Ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco) is the enzyme that fixes carbon dioxide into sugar precursors and is the most abundant protein in nature. However, the assembly pathway for plant Rubisco is not understood. This article describes a protein that is required for proper assembly of maize Rubisco, apparently by allowing the chloroplast-encoded large subunit to fold correctly.
Abstract
Most life is ultimately sustained by photosynthesis and its rate-limiting carbon fixing enzyme, ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco). Although the structurally comparable cyanobacterial Rubisco is amenable to in vitro assembly, the higher plant enzyme has been refractory to such manipulation due to poor understanding of its assembly pathway. Here, we report the identification of a chloroplast protein required for Rubisco accumulation in maize (Zea mays), RUBISCO ACCUMULATION FACTOR1 (RAF1), which lacks any characterized functional domains. Maize lines lacking RAF1 due to Mutator transposon insertions are Rubisco deficient and seedling lethal. Analysis of transcripts and proteins showed that Rubisco large subunit synthesis in raf1 plants is not compromised; however, newly synthesized Rubisco large subunit appears in a high molecular weight form whose accumulation requires a specific chaperonin 60 isoform. Gel filtration analysis and blue native gels showed that endogenous and recombinant RAF1 are trimeric; however, following in vivo cross-linking, RAF1 copurifies with Rubisco large subunit, suggesting that they interact weakly or transiently. RAF1 is predominantly expressed in bundle sheath chloroplasts, consistent with a Rubisco accumulation function. Our results support the hypothesis that RAF1 acts during Rubisco assembly by releasing and/or sequestering the large subunit from chaperonins early in the assembly process.
INTRODUCTION
Ribulose-1,5-bis-phosphate carboxylase/oxygenase (Rubisco) is the major enzyme by which green plants, algae, cyanobacteria, and other autotrophic organisms sequester CO2 into organic compounds via the Calvin-Benson pathway (Andersson and Backlund, 2008). Rubisco catalyzes the photosynthetic carbon reduction and the photorespiratory carbon oxidation reactions of the substrate ribulose-1,5-bisphosphate with CO2 and O2, respectively. The inefficiency of Rubisco in fixing CO2 has a limiting impact on agricultural productivity and in compensation, Rubisco accounts for as much as 20 to 30% and 4 to 9% of total nitrogen compounds in C3 and C4 higher plants, respectively (Feller et al., 2008).
Attempts to improve the catalytic properties of plant Rubisco (reviewed in Parry et al., 2003; Mueller-Cajar and Whitney, 2008) have met with only modest success, which can be traced in part to the lack of a comprehensive knowledge of its biogenesis and the absence of an in vitro reconstitution system. Form I Rubisco, found in higher plants, algae, and cyanobacteria, is a hexadecamer composed of eight large (50-kD) and eight small (13–15 kD) subunits, denoted here as LS and SS, respectively. The genes encoding LS (rbcL) and SS (RBCS) are located in the chloroplast and nuclear genomes, respectively. SS is expressed as a preprotein that is translocated into the chloroplast, where its signal peptide is cleaved prior to its assembly with LS (Nishimura et al., 2008). The two subunits accumulate stoichiometrically in the chloroplast, a phenomenon that is mediated by feedback inhibition of LS synthesis by unassembled subunits (Rodermel et al., 1996; Wostrikoff and Stern, 2007) as well as proteolysis of unassembled SS (Kanevski and Maliga, 1994).
Attempts to delineate the assembly pathway of Form I Rubisco have exploited two major approaches: in vivo assembly of cyanobacterial Rubisco mainly using Escherichia coli cells and in vitro reconstitution of the enzyme via addition of individual components. In the first approach, assembly of Synechococcus PCC 6301 Rubisco in E. coli resulted in a functional enzyme (van der Vies et al., 1986; Tabita, 1999). LS was also expressed alone in this way and shown to have minimal catalytic activity in the octamer form, which could be enhanced by the subsequent addition of SS (Andrews, 1988).
Rubisco assembly requires multiple chaperones. The probable role of chaperonin (Cpn) 60 was first discovered through the copurification of chloroplast Rubisco with a protein homologous to E. coli GroEL (Barraclough and Ellis, 1980). It was subsequently demonstrated that overexpression of E. coli GroEL-ES significantly promoted the assembly and activity of Synechococcus Rubisco in E. coli (Goloubinoff et al., 1989b). In fact, E. coli GroEL-ES and Mg-ATP proved to be the only factors necessary for the reconstitution of a catalytically active Rhodospirillum rubrum Form II Rubisco (Goloubinoff et al., 1989a). However, only recently was Form I Rubisco assembled in vitro (Liu et al., 2010), which required both GroEL-ES and a small chaperone called RBCX (Larimer and Soper, 1993). RBCX appears to play a pivotal role in the solubility of recombinant LS and likely in vivo assembly of functional holoenzyme in Synechococcus strains where the gene is often located within the Rubisco operon (Onizuka et al., 2004; Emlyn-Jones et al., 2006; Saschenbrecker et al., 2007). In maize (Zea mays), rbcX is expressed in leaves (Li et al., 2010); however, the polypeptide remains to be detected in proteomic studies. In Arabidopsis thaliana, no direct evidence for the involvement of one or both RBCX proteins in Rubisco assembly has been documented; however, it was shown that when coexpressed in E. coli, either of the Arabidopsis RBCX proteins could increase the solubility of cyanobacterial Rubisco (Kolesiński et al., 2011). Other than Cpn60, the only chloroplast protein shown to play a direct role in the folding or assembly of plant Rubisco is BUNDLE SHEATH DEFECTIVE2 (BSD2), a DnaJ-like chaperone (Roth et al., 1996; Brutnell et al., 1999) with an unidentified mechanism of action.
Although plant and cyanobacterial Rubisco are both Form I, and the constituent proteins share over 80% amino acid identity (Parry et al., 2003), higher plant Rubisco has proven refractory to manipulation in bacteria. When expressed in E. coli, higher plant SS and LS are insoluble, do not associate with one another to form oligomers, and do not exhibit detectable enzymatic activity (Gatenby et al., 1981, 1987; Gatenby, 1984; Parry et al., 2003). This suggests that additional and possibly plant-specific proteins are required in higher plant Rubisco biogenesis. To identify such proteins, we screened a collection of maize photosynthesis mutants for strains specifically deficient in Rubisco. Here, we report the identification of a chloroplast protein that is essential for Rubisco accumulation in maize.
RESULTS
The raf1-1 Mutant Is Specifically Depleted for Rubisco
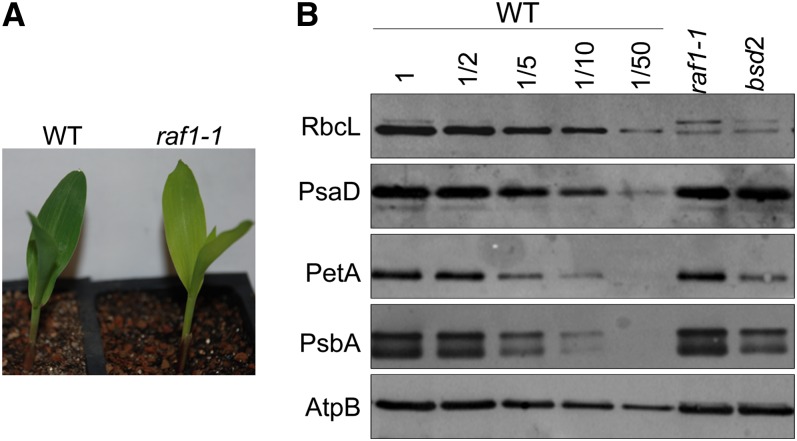
The photosynthetic mutant library (PML) is a collection of ∼2000 maize mutants displaying chlorophyll defects (Stern et al., 2004). These mutants arose in maize lines with active Mutator (Mu) transposons, so it is anticipated that most of the mutations are caused by Mu insertions. Prior screening of the majority of PML mutants by immunoblotting identified a subgroup that had reduced Rubisco but near normal accumulation of the other photosynthetic enzyme complexes that include plastid-encoded subunits (photosystem I, photosystem II, the ATP synthase, and the cytochrome b6f complex). We designated one such mutant raf1-1 (RUBISCO ACCUMULATION FACTOR [RAF]). raf1-1 mutant seedlings were pale green (Figure 1A) and survived on seed reserves for 3 to 4 weeks, as is typical of nonphotosynthetic maize mutants. raf1-1 mutants accumulated <2% of the wild-type level of LS, without a concomitant decrease in representative subunits of photosystem I (PsaD), the cytochrome b6f complex (PetA), photosystem II (PsbA), or the ATP synthase (AtpB). Because the pigmentation and protein profile of raf1-1 mutants were similar to those of a previously described mutant, bsd2 (Roth et al., 1996), we examined BSD2 mRNA by RT-PCR. While the BSD2 transcript is not detectable in the reference bsd2 mutant, it accumulates to the wild-type level in the raf1-1 mutant (see Supplemental Figure 1 online). This made it unlikely that raf1-1 was an allele of bsd2. No other mutants specifically lacking Rubisco have been reported, suggesting that raf1 was a previously unstudied gene.
Figure 1.
A Mu Transposon-Induced Nonphotosynthetic Maize Mutant Specifically Lacks Rubisco.
(A) Wild-type (WT) and homozygous raf1-1 plants at the seedling stage.
(B) Immunoblot analyses of the Rubisco large subunit and other photosynthetic proteins. Total proteins from equal surface area of the seedling leaf tip or dilutions (as indicated) were analyzed by probing with antibodies raised against the proteins indicated at the left.
[See online article for color version of this figure.]
Identification of the raf1 Gene
Genetic crosses had verified that the pale-green growth phenotype and low Rubisco levels cosegregated as a single, recessive Mendelian trait. To identify the Mu insertion underlying this phenotype, DNA was extracted from four raf1-1 seedlings, each from a different ear derived from the original mutant isolate. The Mu insertions in each sample were mapped to the reference maize genome with a high-throughput sequencing method, as recently reported (Williams-Carrier et al., 2010). Six insertion sites were found in all four raf1-1 samples and were therefore candidates for the causal insertion in the raf locus. Of these, locus GRMZM2G457621 stood out as the most promising candidate because it was the only candidate predicted to encode a chloroplast-localized protein and in addition the position of the Mu insertion (∼438 bp downstream of the start codon in the protein-coding region) was expected to cause a strong loss of function. Gene-specific PCR of additional raf1-1 mutant individuals and several +/+ cousins (see Supplemental Figure 2A online) confirmed that this insertion was tightly linked to raf1-1. Furthermore, homozygous raf1-1 plants failed to accumulate mRNA from this locus, as judged by RT-PCR, whereas phenotypically wild-type siblings did accumulate the transcript (see Supplemental Figure 2B online). These data strongly suggested that the Mu insertion in GRMZM2G457621 caused the mutant phenotype.
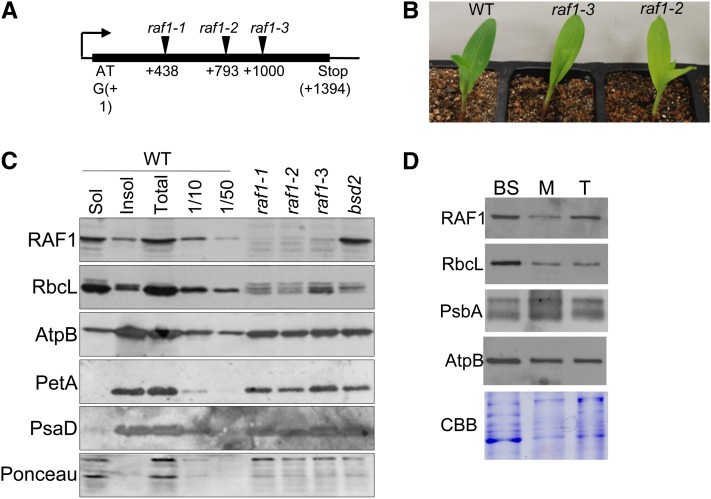
To solidify the assignment of raf1, we sought additional insertions in GRMZM2G457621 through a reverse-genetic screen of the PML collection: Pooled DNA samples representing the complete collection were screened by PCR with a Mu primer in conjunction with a gene-specific primer. Two additional alleles were recovered, which carry insertions 793 and 1000 bp downstream of the start codon (Figure 2A; see Supplemental Figure 3 online). Consistent with the raf1-1 phenotype, mutants homozygous for these insertions were pale green, seedling-lethal, and specifically lacked Rubisco (Figures 2B and 2C), although both the growth phenotype and Rubisco deficiency were slightly less pronounced in raf1-3. Pairwise complementation crosses between plants heterozygous for each of these three alleles produced F1 progeny that segregated ∼25% pale-green seedlings, which specifically lacked Rubisco (see Supplemental Figure 4 online). The failure of these alleles to complement one another proved that GRMZM2G457621 is the raf1 locus.
Figure 2.
Identification of raf1.
(A) GRMZM2G457621 is an intronless locus on chromosome 2. The locations of Mu insertions relative to the translation initiation codon are shown.
(B) Ten-day-old seedlings of the indicated genotypes. WT, the wild type.
(C) Immunoblot analysis of total, soluble, and insoluble proteins extracted from a wild-type leaf tip and total proteins extracted from an equivalent surface area of raf1 leaves. Antibodies used are shown to the left of each panel; the bottom panel is Ponceau-S staining, used to reflect loading.
(D) Immunoblot analysis of bundle sheath–enriched (BS), mesophyll-enriched (M) or total (T) proteins, to detect the proteins shown at the left. CBB, Coomassie blue staining.
[See online article for color version of this figure.]
raf1 Encodes a Previously Unstudied Protein That Is Conserved in the Green Lineage and Localizes to Bundle Sheath Chloroplasts
An antibody raised against recombinant RAF1 detected a protein of ∼47 kD in total leaf extracts from plants with a wild-type raf1 allele, consistent with the predicted size of mature RAF1 (47.5 kD). This protein was missing in raf1 mutants, confirming it to be RAF1 (Figure 2C). The partitioning of RAF1 between soluble and insoluble fractions was similar to that of LS, the implications of which are discussed below.
The algorithms TargetP and iPSORT (Emanuelsson et al., 2000; Bannai et al., 2002) predict that RAF1 localizes to chloroplasts and that raf1 encodes a protein with an N-terminal chloroplast targeting peptide. Proteomic data (Friso et al., 2010) identified RAF1 in the stroma of maize chloroplasts and suggested an approximately threefold higher accumulation in bundle sheath versus mesophyll chloroplasts. However, transcriptome profiling revealed that raf1 transcripts are 12 times more abundant in bundle sheath versus mesophyll cells at the leaf tip (Li et al., 2010). Immunoblot analysis (Figure 2D) showed that the ratio of RAF1 in bundle sheath–enriched versus mesophyll-enriched total cell extracts was similar to that of Rubisco (LS), which is considered to be a bundle sheath–specific protein. This strong enrichment of RAF1 in bundle sheath preparations is most consistent with the transcriptome data.
To investigate the range of organisms encoding RAF1, a BLAST search was performed using the maize RAF1 protein sequence. Numerous homologous proteins were identified, all from other plants or photosynthetic algae or bacteria (examples are given in Supplemental Table 1 online). RAF1 appears to be encoded by single-copy genes in all species examined except Arabidopsis, where it is duplicated. RAF1 does not share overall or segmental homology with any proteins of known structure nor does it contain significant similarity to any known plant protein domain. However, the alignment shown in Supplemental Figure 5 online shows two features of potential interest. First, the Maize Genome Database (http://www.maizegdb.org/) identified the 239– to 298–amino acid region as having similarity to Docking domain A of the erythromycin polyketide synthase superfamily, a domain that is involved in protein–protein interactions (Broadhurst et al., 2003). In addition, we noted that the last four C-terminal amino acids in the plant homologs include three amino acid residues reminiscent of a similarly positioned Hsp70/Hsp90 domain (EEVD) that interacts with tetratricopeptide repeat proteins (Prasad et al., 2010).
Rubisco Subunit Genes Are Normally Transcribed and Translated in raf1 Mutants
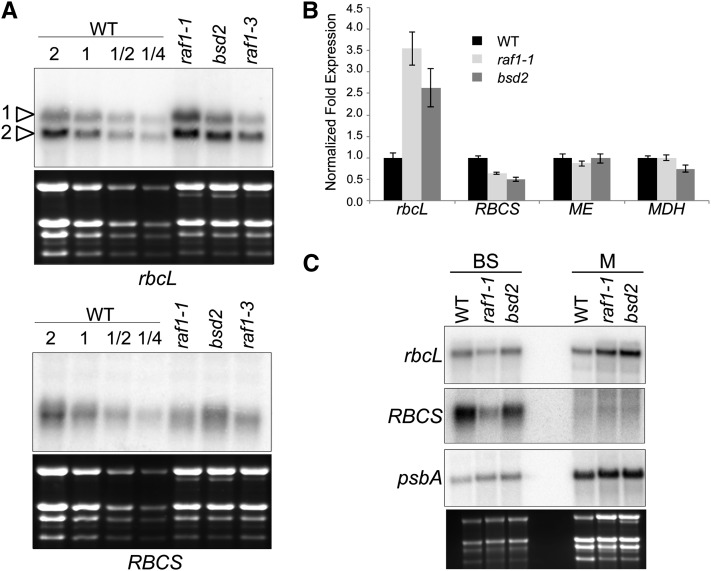
To ascertain whether RAF1 is required for the accumulation of rbcL or RBCS transcripts, gel blot analysis was performed on total RNA from wild-type, raf1, and bsd2 seedling leaves (Figure 3A). Two rbcL transcripts were resolved of which the larger is the primary transcript, and the smaller a processed version (Erion, 1985), which is likely stabilized by the maize ortholog of the RNA binding protein MRL1 (Johnson et al., 2010). While RBCS transcripts appeared to be slightly decreased in raf1-1 mutants, rbcL transcripts accumulated to more than three times the level of the wild type, a result that was confirmed by quantitative RT-PCR (Figure 3B). The expression of malic enzyme (ME) and malate dehydrogenase (MDH) mRNAs, which encode bundle sheath– and mesophyll-specific enzymes, did not change, another indication that the raf1 mutation specifically affects Rubisco. Decreased RBCS mRNA accumulation was also reported in bsd2 mutants (Roth et al., 1996) and in engineered tobacco (Nicotiana tabacum) that produced a reduced amount of Rubisco (Whitney et al., 2009). An increase in rbcL mRNA was previously reported for bsd2 mutants and was ascribed to enhanced stability of rbcL mRNA in mesophyll chloroplasts (Roth et al., 1996; Brutnell et al., 1999). To see if the same phenomenon occurs in raf1 mutants, gel blot analysis was performed on cell type–specific transcripts (Figure 3C). The results indicated that mesophyll cells are the source of rbcL mRNA overaccumulation in raf1 plants, as is the case for bsd2 mutants. Surprisingly, rbcL transcript accumulation was reduced in bundle sheath cells of raf1 compared with the wild type, an experiment which also more clearly revealed the reduction in RBCS mRNA. We assume these are pleiotropic effects related to RAF1’s role in Rubisco assembly or accumulation.
Figure 3.
Accumulation of Rubisco Transcripts in raf1 Mutants.
(A) Equal amounts (or the indicated dilutions) of total RNA from the mid-section of the second leaf of 10-d-old seedlings was analyzed in the indicated genotypes. Arrowheads indicate the bands corresponding to rbcL primary (1) and processed (2) transcripts. WT, the wild type.
(B) Quantitative RT-PCR analysis of rbcL, RBCS, ME (malic enzyme), and MDH (malate dehydrogenase) transcripts. Expression levels are an average of two biological and three technical replicates of each sample, with error bars representing standard errors. The wild-type expression level was set to 1, and samples were normalized to actin mRNA.
(C) RNA gel blot analysis was performed on total RNA isolated from bundle sheath (BS) and mesophyll (M) cells.
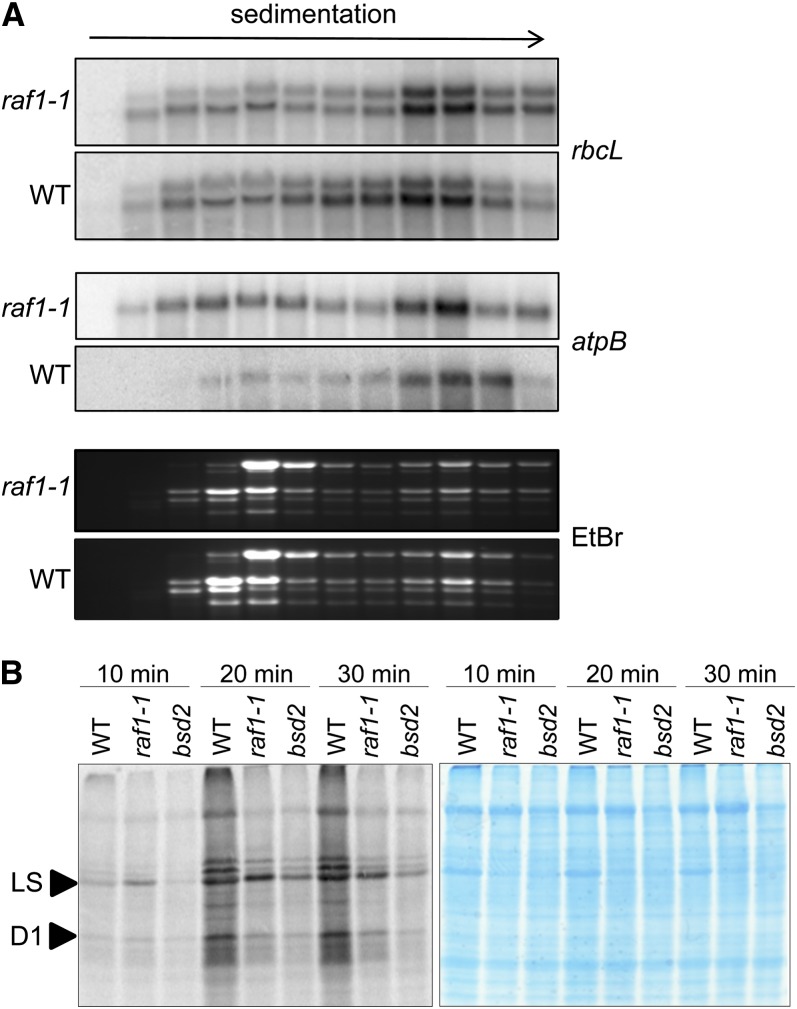
To ascertain whether a defect in the translation of rbcL leads to Rubisco deficiency, the association of rbcL transcripts with polysomes was examined. The distribution of rbcL and a control chloroplast transcript, atpB, was examined among polysome gradient fractions (Figure 4A). The distribution of the rbcL transcript was similar in the wild-type and mutant experiments, providing strong evidence that rbcL translation initiation is not disrupted in raf1 mutants. Interestingly, a larger pool of nonpolysomal atpB mRNA is found in the mutant than in the wild type, even though AtpB protein accumulates to normal levels in the mutant. Thus, the significance of this observation is unclear.
Figure 4.
The rbcL Transcript Is Translated in raf1-1.
(A) Total polysomal extracts from the apical half of the second and third leaves of raf1-1 and wild-type (WT) 12-d-old seedlings were fractionated in 15 to 55% Suc gradients. RNA was extracted from 12 fractions of equal volume and analyzed by gel blot. EtBr, ethidium bromide.
(B) In vivo protein synthesis in 10-d-old leaves of wild-type, raf1-1, and bsd2 seedlings. [35S]Met was incorporated in the presence of cycloheximide, as described in Methods. Total proteins from equal surface areas surrounding the perforations used to introduce the radiolabel were analyzed by SDS-PAGE. The left panel shows autoradiography; the right panel shows Coomassie blue staining of the same gel.
[See online article for color version of this figure.]
To investigate the possibility that a defect in the elongation or termination of LS translation was responsible for the defect in Rubisco biogenesis, we performed in vivo labeling of chloroplast proteins in leaves by uptake of radiolabeled Met in the presence of cycloheximide, an inhibitor of cytosolic translation. Samples of equal surface area were taken at three time points, and total proteins were analyzed by gel electrophoresis. Figure 4B includes a stained gel, where Rubisco deficiency can be clearly visualized for both raf1-1 and bsd2. After 10 min of labeling (at left), however, raf1 exhibited increased incorporation of radiolabel into LS relative to the wild type, with LS being identified as the labeled band that comigrated with stained LS in the same gel. When labeling was continued for a total of 20 or 30 min, the wild type incorporated more label into all proteins than either raf1 or bsd2. When this reduced overall incorporation is taken into account, along with the possible instability of LS in the mutant samples, it is clear that LS synthesis is robust in raf1. We also observed decreased labeling in raf1 of a band migrating just above LS. This is likely to be AtpB, and, if so, its reduced synthesis is in agreement with the polysome data shown in Figure 4A.
RAF1 May Be an LS Chaperone
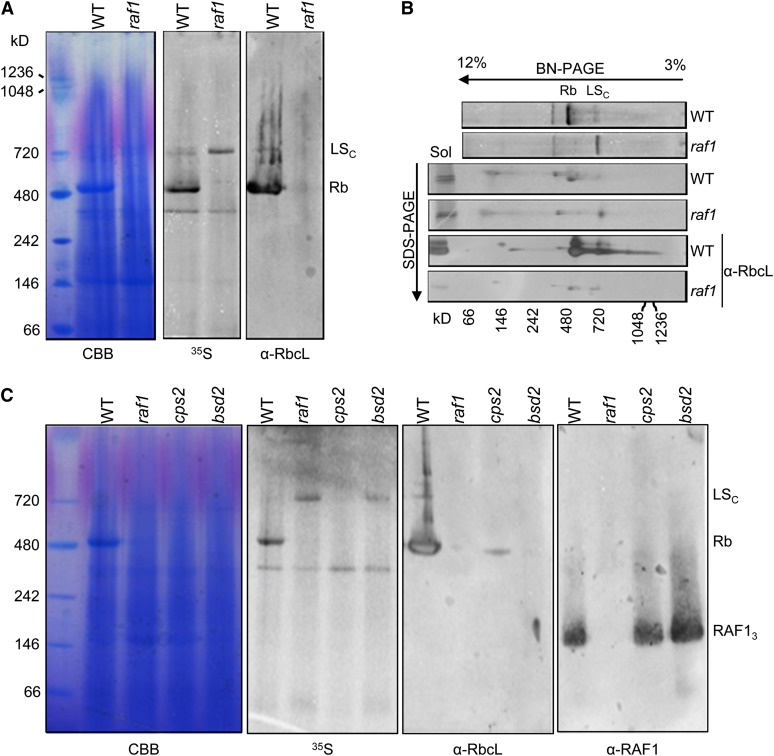
The data presented so far indicate that RAF1 is not required for LS synthesis and that it is required for assembly or stability of Rubisco. We therefore attempted to assess whether the small amount of LS that accumulates in raf1 is indeed assembled into Rubisco holoenzyme. To do so, we took advantage of the fact that LS is robustly labeled in raf1 leaves (Figure 4B). Following labeling in the presence of cycloheximide to impede cytosolic translation, native stromal proteins from the wild type and raf1 were analyzed by Blue Native (BN) gel electrophoresis (Figure 5A) and in a second denaturing dimension (Figure 5B). Figure 5A shows that after 3 h, labeled LS was assembled into Rubisco holoenzyme (Rb; ∼540 kD) in wild-type leaves, whereas the major labeled band in raf1, presumably LS, migrated at ∼800 kD. Based on previous evidence that LS-Cpn60 complexes of this approximate size form in planta (Barraclough and Ellis, 1980; Roy et al., 1982) and a recent study in which the Cpn60 homologs of GroEL were identified in this region of a native gel from Arabidopsis leaf proteins (Peng et al., 2011), we tentatively concluded that newly synthesized LS in raf1 becomes stably complexed with Cpn60; we denote this complex LSC. To visualize the 800-kD complex more clearly, a dilution series of wild-type soluble proteins was separated by BN gel electrophoresis, and an immunoblot was performed for LS (see Supplemental Figure 6 online). To verify that LS was indeed migrating at this position, a second dimension was used (Figure 5B). This showed that the putative LSC complex included a labeled band that comigrates with LS in a denaturing gel, which was verified for the wild type by immunoblotting (lane “Sol”).
Figure 5.
Native Analysis of Newly Synthesized LS.
(A) Leaf proteins of 10-d-old wild-type (WT) and raf1-1/raf1-3 seedlings were labeled for 3 h in vivo with [35S]Met. Total proteins from equal surface areas surrounding the perforations used to introduce the radiolabel were separated in a 3 to 12% native gel, which was analyzed by staining (Coomassie blue [CBB]), autoradiography (35S), and immunoblotting for LS (α-RbcL). LSC marks the migration of the putative LS-chaperone complex, and Rb the position of Rubisco holoenzyme.
(B) BN-PAGE gel lanes (top two rows, 35S shown) were separated in a second dimension 13% SDS-polyacrylamide gel and analyzed by autoradiography (middle two rows) and immunoblotting for LS (bottom two rows).
(C) In vivo–labeled leaf proteins from equal surface areas were analyzed as in (A) from the genotypes given across the top. RAF13 marks the position of the RAF1 trimer.
[See online article for color version of this figure.]
The region corresponding to the position of LSc was excised from the gel and analyzed by liquid chromatography–tandem mass spectrometry (see Supplemental Table 2 online). Several abundant metabolic enzymes were detected, and in the wild-type sample, LS and SS were abundant. Of particular interest was the detection of numerous peptides corresponding to α and β isoforms of maize chloroplast Cpn60, namely, Cpn60β1 (GRMZM2G083716) and one of the maize Cpn60α1 homologs (AC215201.3). Proteomic analysis of maize chloroplasts (Friso et al., 2010) identified four Cpn60 homologs, annotated as Cpn60β1, one Cpn60β4 (GRMZM2G042253) and two Cpn60α1, GRMZM2G434173 and AC215201.3, the latter two sharing 81% amino acid identity. In this same study, the Cpn60β4 was not detectable in BS and Cpn60α1/GRMZM2G434173 was mesophyll abundant. Thus, it was possible that the two Cpn60 homologs we detected are complexed with LS and/or SS in the LSC form.
To obtain genetic evidence that LSc is an LS-Cpn60 complex, we took advantage of the PML collection, where recent deep sequencing of individual mutants had identified an exonic Mu insertion in Cpn60α1/AC215201.3 as the causative mutation in cps2, whose severe Rubisco deficiency was reported previously (Barkan, 1993). Leaf proteins from cps2 and bsd2 mutants were labeled, before analysis by BN gel electrophoresis. Figure 5C shows that after 3 h, LSc was not identifiable in the cps2 mutant, whereas in the bsd2 mutant the labeling of LSc was similar to that observed in the raf1 mutant. Based on this biochemical and genetic evidence, our results are most consistent with RAF1 acting at a postchaperonin step to fold and/or assemble LS into Rubisco.
Figure 5C also shows immunoblot analysis that suggests that the primary form of stromal RAF1 is an ∼143-kD trimer. We also prepared recombinant RAF1 (rRAF1) lacking the predicted 30–amino acid chloroplast transit peptide, which therefore approximates the native form in chloroplasts. When rRAF1 was fractionated by size exclusion chromatography, the vast majority appears as an ∼143-kD trimer, based on the migration of size standards (see Supplemental Figure 7 online). Given the fact that both native and recombinant RAF1 migrate as apparent trimers, we suspect that RAF1 trimers interact weakly and/or transiently with other proteins in vivo, which is consistent with some previously described aspects of cyanobacterial Rubisco assembly (see Discussion).
In Vivo Cross-Linking Stabilizes a RAF1-Containing Complex
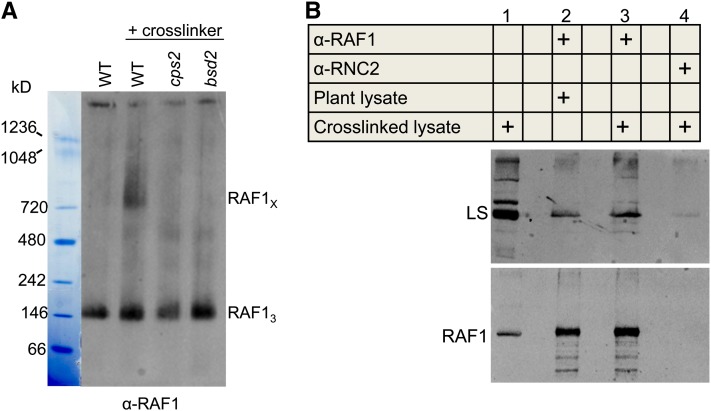
To trap transient or weakly bound complexes formed by RAF1 in plant cells, we optimized and performed reversible in vivo cross-linking in leaves of the wild type and bsd2 and cps2 mutants, by uptake of formaldehyde for 40 min. Total native proteins from equal surface areas were analyzed by BN gel electrophoresis. Immunoblot analysis using the RAF1 antibody revealed the presence of a complex of ∼750 kD, which was exclusively detected in the wild type (Figure 6A). Because both bsd2 and cps2 lack Rubisco, we conclude that the formation of this RAF1-containing complex is Rubisco dependent.
Figure 6.
In Planta Cross-Linking Stabilizes a Complex Formed by RAF1.
(A) Perforated regions of leaves from 10-d-old wild-type (WT), cps2, and bsd2 seedlings were treated with 1.85% formaldehyde for 40 min. Total soluble proteins from equal surface areas surrounding the perforations were extracted and separated in a 3 to 12% native gel, which was analyzed by immunoblotting for RAF1. RAF1X marks the migration of a putative RAF1 complex, and RAF13 the position of the RAF1 trimer. The result is representative of five independent experiments.
(B) Anti-RAF1-protein A affinity beads were used to bind RAF1 from total soluble proteins extracted from cross-linked and non-cross-linked wild-type leaves. Lane 1 was loaded with 0.4% of the input, and the eluates (lanes 2 to 4) were separated in a 13% SDS-polyacrylamide gel and analyzed by immunoblotting for LS and RAF1. To avoid saturating the RAF1 signal, eluates for the RAF1 blot were diluted 120-fold compared with the eluates used for the LS blot. Anti-RNC2-protein A affinity beads were used as a negative control; RNC2 is a nucleolar protein related to RNase III (Comella et al., 2008). Empty lanes were included between those containing samples.
[See online article for color version of this figure.]
To investigate whether the observed protein–protein interactions included one between RAF1 and LS, we prepared an affinity column where anti-RAF1 or a control antibody was conjugated to protein A. This matrix was used to immunoselect RAF1 from total soluble proteins extracted from wild-type plants in the presence or absence of cross-linking. Proteins bound to the column were eluted and the cross-linking reversed using heat, and immunoblot analysis was performed for RAF1 and LS. As shown in Figure 6B, LS was detected at a low background level when the control antibody raised against the nucleolar RNA binding protein RNC2 was used (lane 4). A stronger LS signal was obtained when RAF1 was selected without cross-linking (lane 2), whereas following cross-linking the signal increased further (lane 3). The enhanced selection of LS following cross-linking was consistent in multiple experimental repetitions and indicates that cross-linking stabilizes what is otherwise a somewhat labile interaction.
RAF1 Expression in E. coli Does Not Facilitate Maize Rubisco Assembly
RAF1 is not found in bacteria, and it was possible that previous attempts to express plant Rubisco in E. coli had failed because of this. As a preliminary attempt to test this hypothesis, we expressed maize LS with or without RAF1 and a variety of known or putative maize chaperones, including BSD2, Cpn60, Cpn20, RBCX, and SS. As shown in Supplemental Figure 8 online, the majority of LS remained insoluble even in the presence of RAF1 or RAF1 and BSD2 (lanes 2 to 4). When additional chaperones were added, overall LS expression levels dropped dramatically; however, solubility did not increase. We conclude that RAF1 expression is not sufficient to facilitate assembly of maize Rubisco in E. coli.
DISCUSSION
RAF1, a previously undescribed gene for Rubisco accumulation, was found via a genetic screen for maize mutants that specifically lack Rubisco. The gene was identified by transposon tagging, aided by a high-throughput method for identifying sequences flanking Mu insertions. RAF1 appears to be restricted to the green lineage, joining two other Rubisco-related regulators in this class. One of these is BSD2, mentioned above, which has been hypothesized to act at a co- or posttranslational step. Given its homology to DnaJ chaperones (Brutnell et al., 1999), BSD2 has been hypothesized to be involved in folding or assembly of newly synthesized LS, or in the formation of higher-order oligomers (Nishimura et al., 2008), which suggests a close cooperativity with RAF1. Unlike RAF1, however, BSD2 is found roughly equally in bundle sheath and mesophyll chloroplasts, both at the protein and transcript levels (Friso et al., 2010; Li et al., 2010). Furthermore, RAF1 accumulation is independent of the presence of BSD2 (Figure 2C); therefore, if an association occurs between the two proteins, it is not required for RAF1 stability.
To deduce the stage at which RAF1 is required for Rubisco biogenesis, the transcript abundance for the Rubisco subunit genes and BSD2 was analyzed. We found wild-type levels of BSD2 transcripts and a decrease in RBCS mRNA consistent with previous studies of Rubisco mutants. We also observed increased abundance of rbcL mRNA in mesophyll cells (Figure 3C), as previously reported for bsd2 (Roth et al., 1996; Brutnell et al., 1999). This observation was rationalized in terms of increased rbcL transcript stability in this cell type, stemming from a requirement for BSD2 to deplete ribosomes from rbcL mRNA in mesophyll chloroplasts and thus expose it for degradation. Indeed, instability of the rbcL mRNA rather than reduced rbcL transcription explains its low accumulation in mesophyll cells of wild-type plants (Boinski et al., 1993; Kubicki et al., 1994). Although RAF1 is more abundant in bundle sheath than in mesophyll cells (Figure 2D), this does not exclude its participation in the mechanism proposed for BSD2. Alternatively, the rbcL mRNA phenomenon in raf1 may be a pleiotropic effect.
Inefficient translation of rbcL transcripts was another possible explanation for raf1 Rubisco deficiency. However, neither translation initiation nor elongation is negatively affected in raf1 mutants (Figure 4). Since unassembled LS can repress its own translation through an autoregulatory circuit (Wostrikoff and Stern, 2007), RAF1 appears to be required for folding or assembly of LS into a repression-competent form. RAF1 could also be involved in the association of LS with SS, and we cannot completely rule out that it is required for the stability of fully assembled Rubisco.
To investigate the fate of LS in raf1 mutants, we used native gel electrophoresis of newly labeled proteins (Figure 5A). The data suggest that the newly synthesized LS is trapped in a 800- to 900-kD complex that may include chloroplast chaperonins. A GroEL chaperone-LS complex accumulated when the in vitro synthesis of cyanobacterial LS was conducted in the absence of RBCX (Saschenbrecker et al., 2007) and when the in vitro reconstitution of Rubisco holoenzyme failed due to low affinity of Synechococcus sp PCC6301 RBCX for LS from the same strain (Li et al., 2010). In the latter case, the addition of increasing amounts of high-affinity RBCX led to gradual accumulation of the octameric core of Rubisco, L8, although a fraction of LS remained as a GroEL-bound complex (Li et al., 2010).
In chaperonin complexes, tetradecameric GroEL cooperates with a seven-subunit lid of Cpn10 (GroES in bacteria) to form a protein-folding cage. In chloroplasts, a tandem repeat of two Cpn10 domains is also expressed; this is termed Cpn21 (Bertsch et al., 1992; Hill and Hemmingsen, 2001). Multiple GroEL/Cpn60 isoforms are found in chloroplasts, including those of maize (Friso et al., 2010). Recently, the specificity of Arabidopsis chloroplast Cpn60β4 for the NADH dehydrogenase complex was shown (Peng et al., 2011). Cpn60β4 is a minor chaperonin that forms heterooligomeric complexes with Cpn60α1 and Cpn60β1-3, as shown by mass spectrometry analysis of a native gel band containing the chaperonin complexes. When we conducted a similar analysis of the native gel band corresponding to the LSc region (Figure 5), CPS2 (one of two Cpn60α1 in maize) and Cpn60β1 were the only Cpn60 isoforms detected (see Supplemental Table 2 online). The presence of CPS2 is consistent with our observation that it is required for formation of LSc (Figure 5C).
Labeled LSc was not detected in the cps2 mutant but was found at similar levels in the bsd2 and raf1 mutants. This suggests that BSD2 and RAF1 act subsequent to the formation of the Cpn60-Cpn21/10 chaperonin complex, which is likely to mediate the initial folding of LS in chloroplasts (Figure 7). The majority of obligate chaperonin substrates, including bacterial LS, tend to form kinetically trapped folding intermediates (Hartl and Hayer-Hartl, 2009b), and chaperonins function by performing multiple rounds of binding to such intermediates, maintaining them in an unfolded state inside the cage and releasing them to attempt to fold in the cage or in solution (Weissman et al., 1994; Brinker et al., 2001; Chakraborty et al., 2010). A Ser-to-Phe change at position 112 of Rubisco large subunit in a mutant of tobacco halted the progression of LS from a chaperonin-constituent complex to holoenzyme assembly, consistent with such a pathway (Avni et al., 1989)
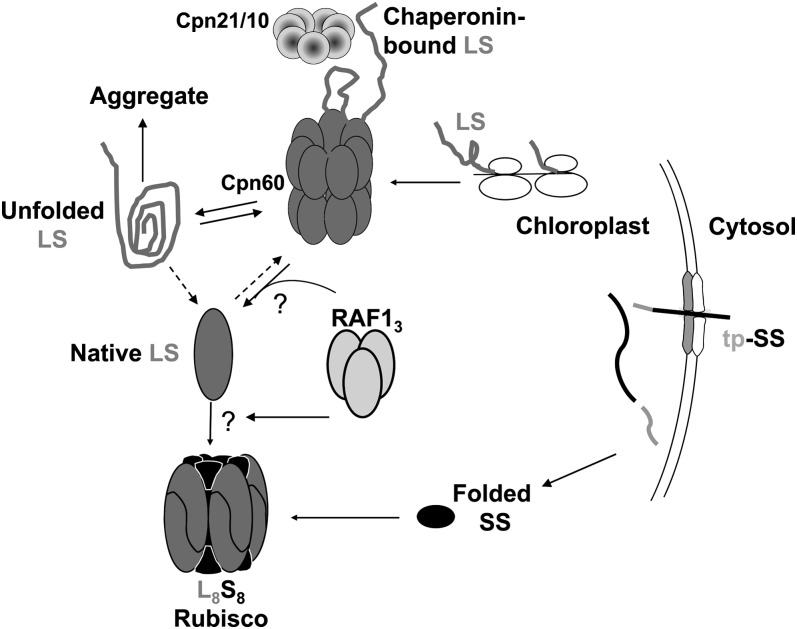
Figure 7.
Model for Role of RAF1 in Rubisco Assembly.
Counterclockwise from top right; newly synthesized LS interacts with the chaperonin complex, which leads to correct folding (Native LS) or aggregation and proteolysis. Trimeric RAF1 would then act to promote the formation of dimeric and/or octameric LS, perhaps in concert with BSD2 (data not shown). In the absence of RAF1, LS would be unable to escape from the chaperonin cycle, ultimately leading to aggregation and proteolysis. “Chaperonin-bound LS” is equivalent to LSc. See Nishimura et al. (2008) for details of other molecular partners involved in Rubisco biogenesis.
As summarized in Figure 7, our results suggest a model in which RAF1 helps assemble LS prior to holoenzyme formation, a role similar to that predicted for RBCX in cyanobacteria. We note that RBCX homologs are encoded in plants and RBCX is transcribed in maize leaves (Li et al., 2010), but unlike RAF1, RBCX was not detected in the maize chloroplast proteome (Friso et al., 2010), as might be expected for a protein with a critical role in assembling the highly abundant Rubisco.
The RBCX-LS interaction during cyanobacterial Rubisco assembly is dynamic, leading to its lability during both gel filtration and native gel electrophoresis (Saschenbrecker et al., 2007). Likewise, while we could detect the RAF1 trimer, no other RAF1-containing complexes were immediately obvious when chloroplast extract was size fractionated. However, in planta cross-linking resulted in the reproducible formation of a larger complex containing RAF1 (Figure 6A). To investigate whether the complex included both RAF1 and LS, immunoselection using the RAF1 antibody was conducted in the presence and absence of cross-linker. The results showed that a very small fraction of the total LS copurifies with RAF1 under these conditions and that cross-linking increases its recovery (Figure 6B). For RbcX, in vitro assembly protocols have been developed (Saschenbrecker et al., 2007; Bracher et al., 2011), which may serve as a guide for further analysis of RAF1’s mechanism of action in Rubisco biogenesis.
The interaction between RAF1 and LS lends further support to our hypothesis that RAF1 is directly required for LS assembly. Because the LSc complex also predominates in the bsd2 mutant, we tentatively propose a postchaperonin role for BSD2, although it may act at a different step than RAF1. As a DnaJ domain-containing protein, BSD2 is a member of a diverse class of cofactors that specify HSP70 function by either directing them to protein clients at precise locations in cells or binding substrates directly, thereby delivering specific clients to HSP70 and directly determining their fate. HSP70 functions at the translational or posttranslational stage (Kampinga and Craig, 2010) by distributing proteins from polysomes to downstream chaperones, such as chaperonins or HSP90 (Langer et al., 1992; Hartl and Hayer-Hartl, 2009a). However, there is no evidence for HSP70 having a role in folding or assembly of Rubisco large subunit nor was it required for in vitro reconstitution of cyanobacterial Rubisco holoenzyme (Liu et al., 2010).
The fact that GroEL-GroES overproduction permits the growth of E. coli cells lacking both of the other two groups of translational chaperones, TF (Trigger factor) and DnaK (Hsp70), along with the observation of polysome association for GroEL-ES, suggested a versatile role for them in cotranslational folding (Ying et al., 2005). This leads us to question whether Hsp70 and the J protein cofactors, such as BSD2, are necessary for LS folding. There is evidence, however, that HSP70 and Cpn60 might individually or cooperatively act on the translocation of Rubisco small subunit across the chloroplast envelope and its subsequent refolding (Kessler and Blobel, 1996; Ivey et al., 2000; Su and Li, 2010). In summary, whether BSD2 regulates Rubisco assembly by direct interaction with either one of the Rubisco subunits or with other chaperones, such as RAF1, Cpn60, or HSP70, remains to be determined. Although our initial attempts to develop a bacterial model for Rubisco assembly were unsuccessful (see Supplemental Figure 7 online), perhaps due to this complexity or specialization of chaperone function, the identification of RAF1 is a further step toward the eventual goal of achieving cell free or bacterial assembly of plant Rubisco.
METHODS
Plant Material and Cloning
The reference raf1-1 allele was identified from the PML collection (http://pml.uoregon.edu/photosyntheticml.html) as described in Results. Phenotypically normal siblings of raf1-1 were self-crossed and outcrossed to inbred A632. The mutation was propagated through multiple rounds of outcrossing and self-pollination. Selfed ears showing cosegregation for pale-green and low Rubisco traits were used as the source of mutant individuals for the cosegregation analysis that identified genetically linked Mu transposon insertions. The method for identification of Mu flanking sequences is published (Williams-Carrier et al., 2010). PCR verification of the genotypes of raf1-1 mutant plants and their wild-type cousins used the gene-specific primer pairs ZmRAF1-FOR1 and ZmRAF1-REV1 to amplify wild-type alleles or one of them with a mixture of Mu-specific primers, EoMu1 and EoMu2, to identify the mutant allele. A list of all primers is presented in Supplemental Table 3 online.
The raf1-2 and raf1-3 alleles were identified in a reverse-genetic screen of the PML collection by amplification of pooled mutant DNAs with ZmRAF1-REV2 and the EoMu primer mix (Williams and Barkan, 2003). ZmRAF1-FOR2, ZmRAF1-REV2, and the EoMu primer mix were used to validate the insertion sites by sequencing and to genotype the plants harboring new alleles prior to complementation crosses. The cps2 mutant was reported previously (Barkan, 1993). The causal insertion in the Cpn60α1 homolog AC215201.3 was identified via the same Illumina-based method as used to identify raf1 (R. Williams-Carrier, S. Belcher, and A. Barkan, unpublished data). All plants were grown in soil under a 16-h light (28°C)/8-h dark (26°C) cycle and harvested between 9 and 12 d after planting.
PCR reactions used Phusion polymerase (Finnzymes) with the GC buffer supplied with the enzyme and the following reaction profile: 98°C for 30 s, followed by 35 cycles of 98°C for 10 s, 60°C for 15 s, and 72°C for 30 s, with a final extension of 72°C for 10 min.
Production and Analysis of rRAF1 and Production of Anti-RAF1 Antisera
The raf1 coding region preceded by glutathione S-transferase was expressed from pGEX 4T-1 (GE Healthcare). The soluble extracted protein was applied to a Glutathione Sepharose 4 Fast Flow (GE Healthcare) column, followed by on-column digestion by thrombin. RAF1 was further purified by Superdex-200 size exclusion chromatography. Antisera were generated by Lampire Biological Laboratories.
Gene Expression Analysis
Total RNA was extracted using Tri-reagent (Molecular Research Center) and 3 to 5 µg were analyzed as described (Cahoon et al., 2004). Gene-specific probes were generated by PCR using the following primer pairs: ZmrbcL-Cod1+ZmrbcL-Rev1, ZmRBCS2-Cod1+ZmRBCS2-Rev1, and ZmPsbA-5′+ZmPsbA-3′. Mesophyll and bundle sheath preparations were as described (Markelz et al., 2003). cDNA synthesis using random hexamers followed by quantitative PCR was performed as described (Hotto et al., 2010). Relative quantification compared with wild-type samples (given a reference value of 1) was achieved after normalization to actin mRNA by the Bio-Rad CFX Manager software. The final data are an average of two biological and three technical replicates. Quantitative PCR used the following primers: Zmqactin-1F and Zmqactin-1R for actin, ZmqLS-1F and ZmqLS-123R for rbcL, ZmqSS-9F and ZmqSS-9R for RBCS1, ZmqME-1F and ZmqME-1R for malic enzyme, and ZmqMDH-1F and ZmqMDH-1R for malate dehydrogenase. Polysomes were analyzed from an extract prepared by grinding 200 mg of tissue in 1 mL of polysome extraction buffer as described (Barkan, 1998).
Protein Analysis
Total protein was extracted from 60 mg of the second leaf as described (Barkan, 1998), and a volume corresponding to 15 to 150 µg of tissue was analyzed using 13% SDS-polyacrylamide gels. To obtain an insoluble fraction, the tissue ground in homogenization buffer was centrifuged at 13,000g for 5 min at 4°C before adding gel loading buffer to both supernatant and pellet. Proteins were analyzed by transfer to polyvinylidene difluoride membranes and chemiluminescence using the ECL Plus Western Blotting Detection System on a Storm Scanner 840 (GE Healthcare). In vivo pulse labeling was performed as described by Barkan (1998) after perforating a band across the top 2-cm segment of the second leaf from 8-d-old seedlings. The labeling mix was prepared from 50 μL of 20 μg/mL cycloheximide in 10 mM NaPhosphate (pH 6.8), 50 μL of [35S]Met (500 μCi), and 2 μL of 10% bromophenol blue. Each labeling experiment was repeated three times.
For BN gel electrophoresis, total soluble proteins were extracted from 100 mg of the second leaf by homogenization in a buffer containing 20 mM Tris-HCl, pH 9.0, 250 mM NaCl, 50 mM NaHCO3, 4 mM MgCl2, and an EDTA-free protease inhibitor cocktail (Roche). After removal of cell debris by centrifugation for 5 min at 13,000g and 4°C, a volume corresponding to 4 mg of tissue was loaded on 3 to 12% bis-Tris 1-mm gels (Invitrogen). The native gel anode buffer was composed of 50 mM bis-Tris and Tricine each at pH 6.8. The dark and light cathode buffers were prepared from the anode buffer with 0.02 and 0.002% Coomassie Brilliant Blue G 250, respectively. BN gels were run in a cold room for 30 min at 150 V before changing the cathode buffer from dark to light and continuing electrophoresis at 250 V. Gels were stained with Instant Coomassie blue (Expedeon) before drying.
Peptide Preparation for Tandem Mass Spectrometry Analysis and Database Searches
For in-gel protein digestion, excised gel bands were washed, reduced, Cys-alkylated, and digested with trypsin (Promega) overnight at 37°C (Shevchenko et al., 2006). Liquid chromatography–tandem mass spectrometry analyses were performed on an integrated Agilent 1100 CapLC, HPLC-Chip column (150 mm, 300A, C18, 160 nL trap) connected in-line to a QTOF-6520 mass spectrometer controlled with MassHunter, Workstation Acquisition Qualitative Analysis (B.04.00). Tandem mass spectrometry spectra were searched with Mascot version 2.2 (Matrix Science) against maize genome release 4a.53 (53,764 models) from http://www.maizesequence.org/ supplemented with the plastid-encoded proteins (111 protein models) and mitochondria-encoded proteins (165 protein models) with the following search parameters: peptide tolerance, 30 mmu; tandem mass spectrometry tolerance, 30 mmu; peptide charge, 1+, 2+, or 3+; trypsin as enzyme allowing up to one missed cleavage; and carboxymethylation on Cys and oxidation on Met as a variable modification.
Formaldehyde Cross-Linking and Coimmunoprecipitation
Introduction of formaldehyde for in vivo cross-linking was performed similarly to the introduction of radiolabeled amino acids for in vivo protein labeling. The cross-linking mix contained 10 mM NaPhosphate, pH 6.8, and 1.85% formaldehyde. For coimmunoprecipitation, 2 g of the cross-linked or 1 g of non-cross-linked leaf tissue was collected from the top 4 cm of the second leaf and used to prepare total soluble protein. Five milliliters of non-cross-linked or 10 mL of cross-linked lysate was filtered through an Express PLUS Membrane (0.22 µm; Millipore) to remove particulates, concentrated to 0.5 mL of non-cross-linked and 1 mL of cross-linked lysate by centrifugation at 4°C in an Ultracel (50- kD cutoff; Millipore), and precleared by incubation with protein A on ice for 30 min. The cross-linked lysate was evenly divided into two samples, which were added to either anti-RAF1 or anti-RNC2-bound protein A affinity beads, prepared as previously described (Watkins et al., 2007), followed by a rotational incubation at 4°C for 90 min. The non-cross-linked lysate was incubated with anti-RAF1-bound protein A affinity beads. The beads were washed 10 times in Tris-buffered saline, pH 7.5, and 0.5% Nonidet P-40, and proteins were eluted from the beads by two sequential incubations in 200 μL of elution buffer (5 mM sodium phosphate, pH 6.8, and 3% SDS). The eluates were pooled, concentrated by methanol/chloroform precipitation, dissolved in 40 μL SDS loading buffer, treated at 100°C for 15 min to reverse the cross-linking, and analyzed by SDS-PAGE (13.5%). To prevent IgG heavy-chain interference with LS and RAF1 detection, the clean-blot IP detection reagent (horseradish peroxidase; Thermo Scientific) was used at a dilution of 1:400.
Accession Number
Protein sequence data from this article can be found in the GenBank/EMBL data libraries under accession number NP_001140763.
Supplemental Material
The following materials are available in the online version of this article.
Supplemental Figure 1. Accumulation of the BSD2 Transcript in raf1.
Supplemental Figure 2. Validation of RAF1 Gene Identification.
Supplemental Figure 3. Gene-Specific PCR Validating the Locations of raf1 Insertions.
Supplemental Figure 4. Complementation Crosses and Protein Analysis.
Supplemental Figure 5. Alignment of RAF1 Homologs.
Supplemental Figure 6. Native Analysis of Native LS from Plant Total Soluble Protein.
Supplemental Figure 7. Analysis of Recombinant RAF1.
Supplemental Figure 8. Coexpression of LS and Rubisco Biogenesis Factors in E. coli.
Supplemental Table 1. Accession Numbers of RAF1 Sequences from Green Species.
Supplemental Table 2. Mass Spectrometry Data Derived from the LSc Gel Region.
Supplemental Table 3. Primers Used in This Work.
Supplementary Material
Acknowledgments
We thank Navid Movahed, in Brian Bothner’s laboratory, Montana State University, for his technical assistance in protein purification, size exclusion chromatography, and mass spectrometry, Tiffany Kroeger (Barkan lab, University of Oregon) for assistance with the reverse-genetic screen, Nicholas Stiffler (Barkan lab, University of Oregon) for bioinformatic analysis of Mu-Illumina sequencing data, and Giulia Friso in Klaas van Wijk’s lab for providing the Mascot search against their assembled maize database. We also thank Kevin Ahern and other staff in Tom Brutnell’s laboratory, Boyce Thompson Institute, for helpful technical discussions. This project was supported by the National Research Initiative Competitive Grants Program Grant 2009-35318-05012 from the USDA National Institute of Food and Agriculture to D.B.S. and by National Science Foundation Grant IOS-0922560 to A.B.
AUTHOR CONTRIBUTIONS
All authors contributed to experimental design and data analysis. L.F., K.W., R.W.-C., and S.B. performed research. L.F., K.W., A.B., and D.B.S. wrote the article.
Glossary
- Rubisco
ribulose-1,5-bis-phosphate carboxylase/oxygenase
- PML
photosynthetic mutant library
- BN
Blue Native
- LS
large (50-kD) subunits
- SS
small (13–15 kD) subunits
References
- Andersson I., Backlund A. (2008). Structure and function of Rubisco. Plant Physiol. Biochem. 46: 275–291 [DOI] [PubMed] [Google Scholar]
- Andrews T.J. (1988). Catalysis by cyanobacterial ribulose-bisphosphate carboxylase large subunits in the complete absence of small subunits. J. Biol. Chem. 263: 12213–12219 [PubMed] [Google Scholar]
- Avni A., Edelman M., Rachailovich I., Aviv D., Fluhr R. (1989). A point mutation in the gene for the large subunit of ribulose 1,5-bisphosphate carboxylase/oxygenase affects holoenzyme assembly in Nicotiana tabacum. EMBO J. 8: 1915–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannai H., Tamada Y., Maruyama O., Nakai K., Miyano S. (2002). Extensive feature detection of N-terminal protein sorting signals. Bioinformatics 18: 298–305 [DOI] [PubMed] [Google Scholar]
- Barkan A. (1993). Nuclear mutants of maize with defects in chloroplast polysome assembly have altered chloroplast RNA metabolism. Plant Cell 5: 389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. (1998). Approaches to investigating nuclear genes that function in chloroplast biogenesis in land plants. Methods Enzymol. 297: 38–57 [Google Scholar]
- Barraclough R., Ellis R.J. (1980). Protein synthesis in chloroplasts. IX. Assembly of newly-synthesized large subunits into ribulose bisphosphate carboxylase in isolated intact pea chloroplasts. Biochim. Biophys. Acta 608: 19–31 [DOI] [PubMed] [Google Scholar]
- Bertsch U., Soll J., Seetharam R., Viitanen P.V. (1992). Identification, characterization, and DNA sequence of a functional “double” groES-like chaperonin from chloroplasts of higher plants. Proc. Natl. Acad. Sci. USA 89: 8696–8700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boinski J.J., Wang J.L., Xu P., Hotchkiss T., Berry J.O. (1993). Post-transcriptional control of cell type-specific gene expression in bundle sheath and mesophyll chloroplasts of Amaranthus hypochondriacus. Plant Mol. Biol. 22: 397–410 [DOI] [PubMed] [Google Scholar]
- Bracher A., Starling-Windhof A., Hartl F.U., Hayer-Hartl M. (2011). Crystal structure of a chaperone-bound assembly intermediate of form I Rubisco. Nat. Struct. Mol. Biol. 18: 875–880 [DOI] [PubMed] [Google Scholar]
- Brinker A., Pfeifer G., Kerner M.J., Naylor D.J., Hartl F.U., Hayer-Hartl M. (2001). Dual function of protein confinement in chaperonin-assisted protein folding. Cell 107: 223–233 [DOI] [PubMed] [Google Scholar]
- Broadhurst R.W., Nietlispach D., Wheatcroft M.P., Leadlay P.F., Weissman K.J. (2003). The structure of docking domains in modular polyketide synthases. Chem. Biol. 10: 723–731 [DOI] [PubMed] [Google Scholar]
- Brutnell T.P., Sawers R.J., Mant A., Langdale J.A. (1999). BUNDLE SHEATH DEFECTIVE2, a novel protein required for post-translational regulation of the rbcL gene of maize. Plant Cell 11: 849–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon A.B., Harris F.M., Stern D.B. (2004). Analysis of developing maize plastids reveals two mRNA stability classes correlating with RNA polymerase type. EMBO Rep. 5: 801–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty K., Chatila M., Sinha J., Shi Q.Y., Poschner B.C., Sikor M., Jiang G.X., Lamb D.C., Hartl F.U., Hayer-Hartl M. (2010). Chaperonin-catalyzed rescue of kinetically trapped states in protein folding. Cell 142: 112–122 [DOI] [PubMed] [Google Scholar]
- Comella P., Pontvianne F., Lahmy S., Vignols F., Barbezier N., Debures A., Jobet E., Brugidou E., Echeverria M., Sáez-Vásquez J. (2008). Characterization of a ribonuclease III-like protein required for cleavage of the pre-rRNA in the 3’ETS in Arabidopsis. Nucleic Acids Res. 36: 1163–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., Brunak S., von Heijne G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Emlyn-Jones D., Woodger F.J., Price G.D., Whitney S.M. (2006). RbcX can function as a rubisco chaperonin, but is non-essential in Synechococcus PCC7942. Plant Cell Physiol. 47: 1630–1640 [DOI] [PubMed] [Google Scholar]
- Erion J. (1985). Characterization of the mRNA transcripts of the maize, ribulose-1,5-bisphosphate carboxylase, large subunit gene. Plant Mol. Biol. 4: 169–179 [DOI] [PubMed] [Google Scholar]
- Feller U., Anders I., Mae T. (2008). Rubiscolytics: Fate of Rubisco after its enzymatic function in a cell is terminated. J. Exp. Bot. 59: 1615–1624 [DOI] [PubMed] [Google Scholar]
- Friso G., Majeran W., Huang M., Sun Q., van Wijk K.J. (2010). Reconstruction of metabolic pathways, protein expression, and homeostasis machineries across maize bundle sheath and mesophyll chloroplasts: Large-scale quantitative proteomics using the first maize genome assembly. Plant Physiol. 152: 1219–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatenby A.A. (1984). The properties of the large subunit of maize ribulose bisphosphate carboxylase/oxygenase synthesised in Escherichia coli. Eur. J. Biochem. 144: 361–366 [DOI] [PubMed] [Google Scholar]
- Gatenby A.A., Castleton J.A., Saul M.W. (1981). Expression in E. coli of maize and wheat chloroplast genes for large subunit of ribulose bisphosphate carboxylase. Nature 291: 117–121 [DOI] [PubMed] [Google Scholar]
- Gatenby A.A., van der Vies S.M., Rothstein S.J. (1987). Co-expression of both the maize large and wheat small subunit genes of ribulose-bisphosphate carboxylase in Escherichia coli. Eur. J. Biochem. 168: 227–231 [DOI] [PubMed] [Google Scholar]
- Goloubinoff P., Christeller J.T., Gatenby A.A., Lorimer G.H. (1989a). Reconstitution of active dimeric ribulose bisphosphate carboxylase from an unfoleded state depends on two chaperonin proteins and Mg-ATP. Nature 342: 884–889 [DOI] [PubMed] [Google Scholar]
- Goloubinoff P., Gatenby A.A., Lorimer G.H. (1989b). GroE heat-shock proteins promote assembly of foreign prokaryotic ribulose bisphosphate carboxylase oligomers in Escherichia coli. Nature 337: 44–47 [DOI] [PubMed] [Google Scholar]
- Hartl F.U., Hayer-Hartl M. (2009a). Converging concepts of protein folding in vitro and in vivo. Nat. Struct. Mol. Biol. 16: 574–581 [DOI] [PubMed] [Google Scholar]
- Hartl F.U., Hayer-Hartl M. (2009b). Converging concepts of protein folding in vitro and in vivo. Nat. Struct. Mol. Biol. 16: 574–581 [DOI] [PubMed] [Google Scholar]
- Hill J.E., Hemmingsen S.M. (2001). Arabidopsis thaliana type I and II chaperonins. Cell Stress Chaperones 6: 190–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotto A.M., Huston Z.E., Stern D.B. (2010). Overexpression of a natural chloroplast-encoded antisense RNA in tobacco destabilizes 5S rRNA and retards plant growth. BMC Plant Biol. 10: 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivey R.A., III, Subramanian C., Bruce B.D. (2000). Identification of a Hsp70 recognition domain within the rubisco small subunit transit peptide. Plant Physiol. 122: 1289–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson X., Wostrikoff K., Finazzi G., Kuras R., Schwarz C., Bujaldon S., Nickelsen J., Stern D.B., Wollman F.A., Vallon O. (2010). MRL1, a conserved Pentatricopeptide repeat protein, is required for stabilization of rbcL mRNA in Chlamydomonas and Arabidopsis. Plant Cell 22: 234–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga H.H., Craig E.A. (2010). The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11: 579–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanevski I., Maliga P. (1994). Relocation of the plastid rbcL gene to the nucleus yields functional ribulose-1,5-bisphosphate carboxylase in tobacco chloroplasts. Proc. Natl. Acad. Sci. USA 91: 1969–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler F., Blobel G. (1996). Interaction of the protein import and folding machineries of the chloroplast. Proc. Natl. Acad. Sci. USA 93: 7684–7689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesiński P., Piechota J., Szczepaniak A. (2011). Initial characteristics of RbcX proteins from Arabidopsis thaliana. Plant Mol. Biol. 77: 447–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicki A., Steinmüller K., Westhoff P. (1994). Differential transcription of plastome-encoded genes in the mesophyll and bundle-sheath chloroplasts of the monocotyledonous NADP-malic enzyme-type C4 plants maize and sorghum. Plant Mol. Biol. 25: 669–679 [DOI] [PubMed] [Google Scholar]
- Langer T., Lu C., Echols H., Flanagan J., Hayer M.K., Hartl F.U. (1992). Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature 356: 683–689 [DOI] [PubMed] [Google Scholar]
- Larimer F.W., Soper T.S. (1993). Overproduction of Anabaena 7120 ribulose-bisphosphate carboxylase/oxygenase in Escherichia coli. Gene 126: 85–92 [DOI] [PubMed] [Google Scholar]
- Li P., et al. (2010). The developmental dynamics of the maize leaf transcriptome. Nat. Genet. 42: 1060–1067 [DOI] [PubMed] [Google Scholar]
- Liu C., Young A.L., Starling-Windhof A., Bracher A., Saschenbrecker S., Rao B.V., Rao K.V., Berninghausen O., Mielke T., Hartl F.U., Beckmann R., Hayer-Hartl M. (2010). Coupled chaperone action in folding and assembly of hexadecameric Rubisco. Nature 463: 197–202 [DOI] [PubMed] [Google Scholar]
- Markelz N.H., Costich D.E., Brutnell T.P. (2003). Photomorphogenic responses in maize seedling development. Plant Physiol. 133: 1578–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Cajar O., Whitney S.M. (2008). Directing the evolution of Rubisco and Rubisco activase: First impressions of a new tool for photosynthesis research. Photosynth. Res. 98: 667–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K., Ogawa T., Ashida H., Yokota A. (2008). Molecular mechanisms of Rubisco biosynthesis in higher plants. Plant Biotechnol. 25: 285–290 [Google Scholar]
- Onizuka T., Endo S., Akiyama H., Kanai S., Hirano M., Yokota A., Tanaka S., Miyasaka H. (2004). The rbcX gene product promotes the production and assembly of ribulose-1,5-bisphosphate carboxylase/oxygenase of Synechococcus sp. PCC7002 in Escherichia coli. Plant Cell Physiol. 45: 1390–1395 [DOI] [PubMed] [Google Scholar]
- Parry M.A., Andralojc P.J., Mitchell R.A., Madgwick P.J., Keys A.J. (2003). Manipulation of Rubisco: The amount, activity, function and regulation. J. Exp. Bot. 54: 1321–1333 [DOI] [PubMed] [Google Scholar]
- Peng L.W., Fukao Y., Myouga F., Motohashi R., Shinozaki K., Shikanai T. (2011). A chaperonin subunit with unique structures is essential for folding of a specific substrate. PLoS Biol. 9: e1001040. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Prasad B.D., Goel S., Krishna P. (2010). In silico identification of carboxylate clamp type tetratricopeptide repeat proteins in Arabidopsis and rice as putative co-chaperones of Hsp90/Hsp70. PLoS ONE 5: e12761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodermel S., Haley J., Jiang C.Z., Tsai C.H., Bogorad L. (1996). A mechanism for intergenomic integration: Abundance of ribulose bisphosphate carboxylase small-subunit protein influences the translation of the large-subunit mRNA. Proc. Natl. Acad. Sci. USA 93: 3881–3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth R., Hall L.N., Brutnell T.P., Langdale J.A. (1996). bundle sheath defective2, a mutation that disrupts the coordinated development of bundle sheath and mesophyll cells in the maize leaf. Plant Cell 8: 915–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy H., Bloom M., Milos P., Monroe M. (1982). Studies on the assembly of large subunits of ribulose bisphosphate carboxylase in isolated pea chloroplasts. J. Cell Biol. 94: 20–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saschenbrecker S., Bracher A., Rao K.V., Rao B.V., Hartl F.U., Hayer-Hartl M. (2007). Structure and function of RbcX, an assembly chaperone for hexadecameric Rubisco. Cell 129: 1189–1200 [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Tomas H., Havlis J., Olsen J.V., Mann M. (2006). In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1: 2856–2860 [DOI] [PubMed] [Google Scholar]
- Stern D.B., Hanson M.R., Barkan A. (2004). Genetics and genomics of chloroplast biogenesis: Maize as a model system. Trends Plant Sci. 9: 293–301 [DOI] [PubMed] [Google Scholar]
- Su P.H., Li H.M. (2010). Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. Plant Cell 22: 1516–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabita F.R. (1999). Microbial ribulose 1,5-bisphosphate carboxylase/oxygenase: A different perspective. Photosynth. Res. 60: 1–28 [Google Scholar]
- van der Vies S.M., Bradley D., Gatenby A.A. (1986). Assembly of cyanobacterial and higher plant ribulose bisphosphate carboxylase subunits into functional homologous and heterologous enzyme molecules in Escherichia coli. EMBO J. 5: 2439–2444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins K.P., Kroeger T.S., Cooke A.M., Williams-Carrier R.E., Friso G., Belcher S.E., van Wijk K.J., Barkan A. (2007). A ribonuclease III domain protein functions in group II intron splicing in maize chloroplasts. Plant Cell 19: 2606–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman J.S., Kashi Y., Fenton W.A., Horwich A.L. (1994). GroEL-mediated protein folding proceeds by multiple rounds of binding and release of nonnative forms. Cell 78: 693–702 [DOI] [PubMed] [Google Scholar]
- Whitney S.M., Kane H.J., Houtz R.L., Sharwood R.E. (2009). Rubisco oligomers composed of linked small and large subunits assemble in tobacco plastids and have higher affinities for CO2 and O2. Plant Physiol. 149: 1887–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams P.M., Barkan A. (2003). A chloroplast-localized PPR protein required for plastid ribosome accumulation. Plant J. 36: 675–686 [DOI] [PubMed] [Google Scholar]
- Williams-Carrier R., Stiffler N., Belcher S., Kroeger T., Stern D.B., Monde R.A., Coalter R., Barkan A. (2010). Use of Illumina sequencing to identify transposon insertions underlying mutant phenotypes in high-copy Mutator lines of maize. Plant J. 63: 167–177 [DOI] [PubMed] [Google Scholar]
- Wostrikoff K., Stern D. (2007). Rubisco large-subunit translation is autoregulated in response to its assembly state in tobacco chloroplasts. Proc. Natl. Acad. Sci. USA 104: 6466–6471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying B.W., Taguchi H., Kondo M., Ueda T. (2005). Co-translational involvement of the chaperonin GroEL in the folding of newly translated polypeptides. J. Biol. Chem. 280: 12035–12040 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.