Abstract
Human respiratory syncytial virus (hRSV) typically affects newborns and young children. Even though it can cause severe and, in some cases, lifelong respiratory infections, there are currently no FDA-approved therapeutics that control this virus. The hRSV F protein facilitates viral fusion, a critical extracellular event that can be targeted for therapeutic intervention by disrupting the assembly of a post-fusion 6-Helix bundle (6HB) within the hRSV F protein. Here we report the development of a fluorescence polarization assay using an engineered hRSV F protein 5-Helix Bundle (5HB). We generated the 5HB and validated its ability to form a 6HB in a fluorescence polarization assay. To test the potential of 5HB as a screening tool, we then investigated a series of truncated peptides derived from the “missing” sixth helix. Using this FP-based 5HB system, we have successfully demonstrated that short peptides can prevent 6HB formation and serve as potential hRSV fusion inhibitors. We anticipate this new 5HB system will provide an effective tool to identify and study potential antivirals to control hRSV infection.
Keywords: hSRV F protein, 5-Helix Bundle, Fluorescence polarization
Introduction
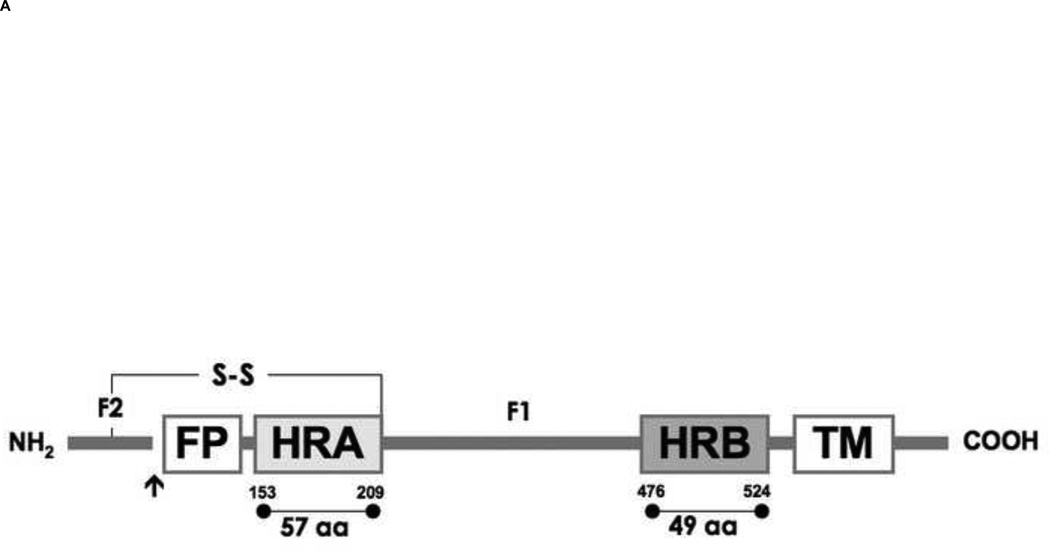
Despite attempts to develop safe, cost-effective treatments to control human respiratory syncytial virus (hRSV)-associated illness [1,2], hRSV remains the leading pathogen causing severe lower respiratory tract infections in infants and children. hRSV infections cause more than 120,000 pediatric hospitalizations and 2,000 deaths in the United States alone with costs of $356–585 million annually [3–7]. Most children are infected with hRSV at least once before the age of 2, and recurrence is common [8]. hRSV has also been an increasingly recognized cause of high morbidity and mortality in the elderly, causing up to 10,000 deaths in individuals over the age of 65 years annually [9,10]. Antiviral drug discovery to control hRSV infections has mainly relied on the screening of chemical libraries or natural products using common virology assays and animal models, yielding limited success [2,11]. The only clinically approved antivirals to date are a nucleoside analog, Ribavirin, and a humanized monoclonal antibody, Synagis. Due to efficacy and cost, these drugs are restricted to high-risk children (babies born at less than 36 weeks or who have heart or lung problems) [12]. With treatment options limited to measures such as supportive care, development of safe and specific agents against hRSV is essential. Viral entry into cells is important to the hRSV infectious cycle. Upon attachment, hRSV uses the fusion protein (F) to cause membrane fusion with the host cells at neutral pH, similar to other paramyxoviruses. In the process of membrane fusion, F refolds to form a stable 6-helix bundle (6HB), structurally characterized by X-ray crystallography [13–16]. In the sequence of the F protein, two hydrophobic, heptad repeats (HRA and HRB) are located adjacent to the fusion peptide and transmembrane domain respectively. HRA and HRB are very far from each other with approximately 250 residues of intervening sequence between them (Fig. 1A). When the F protein is activated, a hydrophobic fusion peptide is exposed and then inserted into the host cell membrane as an anchor. Subsequently, the protein undergoes a dramatic refolding, bringing HRA and HRB together to form the 6HB. This 6HB formation, linked to F protein refolding, is thought to bring the viral and host cell membranes close together, leading to membrane fusion and viral entry [14,15,17–20]. F protein-mediated membrane merger occurs at the cell surface, so the process is theoretically accessible to inhibition by antivirals. Therefore F has been recognized as a potential therapeutic target, yielding numerous peptidic or non-peptidic small molecule fusion inhibitors [1,2,21–31].
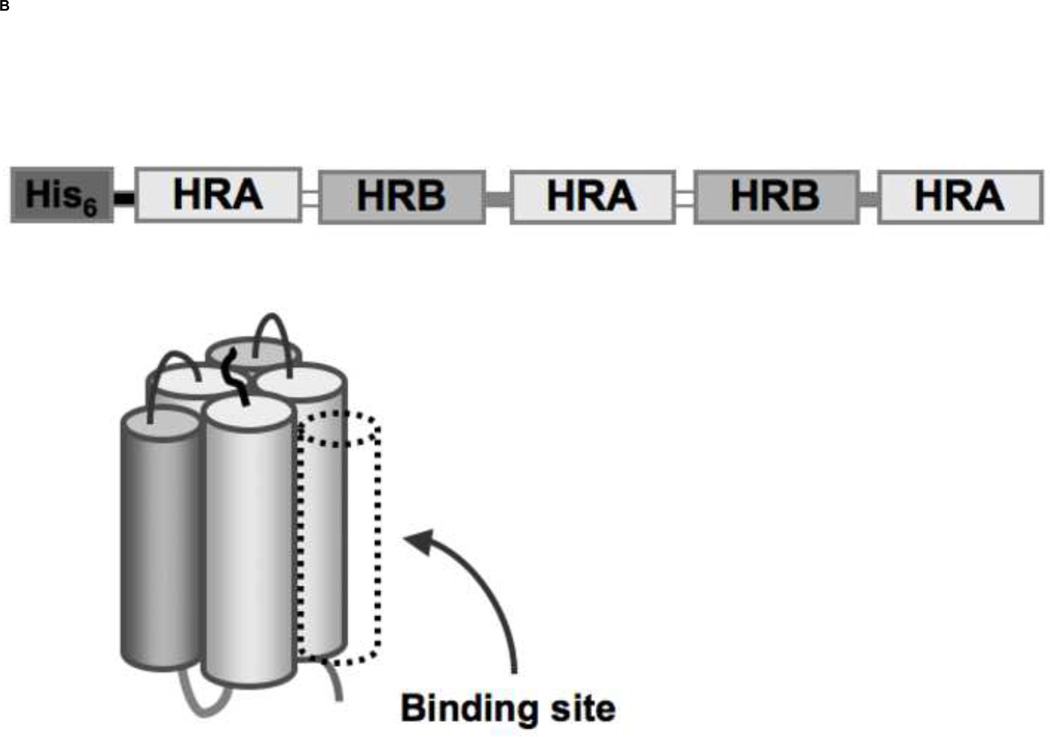
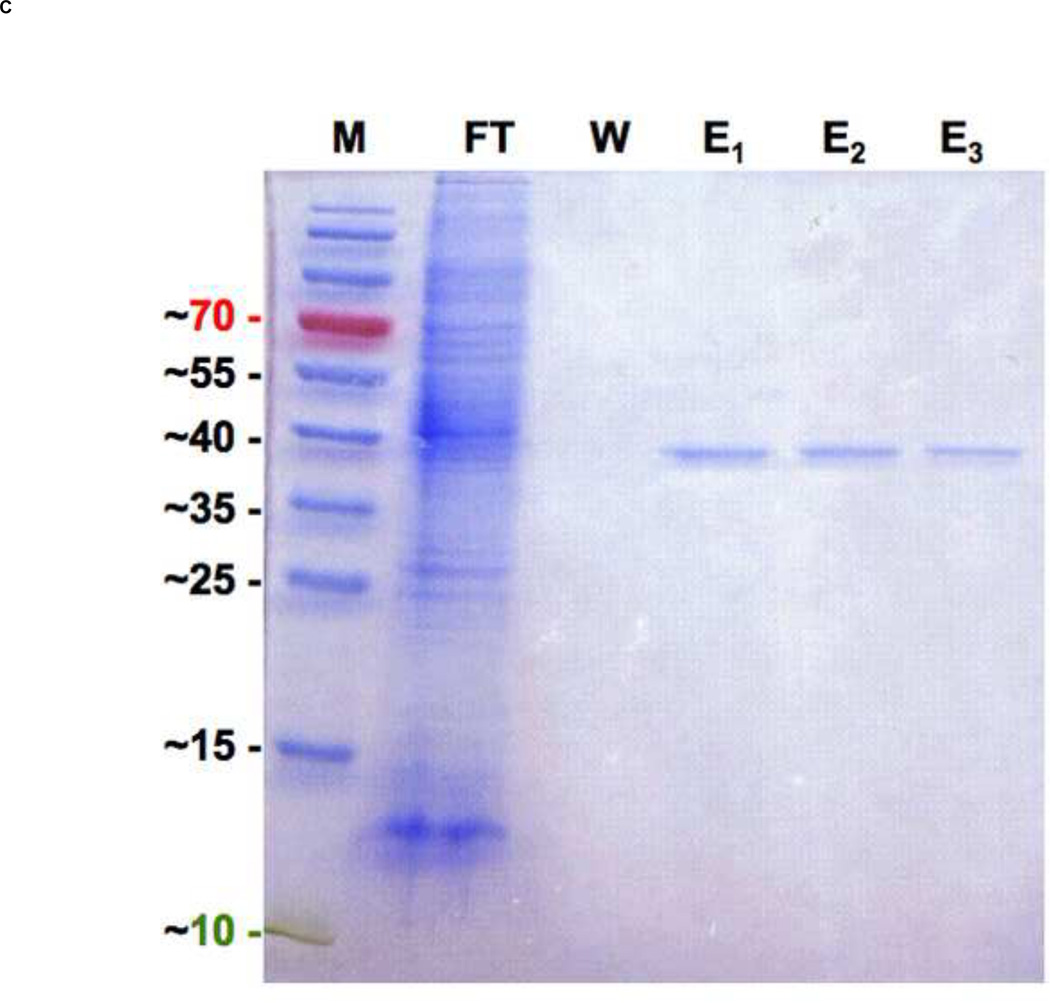
Figure 1. Schematic diagram of hRSV fusion (F) protein and 5-Helix Bundle with a resulting SDS-PAGE of 5HB purification.
(A) F1 and F2 are formed after proteolytic cleavage (arrow) of the precursor protein, F0. The fusion peptide (FP) and transmembrane domain (TM) are indicated. Heptad repeat regions, HRA and HRB are located adjacent to FP and TM respectively. (B) An illustration of the designed 5-Helix Bundle (5HB) and its single-chain polypeptide sequence are shown. A binding site for the missing 3rd HRB and potential fusion inhibitors was shown in dotted line. (C) Purified 5HB samples were analyzed by SDS-PAGE on a 12% non-reducing gel (Molecular marker; M, flow through; FT, wash; W, and elution fractions; E).
To our knowledge, there is no established, simple non-cell based method to screen potential antivirals specifically targeting the hRSV F protein. Previously, the 5-Helix Bundle of the HIV-1 fusion protein gp41 was shown to be a viral entry inhibitor [32], as well as a suitable target for screening small molecule libraries in a high-throughput format [33], suggesting that similar approaches would be applicable to other viruses, like hRSV, that rely on class I viral fusion proteins. We created a 5-Helix Bundle (5HB), variant hRSV F protein, and developed a competitive fluorescence polarization (FP) based assay using the 5HB as a target protein and a fluorescently labeled peptide as a tracer. To validate that the competitive FP-based 5HB assay can provide a reliable screening platform, a series of N- and C- terminally truncated peptides derived from HRB domain of the hRSV F protein were synthesized and tested. Thus, we demonstrate that this simple, low-cost in vitro fluorescence polarization assay can be readily expanded to libraries of peptides, peptidomimetics, or small molecules to rapidly screen potential hRSV fusion inhibitors
Materials & Methods
Protein synthesis
5-Helix Bundle cloning
The 5HB DNA construct is composed of three N57 and two C49 helices, representing residues 126 to 182 (HRA) and 476 to 524 (HRB) of the hRSV F protein, respectively. Each fragment was amplified by PCR using the hRSV strain A2 genome as a template and connected using short linkers: N57 was joined using a linker (PPPELGGP) to C49 to generate a heterodimer, N57-C49; two heterodimers were connected with a short linker (KGSSK); the final N57 was linked after the second C49 via the linker (KGSSK) (Fig. 1B). The engineered gene encoding the 5HB was cloned between the Nde I and BamH III restriction sites of the hexahistidine expression vector pET-15b (Novagen, San Diego, CA, USA). The resulting plasmid carrying the complete 5HB construct was transformed into E.coli strain BL21 (DE3) for protein expression.
5-Helix Bundle expression and purification
Protein was recombinantly expressed in E.coli strain BL21 (DE3) grown to an OD of 0.8 at 600 nm at 37 °C in Luria-Bertani (LB) medium. Protein expression then was induced with 0.5 mM isopropyl-β-D-thiogalctopyranoside (ITPG), and cells were grown for an additional 20 hrs at 20 °C, to enhance the solubility of protein [34]. The cells were harvested by centrifugation at 4500 × g for 15 min, and the resulting cell pellet was resuspended in 20 mM phosphate buffered saline (PBS) and stored at −80 °C. Cells were lysed in lysis buffer (CelLytic™ B cell lysis reagent [cat. no. C8740, Sigma Aldrich, Milwaukee, WI, USA], 20 mM PBS at pH 7.4, 1 mM phenylmethylsulphonyl fluoride [PMSF], protease inhibitor cocktail [Sigma Aldrich, Milwaukee, WI, USA], 1% Triton X-100, 500 mM NaCl and 0.2 mg/ml lysozyme) and incubated for 1 hr at room temperature. Cell lysate was then clarified by centrifugation at 18,000 × g for 30 min. The soluble fraction was immediately incubated with a Nickel-immobilized chelating sepharose fast flow resin (cat. no. 17-0575-02, GE Healthcare, Piscataway, NJ, USA) at room temperature for 30 min with a gentle agitation. The protein-bound resin was washed out with more than 10 column volumes (CV) of a wash buffer (20 mM PBS at pH 7.4, 100 mM imidazole, 1% Triton X-100 and 500 mM NaCl). The 5HB was eluted with an elution buffer (20 mM PBS at pH 7.4, 300 mM imidazole and 500 mM NaCl). The purity of protein was assessed by SDS-PAGE and the protein was used without further purification. Protein concentration was determined by using the BCA protein Assay (cat. no. 23225, Pierce, Rockford, IL, USA). The final yield of soluble 5HB was approximately 1 mg/L of cell culture with batch-to-batch variation.
Circular Dichroism spectroscopy
CD spectra were obtained with a Jasco J-815 spectrophotometer (JASCO, Easton, MD, USA). Sample was prepared in 20 mM PBS, pH 7.4 in concentration of 35 µM. Data were recorded from 195 to 260 nm with a scanning speed of 20 nm/min and a bandwidth at 1.0 nm in a 0.1 cm path-length quartz cell. Each CD spectrum was an average of 3 measurements and corrected for buffer blank obtained under identical conditions. The resulting data was converted to per-residue molar ellipticity units, [Θ] (deg cm2 dmol−1 residue−1), and the secondary structure content was analyzed with the Dichroweb software package. The thermal stability of 5HB was monitored by measuring its molar ellipticity between 0 °C and 85 °C at 222 nm. 2.5 µM of 5HB was used for this study. The rate of temperature change was 2.5 °C /min with a scanning speed of 50 nm/min and a bandwidth of 1.0 nm.
Peptide synthesis
Peptide synthesis reagents were purchased from Applied Biosystems (Foster city, CA, USA) or Sigma-Aldrich (Milwaukee, WI, USA). Resins and Fmoc-protected amino acids were purchased from NovaBioChem (San Diego, CA, USA) or Anaspec (San Jose, CA, USA). Solvents for HPLC were purchased from Fisher Scientific (Pittsburgh, PA, USA). All chemicals were used without additional purification. Fluorescently-labeled peptide (Fl-C35) and truncated peptides (C30, and C35) of 95% purity were commercially obtained (EZBiolab, Carmel, IN, USA and Bio Basic, Markham, ON, Canada) and used without further purification. The remaining truncated peptides (C20, C17 and N15) were synthesized in the laboratory using standard Fmoc chemistry on solid support (preloaded Wang resin, Novabiochem, San Diego, CA, USA) with an ABI 433A automated peptide synthesizer (Applied Biosystems, Foster city, CA, USA). After synthesis, the peptides were cleaved from the resin and deprotected in trifluoroacetic acid (TFA)/water/triisopropylsilane (TIPS)/thionisole (90:5:2.5:2.5 v/v) for 1.5 hr at room temperature. Peptides were purified by preparation RP-HPLC on a C18 column using a linear gradient of 5–99% solvent B in solvent A over 60 min (solvent A is 0.1% (v/v) TFA in water and solvent B is 0.1% (v/v) TFA in acetonitrile). Final purities of synthetic peptides were confirmed to be > 95% by analytical RP-HPLC, and the molecular weight of the purified product was confirmed by electrospray mass spectrometry (ESI) at the Stanford University Mass Spectrometry (SUMS) facility.
FP measurements
FP measurements were performed using a Synergy4 (Biotek, Winooski, VT, USA) plate reader with a tungsten lamp as a light source with an excitation wavelength of 485 nm and an emission wavelength of 530 nm. Fluorescently labeled 35 aa peptide, Fl-C35, was chosen as a tracer due to its inhibitory potency (EC50 of 0.051 µM) [21]. Lyophilized Fl-C35 was dissolved in 20 mM PBS and subsequent dilutions were done in FP buffer (20 mM PBS at pH 7.4, 500 mM NaCl, 0.01% (v/v) Tween-20, and 0.05 mg/ml bovine gamma globulin). Specific controls groups included free Fl-C35 (probe), bound Fl-C35 (Fl-C35 in the presence of 5HB), and FP buffer for every measurement, allowing accurate estimation of specific polarization.
Saturation Binding FP assays
The saturation binding experiments of Fl-C35 to 5HB were performed under the following condition: each well in a black 96-well plate (Corning Inc. Lowell, MA, USA) contained 5 nM Fl-C35 tracer peptide and increasing concentrations (0 to 500 nM) of 5HB in FP buffer in a final volume of 185 µL. The polarization in millipolarization units (mP) was measured after 1 hr incubation at room temperature. Data obtained were analyzed using GraphPad Prism 5.0 (GraphPad, San Diego, CA, USA) to calculate a binding dissociation constant (Kd) by fitting the experimental data using a one-site specific binding model. Experiments were performed in duplicate.
Competitive FP binding assays
Each well in a black 96-well plate (Corning Inc. Lowell, MA, USA) contained 20 nM 5HB and increasing concentrations (0.001 to 200 µM) of each truncated peptide in FP buffer in a final volume of 185 µL. After 1hr incubation at room temperature, 5 nM of Fl-C35 was added and additional 30 min incubation was followed at room temperature. The FP response was measured in duplicate with controls including free Fl-C35, bound Fl-C35, and FP buffer. All experimental data were plotted using GraphPad Prism 5.0 (GraphPad, San Diego, CA, USA). The percentage of inhibition (% Inhibition) was calculated using the following equation:
where mPf is the millipolarization of the free Fl-C35 control, mPb is the millipolarization of the bound Fl-C35 control and mP is the millipolarization of the bound inhibitor to the 5HB.
Results and Discussion
5-Helix Bundle construct design, expression, and purification
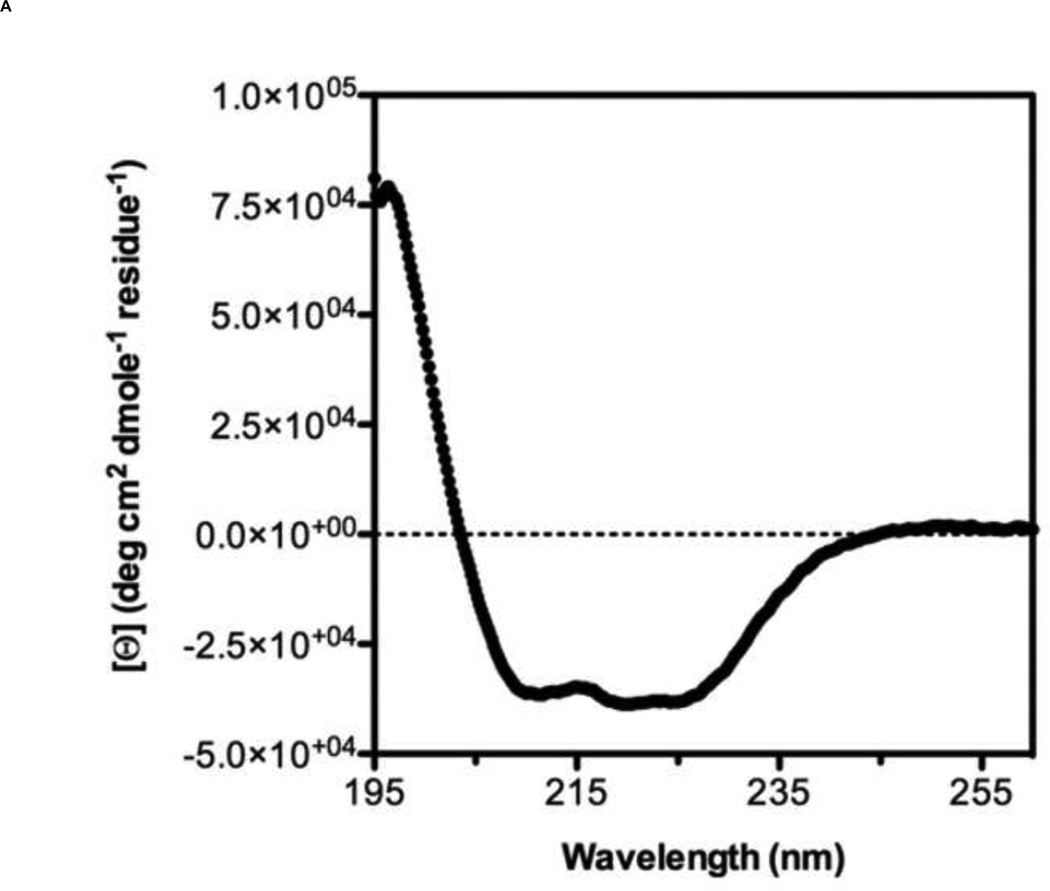
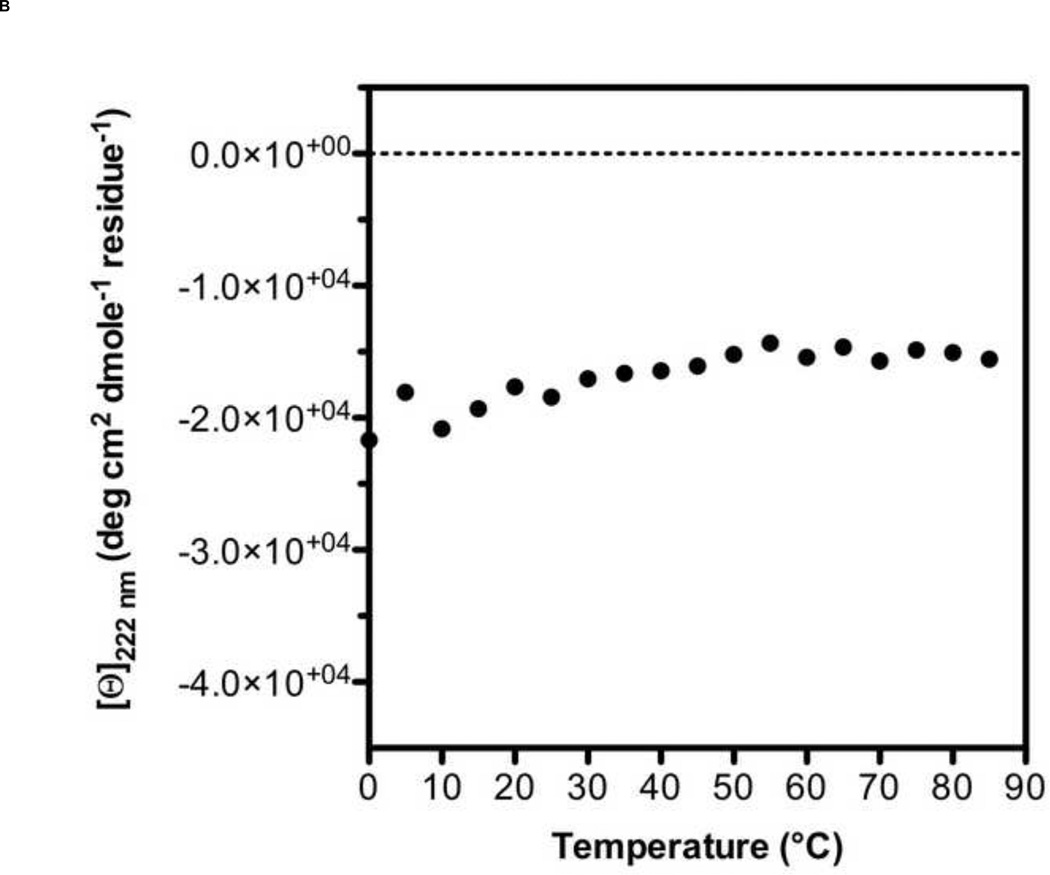
A 5-Helix bundle construct of HIV-1 fusion protein gp41 has been tested as a fusion inhibitor [32] and used as a target protein for screening small molecule libraries [33]. However, analogous constructs for hRSV F have been only tested as fusion inhibitors or vaccine candidates [31,35]. Therefore, we designed a 5-Helix Bundle (5HB) construct to specifically mimic the 6HB that forms during hRSV infection of host cell. The 5HB was generated by connecting three HRA and two HRB helices in an alternating sequence using short peptide linkers. The absence of the 3rd HRB in the 5HB would create a large open binding site for potential fusion inhibitors (Fig. 1B). Soluble 5HB was expressed in BL21 (DE3) cells and purified by metal-affinity chromatography (Fig. 1C). The secondary structure of the 5HB was assessed by Circular Dichroism (CD) spectroscopy (Fig. 2A), showing approximately 90% α-helicity, as calculated using Dichroweb [36,37]. In the range of temperature 0 to 85 °C, the thermal denaturation of the 5HB was not observed, indicating the secondary structure of the 5HB is highly stable (Fig. 2B).
Figure 2. Secondary structure analysis and thermal stability of 5HB by Circular Dichroism spectroscopy.
(A) CD spectrum of 5HB in 10 mM PBS at pH 7.4. (B) The melting curve of 5HB obtained from the ellipticity measurements at 222 nm between 0 °C and 85 °C. ([Θ]: per residue molar ellipticity measured in degrees · cm2 · dmol−1 · residue−1)
Saturation binding FP measurement
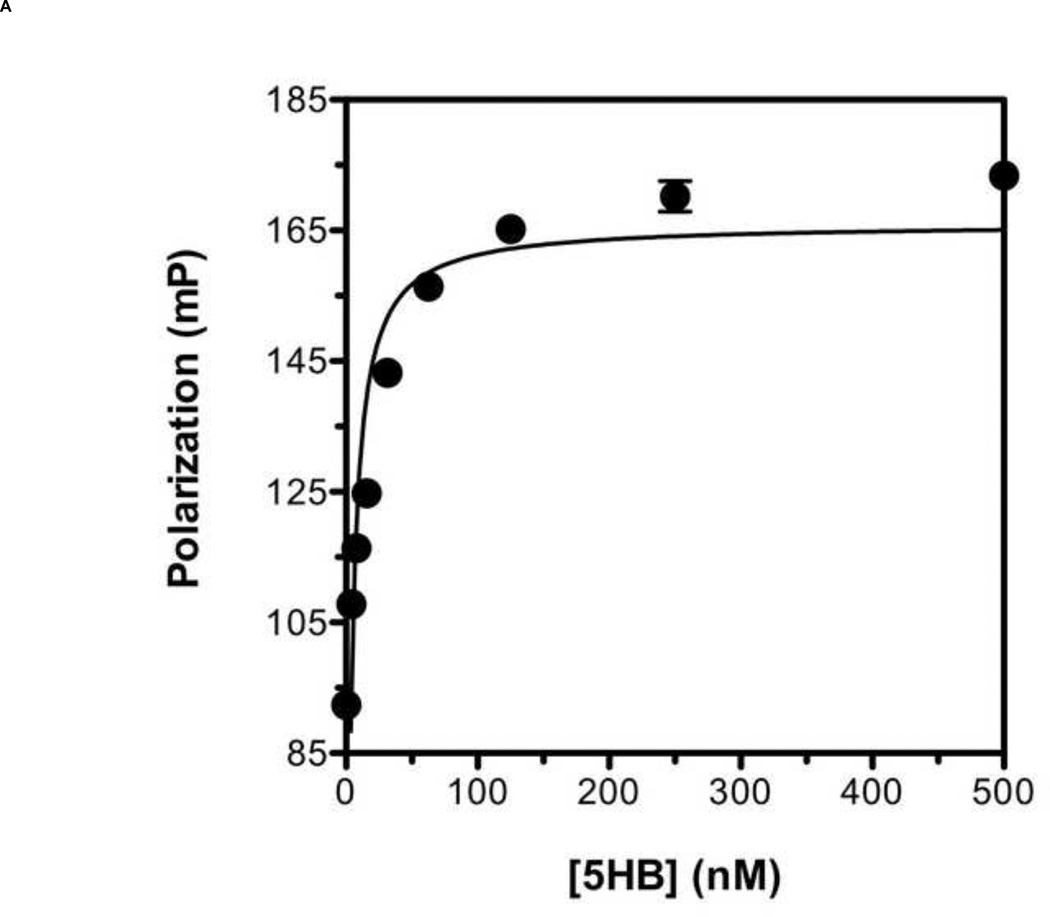
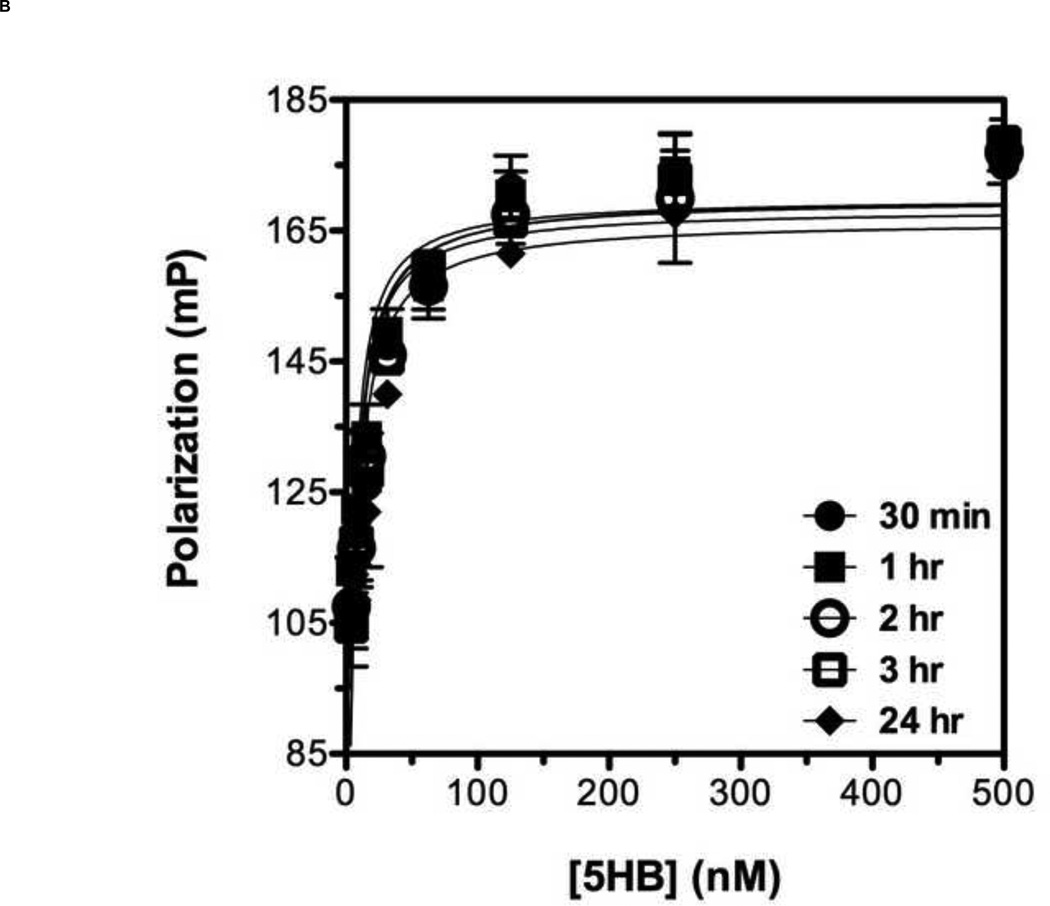
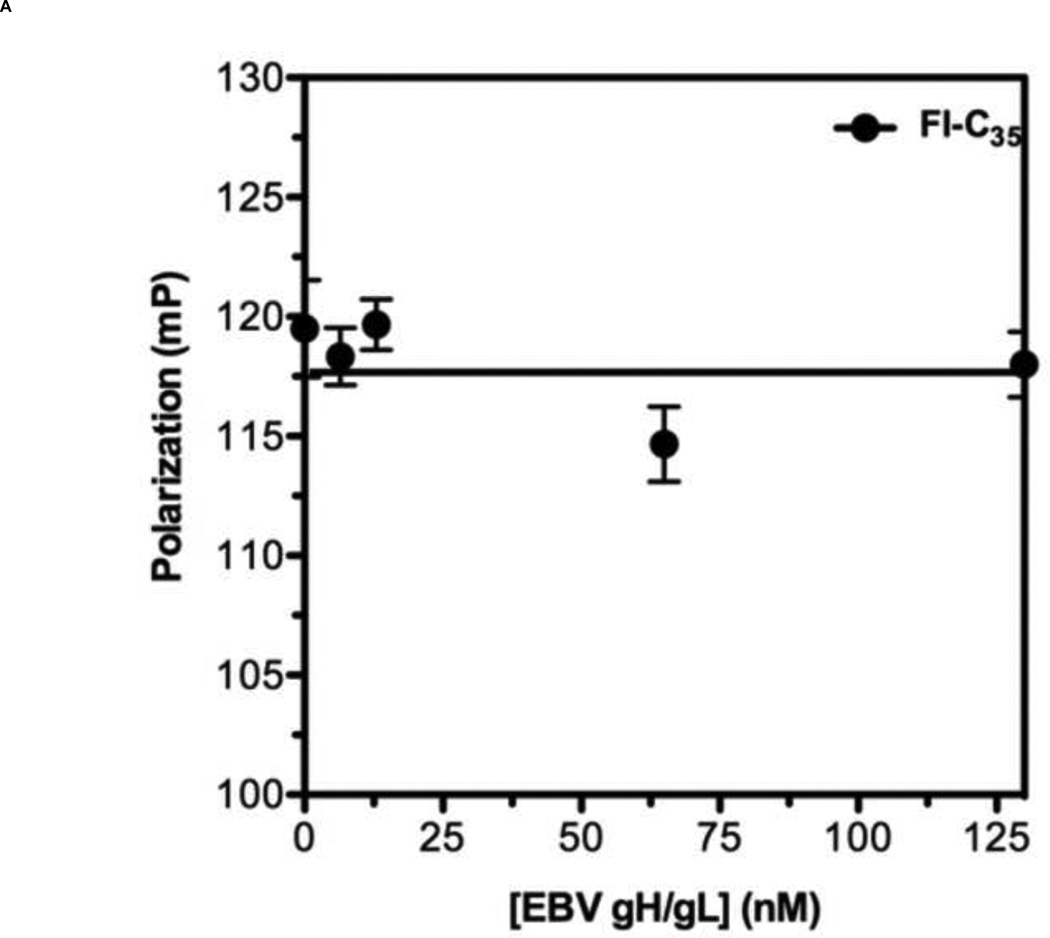
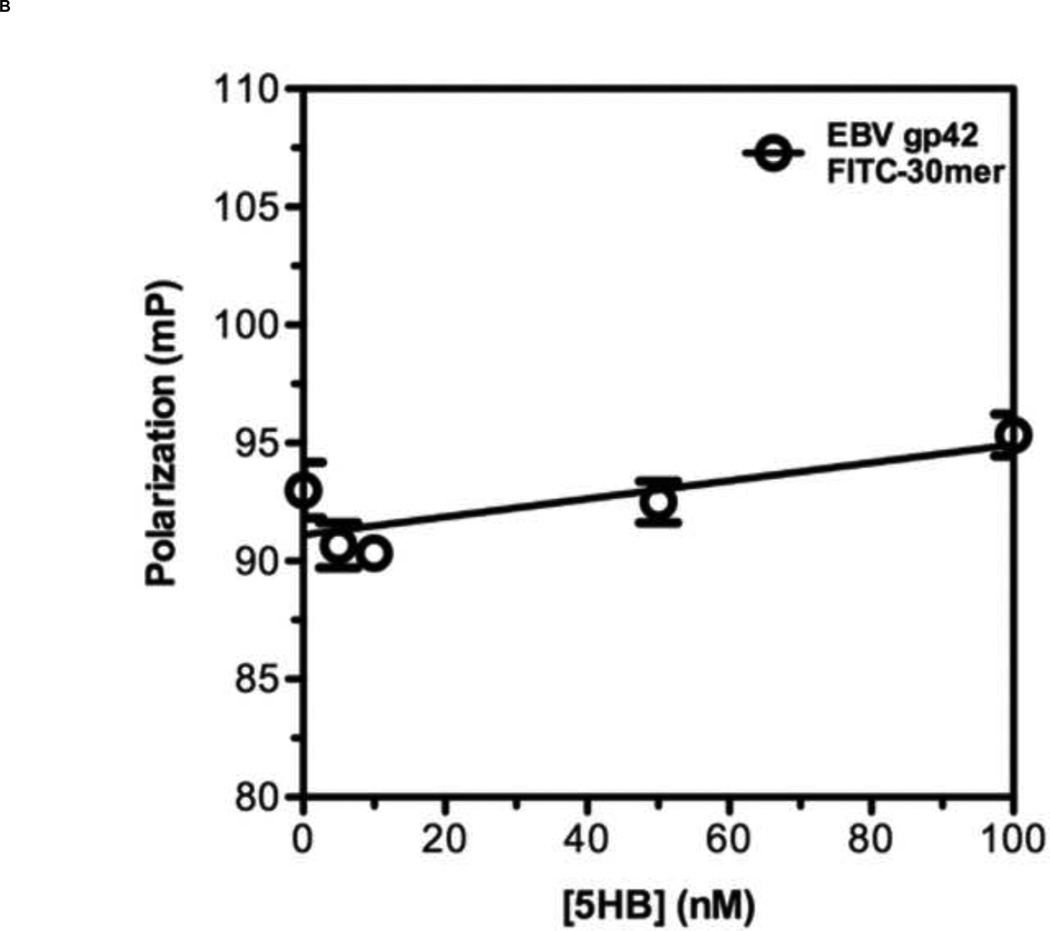
To develop a reliable fluorescence polarization assay, the binding affinity as well as specificity of the probe to the target protein should be high [38]. Previously, it has been shown that a series of overlapping 35-amino acid-long peptides from the conserved HRB domain within various paramyxovirus F proteins could block syncytium formation with EC50 values in the range of 0.015 – 0.25 µM [21]. Taking account of this previous study, we decided to use T-108 (35mer: YDPLVFPSDEFDASISQVN EKINQSLAFIRKSDEL) derived from hRSV F protein as a probe for developing a FP assay for the following reasons: First, the low EC50 value of 0.051 µM suggests that T-108 should bind tightly to the 5HB; Second, T-108 contains two phenylalanine residues (F483 and F488) located at the N-terminus of the HRB region, which engage a deep hydrophobic pocket located at the C-terminus of the HRA helices, known as a antiviral drug target (Fig. 3A; Fig. 3B shows the entire assay system in schematic format) [13,25,26]. This T-108 peptide was labeled with a fluorescein at its N-terminus (Fl-C35). To determine the binding affinity of Fl-C35, we used a fixed concentration of 5 nM Fl-C35 and monitored the FP response of Fl-C35 with increasing concentrations of the 5HB, as shown in Fig. 4A. The FP results were consistent with high affinity binding (Kd = 21 nM). The stability of the FP assay using Fl-C35 and the 5HB is also important as potential use for a high-throughput screening format. We therefore tested the stability of Fl-C35 binding to the 5HB by incubating the plate at room temperature over 24 hrs. Resulting binding curves show that this assay is highly robust (Fig. 4B). The specific binding of Fl-C35 to the 5HB was additionally confirmed in ELISA assays (Resulting data and detailed protocols provided in the supporting information). To further verify the selectivity and specificity of binding Fl-C35 for hRSV F protein, we took advantage of our previous findings of the high affinity binding of a FITC-30mer (30mer: KPNVEVWPVAAPPPVAEQEYGDKEVKLPHW) derived from the Epstein-Barr virus (EBV) gp42 to the EBV gH/gL complex [39,40]. The interaction of the EBV gp42 and the gH/gL complex are known to play key roles in membrane fusion and we previously reported that the EBV gp42-derived FITC-30mer specifically binds to EBV gH/gL protein with an IC50 value of 1.3 nM [40]. To confirm the selectivity and specificity of both the hRSV 5HB and the Fl-C35 probe, we crosstested the EBV gp42-derived FITC-30mer against the hRSV 5HB (Fig. 5A) and our hRSV-derived Fl-C35 against EBV gH/gL complex under the same conditions (Fig. 5B). There was no evidence of nonspecific binding in these controls, indicating the interaction between Fl-C35 and the 5HB is specific and selective, which can provide a solid basis for developing a competitive FP-based 5HB assay.
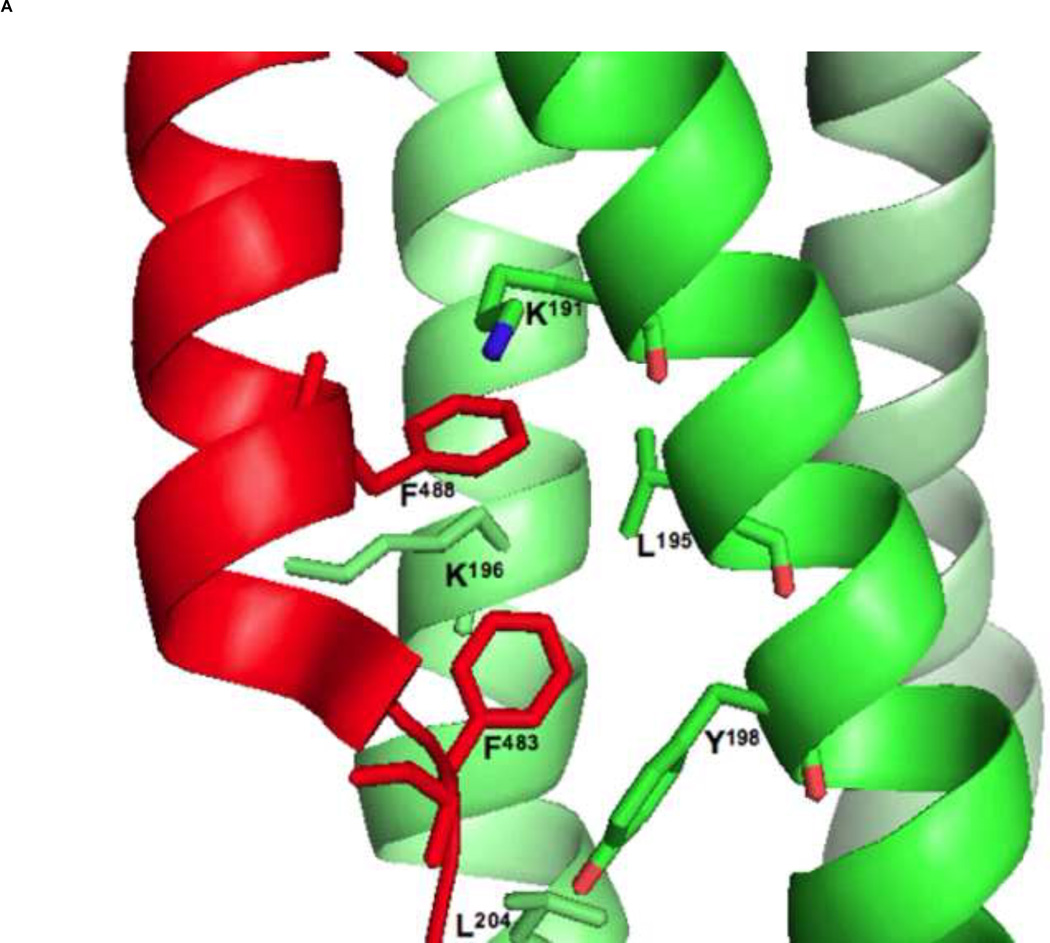
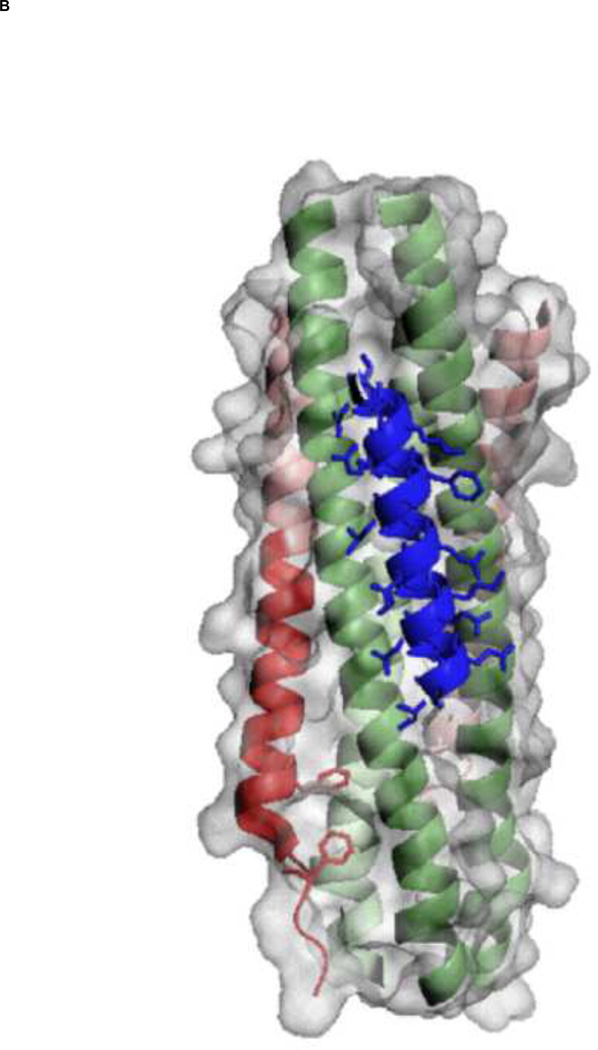
Figure 3. A key interaction between HRA and HRB helices in the 6HB assembly and a 3-D model of C20-bound 5HB.
(A) Two phenylalanine residues of the hRSV F HRB domain (red) play a key role in the interaction with the hRSV F HRA helices (green) by packing into the hydrophobic pocket formed by HRA helices (adapted from [13]). (B) The proposed 5HB model with C20 peptide (blue) and surface area of the 5HB (light gray) are shown. The hydrophobic groove formed by two neighboring HRA helices will provide a wide binding site for future antiviral development. Figures were generated by PyMol software [50].
Figure 4. Binding titration curve of Fl-C35 and its binding stability to the 5HB.
(A) The fluorescence polarization response of Fl-C35 binding to the 5HB was monitored as the concentration of the 5HB increased. The experiment was performed using 5 nM Fl-C35 and the 5HB concentration ranged from 0 to 500 nM. (B) The stability of FL-C35 binding to the 5HB was monitored over a 24 hr period using 5 nM of Fl-C35 in the presence of increasing amount of the 5HB.
Figure 5. Specificity of Fl-C35 to the 5HB.
To confirm the specificity of Fl-C35 to the 5HB, negative controls were tested. (A) 5 nM of Fl-C35 in the presence of increasing amount of the Epstein-Barr virus (EBV) gH/gL, a fusion protein that leads the EBV infection was tested. (B) 5 nM of EBV gp42 FITC-30mer with a wide range of the 5HB in concentration was monitored.
Competitive FP assays
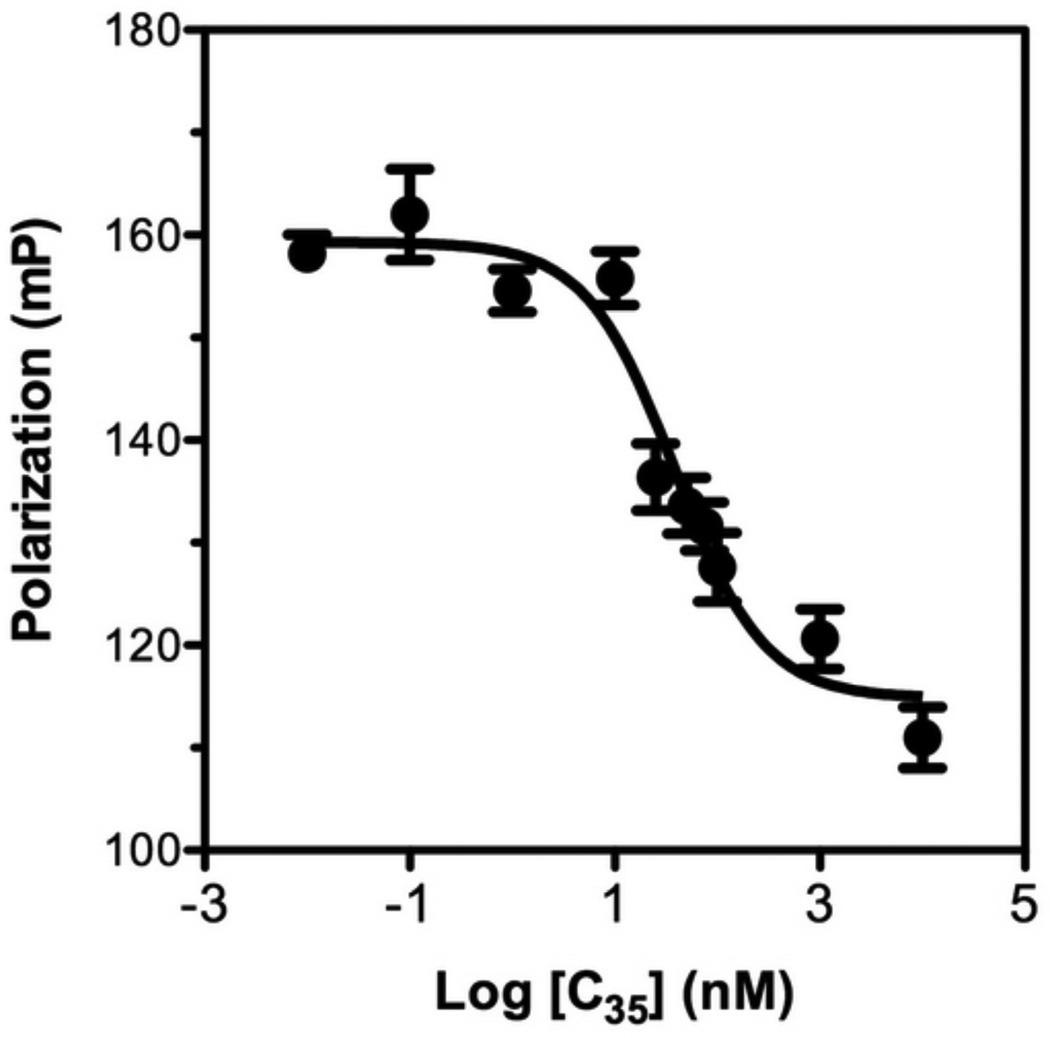
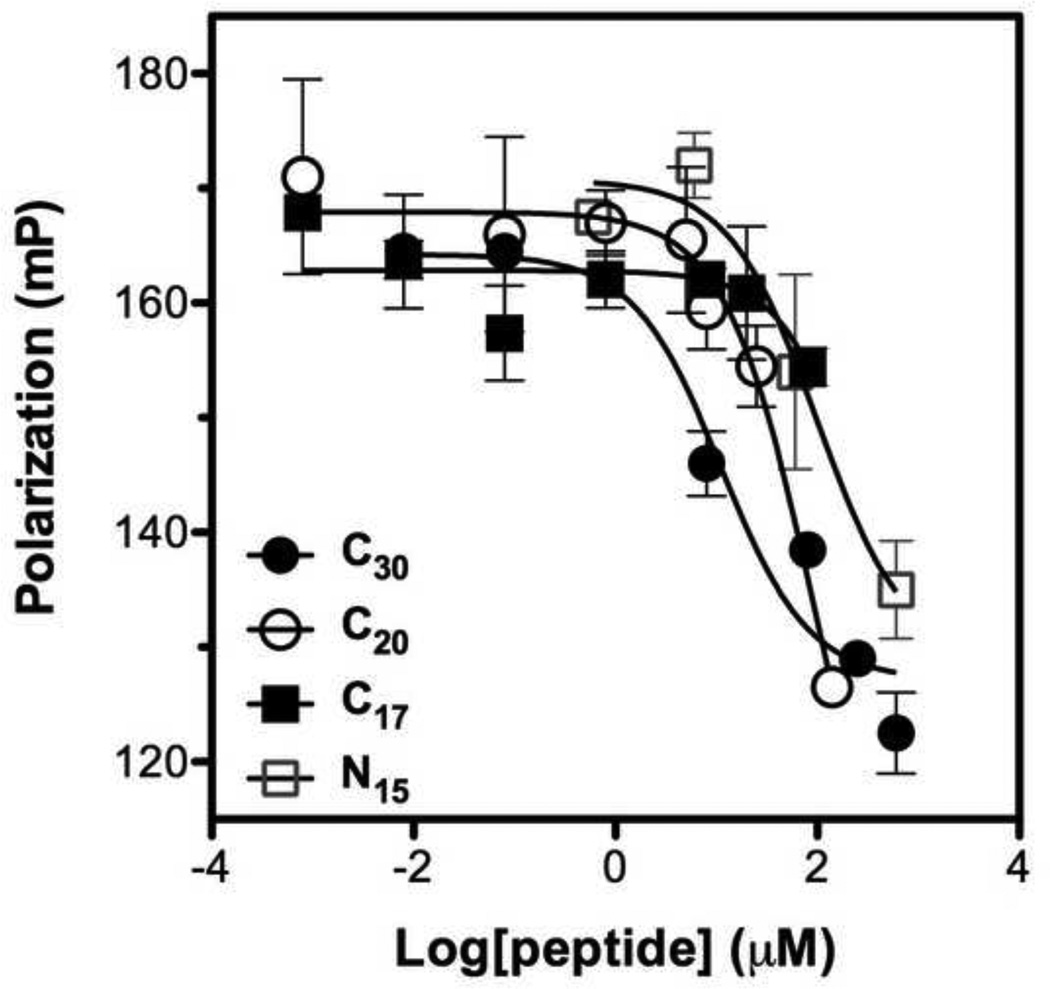
Based on the Kd value observed in the saturation binding FP assay, we established a competitive FP assay to evaluate potential inhibitors based on their ability to displace the Fl-C35 (probe) from the 5HB (target). We first tested unlabelled C35 against Fl-C35 in the presence of the 5HB (Fig. 6). Unlabelled C35 peptide blocked the increase in fluorescence polarization with IC50 of 38.52 nM, competing with Fl-C35 over the binding site on the 5HB. Manufacturing longer bioactive peptides can be problematic due to high-cost, but shorter unstructured peptides can easily lose its efficacy, thereby implying two significant but contradictory criteria of designing peptide therapeutics. We therefore prepared a series of truncated peptides derived from the HRB domain of hRSV F protein (Table 1) and investigated their ability to compete against Fl-C35 using our competitive FP-based 5HB assay (Fig. 7). Truncated peptides tested in this study do not have two phenylalanines that bind to the hydrophobic pocket (Fig. 3A), which have been the focus of small molecule drug discovery effort [1,13,41]. However, in previous work by Lambert and coworkers, the absence of these residues in various 35mer peptides still revealed significant antiviral activity [21]. The results in Fig. 7 and Table 1 summarize our quantitative observations of the binding activity of increasingly truncated peptides, which provide important information of the location as well as length peptides that mediate inhibition. These data clearly suggest that the entire hydrophobic groove on the C-terminus of neighboring HRA helices would be available as a potential drug target for developing hRSV fusion inhibitors (Fig. 3B). Among tested peptides, C20 (IC50 = 14.92 µM) was further investigated for its antiviral activity which showed EC50 > 12.50 µM (Detailed methods provided in the supporting information), denoting that our FP-based assay can be comparable with conventional virological assays. In addition, the reproducibility of measurements for free and bound Fl-C35 controls were examined to calculate the Z’ factor, a parameter for the quality and robustness of the assay [38] according to the guidelines provided by National Institutes of Health (NIH) [42]. The resulting Z’ factor (Z’ > 0.8) suggests that our competitive FP-based assay can be further applied in a high-throughput screening approach.
Figure 6. Competitive fluorescence polarization assay using unlabeled C35 to compete with the binding of FL-C35 to 5HB.
Increasing concentrations of unlabeled C35 were tested in the presence of 5 nM Fl-C35 and 20 nM 5HB.
Table 1. Peptides derived from the HRB sequence of hRSV F protein and tested in this study.
Peptides with longer than 20-amino acid long were commercially obtained and the rest of peptides were synthesized in the laboratory using standard Fmoc chemistry. The purity of peptides is >95% judged by analytical HPLC.
| Peptide | Sequence (amino to carboxy) | % Inhibition (100 µM) |
IC50 (µM) |
|---|---|---|---|
| HRBa | NFYDPLVFPSDEFDASISQVNEKINQSLAFIRKSDELLHNVNAGKSTTN | - | - |
| T-108 | YDPLVFPSDEFDASISQVNEKINQSLAFIRKSDEL | 0.051b | |
| C35 | YDPLVFPSDEFDASISQVNEKINQSLAFIRKSDEL | 100 | 0.038 |
| C30 | VFPSDEFDASISQVNEKINQSLAFIRKSDE | 100 | 6.80 |
| C20 | ISQVNEKINQSLAFIRKSDE | > 90 | 14.92 |
| C17 | VNEKINQSLAFIRKSDE | < 50 | > 100 |
| C13 | INQSLAFIRKSDE | NMc | > 500 |
| C10 | SLAFIRKSDE | NM | > 500 |
| N15 | VFPSDEFDASISQVN | < 50 | > 100 |
HRB sequence from hRSV F protein
EC50 value from Lambert et al. (1996), crude peptide T-108 was analyzed for its ability to prevent cytopathologic effect (CPE) in infectivity assays with hRSV.
NM: Not measurable
Figure 7. Competitive fluorescence polarization assays of N- and C-terminally truncated peptides.
Competitive binding ability of each truncated peptide derived from the hRSV HRB domain was evaluated. Various concentrations of peptides were competed with 5 nM Fl-C35 in the presence of 20 nM 5HB. Symbols represent peptides; C30 , C20
, C20 , C17
, C17 , and N15
, and N15 .
.
Conclusions
In this study, we demonstrated that a peptide-binding assay can be used as a direct screening platform for identifying potential antivirals against hRSV using fluorescence polarization (FP), which has been widely used for a direct, nearly instantaneous measurement of molecular interactions such as protein-protein [43–45], DNA-protein [46], and small molecule-protein [47,48] interactions. Our competitive FP-based assay for the formation of the hRSV 6HB can measure biological activities of short F-derived peptides over a wide range of binding affinity, suggesting that this system is sufficiently sensitive to screen weak binders to the 5HB as potential antiviral candidates. Moreover, this assay could be suitable for a preparatory high throughput screening effort, prior to in vivo assays to prioritize inhibitor candidates. This 5HB construct from the hRSV F protein may prove useful for other viruses within the paramyxovirus family for future drug discovery efforts.
Recently, it has been suggested that a small molecule inhibitor of hRSV F entry may act, not by locking 6HB formation, but by distorting the final 6HB conformation [49]. These studies indicate that the inhibitor (TMC353121) engages both HRA and HRB regions and may thereby enhance 6HB assembly into a non-functional structure. The 5HB system described here may allow quantitative studies of this and other hRSV F fusion inhibitors and help classify the mode of action of these compounds.
Supplementary Material
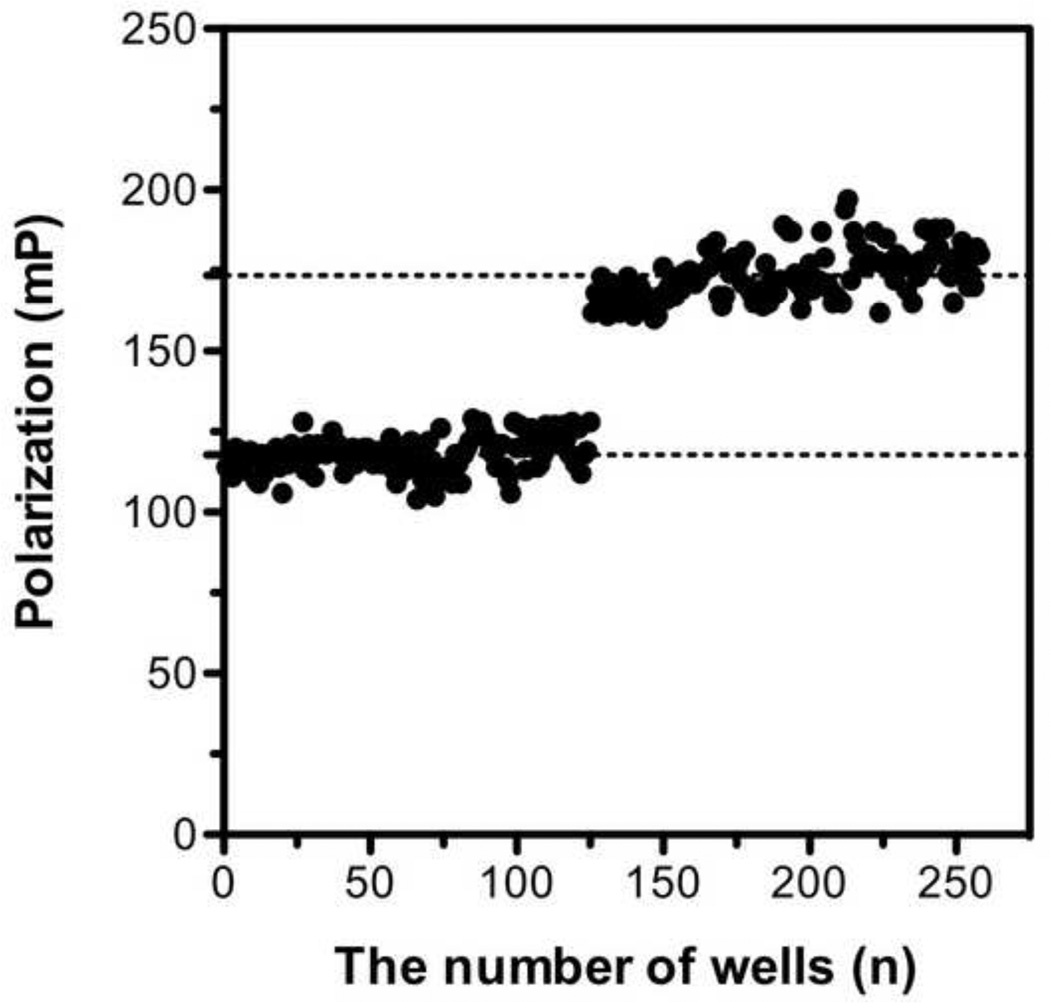
Figure 8. Assay robustness test by FP measurements of free and bound Fl-C35 controls.
FP response for 5nM of Fl-C35 was monitored in the absence and presence of the 5HB construct (20 nM) as negative and positive controls.
Acknowledgments
The project described here was supported in part by National Institutes of Health (NIH) National Institute of Allergy and Infectious Disease (NIAID) Grant R01-AI072666 to A.E.B., NIH Grant R01-GM61050 to T.S.J. and by the Bio-X Interdisciplinary Initiatives Program at Stanford University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Carter M, Cockerill GS. Inhibitors of Respiratory Syncytial Virus. Annual Reports in Medicinal Chemistry. 2008;Vol 43 43:229–245. [Google Scholar]
- 2.Powell KL, Alber D. Development of Antivirals against Respiratory Syncytial Virus. Perspectives in Medical Virology. 2006;14:279–297. [Google Scholar]
- 3.Collins PL, Chanock RM, Murphy BR. Resipiratory syncytial virus. Philadelphia: Lippincott Williams & Wilkins; 2001. [Google Scholar]
- 4.Leader S, Kohlhase K. Recent trends in severe respiratory syncytial virus (RSV) among US infants, 1997 to 2000. Journal of Pediatrics. 2003;143:S127–S132. doi: 10.1067/s0022-3476(03)00510-9. [DOI] [PubMed] [Google Scholar]
- 5.Wyde PR. Respiratory syncytial virus (RSV) disease and prospects for its control. Antiviral Res. 1998;39:63–79. doi: 10.1016/s0166-3542(98)00029-1. [DOI] [PubMed] [Google Scholar]
- 6.Shay DK, Holman RC, Newman RD, Liu LL, Stout JW, Anderson LJ. Bronchiolitis-associated hospitalizations among US children, 1980–1996. JAMA. 1999;282:1440–1446. doi: 10.1001/jama.282.15.1440. [DOI] [PubMed] [Google Scholar]
- 7.Stang P, Brandenburg N, Carter B. The economic burden of respiratory syncytial virus-associated bronchiolitis hospitalizations. Arch Pediatr Adolesc Med. 2001;155:95–96. doi: 10.1001/archpedi.155.1.95. [DOI] [PubMed] [Google Scholar]
- 8.Glezen WP, Taber LH, Frank AL, Kasel JA. Risk of primary infection and reinfection with respiratory syncytial virus. Am J Dis Child. 1986;140:543–546. doi: 10.1001/archpedi.1986.02140200053026. [DOI] [PubMed] [Google Scholar]
- 9.Hashem M, Hall CB. Respiratory syncytial virus in healthy adults: the cost of a cold. Journal of Clinical Virology. 2003;27:14–21. doi: 10.1016/s1386-6532(03)00047-7. [DOI] [PubMed] [Google Scholar]
- 10.Falsey AR, Hennessey PA, Formica MA, Cox C, Walsh EE. Respiratory syncytial virus infection in elderly and high-risk adults. N Engl J Med. 2005;352:1749–1759. doi: 10.1056/NEJMoa043951. [DOI] [PubMed] [Google Scholar]
- 11.Sidwell RW, Barnard DL. Respiratory syncytial virus infections: recent prospects for control. Antiviral Res. 2006;71:379–390. doi: 10.1016/j.antiviral.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 12.Gill MA, Welliver RC. Motavizumab for the prevention of respiratory syncytial virus infection in infants. Expert Opin Biol Ther. 2009;9:1335–1345. doi: 10.1517/14712590903287499. [DOI] [PubMed] [Google Scholar]
- 13.Zhao X, Singh M, Malashkevich VN, Kim PS. Structural characterization of the human respiratory syncytial virus fusion protein core. Proc Natl Acad Sci U S A. 2000;97:14172–14177. doi: 10.1073/pnas.260499197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439:38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin HS, Paterson RG, Wen X, Lamb RA, Jardetzky TS. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc Natl Acad Sci U S A. 2005;102:9288–9293. doi: 10.1073/pnas.0503989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baker KA, Dutch RE, Lamb RA, Jardetzky TS. Structural basis for paramyxovirus-mediated membrane fusion. Mol Cell. 1999;3:309–319. doi: 10.1016/s1097-2765(00)80458-x. [DOI] [PubMed] [Google Scholar]
- 17.Colman PM, Lawrence MC. The structural biology of type I viral membrane fusion. Nat Rev Mol Cell Biol. 2003;4:309–319. doi: 10.1038/nrm1076. [DOI] [PubMed] [Google Scholar]
- 18.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Connolly SA, Leser GP, Yin HS, Jardetzky TS, Lamb RA. Refolding of a paramyxovirus F protein from prefusion to postfusion conformations observed by liposome binding and electron microscopy. Proc Natl Acad Sci U S A. 2006;103:17903–17908. doi: 10.1073/pnas.0608678103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russell CJ, Jardetzky TS, Lamb RA. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J. 2001;20:4024–4034. doi: 10.1093/emboj/20.15.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lambert DM, Barney S, Lambert AL, Guthrie K, Medinas R, Davis DE, Bucy T, Erickson J, Merutka G, Petteway SR., Jr Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc Natl Acad Sci U S A. 1996;93:2186–2191. doi: 10.1073/pnas.93.5.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eckert DM, Kim PS. Design of potent inhibitors of HIV-1 entry from the gp41 N-peptide region. Proc Natl Acad Sci U S A. 2001;98:11187–11192. doi: 10.1073/pnas.201392898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang E, Sun X, Qian Y, Zhao L, Tien P, Gao GF. Both heptad repeats of human respiratory syncytial virus fusion protein are potent inhibitors of viral fusion. Biochem Biophys Res Commun. 2003;302:469–475. doi: 10.1016/s0006-291x(03)00197-9. [DOI] [PubMed] [Google Scholar]
- 24.Douglas JL, Panis ML, Ho E, Lin KY, Krawczyk SH, Grant DM, Cai R, Swaminathan S, Cihlar T. Inhibition of respiratory syncytial virus fusion by the small molecule VP-14637 via specific interactions with F protein. J Virol. 2003;77:5054–5064. doi: 10.1128/JVI.77.9.5054-5064.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cianci C, Langley DR, Dischino DD, Sun Y, Yu KL, Stanley A, Roach J, Li Z, Dalterio R, Colonno R, Meanwell NA, Krystal M. Targeting a binding pocket within the trimer-of-hairpins: small-molecule inhibition of viral fusion. Proc Natl Acad Sci U S A. 2004;101:15046–15051. doi: 10.1073/pnas.0406696101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shepherd NE, Hoang HN, Desai VS, Letouze E, Young PR, Fairlie DP. Modular alpha-helical mimetics with antiviral activity against respiratory syncitial virus. J Am Chem Soc. 2006;128:13284–13289. doi: 10.1021/ja064058a. [DOI] [PubMed] [Google Scholar]
- 27.Rapaport D, Ovadia M, Shai Y. A synthetic peptide corresponding to a conserved heptad repeat domain is a potent inhibitor of Sendai virus-cell fusion: an emerging similarity with functional domains of other viruses. EMBO J. 1995;14:5524–5531. doi: 10.1002/j.1460-2075.1995.tb00239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Young JK, Li D, Abramowitz MC, Morrison TG. Interaction of peptides with sequences from the Newcastle disease virus fusion protein heptad repeat regions. J Virol. 1999;73:5945–5956. doi: 10.1128/jvi.73.7.5945-5956.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shai Y. Functional domains within fusion proteins: prospectives for development of peptide inhibitors of viral cell fusion. Biosci Rep. 2000;20:535–555. doi: 10.1023/a:1010411021326. [DOI] [PubMed] [Google Scholar]
- 30.Debnath AK. Prospects and strategies for the discovery and development of small-molecule inhibitors of six-helix bundle formation in class 1 viral fusion proteins. Curr Opin Investig Drugs. 2006;7:118–127. [PubMed] [Google Scholar]
- 31.Ni L, Zhao L, Qian Y, Zhu J, Jin Z, Chen YW, Tien P, Gao GF. Design and characterization of human respiratory syncytial virus entry inhibitors. Antivir Ther. 2005;10:833–840. [PubMed] [Google Scholar]
- 32.Root MJ, Kay MS, Kim PS. Protein design of an HIV-1 entry inhibitor. Science. 2001;291:884–888. doi: 10.1126/science.1057453. [DOI] [PubMed] [Google Scholar]
- 33.Frey G, Rits-Volloch S, Zhang XQ, Schooley RT, Chen B, Harrison SC. Small molecules that bind the inner core of gp41 and inhibit HIV envelope-mediated fusion. Proc Natl Acad Sci U S A. 2006;103:13938–13943. doi: 10.1073/pnas.0601036103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sorensen HP, Mortensen KK. Soluble expression of recombinant proteins in the cytoplasm of Escherichia coli. Microb Cell Fact. 2005;4:1. doi: 10.1186/1475-2859-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ni L, Zhao L, Gao GF, Qian Y, Tien P. The antibodies directed against N-terminal heptad-repeat peptide of hRSV fusion protein and its analog-5-Helix inhibit virus infection in vitro. Biochem Biophys Res Commun. 2005;331:1358–1364. doi: 10.1016/j.bbrc.2005.04.046. [DOI] [PubMed] [Google Scholar]
- 36.Whitmore L, Wallace BA. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers. 2008;89:392–400. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- 37.Lobley A, Whitmore L, Wallace BA. DICHROWEB: an interactive website for the analysis of protein secondary structure from circular dichroism spectra. Bioinformatics. 2002;18:211–212. doi: 10.1093/bioinformatics/18.1.211. [DOI] [PubMed] [Google Scholar]
- 38.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 39.Kirschner AN, Lowrey AS, Longnecker R, Jardetzky TS. Binding-site interactions between Epstein-Barr virus fusion proteins gp42 and gH/gL reveal a peptide that inhibits both epithelial and B-cell membrane fusion. J Virol. 2007;81:9216–9229. doi: 10.1128/JVI.00575-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu F, Marquardt G, Kirschner AN, Longnecker R, Jardetzky TS. Mapping the N-terminal Residues of EBV gp42 that Bind gH/gL Using Fluorescence Polarization and Cell-based Fusion Assays. J Virol. 2010 doi: 10.1128/JVI.00381-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bonfanti JF, Roymans D. Prospects for the development of fusion inhibitors to treat human respiratory syncytial virus infection. Curr Opin Drug Discov Devel. 2009;12:479–487. [PubMed] [Google Scholar]
- 42.Assay Guidance Manual Version 5.0. Eli Lilly and Company and NIH Chemical Genomics Center; 2009. Dec 8, Available online at: http://www.ncgc.nih.gov/guidance/manual_toc.html. [PubMed] [Google Scholar]
- 43.Park SH, Raines RT. Fluorescence polarization assay to quantify protein-protein interactions. Methods Mol Biol. 2004;261:161–166. doi: 10.1385/1-59259-762-9:161. [DOI] [PubMed] [Google Scholar]
- 44.Knight SM, Umezawa N, Lee HS, Gellman SH, Kay BK. A fluorescence polarization assay for the identification of inhibitors of the p53-DM2 protein-protein interaction. Anal Biochem. 2002;300:230–236. doi: 10.1006/abio.2001.5468. [DOI] [PubMed] [Google Scholar]
- 45.Buchli R, VanGundy RS, Hickman-Miller HD, Giberson CF, Bardet W, Hildebrand WH. Development and validation of a fluorescence polarization-based competitive peptide-binding assay for HLA-A*0201--a new tool for epitope discovery. Biochemistry. 2005;44:12491–12507. doi: 10.1021/bi050255v. [DOI] [PubMed] [Google Scholar]
- 46.Craig JC, Schumacher MA, Mansoor SE, Farrens DL, Brennan RG, Goodman RH. Consensus and variant cAMP-regulated enhancers have distinct CREB-binding properties. J Biol Chem. 2001;276:11719–11728. doi: 10.1074/jbc.M010263200. [DOI] [PubMed] [Google Scholar]
- 47.Mathias U, Jung M. Determination of drug-serum protein interactions via fluorescence polarization measurements. Anal Bioanal Chem. 2007;388:1147–1156. doi: 10.1007/s00216-007-1351-7. [DOI] [PubMed] [Google Scholar]
- 48.Liu Y, Jiang J, Richardson PL, Reddy RD, Johnson DD, Kati WM. A fluorescence polarization-based assay for peptidyl prolyl cis/trans isomerase cyclophilin A. Anal Biochem. 2006;356:100–107. doi: 10.1016/j.ab.2006.04.040. [DOI] [PubMed] [Google Scholar]
- 49.Roymans D, De Bondt HL, Arnoult E, Geluykens P, Gevers T, Van Ginderen M, Verheyen N, Kim H, Willebrords R, Bonfanti JF, Bruinzeel W, Cummings MD, van Vlijmen H, Andries K. Binding of a potent small-molecule inhibitor of six-helix bundle formation requires interactions with both heptad-repeats of the RSV fusion protein. Proc Natl Acad Sci U S A. 2010;107:308–313. doi: 10.1073/pnas.0910108106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, CA, USA: DeLano Scientific LLC.; [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.