This study reveals that, similar to effectors of phytopathogenic bacteria, recognition of filamentous pathogen effectors can be mediated via a host protein that interacts with both the effector and the NB-LRR immunoreceptor.
Abstract
Plant pathogens secrete effector proteins to modulate plant immunity and promote host colonization. Plant nucleotide binding leucine-rich repeat (NB-LRR) immunoreceptors recognize specific pathogen effectors directly or indirectly. Little is known about how NB-LRR proteins recognize effectors of filamentous plant pathogens, such as Phytophthora infestans. AVR2 belongs to a family of 13 sequence-divergent P. infestans RXLR effectors that are differentially recognized by members of the R2 NB-LRR family in Solanum demissum. We report that the putative plant phosphatase BSU-LIKE PROTEIN1 (BSL1) is required for R2-mediated perception of AVR2 and resistance to P. infestans. AVR2 associates with BSL1 and mediates the interaction of BSL1 with R2 in planta, possibly through the formation of a ternary complex. Strains of P. infestans that are virulent on R2 potatoes express an unrecognized form, Avr2-like (referred to as A2l). A2L can still interact with BSL1 but does not promote the association of BSL1 with R2. Our findings show that recognition of the P. infestans AVR2 effector by the NB-LRR protein R2 requires the putative phosphatase BSL1. This reveals that, similar to effectors of phytopathogenic bacteria, recognition of filamentous pathogen effectors can be mediated via a host protein that interacts with both the effector and the NB-LRR immunoreceptor.
INTRODUCTION
A central question in plant pathology is how pathogens are recognized by their hosts and how the subsequent activation of immunity may be evaded or suppressed by pathogens to promote disease. Plants recognize pathogens and mount active defenses to attenuate pathogen progression. First, plant basal defense responses are activated by the perception of pathogen-associated molecular patterns by pattern recognition receptors at the plant cell surface, resulting in pathogen-associated molecular pattern–triggered immunity (Jones and Dangl, 2006). However, pathogens secrete an array of proteins, known as effectors, to suppress these basal defense mechanisms. A second wave of defense acts largely within the cell through the genetically determined recognition of a subset of pathogen effectors, known as avirulence (AVR) proteins. This immune response is mediated by a sophisticated surveillance mechanism that consists of highly specific and structurally conserved plant disease resistance (R) proteins (Jones and Dangl, 2006; van der Hoorn and Kamoun, 2008; Elmore et al., 2011). Each R protein recognizes one or a few corresponding AVR effectors, leading to the activation of effector-triggered immunity (ETI) that often results in a rapid, localized host cell death, termed the hypersensitive response (HR). However, pathogens can also secrete effectors to suppress ETI and promote disease progression (Jones and Dangl, 2006). The fundamental questions in this coevolutionary arms race remain: What are the host targets of effector proteins, why are these proteins targeted, and how do plants sense pathogen effectors? This is particularly relevant for oomycetes and fungi, the filamentous pathogens that cause the most destructive plant diseases. Little is known about the targets of filamentous pathogen effectors and the mechanisms by which these effectors modulate immunity.
Over the past two decades, significant numbers of R genes have been identified. The largest family functions as immune receptors and encode nucleotide binding leucine-rich repeat (NB-LRR) proteins that mediate recognition of pathogen-derived AVR effectors (Eitas and Dangl, 2010; Elmore et al., 2011). Recognition of AVR molecules can be either direct, through interaction with their cognate R proteins, or indirect, through the perception of modifications in host proteins targeted by the AVR effector (Chisholm et al., 2006; Jones and Dangl, 2006; van der Hoorn and Kamoun, 2008; Elmore et al., 2011). In filamentous pathogens, a few instances of direct interaction between an NB-LRR immune receptor and an effector have been confirmed experimentally (Jia et al., 2000; Dodds et al., 2006; Krasileva et al., 2010). Among the oomycetes, one example is the recognition of Hyaloperonospora arabidopsidis ATR1 by Arabidopsis thaliana RPP1, where association of ATR1-RPP1 is dependent on interaction with the LRR domain of the NB-LRR immune receptor (Krasileva et al., 2010). More recently, evidence of direct interaction between Phytophthora infestans effector ipiO (AVRblb1) and the coiled-coil domain of RB (Rpi-blb1) has been reported (Chen et al., 2012). In bacterial systems, effectors are often recognized indirectly through the perception of modifications in an accessory host protein (Elmore et al., 2011). For instance, Pseudomonas syringae AVR proteins AvrB and AvrRpm1 are indirectly recognized by the Arabidopsis NB-LRR protein RPM1 that detects pathogen proteins that mediate phosphorylation of the host target protein RIN4 (Liu et al., 2011). Remarkably, so far, indirect recognition of fungal and oomycete AVR effectors by NB-LRR proteins has not been described even though several cases of direct recognition have been reported (Jia et al., 2000; Dodds et al., 2006; Krasileva et al., 2010).
The study of indirect recognition in well-characterized pathogen systems has led to the development of two models known as the guard and decoy hypotheses. In the guard model, the R protein monitors the status of a host protein, the guardee, which is also required for the virulence function of the effector in the absence of the R protein (van der Hoorn and Kamoun, 2008). By contrast, in the decoy model, the targeted protein acts mainly in effector recognition, mimicking the operative target to activate ETI (van der Hoorn and Kamoun, 2008). The extent to which these models apply to the perception of filamentous pathogen AVR effectors by NB-LRR receptors is unknown. A crucial step is to uncover the host targets of the ever-increasing list of characterized filamentous pathogen AVR effectors. So far, only limited numbers of host targets of these effectors have been described and none have been implicated in perception by NB-LRR proteins (Oliva et al., 2010).
Biotrophic fungi and oomycetes secrete effectors from highly specialized structures, known as haustoria, which form at pathogen-directed invaginations of the host plasma membrane (Panstruga and Dodds, 2009). Once secreted into the extrahaustorial matrix, some effectors translocate into the host cell where they modulate plant immunity (Stassen and Van den Ackerveken, 2011). In oomycetes, such as Phytophthora and downy mildews, the largest class of host-translocated effectors are the RXLR-type proteins, which include all known AVR effectors (Hein et al., 2009; Vleeshouwers et al., 2011). RXLR effectors have a modular architecture with the N-terminal signal peptide and RXLR domain involved in secretion and host translocation and the C-terminal domain carrying the biochemical effector activity (Win et al., 2007). Evolutionary analyses demonstrate that positive selection has acted mainly on the effector domain, probably as a consequence of coevolution with plant targets and/or resistance genes (Win et al., 2007). All Phytophthora genomes examined to date carry a large repertoire of RXLR effector genes. P. infestans, the agent of the economically important potato late blight disease, harbors ∼550 RXLR effector genes, of which seven out of 127 families have been assigned an AVR activity (Haas et al., 2009; Vleeshouwers et al., 2011). These effectors provide a wealth of molecular probes that can be used to unravel novel components of the immune system.
Among the P. infestans AVR effectors, the recently described AVR2 is a canonical RXLR effector protein of 116 amino acids that elicits HR in the presence of the wild potato (Solanum demissum) NB-LRR protein R2 (Gilroy et al., 2011). R2 belongs to a highly diverse gene family located at a major late blight resistance locus on chromosome IV of potato (Solanum tuberosum; Lokossou et al., 2009; Vleeshouwers et al., 2011). Several R2 alleles and orthologs from six different Solanum species have been shown to recognize AVR2 and confer resistance to P. infestans (Champouret, 2010). In P. infestans strain T30-4, Avr2 belongs to a family of 18 genes encoding 13 sequence-divergent members that share similarity throughout the C-terminal effector domains and range from 94 to 118 amino acids (Haas et al., 2009; Champouret, 2010; Vleeshouwers et al., 2011). Several of the AVR2 homologs are specifically recognized inside plant cells by R2 (Champouret, 2010), although genetic analyses of a cross between a virulent and avirulent race has revealed one gene, Avr2 (also known as PITG_22870), as the major contributor to the avirulence phenotype (van der Lee et al., 2001; Gilroy et al., 2011). Strains of P. infestans that are virulent on R2 potatoes either do not express Avr2 and/or express a distinct variant, Avr2-like (referred to here as A2l to avoid confusion with other homologs), that evades perception by R2 (Gilroy et al., 2011). The A2l form was initially isolated from the aggressive clonal lineage genotype 13_A2 (blue13) that has been responsible for destructive potato late blight epidemics in Great Britain since 2007 (Fry et al., 2008; Gilroy et al., 2011). The polymorphic nature of the AVR2 and R2 families, and the occurrence of the latter in Central Mexico, a center of P. infestans diversity, is indicative of coevolutionary balancing selection between P. infestans and Solanum hosts (Champouret, 2010; Vleeshouwers et al., 2011). This system provides a unique opportunity to study differential host recognition and evolution of virulence involving a filamentous pathogen family of host-translocated effectors.
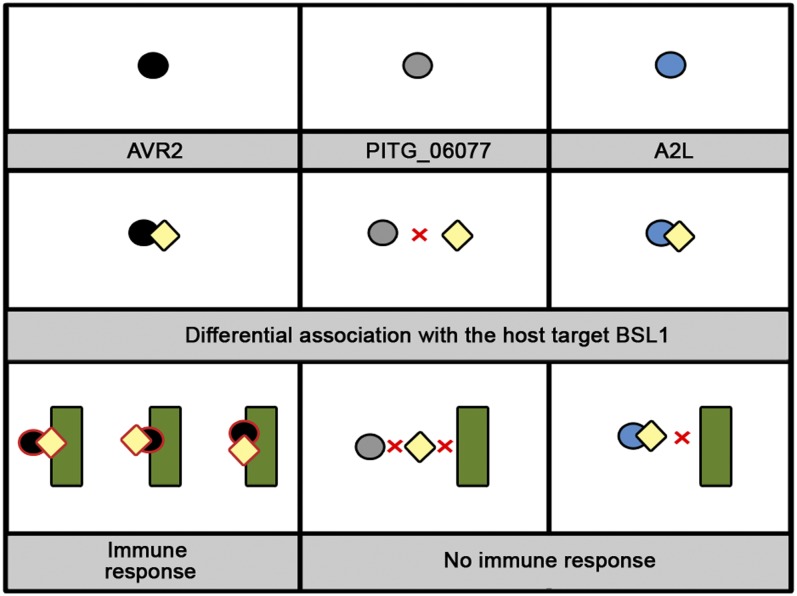
In this study, we used a recently developed in planta coimmunoprecipitation (co-IP) protocol (Win et al., 2011) to identify host targets of AVR2. We discovered that AVR2 associates in planta with BSL1, a kelch repeat domain–containing putative phosphatase. The AVR2–BSL1 interaction was independently confirmed using yeast two-hybrid (Y2H) assays. Not all of the 13 AVR2 family members interacted with BSL1. Remarkably, the five AVR2 homologs that interacted with BSL1 are the only family members that are perceived by R2, pointing to a possible role of BSL1 in R2-mediated recognition. Virus-induced gene silencing (VIGS) experiments demonstrated that BSL1 is required for R2-mediated recognition of AVR2 and resistance to P. infestans. We also discovered that BSL1 associates with R2, but only in the presence of AVR2, possibly through the formation of a ternary complex. Interestingly, A2L, which evades recognition by R2, nevertheless interacts with BSL1. However, unlike AVR2, A2L did not promote the interaction of BSL1 with R2. Our findings show that recognition of the P. infestans AVR2 effector by the NB-LRR protein R2 requires the host protein BSL1. This reveals that, similar to effectors of phytopathogenic bacteria, recognition of filamentous pathogen effectors can be mediated through a host protein that interacts with both the effector and the NB-LRR immunoreceptor (Elmore et al., 2011).
RESULTS
AVR2 Interacts with the Putative Phosphatase BSL1
To identify host targets of AVR2, we used in planta co-IP followed by liquid chromatography–tandem mass spectrometry (LC-MS/MS) (Win et al., 2011). The C-terminal effector domains of each of the 13 AVR2 family members were fused to the FLAG epitope tag at the N terminus in place of the signal peptide and RXLR domain (see Supplemental Figure 1A online). These constructs were expressed in Nicotiana benthamiana leaves using Agrobacterium tumefaciens–mediated transient transformation (agroinfiltration) before being used in co-IPs. Three plant proteins, peroxisomal enzyme catalase 1, thioredoxin peroxidase, and a phosphatase of the BSL family (BSU-LIKE PROTEIN1 [BSL1]), specifically associated with one or more of the eight AVR2 homologs that formed protein complexes detectable by LC-MS/MS (see Supplemental Table 1 and Supplemental Data Set 1 online). To confirm and further study the associations, the three putative target proteins were cloned and fused to green fluorescent protein (GFP). Using co-IPs, we validated the specific in planta association between AVR2 and tomato (Solanum lycopersicum) BSL1 (Sl-BSL1) but not the other two plant proteins (Figure 1A). To determine the extent to which these findings apply to other AVR2 homologs, we also examined PEXRD11, an RXLR effector that was previously shown to activate R2 (Champouret, 2010). PEXRD11 also specifically immunoprecipitated with Sl-BSL1 but not the other two plant proteins (see Supplemental Figure 2 online). Sequence analyses confirmed that Solanaceous (potato and tomato) BSL1s are the putative orthologs of Arabidopsis BSL1, a protein phosphatase involved in brassinosteroid (BR) signal transduction (see Supplemental Figures 3 and 4 and Supplemental Data Set 2 online) (Mora-García et al., 2004; Kim et al., 2009).
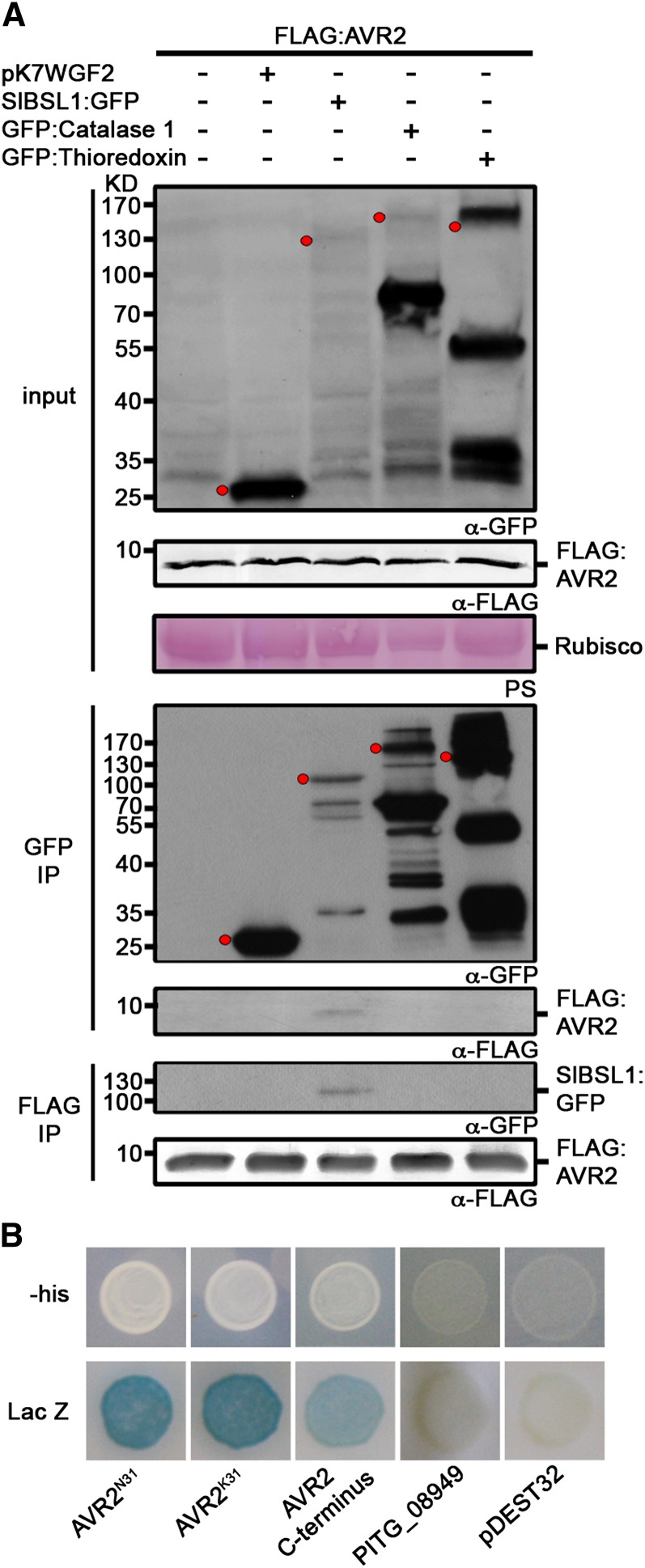
Figure 1.
P. infestans AVR2 Specifically Associates with BSL1.
(A) Immunoblots showing AVR2 specifically coimmunoprecipitates with tomato BSL1 (Sl-BSL1) in planta. FLAG:AVR2 was transiently expressed alone, with the empty vector pK7WGF2 or with GFP-tagged putative target proteins in N. benthamiana. Immunoprecipitates (IP) obtained with anti-FLAG or anti-GFP antiserum and total protein extracts were immunoblotted with appropriate antisera. The expected sizes of the GFP fusion proteins are indicated by red dots in the crude extracts and GFP co-IP probed with anti-GFP antibody. PS, Ponceau stain; Rubisco, ribulose-1,5-bisphosphate carboxylase/oxygenase.
(B) Y2H analysis illustrating that the two allelic variants of AVR2, P. infestans AVR2K31 and AVR2N31, and the C-terminal effector domain of AVR2 (amino acids 66 to 116) interact with potato BSL1 (St-BSL1) in vivo. Both LacZ (blue) and His3 (providing growth on medium lacking His [-his]) reporter genes were activated. Empty vector (pDEST32) was used as a negative control. The RXLR effector PITG_08949, which is closely related to AVR2 across the N-terminal translocation domain but which differs across the C-terminal effector domain due to a likely recombination event (Gilroy et al., 2011), did not interact with St-BSL1 using Y2H analysis.
We also used Y2H analysis to confirm that the potato ortholog of Arabidopsis BSL1, St-BSL1, interacts with AVR2 (Figure 1B). Gilroy et al. (2011) reported that avirulent isolates of P. infestans possess two alleles of AVR2, AVR2K31 and AVR2N31, that differ in a single amino acid in the N terminus at position 31 prior to the effector domain. In Y2H assays, both AVR2 alleles interacted with potato St-BSL1 (Figure 1B). The Y2H assays also confirmed that the C-terminal effector domain of AVR2 (amino acids 66 to 116) is sufficient for interaction with BSL1 (Figure 1B).
In summary, the co-IP and Y2H experiments provided independent evidence that the RXLR effector AVR2 of P. infestans interacts with the plant protein phosphatase BSL1. AVR2 interacts with BSL1 orthologs from the two major host species of P. infestans, tomato, and potato.
AVR2 Interacts with the Putative Phosphatase Domain of BSL1
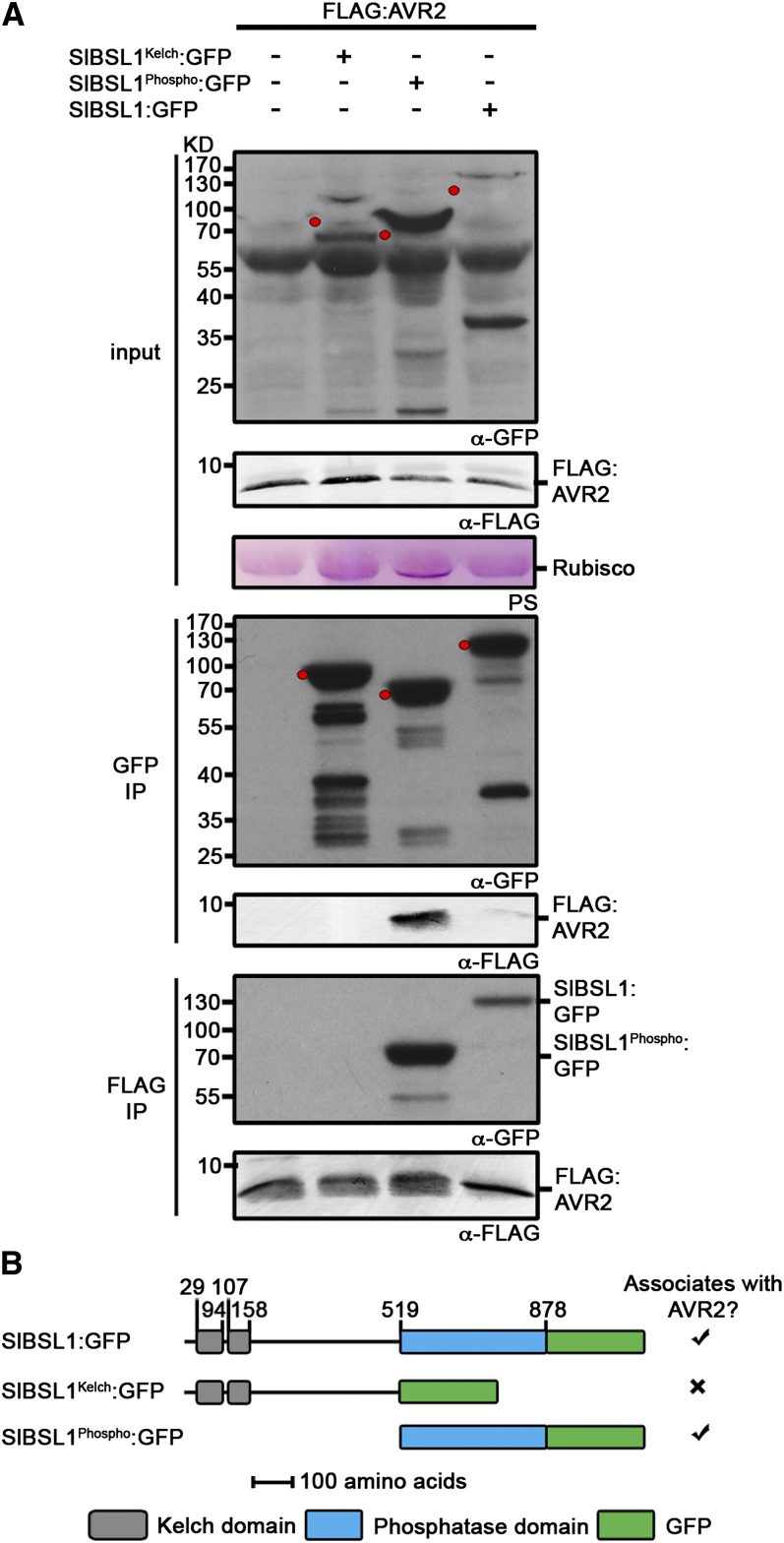
To determine which part of BSL1 interacts with AVR2, we cloned the region upstream of the putative phosphatase domain (amino acids 1 to 519) and the putative phosphatase domain (amino acids 520 to 878) and fused these fragments to the GFP at the C terminus. Using co-IPs, we demonstrated an association between the putative phosphatase domain of Sl-BSL1 and AVR2 in planta (Figure 2). Y2H analysis also illustrated that the putative phosphatase domain of St-BSL1 was sufficient for inducing an interaction with AVR2 in vivo (see Supplemental Figure 5 online).
Figure 2.
Immunoblots Showing AVR2 Specifically Associates with the Putative Phosphatase Domain of Sl-BSL1 in Planta.
(A) FLAG:AVR2 was transiently expressed alone, with SlBSL1Kelch:GFP, SlBSL1Phospho:GFP, or SlBSL1:GFP in N. benthamiana. Immunoprecipitates (IP) obtained with anti-FLAG or anti-GFP antiserum and total protein extracts were immunoblotted with appropriate antisera. The expected sizes of the GFP fusion proteins are indicated by red dots in the crude extracts and GFP co-IP probed with anti-GFP antibody. PS, Ponceau stain; Rubisco, ribulose-1,5-bisphosphate carboxylase/oxygenase.
(B) Schematic illustrating the regions of Sl-BSL1 used to assess interaction with AVR2. Numbers indicate position of the amino acid residues.
AVR2 Localizes around P. infestans Haustoria
To investigate the spatial distribution of AVR2, we expressed a GFP:AVR2 fusion protein (see Supplemental Figure 1B online) in N. benthamiana and assessed the intracellular localization by confocal microscopy. GFP:AVR2 accumulated at the cell periphery and, to a lesser extent, within the cytoplasm and nucleus (see Supplemental Figure 6A online). To investigate this further, we coexpressed GFP:AVR2 with a Remorin red fluorescent protein (RFP) fusion. Remorin is a plant protein that accumulates in lipid rafts of the plasma membrane (Raffaele et al., 2009). GFP:AVR2 and RFP:StRemorin largely colocalized at the plasma membrane in planta (see Supplemental Figure 7 online). However, the AVR2 GFP signal was also detected within the nucleus and at low levels within the cytoplasm, which may be due to partial cleavage of the GFP:AVR2 fusion protein (see Supplemental Figure 6B online). Similar results were obtained from the localization of the AVR2 homolog PEXRD11 in planta (see Supplemental Figure 7 online).
To investigate the intracellular localization of AVR2 during infection, we inoculated N. benthamiana leaves expressing GFP:AVR2 with the P. infestans strain 88069td that expresses an RFP marker (Bozkurt et al., 2011; Chaparro-Garcia et al., 2011). Infected plant cells were identified under bright-field microscopy by the presence of P. infestans haustoria. Remarkably, we observed that the GFP:AVR2 signal was altered in infected cells with preferential accumulation around haustoria (perihaustorial localization) (Figure 3A).
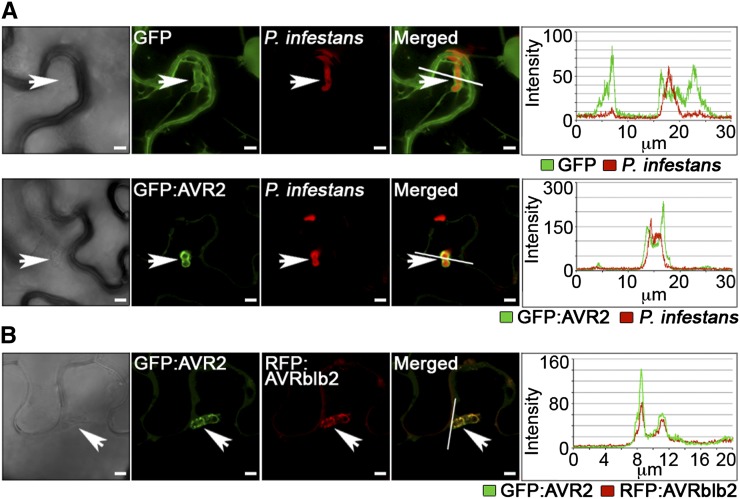
Figure 3.
P. infestans AVR2 Accumulates around Haustoria and Colocalizes with the Perihaustorial-Localized Effector AVRblb2 in Planta.
(A) P. infestans (red)–infected N. benthamiana cells preferentially accumulated transiently expressed GFP:AVR2 (green) around haustoria (arrowheads) when compared with the localization of GFP alone.
(B) P. infestans–infected cells accumulated GFP:AVR2 and RFP:AVRblb2 at sites of haustorial penetration (arrowheads), illustrated by overlapping peaks of fluorescence intensity.
Pictures were taken at 4 DAI. Bars + 5 µm.
To investigate further the localization of AVR2 during infection, we coexpressed AVR2 with the P. infestans plasma membrane–associated perihaustorial effector AVRblb2 (Bozkurt et al., 2011). GFP:AVR2 and RFP:AVRblb2 fusion proteins were coexpressed in N. benthamiana and their spatial distribution assessed by confocal microscopy. The GFP:AVR2 and RFP:AVRblb2 fluorescent signals overlapped in planta, supporting that AVR2 localizes, at least partially, to the plasma membrane (see Supplemental Figure 8 online). When leaves expressing GFP:AVR2 and RFP:AVRblb2 were inoculated with P. infestans, the two signals preferentially coaccumulated around haustoria, further supporting perihaustorial localization of AVR2 (Figure 3B).
BSL1 Colocalizes with AVR2 in Planta and during Infection
To examine the spatial distribution of BSL1 in planta, we expressed a SlBSL1:GFP fusion protein in N. benthamiana. The fluorescent signal for SlBSL1:GFP was excluded from the nucleus and detected at the cell periphery and within the cytoplasm (Figure 4A). To determine whether Sl-BSL1 localized at the plasma membrane, we coexpressed SlBSL1:GFP with RFP:StRemorin in N. benthamiana. The two fluorescent signals largely overlapped, indicating partial plasma membrane association of Sl-BSL1, as has been described previously for At-BSL1 (Benschop et al., 2007; Kim et al., 2009). However, following salt-induced plasmolysis, the signals were quite distinct, indicating that Sl-BSL1 only partially localized at the plasma membrane in planta (see Supplemental Figure 7 online).
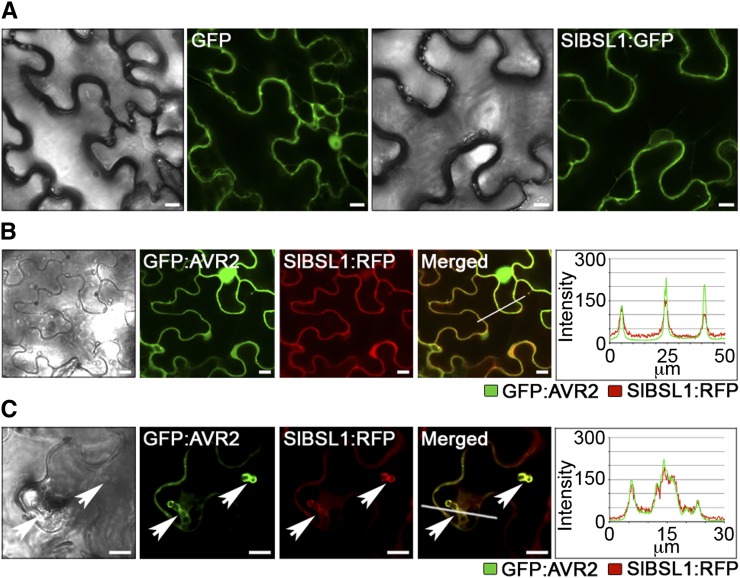
Figure 4.
Sl-BSL1 and P. infestans AVR2 Colocalize and Accumulate around Haustoria in Planta.
(A) Transient expression of SlBSL1:RFP in N. benthamiana reveals that Sl-BSL1 is excluded from the nucleus and localizes at the cell periphery and within the cytoplasm.
(B) Transient expression of GFP:AVR2 and SlBSL1:RFP fusion proteins revealed similar patterns of localization for the two markers, accumulating at the cell periphery, as shown by overlapping peaks in fluorescence intensity. GFP:AVR2 was also detected in the nucleus.
(C) GFP:AVR2 and SlBSL1:RFP localize strongly at sites of P. infestans haustorial penetration in N. benthamiana.
Pictures were taken at 3 DAI. Bars + 10 µm.
To determine whether AVR2 and Sl-BSL1 colocalize in planta, we coexpressed GFP:AVR2 and SlBSL1:RFP fusion proteins in N. benthamiana. We observed a clear overlap in the two fluorescent protein signals in planta (Figure 4B). When N. benthamiana leaves expressing GFP:AVR2 and SlBSL1:RFP were inoculated with P. infestans, the two fluorescent signals colocalized at the sites of haustorial penetration indicating perihaustorial localization of both proteins (Figure 4C).
Similar results were obtained from the localization of the AVR2 homolog PEXRD11 in planta. We observed perihaustorial accumulation of GFP:PEXRD11 and colocalization with AVRblb2 around haustoria in infected N. benthamiana cells (see Supplemental Figure 9 online). GFP:PEXRD11 also colocalized with SlBSL1:RFP in planta and at haustoria in cells infected with P. infestans (see Supplemental Figure 10 online).
To investigate the potential interaction of AVR2 and BSL1 at the observed sites of colocalization, we used bimolecular fluorescence complementation. The N terminus of yellow fluorescent protein (YFP; YN) was fused to full-length AVR2N31 or AVR2K31 (minus the signal peptide region) and coexpressed with the C terminus of YFP fused to St-BSL1 in N. benthamiana. Using confocal microscopy, fluorescence was detected at the cell periphery and in the cytoplasm, but excluded from within the nucleus, supporting interaction between AVR2 and BSL1 at these sites (see Supplemental Figure 11 online).
AVR2 Homologs That Are Recognized by R2 Associate with BSL1
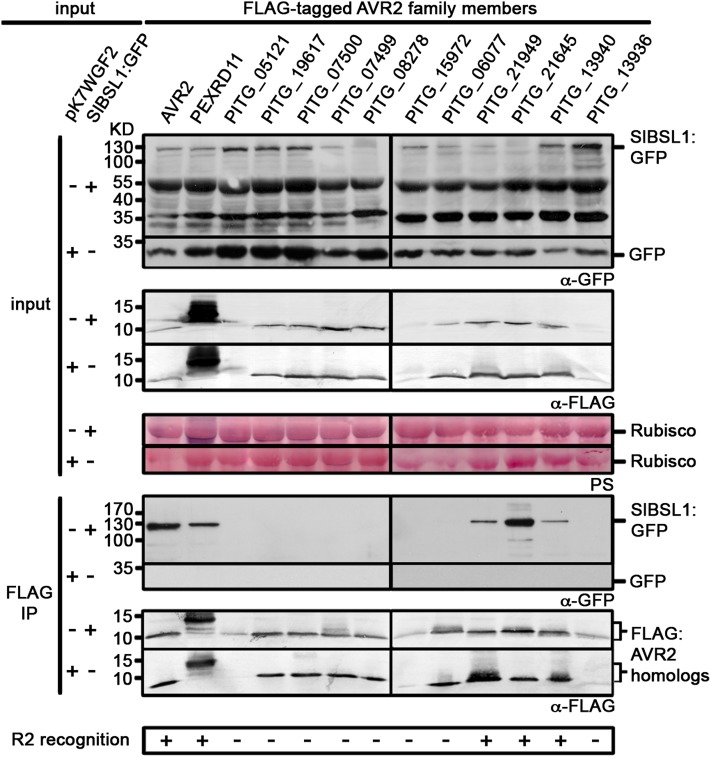
AVR2 belongs to a large family of RXLR effectors from P. infestans with highly divergent amino acid sequences (ranging from 28 to 98% in identity between family members) that are differentially recognized by wild potato R2 protein (Haas et al., 2009; Champouret, 2010). Therefore, we extended our study to determine if BSL1 could associate with other members of this family. AVR2 and PEXRD11 belong to RXLR families 7 and 32 according to the classification of Haas et al. (2009). Of these families, 13 RXLR effectors have divergent sequences yet share similarity to AVR2 in their C-terminal domains (Champouret, 2010). We coexpressed these family members as FLAG-tagged fusion proteins with SlBSL1:GFP in N. benthamiana and performed co-IPs using an anti-FLAG antibody. Co-IP analysis was inconclusive for three family members (PITG_05121, PITG_15972, and PITG_13936) as these FLAG-tagged fusion proteins displayed reduced stability in planta. Of the remaining 10 family members, we found that only five associated with Sl-BSL1 (Figure 5). Remarkably, four AVR2 homologs that associate with Sl-BSL1 (AVR2, PEXRD11, PITG_21949, and PITG_21645) are the same effectors that were previously shown to activate R2 (Champouret, 2010). A fifth AVR2 homolog, PITG_13940, also associated with Sl-BSL1 and was found to activate R2 in our assays, when infiltrated at higher concentrations than used by Champouret (2010) (see Supplemental Figure 12 online). This shows a correlation between the AVR2 family members that associate with BSL1 and their recognition by R2 and raises the possibility that BSL1 plays a role in R2 recognition of AVR2.
Figure 5.
Immunoblots Showing That Five of the 13 AVR2 Family Members Associate with Sl-BSL1 and Activate R2-Mediated Recognition.
FLAG-tagged protein fusions of AVR2 family members were transiently expressed with pK7WGF2 or SlBSL1:GFP in N. benthamiana. Immunoprecipitates obtained with anti-FLAG or anti-GFP antiserum and total protein extracts were immunoblotted with appropriate antisera. IP, immunoprecipitate; PS, Ponceau stain.
[See online article for color version of this figure.]
BSL1 Is Specifically Required for R2-Mediated Recognition and Resistance
To determine the extent to which BSL1 is involved in AVR2-R2 recognition, we performed VIGS of Nb-BSL1 (see Supplemental Figure 13 online) in N. benthamiana. To reduce the potential for off-target silencing, we selected a 290-bp fragment of the kelch repeat domain and a 300-bp region of the phosphatase domain of Nb-BSL1 and expressed them in the tobacco rattle virus (TRV) VIGS-based silencing system (see Supplemental Figure 13A online) (Ratcliff et al., 1999). While sequences encoding these regions are found in other genes, the combination of kelch-repeat and phosphatase domains is only found in members of the BSL family.
Plants expressing the TRV:NbBSL1 constructs did not show any marked phenotypic alterations and were comparable to the TRV:GFP control plants (see Supplemental Figure 13B online). Using quantitative RT-PCR, we observed a reduction in the accumulation of Nb-BSL1 transcripts only in plants infected with the two TRV:NbBSL1 constructs. To ensure that silencing of BSL1 was specific and did not affect transcript levels of related genes, we monitored the expression of BSL2, the closest paralog based on the recently released genome sequence draft of N. benthamiana (http://solgenomics.net/organism/Nicotiana_benthamiana/genome; see Methods). Unlike the observed decrease in BSL1 transcripts, TRV:NbBSL1 inoculations failed to affect the accumulation of BSL2 transcripts (see Supplemental Figures 13C and 13D online).
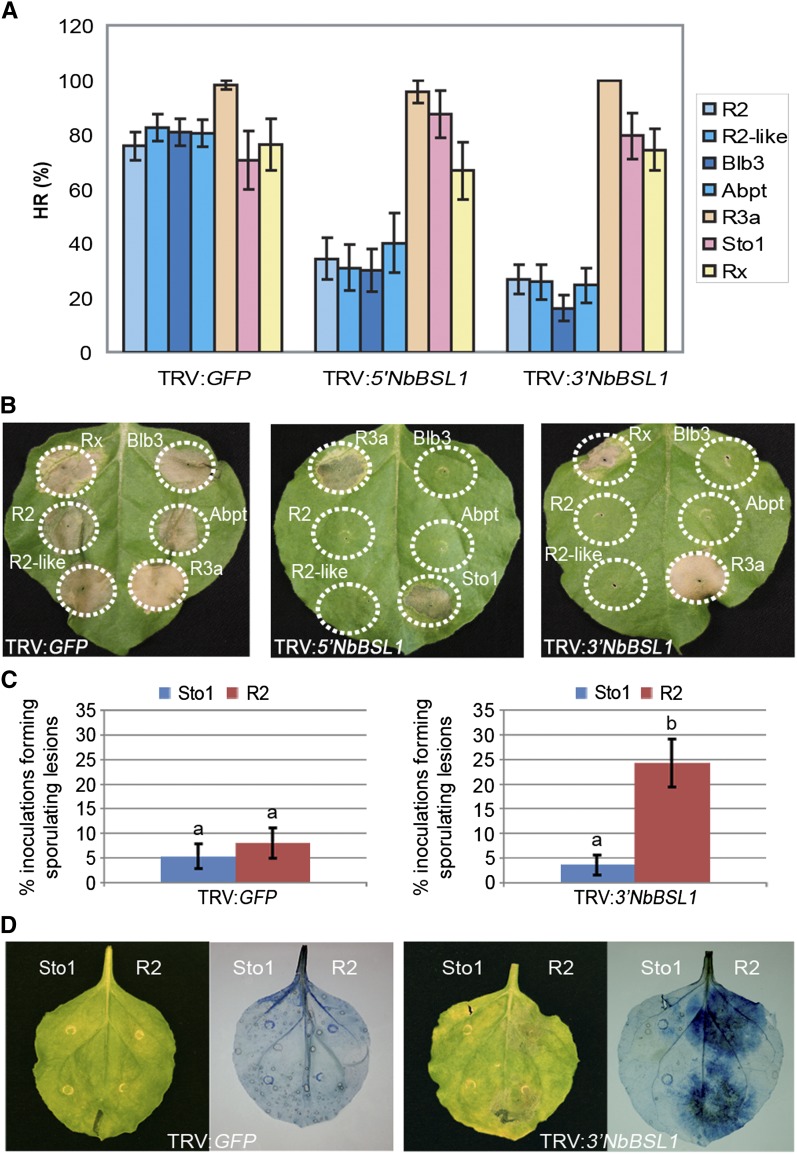
We coexpressed AVR2 with R2 and three additional R2 family members (R2-like, Rpi-abpt, and Rpi-blb3) that have the same recognition specificity (Lokossou et al., 2009) in BSL1-silenced plants and monitored development of the HR. We found that BSL1 silencing, using either TRV:NbBSL1 construct, significantly reduced recognition of AVR2 by all four R2 orthologs (Figure 6A). To ensure that the observed reduction in HR was not due to pleotropic effects from silencing of BSL1, we also confirmed that R2 was stable in BSL1-silenced plants (see Supplemental Figure 13E online).
Figure 6.
BSL1 Expression Is Required for R2-Mediated Recognition and Resistance.
(A) and (B) VIGS of Nb-BSL1 specifically perturbed R2-mediated hypersensitivity (HR). AVR-R combinations were coexpressed (only the R proteins are indicated in the figure) in N. benthamiana using agroinfiltration. HR (%) indicates the percentage of infiltration sites showing a confluent zone of cell death.
(C) and (D) VIGS of Nb-BSL1 inhibited R2-mediated resistance. Growth of P. infestans (also visualized by trypan blue staining) was significantly increased on BSL1-silenced plants transiently expressing R2 when compared with Rpi-sto1 expression. Letter “b” indicates a significant difference (P < 0.001) in the mean number of plants that show infection following R2 treatment, compared with Sto1 treatment (“a”) on TRV:3′NbBSL1 plants, using logistical regression analysis. Error bars represent ± se.
To determine whether BSL1 silencing affects other AVR–R interactions, we coexpressed three additional AVR-R combinations, AVR3a/R3a (Armstrong et al., 2005), IPI-O/Rpi-sto1 (Champouret et al., 2009), and PVX-CP/Rx (Moffett et al., 2002), in leaves of BSL1-silenced N. benthamiana. Silencing of BSL1 did not affect the HR triggered by these other AVR–R interactions (Figures 6A and 6B). Also, we did not observe cell death following expression of either effectors (AVR3a, PVX-CP, and ipiO1) or corresponding resistance proteins (R3a, Rx, and Sto1) alone in BSL1-silenced plants (see Supplemental Figure 13F online). In conclusion, BSL1 is required specifically for the recognition of AVR2 by R2 among the AVR-R pairs we examined.
To determine whether silencing of BSL1 perturbs R2-mediated resistance to P. infestans, we infected BSL1-silenced plants transiently expressing R2 with P. infestans (Figures 6C and 6D). First, we expressed R2 and, as a control, the resistance protein Rpi-sto1 in each half of the same leaf (total 60 leaves each) from BSL1-silenced and TRV:GFP plants through agroinfiltration. Next, we infected these leaves with the P. infestans strain 88069, which expresses recognized forms of both Avr effector genes (Avr2 and Avrblb1). In leaf panels that transiently expressed R2, we observed a significant increase in the number of sites showing late blight lesions in the BSL1-silenced plants compared with TRV:GFP plants (Figures 6C and 6D). By contrast, in leaf panels expressing Rpi-sto1, the number of sites with sporulating lesions remained low in both BSL1-silenced and TRV:GFP plants, indicating Rpi-sto1–mediated resistance was not affected. We conclude that silencing of BSL1 specifically compromises R2-mediated resistance to P. infestans. However, silencing of BSL1 did not alter the compatible interaction with P. infestans in the absence of R2 (see Supplemental Figure 14 online).
AVR2 Promotes the Interaction between BSL1 and R2
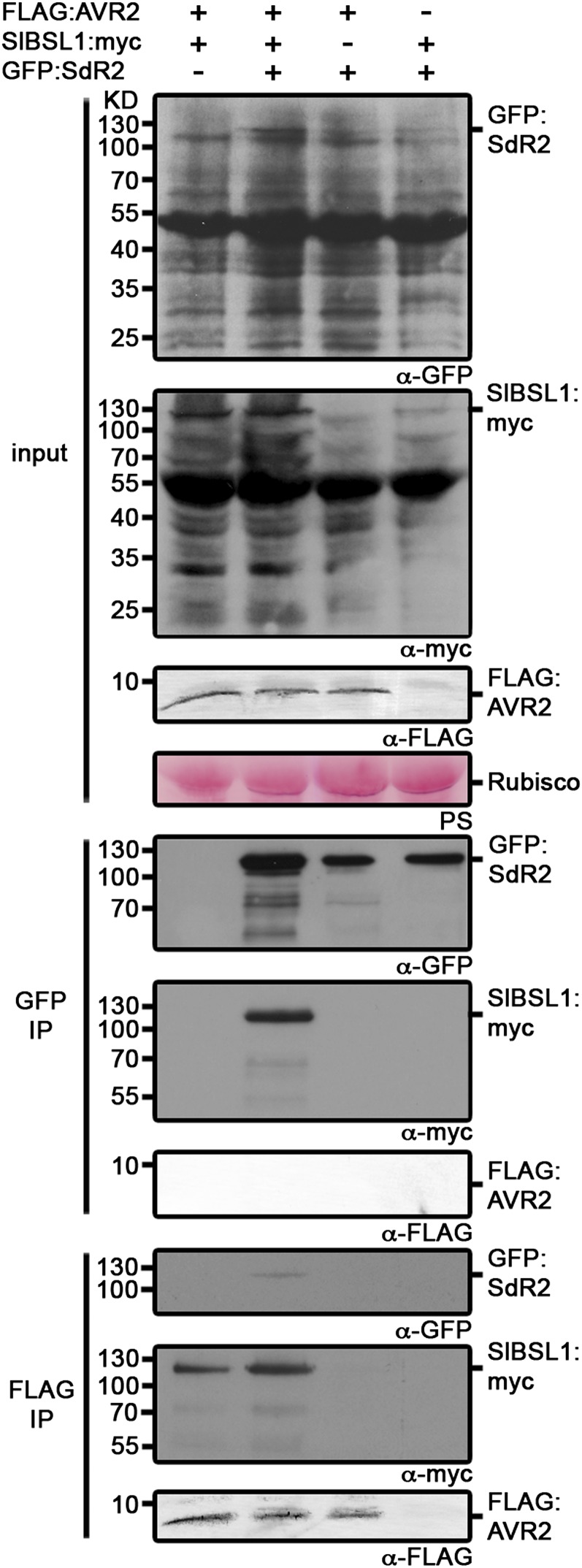
Given that BSL1 is required for R2-mediated recognition of AVR2, a logical step was to investigate whether these proteins associate in planta. To elucidate this, we performed co-IP experiments involving a GFP-tagged R2 protein (GFP:SdR2), FLAG-tagged AVR2 protein (FLAG:AVR2), and a myc-tagged BSL1 protein (SlBSL1:myc) in different combinations (Figure 7). These constructs were expressed in N. benthamiana using agroinfiltration. Co-IPs with anti-GFP antibodies resulted in the recovery of Sl-BSL1 only in the presence of AVR2 expression (Figure 7). The same co-IPs failed to recover AVR2. However, co-IPs performed with anti-FLAG antibodies revealed a weak GFP reacting signal corresponding to R2 (Figure 7). This indicates that, although we have obtained no evidence for an exclusive direct interaction between AVR2 and R2, these two proteins may associate through a ternary complex with BSL1 that forms in planta (Figure 7). This observation was also confirmed by Y2H analysis, which failed to demonstrate a direct interaction between AVR2 and R2 (see Supplemental Figure 15 online).
Figure 7.
Immunoblots Showing That AVR2 Expression Is Required for Interaction of Sl-BSL1 with R2.
FLAG:AVR2, SlBSL1:myc, and GFP:SdR2 were transiently expressed in combination in N. benthamiana. Immunoprecipitates obtained with anti-FLAG or anti-GFP antiserum, and total protein extracts were immunoblotted with appropriate antisera. IP, immunoprecipitate; PS, Ponceau stain; Rubisco, ribulose-1,5-bisphosphate carboxylase/oxygenase.
[See online article for color version of this figure.]
The Stealthy A2L Variant Associates with BSL1
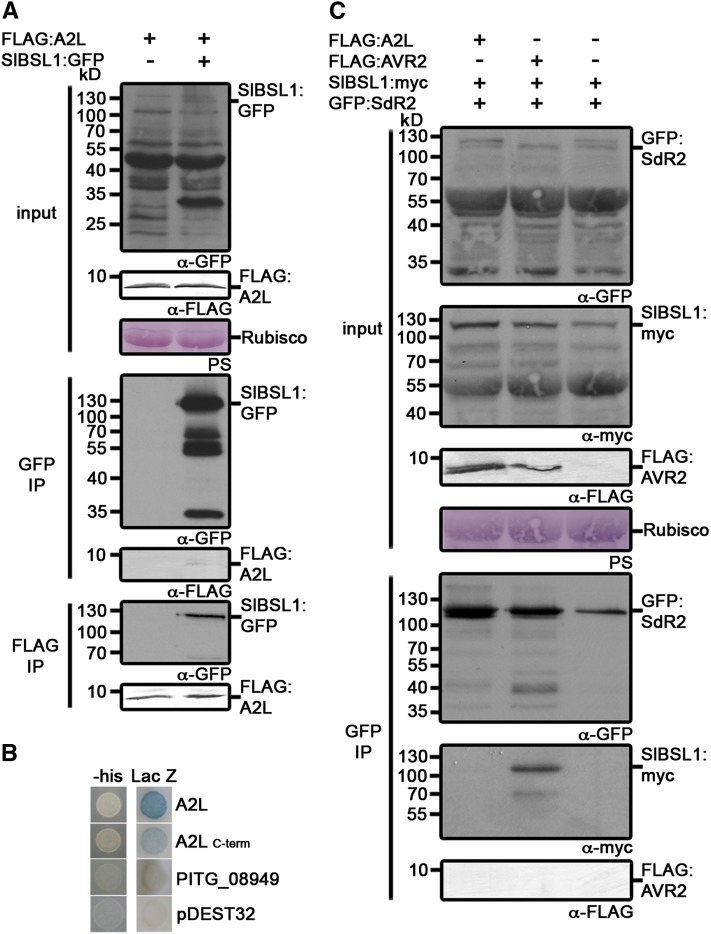
Strains of P. infestans, such as those from the virulent clonal lineage 13_A2, that are virulent on R2 potatoes, express the A2L variant that evades perception by R2 (Gilroy et al., 2011). To determine whether A2L interacts with BSL1, we expressed a FLAG-tagged C-terminal region of A2L (amino acids 66 to 116) and SlBSL1:GFP in N. benthamiana by agroinfiltration. Co-IP analysis revealed that the FLAG:A2L protein associates with Sl-BSL1 (Figure 8A). We also demonstrated the direct interaction of the full-length (minus signal peptide) and C-terminal domain (amino acids 66 to 116) of A2L with St-BSL1 by Y2H analysis (Figure 8B).
Figure 8.
A2L Associates with BSL1 but Does Not Promote an Interaction between BSL1 and R2.
(A) Immunoblots showing A2L coimmunoprecipitates with tomato BSL1 in planta. FLAG:A2L was transiently expressed alone or with SlBSL1:GFP in N. benthamiana. Immunoprecipitates obtained with anti-FLAG or anti-GFP antiserum and total protein extracts were immunoblotted with appropriate antisera. IP, immunoprecipitate; Rubisco, ribulose-1,5-bisphosphate carboxylase/oxygenase.
(B) Y2H analysis demonstrating that A2L (excluding the signal peptide) and the C-terminal effector domain of A2L (amino acids 66 to 116) interact with potato BSL1 (St-BSL1) in vivo. Both LacZ (blue color) and His3 (providing growth on medium lacking His [-his]) reporter genes were activated. Empty vector (pDEST32) and effector PITG_08949 (Gilroy et al., 2011) acted as negative controls (no reporter activity).
(C) Immunoblots showing A2L does not promote interaction of Sl-BSL1 with R2. FLAG:A2L, SlBSL1:myc, and GFP:SdR2 were transiently expressed in combination in N. benthamiana. Immunoprecipitates obtained with anti-FLAG or anti-GFP antiserum and total protein extracts were immunoblotted with appropriate antisera. PS, Ponceau stain.
A2L Does Not Promote the Interaction between BSL1 and R2
Given that AVR2 promotes the association of BSL1 with R2, we investigated whether the virulent form A2L could also promote this association. We coexpressed FLAG:A2L, SlBSL1:myc, and GFP:SdR2 in N. benthamiana by agroinfiltration and performed co-IPs as described earlier for AVR2. Co-IPs with GFP antibodies did not recover Sl-BSL1 in the presence of A2L, in sharp contrast with the AVR2 co-IPs (Figure 8C). Therefore, although A2L can associate with BSL1, it differs from AVR2 by not promoting the interaction of BSL1 with R2.
DISCUSSION
Plant NB-LRR receptors can recognize specific pathogen effectors by direct binding or indirectly via plant proteins that are targeted by the effectors. So far, all cases of indirect recognition by NB-LRR proteins have involved bacterial and viral plant pathogens, although these immune receptors can perceive pathogens as diverse as fungi, oomycetes, nematodes, and insects (Rafiqi et al., 2009). Here, we show using co-IP and Y2H that AVR2 and R2 may associate through a ternary complex with the host protein BSL1. Our localization studies indicate that AVR2 and BSL1 share overlapping spatial distributions, which further supports their potential for interaction. We found that the putative plant phosphatase BSL1 is required for R2-mediated perception of the P. infestans RXLR effector AVR2 and resistance to an avirulent strain of the late blight pathogen. AVR2 associates with BSL1 and mediates the interaction of BSL1 with R2 in planta. Strains of P. infestans that are virulent on R2 potatoes express an unrecognized form, A2L (Gilroy et al., 2011). A2L also interacts with BSL1 but does not promote the association of BSL1 with R2.
Our objective was to understand how members of the P. infestans AVR2 effector family differentially activate R2-mediated hypersensitivity. Our findings point to a model in which any of an AVR2-modified BSL1, a BSL1-modified AVR2, or a AVR2-BSL1 complex interacts with and activates R2 (Figure 9). The stealthy effector A2L still interacts with BSL1, but this association does not lead to R2 activation. One possibility is that AVR2 induces conformational and/or biochemical changes in BSL1 and that the modified BSL1 protein can now associate with the R2 receptor to activate defense responses. Similar mechanisms of action have been reported for the recognition of bacterial effectors by NB-LRR proteins (Collier and Moffett, 2009; Elmore et al., 2011). For instance, P. syringae AvrB and AvrRpm1 promote the phosphorylation of the accessory host protein RIN4, which is perceived by the NB-LRR protein RPM1, resulting in its activation (Chung et al., 2011). By contrast, RPS2 perceives the proteolytic cleavage of RIN4 by P. syringae effector AvrRpt2 (Belkhadir et al., 2004). Future work will determine the nature of any potential conformational and/or biochemical modifications of BSL1 that could be triggered by AVR2 and how the modified BSL1 promotes interaction with R2 and activates R2 immune signaling.
Figure 9.
Models Illustrating the Potential Link between Association of AVR2 Family Members with BSL1 and R2-Mediated Recognition and Resistance.
Activation of R2 by AVR2 could be mediated by an AVR2-modifed BSL1, BSL1-modified AVR2, or AVR2-BSL1 complex (left to right). Circles, corresponding effectors; yellow diamonds, BSL1; green rectangles, R2. Potential modifications are indicated by red highlighting.
The association of AVR2 with BSL1, the requirement of AVR2 for association of R2 with BSL1, and our inability to detect direct interaction between R2 and AVR2 are compatible with indirect recognition, with BSL1 acting as an intermediary host protein that interacts with both the effector protein and NB-LRR immune receptor. To date, the only reported case of indirect recognition for filamentous plant pathogens is the recognition of the Cladosporium fulvum apoplastic effector Avr2 by tomato Cf-2. C. fulvum Avr2 selectively inhibits two papain-like Cys proteases, RCR3 and PIP1, which are transcriptionally upregulated by the salicylic acid–regulated defense pathway (Rooney et al., 2005; Shabab et al., 2008). Although the interaction of the C. fulvum Avr2 with RCR3 has been demonstrated (Rooney et al., 2005), there is no evidence for association of RCR3 or PIP1 with Cf-2. Therefore, our study represents a clear example of a host protein that associates with both a filamentous plant pathogen effector and its corresponding NB-LRR immune receptor.
Our interpretation of BSL1-mediated indirect recognition of AVR2 contrasts sharply with examples of direct perception of filamentous pathogen effectors by NB-LRR proteins. Examples include the perception of Magnaporthe oryzae AVR-Pita effector by the rice (Oryza sativa) Pi-ta protein, flax rust Melampsora lini AvrL567 by flax L proteins, H. arabidopsidis ATR1 by Arabidopsis RPP1, and P. infestans effector ipiO (Avr-blb1) by the coiled-coil domain of RB (Blb1) (Jia et al., 2000; Dodds et al., 2006; Krasileva et al., 2010; Chen et al., 2012). By contrast, in bacterial systems, effectors are often recognized indirectly with only one case of direct perception of a bacterial effector by an NB-LRR receptor reported (Deslandes et al., 2003), indicating that the predominant mechanism of plant perception for different classes of pathogens may differ.
The host target of AVR2, BSL1, and its close homolog in Arabidopsis, BSU1, have been shown to promote BR signal transduction actively (Kim et al., 2009). Proteomic analysis in Arabidopsis showed that BSL1 is a plasma membrane protein, and live-cell imaging revealed that BSL1 is also observed in the cytoplasm (Benschop et al., 2007; Kim et al., 2009). This supports a potential role and site of action for BSL1 in upstream BR signaling events at the cell surface (Kim et al., 2009). Our results indicate that solanaceous BSL1 may also associate with the plasma membrane and cytoplasm in N. benthamiana (Figure 4). Similar to the Arabidopsis T-DNA knockouts of BSL1, which are indistinguishable from wild-type plants due to the functional redundancy between members of the BSL family (Mora-García et al., 2004), silencing of BSL1 in N. benthamiana revealed no striking developmental phenotype (see Supplemental Figure 13B online). Whether or not BSL1 is a positive regulator of BR signaling in solanaceous plants remains to be determined. In addition, silencing of BSL1 had no impact upon the compatible interaction between N. benthamiana and P. infestans. However, given the documented functional redundancy between BSL family members in the context of BR signal transduction in Arabidopsis, this may also explain an unaltered virulence phenotype when silencing BSL1 alone in this study. Elucidating the function of BSL1 and how it is perturbed by AVR2, and, indeed, whether AVR2 interacts with and modifies other members of the BSL family, will be necessary to establish whether BSL1 is acting as a guardee or decoy during recognition of P. infestans by R2.
Late blight is the most destructive disease of potato, the third most important food crop in the world. P. infestans continues to cost modern agriculture billions of dollars annually and is a critical constraint for subsistence potato farming. The most sustainable strategy to manage late blight is to breed broad-spectrum disease resistance into potato. The R2 gene has been exploited in agriculture (Gilroy et al., 2011; Vleeshouwers et al., 2011). But although virulent races of the pathogen, including the epidemic clonal lineage blue13, have emerged, R2 potatoes have been reported to confer a degree of resistance to local P. infestans populations in China, Russia, France, and The Netherlands (Vleeshouwers et al., 2011). Our gained understanding of the mechanism of recognition of P. infestans by R2 should prove helpful in managing and improving R2-mediated late blight resistance. For example, synthetic R2 mutant genes with expanded pathogen recognition specificities could be generated to enhance the spectrum and durability of late blight resistance or to extend recognition to effectors from other pathogens that may also target and modify BSL1 (Vleeshouwers et al., 2011).
METHODS
Plasmid Construction and Preparation
All primers used in this study are listed in Supplemental Table 2 online. The C-terminal effector domain of each Avr2 family member and A2l were custom synthesized with an N-terminal FLAG-tag in place of the signal peptide and RXLR-ERR region, or for PEXRD11 just the signal peptide region, by Genscript and subcloned into the Tobacco mosaic virus–based Agrobacterium tumefaciens binary vector pTRBO (Lindbo, 2007) (see Supplemental Figure 1A online). All Gateway cloning was performed following the manufacturer’s instructions (Invitrogen). GFP:Avr2 and GFP:PexRd11 constructs were generated as follows: The full-length Avr2 gene (excluding the N-terminal signal peptide region; see Supplemental Figure 1B online) and the C-terminal domain of PexRd11 (following the RXLR-EER motif; see Supplemental Figure 1B online) were generated by PCR using primer combinations 5AVR2/3AVR2 or 5RD11/3RD11 and genomic DNA from Phytophthora infestans isolate 88069. These amplicons were then cloned into the entry vector pENTR/D-TOPO (Invitrogen) and resulting clones confirmed by sequencing. The entry clone inserts were then introduced into pK7WGF2 (Karimi et al., 2002) by Gateway LR recombination (Invitrogen). Initially constructs were designed both with and without the RXLR region. However, we now know that the RXLR domain has no impact on the observed localization patterns.
BSL1, Catalase 1, and thioredoxin peroxidase were cloned into pENTR/D-TOPO (Invitrogen) using the PCR products amplified from tomato (Solanum lycopersicum), Nicotiana benthamiana, or Nicotiana tabacum cDNA, respectively, using primers 5BSL1/3BSL1, 5Cat1/3Cat1, or 5Thper/3Thper. GFP fusions of the three amplicons were generated by Gateway LR recombination of pENTR:BSL1 with pK7FWG2 and pENTR:Catalase 1 or pENTR:thioredoxin with pK7WGF2 (Karimi et al., 2002). RFP and 4xmyc fusions of BSL1 were generated by Gateway LR recombination of pENTR:BSL1 with pGWB554 (BSL1:RFP) or pGWB17 (BSL1:myc) (Nakagawa et al., 2007). The region upstream of the BSL1 phosphatase domain and the phosphatase domain were cloned into pENTR/D-TOPO (Invitrogen) using the PCR products amplified from tomato (Solanum lycopersicum), using primers 5BSL1/3BSL1_kel or 5BSL1_phos/3BSL1. GFP fusions of the two amplicons were generated by Gateway LR recombination of pENTR:BSL1Kelch and pENTR:BSL1Phospho with pK7FWG2.
RFP:AVRblb2 was previously described (Bozkurt et al., 2011). RFP:StRemorin was generated by cloning the PCR amplicon produced using potato (Solanum tuberosum) cDNA as a template with primers FStrem13/RStrem13. The amplicon was then used as a template in a second PCR with attB-specific primers and the amplicon cloned into pDONR201. The resulting entry clone insert was then introduced into pGWB555 by Gateway LR recombination.
GFP:SdR2 and YFP:SdR2 were generated by cloning the PCR amplicon produced using pKGW-MG:R2 as a template (Lokossou et al., 2009) with primers 5Nterm_R2/3Nterm_R2 into the entry vector pENTR/D-TOPO. The resulting entry clone insert was then introduced into pK7WGF2 or pB7WGY2 (Karimi et al., 2002) by Gateway LR recombination and into pDEST32 and pDEST22 vectors for Y2H analyses.
The Y2H DNA binding domain and activation domain fusions of AVR2 and an activation domain fusion of BSL1 were generated as follows. First, pDNR221 entry clones containing AVR2K31, AVR2N31, A2L, the C-terminal effector domain of both AVR2 and A2L (amino acids 66 to 116), or the control RXLR effector PITG_08949 (Gilroy et al., 2011) were recombined with pDEST32 and pDEST22 using Gateway LR recombination. Next, St-BSL1 was PCR amplified from potato cDNA using primers 5StBSL1-1/3StBSL1-1, which contain partial attB sites. The amplicon was then used as template in a second PCR with primers 5StBSL1-2/3StBSL1-2 to complete the attB recombination sites, before cloning into the entry vector pDON201 followed by Gateway LR recombination into pDEST22. The phosphatase domain of St-BSL1 was amplified and also cloned into pDEST22 using primers 5StBSL1-CT/3StBSL1-1.
Constructs used for VIGS of BSL1 were generated as follows. A 290- and 300-bp fragment of BSL1 was PCR amplified from N. benthamiana cDNA using primers 5BSL1-290/3BSL1-290 and 5BSL1-300/3BSL1-300, respectively (see Supplemental Figure 13A online). Each fragment was then cloned into the TRV-based silencing vector (Ratcliff et al., 1999).
YN:AVR2N31, YN:AVR2K31, and YN:PITG_08949 were generated by recombining pDNR221 entry clones containing AVR2K31, AVR2N31, or the control RXLR effector PITG_08949 (Gilroy et al., 2011) with vector CL112 (Bos et al., 2010) using Gateway LR recombination. YC:StBSL1 was generated by recombining the pDON201 entry clone containing St-BSL1 into vector CL113 (Bos et al., 2010) using Gateway LR recombination.
Transient Gene Expression Assays
In planta transient expression by agroinfiltration was performed according to methods described previously (Bos et al., 2006). For each of the Agrobacterium strains, a final OD600 of 0.3 to 0.4 in agroinfiltration medium (10 mM MES, 10 mM MgCl2, and 150 μM acetosyringone, pH 5.6) was used. For the transient coexpression assays, Agrobacterium strains carrying the plant expression constructs were mixed in a 1:1 ratio in agroinfiltration medium to a final OD600 of 0.4 to 0.6. GFP:SdR2 induced a delayed HR when coinfiltrated with AVR2; therefore, co-IPs were conducted with tissue not displaying HR.
Co-IP Experiments and Immunoblot Analysis
For co-IP experiments, total proteins were extracted from N. benthamiana leaves 4 d after agroinfiltration and peptides prepared for LC-MS/MS as described previously (Win et al., 2011). Immunoblot analyses were performed on SDS-PAGE separated proteins as described elsewhere (Oh et al., 2009). Monoclonal FLAG M2-alkaline phosphatase antibodies (Sigma-Aldrich) were used at 1:10,000 dilution and blots developed using the AP conjugate substrate kit (Bio-Rad). The α-GAL4 activation domain monoclonal antibody (Clontech) was used at 1:6000 to detect Sd-R2 protein expression and stability in yeast cells. Polyclonal GFP antibodies (Invitrogen) at 1:4000 and polyclonal myc-antibodies at 1:4000 (Santa Cruz Biotechnology) were used as primary antibodies, and anti-rabbit polyclonal antibody conjugated to horseradish peroxidase (Sigma-Aldrich) was used as a secondary antibody at a dilution of 1:12,000. Protein bands on immunoblots were detected using ECL substrate (Thermo Scientific Pierce) and exposed on Amersham Hyperfilm ECL (GE Healthcare).
Y2H Analyses
pDEST22:StBSL1 was cotransformed with the pDEST32 constructs containing AVR2K31, AVR2N31, A2L, the C-terminal effector domain of both AVR2 and A2L (amino acids 66 to 116), the control RXLR effector PITG_08949, or pDEST32 alone into the yeast strain MaV203. pDEST22:StBSL1phospho (phosphatase domain) was cotransformed with pDEST32 constructs containing AVR2K31, AVR2N31, the C-terminal effector domain of AVR2 (amino acids 66 to 116), and the control RXLR effector PITG_08949. pDEST22:SdR2 and pDEST32:SdR2 were cotransformed with appropriate effector forms in the corresponding pDEST22 or pDEST32 vectors (as above). Transformed cells were plated out on synthetic complete media lacking Leu and Trp. Colonies were picked from these plates to test interaction in reporter gene assays (the same medium for the LacZ assay and the same medium but lacking His for the HIS3 assay), using the ProQuest system (Invitrogen), following the manufacturer’s protocol.
VIGS
VIGS was performed in N. benthamiana as described by Peart et al. (2002). A TRV construct expressing GFP was used as a control (Gilroy et al., 2007). To confirm silencing RT-PCR was performed on BSL1- and GFP-silenced plant material 2 to 4 weeks after infection with TRV constructs, to PCR amplify either Nb-BSL1 using primers 5BSL1-RT/3BSL1-RT or Nb-BSL2 using primers 5BSL2-RT/3BSL2-RT. Nb-BSL2 was identified as the closest paralog based on a TBLASTN search of the recently released genome sequence draft of N. benthamiana (http://solgenomics.net/organism/Nicotiana_benthamiana/genome) with Arabidopsis BSL family members At-BSU1, At-BSL2, and At-BSL3. To assess the stability of YFP:SdR2 in BSL1-silenced plants, YFP:SdR2 was transiently expressed in plants expressing each TRV construct and in unsilenced plants. Total proteins were extracted 3 d after infection (DAI) and immunoblot analysis undertaken as described previously (Gilroy et al., 2011).
HR Assays
Following VIGS, transgenes were delivered into N. benthamiana leaves by agroinfiltration 2 to 3 weeks after infiltration with the TRV constructs. Agrobacterium cultures were resuspended in agroinfiltration medium at a final concentration of OD600 0.4 for those delivering the effector transgenes and OD600 0.75 for those delivering the R genes and results recorded and photographed 5 d after infiltration. Following agroinfiltration, HR was monitored as described previously (Gilroy et al., 2011) with six independent biological experiments, each with three leaves per plant, from four plants.
To assess the ability of PITG_13940 to induce R2-mediated HR, pTRBO:AVR2, pTRBO:PEXRD11, and pTRBO:PITG_13940 were transiently coexpressed in N. benthamiana with pKGW-MG:R2 (Lokossou et al., 2009) using agroinfiltration.
Agrobacterium Transient Assay
Agrobacterium transient assay was performed on N. benthamiana plants following VIGS 3 weeks after initial infiltration with the TRV constructs. First, Agrobacterium cultures were resuspended in agroinfiltration medium at a final concentration of OD600 0.3 and used for transient expression in planta by agroinfiltration of the VIGS plants. After 2 d, each infiltration site was inoculated with zoospores from P. infestans isolate 88069 as described previously (Bos et al., 2010). Three biological replicates of 20 leaves each were assessed for late blight lesions 6 d after infection (DAI) for each VIGS construct. The percentage of inoculation sites that formed sporulating lesions were compared with the total number of sites inoculated. Leaves were photographed at 5 to 6 DAI. Trypan blue staining was performed as described previously by Gilroy et al. (2007).
P. infestans Infection Assays for Microscopy
Assays to observe the localization of AVR2, PEXRD11, and BSL1 during infection were performed as follows: First GFP:Avr2, GFP:PexRD11, and/or BSL1:RFP were transiently expressed under the 35S promoter in N. benthamiana leaves by agroinfiltration. At 48 h after infiltration, the infiltrated leaves were detached and inoculated with the P. infestans 88069 or 88069td (100,000 zoospores/mL) strains on six spots for each leaf. Localization of the fluorescent proteins at sites of P. infestans haustoria formation was recorded using confocal microscopy 2 to 4 DAI.
Confocal Microscopy
Patches of N. benthamiana leaves were cut and mounted in water and analyzed on a Leica DM6000B/TCS SP5 confocal microscope (Leica Microsystems CMS) with the following excitation wavelengths: GFP, 488 nm; RFP, 561 nm. Scanning was performed in sequential mode to prevent signal bleed-through. For bimolecular fluorescence complementation, a Leica SP2 confocal laser scanning microscope on a DM6000 microscope fitted with a FI/RH filter block and water dipping lenses (HCX APO L10×/0.30 W U-V-1, L20×/0.50 W U-V-1, L40×/0.80 W U-V-1, or L63×/0.90 W U-V-1) was used. YFP fluorescence was collected using excitation at 514 nm with emission collected at 530 to 575 nm using the L40×/0.80 W U-V-1 lens.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: Avr2 (PITG_22870, XM_002902940.1 and PITG_08943, XM_002902939.1), PexRd11 (PITG_13930, XM_002899550.1), PITG_05121 (XM_002998776.1), PITG_19617 (XM_002996892.1), PITG_07500 (XM_002904456.1), PITG_07499 (XM_002904455.1), PITG_08278 (XM_002903638.1), PITG_15972 (XM_002898139.1), PITG_06077 (XM_002998224.1), PITG_21949 (XM_002996830.1), PITG_21645 (XM_002894729.1), PITG_13940 (XM_002899557.1), PITG_13936 (XM_002899553.1), S. lycopersicum BSL1 (JQ886089), N. tabacum Catalase 1 (JQ886090), N. tabacum Thioredoxin peroxidase (JQ886091), Solanum demissum R2 (FJ536325.1), S. tuberosum BSL1 (JQ950742) and BSL2 JQ950743), N. benthamiana BSL1 (JQ950744), and N. benthamiana BSL2 (JQ969018).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Schematic Representation of AVR2 Family Fusion Constructs Used in Co-IPs.
Supplemental Figure 2. Immunoblots Showing That P. infestans PEXRD11 Specifically Associates with Sl-BSL1 in Planta.
Supplemental Figure 3. Sequence Analysis Confirms That BSL1 from Solanum tuberosum (Potato), Solanum lycopersicum (Tomato), and Nicotiana benthamiana Are Orthologs of Arabidopsis BSL1.
Supplemental Figure 4. Arabidopsis, Solanum tuberosum (Potato), and Solanum lycopersicum (Tomato) BSL1 Full-Length Amino Acid Sequences Are Highly Conserved.
Supplemental Figure 5. Y2H Analysis Illustrating That P. infestans AVR2K31, AVR2N31, and the C-Terminal Effector Domain of AVR2 (Amino Acids 66 to 116) Interact with the Phosphatase Domain (Amino Acids 520 to 881) of Potato BSL1 (St-BSL1) in Vivo.
Supplemental Figure 6. Localization and Stability of P. infestans AVR2 in Planta.
Supplemental Figure 7. P. infestans AVR2 and PEXRD11 Colocalize with the Plasma Membrane–Associated Protein Remorin in Planta.
Supplemental Figure 8. P. infestans AVR2 and PEXRD11 Colocalize with the P. infestans Plasma Membrane–Associated Effector AVRblb2 in Planta.
Supplemental Figure 9. P. infestans PEXRD11 Accumulates around Haustoria and Colocalizes with the Perihaustorial-Localized Effector AVRblb2 in Planta.
Supplemental Figure 10. Tomato BSL1 and P. infestans PEXRD11 Colocalize in Planta and Accumulate around Haustoria.
Supplemental Figure 11. AVR2 Is in Close Proximity to St-BSL1 at the Cell Periphery and within the Cytoplasm.
Supplemental Figure 12. PITG_13940 Activates R2-Mediated Hypersensitivity (HR).
Supplemental Figure 13. Virus-Induced Gene Silencing of BSL1 in N. benthamiana.
Supplemental Figure 14. VIGS of BSL1 in N. benthamiana Did Not Affect Colonization by P. infestans.
Supplemental Figure 15. Y2H Analysis Failed to Demonstrate a Direct Interaction between AVR2 and R2.
Supplemental Table 1. Plant Proteins That Specifically Associated with AVR2 Family Effectors after Coimmunoprecipitation as Identified by Mass Spectrometry.
Supplemental Table 2. Details of Primers Used in This Study.
Supplemental Data Set 1. Peptide Matches from Mascot Spectra Searches That Identified Plant Proteins Specifically Associated with AVR2 Family Effectors in Coimmunoprecipitations.
Supplemental Data Set 2. Text File of the Alignment Used for the Phylogenetic Analysis Shown in Supplemental Figure 3.
Supplementary Material
Acknowledgments
We thank Matthew Smoker (The Sainsbury Laboratory [TSL]), Jodie Pike (TSL), Jan Sklenar (TSL), and Alex Jones (TSL) for technical assistance, Katrin Mackenzie (BioSS) for statistical analyses, Leighton Pritchard (James Hutton Institute [JHI]) for bioinformatic analysis of the BSL gene family, Miles Armstrong (JHI) for assistance with Y2H analyses, and Stephen Whisson (JHI) for providing P. infestans 88069td. This project was funded by the Gatsby Charitable Foundation, a Leverhulme Early Career Fellowship, Marie Curie FP7-PEOPLE-2007-2-1-IEF, Deutsche Forschungsgemeinschaft Grant SCHO1347/1-1, the Rural and Environmental Science and Analytical Services department of the Scottish Government, and the Biotechnology and Biological Sciences Research Council.
AUTHOR CONTRIBUTIONS
D.G.O.S., S.B., J.W., S.S., P.R.J.B., E.M.G., and S.K. designed research. D.G.O.S., S.B., J.W., S.S., I.H., T.O.B., and E.M.G. performed research. D.G.O.S., S.B., J.W., S.S., N.C., V.G.A.A.V., P.R.J.B., E.M.G., and S.K. analyzed data. D.G.O.S., S.B., P.R.J.B., and S.K. wrote the article, and all authors commented on the article before submission.
Glossary
- RXLR
to be defined
- ETI
effector-triggered immunity
- HR
hypersensitive response
- NB-LRR
nucleotide binding leucine-rich repeat
- Y2H
yeast two-hybrid
- co-IP
coimmunoprecipitation
- LC-MS/MS
liquid chromatography–tandem mass spectrometry
- GFP
green fluorescent protein
- BR
brassinosteroid
- RFP
red fluorescent protein
- VIGS
virus-induced gene silencing
- TRV
tobacco rattle virus
- YFP
yellow fluorescent protein
- DAI
days after infection
References
- Armstrong M.R., et al. (2005). An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc. Natl. Acad. Sci. USA 102: 7766–7771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir Y., Nimchuk Z., Hubert D.A., Mackey D., Dangl J.L. (2004). Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1. Plant Cell 16: 2822–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benschop J.J., Mohammed S., O’Flaherty M., Heck A.J., Slijper M., Menke F.L. (2007). Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol. Cell. Proteomics 6: 1198–1214 [DOI] [PubMed] [Google Scholar]
- Bos J.I., et al. (2010). Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc. Natl. Acad. Sci. USA 107: 9909–9914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos J.I., Kanneganti T.D., Young C., Cakir C., Huitema E., Win J., Armstrong M.R., Birch P.R., Kamoun S. (2006). The C-terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a-mediated hypersensitivity and suppress INF1-induced cell death in Nicotiana benthamiana. Plant J. 48: 165–176 [DOI] [PubMed] [Google Scholar]
- Bozkurt T.O., Schornack S., Win J., Shindo T., Ilyas M., Oliva R., Cano L.M., Jones A.M., Huitema E., van der Hoorn R.A., Kamoun S. (2011). Phytophthora infestans effector AVRblb2 prevents secretion of a plant immune protease at the haustorial interface. Proc. Natl. Acad. Sci. USA 108: 20832–20837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champouret N. (2010). Functional Genomics of Phytophthora infestans Effectors and Solanum Resistance Genes. PhD dissertation (Wageningen, The Netherlands: Wageningen University), p. 162
- Champouret N., Bouwmeester K., Rietman H., van der Lee T., Maliepaard C., Heupink A., van de Vondervoort P.J.I., Jacobsen E., Visser R.G.F., van der Vossen E.A.G., Govers F., Vleeshouwers V.G. (2009). Phytophthora infestans isolates lacking class I ipiO variants are virulent on Rpi-blb1 potato. Mol. Plant Microbe Interact. 22: 1535–1545 [DOI] [PubMed] [Google Scholar]
- Chaparro-Garcia A., Wilkinson R.C., Gimenez-Ibanez S., Findlay K., Coffey M.D., Zipfel C., Rathjen J.P., Kamoun S., Schornack S. (2011). The receptor-like kinase SERK3/BAK1 is required for basal resistance against the late blight pathogen Phytophthora infestans in Nicotiana benthamiana. PLoS ONE 6: e16608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu Z., Halterman D.A. (2012). Molecular determinants of resistance activation and suppression by Phytophthora infestans effector IPI-O. PLoS Pathog. 8: e1002595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm S.T., Coaker G., Day B., Staskawicz B.J. (2006). Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- Chung E.H., da Cunha L., Wu A.J., Gao Z., Cherkis K., Afzal A.J., Mackey D., Dangl J.L. (2011). Specific threonine phosphorylation of a host target by two unrelated type III effectors activates a host innate immune receptor in plants. Cell Host Microbe 9: 125–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier S.M., Moffett P. (2009). NB-LRRs work a “bait and switch” on pathogens. Trends Plant Sci. 14: 521–529 [DOI] [PubMed] [Google Scholar]
- Deslandes L., Olivier J., Peeters N., Feng D.X., Khounlotham M., Boucher C., Somssich I., Genin S., Marco Y. (2003). Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. USA 100: 8024–8029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds P.N., Lawrence G.J., Catanzariti A.M., Teh T., Wang C.I., Ayliffe M.A., Kobe B., Ellis J.G. (2006). Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. USA 103: 8888–8893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitas T.K., Dangl J.L. (2010). NB-LRR proteins: Pairs, pieces, perception, partners, and pathways. Curr. Opin. Plant Biol. 13: 472–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore J.M., Lin Z.J., Coaker G. (2011). Plant NB-LRR signaling: Upstreams and downstreams. Curr. Opin. Plant Biol. 14: 365–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry W.E., Gruenwald N.J., Cooke D.E.L., McLeod A., Forbes G.A., Cao K. (2008). Population genetics and population diversity of Phytophthora infestans. In Oomycete Genetics and Genomics, K. Lamour and S. Kamoun, eds (Hoboken, NJ: John Wiley & Sons), pp. 139–164
- Gilroy E.M., et al. (2011). Presence/absence, differential expression and sequence polymorphisms between PiAVR2 and PiAVR2-like in Phytophthora infestans determine virulence on R2 plants. New Phytol. 191: 763–776 [DOI] [PubMed] [Google Scholar]
- Gilroy E.M., et al. (2007). Involvement of cathepsin B in the plant disease resistance hypersensitive response. Plant J. 52: 1–13 [DOI] [PubMed] [Google Scholar]
- Haas B.J., et al. (2009). Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461: 393–398 [DOI] [PubMed] [Google Scholar]
- Hein I., Gilroy E.M., Armstrong M.R., Birch P.R. (2009). The zig-zag-zig in oomycete-plant interactions. Mol. Plant Pathol. 10: 547–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y., McAdams S.A., Bryan G.T., Hershey H.P., Valent B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19: 4004–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. (2006). The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kim T.W., Guan S., Sun Y., Deng Z., Tang W., Shang J.X., Sun Y., Burlingame A.L., Wang Z.Y. (2009). Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 11: 1254–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasileva K.V., Dahlbeck D., Staskawicz B.J. (2010). Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell 22: 2444–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindbo J.A. (2007). TRBO: A high-efficiency tobacco mosaic virus RNA-based overexpression vector. Plant Physiol. 145: 1232–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Elmore J.M., Lin Z.J., Coaker G. (2011). A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe 9: 137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokossou A.A., Park T.H., van Arkel G., Arens M., Ruyter-Spira C., Morales J., Whisson S.C., Birch P.R., Visser R.G., Jacobsen E., van der Vossen E.A. (2009). Exploiting knowledge of R/Avr genes to rapidly clone a new LZ-NBS-LRR family of late blight resistance genes from potato linkage group IV. Mol. Plant Microbe Interact. 22: 630–641 [DOI] [PubMed] [Google Scholar]
- Moffett P., Farnham G., Peart J., Baulcombe D.C. (2002). Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J. 21: 4511–4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora-García S., Vert G., Yin Y., Caño-Delgado A., Cheong H., Chory J. (2004). Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 18: 448–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Oh S.K., et al. (2009). In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi-blb2. Plant Cell 21: 2928–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva R., et al. (2010). Recent developments in effector biology of filamentous plant pathogens. Cell. Microbiol. 12: 1015. [DOI] [PubMed] [Google Scholar]
- Panstruga R., Dodds P.N. (2009). Terrific protein traffic: The mystery of effector protein delivery by filamentous plant pathogens. Science 324: 748–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart J.R., et al. (2002). Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc. Natl. Acad. Sci. USA 99: 10865–10869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele S., et al. (2009). Remorin, a solanaceae protein resident in membrane rafts and plasmodesmata, impairs potato virus X movement. Plant Cell 21: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiqi M., Bernoux M., Ellis J.G., Dodds P.N. (2009). In the trenches of plant pathogen recognition: Role of NB-LRR proteins. Semin. Cell Dev. Biol. 20: 1017–1024 [DOI] [PubMed] [Google Scholar]
- Ratcliff F.G., MacFarlane S.A., Baulcombe D.C. (1999). Gene silencing without DNA. rna-mediated cross-protection between viruses. Plant Cell 11: 1207–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney H.C., Van’t Klooster J.W., van der Hoorn R.A., Joosten M.H., Jones J.D., de Wit P.J. (2005). Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science 308: 1783–1786 [DOI] [PubMed] [Google Scholar]
- Shabab M., Shindo T., Gu C., Kaschani F., Pansuriya T., Chintha R., Harzen A., Colby T., Kamoun S., van der Hoorn R.A. (2008). Fungal effector protein AVR2 targets diversifying defense-related cys proteases of tomato. Plant Cell 20: 1169–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stassen J.H., Van den Ackerveken G. (2011). How do oomycete effectors interfere with plant life? Curr. Opin. Plant Biol. 14: 407–414 [DOI] [PubMed] [Google Scholar]
- van der Hoorn R.A., Kamoun S. (2008). From guard to decoy: A new model for perception of plant pathogen effectors. Plant Cell 20: 2009–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lee T., Robold A., Testa A., van ’t Klooster J.W., Govers F. (2001). Mapping of avirulence genes in Phytophthora infestans with amplified fragment length polymorphism markers selected by bulked segregant analysis. Genetics 157: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vleeshouwers V.G., Raffaele S., Vossen J., Champouret N., Oliva R., Segretin M.E., Rietman H., Cano L.M., Lokossou A., Kessel G., Pel M.A., Kamoun S. (2011). Understanding and exploiting late blight resistance in the age of effectors. Annu. Rev. Phytopathol. 49: 25.21–25.25 [DOI] [PubMed] [Google Scholar]
- Win J., Kamoun S., Jones A.M. (2011). Purification of effector-target protein complexes via transient expression in Nicotiana benthamiana. Methods Mol. Biol. 712: 181–194 [DOI] [PubMed] [Google Scholar]
- Win J., Morgan W., Bos J., Krasileva K.V., Cano L.M., Chaparro-Garcia A., Ammar R., Staskawicz B.J., Kamoun S. (2007). Adaptive evolution has targeted the C-terminal domain of the RXLR effectors of plant pathogenic oomycetes. Plant Cell 19: 2349–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.