Abstract
Recently, heterozygous mutations in GATA6 have been found in neonatal diabetic patients with failed pancreatic organogenesis. To investigate the roles of GATA4 and GATA6 in mouse pancreas organogenesis, we conditionally inactivated these genes within the pancreas. Single inactivation of either gene did not have a major impact on pancreas formation, indicating functional redundancy. However, double Gata4/Gata6 mutant mice failed to develop pancreata, died shortly after birth, and displayed hyperglycemia. Morphological defects in Gata4/Gata6 mutant pancreata were apparent during embryonic development, and the epithelium failed to expand as a result of defects in cell proliferation and differentiation. The number of multipotent pancreatic progenitors, including PDX1+ cells, was reduced in the Gata4/Gata6 mutant pancreatic epithelium. Remarkably, deletion of only 1 Gata6 allele on a Gata4 conditional knockout background severely reduced pancreatic mass. In contrast, a single WT allele of Gata4 in Gata6 conditional knockout mice was sufficient for normal pancreatic development, indicating differential contributions of GATA factors to pancreas formation. Our results place GATA factors at the top of the transcriptional network hierarchy controlling pancreas organogenesis.
Introduction
In vertebrates, the GATA zinc finger transcription factor family comprises 6 members that recognize the consensus-binding site A/T-GATA-A/G in the promoter regions of multiple genes (1). Studies have shown that GATA1, GATA2, and GATA3 are involved in hematopoietic cell differentiation, whereas GATA4, GATA5, and GATA6 control specification and differentiation of mesoderm- and endoderm-derived cell types (2, 3). In humans, GATA4 and GATA6 mutations have been reported to cause congenital heart diseases (4–7), highlighting the crucial roles of these factors in embryonic development. More recently, genome sequencing of 27 neonatal diabetic patients with pancreatic agenesis or severe pancreas hypoplasia revealed that half of the patients had spontaneous heterozygous mutations in the GATA6 gene (8). These findings add GATA6 to the short list of genes, including PDX1 and PTF1A, identified to date whose inactivation causes pancreatic agenesis in humans and indicates a role for GATA6 in pancreas morphogenesis.
Studies in mice have shown that GATA6 is broadly coexpressed with GATA4 in the early foregut endoderm and later throughout the nascent pancreatic epithelium (9). As embryonic development proceeds and pancreatic cells differentiate into endocrine and exocrine lineages, Gata4 and Gata6 expression become mutually exclusive. Gata4 is expressed in digestive enzyme–producing acinar cells, whereas Gata6 expression is restricted to endocrine islets (10). The dynamic expression pattern of Gata4 and Gata6 during pancreatic development suggests a functional role for these transcription factors in pancreas development. Efforts to elucidate the role of GATA4 and GATA6 in pancreas formation have been hampered by the early embryonic lethality of Gata4- and Gata6-null mice. Gata4-null mice die at E8.5, exhibiting defects in the heart tube, extraembryonic endoderm, and ventral foregut morphogenesis (11, 12). Similarly, Gata6-null mice do not develop beyond gastrulation due to defects in extraembryonic tissues and die in utero (13).
Several strategies have been used to circumvent the early embryonic lethality of Gata4- and Gata6-null mice. By using tetraploid complementation approaches, it has been shown that Gata4–/– or Gata6–/– mutant embryos fail to induce the pancreatic developmental program (14). However, it remains unclear whether these phenotypes reflect a cell-autonomous role of GATA factors in pancreas development. In other studies, transgenic mice were engineered to overexpress GATA4 or GATA6 protein fused to the repressor domain of the Drosophila engrailed protein in the Pdx1 expression domain. These mice exhibited a total absence or severe reduction of pancreatic mass (9). Together, these studies point to an autonomous role of GATA factors in embryonic pancreas development.
To determine whether GATA4 and GATA6 play a functional role in pancreas formation, we have conditionally inactivated these genes in the pancreatic progenitor domain via Cre/lox technology. While the lack of either GATA4 or GATA6 activity appears not to have a major impact in pancreas morphogenesis, the inactivation of both Gata4 and Gata6 in the pancreatic primordia leads to pancreatic agenesis, indicating a functional redundancy for these transcription factors during pancreas formation. The pancreatic epithelium fails to grow, and pancreatic cell proliferation is reduced in Gata4/Gata6 double-mutant mice. Cell commitment toward endocrine and exocrine lineages is also severely impaired, and expression of several multipotent pancreatic progenitor markers, including Pdx1, is reduced in the absence of GATA4 and GATA6 activity. Remarkably, deletion of only 1 allele of Gata6 in a Gata4 conditional knockout background results in severe pancreas hypoplasia. In contrast, a single allele of Gata4 in the absence of Gata6 WT alleles is sufficient for normal pancreatic development, indicating that GATA4 and GATA6 are not completely equivalent in regulating pancreas formation. We demonstrate that GATA4 and GATA6 transcription factors bind to 2 conserved GATA sites in the conserved area III of Pdx1 by ChIP and EMSA. Furthermore, these GATA sites are required for Pdx1 promoter activity in transgenic mice. Together, our results demonstrate a crucial role for GATA factors in pancreas formation and identify Pdx1 as a direct target of GATA factors.
Results
Single inactivation of Gata4 and Gata6 does not affect pancreas formation.
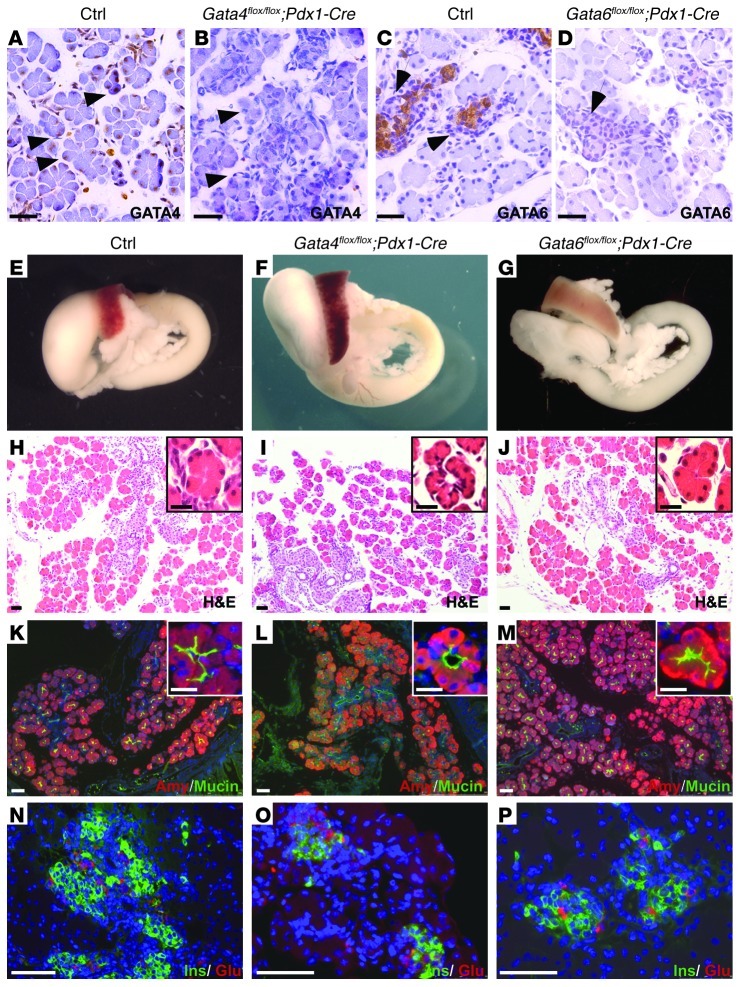
To investigate the role of GATA4 and GATA6 in pancreas development, mice with a conditional (flox) allele of Gata4 (Gata4flox/flox) or Gata6 (Gata6flox/flox) were crossed to a transgenic mouse line that expresses Cre recombinase under the control of the pancreatic and duodenal homeobox gene 1 (Pdx1) promoter (Pdx1-Cre mice) (15). Pdx1 is expressed early in the multipotent pancreatic progenitors that give rise to all pancreatic cell types, thus allowing gene inactivation in the entire pancreas (15). Expression analysis of Cre activity in Pdx1-Cre;Rosa26R transgenic mice revealed robust lacZ staining in the pancreatic epithelium as early as E9.5 (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI63240DS1). Gata4flox/flox and Gata6flox/flox mice have been used previously to successfully analyze the role of these transcription factors in other organs, such as heart and lung (16, 17). Gata4flox/flox;Pdx1-Cre and Gata6flox/flox;Pdx1-Cre mice were born at the expected Mendelian frequency and appeared overtly normal. Immunohistochemical staining demonstrated an efficient recombination of Gata4 and Gata6 floxed alleles by Cre recombinase in pancreatic tissue (Figure 1, A–D). Gross morphological examination and histological analysis by H&E staining of newborn pancreata did not reveal obvious defects in either Gata4flox/flox;Pdx1-Cre or Gata6flox/flox;Pdx1-Cre mice when compared with control littermates (Figure 1, E–J). Occasional, mild ductal dilation in acinar cells of Gata4flox/flox;Pdx1-Cre newborn pancreata was detected (Figure 1I). However, this phenotype was not observed in adult mice (data not shown), indicating a transient role for GATA4 in early exocrine formation. Immunohistochemical analysis for different epithelial cell lineages (acinar, ductal, and islet) within the pancreas revealed that pancreatic cell differentiation was unaffected in Gata4flox/flox;Pdx1-Cre and Gata6flox/flox;Pdx1-Cre mutant mice (Figure 1, K–P). Furthermore, Gata4flox/flox;Pdx1-Cre and Gata6flox/flox;Pdx1-Cre mice exhibited normal glucose tolerance to intraperitoneal glucose load (Supplemental Figure 2). Taken together, our results show that pancreas morphogenesis is not affected by inactivation of either GATA4 or GATA6 alone.
Figure 1. Single inactivation of Gata4 and Gata6 does not affect pancreas formation.
(A–D) Immunohistochemical analyses show strong expression of GATA4 in acinar cells (arrowheads in A) and GATA6 in endocrine cells (arrows in C) in pancreatic sections of control mice at P1. Loss of GATA4 (B, arrowheads) and GATA6 (D, arrows) in newborn conditional mutant mice is confirmed by immunohistochemical analysis. (E–G) Gross morphology of neonatal WT and conditional mutant guts. H&E-stained sections of newborn control (H), Gata4 (I), and Gata6 (J) conditional knockout pancreata does not reveal major defects in pancreas architecture. Mature acinar (amylase), ductal (mucin) (K–M), and islet (insulin and glucagon) markers (N–P) are normally expressed in single Gata4 and Gata6 conditional knockout mice. Insets in H–M show higher magnification of acinar cells. Mild ductal dilation in acinar cells of Gata4flox/flox;Pdx1-Cre is observed (I and L, insets). Nuclei are counterstained with DAPI in K–P. Scale bars: 50 μm; 25 μm (insets).
Pancreatic agenesis in GATA4 and GATA6 double-mutant mice.
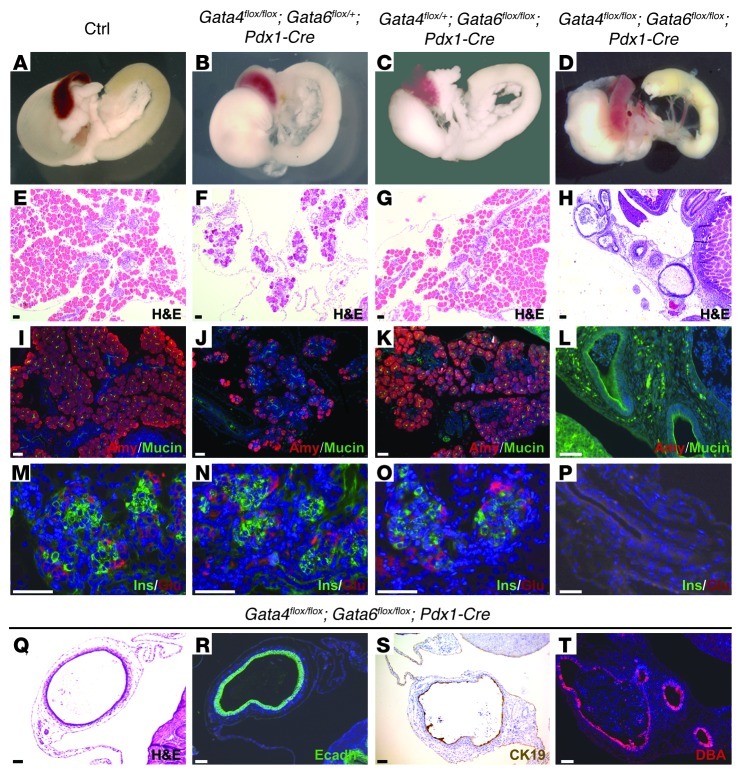
Previous studies have shown that Gata4 and Gata6 have overlapping expression patterns at early stages of pancreas formation, when the pancreatic epithelium comprises mainly multipotent pancreatic progenitor cells (MPCs) (9) (see also Supplemental Figure 4). Since pancreas formation is unaffected in individual Gata4 and Gata6 conditional knockout mice, we reasoned that these transcription factors might have redundant roles during pancreas development. To test this hypothesis, we generated double Gata4flox/flox;Gata6flox/flox;Pdx1-Cre mutant mice (double-mutant mice hereafter). Double-mutant mice displayed growth retardation and hyperglycemia and died shortly after birth (data not shown). Dissection of newborn double-mutant mice revealed a near total absence of pancreatic tissue (Figure 2D). Close histological analysis of the pancreatic remnant revealed the presence of epithelial cysts and abundant stroma (Figure 2, H and Q), while exocrine and endocrine tissue were almost completely absent (Figure 2, L and P). Cysts comprised epithelial cells that express mature ductal markers, including cytokeratin 19 (Figure 2S), and react with lectin Dolichos biflorus agglutinin (DBA) (Figure 2T). Although the majority of double-mutant mice displayed pancreatic agenesis, the degree of pancreatic tissue loss was variable among individual mutant pups, with approximately 20% of double-mutant mice displaying severe pancreatic hypoplasia. Histological analysis of hypoplastic pancreata revealed well-differentiated tissue (Supplemental Figure 3). However, GATA4 and GATA6 expression was still observed in these cells, indicating inefficient excision of Gata4 and Gata6 floxed alleles, perhaps relating to the number of floxed alleles requiring recombination (Supplemental Figure 2 and data not shown). Pdx1-Cre activity in pancreas is reported to be mosaic (15), and thus it is conceivable that unrecombined Gata4+Gata6+ progenitor cells might have partially repopulated the pancreas in double-mutant mice. The phenotype of the double-mutant mice suggests that loss of both GATA4 and GATA6 confers a competitive disadvantage to developing pancreatic cells. A dramatic reduction in pancreatic mass was also observed in Gata4flox/flox;Gata6flox/+;Pdx1-Cre mutant mice (Figure 2, B and F), indicating that pancreas formation is sensitive to reduced Gata6 dosage. Gata4flox/flox;Gata6flox/+;Pdx1-Cre mutant mice displayed severe acinar cell loss, and the architecture of remaining acini appeared impaired (Figure 2, F and J). Interestingly, tissue architecture of Gata4flox/+;Gata6flox/flox;Pdx1-Cre mutant pancreata was completely normal (Figure 2, C, G, K, and O), indicating that GATA4 and GATA6 are not completely equivalent in regulating pancreas formation. Together, our results indicate that pancreatic morphogenesis requires GATA4 and GATA6 activity and that these transcription factors play redundant functions in this process.
Figure 2. Pancreatic agenesis in Gata4/Gata6 double mutant.
Gross appearance of neonatal WT and conditional mutant guts (A–D) and pancreatic sections stained with H&E (E–H) reveal the abnormal morphology of double-mutant pancreata at P1. Gata4flox/flox;Gata6flox/+;Pdx1-Cre mice show pancreatic hypoplasia with scarcity of acinar cells (B and F). Gata4flox/+;Gata6flox/flox;Pdx1-Cre mice display normal pancreatic mass and architecture (C and G). Immunohistochemical analysis shows reduced expression of the acinar marker, amylase, in Gata4flox/flox;Gata6flox/+;Pdx1-Cre pancreatic sections (J) compared with Gata4flox/+;Gata6flox/flox;Pdx1-Cre (K) and control littermates (I). The double-mutant pancreatic remnant displays cystic structures surrounded by abundant stroma (H, L, P, and Q). The cystic structures express mucin (L) and cytokeratin 19 (S) and react with DBA lectin (T), which are markers of differentiated ductal cells. Immunostaining for E-cadherin confirms the epithelial nature of the cysts (R). Insulin and glucagon staining reveals normal differentiation of the endocrine lineage in Gata4flox/flox;Gata6flox/+;Pdx1-Cre (N) and Gata4flox/+;Gata6flox/flox;Pdx1-Cre (O) mutant mice in comparison with control mice (M). In contrast, Gata4/Gata6 double-mutant mice lack endocrine cells (P). Counterstaining with DAPI was performed to reveal nuclei. Scale bars: 50 μm.
Pancreatic epithelium of Gata4/Gata6 double-mutant embryos fails to expand.
To delineate the role of GATA4 and GATA6 during pancreas development, we characterized the pancreatic morphology of double-mutant mice at different developmental stages. During the early stages of pancreatic development, when bud formation occurs (E11.5), Gata4 and Gata6 expression completely overlapped in the pancreatic epithelium (Supplemental Figure 4). However, no major morphological changes in pancreatic epithelia of the double-mutant mice were observed (Supplemental Figure 4). Efficient inactivation of the Gata4 and Gata6 genes by Cre recombinase at this developmental stage was confirmed by immunohistochemistry (Supplemental Figure 4). By E13.5, the pancreatic epithelium expands and undergoes extensive branching to form a network of tubules. Concomitantly, a massive wave of endocrine and acinar cell differentiation, known as the secondary transition, begins (18, 19). At this embryonic stage, Gata6 is homogeneously expressed throughout the pancreatic epithelium (Supplemental Figure 4). In contrast, Gata4 displays a very specific expression pattern, restricted to the tips of the branching ductal epithelium, which will further adopt an acini fate (Supplemental Figure 4). Histological analysis of double-mutant embryos revealed a smaller and disorganized epithelium compared with Gata4flox/flox;Gata6flox/+;Pdx1-Cre, Gata4flox/+;Gata6flox/flox;Pdx1-Cre, and control embryos, suggesting that epithelial expansion was disrupted (Figure 3, A–D). Furthermore, immunohistochemical staining for mucin on whole-mount pancreata revealed a significant reduction in the pancreatic epithelial area of the double mutant when compared with control littermates (Supplemental Figure 5). Consistent with these data, pancreatic epithelial morphogenesis was severely affected in double-mutant embryos at E15.5 (Supplemental Figure 4). These results indicate that pancreatic agenesis in newborn double-mutant mice is a consequence of defective pancreatic morphogenesis during early pancreatic development.
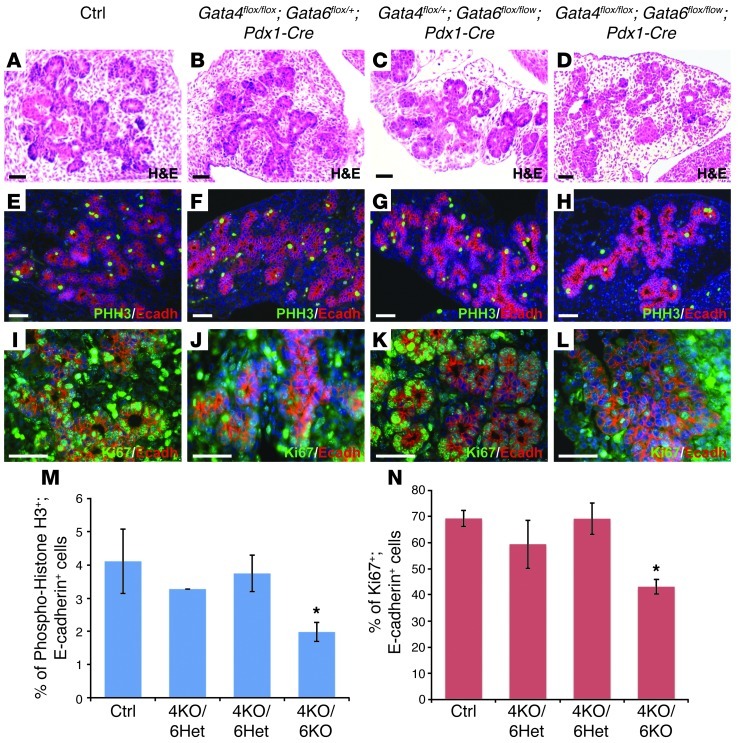
Figure 3. Pancreatic epithelial expansion is impaired in the absence of GATA4 and GATA6 activity.
The pancreatic epithelia of control, Gata4flox/flox;Gata6flox/+;Pdx1-Cre, and Gata4flox/+;Gata6flox/flox;Pdx1-Cre embryos at E13.5 display normal morphology (A–C), whereas double-mutant pancreatic epithelium appears disorganized and reduced in epithelial area (D). Immunohistochemistry analysis of the mitotic marker phospho-histone H3 (PHH3) reveals a significant reduction in proliferating pancreatic epithelial cells in the double-mutant (H) compared with littermate embryos at E13.5 (E–G). Immunostaining with another proliferation marker, Ki67, confirms the reduction in proliferation of E13.5 pancreatic epithelial cells in the double mutant compared with littermates (I–L). Counterstaining with DAPI was performed to reveal nuclei. Quantification of proliferating cells, measured as the number of PHH3 and Ki67-positive cells (M and N, respectively) per E-cadherin–positive cells. *P < 0.05. Scale bars: 50 μm.
Defects in proliferation of pancreatic progenitors in Gata4/Gata6 double-mutant embryos.
During pancreatic bud growth, extensive proliferation of the pancreatic epithelial cells occurs. To determine whether pancreatic growth arrest in double-mutant mice was due to defects in proliferation of pancreatic epithelial cells, we performed immunohistochemical analyses for proliferation markers at E13.5. Quantification for the mitotic marker phospho-histone H3 revealed a 50% reduction in proliferating cell number in the pancreatic epithelium of the double-mutant embryos when compared with control, Gata4flox/flox;Gata6flox/+;Pdx1-Cre, and Gata4flox/+;Gata6flox/flox;Pdx1-Cre littermates (Figure 3, E–H, and M). These results were confirmed using another proliferation marker, Ki67, a protein present during all active phases of the cell cycle. Based on Ki67 staining, a high percentage of proliferative pancreatic cells was found in control, Gata4flox/flox;Gata6flox/+;Pdx1-Cre, and Gata4flox/+;Gata6flox/flox;Pdx1-Cre littermate embryos (Figure 3, I–K, and N). In contrast, double-mutant mice showed a decreased number of Ki67-positive cells in the pancreatic epithelium compared with control mice (Figure 3, I, L, and N). Although not statistically significant, we also observed a decreasing trend in proliferation of pancreatic epithelial cells in Gata4flox/flox;Gata6flox/+;Pdx1-Cre embryos compared with control mice that might partially explain the hypoplastic phenotype of these mice at neonatal stages (Figure 3, F, J, M, and N). These results indicate that GATA4 and GATA6 are necessary for the normal proliferation rate of the pancreatic cells to sustain embryonic epithelial growth.
Endocrine and exocrine differentiation is arrested in the absence of GATA4 and GATA6 activity.
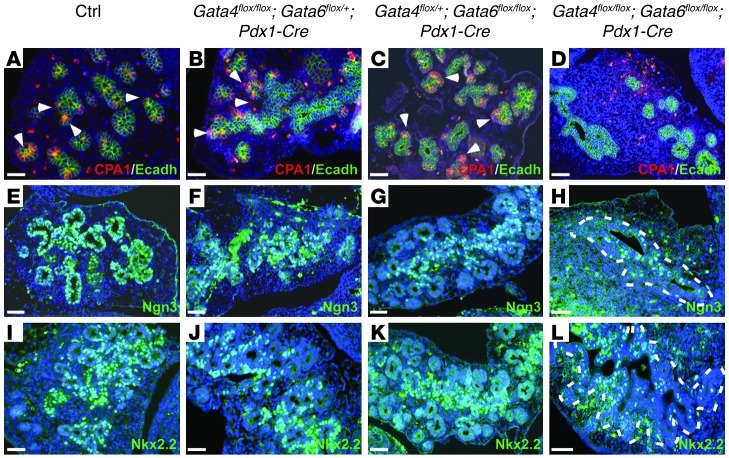
Our previous results indicated that pancreatic epithelial growth and expansion were impaired in the absence of GATA4 and GATA6 function at E13.5. The epithelial remodeling that occurs during this period appears to be intimately connected to the formation of different progenitor domains in the progenitor ductal epithelium. Different pancreatic lineage progenitors display a characteristic distribution within the pancreatic epithelium at this embryonic stage (20). Multipotent progenitor cells are found at the tips of the branching pancreatic epithelium, while ductal-endocrine progenitor cells are located along the trunk (20–22). The tip and trunk domains can be easily identified by the expression of Carboxypeptidase A1 (Cpa1) and Neurogenin 3 (Ngn3), respectively (20, 23). Cpa1 and Ngn3 expression were analyzed by immunohistochemistry to determine whether the establishment of different pancreatic cell lineages within MPCs was affected in double-mutant mice. As expected, cells stained for Cpa1 were observed at the tips of the pancreatic epithelium, and Ngn3+ cells were located in the epithelial trunk in control, Gata4flox/flox;Gata6flox/+;Pdx1-Cre, and Gata4flox/+;Gata6flox/flox;Pdx1-Cre embryos (Figure 4, A–C, and E–G). In stark contrast, the pancreatic epithelium of the double-mutant embryos lacked clear tip-trunk structures, and Cpa1+ cells were totally absent in Gata4/Gata6 mutant pancreatic epithelium (Figure 4D). Indeed, no clear acinar morphology could be observed in double-mutant pancreas by H&E staining (Figure 3D). Endocrine commitment was affected as well in double-mutant embryos, as only a low number of Ngn3-expressing cells were found in the epithelial trunk (Figure 4H). In concordance with this observation, the number of cells expressing the transcription factor Nkx2.2, an endocrine progenitor marker regulated by Ngn3 (24), was also dramatically decreased in the double-mutant pancreata compared with littermate embryos (Figure 4, I and L). Thus, differentiation of pancreatic multipotent progenitor cells toward acinar and endocrine lineages requires GATA4 and GATA6 function.
Figure 4. Endocrine and acinar differentiation are compromised in Gata4/Gata6 double-mutant embryos.
The enzyme Carboxipeptidase A1 (Cpa1) is expressed in the multipotent progenitor cell population located at the tip of the E13.5 branching epithelium in control, Gata4flox/flox;Gata6flox/+;Pdx1-Cre, and Gata4flox/+;Gata6flox/flox;Pdx1-Cre mice (A–C, arrowheads). In stark contrast, Cpa1+ cells were not detected in the double-mutant pancreatic epithelium (D). The proendocrine markers Ngn3 (E–G) and Nk2.2 (I–K) were mainly expressed in the epithelial trunk of control, Gata4flox/flox;Gata6flox/+;Pdx1-Cre, and Gata4flox/+;Gata6flox/flox;Pdx1-Cre embryos at E13.5. On the contrary, endocrine differentiation is disrupted in the double-mutant embryos as the number of cells expressing Ngn3 (H) and Nkx2.2 (L) are reduced compared with littermate embryos. The pancreatic epithelium is outlined in white in H and L. Counterstaining with DAPI was performed to reveal nuclei. Scale bars: 50 μm.
GATA4 and GATA6 are required to maintain the pancreatic progenitor cell pool.
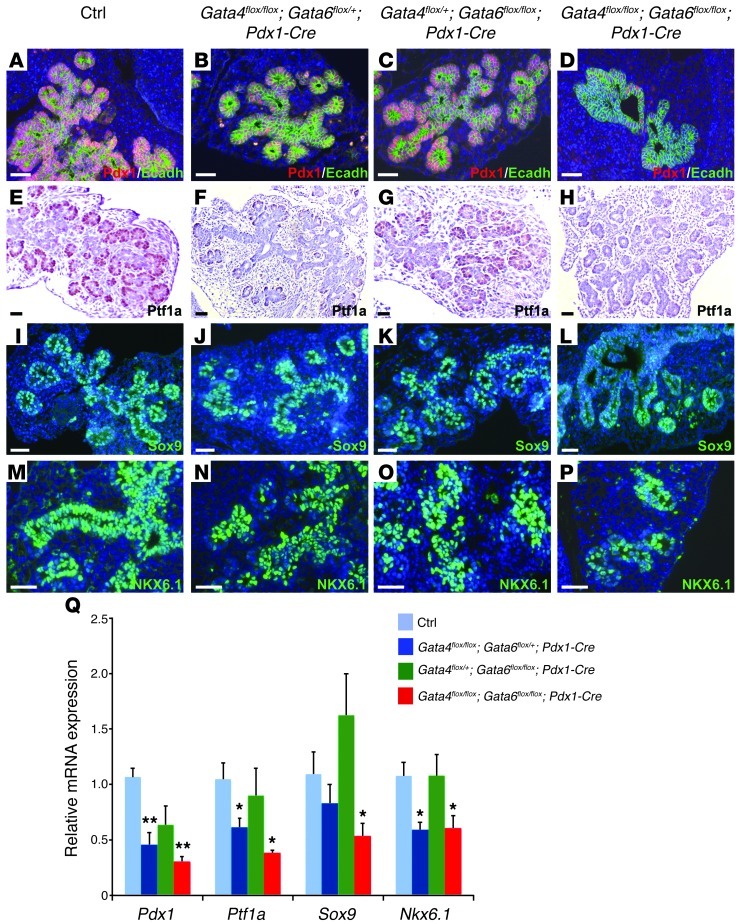
Exocrine and endocrine progenitors failed to form properly in Gata4/Gata6 mutant mice, raising the question of whether MPC formation and/or identity might be affected as well. To test this hypothesis, we decided to analyze the expression of several transcription factors that define MPC identity, including Pdx1, Ptf1a, Sox9, and Nkx6.1, in double-mutant embryos at E13.5 (22, 23, 25, 26). MPCs of control and Gata4flox/+;Gata6flox/flox;Pdx1-Cre mice displayed normal distribution and expression levels of progenitor markers (Figure 5, A, C, E, G, I, K, M, and O). In contrast, the numbers of Pdx1-, Ptf1a-, Sox9-, and Nkx6.1-expressing cells in double-mutant embryos were dramatically reduced compared with control and Gata4flox/+;Gata6flox/flox;Pdx1-Cre littermate embryos (Figure 5, D, H, L, and P). The decrease of Pdx1- and Ptf1a-expressing cells was particularly remarkable, as only a few positive cells could be observed in the epithelium of the double-mutant embryos (Figure 5, D and H). The reduced pool of cells expressing MPC markers in double-mutant embryos led to an overall decrease in the expression of progenitor markers (Figure 5Q). A decrease in the number of cells expressing Pdx1, Ptf1a, and Nkx6.1 was also found in Gata4flox/flox;Gata6flox/+;Pdx1-Cre, consistent with these mice displaying defects in pancreas formation at birth (Figure 5, B, F, J, and N). These results indicate that GATA4 and GATA6 activity are required to maintain normal numbers of MPCs during pancreatic development.
Figure 5. Reduced number of MPCs in Gata4/Gata6 double-mutant mice.
Control embryos show strong expression of all multipotent pancreatic progenitor markers, Pdx1, Ptf1a, Sox9, and Nkx6.1 at E13.5 (A, E, I, and M, respectively). Similarly, Gata4flox/+;Gata6flox/flox;Pdx1-Cre embryos display normal distribution and expression levels (C, G, K, and O). However, Gata4flox/flox;Gata6flox/+;Pdx1-Cre mice show a significant decrease of cell numbers expressing Pdx1, Ptf1, and Nkx6.1 (B, F, and N) and a moderate reduction in the number of Sox9+ cells (J). The reduced number of cells expressing all pancreatic progenitor markers is even more dramatic in the double mutant (D, H, L, and P). (Q) qPCR analysis of multipotent pancreatic progenitor markers in E13.5 pancreata. *P < 0.02; **P < 0.001. Scale bars: 50 μm.
GATA4 and GATA6 bind to 2 conserved GATA sites in the Pdx1 promoter region.
The impairment in pancreatic epithelial growth and cell differentiation observed in Gata4/Gata6 mutant mice is reminiscent of that in Pdx1- and Ptf1a-null mice (25, 27). This observation, in combination with the dramatic reduction in the number of cells expressing both Pdx1 and Ptf1a in the absence of GATA4 and GATA6 activity, prompted us to investigate whether GATA factors might regulate Ptf1a and Pdx1 expression in pancreatic progenitors. Previous studies have identified a 5′ distal enhancer region that controls the expression of Ptf1a in both dorsal and ventral pancreatic buds as early as E10.5 (28). Our bioinformatics analyses did not identify any conserved GATA site within these regulatory sequences, suggesting that the activity of this enhancer during pancreatic development might be independent of GATA factors. In spite of this, we cannot discard a role for GATA factors in the transcriptional regulation of Ptf1a through different regulatory sequences. Indeed, the relatively nonconserved proximal promoter contains several GATA sites that were able to bind recombinant GATA4 and GATA6 proteins (Supplemental Figure 6). However, this region has only so far been associated with the transcriptional control of Ptf1a at later stages of pancreas development (28).
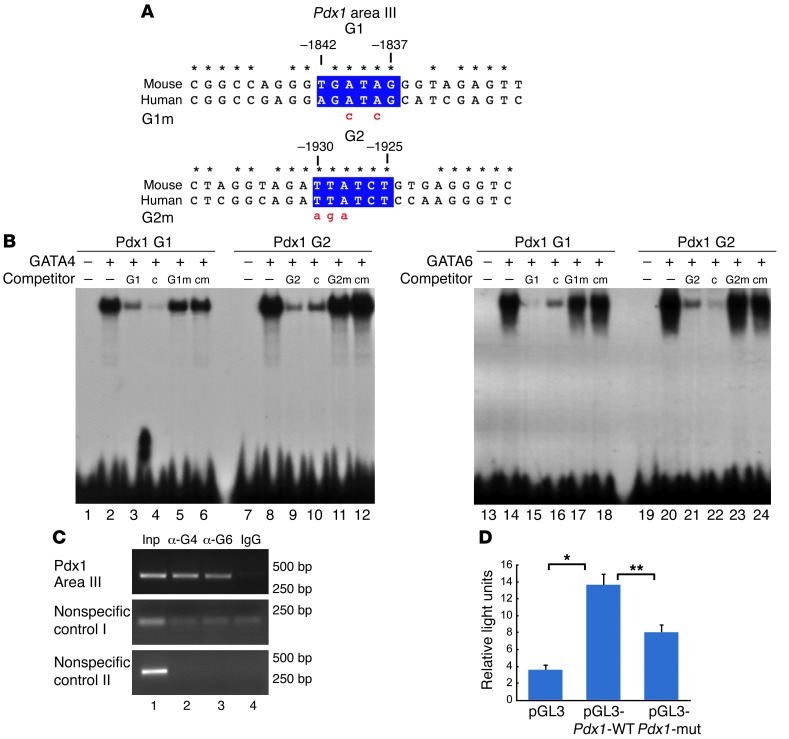
The transcriptional regulation of Pdx1 has been extensively studied, and several regulatory sequences or enhancers in the 5′ conserved region of the Pdx1 promoter have been identified (29, 30). These enhancers, known as areas I, II, III, and IV, are bound by transcription factors, including HNF1a, Foxa2, Foxa1, HNF6, Pax6, MafA, and Ptf1, to direct Pdx1 expression in pancreatic progenitors and/or in adult β cells (29, 31–37). Areas I and II direct Pdx1 expression in pancreatic endocrine cells, but not exocrine cells, whereas the conserved area III controls the expression of Pdx1 in pancreatic progenitor cells (37). With this consideration, we analyzed the conserved enhancer areas in the Pdx1 locus for candidate GATA sites. Interestingly, 2 perfect and conserved candidate GATA-binding sites were found in area III of the previously identified Pdx1 enhancer (Figure 6A). To determine whether these 2 putative sites, called G1 and G2, represent bona fide sites for GATA factors in the Pdx1 enhancer, we assessed GATA4 and GATA6 protein binding (Figure 6B). First, we performed EMSA using recombinant proteins. Recombinant GATA4 protein bound efficiently to labeled oligos encompassing the Pdx1 G1 and Pdx1 G2 GATA sites (Figure 6B, lanes 2 and 8). The binding to G1 and G2 sites was specific. as it was competed off by excess, unlabeled self probe (Figure 6B, lanes 3 and 9) and by excess, unlabeled GATA control probe (Figure 6B, lanes 4 and 10). In contrast, binding of GATA4 to G1 and G2 sites was not competed off by unlabeled mutant versions of the G1 and G2 sites, G1m and G2m (Figure 6, A and B, lanes 5 and 11), or GATA mutant control site (Figure 6B, lanes 6 and 12). Similarly to GATA4, recombinant GATA6 protein strongly bound to both G1 and G2 sites of the Pdx1 enhancer (Figure 6B, lanes 14 and 20). This binding was specific, as it was competed off by an excess of unlabeled self probe (Figure 6B, lanes 15 and 21), but not by an excess of unlabeled mutant probes (Figure 6B, lanes 17 and 23). In addition, we found 2 conserved GATA sites in conserved area I that were able to bind GATA4 and GATA6 in EMSA experiments (Supplemental Figure 7), suggesting that other GATA sites may also participate in the transcriptional regulation of Pdx1 at later stages of pancreas development.
Figure 6. GATA4 and GATA6 bind to the Pdx1 conserved area III in vitro and in pancreatic cell line.
(A) Highly conserved region in the cis-regulatory area III of Pdx1. Two conserved GATA sites, as revealed by bioinformatics analysis, are shown in blue boxes. Numbers indicate the position of the GATA sites relative to the Pdx1 translational start site. Point mutations introduced into GATA sites, G1m and G2m, are indicated in red lowercase. Asterisks denote nucleotides that have been perfectly conserved between mouse and human. (B) Recombinant GATA4 and GATA6 proteins are able to bind to G1 and G2 GATA sites of the Pdx1 enhancer as shown by EMSA. Competition experiments were performed by adding excess unlabeled probes of G1, G2, or control (denoted as c in competitor row) GATA sites, and the corresponding mutant versions (G1m, G2m, or cm) to the binding reaction. (C) ChIP experiments performed in mouse pancreatic ductal cells (mPAC cells) using specific GATA4 and GATA6 antibodies (lanes 2, 3, respectively) and nonspecific anti-IgG (lane 4) show that anti-GATA4 and anti-GATA6 antibodies are able to immunoprecipitate the GATA sites of the Pdx1 conserved area III, but not nonspecific genomic regions. Lane 1 contains PCR products from input DNA (Inp) amplified prior to immunoprecipitation. Sizes of the PCR products in bp are shown on the right. (D) A WT Pdx1 promoter-luciferase construct (pGL3-Pdx1-WT) is significantly activated by endogenous factors present in mPAC cells compared with the activity of the empty reporter pGL3 vector. Mutations in the GATA sites of Pdx1 (pGL3-Pdx1-mut) significantly attenuate the luciferase activity. *P = 0.002; **P = 0.005.
To investigate whether GATA4 and GATA6 could bind to conserved area III of the Pdx1 cis-regulatory sequences in pancreatic cells, we performed a ChIP assay in a murine pancreatic ductal cell line, mPAC, that endogenously expresses GATA4, GATA6, and PDX1 (data not shown). Anti-GATA4 antibody was able to specifically immunoprecipitate DNA fragments encompassing the G1 and G2 sites of the Pdx1 conserved area III, but not nonspecific genomic regions (Figure 6C, lane 2). Incubation of sheared DNA with anti-GATA6 also resulted in specific immunoprecipitation of DNA fragments containing the GATA sites of the Pdx1 enhancer (Figure 6C, lane 3). The addition of nonspecific IgG to the reaction did not result in the immunoprecipitation of the Pdx1 conserved area III or nonspecific genomic regions (Figure 6C, lane 4), confirming the specificity of the immunoprecipitation by the corresponding GATA antibodies. These results demonstrate that endogenous GATA4 and GATA6 bind to the GATA sites in the endogenous Pdx1 enhancer.
The observation that GATA4 and GATA6 bound to the Pdx1 conserved area III by EMSA and in mPAC pancreatic cells suggests that GATA4 and GATA6 transcription factors are required to transcriptionally regulate Pdx1 expression. Therefore, we examined the requirement of the G1 and G2 sites to activate the Pdx1 promoter in mPAC cells. A reporter construct containing the conserved area III of Pdx1 was fused to the luciferase gene to generate the pGL3-Pdx1-WT reporter plasmid. Likewise, the 2 conserved GATA sites were mutated to generate the pGL3-Pdx1-mut reporter plasmid. The luciferase activity of these plasmids was measured in transiently transfected mPAC cells. mPAC cells transfected with pGL3-Pdx1-WT display significantly higher luciferase activity compared with mPAC cells transfected with the parent reporter vector pGL3-basic, indicating that the Pdx1 enhancer region is activated by endogenous factors in mPAC cells (Figure 6D). Activation of the Pdx1 promoter significantly decreased when the 2 GATA sites in area III were mutated (Figure 6D). Taken together, these results demonstrate that GATA factors are required for the transcriptional regulation of Pdx1 in at least some pancreatic cell lines in vitro, suggesting that they may function through these sites in the MPCs.
Pdx1 transcriptional activity depends on GATA sites in vivo.
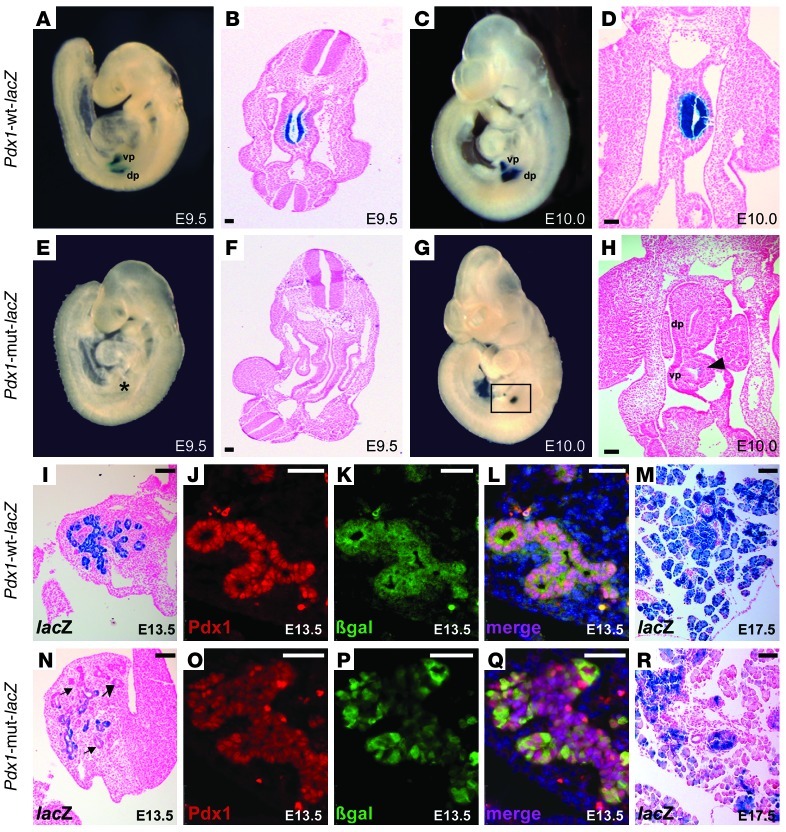
Previous studies have reported that a region 4.6 kb upstream of the Pdx1 translational start site containing conserved areas I, II, and III is sufficient to faithfully recapitulate endogenous Pdx1 expression throughout development in transgenic mice (38, 39). We generated transgenic mice harboring these upstream sequences of the Pdx1 promoter fused to the lacZ reporter gene, which we refer to as Pdx1-WT-lacZ. To test the requirement of the GATA sites for the Pdx1 enhancer activity in vivo, we introduced mutations in the GATA sites of Pdx1 area III to generate the Pdx1-mut-lacZ transgene. The introduced mutations were identical to those used in EMSA analyses that abolished the binding of GATA4 and GATA6 recombinant proteins (Figure 6, A and B). We generated 3 stable transgenic lines for the Pdx1-WT-lacZ construct and 2 stable transgenic lines for the Pdx1-mut-lacZ construct. The analysis of the lacZ expression by X-gal staining was performed in at least 6 transgenic embryos from each founder at different developmental stages. We observed that the lacZ expression pattern of the transgenic embryos was temporally and spatially very consistent among the founders from each construct. As expected, the 4.6 kb cis-regulatory region was sufficient to recapitulate the endogenous expression of Pdx1 in transgenic embryos from early embryonic stages. At E9.5, β-gal activity was observed in both the dorsal and ventral pancreatic buds in Pdx1-WT-lacZ transgenic embryos (Figure 7, A and B). In contrast, no X-gal staining was observed in any Pdx1-mut-lacZ transgenic embryo (Figure 7, E and F). By E10.0, very robust X-gal staining was observed in the pancreatic foregut of Pdx1-WT-lacZ transgenic embryos (Figure 7, C and D), while only a few lacZ-positive cells were detected in the pancreatic epithelium of Pdx1-mut-lacZ embryos (Figure 7, G and H). At midgestation, around E13.5, the Pdx1-WT-lacZ transgene continued to be active in the branching epithelial tree of the pancreas (Figure 7I). The β-gal expression directed by the Pdx1-WT-lacZ transgene completely mirrored the endogenous expression of Pdx1 (Figure 7, J–L). In sharp contrast, mutations in Pdx1 area III GATA sites resulted in a dramatic decrease in the Pdx1-mut-lacZ transgene activity in the pancreatic epithelium (Figure 7N). Immunohistochemical analysis confirmed the reduction of β-gal expression in Pdx1-mut-lacZ embryos (Figure 7, O–Q). At later stages of development, the activity of the Pdx1-mut-lacZ transgene was even more diminished compared with the activity of the Pdx1-WT-lacZ transgene (Figure 7M). Furthermore, X-gal staining was observed only in scattered cells of Pdx1-mut-lacZ pancreatic embryos (Figure 7R). These results indicate that GATA sites are required for Pdx1 enhancer activity during pancreas development and place GATA factors upstream of Pdx1 in the regulatory network controlling pancreas formation.
Figure 7. Conserved GATA sites in area III are required for Pdx1 enhancer activation in vivo.
Whole-mount (A, C, E, and G) and transversal sections (B, D, F, and H) of representative Pdx1-WT-lacZ and Pdx1-mut-lacZ transgenic embryos stained with X-gal. β-gal activity in both dorsal and ventral pancreatic buds is first observed in Pdx1-WT-lacZ embryos at E9.5 (A and B). In contrast, no X-gal staining is observed in the pancreatic buds of Pdx1-mut-lacZ embryos (E and F, asterisk). By E10.0, β-gal activity in the Pdx1-mut-lacZ embryos (square in G, arrowhead in H) is dramatically reduced compared with Pdx1-WT-lacZ embryos (C and D). At E13.5, all the pancreatic epithelial cells in Pdx1-WT-lacZ embryos show homogeneous β-gal activity (I). Similarly, strong X-gal staining is observed in most of the pancreatic cells of Pdx1-WT-lacZ embryos at E17.5 (M). In contrast, lacZ expression is markedly reduced in the pancreatic epithelium of Pdx1-mut-lacZ embryos both at E13.5 (arrows in N) and at E17.5 (R). Immunofluorescence staining at E13.5 reveals a complete overlapping expression pattern of Pdx1 and β-gal in Pdx1-WT-lacZ pancreas (J–L), while only a fraction of Pdx1+ cells express β-gal in Pdx1-mut-lacZ embryos (O–Q). dp, dorsal pancreas; vp, ventral pancreas. Scale bars: 50 μm.
Discussion
Pancreas specification and differentiation follow a well-orchestrated cascade of transcription factors that control the pancreatic genetic program in a spatiotemporal fashion. Inactivation of several of these transcription factors results in pancreatic hypoplasia or pancreatic agenesis in humans and mice (26, 27, 40, 41), reflecting their critical role in pancreas formation. In the rare examples of patients with pancreatic agenesis in which the affected gene has been identified, neonatal diabetes mellitus has been associated with homozygous mutations in the coding region of PDX1 and PTF1A genes (42, 43).
More recently, heterozygous mutations in GATA6 have been found in neonatal diabetic patients associated with pancreas agenesis and other extrapancreatic disorders, suggesting a possible functional role of this gene in human pancreas formation (8). In agreement with these observations, our studies performed in mice have established the GATA family transcription factors as critical regulators of pancreatic development. In contrast with humans, single inactivation of Gata6 (or Gata4) in the mouse pancreatic progenitor cells does not have an obvious effect on pancreas formation. The discrepancy between human and mouse GATA6 function could be related to the previously suggested role for GATA transcription factors in earlier pancreas specification (9). GATA6 mutations in humans could cause a failure in endoderm specification toward pancreatic fate, resulting in pancreas agenesis. Gene inactivation by Pdx1-directed Cre expression in Gata6 mutant mice is achieved after the pancreas has been specified, and therefore it can be argued that a possible role for GATA6 (or GATA4) in pancreas specification was not addressed in our study. However, inactivation of Gata4 and Gata6 in the prepancreatic endoderm using the FoxA3-Cre line results in proper pancreatic specification, suggesting that GATA factors might not be required to initiate the pancreatic program (44). The discrepancy between the mouse and human phenotype of GATA6 deficiency might also be explained by differences in the pancreatic genetic program between species. The spatiotemporal pattern of GATA factor expression during human pancreas development has not been reported. Thus, it remains to be determined whether GATA4 and GATA6 expression overlap and are functionally redundant in early human pancreatic formation, as is true in the mouse.
Based on the overlapping expression of GATA4 and GATA6 in identical cell compartments of different tissues during mouse embryonic development and their ability to recognize the same binding sites in the regulatory sequences of target genes, a certain degree of functional redundancy between these 2 transcription factors could have been anticipated. For example, at the onset of cardiac development, GATA4 and GATA6 are coexpressed, and thus, it was not entirely unexpected when functional studies in mice showed that GATA4 and GATA6 had redundant roles in controlling cardiomyocyte cell differentiation (45). More recently, it has been reported that GATA4 and GATA6 also share common functions in regulating cell proliferation and differentiation in proximal intestinal cells (46). Similar to these studies, our results indicate that these 2 GATA transcription factors are also functionally complementary during early stages of pancreas formation.
The pancreatic agenesis phenotype of the Gata4/Gata6 mutant mice is the result of defects that occur during the early stages of pancreas formation. We have observed defects in pancreatic epithelial morphology of double-mutant mice as early at E13.5. At that developmental stage, there is a marked reduction in the number of proliferating epithelial cells. Xuan et al. have reported that cell proliferation is decreased even at earlier embryonic stages in Gata4/Gata6 mutant mice, indicating that GATA4 and GATA6 function is required from the early post-bud stages of pancreas development (44). The exact mechanism by which the loss of GATA4 and GATA6 results in arrested pancreatic development needs to be investigated. However, it is tempting to speculate that a critical number of pancreatic progenitors are required to ensure the progression of the developmental program. Thus, diminishing the size of the pancreatic progenitor pool (as in Gata4/Gata6 mutant mice) could compromise the ability to move forward in the organogenesis program and ultimately result in the loss of pancreatic mass after birth. An attractive model for the expansion of MPCs would involve GATA, Pdx1, Ptf1a, and likely other early pancreatic transcription factors working in a cross-regulatory network to engage the proper level of progenitor cell proliferation necessary to promote the progression of pancreas organogenesis.
Although several transcription factors have been identified for controlling Pdx1 expression in adult β cells, less is known about the transcription factors that directly regulate the expression of Pdx1 in the early stages of pancreas development. Hnf6, Ptf1a, and more recently, FoxA1/FoxA2 have been identified as direct regulators of early Pdx1 expression (35, 37, 41). Our in vivo transgenic analysis adds GATA factors to the short list of identified transcription factors that control Pdx1 expression during early pancreatic development. The 2 conserved GATA sites we identified in our studies are located in area III of the Pdx1 regulatory sequence, an area that has been proposed to mediate early, pancreas-wide Pdx1 expression (37). Consistent with this, these GATA sites are required for Pdx1 expression in the early pancreatic bud. Interestingly, they seem to be required for maintenance of Pdx1 expression at later stages of pancreatic formation as well. However, these mutations do not completely abolish Pdx1 expression, and thus it is possible that additional GATA sites might account for the lower β-gal activity observed in Pdx1-mut-lacZ transgenic embryos.
An interesting observation from our analyses of different combinations of Gata4 and Gata6 floxed alleles is that, although both transcription factors are required for proper pancreas formation, GATA4 is more competent than GATA6 in supporting pancreas development. Gata4flox/+;Gata6flox/flox;Pdx1-Cre mutant pancreata appear completely normal. In contrast, although a pancreas is formed in Gata4flox/flox;Gata6flox/+;Pdx1-Cre mutant mice, a severe loss of pancreatic acinar mass and disrupted pancreatic morphology is observed. The more severe phenotype observed in Gata4flox/flox;Gata6flox/+;Pdx1-Cre embryos compared with the single Gata4-knockout mice suggests that GATA6 might also contribute to exocrine development before its expression becomes restricted to islet and ductal cells. The partial redundancy of GATA4 and GATA6 indicates that they have common and distinct functions in regulating gene expression during pancreatic development. It will be interesting to determine whether different DNA-binding affinities or recruitment of specific coactivators might explain the differences in GATA4 and GATA6 activities in pancreas development.
In summary, we have demonstrated a critical role for GATA4 and GATA6 transcription factors in pancreas formation. Our results place GATA factors at the top of the transcriptional network hierarchy controlling pancreas organogenesis. The direct regulation of Pdx1 expression by GATA factors offers a model that explains the pathogenesis of congenital pancreatic anomalies in human. Advances in our current understanding of pancreatic islet formation have sparked a great interest in the development of cell-based therapies for diabetes. Our data here suggest that manipulation of GATA factor activities should be now considered when devising strategies to generate β cells, either in vitro or in vivo.
Methods
Mice.
Gata4flox/flox, Gata6flox/flox, and Pdx1-Cre mice have been previously described (14, 15, 23, 47). The strategy for genotyping Gata6 WT and floxed alleles has been previously described (47). Gata4 WT and floxed alleles were detected by PCR using the following primers: 5′-CCCAGTAAAGAAGTCAGCACAAGGAAAC-3′and 5′-AGACTATTGATCCCGGAGTGAACATT-3′. Cre transgene was detected by PCR using the following primers: 5′-TGCCACGACCAAGTGACAGC-3′ and 5′-CCAGGTTACGGATATAGTTCATG-3′.
Reporter transgenic constructs.
A 4,660-bp SpeI/EcoRV fragment containing 4620 bp upstream and 40 bp downstream of the transcriptional start site from the mouse Pdx1 promoter region was cloned into SpeI-SmaI sites of the AUG–β-gal lacZ reporter plasmid (48) to generate the Pdx1-lacZ transgene. GATA site mutations in the Pdx1 promoter (Pdx1-mut) were generated by PCR using the following plus strand sequence: G1m, 5′-CCACTAGGTAGAAGATCTGTGAGGGTCAACAC-3′; G2m, 5′-CACTCGCGGCCGAGGAGCTCGCATCGAGTCCCTGGC-3′. Mutagenesis was confirmed by sequencing on both strands.
Pdx1-WT-lacZ and Pdx1-mut-lacZ transgenic reporter fragments were excised from the plasmid backbone with SpeI/HindIII, gel purified, and suspended in 10 mM Tris-HCl, 0.1 mM EDTA (pH 7.4), for pronuclear injection as described previously (49). Injected embryos were implanted into pseudopregnant FVB females, and embryos were allowed to develop to adulthood for establishment of stable transgenic lines. Three stable transgenic lines harboring the Pdx1-WT-lacZ construct and 2 stable transgenic lines harboring the Pdx1-mut-lacZ construct were generated. Each transgenic line was bred to C57BL/6 WT mice, and the embryos or tissues from these crosses were stained with X-gal to visualize β-gal, as described previously (50).
Histology, immunohistochemistry, and immunofluorescence.
Whole embryos or dissected pancreata collected at embryonic or adult stages were fixed in 4% paraformaldehyde in PBS overnight at 4°C, dehydrated with ethanol and xylene, and embedded in paraffin. Sections were cut at a thickness of 6 μm with a Leica DM6000B microtome. Sections were dewaxed through a series of xylene and ethanol washes. Embryos or tissues were counterstained with Nuclear Fast Red or H&E to visualize structures as previously described (49, 51). Prior to incubation with antibodies, paraffin sections were treated for immunohistochemistry and immunofluorescence as described previously (52).
The following primary antibodies were used at the indicated dilutions: mouse anti-GATA4 (1:100; Sc-25310, Santa Cruz Biotechnology Inc.); rabbit anti-GATA6 (1:50; Sc-9055, Santa Cruz Biotechnology Inc.); guinea pig anti-Pdx1 (1:200; ab47308, Abcam); rabbit anti-Sox9 (1:800; AB5535, Millipore); mouse anti–E-cadherin (1:100; 610186, BD Biosciences); mouse anti-amylase (1:200; Sc-46657, Santa Cruz Biotechnology Inc.); hamster anti-mucin (1:300; HM-1630, Thermo Scientific), rabbit anti–β-gal (1:500; 559762, MP Biochemicals); rabbit anti-Ki67 (1:100; RM-9106-S, Thermo Scientific); rabbit anti–phospho-histone H3 (1:500; 06-570, Millipore); biotinylated DBA (1:300; B-1035, Vector Laboratories); rabbit anti–carboxypeptidase 1 (1:800; 1810-0006, AbD Serotec); rabbit anti-Ptf1a (1:2000; AB2153 [ref. 53], Beta Cell Biology Consortium), rabbit anti-glucagon (1:200; 2760, Cell Signaling), and mouse anti-insulin (1:500; I2018, Sigma-Aldrich). The following primary antibodies were obtained from the Developmental Studies Hybridoma Bank (DSHB) developed under the auspices of the National Institute of Child Health and Human Development (NICHD) and maintained by The University of Iowa, Department of Biology, Iowa City, Iowa, USA: mouse anti-Ngn3 (1:200; DSHB, F25A1B3, developed by Ole D. Madsen, Hagedorn Research Institute, Gentofte, Denmark); rat anti–cytokeratin 19 (1:200; DSHB, TROMA III, developed by Rolf Kemler, Max-Planck Institute of Immunobiology and Epigenetics, Freiburg, Germany); mouse anti-Nkx2.2 (1:25; DSHB, 74.5A5, developed by Thomas M. Jessell and Susan Brenner-Morton, Columbia University, New York, New York, USA); mouse anti-Nkx6.1 (1:50; DSHB, F55A10, developed by Ole D. Madsen). Rabbit, rat, guinea pig, hamster, and mouse Alexa Fluor 568 and Alexa Fluor 488 (Invitrogen) secondary antibodies were used at 1:300 dilutions. For Nkx6.1 staining, signal from the secondary antibody was amplified using the TSA Fluorescein System (PerkinElmer). Counterstaining with DAPI was performed to visualize nuclei. The slides were analyzed using epifluorescence microscopy (Leica AF6000). Immunoperoxidase staining for the peroxidase substrate diaminobenzidine (DAB) was performed using the Vectastain Elite ABC Kit (Vector Laboratories).
EMSA.
DNA-binding reactions were performed as described previously (54). Gata4 and Gata6 cDNAs were transcribed and translated using the TNT Coupled Transcription-Translation System (Promega) as described in the manufacturer’s directions. GATA4 and GATA6 proteins were generated from pCITE-GATA4 and pCITE-GATA6 plasmids, which have been described previously (55). The sense strand sequence of the mouse Pdx1 GATA sites used for EMSA were as follows: G1, 5′-GGGCCACTAGGTAGATTATCTGTGAGGGTCAACAC-3′; G2, 5′-GGACTCGCGGCCGAGGAGATAGCATCGAGTCCCTGGC-3′; G3–G4, 5′-GGGTACCTCCAGTATCAGGGAGGACTATCAGGACGTCC-3′. Control and mutant control oligos for GATA sites have previously been described (55). The sense strand sequences for the G1 and G2 Gata mutant sites (G1m and G2m) were the same as for the mutagenic primers described above, and for G3m–G4m, the sequence was as follows: 5′-GTACCTCCAGGATCCGGGAAGACTCTCAGGACG- 3′.
The sense strand sequences of the mouse Ptf1a GATA sites used for EMSA were as follows: G1, 5′-GGCATCAGGATTTTGATAAGGTTTTGAAATTTGG-3′; G1m, 5′-CATCAGGATTTTGCCCGGGTTTTGAAATTTGG-3′; G2–G3, 5′-GCACTTATTTGGATAATTATATCAGCATTTAGG-3′; G2m–G3m,5′-CACTTATTTGGTATAATCGATCAGCATTTAGG-3′.
ChIP.
ChIP assays were performed using Dynabeads Protein A (100.01D, Invitrogen) following the recommendations of the manufacturer, with some modifications. Briefly, mPAC mouse pancreatic cells were grown in DMEM supplemented with 10% FBS on a 10-cm plate to approximately 1.5 × 106 cells (56). At that density, cells were treated with 1% formaldehyde at 37°C for 10 minutes to crosslink protein-DNA complexes. Sheared DNA from mPAC cells was incubated with Dynabeads Protein A plus 4 μg of mouse anti-GATA4 antibody (Sc- 25310, Santa Cruz Biotechnology Inc.) or rabbit anti-GATA6 antibody (Sc-9055, Santa Cruz Biotechnology Inc.) for 1 hour at 4°C. The same amount of mPAC sheared DNA was incubated with 4 μg of nonspecific IgG as a negative control. The DNA fragments were collected after the incubation using a DynaMag-2 (123.21D, Invitrogen). Following incubation in 200 mM NaCl at 65°C overnight to reverse the crosslinks, the DNA was recovered by phenol-chloroform extraction and subjected to PCR. The following primers were used to detect the fragment encompassing both GATA sites of the Pdx1 promoter: 5′-TGACCAGGTGAAGGAAGGTCC-3′ and 5′-AGCCACCTGTGCCCGTCAAGG-3′. The primers used to detect the second exon of Cyclin D2 as nonspecific control have previously been described (54). The following primers were used to detect the 3′ UTR region of Pdx1 as nonspecific control II: 5′-CTTTCTGGATGCTGCTCACAG-3′; 5′-TAATGTAGATTTGTCTCATCC-3′.
Luciferase reporter plasmids, cell culture, and transfection.
A 1,927-bp fragment containing 1,886 bp upstream and 41 bp downstream of the transcriptional start site from the mouse Pdx1 promoter region with either the WT GATA sequences or the GATA mutations, described above, was cloned in the pGL3-basic reporter vector (Promega) to generate pGL3-Pdx1-WT or pGL3-Pdx1-mut reporter plasmids. mPAC cells were maintained in DMEM supplemented with 10% FBS. Cells were cultured in 24-well plates at a density of 80,000 cells per well and transfected using Lipofectamine 2000 (Promega) following the manufacturer’s recommendations. Each well was transfected with 0.5 μg of pGL3-basic, pGL3-Pdx1-WT, or pGL3-Pdx1-mut reporter plasmids. The firefly luciferase activity was normalized to renilla luciferase activity by cotransfecting each well with 5 ng of pRL-renilla reporter vector (Promega). Cells were harvested 48 hours after transfection, and the luciferase activity was measured using a Dual Luciferase Kit Assay (Promega) and detected in a luminometer (Glomax 20/20, Promega). At least 3 independent triplicate sets of transfections were performed with each construct.
Quantitative RT-PCR.
Dissected pancreata from 3 E13.5 embryos of control (Pdx1-Cre–), Gata4flox/flox;Gata6flox/+;Pdx1-Cre, Gata4flox/+;Gata6flox/flox;Pdx1-Cre, and Gata4flox/flox;Gata6flox/flox;Pdx1-Cre mice were pooled to obtain RNA. Total RNA was isolated using RNeasy Plus Micro Kit (74034, QIAGEN), and cDNA was prepared using QuantiTect Reverse Transcription Kit (205311, QIAGEN). Quantitative PCR (qPCR) analysis was performed using FastStart Universal SYBR Green Master (04913850001, Roche), and the following primers were used: Ptf1a F, 5′-GCACTCTCTTTCCTGGACTGA-3′; Ptf1a R, 5′-TCCACACTTTAGCTGTACGGA-3′; Pdx1 F, 5′-AGCTCCCTTTCCCGTGGATGAAAT-3′; Pdx1 R, 5′-TAGGCAGTACGGGTCCTCTTGTTT-3′; Nkx2.2 F, 5′-TCGCTCTCCCCTTTGAACTTT-3′; Nkx2.2 R, 5′-GTTAACGTTGGGATGGTTTGG-3′; Sox9 F, 5′-GCAGACCAGTACCCGCATCT-3′; Sox9 R, 5′-TTCAGCAGCCTCCAGAGCTT-3′; β-actin F, 5′-TCCTGTGGCATCCACGAAACTACA-3′; β-actin R, 5′-ACCAGACAGCACTGTGTTGGCATA-3′. For detection of Pdx1, Ptf1, Sox9, and Nkx6.1, 10 ng of total cDNA was used per reaction, and each reaction was done in triplicate in 3 independent experiments. Gene expression levels were normalized to β-actin. The gene expression changes in mutant mice were determined using the comparative ΔΔCt method (57).
Quantitation of proliferative cells.
The number of proliferating cells were determined by counting phospho-histone H3– or Ki67-labeled cells within the pancreatic epithelium, delimitated by E-cadherin+ cells, in a series of sections from at least 3 pancreatic embryos of each genotype: control, Gata4flox/flox;Gata6flox/+;Pdx1-Cre (4KO/6Het), Gata4flox/+;Gata6flox/flox;Pdx1-Cre (4Het/ 6KO), and Gata4flox/flox;Gata6flox/flox;Pdx1-Cre (4KO/6KO).
Statistics.
All values are expressed as mean ± SEM. Statistical analyses were performed using a 2-tailed Student’s t test. P < 0.05 was considered statistically significant.
Study approval.
All experiments using animals complied with institutional guidelines and were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Sevilla, Sevilla, Spain.
Supplementary Material
Acknowledgments
We thank David Cano, Benoit Gauthier, and Janet Lau for their helpful comments on the manuscript. We also thank Antonio Cárdenas and Raquel Araujo for their technical assistance and María del Pilar González Sánchez and Ana Morilla Camacho from CPYEA for the generation of transgenic mice. We also thank Paloma Domínguez (CABIMER) for her assistance with the confocal microscope. M. Carrasco was supported by a predoctoral fellowship from the Spanish Ministry of Education. I. Delgado was supported by a contract from Consejería de Salud, Junta de Andalucía (PI-0008). This work was supported by grants from ISCIII cofunded by Fondos FEDER (PI08/0018, PI11/01125 to A. Rojas) (RD06/0010/0025 and PI10/00964 to B. Soria), Consejería de Economía, Innovación y Ciencia (P10.CTS.6505 to B. Soria), and from Consejería de Salud, Junta de Andalucía (PI-0008/2009 to A. Rojas) (PI-022/2008 to F. Martín).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(10):3504–3515. doi:10.1172/JCI63240.
See the related Commentary beginning on page 3469.
References
- 1.Patient RK, McGhee JD. The GATA family (vertebrates and invertebrates). Curr Opin Genet Dev. 2002;12(4):416–422. doi: 10.1016/S0959-437X(02)00319-2. [DOI] [PubMed] [Google Scholar]
- 2.Bresnick EH, Lee HY, Fujiwara T, Johnson KD, Keles S. GATA switches as developmental drivers. J Biol Chem. 2010;285(41):31087–31093. doi: 10.1074/jbc.R110.159079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Molkentin JD. The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J Biol Chem. 2000;275(50):38949–38952. doi: 10.1074/jbc.R000029200. [DOI] [PubMed] [Google Scholar]
- 4.Garg V, et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003;424(6947):443–447. doi: 10.1038/nature01827. [DOI] [PubMed] [Google Scholar]
- 5.Lin X, et al. A novel GATA6 mutation in patients with tetralogy of Fallot or atrial septal defect. J Hum Genet. 2010;55(10):662–667. doi: 10.1038/jhg.2010.84. [DOI] [PubMed] [Google Scholar]
- 6.Maitra M, Koenig SN, Srivastava D, Garg V. Identification of GATA6 sequence variants in patients with congenital heart defects. Pediatr Res. 2010;68(4):281–285. doi: 10.1203/PDR.0b013e3181ed17e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kodo K, Yamagishi H. GATA transcription factors in congenital heart defects: a commentary on a novel GATA6 mutation in patients with tetralogy of Fallot or atrial septal defect. J Hum Genet. 2010;55(10):637–638. doi: 10.1038/jhg.2010.105. [DOI] [PubMed] [Google Scholar]
- 8.Allen HL, et al. GATA6 haploinsufficiency causes pancreatic agenesis in humans. Nat Genet. 2011;44(1):20–22. doi: 10.1038/ng.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Decker K, Goldman DC, Grasch CL, Sussel L. Gata6 is an important regulator of mouse pancreas development. Dev Biol. 2006;298(2):415–429. doi: 10.1016/j.ydbio.2006.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ketola I, et al. Transcription factor GATA-6 is expressed in the endocrine and GATA-4 in the exocrine pancreas. Mol Cell Endocrinol. 2004;226(1–2):51–57. doi: 10.1016/j.mce.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 11.Kuo CT, et al. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997;11(8):1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- 12.Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11(8):1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- 13.Koutsourakis M, Langeveld A, Patient R, Beddington R, Grosveld F. The transcription factor GATA6 is essential for early extraembryonic development. Development. 1999;126(9):723–732. doi: 10.1242/dev.126.9.723. [DOI] [PubMed] [Google Scholar]
- 14.Watt AJ, Zhao R, Li J, Duncan SA. Development of the mammalian liver and ventral pancreas is dependent on GATA4. BMC Dev Biol. 2007;7:37. doi: 10.1186/1471-213X-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hingorani SR, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4(6):437–450. doi: 10.1016/S1535-6108(03)00309-X. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, et al. A Gata6-Wnt pathway required for epithelial stem cell development and airway regeneration. Nat Genet. 2008;40(7):862–870. doi: 10.1038/ng.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oka T, et al. Cardiac-specific deletion of Gata4 reveals its requirement for hypertrophy, compensation, and myocyte viability. Circ Res. 2006;98(6):837–845. doi: 10.1161/01.RES.0000215985.18538.c4. [DOI] [PubMed] [Google Scholar]
- 18.Jorgensen MC, Ahnfelt-Ronne J, Hald J, Madsen OD, Serup P, Hecksher-Sorensen J. An illustrated review of early pancreas development in the mouse. Endocr Rev. 2007;28(6):685–705. doi: 10.1210/er.2007-0016. [DOI] [PubMed] [Google Scholar]
- 19.Pan FC, Wright C. Pancreas organogenesis: from bud to plexus to gland. Dev Dyn. 2011;240(3):530–565. doi: 10.1002/dvdy.22584. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Q, Law AC, Rajagopal J, Anderson WJ, Gray PA, Melton DA. A multipotent progenitor domain guides pancreatic organogenesis. Dev Cell. 2007;13(1):103–114. doi: 10.1016/j.devcel.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 21.Solar M, et al. Pancreatic exocrine duct cells give rise to insulin-producing beta cells during embryogenesis but not after birth. Dev Cell. 2009;17(6):849–860. doi: 10.1016/j.devcel.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 22.Schaffer AE, Freude KK, Nelson SB, Sander M. Nkx6 transcription factors and Ptf1a function as antagonistic lineage determinants in multipotent pancreatic progenitors. Dev Cell. 2010;18(6):1022–1029. doi: 10.1016/j.devcel.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu G, Dubauskaite J, Melton DA. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129(10):2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- 24.Watada H, Scheel DW, Leung J, German MS. Distinct gene expression programs function in progenitor and mature islet cells. J Biol Chem. 2003;278(19):17130–17140. doi: 10.1074/jbc.M213196200. [DOI] [PubMed] [Google Scholar]
- 25.Kawaguchi Y, Cooper B, Gannon M, Ray M, MacDonald RJ, Wright CV. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat Genet. 2002;32(1):128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- 26.Seymour PA, et al. From the Cover: SOX9 is required for maintenance of the pancreatic progenitor cell pool. Proc Natl Acad Sci U S A. 2007;104(6):1865–1870. doi: 10.1073/pnas.0609217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jonsson J, Ahlgren U, Edlund T, Edlund H. IPF1, a homeodomain protein with a dual function in pancreas development. Int J Dev Biol. 1995;39(5):789–798. [PubMed] [Google Scholar]
- 28.Masui T, Swift GH, Hale MA, Meredith DM, Johnson JE, Macdonald RJ. Transcriptional autoregulation controls pancreatic Ptf1a expression during development and adulthood. Mol Cell Biol. 2008;28(17):5458–5468. doi: 10.1128/MCB.00549-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerrish K, et al. Pancreatic beta cell-specific transcription of the pdx-1 gene. The role of conserved upstream control regions and their hepatic nuclear factor 3beta sites. J Biol Chem. 2000;275(5):3485–3492. doi: 10.1074/jbc.275.5.3485. [DOI] [PubMed] [Google Scholar]
- 30.Gerrish K, Van Velkinburgh JC, Stein R. Conserved transcriptional regulatory domains of the pdx-1 gene. Mol Endocrinol. 2004;18(3):533–548. doi: 10.1210/me.2003-0371. [DOI] [PubMed] [Google Scholar]
- 31.Marshak S, Benshushan E, Shoshkes M, Havin L, Cerasi E, Melloul D. Functional conservation of regulatory elements in the pdx-1 gene: PDX-1 and hepatocyte nuclear factor 3beta transcription factors mediate beta-cell-specific expression. Mol Cell Biol. 2000;20(20):7583–7590. doi: 10.1128/MCB.20.20.7583-7590.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerrish K, Cissell MA, Stein R. The role of hepatic nuclear factor 1 alpha and PDX-1 in transcriptional regulation of the pdx-1 gene. J Biol Chem. 2001;276(51):47775–47784. doi: 10.1074/jbc.M109244200. [DOI] [PubMed] [Google Scholar]
- 33.Ben-Shushan E, Marshak S, Shoshkes M, Cerasi E, Melloul D. A pancreatic beta -cell-specific enhancer in the human PDX-1 gene is regulated by hepatocyte nuclear factor 3beta (HNF-3beta ), HNF-1alpha, and SPs transcription factors. J Biol Chem. 2001;276(20):17533–17540. doi: 10.1074/jbc.M009088200. [DOI] [PubMed] [Google Scholar]
- 34.Samaras SE, Cissell MA, Gerrish K, Wright CV, Gannon M, Stein R. Conserved sequences in a tissue-specific regulatory region of the pdx-1 gene mediate transcription in Pancreatic beta cells: role for hepatocyte nuclear factor 3 beta and Pax6. Mol Cell Biol. 2002;22(13):4702–4713. doi: 10.1128/MCB.22.13.4702-4713.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacquemin P, Lemaigre FP, Rousseau GG. The Onecut transcription factor HNF-6 (OC-1) is required for timely specification of the pancreas and acts upstream of Pdx-1 in the specification cascade. Dev Biol. 2003;258(1):105–116. doi: 10.1016/S0012-1606(03)00115-5. [DOI] [PubMed] [Google Scholar]
- 36.Samaras SE, Zhao L, Means A, Henderson E, Matsuoka TA, Stein R. The islet beta cell-enriched RIPE3b1/Maf transcription factor regulates pdx-1 expression. J Biol Chem. 2003;278(14):12263–12270. doi: 10.1074/jbc.M210801200. [DOI] [PubMed] [Google Scholar]
- 37.Wiebe PO, et al. Ptf1a binds to and activates area III, a highly conserved region of the Pdx1 promoter that mediates early pancreas-wide Pdx1 expression. Mol Cell Biol. 2007;27(11):4093–4104. doi: 10.1128/MCB.01978-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gannon M, Gamer LW, Wright CV. Regulatory regions driving developmental and tissue-specific expression of the essential pancreatic gene pdx1. Dev Biol. 2001;238(1):185–201. doi: 10.1006/dbio.2001.0359. [DOI] [PubMed] [Google Scholar]
- 39.Stoffers DA, Heller RS, Miller CP, Habener JF. Developmental expression of the homeodomain protein IDX-1 in mice transgenic for an IDX-1 promoter/lacZ transcriptional reporter. Endocrinology. 1999;140(11):5374–5381. doi: 10.1210/en.140.11.5374. [DOI] [PubMed] [Google Scholar]
- 40.Krapp A, et al. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes Dev. 1998;12(23):3752–3763. doi: 10.1101/gad.12.23.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao N, LeLay J, Vatamaniuk MZ, Rieck S, Friedman JR, Kaestner KH. Dynamic regulation of Pdx1 enhancers by Foxa1 and Foxa2 is essential for pancreas development. Genes Dev. 2008;22(24):3435–3448. doi: 10.1101/gad.1752608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sellick GS, et al. Mutations in PTF1A cause pancreatic and cerebellar agenesis. Nat Genet. 2004;36(12):1301–1305. doi: 10.1038/ng1475. [DOI] [PubMed] [Google Scholar]
- 43.Stoffers DA, Ferrer J, Clarke WL, Habener JF. Early-onset type-II diabetes mellitus (MODY4) linked to IPF1. Nat Genet. 1997;17(2):138–139. doi: 10.1038/ng1097-138. [DOI] [PubMed] [Google Scholar]
- 44.Xuan S, et al. Pancreas-specific deletion of mouse Gata4 and Gata6 causes pancreatic agenesis. . J Clin Invest. 2012;122(10):3516–3528. doi: 10.1172/JCI63352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao R, Watt AJ, Battle MA, Li J, Bondow BJ, Duncan SA. Loss of both GATA4 and GATA6 blocks cardiac myocyte differentiation and results in acardia in mice. Dev Biol. 2008;317(2):614–619. doi: 10.1016/j.ydbio.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beuling E, et al. GATA factors regulate proliferation, differentiation, and gene expression in small intestine of mature mice. Gastroenterology. 2011;140(4):1219–1229. doi: 10.1053/j.gastro.2011.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sodhi CP, Li J, Duncan SA. Generation of mice harbouring a conditional loss-of-function allele of Gata6. BMC Dev Biol. 2006;6:19. doi: 10.1186/1471-213X-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McFadden DG, Charite J, Richardson JA, Srivastava D, Firulli AB, Olson EN. A GATA-dependent right ventricular enhancer controls dHAND transcription in the developing heart. Development. 2000;127(24):5331–5341. doi: 10.1242/dev.127.24.5331. [DOI] [PubMed] [Google Scholar]
- 49.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating The Mouse Embryo. Plainview, New York, USA: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 50.Dodou E, Xu SM, Black BL. mef2c is activated directly by myogenic basic helix-loop-helix proteins during skeletal muscle development in vivo. Mech Dev. 2003;120(9):1021–1032. doi: 10.1016/S0925-4773(03)00178-3. [DOI] [PubMed] [Google Scholar]
- 51.Anderson JP, et al. HRC is a direct transcriptional target of MEF2 during cardiac, skeletal, and arterial smooth muscle development in vivo. Mol Cell Biol. 2004;24(9):3757–3768. doi: 10.1128/MCB.24.9.3757-3768.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rojas A, Schachterle W, Xu SM, Martín F, Black BL. Direct transcriptional regulation of Gata4 during early endoderm specification is controlled by FoxA2 binding to an intronic enhancer. Dev Biol. 2010;346(2):346–355. doi: 10.1016/j.ydbio.2010.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hald J, Sprinkel AE, Ray M, Serup P, Wright C, Madsen OD. Generation and characterization of Ptf1a antiserum and localization of Ptf1a in relation to Nkx6.1 and Pdx1 during the earliest stages of mouse pancreas development. J Histochem Cytochem. 2008;56(6):587–595. doi: 10.1369/jhc.2008.950675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rojas A, Kong SW, Agarwal P, Gilliss B, Pu WT, Black BL. GATA4 is a direct transcriptional activator of cyclin D2 and Cdk4 and is required for cardiomyocyte proliferation in anterior heart field-derived myocardium. Mol Cell Biol. 2008;28(17):5420–5431. doi: 10.1128/MCB.00717-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rojas A, De Val S, Heidt AB, Xu SM, Bristow J, Black BL. Gata4 expression in lateral mesoderm is downstream of BMP4 and is activated directly by Forkhead and GATA transcription factors through a distal enhancer element. Development. 2005;132(15):3405–3417. doi: 10.1242/dev.01913. [DOI] [PubMed] [Google Scholar]
- 56.Yoshida T, Hanahan D. Murine pancreatic ductal adenocarcinoma produced by in vitro transduction of polyoma middle T oncogene into the islets of Langerhans. Am J Pathol. 1994;145(3):671–684. [PMC free article] [PubMed] [Google Scholar]
- 57.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.