In response to previous commentaries, we report here that different CLAVATA3 (CLV3) peptides have strikingly different activities in immune response gene activation and root growth repression, which are mediated by the FLAGELLIN SENSING2 (FLS2) and CLAVATA2 (CLV2) receptors, respectively (Figures 1 and 2). FLS2-dependent immune response gene activation triggered by the modified (hydroxylated at Pro-4 and -7) 12–amino acid CLV3 peptide (MCLV3p) was consistently observed in six independent batches (Figures 1 to 5; see Supplemental Figure 1 online). In addition to differential FLS2 binding and distinctive FLS2-mediated whole-seedling growth suppression induced by flg22 but not by MCLV3p (Figure 2), we also discovered that MCLV3p and flg22 exhibit distinct pH profiles in activating multiple immune response genes (Figure 5; see Supplemental Figures 2 and 3 online). These results and peptide analysis by sensitive mass spectrometry (Figure 4) make it unlikely that the defense-related responses we observe with MCLV3p are a consequence of flg22 or flg22-type peptide contamination in our MCLV3p preparations. Finally, we confirm that bacterial infection occurs inside the shoot apical meristem of clv3 and fls2 seedlings using confocal imaging and scanning electron microscopy (Table 1; Figure 7; see Supplemental Figures 4 and 5 online).
Figure 1.
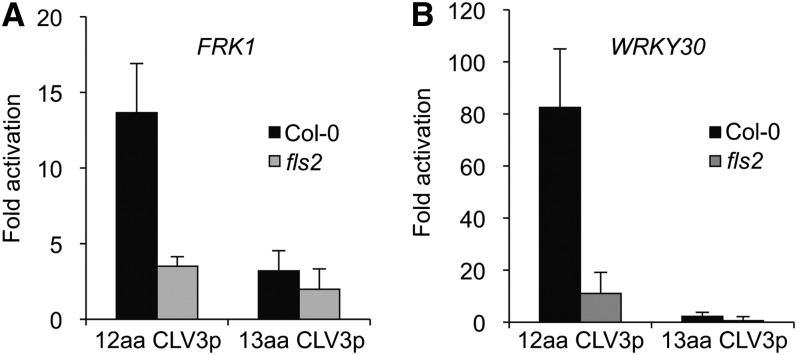
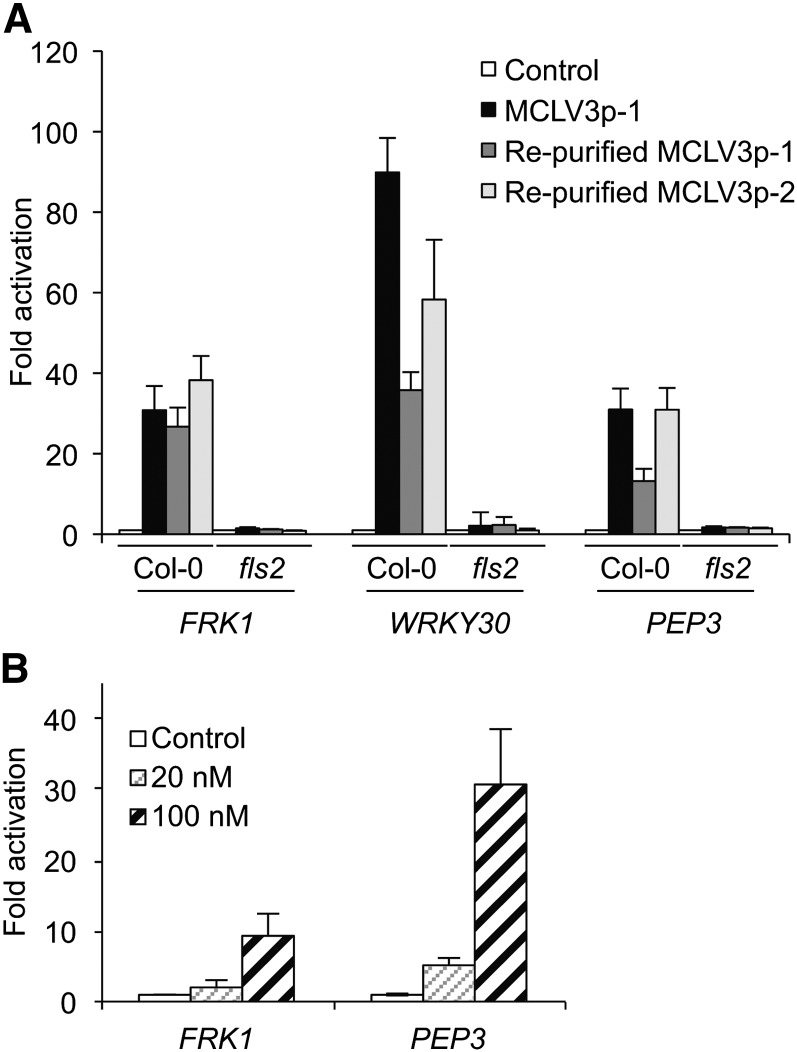
Differential Immune Response Marker Gene Activation by the 12–Amino Acid CLV3p and the 13–Amino Acid CLV3p.
Immune response marker genes, FRK1 (At2g19190) (A) and WRKY30 (At5g24110) (B), were activated by the 12–amino acid (aa) CLV3p but not the 13–amino acid CLV3p. Seven-day-old seedlings of Col-0 and the fls2 mutant grown in liquid culture were analyzed by qRT-PCR after treatment with 20 μM peptides for 1 h. The expression of each gene was normalized by ACT2 (At3g18780). Error bars indicate sd (n = 3).
Figure 2.
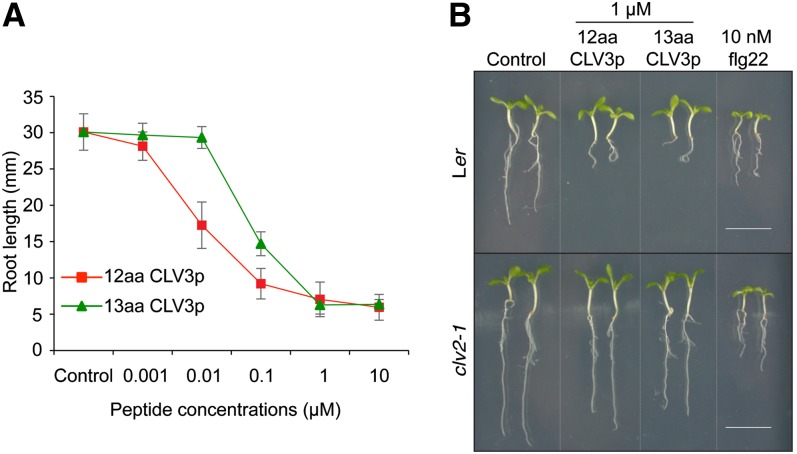
Differential CLV3p and flg22 Activities in Seedling Assays.
(A) Root growth inhibition by the 12–amino acid CLV3p and the 13–amino acid CVL3p. Seedlings were grown in a liquid medium without (Control) or with various concentrations of the 12–amino acid CLV3p (square) or the 13–amino acid CLV3p (triangle). Primary root length was measured after 14 d. Error bars indicate sd (n = 10).
(B) Flg22 but not CLV3p triggers whole-seedling growth suppression. Ler (top panel) or clv2-1 (bottom panel) seedlings were grown in a liquid medium with 1 μM 12–amino acid (aa) CLV3p, 1 μM 13–amino acid CLV3p, or 10 nM flg22 for 9 d. The differential flg22 and CLV3p activities in whole-seedling suppression are best illustrated in the clv2 mutant. Bars = 1 cm.
Figure 5.
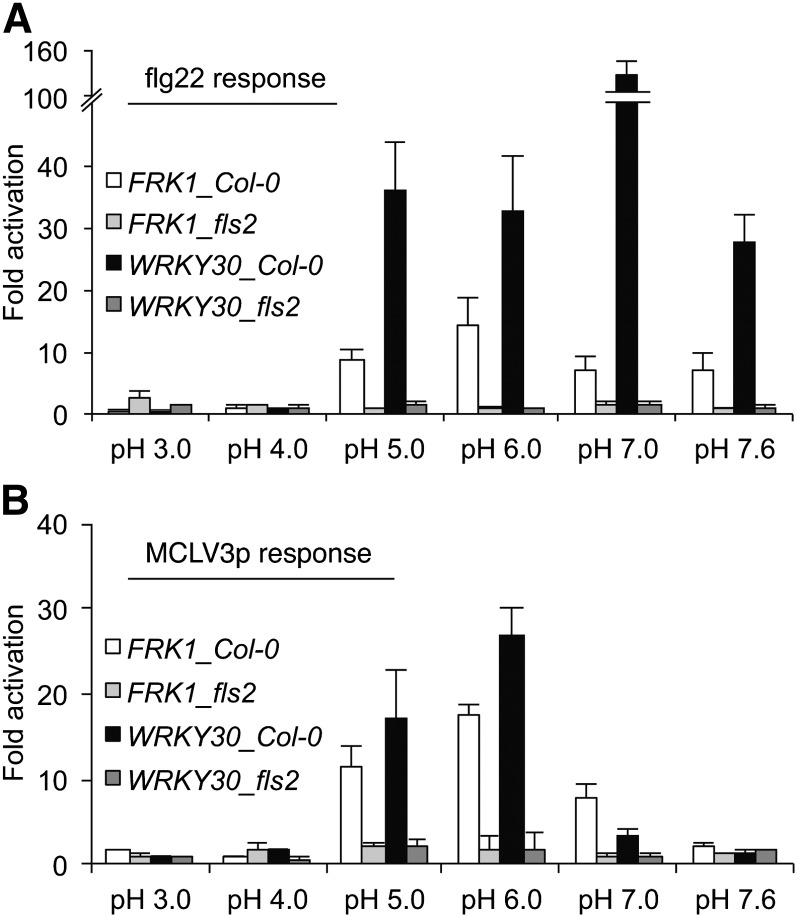
Flg22 and MCLV3p Exhibit Distinct pH Optima in Immune Response Marker Gene Activation.
Immune response marker gene activation by 1 nM flg22 (A) or 1 μM MCLV3p (B) in Col-0 and fls2 seedlings grown in liquid culture. Seven-day-old seedlings grown in liquid medium were treated with 1 nM flg22 (A) or 1 μM MCLV3p (B) for 1 h in various pH ranges. The expression of each gene was normalized by ACT2. Relative fold activation was normalized by the control level without peptide treatment in each pH. Error bars indicate sd (n = 3).
Figure 4.
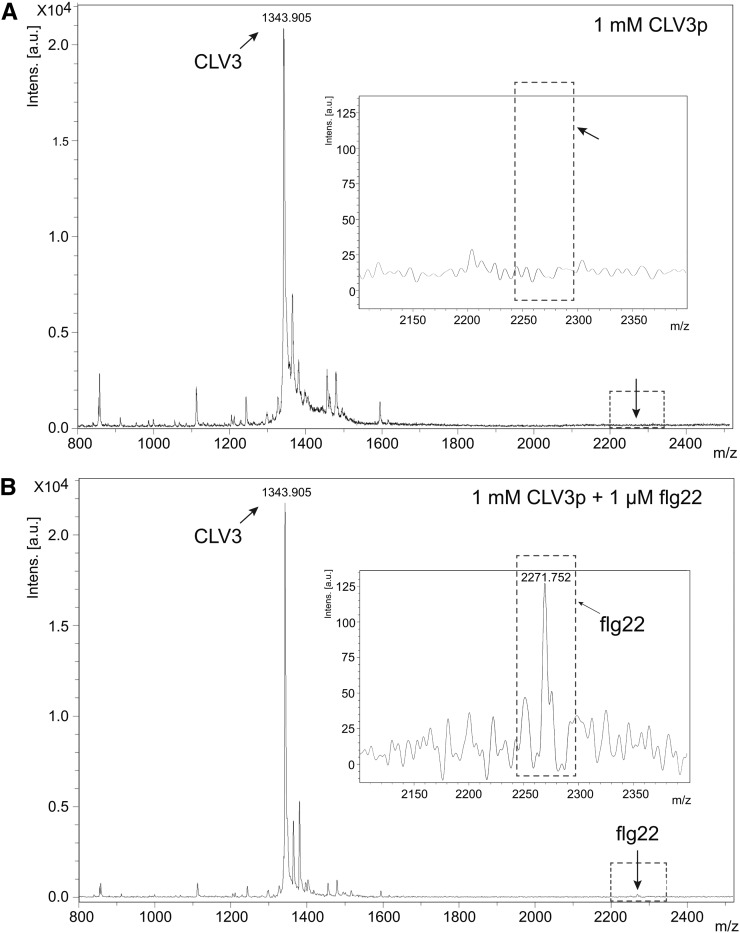
Analysis of CLV3p and flg22 by MALDI-TOF MS.
(A) Mass spectrum of 0.5 nM CLV3p (0.5 μL spotted from a 1 mM solution).
(B) Mass spectrum of 0.5 nanomole of CLV3p mixed with 0.5 pM of flg22 (0.5 μL spotted from a solution of 1 mM CLV3p containing 1 μM flg22).
Insets show the magnified portion of the spectrum in the region of flg22 mass (mass range 2100 to 2400). Arrows represent the same position in (A) and (B). a.u., arbitrary units; m/z, mass-to-charge ratio.
Table 1.
Infection Rate of Pst DC3000-GFP in the SAM
| Genotype | Total Seedlings | Available SAM | Infected SAM |
| Ler | 185 | 61 | 0 |
| fls2-24 | 151 | 45 | 21 |
| clv1-1 | 105 | 48 | 0 |
| clv2-1 | 108 | 44 | 0 |
| clv3-2 | 258 | 122 | 40 |
Seedlings grown in liquid medium were cocultured with Pst DC3000-GFP as described in Methods. Available SAM, the number of intact SAM that were successfully analyzed by a confocal microscopy; infected SAM, the number of SAM with bacteria observed in or near the inner SAM region.
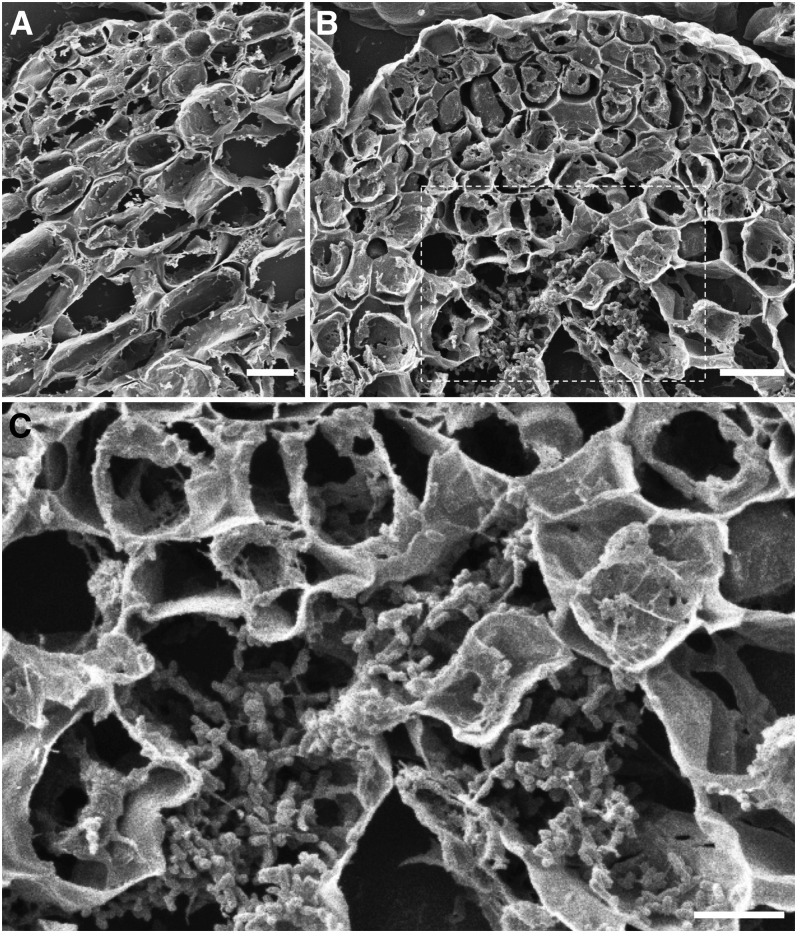
Figure 7.
Analysis of Pst DC3000 Infection in the SAM Regions of Seedlings Grown in Liquid Culture by Scanning Electron Microscopy.
Scanning electron microscopy analyses were performed with tissue cross sections.
(A) Immune protection of the Ler SAM area. A cross section through the SAM area is shown. Multiple cell layers below the SAM did not reveal any Pst DC3000 bacteria at 4 DAC. Bacteria were observed outside the dome.
(B) Compromised immune protection in the clv3 SAM area. Pst DC3000 colonized in the SAM and the cells below the SAM (4 DAC). Bars in (A) and (B) = 10 μm.
(C) Magnified image of the selected regions (white dot box) in (B) showing bacterial colonization. Bar = 5 μm.
The Commentaries by Mueller et al. (2012) and Segonzac et al. (2012) challenge the results reported by Lee et al. (2011), in which we proposed that in addition to its role in development, the signaling peptide CLV3p secreted from stem cells activates the Arabidopsis thaliana innate immune response in the shoot apical meristem (SAM) in a manner that is dependent on the flagellin receptor FLS2. The Mueller et al. (2012b) Commentary suggests that our results are most likely the consequence of flagellin-derived peptide flg22 contamination in our MCLV3p preparations. As discussed in more detail below, new data, which demonstrate differential CLV3p-FLS2 and flg22-FLS2 responses under different experimental conditions, make it unlikely that flg22 contamination is responsible for the FLS2-dependent responses we observe with CLV3p. It is likely that the significant experimental differences between our studies and the studies of Mueller et al. (2012b) and Segonzac et al. (2012) may account for the different results obtained in different laboratories.
In contrast with our work reported in Lee et al. (2011), which used the 12–amino acid MCLV3p, in the Commentary by Segonzac et al. (2012), a 13–amino acid CLV3p was tested. The 12–amino acid MCLV3p used in our experiments and an arabinosylated derivative of the 13–amino acid MCLV3p used by Segonzac et al. (2012) have been identified as endogenous CLV3 peptides by sensitive mass spectrometry methods (Kondo et al., 2006; Ohyama et al., 2009). Importantly, the 13–amino acid nonarabinosylated CLV3p showed 10-fold weaker binding to the CLV1 receptor than the 12–amino acid nonarabinosylated MCLV3p (Ohyama et al., 2009). In our preparation to respond to the Segonzac et al. (2012) Commentary, we ordered new batches of the 12–amino acid CLV3p as well as the 13–amino acid CLV3p used by Segonzac et al. (2012) from the same commercial source as reported by Segonzac et al. (2012). We then compared these two peptide preparations side by side in a variety of assays. The key result is that the 12–amino acid CLV3p but not the 13–amino acid CLV3p significantly activated immune response marker genes through FLS2 in seedlings (Figure 1). On the other hand, both the 12–amino acid and the 13–amino acid CLV3 peptides inhibited root growth mediated by the CLV2 receptor (Figure 2) (Kondo et al., 2006; Miwa et al., 2008; Lee et al., 2011). Strikingly, however, flg22 but not the 12–amino acid CLV3p triggered whole-seedling growth suppression in the clv2 mutant, suggesting differential flg22 and MCLV3p activities in different growth responses mediated by different receptors (Figure 2B) (Lee et al., 2011). If MCLV3p activation of immune responses was due to flg22 contamination, MCLV3p should have suppressed whole-seedling growth similarly to flg22. Thus, the discrepancies between our results and the results reported by Segonzac et al. (2012) are most likely due to the simple fact that Segonzac et al. (2012) did not test the same CLV3 peptide that was used in our experiments.
In contrast with Segonzac et al. (2012), Mueller et al. (2012b) tested the same MCLV3p peptide that was used in our experiments. What then accounts for the failure of Mueller et al. (2012b) to detect FLS2-dependent MCLV3p-mediated activation of defense response genes? One possibility is that Mueller et al. (2012b) used a different biological assay than the ones we used to monitor FLS2-mediated signaling. Using an FRK1-LUC reporter, Mueller et al. (2012b) observed extremely low and nonspecific MCLV3p activity, which led them to conclude that MCLV3p could not activate an immune response. The failure of Mueller et al. (2012b) to replicate our results primarily may be due to the fact that FRK1 is not a very sensitive reporter for FLS2-mediated early immune activation (Segonzac et al. [2012] observed only twofold FRK1 activation by flg22 in 1 h shown in their Figures 1C and 2B). We think the best way to resolve the discrepancy between our data and the data of Mueller et al. (2012b) would be to order new batches of MCLV3p and then test them in both laboratories using the same assays that employ several sensitive immune response genes (Figures 1, 3, and 5; see Supplemental Figures 1 to 3 online).
Figure 3.
Sensitive MCLV3p Activation of Immune Response Marker Genes after Repurification.
(A) Repurified MCLV3p from two independent batches activated immune marker genes, FRK1, WRKY30, and PEP3 (At5g64905), in Col-0 seedlings grown in liquid culture. The treatment was without (Control) or with 1 μM MCLV3p for 1 h.
(B) Immune response marker gene activation by nanomolar range of MCLV3p in the mesophyll protoplast assay. Protoplasts were collected after treatment without (Control) or with 20 or 100 nM of MCLV3p for 1 h.
Expression of each gene was analyzed by qRT-PCR and normalized by ACT2. Error bars indicate sd (n = 3).
In their Commentary, Mueller et al. (2012b) share their experience with inadvertent cross-contamination in commercial peptide preparations and suggest sound experimental precautions in handling synthetic peptides. As clearly demonstrated by Mueller et al., (2012b) their unexpected experimental problem due to flg22-type peptide contamination was resolved after a second batch of INFLORESCENCE DEFICIENT IN ABSCISSION-LIKE peptide had been ordered and tested, illustrating the importance of testing independently synthesized batches of synthetic peptides. For the following reasons, however, we think that it is highly unlikely that flg22 contamination of our MCLV3p preparations can explain the results reported by Lee et al. (2011).
First, in accordance with the recommendations of Mueller et al. (2012), in the course of our initial studies on CLV3p-FLS2 signaling spanning five years, and in the course of preparing this rebuttal, we ordered and tested four different batches of the 12–amino acid MCLV3p and two additional batches of Tyr-MCLV3p (adding Tyr in front of the first Arg of MCLV3p for 125I-labeling) for FLS2 binding analyses from four different suppliers. All six of these independent batches of 12–amino acid MCLV3p and Tyr-MCLV3p preparations activated immune response marker genes in an FLS2-dependent manner (Figures 1, 3, and 5; see Supplemental Figures 1 to 3 online) (Lee et al., 2011). Unlike adding His at the C terminus in the 13–amino acid CVL3p, the additional Tyr at the N terminus did not abolish MCLV3p activity in activating immune marker genes (see Supplemental Figure 1 online).
The second reason why we think that flg22 contamination is not the explanation for our results is that two MCLV3p preparations synthesized in house at the Massachusetts General Hospital (MGH) peptide core facility were subjected to HPLC purification and retested (Figure 3A). These MCLV3p preparations initially had not been HPLC purified because they were 93 to 97% pure, and the procedures used by the MGH core facility are designed to eliminate cross-contamination of peptides (Shimizu et al., 2000). Importantly, these two MCVL3p preparations showed similar characteristic activities in immune response gene activation before and after HPLC purification (Figure 3A). In the more sensitive mesophyll protoplast assay, MCLV3p activated immune response marker genes at low concentrations (20 to 100 nM) as reported previously for the SAM tissues, consistent with the Kd value of 34.7 nM for FLS2 binding (Figure 3B) (Lee et al., 2011). Importantly, we previously reported that immune marker genes were expressed at lower levels in the SAM tissues of clv3 seedlings compared with those in the SAM tissues of Landsberg erecta (Ler) wild-type seedlings. Application of 10 to 60 nM MCLV3p restored immune gene expression in the SAM of clv3 seedlings (Lee et al., 2011). Although HPLC sample loops and columns can be potential sources of contamination with other peptides, the retention time of flg22 on our reverse-phase HPLC is distinct from the retention time of MCLV3p. Moreover, the mass of flg22 was never detected in any of the MCLV3p samples (unpurified, purified, and side fractions) analyzed by mass spectrometry. We further developed a sensitive matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry (MALDI-TOF MS) method to show that the MCLV3p preparation used in these experiments is free of flg22 contamination at a 1:1000 ratio based on parallel analysis between MCLV3p alone and a mix of flg22 and MCLV3p (Figure 4).
The third reason we think that flg22 contamination is unlikely is because we have discovered in the course of preparing this rebuttal that MCLV3p and flg22 have very different pH optima. Because the positively charged N-terminal Arg and the C-terminal His-His residues are essential for the 12–amino acid MCLV3p activity (Kondo et al., 2006, 2008; Betsuyaku et al., 2011; Lee et al., 2011) and for transgenic CLV3 complementation in clv3 (Song et al., 2012), we hypothesized that CLV3p-FLS2 interactions may be pH dependent. Indeed, it has been reported that the processing of the CLV3 precursor proteins in vitro is pH dependent (Ni et al., 2011). Interestingly, FLS2-dependent MCLV3p activity is most active at pH 5 to 6 but is abolished at pH 7.6. This reflects the physiological apoplastic environment where the pH is reported to be 5 to 6 (Bibikova et al., 1998), whereas flg22 activates similar responses from pH 5.0 to 7.6 and peaks at pH 7.0 (Figure 5; see Supplemental Figure 2 online). As a control, although 1 μM MCLV3p did not significantly activate immune response genes at pH 7, addition of 1 nM flg22 to 1 μM MCLV3p resulted in strong activation. This latter experiment supports the conclusion that the MCLV3p peptide preparation was not contaminated with flg22 and moreover shows that an excess of MCLV3p does not block the activity of flg22 (see Supplemental Figure 3 online). Thus, carrying out experiments at relatively high pH (7 to 7.6) can be used to distinguish the activities of flg22 and MCLV3p. In summary, the observation that MCLV3p activated immune gene expression at low but not high pH suggests that the immune gene activation is not a consequence of flg22 contamination.
Another argument used by Mueller et al. (2012b) to suggest that our results were a consequence of flg22 contamination is that they found that excess amounts of CLV3p do not compete with 125I-Tyr-flg22 for binding to FLS2. It is important to note that the binding and competition assays performed by Lee et al. (2011) and by Mueller et al. (2012b) were performed under different experimental conditions. Lee et al. conducted binding and competition experiments using 125I-Tyr-MCLV3p, not 125I-Tyr-flg22, as was the case with Mueller et al. (2012). Further molecular understanding will require biochemical, structural, and genetic mapping of the precise functional peptide–leucine-rich repeat interactions for each peptide ligand with FLS2 (Mueller et al., 2012a; Shinohara et al., 2012).
In their Commentary, Segonzac et al. (2012) report that they were not able to observe the characteristically enlarged vegetative SAM and Pseudomonas syringae pv tomato (Pst) DC3000-GFP (for green fluorescent protein) infection in the clv3 seedlings (Medford et al., 1992; Bowman, 1993; Fiers et al., 2006; Kinoshita et al., 2010; Lee et al., 2011) and argue from first principles that it would not be possible for P. syringae to infect the SAM. However, Segonzac et al. (2012) used very different conditions for growing seedlings when they attempted to repeat our bacterial infection experiments. We tested the growth conditions described in their Commentary, which involve the use of 24-well plates, and found that the seedlings were stressed and exhibited delayed development compared with ours, which might explain the severely retarded shoot development in their CLV3p-treated seedlings and the relatively flat SAM they observed even in clv3 seedlings. Furthermore, our seedlings were grown in liquid medium at higher temperature (27°C compared with 22°C) and constant light (compared with 16-h light) during bacterial infection, which likely explains the more advanced SAM development in wild-type and clv3 seedlings that we observe (Medford et al., 1992; Bowman, 1993; Fiers et al., 2006; Kinoshita et al., 2010; Lee et al., 2011) but was not observed by Segonzac et al. (2012).
In the preparation of this rebuttal, we repeated our infection experiments using many more seedlings (807 total) to ensure the reproducibility of our original observations (Lee et al., 2011). As shown in Table 1, Pst DC3000-GFP cells were consistently observed only in the SAM of clv3 and fls2 seedlings grown in liquid culture but not in those of Ler, clv1, and clv2. By staining the plant cell membrane with FM4-64 to mark plant cells on the epidermal cell layer and provide a red color contrast, green Pst DC3000-GFP images inside the SAM are clearly visible through optical sections using a confocal laser scanning spectral microscope (Leica TCS-NT confocal laser scanning spectrophotometer) (see Supplemental Figure 4 online). Unlike the stable transgenic line expressing a plasma membrane–localized dtTOMATO fusion used by Segonzac et al. (2012), the red dye FM4-64 became defuse during the lengthy procedure to identify the infected SAM domes (see Methods). When stained only briefly with FM4-64, we observed clear cellular definition with no cell damages in our SAM samples. Because the length of bacterial cells (∼1.5 μm) is much shorter than the serial optical sections when we examined the SAM dome (scale bars are shown in all figures), by setting the starting focus point at the surface of the SAM defined by the FM4-64–stained epidermal cell layer, the Pst DC3000-GFP cells on the SAM surface or outside would disappear quickly after just a few micrometers scanning into the SAM through confocal optical sections (see Supplemental Figure 4 online). The method allowed us to confidently identify Pst DC3000-GFP cells inside the SAM even with relatively low cellular details. Quantification of infected Pst DC3000-GFP in the dissected SAM from 10 randomly pooled seedlings was determined by quantitative PCR using specific GFP primers (Figure 6).
Figure 6.
Quantification of Pst DC3000-GFP in the SAM.
Bacteria harboring the GFP gene were quantified by quantitative PCR using specific GFP primers and normalized with Arabidopsis ACT2 gene in the SAM tissues. Cocultivated SAM tissues were harvested from seedlings grown in liquid culture at 4 DAC. Error bars indicate sd (n = 3).
As the SAM inner tissues are not fully accessible to confocal microscopy–based imagining, we developed a scanning electron microscopy method to generate cross sections of the SAM dome from 4 d after infection seedlings to directly visualize colonized Pst DC3000 cells (Plotnikova et al., 2000). We observed clear bacterial colonization of cells very close to the SAM in clv3 but not Ler wild-type seedlings, in which we only observed invaded cells many cell layers away from the SAM. These data suggest that CLV3p-based immune protection in the wild-type SAM extends multiple cell layers below the SAM (Figure 7; see Supplemental Figure 5 online). However, in cotyledons and leaves distant from the SAM, the virulent bacterial pathogen successfully colonized in both wild-type and clv3 seedlings (Lee et al., 2011).
As Segonzac et al. (2012) report that they could not identify the SAM in their seedlings, we present an example of the seedling images showing the visible SAM in Ler and clv3 after removing the cotyledons and petioles using a pair of forceps and gently placing a cover glass on top of the seedling without damaging tissues (see Supplemental Figure 6 online). We also provide detailed methods for our bacterial infection experiments (see Methods). Although the experimental procedure is tedious and lengthy, with patience and practice, we were able to observe the SAM not masked by leaf primordia (see Supplemental Figure 6 online) in ∼30 to 47% wild-type and mutant seedlings screened under a dissecting microscope. These seedlings with clearly visible SAM were then further examined for the presence of Pst DC300-GFP by optical sectioning using a confocal laser scanning spectral microscope (Table 1).
Finally, a separate set of experiments were performed by Segonzac et al. (2012), which showed that in vivo dexamethasone-induced expression of CLV3 in transgenic plants did not activate immune response genes. Based on these data, they argue that CLV3-derived peptides cannot activate immune responses in planta. However, it is possible that the whole seedlings analyzed by Segonzac et al. (2012) secrete only the 13–amino acid peptide as reported (Ohyama et al., 2009), which, as we show in this rebuttal, is able to complement the CLV1/2-mediated developmental phenotype, but fails to activate immune response genes (Figure 1). As the physiological conditions and sensitivity for MCLV3p and flg22 activation of immune response genes are different and MCLV3p-FLS2 and MCLV3p-CLV1/2 signaling pathways are distinct (Lee et al., 2011), additional experiments will need to be conducted with the dexamethasone-inducible CLV3 lines, especially in the SAM.
The biological relevance, if any, of our finding that the SAM of clv3 and fls2 mutants can be infected with P. syringae using liquid-grown Arabidopsis seedlings has not been definitively established. However, P. syringae tends to be seed borne and is dispersed via rain splash (Hirano and Upper, 1990), and it is possible that young seedlings growing in rainwater could be potentially susceptible to bacterial infection through an unconventional entrance in the SAM, explaining the importance of CLV3-mediated activation of immunity. It is not surprising that P. syringae growth is slower in the SAM than in cotyledons and leaves, as different pathogens have presumably evolved and adapted to different organs or tissues as their preferred proliferation niche (Hirano and Upper, 1990; Alfano and Collmer, 1996; Blander and Sander, 2012; Medzhitov et al., 2012). Our data suggest that the Arabidopsis SAM displays an FLS2-dependent innate immune response against P. syringae, which is only revealed in the clv3 and fls2 mutants under liquid growth conditions. As a critical reservoir of stem cells for organogenesis, the protection of SAM from infection and stress is critical to ensure plant survival and longevity, especially when the SAM is in vulnerable conditions or damaged. It will be interesting and important to further explore the molecular details of SAM immunity.
In summary, our findings for differential MCLV3p and flg22 signaling responses make the possibility of flg22 contamination of MCLV3 preparations unlikely and provide evidence for bacterial infection close to the SAM of clv3 mutants with compromised innate immunity as reported by Lee et al., (2011). In future experiments, it will be interesting to unravel the molecular mechanisms of differential pathogen defense and tolerance in different plant organs and tissues, including the SAM.
METHODS
Plant Materials and Growth Conditions
Columbia-0 (Col-0) and Ler were used as wild-type Arabidopsis thaliana plants in this study. The fls2 (Salk_141277) mutant is in Col-0 background (Shan et al., 2008), and clv1-1, clv2-1, clv3-2, and fls2-24 are in Ler background (Koornneef et al., 1983; Clark et al., 1995; Gómez-Gómez and Boller, 2000). For liquid culture of wild-type and mutant seedlings, seeds were germinated and grew in six-well plates containing 1 mL of liquid medium (0.5×Murashige and Skoog and 0.5% Suc, pH 5.8 adjusted with KOH). Ten seedlings were grown at 23°C, 65% humidity, and 75 μmol m−2 s−1 light intensity under 12-h-light/12-h-dark photoperiod condition for 7 d (for quantitative RT-PCR [qRT-PCR]), 9 d (for seedling growth inhibition assay), or 14 d (for root growth inhibition assay). The seedling growth condition for bacterial infection assays is provided in detail below.
Peptides
Two independent batches of MCLV3p (12 amino acids, RTVPhSGPhDPLHH) were synthesized in different years at the MGH Peptide/Protein Core Facility (Charlestown, MA). Ph is Hyp. Peptides were synthesized on an automated peptide synthesizer (Model Apex 396; AAPPTEC) with Fmoc solid-phase chemistry using 2-(6-chloro-1-H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate in the presence of N-N-diisopropylethylamine as an activator. Following the assembly of the peptide chain, the dried resins were cleaved and deprotected with cocktail containing 1% 3,6-dioxa-1,8-octanediol/2.5% deionized water/2.5% triisopropylsilane/94% trifluoroacetic acid (TFA) for 2 to 2.5 h at room temperature. Crude peptides were precipitated and washed twice in cold methyl tert-butyl ether. Precipitates were then dissolved in 20% acetonitrile/0.01% TFA in deionized water and freeze-dried.
CLV3 peptides were purified on an Applied Biosystems micropreparative (0.1 to 2 mg of sample load; 0.4 mL flow rate) HPLC using a Vydac, C-18, 0.21 × 15-cm column and a linear gradient of 5 to 95% B (buffer A = 0.06% TFA in deionized water and buffer B = 0.05% TFA in 80% acetonitrile) in 9 min. Following each run, the HPLC sample loops and the columns were thoroughly flushed and a blank run was performed before the purification of the next sample.
Peptides were analyzed on a Beckman Coulter System Gold HPLC using a Vydac, reverse-phase C-18, 0.46 × 15-cm column. A 5 to 65% B (buffer A = 0.1% TFA in deionized water and buffer B = 0.1% TFA in acetonitrile) linear gradient with a flow of 1 mL/min in 30 min was used. The molecular weight of peptides was confirmed by MALDI-TOF MS on an Applied Biosystems Voyager DE instrument using α-cyano-4-hydroxycinnamic acid matrix.
Note: The first two batches of the 12–amino acid MCLV3p were synthesized 1 year apart at the MGH Endocrine Unit protein/peptide core facility that specializes in the synthesis of peptide hormones. The design of this core facility includes meticulous efforts to eliminate cross-contamination of peptides. For example, in an important study, the biological activity of an N-terminal 14–amino acid peptide of parathyroid hormone was confirmed to be free of any contamination with a longer 34–amino acid peptide (Shimizu et al., 2000). The two batches of MCLV3p are free of flg22 or flg22-type contamination based on HPLC, MALDI-TOF MS, and peptide sequencing analyses. The further purified 12–amino acid MCLV3p shows consistent activity and higher efficacy in triggering characteristic immune response marker genes in an FLS2-dependent manner (Figure 3A). The third batch of the 12–amino acid MCLV3p was ordered from GenScript. Two independent batches of Tyr-MCLV3p were requested from Phoenix Pharmaceuticals. New batches of the 12–amino acid CLV3p and the 13–amino acid CLV3p were purchased from Biomatik. The 12–amino acid CLV3p used in this study is identical to the endogenous modified CLV3 peptide identified by in situ MALDI-TOF MS analysis (Kondo et al., 2006), and the 13–amino acid CLV3p (RTVPhSGPhDPLHHH) contains an additional His residue at the C terminus without arabinosylation at the 7th Hyp residue (Ohyama et al., 2009).
MALDI-TOF MS
MALDI-TOF-MS (Karas and Hillenkamp, 1988) analyses were performed on a Microflex instrument (Bruker Daltonics) that was operated at an accelerated voltage of 20 kV for detection of positive ions in linear mode. The super-dihydroxybenzoic acid matrix (Sigma-Aldrich) consisting of a 9:1 mixture of 2,5-dihydroxybenzoic acid and 2-hydroxy-5-methoxybenzoic acid was used. The matrix solution (10 mg/mL) was prepared in 33% acetonitrile/0.1% TFA in water. Solutions of varying concentrations of pure CLV3p, pure flg22, and the mixture of these two peptides were prepared in Milli-Q water. We mixed 0.5 μL of matrix and 0.5 μL of sample solution aliquots directly on the spots of a ground steel target (Bruker Daltonics) and air dried at room temperature.
Flg22 could be reliably detected in a mixture of 1 mM CLV3p and 1 μM flg22 in a 1:1000 ratio. Due to the use of linear mode of detection, masses observed were within 1 to 2 mass units of the calculated monoisotopic [M+H]+ masses of 1344.65 for CLV3p and 2272.20 for flg22, respectively.
MALDI-TOF MS with sDHB matrix was helpful in the detection of 1 μM flg22 mixed with 1 mM CLV3p. Under the same conditions of detection, flg22 contamination was not seen in the 1 mM solution of CLV3p.
qRT-PCR Analysis
Total RNA was isolated from Arabidopsis seedlings with TRIzol reagent (Invitrogen). First-strand cDNA was synthesized from 1 μg of total RNA with M-MLV reverse transcriptase (Promega). All qRT-PCR analyses were performed by CFX96 real-time PCR detection system with iQ SYBR green supermix (Bio-Rad). ACTIN2 (ACT2, At3g18780) was used as a control gene.
For the assay of pH-dependent gene expression, 7-d-old seedlings were preincubated for 30 min in a 10-fold diluted citric acid-Na2HPO4 buffer before adding peptides. A citric acid-Na2HPO4 buffer solution covers the wide range from pH 2.6 to 7.6 (information from the buffer reference center of Sigma-Aldrich).
Bacterial Infection Assay in the SAM and Confocal Microscopy
Nine seeds were sowed in 1 mL liquid medium (0.5× Murashige and Skoog and 0.5% Suc, pH 5.8 adjusted with KOH) in six-well plates and grown under constant light (50 to 65 μmol m−2 s−1) at 25 to 27°C without shaking for 2 d. Several fresh colonies of Pst DC3000-GFP were inoculated in King's B liquid medium (50 μg/mL of rifampicin and 15 μg/mL of tetracyclin) with shaking at 28°C. Overnight cultured Pst DC3000-GFP was washed twice with water and diluted to OD600 = 0.02. Diluted Pst DC3000-GFP (50 μL) was added into 1 mL of liquid medium with 2-d-old seedlings. Plants and bacteria were cocultivated with gentle shaking (50 rpm) for 4 d under constant light.
For observation of bacteria-infected SAM with the membrane staining dye FM4-64 (Molecular Probes), cocultivated seedlings were washed with water twice without the ethanol wash step (Lee et al., 2011). Seedlings were placed on a glass slide in several drops of water, and cotyledons, roots, and visible leaves were removed under a stereomicroscope (Wild M5A; Wild Heerbrugg) or using a magnifying glasses (OptiVISOR; Donegan Optical Company). Fine forceps (Fine Science Tools) were used to remove tissues but leave the area around the SAM intact (see Supplemental Figure 6 online). Then, prepared seedlings with intact shoot apex were stained in 18 μM FM4-64 for 5 min on the ice in the dark (Bolte et al., 2004). FM4-64 dye was prepared in Hank’s Balanced Salt Solution buffer without calcium, magnesium, and phenol red. Stained tissues were placed on a glass slide in 200 μL of 1×Hank’s Balanced Salt Solution, and a cover slip was laid gently on top of the five to 10 seedlings. The cover slip was brought closer to the glass slide by removing the solution with Kimwipes (very gently squashed without pressing the cover slip). Intact SAM structures were visible in 30 to 33% of Ler and fls2-24 seedlings and 41 to 47% of clv1-1, clv2-1, and clv3-2. The SAM infected by Pst DC3000-GFP was only observed in clv3 and fls2 seedlings. For imaging analysis with the confocal laser scanning microscope (Leica TCS-NT), GFP and FM4-64 were excited with an air-cooled argon laser at 488 nm and fluorescence was detected at 510 to 540 nm (GFP) and 635 to 680 nm (FM4-64).
For the quantitative PCR analysis of bacterial infection, diluted PstDc3000-GFP (OD600 = 0.5, 50 μL) was cocultivated with wild-type or mutant seedlings for 4 d. After washing seedlings with 70% ethanol twice and rinsing them with water twice, 10 SAM tissues, after removing cotyledons, visible leaves, hypocotyl, and roots by a pair of fine forceps, were harvested and ground in 100 μL of water using a blue pestle and hand grinder (Sigma-Aldrich). Control experiments were conducted at 0 d after cocultivation (DAC) to determine nonspecific bacterial attachment 1 h after cocultivation. Bacterial growth was detected by DNA PCR using the specific GFP primers. The GFP PCR was normalized using the Arabidopsis ACT2 gene in the SAM tissues.
Paraffin Embedding and Scanning Electron Microscopy Analysis of Bacteria-Infected SAM Tissues
Two-day-old Ler and clv3 seedlings were infected with P. syringae pv tomato DC3000 for 4 d. Seedling samples were washed with 70% ethanol and rinsed with water twice. After removing visible roots, cotyledons, and true leaves, the SAM tissue samples were fixed in 4% (w/v) paraformaldehyde/4% (v/v) DMSO at 4°C for overnight. Then, fixed samples were washed in phosphate buffer, pH 7.2, and passed through ethanol series (30, 50, 70, and 80% for 1 h in each step) at 4°C and stained with 0.1% Eosin Y (Sigma-Aldrich) in 95% ethanol at 4°C for overnight. Ethanol was changed through histoclear series (75% ethanol: 25% histoclear, 50% ethanol: 50% histoclear, 25% ethanol: 75% histoclear, 100% histoclear, 100% histoclear for 1 h in each step at room temperature) (National Diagnostics). Histoclear was then gradually changed with melted paraffin (Fisher Scientific) in a 60°C chamber. Replacement of freshly melted paraffin was performed for 4 d. Paraffin-embedded tissues were poured into the mold and adjusted in appropriated position. Section was performed with a rotary microtome (Leica RM2255) at 8-μm thickness.
Section ribbons of SAM tissues were placed to prewarm water on round cover glasses (diameter = 12 mm) coated with poly-l-Lys, and incubated on a slide warmer (Fisher Scientific) at 42°C for overnight. After removing paraffin in the 100% histoclear for 10 min twice, the apical meristem cross sections attached to the cover glasses were postfixed in 4% paraformaldehyde/1% glutaraldehyde solution in phosphate buffer, pH 7.2, at 4°C for overnight, then washed in phosphate buffer, pH 7.2, and dehydrated in ethanol series (30, 50, 70, 96, and 100% for 15 min twice in each step). The cover glasses with apical meristem cross sections were dried at critical point in a Samdri-795 semiautomatic critical point dryer. The dried apical meristems were mounted on metal stubs (diameter =1.2 mm) and covered with 600A layer of chromium in a Gatan High Resolution Ion Beam Coater 681. The coated samples were analyzed under a JEOL 7401F scanning electron microscope.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: FRK1 (At2g19190), WRKY30 (At5g24110), PEP3 (At5g64905), FAD (At1g26380), WAK2 (At1g79680), and ACT2 (At3g18780).
Supplementary Material
Acknowledgments
We thank Fred M. Ausubel and Thomas Boller for discussions, Andrew F. Bent for comments, Ok-Kyung Chah for technical assistance, and Richard F. Cook of the Massachusetts Institute of Technology Biopolymers Facility for access to the Bruker Daltonics Microflex mass spectrometry instrument. This work was supported by the National Institutes of Health (R01GM060493 and R01GM070567) and the National Science Foundation (IOS-0618292) to J.S.
AUTHOR CONTRIBUTIONS
H.L. and J.S. designed the experiments. H.L. carried out the experiments and prepared the data. A.K. synthesized MCLV3 and flg22 peptides. A.K. and X.-C.Z. analyzed CLV3 and flg22 peptides by MS analysis. J.M.P. performed the scanning electron microscopy analysis. H.L. and J.S. wrote the article.
REFERENCES
- Alfano J.R., Collmer A. (1996). Bacterial pathogens in plants: Life up against the wall. Plant Cell 8: 1683–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betsuyaku S., Takahashi F., Kinoshita A., Miwa H., Shinozaki K., Fukuda H., Sawa S. (2011). Mitogen-activated protein kinase regulated by the CLAVATA receptors contributes to shoot apical meristem homeostasis. Plant Cell Physiol. 52: 14–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova T.N., Jacob T., Dahse I., Gilroy S. (1998). Localized changes in apoplastic and cytoplasmic pH are associated with root hair development in Arabidopsis thaliana. Development 125: 2925–2934 [DOI] [PubMed] [Google Scholar]
- Blander J.M., Sander L.E. (2012). Beyond pattern recognition: Five immune checkpoints for scaling the microbial threat. Nat. Rev. Immunol. 12: 215–225 [DOI] [PubMed] [Google Scholar]
- Bolte S., Talbot C., Boutte Y., Catrice O., Read N.D., Satiat-Jeunemaitre B. (2004). FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J. Microsc. 214: 159–173 [DOI] [PubMed] [Google Scholar]
- Bowman J. (1993). Early development of the apical meristem of WT A. thaliana. In Arabidopsis: An Atlas of Morphology and Development. J. Bowman, ed, pp. 18–21 Springer-Verlag New York, Inc., New York
- Clark S.E., Running M.P., Meyerowitz E.M. (1995). CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121: 2057–2067 [Google Scholar]
- Fiers M., Golemiec E., van der Schors R., van der Geest L., Li K.W., Stiekema W.J., Liu C.-M. (2006). The CLAVATA3/ESR motif of CLAVATA3 is functionally independent from the nonconserved flanking sequences. Plant Physiol. 141: 1284–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L., Boller T. (2000). FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Hirano S.S., Upper C.D. (1990). Population biology and epidemiology of Pseudomonas syringae. Annu. Rev. Phytopathol. 28: 155–177 [Google Scholar]
- Karas M., Hillenkamp F. (1988). Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal. Chem. 60: 2299–2301 [DOI] [PubMed] [Google Scholar]
- Kinoshita A., Betsuyaku S., Osakabe Y., Mizuno S., Nagawa S., Stahl Y., Simon R., Yamaguchi-Shinozaki K., Fukuda H., Sawa S. (2010). RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 137: 3911–3920 [DOI] [PubMed] [Google Scholar]
- Kondo T., Nakamura T., Yokomine K., Sakagami Y. (2008). Dual assay for MCLV3 activity reveals structure-activity relationship of CLE peptides. Biochem. Biophys. Res. Commun. 377: 312–316 [DOI] [PubMed] [Google Scholar]
- Kondo T., Sawa S., Kinoshita A., Mizuno S., Kakimoto T., Fukuda H., Sakagami Y. (2006). A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313: 845–848 [DOI] [PubMed] [Google Scholar]
- Koornneef M., van Eden J., Hanhart C.J., Stam P., Braaksma F.J., Feenstra W.J. (1983). Linkage map of Arabidopsis thaliana. J. Hered. 74: 265–272 [Google Scholar]
- Lee H., Chah O.-K., Sheen J. (2011). Stem-cell-triggered immunity through CLV3p-FLS2 signalling. Nature 473: 376–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medford J.I., Behringer F.J., Callos J.D., Feldmann K.A. (1992). Normal and abnormal development in the Arabidopsis vegetative shoot apex. Plant Cell 4: 631–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R., Schneider D.S., Soares M.P. (2012). Disease tolerance as a defense strategy. Science 335: 936–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa H., Betsuyaku S., Iwamoto K., Kinoshita A., Fukuda H., Sawa S. (2008). The receptor-like kinase SOL2 mediates CLE signaling in Arabidopsis. Plant Cell Physiol. 49: 1752–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller K., Bittel P., Chinchila D., Jehle A.K., Albert M., Boller T., Felix G. (2012a). Chimeric FLS2 receptors reveal the basis for differential flagellin perception in Arabidopsis and tomato. Plant Cell 24:2213–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller K., Chinchilla D., Albert M., Jehle A.K., Kalbacher H., Boller T., Felix G. (2012b). The flagellin receptor FLS2 is blind to peptides derived from CLV3 or Ax21 but perceives traces of contaminating flg22. Plant Cell 24:3193–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J., Guo Y., Jin H., Hartsell J., Clark S.E. (2011). Characterization of a CLE processing activity. Plant Mol. Biol. 75: 67–75 [DOI] [PubMed] [Google Scholar]
- Ohyama K., Shinohara H., Ogawa-Ohnishi M., Matsubayashi Y. (2009). A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat. Chem. Biol. 5: 578–580 [DOI] [PubMed] [Google Scholar]
- Plotnikova J.M., Rahme L.G., Ausubel F.M. (2000). Pathogenesis of the human opportunistic pathogen Pseudomonas aeruginosa PA14 in Arabidopsis. Plant Physiol. 124: 1766–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segonzac C., Nimchuk Z.L., Beck M., Tarr P.T., Robatzek S., Meyerowitz E., Zipfel C. (2012). The shoot apical meristem regulatory peptide CLV3 does not activate innate immunity. Plant Cell 24:3186–3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L., He P., Li J., Heese A., Peck S.C., Nürnberger T., Martin G.B., Sheen J. (2008). Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4: 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu M., Potts J.T., Jr, Gardella T.J. (2000). Minimization of parathyroid hormone. Novel amino-terminal parathyroid hormone fragments with enhanced potency in activating the type-1 parathyroid hormone receptor. J. Biol. Chem. 275: 21836–21843 [DOI] [PubMed] [Google Scholar]
- Shinohara H., Moriyama Y., Ohyama K., Matsubayashi Y. (2012). Biochemical mapping of a ligand-binding domain within Arabidopsis BAM1 reveals diversified ligand recognition mechanisms of plant LRR-RKs. Plant J. 70: 845–854 [DOI] [PubMed] [Google Scholar]
- Song X.-F., Yu D.-L., Xu T.-T., Ren S.-C., Guo P., Liu C.-M. (2012). Contributions of individual amino acid residues to the endogenous CLV3 function in shoot apical meristem maintenance in Arabidopsis. Mol. Plant 5: 515–523 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.