzel R, McDonald C, Shrive AK, Moore GR, Coggins JR, Sawyer L, Kleanthous C. Re-evaluating the role of His-143 in the mechanism of type I dehydroquinase from Escherichia coli using two-dimensional 1H,13C NMR. J. Biol. Chem. 1998;273:9602–9607. doi: 10.1074/jbc.273.16.9602. [DOI] [PubMed] [Google Scholar]
- 71.Kwak JE, Lee JY, Han BW, Moon J, Sohn SH, Suh SW. Crystallization and preliminary X-ray crystallographic analysis of type II dehydroquinase from Helicobacter pylori. Acta Cryst. D. 2001;57:279–280. doi: 10.1107/s0907444900016267. [DOI] [PubMed] [Google Scholar]
- 72.Ingold CK. Structure and Mechanism in Organic Chemistry. 2nd edn Cornell University Press; Ithaca, New York: 1969. [Google Scholar]
- 73.Jaccard P. La distribution de la flore dans la zone alpine. Rev. Gen. Sci. Pures Appl. 1907;18:961–967. [Google Scholar]
- 74.Durbin R, Eddy SR, Krogh A, Mitchison G. Biological Sequence Analysis. Cambridge University Press; Cambridge: 1998. [Google Scholar]
- 75.Waterman MS. Introduction to Computational Biology: Maps, Sequences, and Genomes: Interdisciplinary Statistics. Chapman and Hall/CRC; Boca Raton, Florida: 1995. [Google Scholar]
- 76.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 77.Smith TF, Waterman MS. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.