Abstract
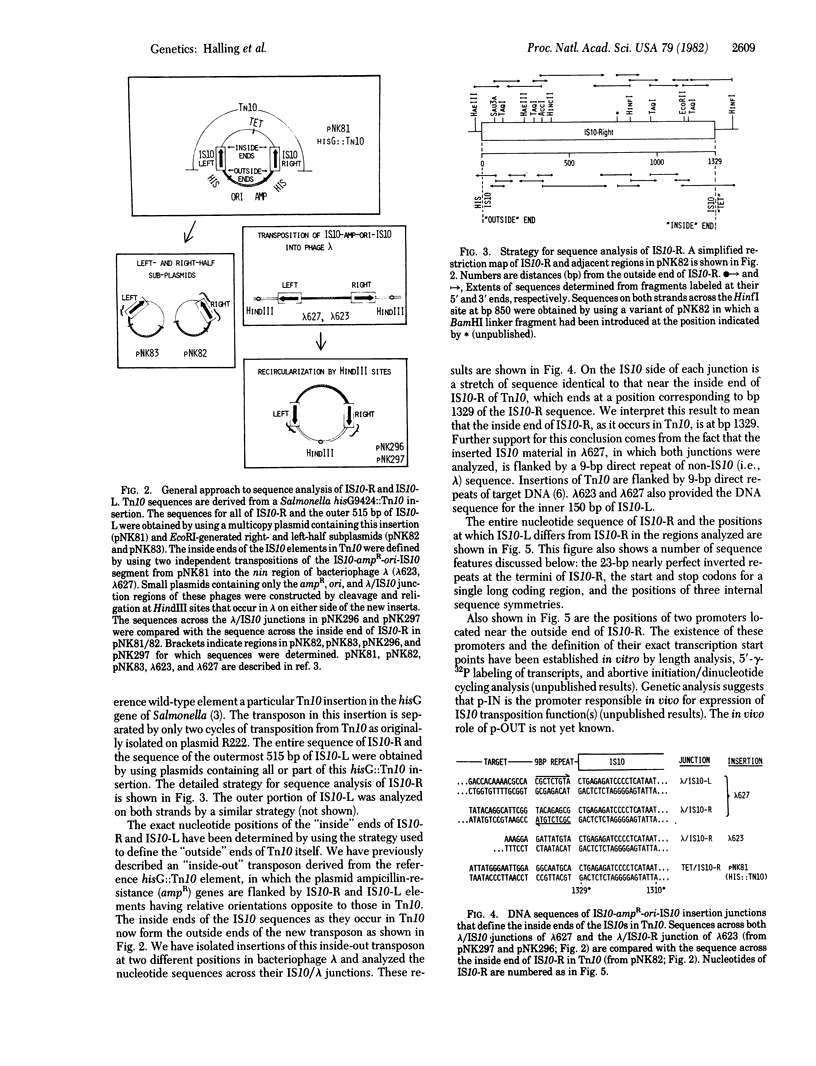
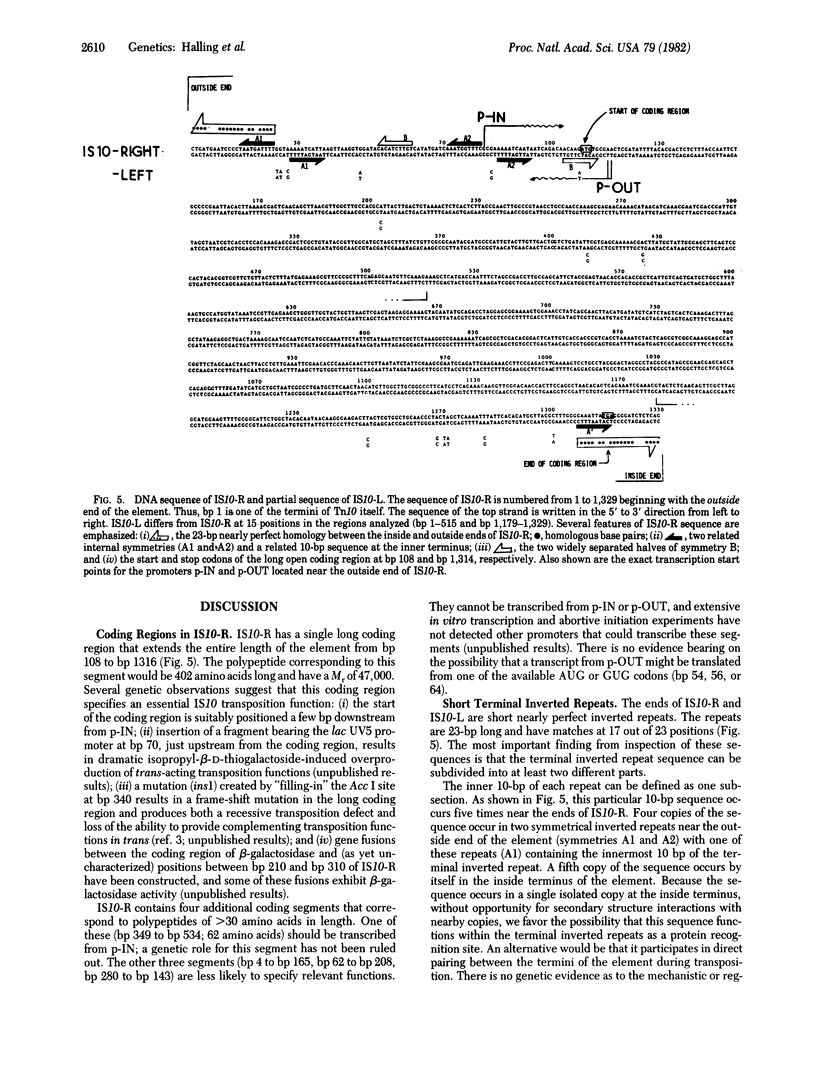
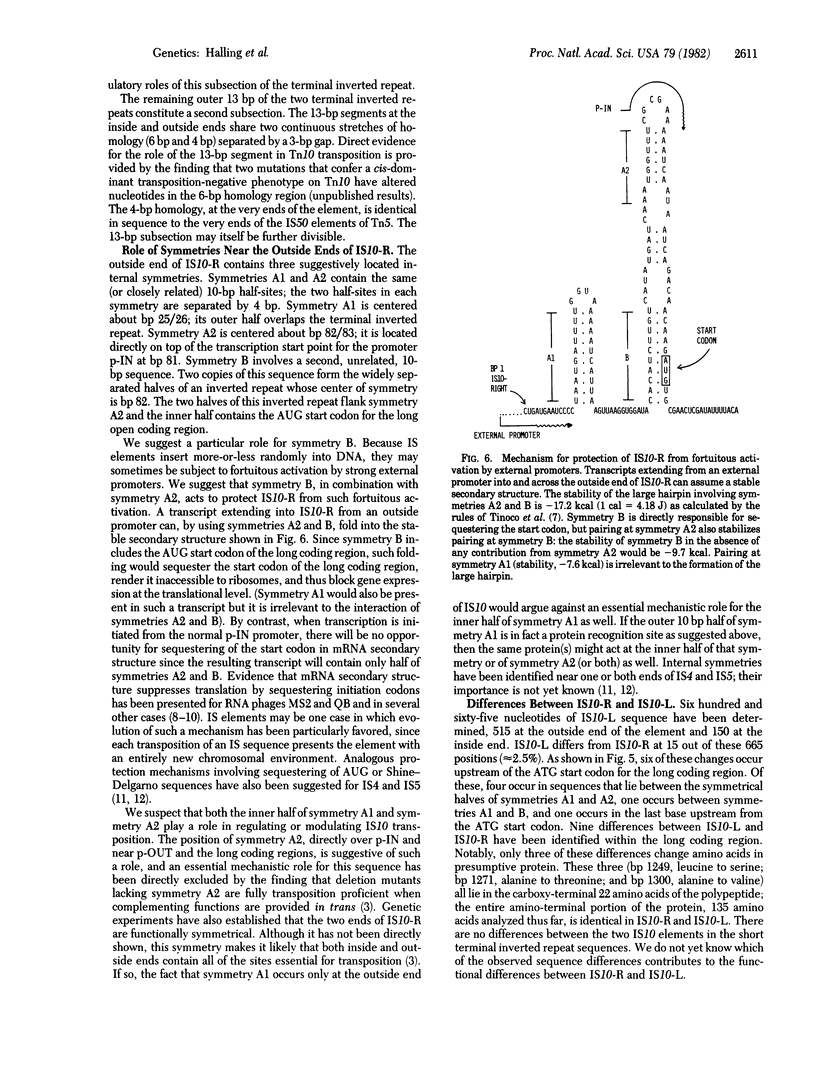
Tn10 is 9,300 base pairs long and has inverted repeats of an insertion sequence (IS)-like sequence (IS10) at its ends. IS10-right provides all of the Tn10-encoded functions used for normal Tn10 transposition. IS10-left can also provide these functions but at a much reduced level. We report here the complete nucleotide sequence of IS10-right and a partial sequence of IS10-left. From our analysis of this information, we draw the following conclusions. (i) IS10-right is 1,329 base pairs long. Like most IS elements, it has short (23-base pair) nearly perfect inverted repeats at its termini. We can divide these 23-base pair segments into at least two functionally distinct parts. IS10-right also shares with other elements the presence of a single long coding region that extends the entire length of the element. Genetic evidence suggests that this coding region specifies an essential IS10 transposition function. A second, overlapping, coding region may or may not be important. (ii) The “outside” end of IS10-right contains three suggestively positioned internal symmetries. Two of these (A1 and A2) are nearly identical in sequence. Symmetry A1 overlaps the terminal inverted repeat; symmetry A2 overlaps the promoter shown elsewhere to be responsible for expression of IS10 functions and lies very near a second characterized promoter that directs transcription outward across the end of IS10. Symmetries A1 and A2 may play a role in modulation of Tn10 activity and are likely to function at least in part as protein recognition sites. We propose that the third symmetry (B) acts to prevent fortuitous expression of IS10 functions from external promoters. The transcripts from such promoters can assume a stable secondary structure in which the AUG start codon of the long coding region is sequestered in a region of double-stranded mRNA formed by pairing between the two halves of symmetry B. (iii) IS10-left differs from IS10-right at many nucleotide positions in both the presumptive regulatory region and the long coding region. The available evidence suggests that Tn10 may be older than other analyzed drug-resistance transposons and thus have had more time to accumulate mutational changes.
Keywords: transposon genetic organization, insertion sequence genetic organization, inverted repeat, regulation of transposition, transposon evolution
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Auerswald E. A., Ludwig G., Schaller H. Structural analysis of Tn5. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):107–113. doi: 10.1101/sqb.1981.045.01.019. [DOI] [PubMed] [Google Scholar]
- Backman K., Ptashne M., Gilbert W. Construction of plasmids carrying the cI gene of bacteriophage lambda. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4174–4178. doi: 10.1073/pnas.73.11.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J., Davis M. A., Roberts D. E., Takeshita K., Kleckner N. Genetic organization of transposon Tn10. Cell. 1981 Jan;23(1):201–213. doi: 10.1016/0092-8674(81)90285-3. [DOI] [PubMed] [Google Scholar]
- Iserentant D., Fiers W. Secondary structure of mRNA and efficiency of translation initiation. Gene. 1980 Apr;9(1-2):1–12. doi: 10.1016/0378-1119(80)90163-8. [DOI] [PubMed] [Google Scholar]
- Klaer R., Kühn S., Tillmann E., Fritz H. J., Starlinger P. The sequence of IS4. Mol Gen Genet. 1981;181(2):169–175. doi: 10.1007/BF00268423. [DOI] [PubMed] [Google Scholar]
- Kleckner N., Chan R. K., Tye B. K., Botstein D. Mutagenesis by insertion of a drug-resistance element carrying an inverted repetition. J Mol Biol. 1975 Oct 5;97(4):561–575. doi: 10.1016/s0022-2836(75)80059-3. [DOI] [PubMed] [Google Scholar]
- Kleckner N. DNA sequence analysis of Tn10 insertions: origin and role of 9 bp flanking repetitions during Tn10 translocation. Cell. 1979 Apr;16(4):711–720. doi: 10.1016/0092-8674(79)90087-4. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- Lodish H. F. Secondary structure of bacteriophage f2 ribonucleic acid and the initiation of in vitro protein biosynthesis. J Mol Biol. 1970 Jun 28;50(3):689–702. doi: 10.1016/0022-2836(70)90093-8. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Oka A., Sugisaki H., Takanami M. Nucleotide sequence of the kanamycin resistance transposon Tn903. J Mol Biol. 1981 Apr 5;147(2):217–226. doi: 10.1016/0022-2836(81)90438-1. [DOI] [PubMed] [Google Scholar]
- Ross D. G., Swan J., Kleckner N. Physical structures of Tn10-promoted deletions and inversions: role of 1400 bp inverted repetitions. Cell. 1979 Apr;16(4):721–731. doi: 10.1016/0092-8674(79)90088-6. [DOI] [PubMed] [Google Scholar]
- Rothstein S. J., Reznikoff W. S. The functional differences in the inverted repeats of Tn5 are caused by a single base pair nonhomology. Cell. 1981 Jan;23(1):191–199. doi: 10.1016/0092-8674(81)90284-1. [DOI] [PubMed] [Google Scholar]
- Shapiro J. A. Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1933–1937. doi: 10.1073/pnas.76.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steege D. A. 5'-Terminal nucleotide sequence of Escherichia coli lactose repressor mRNA: features of translational initiation and reinitiation sites. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4163–4167. doi: 10.1073/pnas.74.10.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]


