This study characterizes the temperature-sensitive Arabidopsis thaliana lig mutant, which exhibits ectopic lignin deposition and growth defects at high temperatures. It finds that impairment of N-glycan synthesis in the mutant likely activates lignin biosynthesis, probably via endoplasmic reticulum stress and the unfolded protein response.
Abstract
To study the regulatory mechanisms underlying lignin biosynthesis, we isolated and characterized lignescens (lig), a previously undescribed temperature-sensitive mutant of Arabidopsis thaliana that exhibits ectopic lignin deposition and growth defects under high-temperature conditions. The lig mutation was identified as a single base transition in GNA1 encoding glucosamine-6-phosphate N-acetyltransferase (GNA), a critical enzyme of UDP-N-acetylglucosamine (UDP-GlcNAc) biosynthesis. lig harbors a glycine-to-serine substitution at residue 68 (G68S) of GNA1. Enzyme activity assays of the mutant protein (GNA1G68S) showed its thermolability relative to the wild-type protein. The lig mutant exposed to the restrictive temperature contained a significantly smaller amount of UDP-GlcNAc than did the wild type. The growth defects and ectopic lignification of lig were suppressed by the addition of UDP-GlcNAc. Since UDP-GlcNAc is an initial sugar donor of N-glycan synthesis and impaired N-glycan synthesis is known to induce the unfolded protein response (UPR), we examined possible relationships between N-glycan synthesis, UPR, and the lig phenotype. N-glycans were reduced and LUMINAL BINDING PROTEIN3, a typical UPR gene, was expressed in lig at the restrictive temperature. Furthermore, treatment with UPR-inducing reagents phenocopied the lig mutant. Our data collectively suggest that impairment of N-glycan synthesis due to a shortage of UDP-GlcNAc leads to ectopic lignin accumulation, mostly through the UPR.
INTRODUCTION
Lignin is a complex heteropolymer derived mainly from three hydroxycinnamyl alcohol monomers (or monolignols) named p-coumaryl, coniferyl, and sinapyl alcohols. The monolignols are synthesized from Phe through the general phenylpropanoid pathway, followed by the lignin-specific pathway. Monolignols are transported to cell walls, where they are oxidatively polymerized by various classes of enzymes, such as peroxidases and laccases, to form lignin. Lignin is produced in association with xylem differentiation and is deposited into the secondary cell walls of tracheary elements and fiber cells. In addition to contributing to water conductance through vascular networks, lignin provides mechanical support for the plant body. Lignin synthesis, which is also induced by various stresses (e.g., wounding and pathogen infection), constitutes an important part of the plant defense system (Boerjan et al., 2003).
Much of our knowledge on the developmental regulation of lignin synthesis has come from studies of tracheary element differentiation. During tracheary element differentiation, a set of genes for the enzymes that catalyze lignin biosynthesis are expressed together with genes related to secondary cell wall formation (Demura et al., 2002; Milioni et al., 2002). Expression of these genes is activated directly by a specific kind of MYB transcription factor (Patzlaff et al., 2003; Zhou et al., 2009; Zhao and Dixon, 2011). NAC domain proteins, such as VASCULAR-RELATED NAC-DOMAIN6 (VND6) and VND7 in Arabidopsis thaliana, which can direct the whole program of cell differentiation (Kubo et al., 2005; Yamaguchi and Demura, 2010), are key regulators that act upstream of the MYB factors.
Environmental stresses induce lignin production through variable and complicated signaling cascades that involve Ca2+ ions, reactive oxygen species, jasmonic acid, and other signal molecules (Mahalingam and Fedoroff, 2003; Pauwels et al., 2008). Under the influence of these signaling cascades, the expression of genes involved in lignin synthesis is stimulated at the transcription level (Zhao and Dixon, 2011). This may be attributed, at least partly, to the downregulation of repressor-type MYB transcription factors (Jin et al., 2000; Cheong et al., 2002).
Interestingly, the integrity of cell walls is another factor that regulates lignin biosynthesis, as shown by molecular-genetic and physiological analyses in Arabidopsis. Using the ectopic lignin deposition1 mutant, which has a point mutation in the CELLULOSE SYNTHASE3 gene, and isoxaben, an inhibitor of cellulose synthesis, Caño-Delgado et al. (2000, 2003) demonstrated that cellulose synthesis defects cause deposition of a large amount of lignin over normally nonlignified tissues. Such ectopic lignin deposition was also observed in plants treated with thaxtomin A, which perturbs cell wall structure (Bischoff et al., 2009). Recent studies have indicated that the lignification induced by cell wall damage is mediated by a receptor-like kinase, THESEUS1 (Hématy et al., 2007), and regulated by reactive oxygen species– and jasmonic acid–dependent processes (Denness et al., 2011).
Many temperature-sensitive mutants of Arabidopsis have been isolated using in vitro organogenesis as an index phenotype (Yasutani et al., 1994; Konishi and Sugiyama, 2003; Sugiyama, 2003). In the same screening described by Konishi and Sugiyama (2003), we identified a mutant that exhibits severe growth defects associated with ectopic lignin deposition and designated it lignescens (lig). Here, we report the phenotypic characterization of the lig mutant and the identification of LIG as GNA1, which encodes glucosamine-6-phosphate N-acetyltransferase (GNA). We further show several lines of evidence that suggest that impaired synthesis of N-glycans resulting from a shortage of UDP-N-acetylglucosamine (UDP-GlcNAc) is responsible for the lig phenotype. Our findings add to the present body of knowledge on the regulatory networks underlying lignin biosynthesis.
RESULTS
Isolation of lig as a Novel Temperature-Sensitive Mutant
In a screen of a mutagenized population of Arabidopsis for temperature-sensitive mutants with defects in adventitious root formation (Konishi and Sugiyama, 2003), we identified a mutant in which the adventitious roots ceased to grow and large amounts of lignin accumulated soon after exposure to high-temperature conditions. Initial genetic characterization of this mutant showed that its temperature sensitivity is a recessive and monogenic trait (see Supplemental Table 1 online). With reference to the nature of abnormal lignification, this mutant was designated lig.
Temperature-Dependent Growth Defect and Ectopic Lignin Deposition in the lig Mutant
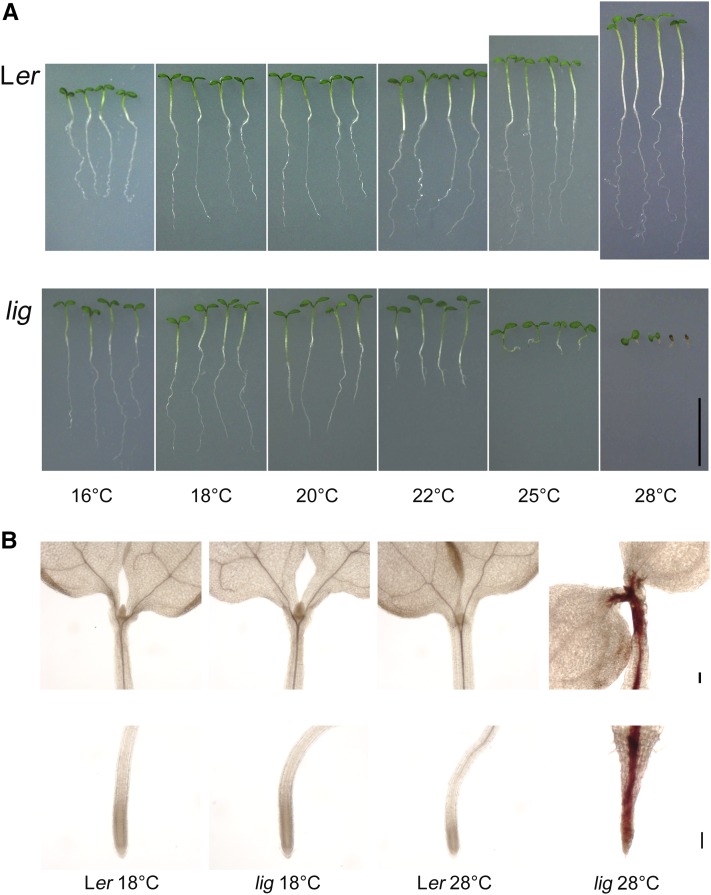
Overall growth was compared between wild-type and lig seedlings cultured at various temperatures (Figure 1A). At 22°C, the roots of the lig seedlings were obviously shorter than those of the wild type (Figure 1A). At 25 and 28°C, elongation growth of the lig seedlings was severely inhibited in both the hypocotyls and roots (Figure 1A).
Figure 1.
Temperature-Dependent Growth Defect and Lignin Accumulation in the lig Seedlings.
(A) Wild-type (Ler) and lig mutant seedlings were cultured at various temperatures for 7 d. Bar = 10 mm.
(B) Shoots and primary root tips of 5-d-old seedlings stained with phloroglucinol-HCl to detect lignin. Bars = 100 µm.
Wild-type and lig seedlings grown at 18 and 28°C were stained with phloroglucinol-HCl to visualize lignin deposition. In the lig seedlings stunted at 28°C, lignin deposition was pronounced within and around the stele tissues of hypocotyls and roots (Figure 1B). In these seedlings, unusual lignin deposition was also observed in the epidermis near the shoot apical meristem (Figure 1B). At 18°C, however, the lig seedlings exhibited the same level and distribution of lignin deposition as the wild type, with lignin being confined to the vascular bundle (Figure 1B). These observations showed that the lig mutation causes temperature-dependent growth defects and ectopic lignin deposition in seedlings.
The deleterious effects of the lig mutation were alleviated at lower temperatures but not eliminated even at 18°C, which was apparent after culture for a longer period. Compared with the wild type, lig mutant plants cultured at 18°C exhibited retarded vegetative growth and delayed flowering (Figure 2). Additionally, the inflorescence stems and siliques of reproductive-stage lig plants grown at 18°C were shorter and thicker than those of the wild type, but did not exhibit ectopic lignin deposition (Figure 2).
Figure 2.
Morphology of lig Mutant Plants Grown at 18°C.
Wild-type (Ler) and lig mutant plants cultured at 18°C for 31 d (rosette leaves; [A] and [B]) and 70 d (inflorescence stems and siliques; [C] to [E]).
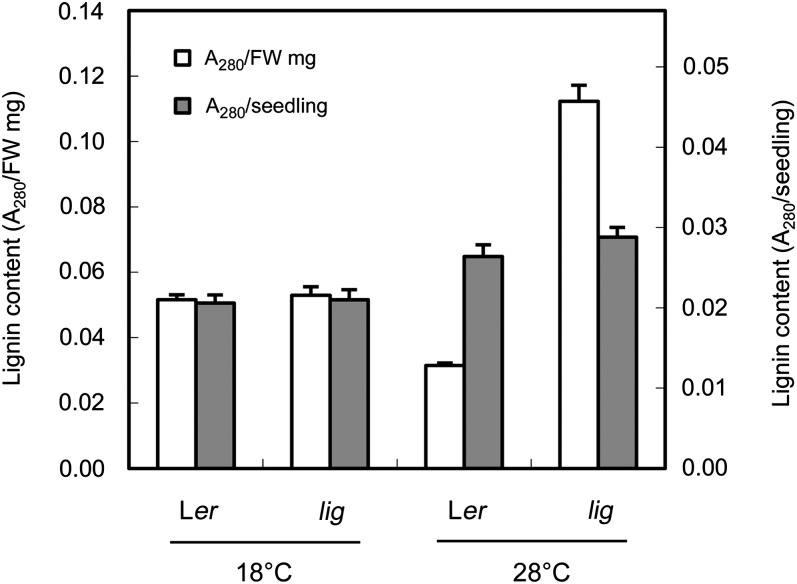
Increased Lignin Content of the lig Seedlings Cultured at 28°C
Quantification of lignin content using the acetyl bromide method (Morrison, 1972) demonstrated that lig seedlings contained more lignin than the wild type when cultured at 28°C but not when cultured at 18°C (Figure 3). On a fresh weight basis, the lignin content was approximately threefold higher in lig seedlings than in wild-type seedlings after culture at 28°C for 5 d (Figure 3). These results indicate that the mutation in lig not only disrupts the spatial control of lignin deposition but also induces active lignin biosynthesis.
Figure 3.
Increased Lignin Content in lig Seedlings.
The lignin content of wild-type (Ler) and lig seedlings grown at 18 or 28°C for 5 d was analyzed using the acetyl-bromide method. Vertical bars represent se values for three determinations. FW, fresh weight.
Lignin Deposition in the Root Elongation Zone of lig Seedlings Exposed to 28°C
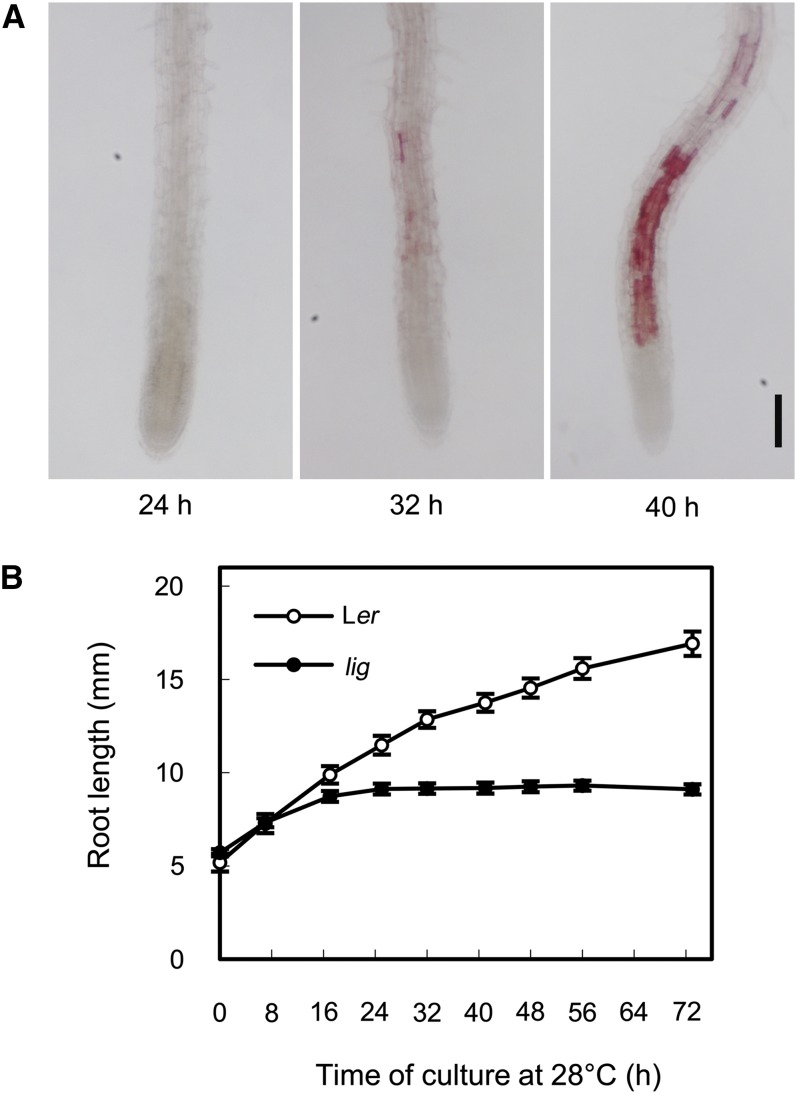
We next examined the time course of root growth and lignin deposition in lig seedlings that had undergone a temperature shift from 18 to 28°C. Thirty-two hours after the temperature shift, lignin deposition became evident in several cells of the root elongation zone, and within the next 8 h, lignin deposition was expanded to the whole root elongation zone but excluded from the root apical meristem (Figure 4A). Thus, the mechanism of lignin induction in the lig mutant appeared to be related to cell elongation and not cell division. In lig seedlings, retardation of root growth was detectable as early as 16 h after the temperature shift, and root growth ceased almost completely within the next 8 h (Figure 4B). These observations indicate that the growth defect precedes ectopic lignin deposition in the lig mutant.
Figure 4.
Time Course of Lignin Accumulation and Root Growth in lig Seedlings Subjected to a Temperature Shift.
Seedlings were cultured at 18°C for 5 d and then at 28°C.
(A) Primary root tips of the lig seedlings were collected 24, 32, and 40 h after the temperature shift and stained with phloroglucinol-HCl for detection of lignin. Bar = 100 µm.
(B) Changes in the primary root length of the wild-type (Ler) and lig seedlings after temperature shift. Vertical bars represent se values for 19 to 23 seedlings.
Effect of the Inhibition of Lignin Deposition on Root Growth of the lig Mutant
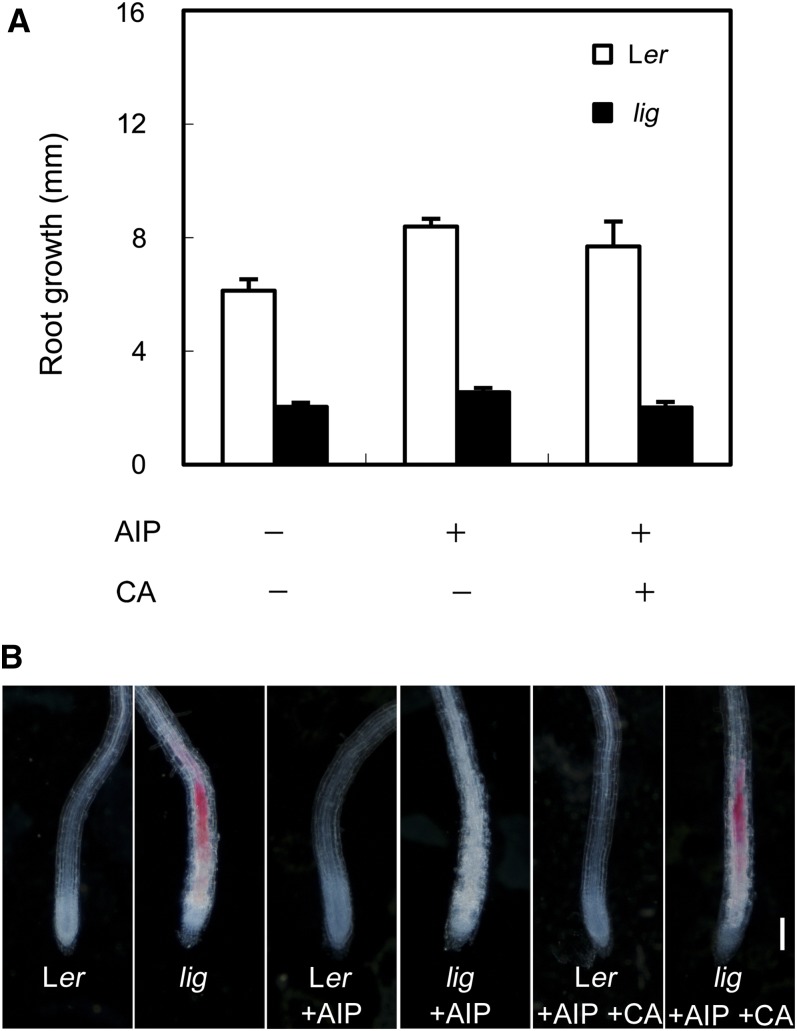
As lignification decreases the extensibility of cell walls, one might speculate that the defect in root elongation in the lig mutant is the result of ectopic lignin deposition in the elongation zone. To test this possibility, we examined the effect of 2-aminoindan-2-phosphonic acid (AIP), which is known to inhibit Phe ammonia-lyase and thereby lignin biosynthesis (Zoń and Amrhein, 1992), on root growth of the lig mutant. AIP treatment did not rescue the root growth defect of lig seedlings at 28°C, although ectopic lignin deposition in these roots was entirely suppressed (Figure 5). This result demonstrated that the root growth defect in the lig mutant is not merely the consequence of ectopic lignin deposition. When AIP was given together with coniferyl alcohol (CA), a kind of monolignol, lignin deposition occurred in the root elongation zone of the lig mutant but not of the wild type (Figure 5B). This result shows that monolignols are not constitutively polymerized into lignin in the elongation zone but that this activity was induced in lig.
Figure 5.
Effect of AIP Treatment on Root Growth and Ectopic Lignin Deposition in lig Seedlings.
Seedlings of the wild type (Ler) and lig mutant were cultured on solidified germination medium containing no additives, 10 µM AIP, or 10 µM AIP and 100 µM CA for 3 d at 28°C after 5 d of culture at 18°C.
(A) Growth of primary roots during 3 d of culture at 28°C. Vertical bars represent se values for four to six seedlings.
(B) Primary root tips stained with phloroglucinol-HCl for detection of lignin. Bar = 100 µm.
Enhanced Hydrogen Peroxide Production and Peroxidase Activity in the Root Elongation Zone of the lig Mutant
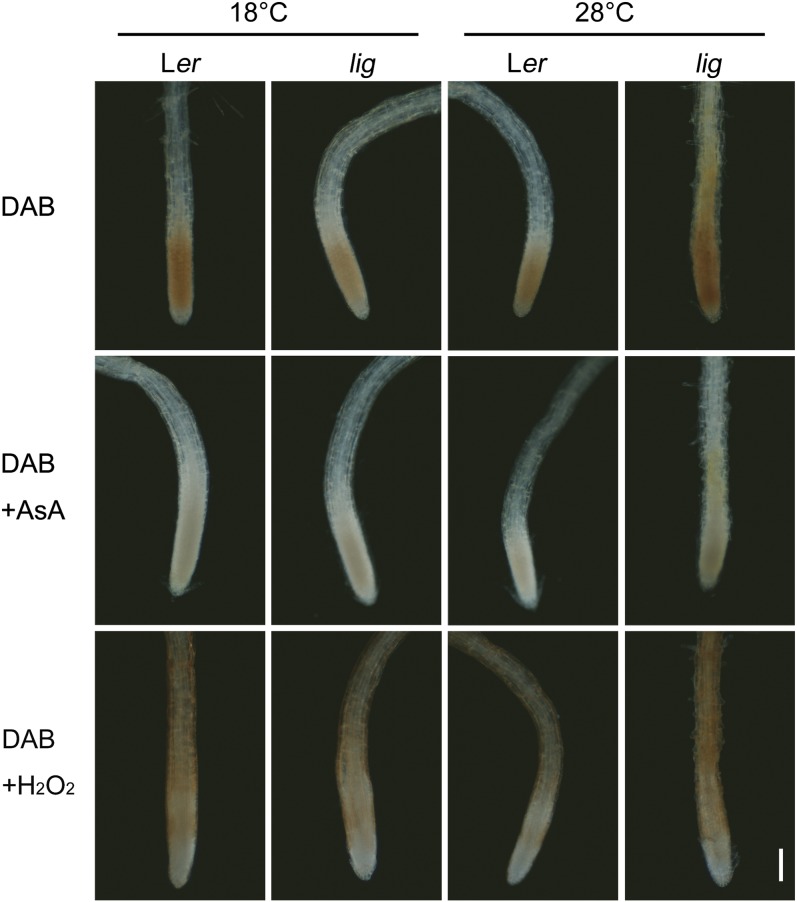
Hydrogen peroxide (H2O2) is reduced by peroxidases and, in turn, oxidizes monolignols during lignin polymerization (Boerjan et al., 2003). Here, we examined H2O2 production and peroxidase activity in the root tissues using the 3,3′-diaminobenzidine (DAB) staining method (Thordal-Christensen et al., 1997). DAB staining in the absence of added H2O2 showed that the endogenous level of H2O2 is elevated in the root elongation zone of the lig mutant (Figure 6). In the elongation zone of the lig mutant, enhanced peroxidase activity was also observed by DAB staining with excess H2O2 (Figure 6).
Figure 6.
Detection of H2O2 Production and Peroxidase Activity in the Root Tip Region using DAB Staining.
Seedlings of the wild type (Ler) and lig mutant that had been grown at 18°C for 5 d and then at 28°C for 32 h were subjected to DAB staining. DAB staining procedures in the absence or presence of added H2O2 were employed to detect endogenous H2O2 and peroxidase activity, respectively. Root tips incubated with DAB and ascorbic acid (AsA) are shown as negative controls. Bar = 100 µm.
Temperature-Dependent Expression of Stress-Responsive Monolignol Biosynthesis Genes in the lig Mutant
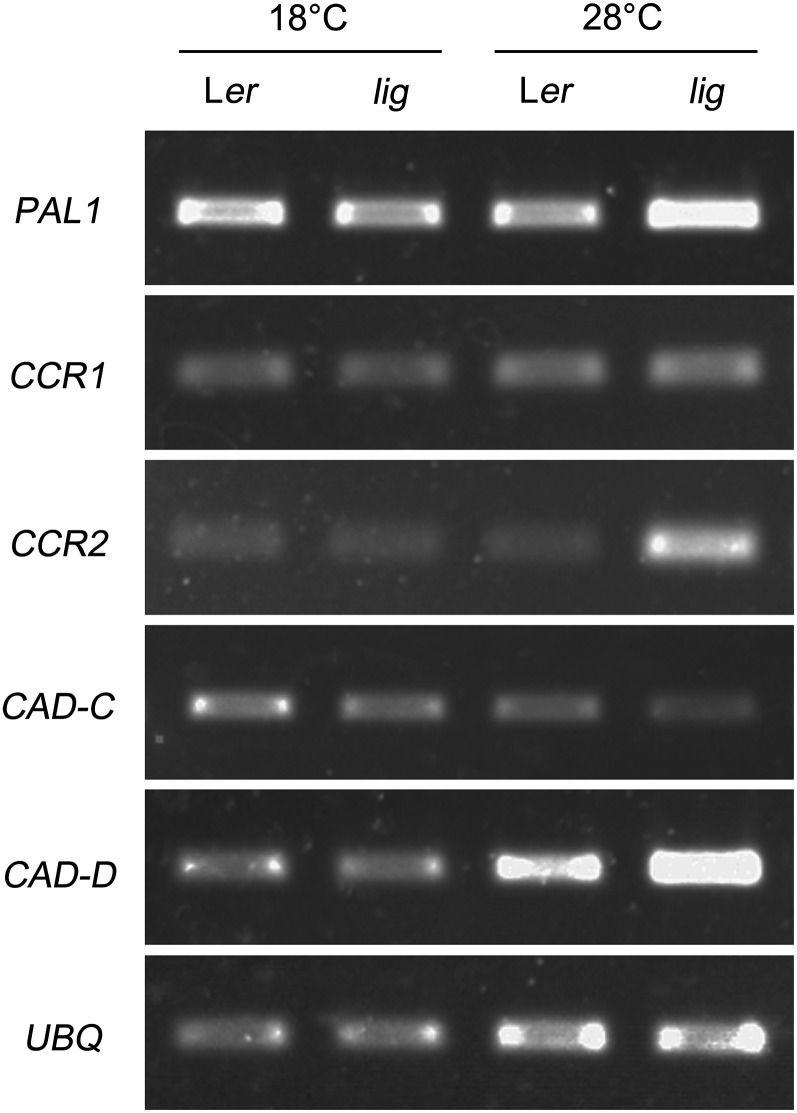
The expression levels of monolignol biosynthesis genes, such as PAL1 encoding Phe ammonia-lyase; CCR1 and CCR2, encoding cinnamoyl-CoA reductase; and CAD-C and CAD-D, encoding cinnamyl alcohol dehydrogenase, were assessed under different temperature conditions by RT-PCR analysis. The expression levels of PAL1, CCR2, and CAD-D were increased in lig seedlings grown at 28°C, but CCR1 and CAD-C expression was unaltered (Figure 7). It has been shown that PAL1, CCR2, and CAD-D are strongly induced by pathogen infection, while CCR1 and CAD-C are not (Giacomin and Szalay, 1996; Lauvergeat et al., 2001; Tronchet et al., 2010). Therefore, this result suggests that stress-responsive lignin biosynthesis is stimulated in the lig mutant at 28°C.
Figure 7.
Expression Analysis of Monolignol Biosynthesis Genes.
Total RNAs were prepared from wild-type (Ler) and lig mutant seedlings grown at 18 or 28°C for 5 d, and the expression of PAL1, CCR1, CCR2, CAD-C, and CAD-D was analyzed by RT-PCR. Amplification of UBQ is shown as a loading control. Similar experiments were performed four times with similar results.
Identification of LIG as a Gene Encoding Glucosamine-6-Phosphate Acetyltransferase
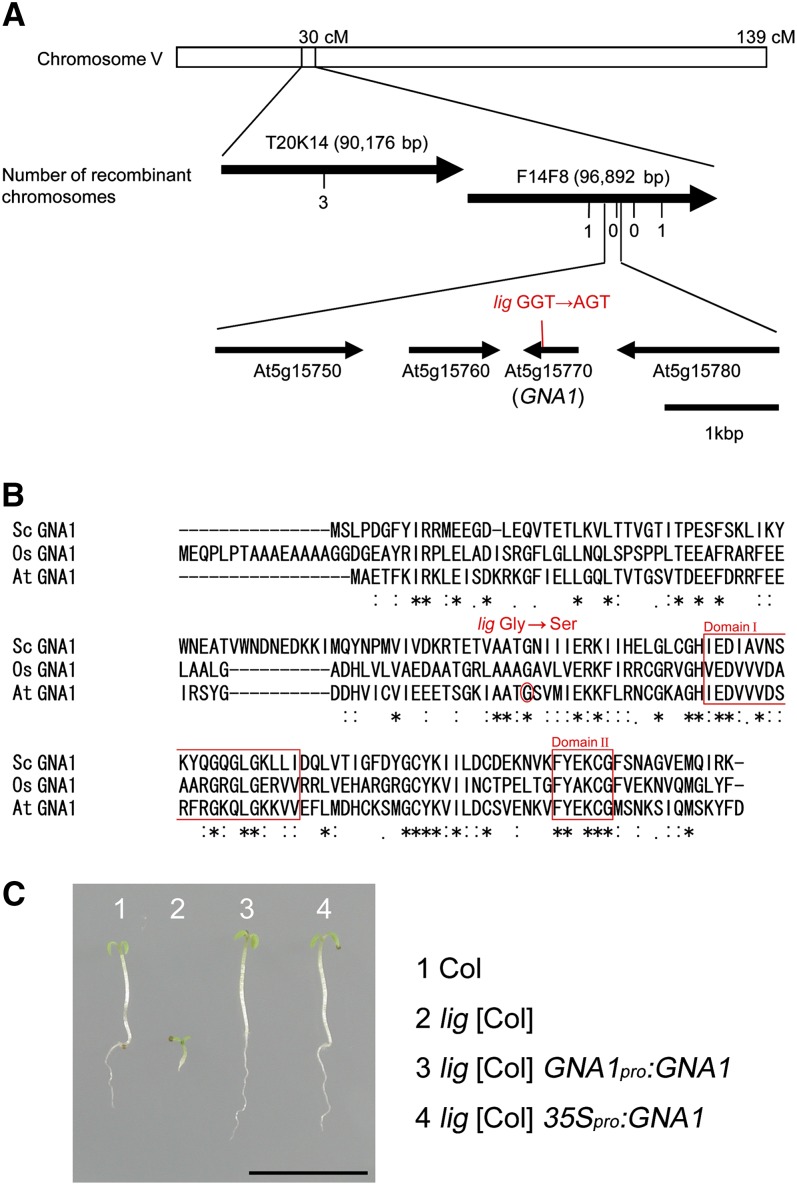
The lig mutation was localized to a 25-kb region at the 30-centimorgan position of chromosome V by chromosome mapping (Figure 8A). At5g15770, which resides in this region, was found to contain a missense mutation that was linked to the lig phenotype. At5g15770 encodes a putative GNA and is referred to as GNA1. The missense mutation in GNA1 of the lig mutant is a G-to-A transition that causes a single amino acid substitution from Gly to Ser at residue 68 (Figure 8B). This Gly residue is conserved among GNAs, such as that from Saccharomyces cerevisiae (Sc GNA1) and rice (Oryza sativa; Os GNA1) (Figure 8B), but is not included in either of two short regions common to several types of acetyltransferases (domain I and domain II; Mio et al., 1999). The Arabidopsis genome contains a single copy of the GNA1 gene and no obvious paralogs.
Figure 8.
Positional Cloning of LIG.
(A) Chromosomal position of the lig mutation. Thick arrows represent BAC clones around the LIG locus on chromosome V. The number of recombination events between DNA polymorphism markers and the LIG locus is given beneath the BAC clones. Thin arrows show genes (open reading frames) predicted to occur at or near the position of the lig mutation. cM, centimorgans.
(B) Alignment of the amino acid sequences of the Arabidopsis GNA1 (At GNA1) with Os GNA1 and Sc GNA1. Asterisks, double dots, and single dots represent identity, strong similarities, and weak similarities, respectively. The position of the lig mutation is marked with a circle. Domains I and II are boxed.
(C) Complementation of the lig phenotype by the introduction of wild-type GNA1. Seedlings of Col, lig [Col], lig [Col] carrying GNA1pro:GNA1, and lig [Col] carrying 35Spro:GNA1 were grown at 28°C for 5 d. Bar = 1 cm.
[See online article for color version of this figure.]
The lig mutant was transformed with GNA1pro:GNA1 or 35Spro:GNA1 to express the wild-type GNA1 transgene under its own promoter or the cauliflower mosaic virus 35S promoter, respectively. The lig phenotypes were complemented by these genes, as seen in images of these seedlings grown at 28°C (Figure 8C). Thus, LIG corresponds to GNA1.
Thermolability of GNA1G68S
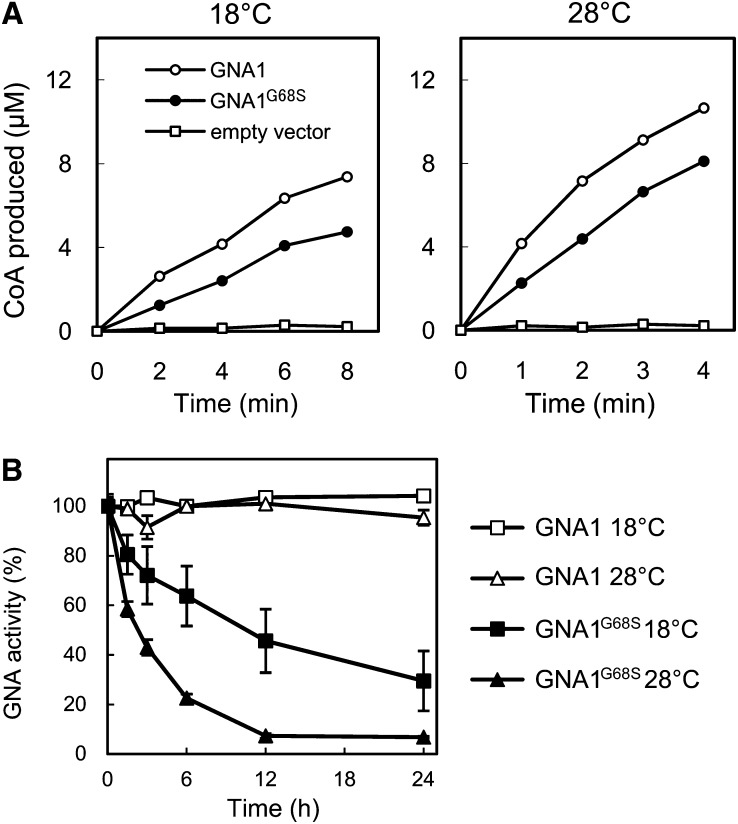
GNA catalyzes the generation of N-acetylglucosamine-6-phosphate and CoA from glucosamine-6-phosphate and acetyl-CoA. Wild-type GNA1 and the lig mutant GNA1 (GNA1G68S) were prepared as recombinant proteins in Escherichia coli, and their GNA enzyme activities were examined at 18 and 28°C by measuring CoA production. Considerable GNA activity was detected for wild-type GNA1 and GNA1G68S, at both 18 and 28°C, although the activity of GNA1G68S was a little lower than that of the wild type (Figure 9A). However, when these proteins were incubated at 18 or 28°C for up to 24 h, GNA1G68S was found to be much more labile than wild-type GNA1. Particularly, the enzyme activity of GNA1G68S disappeared rapidly at 28°C (Figure 9B). The thermolability of GNA1G68S thus reasonably accounts for the temperature-sensitive nature of the lig mutant.
Figure 9.
Enzyme Activities of Recombinant GNA1 and GNA1G68S Proteins.
(A) Time-course analysis of CoA production catalyzed by the GNA activity of GNA1 and GNA1G68S at 18 and 28°C.
(B) Effect of incubation at 18 or 28°C for an extended period of time on the GNA activity of GNA1 or GNA1G68S. The enzyme activities were measured at 28°C after incubation for the indicated periods. Vertical bars represent se values for three measurements.
Deficiency of UDP-GlcNAc in the lig Mutant
The acetylation of glucosamine-6-phosphate by GNA is a critical step in the biosynthesis of UDP-GlcNAc (Mio et al., 1999). Therefore, we speculated that the growth defect and ectopic lignin deposition in the lig mutant may result from a deficiency of UDP-GlcNAc. We examined this possibility by comparing UDP-GlcNAc content between lig and the wild type and by adding UDP-GlcNAc to the lig mutant. We first applied the methods of Nakajima et al. (2010) and Carpita and Delmer (1981) for HPLC analysis of nucleotide sugars, but neither of them gave a resolution sufficiently high to isolate UDP-GlcNAc. Then we modified the latter method by changing the pH of the buffers and finally succeeded in separation and quantification of UDP-GlcNAc (see Supplemental Figure 1 online). When grown at 18°C, the lig seedlings contained a rather higher amount of UDP-GlcNAc than the wild type (Table 1). In the lig seedlings exposed to 28°C for 3 d, however, the content of UDP-GlcNAc was decreased to about one-third of the wild-type level (Table 1).
Table 1. Effect of the lig Mutation and the Addition of GlcNAc and GalNAc on the Contents of UDP-GlcNAc and UDP-GalNAc.
| Temperature | Addition |
UDP-GlcNAc (nmol/g) |
UDP-GalNAc (nmol/g) |
||
|---|---|---|---|---|---|
| Ler | lig | Ler | lig | ||
| 18°C, 8 d | None | 10.2 ± 0.5 | 14.5 ± 2.0 | 4.3 ± 0.4 | 5.7 ± 2.6 |
| 18°C, 5 d/28°C, 3 d | None | 9.4 ± 0.5 | 3.1 ± 0.6* | 3.5 ± 0.4 | 1.9 ± 0.5 |
| 18°C, 5 d/28°C, 4 d | GlcNAc | 25.0 ± 3.5 | 29.9 ± 8.5 | 7.8 ± 1.4 | 9.7 ± 3.7 |
| 18°C, 5 d/28°C, 4 d | GalNAc | 51.1 ± 13.3 | 55.8 ± 15.5 | 17.3 ± 8.1 | 19.9 ± 4.5 |
Seedlings of the wild type (Ler) and lig mutant that had been grown at 18°C for 8 d, or at 18°C for 5 d and then at 28°C for 3 d or 4 d. GlcNAc and GalNAc were administered at the final concentration of 10 µM. Nucleotide sugars were extracted from the root tissues and analyzed by HPLC for measurement of UDP-GlcNAc and UDP-GalNAc. The data represent average values per gram fresh weight obtained from three (top two rows) or two (bottom two rows) biological repeats with the mean se. An asterisk indicates a significant difference from the wild-type value under the same condition (Student’s t test, P < 0.01).
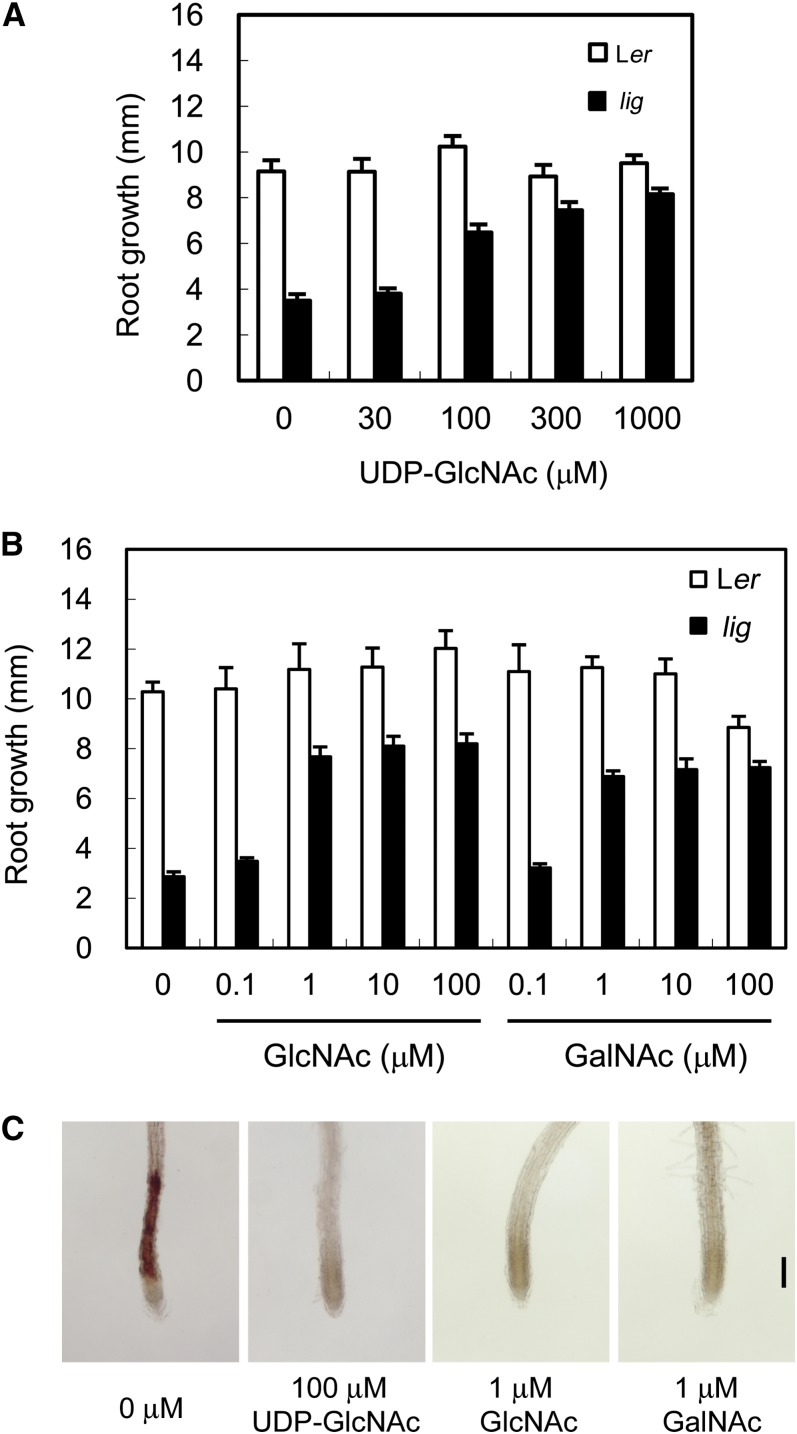
In the presence of 100 µM or higher concentrations of UDP-GlcNAc, the root growth of lig at 28°C was recovered (Figure 10A). In addition, 1 µM GlcNAc or 1 µM N-acetylgalactosamine (GalNAc) restored the lig phenotypes as well (Figure 10B). Ectopic lignin deposition was also suppressed by the addition of UDP-GlcNAc, GlcNAc, or GalNAc (Figure 10C). Application of nonacetylated glucosamine or glucosamine-6-phosphate instead of these acetylated sugars did not alleviate the growth defect of the lig mutant (see Supplemental Figure 2 online). Administration of GlcNAc or GalNAc markedly increased the UDP-GlcNAc content both in the wild type and in lig (Table 1), indicating that the acetylated glucosamine or galactosamine derivatives can directly or indirectly supply UDP-GlcNAc without a requirement for GNA activity. The results obtained here suggest that the lig phenotypes are caused by a deficiency of UDP-GlcNAc.
Figure 10.
Effects of Acetylated Sugars on Root Growth and Ectopic Lignin Deposition in the lig Mutant.
Seedlings of the wild type (Ler) and lig mutant were cultured on solidified germination medium containing various concentrations of UDP-GlcNAc, GlcNAc, or GalNAc for 3 d at 28°C, after 6 d of culture at 18°C.
(A) and (B) Growth of primary roots during 3 d of culture at 28°C in the presence of UDP-GlcNAc (A) or GlcNAc or GalNAc (B). Vertical bars represent se values for six to 15 seedlings.
(C) Primary root tips of lig seedlings stained with phloroglucinol-HCl for detection of lignin. Bar = 100 µm.
Decrease of N-Linked Glycans in the lig Mutant
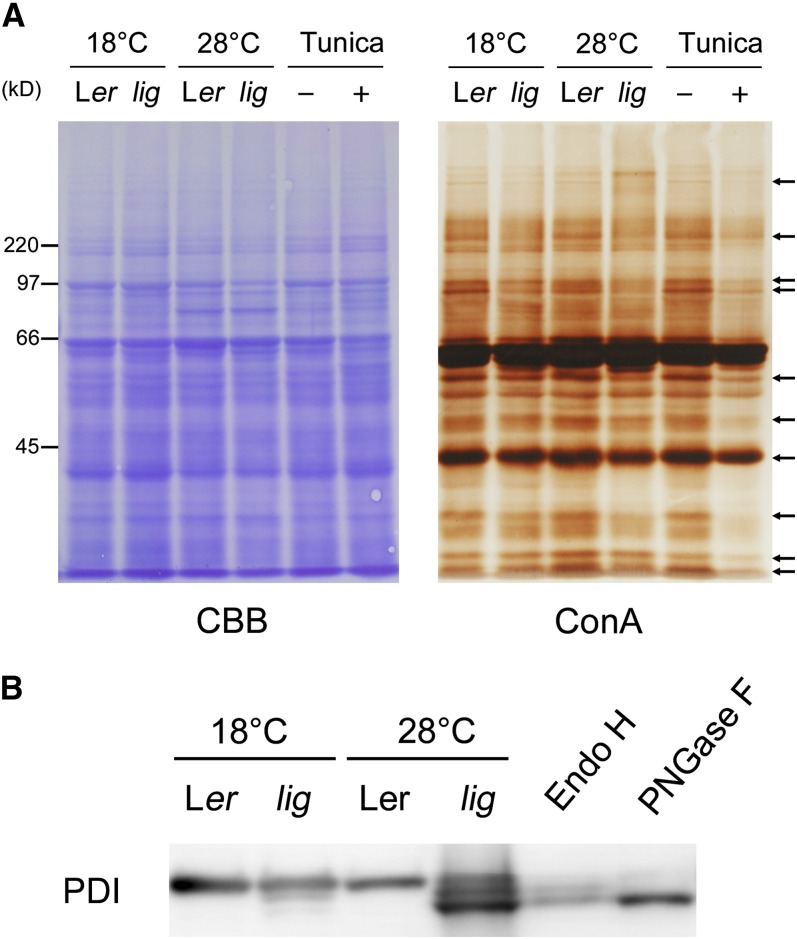
UDP-GlcNAc serves as a donor of GlcNAc in the biosynthesis of various glycans. Among these glycans, N-linked glycans are likely to be particularly vulnerable to the UDP-GlcNAc deficiency because N-glycan synthesis requires GlcNAc as the initial sugar moiety (Pattison and Amtmann, 2009). For this reason, we examined whether the lig mutation affected N-linked glycan content. SDS-PAGE followed by lectin staining with concanavalin A (ConA), a lectin that recognizes branches of oligomannose chains widely present in N-glycans on glycoproteins, was used to monitor N-linked glycan content. In the lig mutant, the intensity of ConA staining for many of the glycoprotein bands was weaker than it was in the wild type (Figure 11A). Differences in ConA staining between the lig mutant and the wild type were detectable in samples prepared from the 18°C culture but were slightly more obvious in the samples from the 28°C culture. Treatment of the wild-type seedlings with tunicamycin, an inhibitor of the first step of N-glycan synthesis (Hori and Elbein, 1981), strikingly reduced ConA staining, confirming that the ConA staining method effectively detects N-glycans.
Figure 11.
Effect of the lig Mutation on the Electrophoretic Profiles of N-Glycoproteins.
(A) Proteins were extracted from the roots of seedlings of the wild type (Ler) and lig mutant grown for 5 d at 18°C and then for 3 d at 18 or 28°C, and also from the roots of wild-type seedlings cultured with 100 ng/mL tunicamycin for 3 d after 5 d of tunicamycin-free culture at 18°C. The protein samples were separated by SDS-PAGE and subjected to Coomassie blue (CBB) staining for total protein detection or lectin staining with ConA for N-glycan detection. Arrows indicate representative bands that are weaker in lig than the wild type at 28°C.
(B) Immunoblot analysis with the anti-PDI antibody. Proteins were extracted from the seedlings of the wild type (Ler) and lig mutant grown for 6 d at 18 or 28°C. A portion of the protein sample prepared from the wild type (Ler, 28°C) was incubated in the presence of Endo H or PNGase F for digestion of N-glycans before SDS-PAGE. Each lane contains proteins equivalent to the extract from 10 mg (fresh weight) of seedlings.
[See online article for color version of this figure.]
For closer examination of the status of N-glycans, we focused on protein disulfide isomerase (PDI), an endoplasmic reticulum (ER)-resident protein that is normally N-glycosylated at two Asn residues and has been used as a model protein for N-glycosylation analysis (Kajiura et al., 2010). Immunoblot analysis indicated that, while PDI of the wild-type plants migrated in electrophoresis as a single band, the lig mutant cultured at 28°C had two additional bands of PDI with higher mobility (Figure 11B). These two bands were judged to represent the PDI protein lacking one or two N-glycan chains, since they were very similar in electrophoretic mobility with digestion products of PDI obtained by treatment with endoglycosidase H (Endo H) or peptide:N-glycosidase F (PNGase F) (Figure 11B). These results showed that N-glycans were decreased in the lig mutant.
The electrophoretic mobility of another glycoprotein, PYK10, which resides in the ER body (Matsushima et al., 2003), was also tested in the presence of the lig mutation. In this protein, however, we could not detect mobility changes indicative of loss of N-glycan chains (see Supplemental Figure 3 online). This result is suggestive of variation in the susceptibility of N-glycosylation of different glycoproteins to the lig mutation.
Unfolded Protein Response Induced by the lig Mutation
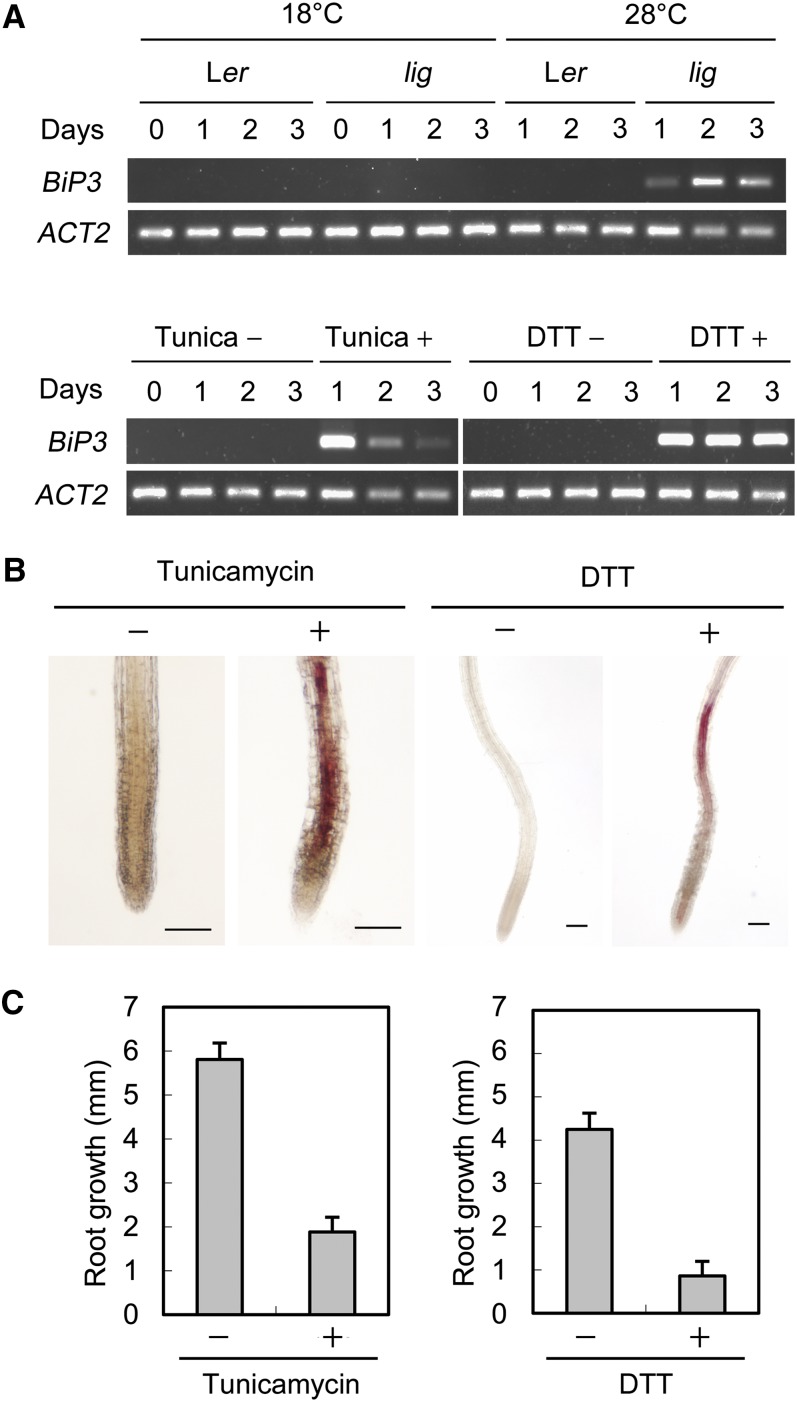
Impairment of protein N-glycosylation is known to cause ER stress leading to the unfolded protein response (UPR) (Urade, 2007). We examined if the UPR would occur in the lig mutant by monitoring expression of LUMINAL BINDING PROTEIN3 (BiP3), a typical UPR gene often used as an indicator of the UPR (Iwata and Koizumi, 2005). BiP3 expression was highly elevated in the lig mutant after exposure to the restrictive temperature (Figure 12A), a result that indicates that the lig mutation induces the UPR in a temperature-dependent manner.
Figure 12.
Relationship between the UPR and Ectopic Lignin Deposition in the lig Mutant.
(A) RT-PCR analysis of BiP3 expression. Total RNA was isolated from seedlings of the wild-type (Ler) and lig mutant grown at 18 or 28°C for the indicated periods after 7 d of culture at 18°C and subjected to RT-PCR analysis. RNA samples were also prepared from the wild-type seedlings grown at 18°C with or without UPR-inducing reagents (100 ng/mL tunicamycin or 1 mM DTT) for the indicated periods after 7 d of culture without these reagents at 18°C. Amplification of ACT2 is shown as a loading control.
(B) and (C) Wild-type (Ler) seedlings were grown at 18°C for 5 d and then treated with 100 ng/mL tunicamycin or 1 mM DTT for 3 d. Experiments for tunicamycin treatment and DTT treatment were separately conducted.
(B) Primary root tips stained with phloroglucinol-HCl for detection of lignin. Bars = 100 µm.
(C) Growth of primary roots during 3 d of culture with or without tunicamycin or DTT. Vertical bars represent se values for eight to 10 seedlings.
Phenocopy of the lig Mutant by Tunicamycin and DTT
Tunicamycin induces the UPR through inhibition of protein N-glycosylation (Martínez and Chrispeels, 2003). DTT, a reducing agent that disrupts disulfide bonds, also induces the UPR, but unlike tunicamycin, without preventing N-glycosylation (Martínez and Chrispeels, 2003). Wild-type seedlings treated with these UPR inducers were inspected for elongation growth and lignin deposition. Both tunicamycin and DTT induced severe growth defects and ectopic lignin deposition (Figures 12B and 12C), resulting in plants that resembled the lig mutant. Thus, tunicamycin treatment and DTT treatment phenocopied the lig mutant. This result further supports the connection between the lig phenotype and the UPR.
DISCUSSION
Deficiency in UDP-GlcNAc Synthesis Causes Growth Retardation and Ectopic Lignin Deposition
The lig mutant exhibited severe growth retardation and abnormal lignin deposition under high temperature conditions (i.e., 28°C; Figure 1). Positional cloning identified the LIG gene as GNA1, a single-copy gene encoding GNA (Figure 8). GNA catalyzes the acetylation of glucosamine-6-phosphate, which is crucial for UDP-GlcNAc biosynthesis in S. cerevisiae (Mio et al., 1999; Milewski et al., 2006). Enzymological analysis of recombinant GNA1 proteins confirmed the GNA activity of GNA1 and attributed the temperature-dependent defects of the lig mutant to the thermolability of the mutated GNA1 (Figure 9). Quantification of UDP-GlcNAc content showed a significant reduction of UDP-GlcNAc in the lig mutant at 28°C (Table 1). Furthermore, the addition of UDP-GlcNAc suppressed both the growth defect and ectopic lignin deposition of lig seedlings grown at 28°C (Figure 10). These findings strongly suggest that the shortage of UDP-GlcNAc due to the genetic lesion of GNA results in the lig phenotype at 28°C.
The only plant GNA mutant that has been reported to date is gna1 of O. sativa, which carries a T-DNA insertion in the rice GNA gene, Os GNA1 (Jiang et al., 2005). Like the lig mutant, this mutant is deficient in UDP-GlcNAc; however, in contrast with lig, it exhibits more severe root growth defects at lower temperatures (25°C) than at higher temperatures (32°C). Unfortunately, it is not known why the rice gna1 mutant is cold sensitive, and lignin-related phenotypes have not been described for gna1. A detailed comparison of gna1 with lig would provide further insight into the physiological impacts of the UDP-GlcNAc deficiency.
Arabidopsis Can Use GlcNAc and GalNAc for UDP-GlcNAc Synthesis
The lig phenotypes were suppressed by the addition of GlcNAc, GalNAc, or UDP-GlcNAc (Figure 10). We measured the UDP-GlcNAc content after treatment with GlcNAc or GalNAc and found it strikingly increased (Table 1). These results strongly suggest that Arabidopsis contains a metabolic pathway(s) in which UDP-GlcNAc is synthesized from GlcNAc and GalNAc independently of GNA activity. Mammals have been reported to possess GlcNAc kinases, which phosphorylate GlcNAc to supply GlcNAc-6-phosphate for UDP-GlcNAc synthesis (Hinderlich et al., 1998, 2000). By conducting a BLAST search, we identified At1g30540 as a putative homolog of the mammalian GlcNAc kinase genes in the Arabidopsis genome. This GlcNAc kinase gene homolog might enable Arabidopsis to synthesize UDP-GlcNAc from GlcNAc. In Candida albicans, a kind of hexokinase was shown to function as a GlcNAc kinase (Mio et al., 2000; Yamada-Okabe et al., 2001). Arabidopsis has three hexokinase genes and three hexokinase-like genes (Karve et al., 2008). Proteins encoded by these genes are also candidates for enzymes that phosphorylate GlcNAc. Although the metabolic fate of GalNAc in plants is not clear, considering that epimerases that can catalyze the conversion between Gal and Glc moieties of sugar derivatives are known to exist in various organisms (Allard et al., 2001), it is likely that epimerization occurs on GalNAc or on GalNAc-derived metabolites to produce a UDP-GlcNAc synthesis precursor(s).
GlcNAc and GalNAc were much more effective at restoring the phenotype of the lig mutant than was UDP-GlcNAc (Figure 10). In general, nucleotide sugars cannot substantially move across cell membranes without the aid of transporters. The lower ability of UDP-GlcNAc to restore the mutant phenotype might be explained by its low membrane permeability and lack of efficient UDP-GlcNAc transport systems.
De Novo Lignin Synthesis Is Possibly Triggered by the Stress Response in the Elongation Zone of the lig Mutation
Since root growth of the lig mutant was not recovered by the inhibition of lignin biosynthesis (Figure 5), abnormal lignin deposition is not responsible for the growth defect. In the lig seedlings grown at 28°C after an initial culture at 18°C, root growth ceased within 24 h after the increase in temperature and then ectopic lignin deposition started to occur (Figure 4). This observation rather implicates impaired root growth in the induction of ectopic lignin deposition.
We found that the expression levels of monolignol biosynthesis genes PAL1, CCR2, and CAD-D are elevated in lig seedlings grown at 28°C (Figure 7). DAB staining experiments indicated that H2O2 production and peroxidase activity, which together polymerize monolignols to form lignin, are enhanced in the elongation zone of the lig mutant after exposure to 28°C (Figure 6). The occurrence of lignin-forming activity in the elongation zone was also demonstrated by treatment with AIP and CA (Figure 5). These results collectively indicate that an entire system required for de novo lignin biosynthesis is induced in the lig mutant in association with the defect of elongation growth.
Expression of PAL1, CCR2, and CAD-D was previously shown to be triggered by pathogen infection (Giacomin and Szalay, 1996; Lauvergeat et al., 2001; Tronchet et al., 2010). It is well known that H2O2 is generated upon biotic and abiotic stresses (Jaspers and Kangasjärvi, 2010; Torres, 2010). Therefore, the elevated expression of monolignol biosynthesis genes and H2O2 production in the lig mutant may be induced as responses to some stress related to the growth defect.
Impairment in N-Glycan Synthesis and Resultant UPR Lead to Ectopic Lignin Deposition in the lig Mutant
All N-linked glycans have two GlcNAc moieties at their attachment site to proteins. UDP-GlcNAc is an indispensable donor of these GlcNAcs during N-glycan biosynthesis. Hence, the shortage of UDP-GlcNAc is expected to interfere with normal N-glycan synthesis. In accordance with this, the level of N-glycans associated with glycoproteins was shown to be decreased in the lig mutant, which is deficient in UDP-GlcNAc due to a genetic lesion in GNA (Figure 11). Tunicamycin, an inhibitor of N-linked glycan synthesis, resulted in growth retardation and ectopic lignin deposition, features that were also characteristic of lig (Figures 12B and 12C). These findings suggest that impairment of N-glycan synthesis is a major cause of the lig phenotype.
N-linked glycans of glycoproteins are very important for their proper folding. When N-glycosylation is interfered with, misfolded proteins are increased in the ER lumen, which elicits ER stress and consequently induces the UPR (Liu and Howell, 2010). In the lig mutant, the UPR was shown to occur at the restrictive temperature, consistent with the impairment of N-glycan synthesis (Figure 12A). Additionally, we found that treatment with DTT, which induces the UPR by a mechanism unrelated to N-glycan synthesis, phenocopies the lig mutant (Figures 12B and 12C). We can reasonably speculate from all these results that the ectopic lignin accumulation caused by the lig mutation is largely mediated by the UPR.
There have been several reports that ectopic lignin deposition is related to cell wall damage in cellulose-deficient mutants and in plants treated with a cellulose synthesis inhibitor (Caño-Delgado et al., 2000, 2003; Bischoff et al., 2009). Notably, tunicamycin treatment was reported to induce radial swelling and callose deposition in the root tip, which are characteristic of cellulose-deficient phenotypes (Lukowitz et al., 2001). We therefore suspect that cell wall damage may lie somewhere, possibly downstream or in parallel to the UPR, in the pathway from the impairment of N-glycan synthesis toward the ectopic lignin deposition.
UDP-GlcNAc is involved not only in N-glycan synthesis but also in the synthesis of other glycoconjugates, including glycosylphosphatidylinositol (GPI) anchors. Cellulose-deficient phenotypes were observed in a mutant defective in GPI anchor synthesis (Gillmor et al., 2005). Thus, the possibility also cannot be excluded that, in addition to the defects in N-glycan biosynthesis, defects in GPI anchors might cause cell wall damage that leads to the induction of lignin synthesis in the lig mutant.
In summary, through analyses of the lig mutant, we have shown that a shortage of UDP-GlcNAc and the subsequent impairment of N-glycan synthesis may activate lignin biosynthesis, probably via ER stress and the UPR. It seems possible that a type of stress response to cell wall damage is also involved in the activation of lignin biosynthesis in the lig mutant. The temperature-sensitive nature of the lig mutant resulting from the thermolability of GNA offers a unique opportunity to study further the physiological roles of UDP-GlcNAc and the molecular mechanisms underlying stress-induced lignification.
METHODS
Plant Materials
From an ethyl methanesulfonate–mutagenized population of the Landsberg erecta (Ler) ecotype of Arabidopsis thaliana, lig was isolated as a temperature-sensitive mutant defective in adventitious root formation by the same screening as described by Konishi and Sugiyama (2003). Before detailed characterization, the lig mutant was genetically purified by crossing three times with Ler, followed by self-propagation. In this study, the Ler strain was used as the wild type, unless otherwise indicated.
In the complementation analysis, we used lig [Col], a lig mutant line of which the genetic background was partially replaced by crossing with the Columbia (Col) ecotype. lig [Col] was established from F3 lines that were homozygous for the lig mutation and did not exhibit the erecta phenotypes. Whereas the original lig line was largely incompetent for plant transformation, lig [Col] allowed a considerably higher efficiency of transformation.
Plant Culture Conditions
Seedlings were aseptically and vertically grown on germination medium solidified with agar (GMA) under continuous light (15 to 25 µmol/m2/s). GMA is Murashige and Skoog medium (Murashige and Skoog, 1962) supplemented with 10 g/L Suc, buffered to pH 5.7 with 0.5 g/L MES, and solidified with 15 g/L agar. For treatment with AIP, acetylated sugars, DTT, or tunicamycin, seedlings were cultured on GMA that contained these reagents.
For long-term culture, plants were grown on a 1:1 mixture (v/v) of potting compost (Hanasaki-baido; Takii Shubyo) to vermiculite under continuous light (30 to 40 µmol/m2/s).
Induction of Adventitious Root Formation
Hypocotyl segments excised from seedlings were cultured on root-inducing medium at 28°C under continuous light (15 to 25 µmol/m2/s). Root-inducing medium was Gamborg’s B5 medium (Gamborg et al., 1968) supplemented with 20 g/L Glc and 0.5 mg/L indole-3-butyric acid, buffered to pH 5.7 with 0.5 g/L MES, and solidified with 2.5 g/L gellan gum. The ability to produce adventitious roots under these conditions was used as an index phenotype in mutant isolation and chromosome mapping.
Phloroglucinol-HCl Staining
Seedlings were fixed and cleared in 70% ethanol overnight at 4°C. After rinsing with water, the seedlings were stained with 1% (w/v) phloroglucinol in 20% (w/v) HCl (Nakano and Meshitsuka, 1992) and then observed under a light microscope (Labophoto-2; Nikon).
Measurement of Lignin Content
Lignin content was determined according to the method of Morrison (1972), with some modifications. Ten seedlings stored at −70°C were homogenized in ice-cold 95% ethanol using a plastic pestle and then using an ultrasonic disruptor (UD200; Tomy Seiko). The homogenate was centrifuged at 3000g for 5 min. The resultant pellet was washed three times with 95% ethanol and twice with a 1:2 (v/v) mixture of ethanol to hexane and allowed to air-dry at 36°C overnight. The dried pellet was ultrasonically homogenized in acetic acid and centrifuged at 1000g for 5 min. The pellet was resuspended in 25% acetyl bromide in acetic acid and centrifuged at 1000g for 5 min. To each sample, 300 μL 25% acetyl bromide in acetic acid was added. After heating to 70°C for 30 min, 270 μL 2 M NaOH, 1.5 mL acetic acid, 30 μL 7.5 M hydroxylamine hydrochloride, and 900 μL acetic acid were added to each sample. The samples were centrifuged at 1000g for 5 min, and the absorbance of the supernatant was measured at 280 nm to determine the lignin content.
DAB Staining
H2O2 was detected according to the method of Thordal-Christensen et al. (1997), with some modifications. Seedlings were placed in 10 mM sodium-acetate buffer, pH 5.5, that contained 1 mg/mL DAB for 30 min at room temperature. The seedlings were washed with water and cleared by boiling for 10 min in 95% ethanol. The samples were then observed under a light microscope. As a negative control, 10 mM ascorbic acid was added to the DAB solution before the seedlings were immersed. For the histochemical detection of peroxidase activity, seedlings were placed in 10 mM sodium-acetate buffer, pH 5.5, that contained 1 mg/mL DAB and 0.1 mM H2O2 for 5 min at room temperature.
RT-PCR Analysis of Gene Expression
Total RNA was isolated from seedlings according to the method of Verwoerd et al. (1989), treated with DNase I (Takara), and then reverse transcribed with Super-Script II reverse transcriptase (Invitrogen). The first-strand cDNA was used as a template for PCR amplification of cDNA fragments. PCR reactions were performed using PrimeSTAR GXL DNA Polymerase (Takara) with gene-specific primers (see Supplemental Table 2 online). Denaturation took place at 98°C for 10 s, annealing at 56°C for 15 s, and elongation at 68°C for 1 min. Cycle numbers are listed in Supplemental Table 2 online. The cycle numbers used here produced the exponential phase of PCR amplification, which was verified by performing the PCRs for various numbers of cycles. The PCR products were electrophoresed on a 1% agarose-ME (Nacalai Tesque), and the gel was stained with ethidium bromide.
Chromosome Mapping
The lig mutant was crossed to the wild-type Col ecotype, and the F2 progeny were used for chromosome mapping. F2 plants were tested for their ability to produce adventitious roots at 28°C and for DNA polymorphisms. In some cases, the adventitious rooting ability of F3 plants derived from each individual F2 plant was tested to genotype the parental F2 plants for the lig mutation. The chromosomal location of lig was determined on the basis of linkage between the lig mutation and the polymorphic alleles.
Sequence Alignment
Sequence alignment was performed using ClustalW (version 1.83) at the DNA Data Bank of Japan using default parameters.
Complementation Test
To construct GNA1pro:GNA1, the −1653/ +450 region (+1 = the first base of the translation initiation codon, +448 = the first base of the stop codon) of GNA1 was amplified by PCR from the Ler genomic DNA, cloned into the pENTR/D-TOPO vector (Invitrogen), and then transferred to the Gateway binary vector pGWB1 (Nakagawa et al., 2007). 35Spro:GNA1 was similarly constructed from the +1/+450 region fragment of the GNA1 gene in the binary vector pGWB2 (Nakagawa et al., 2007). The constructs were introduced into the lig [Col] plants via Agrobacterium tumefaciens. Plant transformation was performed according to the floral dip method by Clough and Bent (1998).
Preparation of GNA1 Recombinant Proteins
DNA fragments of the coding region of GNA1 (+4 to +450) were PCR amplified from genomic DNA isolated from wild-type Ler and the lig mutant and cloned between the BamHI and PstI sites of the pQE30 vector (Qiagen) to construct plasmids for production of His-tagged GNA1 and GNA1G68S proteins. These plasmids were transfected into Escherichia coli M15. The transformed bacterial cells were grown at 37°C until OD600 reached 0.6, and the cultures were supplemented with 1 mM isopropyl-β-D-thiogalactoside and incubated for another 5 h at 18°C. The cultured cells were harvested and resuspended in equilibration/wash buffer, pH 7.0, containing 50 mM sodium phosphate and 300 mM NaCl. After incubation with 0.75 mg/mL lysozyme at 4°C for 20 min, the cells were disrupted by ultrasonic treatment. The cell lysates were centrifuged at 12,000g for 20 min at 4°C. The His-tagged recombinant proteins were purified from the supernatant using TALON metal affinity resin (Clontech) column chromatography. The purity of the recombinant proteins was checked by SDS-PAGE.
GNA Enzyme Assay
GNA enzyme activity was determined according to the method by Mio et al. (1999) with some modifications. Enzyme reactions were performed in 50 μL of a reaction mixture containing 50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 200 µM glucosamine-6-phosphate, 200 µM acetyl-CoA, 10% glycerol, and ∼0.1 µg purified recombinant GNA1 protein. The reaction mixture was mixed on ice and then incubated at 18 or 28°C for the indicated periods. The reaction was terminated by adding 50 μL 50 mM Tris-HCl, pH 7.5, buffer containing 6.4 M guanidine hydrochloride. To detect the thiol group of CoA produced by the GNA reaction, 50 μL 50 mM Tris-HCl, pH 7.5, buffer containing 1 mM EDTA and 0.1 mM 5,5′-dithiobis(2-nitrobenzoic acid) was added, and the absorbance of the solution was measured at 412 nm. For thermolability analysis, recombinant GNA1 proteins (0.02 µg/µL) were incubated in the equilibration/wash buffer containing 10% glycerol for various periods of time at 18 or 28°C before enzyme assay.
Determination of UDP-GlcNAc and UDP-GalNAc by HPLC
Roots of seedlings (27 to 130 mg) were frozen with liquid nitrogen and stored at −70°C. Frozen samples were disrupted with a bead-type cell disrupter (MS-100; Tomy Seiko) and then homogenized with an ultrasonic disrupter (UD-200; Tomy Seiko) in 1 mL extraction buffer containing 75% ethanol and 25% sodium-phosphate buffer saline (10 mM sodium phosphate and 0.15 M NaCl, pH 7.4) on ice. After addition of cytidine-5′-monophospho-N-acetyl-d-neuraminic acid, which plants usually do not contain, as an internal standard, nucleotide sugars were purified with Envi-Carb SPE column (250 mg/6 mL column; Supelco) according to of Nakajima et al. (2010), lyophilized, and stored at −70°C.
HPLC analysis of nucleotide sugars was performed based on the method of Carpita and Delmer (1981) with several modifications. The lyophilized sample was dissolved in 200 to 250 μL distilled water, and a 20 to 50 μL aliquot was fractionated by gradient HPLC at 40°C on an anion exchange column (Partisil-10 SAX, 4.6 × 250 mm; GL Science) with a HPLC system (L-6210 intelligent pump, L-4200H UV-VIS detector, 655A-52 column oven, D-2500 chromatographic-data processor; Hitachi) using a mixture of solvent A (5 mM KH2PO4, pH 3.1) and solvent B (0.6 M KH2PO4, pH 3.6). The pH of solvents was adjusted with phosphoric acid at room temperature. The gradient condition was 0 to 12% B for 30 min, 12 to 40% B for 5 min, 40 to 100% B for 5 min, 100% B for 10 min, and then 0% B for 10 min for reequilibration; the solvent flow rates were 1 mL/min for the first 50 min and 2 mL/min for the later 10 min for reequilibration. The eluate was monitored by absorbance at 254 nm. The concentration of UDP-GlcNAc and UDP-GalNAc were estimated by comparison with the peak heights detected by absorbance at 254 nm during HPLC of authentic samples of known concentrations and internal standards.
Detection of N-Glycosylated Proteins
Roots of seedlings were homogenized in 50 mM Tris-HCl, pH 7.2, on ice using a plastic pestle and then an ultrasonic disruptor (UD200; Tomy Seiko). After centrifugation at 12,000g for 5 min at 4°C, the supernatant was collected. The supernatant was mixed with 1.2 volumes of methanol and 0.3 volume of chloroform and centrifuged at 12,000g for 5 min at 4°C. The pellet was rinsed with methanol, dried at room temperature for 10 min, resuspended in SDS-PAGE sample buffer containing β-mercaptoethanol, and heated at 95°C for 5 min. Proteins were separated by SDS-PAGE using a 10% acrylamide gel and electroblotted onto a polyvinylidene difluoride (PVDF) membrane (NIPPON Gene).
For N-glycan detection with ConA, the PVDF membrane was washed four times in 10 mM Tris-HCl, pH 7.2, buffer containing 150 mM NaCl and 0.05% Tween 20 (buffer A) for 10 min, and then incubated for 1 h in buffer A containing 50 µg/mL ConA (Sigma-Aldrich) at 25°C. After another 10-min wash with buffer A, the membrane was incubated for 1 h in buffer A containing 50 µg/mL horseradish peroxidase (Wako). The membrane was washed three times by immersion in buffer A for 10 min, rinsed with 15 mM sodium-phosphate buffer, pH 7.0, and then transferred to 15 mM sodium-phosphate buffer, pH 7.0, containing 1 mg/mL DAB. Five minutes later, H2O2 was added to a final concentration of 5 mM to detect peroxidase activity.
For detection of PDI and PYK10 by immunoblotting, the PVDF membranes were incubated for 1 h at 37°C with 5% (w/v) skim milk in 100 mM maleic acid buffer, pH 7.5, containing 150 mM NaCl and 0.3% Tween 20, and then incubated with anti-PDI antibody (Kajiura et al., 2010) and anti-PYK10 antibody (Matsushima et al., 2003), respectively, in the same buffer for 1 h at 37°C. After washing four times with 100 mM maleic acid buffer, the membrane was incubated with alkaline phosphatase–conjugated mouse monoclonal anti-rabbit IgG (Sigma-Aldrich) in 100 mM maleic acid buffer containing 0.1% skim milk for 1 h at 37°C. After three washes with 100 mM maleic acid buffer, the membrane was placed in 100 mM Tris-HCl buffer, pH 9.5, containing 0.1 M NaCl. Signals were detected with CDP-star ready-to-use (Roche) and a luminescent image analyzer (LAS-1000 UV mini; Fujifilm).
Digestion of N-glycans was performed using Endo H (New England Biolabs) and PNGase F (New England Biolabs) according to the manufacturer’s protocol. Briefly, methanol-chloroform purified proteins were resuspended in denaturing buffer containing 0.5% SDS and 40 mM DTT, heated for 10 min at 100°C, and then treated with Endo H (New England Biolabs) and PNGase F (New England Biolabs).
Accession Numbers
Locus information and sequence data from this article can be found in the Arabidopsis Genome Initiative or in the GenBank/EMBL data libraries under the following accession numbers: GNA1, At5g15770; BiP3, At1g09080; CAD-C, At3g19450; CAD-D, At4g34230; CCR1, At1g15950; CCR2, At1g80820; PAL1, At2g37040; UBQ, At4g05320; Os GNA1, AAV40998; and Sc GNA1, BAA36495.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Anion Exchange HPLC Elution Profiles for UDP-GlcNAc and UDP-GalNAc Analyses in the lig Mutant and Wild-Type Seedlings.
Supplemental Figure 2. Effects of Treatment with Nonacetylated Glucosamine or Glucosamine-6-Phosphate on Root Growth and Ectopic Lignin Deposition in the lig Mutant.
Supplemental Figure 3. Immunoblot Analysis with the Anti-PYK10 Antibody.
Supplemental Table 1. Segregation of the Adventitious Root Phenotype in the Progeny of the Mutant Line Backcrossed with the Wild-Type Ler.
Supplemental Table 2. Primers Used in the RT-PCR Analysis.
Supplementary Material
Acknowledgments
We thank Jerzy Zoń (Wroclaw University of Technology), Tsuyoshi Nakagawa (Shimane University), Kazuko Nishimura (Kyoto University), and Kazuhito Fujiyama (Osaka University) for providing AIP, pGWB vectors, anti-PYK10 antibody, and anti-PDI antibody, respectively. We thank Masahiro Inouhe (Ehime University) for helpful discussion and use of facilities. We also thank Aya Yashiki, Hiroko Kimura, and Mika Tanimoto (Ehime University) for research assistance. This work was supported in part by Grants-in-Aid from the Ministry of Sports, Culture, Science, and Technology of Japan (RFTF00L01605 and 19060001).
AUTHOR CONTRIBUTIONS
M.S. and Y.S. designed the project. M.N. carried out most of the experiments. M.S. isolated the lig mutant and performed map-based cloning. J.D. contributed to initial characterization of lig and chromosome mapping. H.U. and T.G. contributed to the phenotypic analysis and RT-PCR analysis, respectively. M.N., M.S., and Y.S. wrote the article.
Glossary
- GNA
glucosamine-6-phosphate N-acetyltransferase
- UDP-GlcNAc
UDP–N-acetylglucosamine
- GlcNAc
N-acetylglucosamine
- GalNAc
N-acetylgalactosamine
- UDP-GalNAc
UDP–N- acetylgalactosamine
- AIP
2-aminoindan-2-phosphonic acid
- CA
coniferyl alcohol
- H2O2
hydrogen peroxide
- DAB
3,3′-diaminobenzidine
- ConA
concanavalin A
- PDI
protein disulfide isomerase
- ER
endoplasmic reticulum
- Endo H
endoglycosidase H
- PNGase F
peptide:N-glycosidase F
- UPR
unfolded protein response
- GPI
glycosylphosphatidylinositol
- Ler
Landsberg erecta
- Col
Columbia
- GMA
germination medium solidified with agar
- PVDF
polyvinylidene difluoride
References
- Allard S.T.M., Giraud M.-F., Naismith J.H. (2001). Epimerases: Structure, function and mechanism. Cell. Mol. Life Sci. 58: 1650–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff V., Cookson S.J., Wu S., Scheible W.-R. (2009). Thaxtomin A affects CESA-complex density, expression of cell wall genes, cell wall composition, and causes ectopic lignification in Arabidopsis thaliana seedlings. J. Exp. Bot. 60: 955–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W., Ralph J., Baucher M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Caño-Delgado A.I., Metzlaff K., Bevan M.W. (2000). The eli1 mutation reveals a link between cell expansion and secondary cell wall formation in Arabidopsis thaliana. Development 127: 3395–3405 [DOI] [PubMed] [Google Scholar]
- Caño-Delgado A.I., Penfield S., Smith C., Catley M., Bevan M. (2003). Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J. 34: 351–362 [DOI] [PubMed] [Google Scholar]
- Carpita N.C., Delmer D.P. (1981). Concentration and metabolic turnover of UDP-glucose in developing cotton fibers. J. Biol. Chem. 256: 308–315 [PubMed] [Google Scholar]
- Cheong Y.H., Chang H.-S., Gupta R., Wang X., Zhu T., Luan S. (2002). Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol. 129: 661–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demura T., et al. (2002). Visualization by comprehensive microarray analysis of gene expression programs during transdifferentiation of mesophyll cells into xylem cells. Proc. Natl. Acad. Sci. USA 99: 15794–15799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denness L., McKenna J.F., Segonzac C., Wormit A., Madhou P., Bennett M., Mansfield J., Zipfel C., Hamann T. (2011). Cell wall damage-induced lignin biosynthesis is regulated by a reactive oxygen species- and jasmonic acid-dependent process in Arabidopsis. Plant Physiol. 156: 1364–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamborg O.L., Miller R.A., Ojima K. (1968). Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell Res. 50: 151–158 [DOI] [PubMed] [Google Scholar]
- Giacomin L.T., Szalay A.A. (1996). Expression of a PAL1 promoter luciferase gene fusion in Arabidopsis thaliana in response to infection by phytopathogenic bacteria. Plant Sci. 116: 59–72 [Google Scholar]
- Gillmor C.S., Lukowitz W., Brininstool G., Sedbrook J.C., Hamann T., Poindexter P., Somerville C. (2005). Glycosylphosphatidylinositol-anchored proteins are required for cell wall synthesis and morphogenesis in Arabidopsis. Plant Cell 17: 1128–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hématy K., Sado P.-E., Van Tuinen A., Rochange S., Desnos T., Balzergue S., Pelletier S., Renou J.-P., Höfte H. (2007). A receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis. Curr. Biol. 17: 922–931 [DOI] [PubMed] [Google Scholar]
- Hinderlich S., Berger M., Schwarzkopf M., Effertz K., Reutter W. (2000). Molecular cloning and characterization of murine and human N-acetylglucosamine kinase. Eur. J. Biochem. 267: 3301–3308 [DOI] [PubMed] [Google Scholar]
- Hinderlich S., Nöhring S., Weise C., Franke P., Stäsche R., Reutter W. (1998). Purification and characterization of N-acetylglucosamine kinase from rat liver—Comparison with UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase. Eur. J. Biochem. 252: 133–139 [DOI] [PubMed] [Google Scholar]
- Hori H., Elbein A.D. (1981). Tunicamycin inhibits protein glycosylation in suspension cultured soybean cells. Plant Physiol. 67: 882–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata Y., Koizumi N. (2005). An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proc. Natl. Acad. Sci. USA 102: 5280–5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspers P., Kangasjärvi J. (2010). Reactive oxygen species in abiotic stress signaling. Physiol. Plant. 138: 405–413 [DOI] [PubMed] [Google Scholar]
- Jiang H., Wang S., Dang L., Wang S., Chen H., Wu Y., Jiang X., Wu P. (2005). A novel short-root gene encodes a glucosamine-6-phosphate acetyltransferase required for maintaining normal root cell shape in rice. Plant Physiol. 138: 232–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Cominelli E., Bailey P., Parr A., Mehrtens F., Jones J., Tonelli C., Weisshaar B., Martin C. (2000). Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 19: 6150–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiura H., Seki T., Fujiyama K. (2010). Arabidopsis thaliana ALG3 mutant synthesizes immature oligosaccharides in the ER and accumulates unique N-glycans. Glycobiology 20: 736–751 [DOI] [PubMed] [Google Scholar]
- Karve A., Rauh B.L., Xia X., Kandasamy M., Meagher R.B., Sheen J., Moore B.D. (2008). Expression and evolutionary features of the hexokinase gene family in Arabidopsis. Planta 228: 411–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi M., Sugiyama M. (2003). Genetic analysis of adventitious root formation with a novel series of temperature-sensitive mutants of Arabidopsis thaliana. Development 130: 5637–5647 [DOI] [PubMed] [Google Scholar]
- Kubo M., Udagawa M., Nishikubo N., Horiguchi G., Yamaguchi M., Ito J., Mimura T., Fukuda H., Demura T. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19: 1855–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauvergeat V., Lacomme C., Lacombe E., Lasserre E., Roby D., Grima-Pettenati J. (2001). Two cinnamoyl-CoA reductase (CCR) genes from Arabidopsis thaliana are differentially expressed during development and in response to infection with pathogenic bacteria. Phytochemistry 57: 1187–1195 [DOI] [PubMed] [Google Scholar]
- Liu J.X., Howell S.H. (2010). Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell 22: 2930–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W., Nickle T.C., Meinke D.W., Last R.L., Conklin P.L., Somerville C.R. (2001). Arabidopsis cyt1 mutants are deficient in a mannose-1-phosphate guanylyltransferase and point to a requirement of N-linked glycosylation for cellulose biosynthesis. Proc. Natl. Acad. Sci. USA 98: 2262–2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalingam R., Fedoroff N. (2003). Stress response, cell death and signalling: The many faces of reactive oxygen species. Physiol. Plant. 119: 56–68 [Google Scholar]
- Martínez I.M., Chrispeels M.J. (2003). Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 15: 561–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima R., Kondo M., Nishimura M., Hara-Nishimura I. (2003). A novel ER-derived compartment, the ER body, selectively accumulates a β-glucosidase with an ER-retention signal in Arabidopsis. Plant J. 33: 493–502 [DOI] [PubMed] [Google Scholar]
- Milewski S., Gabriel I., Olchowy J. (2006). Enzymes of UDP-GlcNAc biosynthesis in yeast. Yeast 23: 1–14 [DOI] [PubMed] [Google Scholar]
- Milioni D., Sado P.-E., Stacey N.J., Roberts K., McCann M.C. (2002). Early gene expression associated with the commitment and differentiation of a plant tracheary element is revealed by cDNA-amplified fragment length polymorphism analysis. Plant Cell 14: 2813–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mio T., Kokado M., Arisawa M., Yamada-Okabe H. (2000). Reduced virulence of Candida albicans mutants lacking the GNA1 gene encoding glucosamine-6-phosphate acetyltransferase. Microbiology 146: 1753–1758 [DOI] [PubMed] [Google Scholar]
- Mio T., Yamada-Okabe T., Arisawa M., Yamada-Okabe H. (1999). Saccharomyces cerevisiae GNA1, an essential gene encoding a novel acetyltransferase involved in UDP-N-acetylglucosamine synthesis. J. Biol. Chem. 274: 424–429 [DOI] [PubMed] [Google Scholar]
- Morrison I.M. (1972). A semi-micro method for the determination of lignin and its use in predicting the digestibility of forage crops. J. Sci. Food Agric. 23: 455–463 [DOI] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15: 473–497 [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Nakajima K., Kitazume S., Angata T., Fujinawa R., Ohtsubo K., Miyoshi E., Taniguchi N. (2010). Simultaneous determination of nucleotide sugars with ion-pair reversed-phase HPLC. Glycobiology 20: 865–871 [DOI] [PubMed] [Google Scholar]
- Nakano J., Meshitsuka G. (1992). The detection of lignin. In Methods in Lignin Chemistry, S.Y. Lin and C.W. Dence, eds (New York: Springer-Verlag), pp. 23–32.
- Pattison R.J., Amtmann A. (2009). N-glycan production in the endoplasmic reticulum of plants. Trends Plant Sci. 14: 92–99 [DOI] [PubMed] [Google Scholar]
- Patzlaff A., McInnis S., Courtenay A., Surman C., Newman L.J., Smith C., Bevan M.W., Mansfield S., Whetten R.W., Sederoff R.R., Campbell M.M. (2003). Characterisation of a pine MYB that regulates lignification. Plant J. 36: 743–754 [DOI] [PubMed] [Google Scholar]
- Pauwels L., Morreel K., De Witte E., Lammertyn F., Van Montagu M., Boerjan W., Inzé D., Goossens A. (2008). Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc. Natl. Acad. Sci. USA 105: 1380–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama M. (2003). Isolation and initial characterization of temperature-sensitive mutants of Arabidopsis thaliana that are impaired in root redifferentiation. Plant Cell Physiol. 44: 588–596 [DOI] [PubMed] [Google Scholar]
- Thordal-Christensen H., Zhang Z., Wei Y., Collinge D.B. (1997). Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 11: 1187–1194 [Google Scholar]
- Torres M.A. (2010). ROS in biotic interactions. Physiol. Plant. 138: 414–429 [DOI] [PubMed] [Google Scholar]
- Tronchet M., Balagué C., Kroj T., Jouanin L., Roby D. (2010). Cinnamyl alcohol dehydrogenases-C and D, key enzymes in lignin biosynthesis, play an essential role in disease resistance in Arabidopsis. Mol. Plant Pathol. 11: 83–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urade R. (2007). Cellular response to unfolded proteins in the endoplasmic reticulum of plants. FEBS J. 274: 1152–1171 [DOI] [PubMed] [Google Scholar]
- Verwoerd T.C., Dekker B.M.M., Hoekema A. (1989). A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 17: 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada-Okabe T., Sakamori Y., Mio T., Yamada-Okabe H. (2001). Identification and characterization of the genes for N-acetylglucosamine kinase and N-acetylglucosamine-phosphate deacetylase in the pathogenic fungus Candida albicans. Eur. J. Biochem. 268: 2498–2505 [DOI] [PubMed] [Google Scholar]
- Yamaguchi M., Demura T. (2010). Transcriptional regulation of secondary wall formation controlled by NAC domain proteins. Plant Biotechnol. 27: 237–242 [Google Scholar]
- Yasutani I., Ozawa S., Nishida T., Sugiyama M., Komamine A. (1994). Isolation of temperature-sensitive mutants of Arabidopsis thaliana that are defective in the redifferentiation of shoots. Plant Physiol. 105: 815–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Dixon R.A. (2011). Transcriptional networks for lignin biosynthesis: More complex than we thought? Trends Plant Sci. 16: 227–233 [DOI] [PubMed] [Google Scholar]
- Zhou J., Lee C., Zhong R., Ye Z.-H. (2009). MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21: 248–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoń J., Amrhein N. (1992). Inhibitors of phenylalanine ammonia-lyase: 2-aminoindan-2-phosphonic acid and related compounds. Liebigs Ann. Chem. 1992: 625–628 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.