Background: Salmonella utilize ZirTS to decrease virulence for increased host transmission.
Results: ZirS is an immunoglobulin fold that binds the previously unknown zir protein ZirU, and both interact with the extracellular domains of ZirT.
Conclusion: The zir antivirulence pathway is an immunoglobulin adhesion system that assembles in the extracellular environment.
Significance: Understanding antivirulence completes the picture of host-microbe interactions for a full model of pathogenesis.
Keywords: Mass Spectrometry (MS), Membrane Proteins, Microbial Pathogenesis, Nuclear Magnetic Resonance, Structural Biology, Antivirulence, SILAC, Salmonella, ZirS, ZirT
Abstract
The co-evolutionary relationship between pathogen and host has led to a regulatory cycle between virulence factors needed for survival and antivirulence factors required for host transmission. This is exemplified in Salmonella spp. by the zirTS antivirulence genes: a secretion pathway comprised of the outer membrane transporter ZirT, and its secreted partner, ZirS. ZirTS act within the gastrointestinal tract to function as a virulence modulator and during Salmonella shedding in anticipation of a new host. Together, ZirT and ZirS decrease virulence by lowering bacterial colonization at systemic sites through an unknown mechanism. To understand this mechanism, we have probed the zirTS pathway both structurally and biochemically. The NMR derived structural ensemble of the C-terminal domain of ZirS reveals an immunoglobin superfamily fold (IgSF). Stable isotope labeling by amino acids in cell culture experiments show that the ZirS IgSF domain interacts with its transporter ZirT, and reveal a new protein interaction partner of the pathway, a protein encoded adjacent to zirTS that we have designated as ZirU. ZirU is secreted by ZirT and is also a predicted IgSF. Biochemical analysis delineates ZirT into an N-terminal porin-like β domain and C-terminal extracellular soluble IgSF domain, whereas biophysical characterization suggests that the transporter undergoes self-association in a concentration-dependent manner. We observe that ZirS and ZirU directly interact with each other and with the extracellular domains of ZirT. Here we show that the zir antivirulence pathway is a multiprotein immunoglobulin adhesion system consisting of a complex interplay between ZirS, ZirT, and ZirU.
Introduction
Pathogenic microrganisms have the dual challenge of both adapting to survive within the environment of their host and mediating their transmission to new hosts. Such organisms employ a number of virulence genes, which are often encoded together on a localized genetic segment or pathogenicity island (1, 2). These factors are readily identifiable as their disruption reduces or abolishes the severity of the disease phenotype of the infecting species. Prominent examples include the type III secretion system, found in Gram-negative bacteria such as Salmonella and Yersinia (3), and the cytotoxin-associated genes (cags) that constitute the type IV secretion system in the Gram-negative bacteria H. pylori (4). In both cases these genes encode proteins that interact directly within eukaryotic cells, gained through co-evolution with their mammalian hosts (4, 5). These virulence factors frequently mimic the function of host proteins to take control of cellular signaling events (6, 7) allowing the bacteria to evade the immune system and replicate.
Contingent with virulence factors, pathogens also employ genes that encode for antivirulence function(s). Similar to virulence proteins, antivirulence proteins can be identified by their disruption, but instead result in a more severe disease or hypervirulence phenotype. These systems necessarily evolve with their virulence counterparts to facilitate the transmission to a new host by prolonging host survival (8–10), or are critical for survival in another part of the pathogen life cycle (11). Although as a field antivirluence is relatively new, examples are readily appearing with the availability of pathogen genomes and bioinformatic techniques (8). Antivirulence proteins have been characterized in Leishmania spp., protozoan parasites that cause skin lesions and eventual death (10), and in Salmonella spp., a Gram-negative enteropathogenic bacteria that is a common cause of gastroenteritis and the causative agent of typhoid fever (12–15). In each case, the antivirulence proteins act to decrease population growth of the pathogen during specific points of infection, effectively increasing the levels of host survival required to allow for efficient transmission. In Salmonella spp., two characterized examples, sciS (12) and the zirTS pathway (13), highlight the pathogenic importance of antivirulence.
Upon ingestion of contaminated food or drinking water, Salmonella enterica serovar Typhimurium (Salmonella typhimurium) localize to the intestinal mucosa and phagocytotic cells of their human hosts where they internalize and replicate by virtue of their type III secretion systems (3, 14). Here the bacteria cause a self-limiting gastroenteritis, with diarrheal symptoms resulting when newly replicated S. typhimurium induce shedding of the intestinal mucosal lining. This ultimately leads to the release of the pathogen into the environment for transmission to new hosts (16). The gene sciS has been shown to be intimately involved in these replication events, where it regulates Salmonella division within macrophages (12). The deletion of sciS results in an increase of bacterium in macrophages infected in vitro, and an 8-fold increase of lethality in mice infected intragastrically (12). Together this suggests that sciS attenuates virulence at the late stages of infection (12).
The zirTS pathway also regulates virulence by attenuating population growth, but instead influences the early stages of infection (13). Deletion of zirTS genes results in a hypervirulence phenotype, with the median survival time of BALB/c mice dropping from 8.5 to 6 days, and wild-type Salmonella outcompeting deletion strains by 4-fold as observed by competitive index experiments (13). Additionally, it was found that zirTS is expressed throughout the gastrointestinal tract rather than at systemic sites, and is highly expressed during bacterial shedding in fecal pellets at 8 weeks postinfection (13). Furthermore, the system was shown to be negatively regulated by OxyR, a transcription factor that controls the expression of genes in response to oxidative stress (17), and also by the presence of zinc in the media. As Salmonella spp. are primarily transmitted by the ingestion of contaminated fecal matter, ZirTS is expressed in the anticipation of infection of a new host to regulate bacterial virulence upon colonization of the intestinal tract (13).
The zirTS genes are located on a horizontally acquired genomic island that is conserved in pathogenic strains of Salmonella (13, 18). ZirS (zinc regulated secreted) is a 31-kDa protein of unknown function that is secreted by ZirT (zinc regulated transporter), a 72-kDa outer membrane protein (19) with sequence homology to Invasin/Intimin proteins involved in bacterial-host adhesion (13). ZirS and ZirT are predicted to constitute a secretion system with aspects reminiscent of type V two-partner secretion systems (20). However, ZirS lacks the N-terminal two-partner secretion system domain typically necessary for targeting of the secreted component to the cognate transporter in other type V systems. Additionally, ZirS has sequence homology (24% identity) with a putative human zinc finger protein (NP_065798), and the open reading frame adjacent to ZirS (STM1667) is a predicted thiol peroxidase (13). In light of the negative regulation of the pathway by OxyR and zinc, these previous observations suggested a potential requirement for specific environmental regulatory cues for ZirTS function.
To elucidate the molecular details governing the mechanism of action and regulation of the zirTS antivirulence pathway in Salmonella, we have undertaken a structural approach using NMR spectroscopy and computational modeling, coupled with biochemical and biophysical analysis. We characterize features of the known components of the zirTS pathway, ZirS and ZirT, and identify an additional protein component that directly interacts with the system, named herein as ZirU. Our results show that this secretion pathway is divergent from type V or other secretion, describes the zir antivirulence pathway as a multiprotein immunoglobulin adhesion system, and offers mechanistic insight into an antivirulence mechanism with a new role for the Ig-fold in bacterial pathogenesis.
EXPERIMENTAL PROCEDURES
Expression and Purification of ZirS-IgSF, ZirT-IgSF, and ZirU
ZirS(136–276) was expressed as an N-terminal GST-tagged protein in pGEX6p1 (GE Healthcare) with restriction sites BamHI and Notl. Isotopically labeled samples were prepared with Escherichia coli BL21(DE3) grown at 37 °C in M9 minimal medium containing 1 g/liter of 15NH4Cl and/or 3 g/liter of [13C6]glucose. At A600 >0.6 the cells were induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside and the temperature lowered to 20 °C. After 20 h the cells were harvested, lysed by an Avestin homogenizer, and the lysate cleared by centrifugation. GST-ZirS was purified by passing the lysate over Q-Sepharose followed by glutathione-Sepharose at 4 °C. The material was then digested overnight during dialysis with 3C protease and GST was removed using a Mono Q column. The ZirS material was concentrated and further purified by gel filtration using an SD75 column (GE Healthcare) equilibrated in 50 mm Tris, pH 7.1, 50 mm NaCl, 0.1 mm Tris(2-carboxyethyl)phosphine, and 0.1 mm PMSF. All steps were carried out at 4 °C. ZirT(430–660) and ZirU(38–281) were purified as N-terminal GST-tagged constructs as per ZirS, but GST was not removed. The final buffer was 50 mm Tris, pH 7.5, 100 mm NaCl, 0.1 mm Tris(2-carboxyethyl)phosphine.
Expression and Purification of ZirT
ZirT(1–660) was first cloned into pET52b (Novagen) with restriction sites Ncol and Sacl, then subcloned with the His tag and thrombin site into pBAD (Invitrogen). The vector was transformed into S. typhimurium lacking InvG and ZirT expression induced with 0.1% arabinose overnight at 20 °C. Cells were harvested by centrifugation, lysed with an Avestin homogenizer, and inclusion bodies were removed at 7,000 rpm for 20 min. Membranes were harvested by ultracentrifugation at 100,000 × g for 1 h and resuspended in 50 mm HEPES, pH 7.5, 200 mm NaCl, 10% glycerol, 0.1 mm PMSF, 0.1 mm Tris(2-carboxyethyl)phosphine. ZirT was extracted from membranes in the same buffer plus 1% N,N-dimethyldodecylamine N-oxide (LDAO) (Anatrace) and unextracted material was removed by ultracentrifugation. ZirT was purified by nickel-Sepharose in buffer with 0.2% N,N-dimethyldodecylamine N-oxide followed by gel filtration with Superose-6 in buffer plus 0.8% β-octyl glucoside (Anatrace).
NMR Spectroscopy
NMR spectra were recorded at 25 or 35 °C on Varian Unity 500 and cryoprobe-equipped Inova 600 MHz spectrometers. Final ZirS samples were composed of 0.5 mm protein in 50 mm Tris, pH 7.1, 50 mm NaCl, 0.1 mm Tris(2-carboxyethyl)phosphine, and 0.1 mm PMSF with the addition of 5% D2O. Signals from the 1H, 13C, and 15N nuclei of ZirS were detected and assigned by standard heteronuclear NMR experiments (21). 15N NOE relaxation data for ZirS were recorded using a 600 MHz spectrometer at 20 °C following established methods (22). The NMR data were processed with NMRpipe (23) and analyzed with Sparky.2
Structure Calculation
The ZirS structural ensemble was calculated using CYANA, as summarized in supplemental Table S1 (24, 25). Backbone dihedral angles were predicted from chemical shifts by TALOS+ (26). Distance restraints were derived from a 100-ms mixing time simultaneous three-dimensional 13C/15N-edited NOESY-HSQC recorded with a 600 MHz spectrometer at 25 °C (27) and assigned automatically. Hydrogen bond restraints were based on diagnostic NOE patterns. The calculation consisted of 7 cycles of refinement with 100 iterations per cycle (25). The 10 structures with the lowest calculated target function were chosen as a representative ensemble and refined in water using CNS (28, 29).
Computational Modeling
Structural models for the ZirS N-terminal domain were calculated ab initio using the server Robetta (30) and manually with Rosetta (31) imposing a disulfide restraint. Residues 44–146 of ZirS were chosen to model due to secondary structure predictions with Jpred (32). The forced disulfide constraint resulted in poorly packed folds consisting of isolated β-hairpins, whereas the Robetta server yielded folded domains that were analyzed by the Dali server (33). ZirT(430–660) was modeled by the server Phyre2 (34) and with the program Modeller (35). Both constructed highly similar homology models with Invasin (Protein Data Bank code 1cwv) as the template.
β Domain Refolding
ZirT porin-like β domain constructs were overexpressed as C-terminal His-tagged constructs in the vector pET52b and purified as inclusion bodies. A concentration of 0.15 mg/ml of purified material was resuspended in 20 mm potassium phosphate, pH 8, and 2% SDS at room temperature. Refolding was induced with the addition of 1.5 m 2-methyl-2,4-pentanediol (MPD)3 and monitored by SDS-PAGE. The secondary structure was measured by circular dichroism spectroscopy after concentration to 4.2 mg/ml (Jasco J-810). The CD data were de-convoluted using Dichroweb (36) and compared with Jpred (32) secondary structure predictions.
ZirT Topology Assay
ZirT was expressed as described above. After 1 h of induction at 37 °C, 1 ml of cells were harvested, washed 3 times in PBS to remove arabinose, and then resuspended in 500 μl. Thrombin was added at a total of 0, 8, or 80 μg and incubated for 1 or 2 h. The reaction was stopped with the addition of PBS and 1 mm PMSF and washed 3 times in the same buffer. The cells were resuspended in a final volume of 300 μl lysed by sonication and membranes were harvested. Samples were analyzed by Western blot with anti-His tag (Cedarlane) and developed by Immobilon Western Chemilumiescent HRP substrate (Millipore). Quantification was performed with Genesnap (SYNGENE) and graphed with GraphPad Prism.
ZirT Oligomerization
ZirT was concentrated to ∼3 mg/ml for gel filtration as described previously. Immediately after gel filtration, samples were assayed by dynamic light scattering (DynaPro Wyatt). Samples were microcentrifuged at maximum for 5 min before and measured at 0.4 and 1 mg/ml. Data were analyzed with a model consisting of an isotropic sphere and an aqueous solution plus 10% glycerol. Each concentration consisted of at least seven individual measurements contributing to the observed distribution.
ZirU Secretion Assay
ZirU(1–281) and ZirS(1–276) were cloned into the vector pWSK29 with a C-terminal 2HA tag and transformed into Salmonella strain SL1344 or SL1344ΔzirT (13). For samples containing ZirT, ZirT was added in trans on the vector pBR322. Expression and secretion were monitored in the pellet and media by Western blot with anti-HA and anti-DnaK antibody and goat anti-mouse HRP secondary antibody for detection. All media fractions were sterile filtered before visualization.
Metal Binding Analysis
The presence of metals was tested by inductively coupled plasma mass spectrometry (ICP-MS) with a NexION 3000 ICP mass spectrometer (PerkinElmer Life Sciences) and analyzed by the NexION software program. GST was used as a negative control and the zinc binding of New Delhi β-lactamase (kindly supplied by Dustin King) as a positive control. Purified GST-ZirS(136–276), GST-ZirU(38–281), and ZirT-Histag(430–660) were assayed. All proteins were diluted to 5 μm in 20 mm HEPES, pH 7.5, 100 mm NaCl, and then diluted 1/10 in an internal standard (10 μg/liter Sc45, 1% nitric acid, Inorganic Ventures). Each sample was measured three times and the average abundance was recorded.
Stable Isotope Labeling with Amino Acids in Cell Culture (SILAC)
Salmonella and HeLa cells were grown in minimal media supplemented with isotopically labeled arginine (13C6) and lysine (D6) for “heavy” extracts and unlabeled amino acids for “light” extracts. For bait, 40 pmol of GST-ZirS IgSF was bound to glutathione beads and an equivalent molar ratio of GST alone as the control. The co-precipitation was performed according to standard methods (37) and proteins were digested with endopeptidase LysC and trypsin (mass spectroscopy grade, Promega) as described (PMID 12724530). Digested peptides were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) on a LTQ-OrbitrapXL (ThermoFisher, Bremen, Germany) on-line coupled to an Agilent 1100 Series nanoflow HPLC using nanospray ionization source (Proxeon Biosystems) holding columns packed into 15-cm long, 75-μm inner diameter fused silica emitters (8-μm-diameter opening, pulled on a P-2000 laser puller from Sutter Instruments) using 3-μm diameter ReproSil Pur C18 beads. Buffer A consisted of 0.5% acetic acid, and buffer B consisted of 0.5% acetic acid and 80% acetonitrile. Gradients were run from 6% B to 30% B over 60 min, then 30% B to 80% B in the next 10 min, held at 80% B for 5 min, and then dropped to 6% B for another 15 min to recondition the column. The LTQ-OrbitrapXL was set to acquire a full-range scan at 60,000 resolution from 350 to 1500 Thompsons in the Orbitrap and to simultaneously fragment the top five peptide ions in each cycle in the LTQ.
Protein identification and quantification were done using Proteome Discover (version 1.2, ThermoFisher) and Mascot (version 2.3, Matrix Science) to search against the International Protein Index Human (version 3.69, 149,771 sequences, common serum contaminants were added and all reversed sequences were concatenated) data base with the following criteria: electrospray ionization-ion trap fragmentation characteristics, tryptic specificity with up to one missed cleavages; ±10 parts per million and ±0.6 Da accuracy for MS and MS/MS measurements, respectively; cysteine carbamidomethylation as a fixed modification; N-terminal protein acetylation, methionine oxidation, duplex ([2H4]Lys, [13C6]Arg) SILAC modifications as appropriate; peptide false discovery rate was set at 1%. Quantitation was done using a mass precision of 2 ppm (three times the mass precision is used to create extracted ion chromatograms). After extracting each ion chromatogram, Proteome Discoverer runs several filters to check for, among other things, interfering peaks and the expected isotope pattern, and peptides that do not meet all the criteria are not used in calculating the final ratio for each protein. Analytical variability of SILAC data in the types of experiments performed here is typically <30% on average and biological variability was addressed in these experiments by performing at least three independent replicates of each experiment.
Far Western
Purified bait proteins were run on an SDS-PAGE and transferred to nitrocellulose. Full-length ZirS-HA and ZirU-HA were cloned as described previously (13) and expressed in S. typhimurium strain SL1344. Either solubilized pellet or media containing ZirS-HA or ZirU-HA was used to blot the nitrocellulose matrix containing bait proteins. For controls, no HA-labeled protein was added. After binding the matrix was washed at least three times and then incubated with anti-HA antibody (Cedar lane) followed by three more washes to remove unbound anti-HA antibody. In all steps the buffer was PBS. Experiments with controls were visualized by exposure on the same film.
RESULTS
ZirS Structure
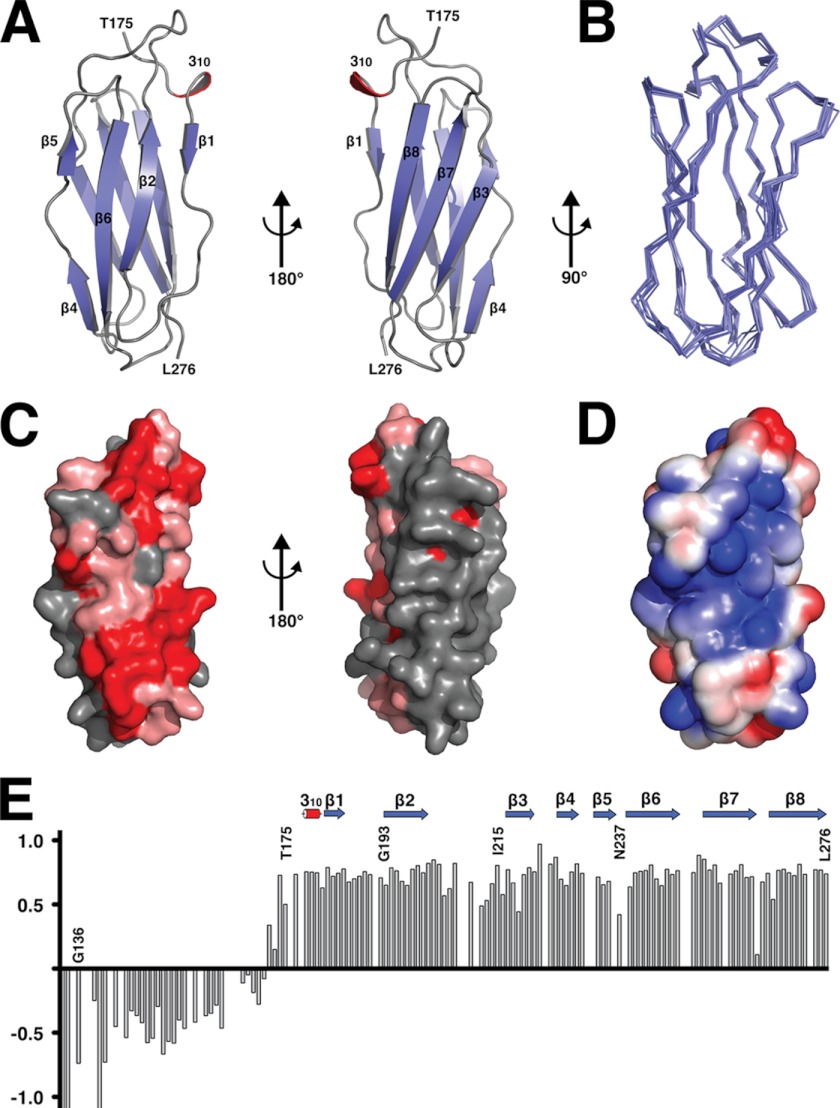
The NMR spectroscopically derived solution state structure of ZirS consists of eight β-strands and one 310 helix that fold into the canonical β-sandwich of the immunoglobulin superfamily (IgSF) (Fig. 1A) (38). The ZirS structural ensemble is displayed in Fig. 1B, the statistics of the calculation are summarized in supplemental Table S1, and the assigned 15N-HSQC spectrum in Fig. 2A. The ZirS mean structure was submitted to the Dali server search engine (33), and showed the highest similarity to the Invasin subunit D3 of Yersinia pseudotuberculosis (39) with a Z-score of 9.9 and a root mean square deviation of 2.5 Å over 92 common backbone atoms. ZirS is also structurally related to Gelation Factor (Z-score 8.4), Intimin (Z-score 7.8), and Filamin (Z-score 7.5), as well as to a number of other IgSF members such as Complement C3 and C5 (Z-score 6.5) and myosin-binding proteins (Z-score 5.8).
FIGURE 1.
Structure of the ZirS C-terminal IgSF domain. A, a ribbon diagram of a low energy representative of the structural ensemble of ZirS residues 175–276 showing two rotated views. β-Sheets are colored in blue and the single 310 helix in red. B, overlay of the 10-member ensemble of the lowest energy ZirS structures. C, the surface representation of the structures displayed in panel A, colored for sequence conservation by the Consurf server. Red is highly conserved, pink shows areas of greater than average conservation, and gray areas of no conservation (Consurf score of 5 or less). D, the left-most surface in panel C colored by electrostatic potential generated by APBS (Adaptive Poisson-Boltzmann Solver) and the PARSE force field. The potential is contoured from −2 kT/e (red) to 2 kT/e (blue). E, 1H-15N heteronuclear NOE relaxation data for ZirS recorded with a 600 MHz spectrometer at 20 °C. Decreasing NOE values result from increasing mobility of the 15N-1HN bond vector on the sub-nanosecond time scale. Missing data points correspond to prolines or residues with overlapping signals. Graphics were generated using PyMOL (Schrödinger, L., The PyMOL Molecular Graphics System, version 1.3).
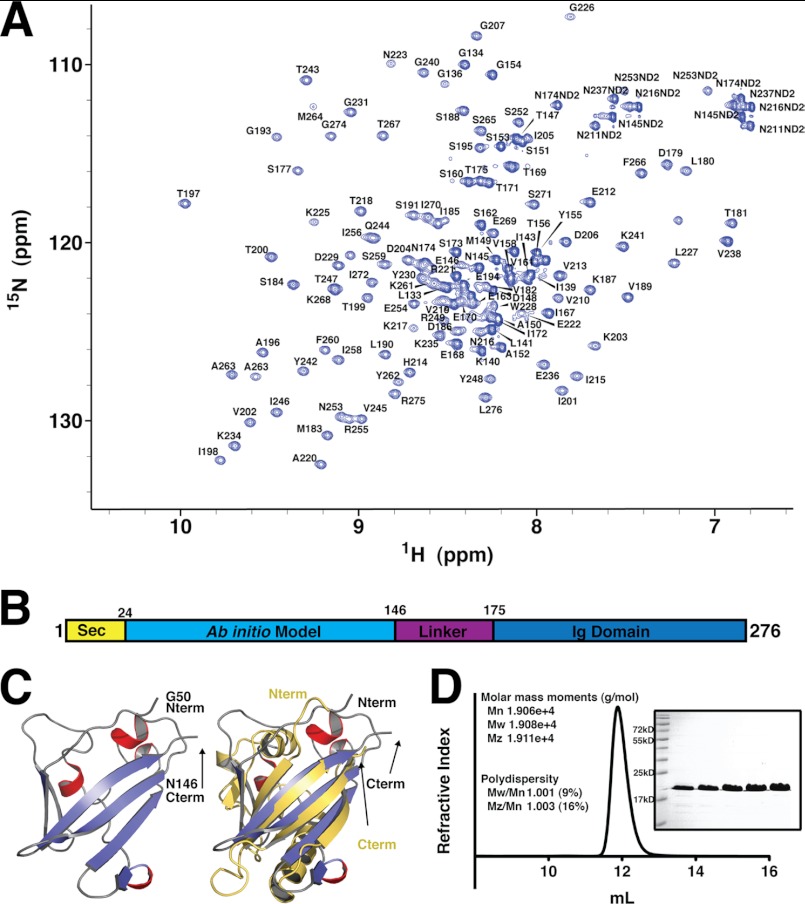
FIGURE 2.
ZirS domain structure and N-terminal ab initio model. A, assigned 15N-HSQC of ZirS C-terminal IgSF domain residues 136–276. B, domain map of ZirS based on the ZirS IgSF structure, expression trials, and ab initio model. C, an ab initio model of ZirS(50–146) generated by Rosetta. α-Helices are colored in red and β-strands in blue with an overlay of the model with an HTR-like modeling protein (PDB code 3fc7) in yellow. D, static gel-filtration coupled light scattering of ZirS(136–276) in the NMR buffer. The inset is the SDS-PAGE gel of purified ZirS.
The β-strands of the ZirS Ig domain fold to form two distinct faces on opposite sides of the molecule, with strands 1, 2, 4, 5, and 6 forming one β-sheet face and strands 3, 7, and 8 creating an opposing β-sheet face (Fig. 1A). When analyzed for surface and sequence conservation with the Consurf server (40) and PSI-Blast (41), one face of ZirS is highly conserved, whereas the other is variable (Fig. 1C). Given that the Ig domain of ZirS behaves as a monomer in solution (Fig. 2D) the former surface is not conserved for oligomerization of the IgSF domain itself, and thus is likely the binding site for a biological interaction partner. Furthermore, a computational analysis with Adaptive Poisson-Boltzmann Solver (42) revealed that the conserved surface of ZirS is an electropositive groove (comprised mainly of lysines and the polar side chains of Ser or Thr) flanked by two protruding areas, which tend toward a more negative potential (Fig. 1D and supplemental Fig. S1B) suggesting a largely electrostatic binding mode.
The observed ZirS IgSF domain spans residues 175–276 of the purified construct (supplemental Fig. S1). Residues 136–174 of the NMR ensemble were omitted from Fig. 1 for clarity as they do not fold into an ordered structure. Although readily detectable, these 38 residues are disordered and mobile on a nanosecond-picosecond time scale as indicated by their random coils 1HN-15N chemical shifts (Fig. 2A) and by 1H-15N heteronuclear NOE measurements (Fig. 1E). DISOPRED (43) does not flag these residues as intrinsically disordered, however, Jpred (32) fails to confidently predict any regular secondary structure for residues 150–174. These residues (150–174) may function as a flexible linker between the C-terminal IgSF domain of ZirS and the N-terminal domain of the protein (Fig. 2C), a common feature in linked-domain proteins (44).
Despite our efforts, attempts to overexpress the ZirS N-terminal domain for structural studies were unsuccessful (supplemental Fig. S1A). To gain insight into this domain, a structural model was calculated ab initio, using Rosetta (30, 31). Only the lowest energy model was a fully folded globular domain, which predicted an α/β structure (Fig. 2, B and C). Submission to the Dali server showed structural homology to members of the PAS-domain family (top Z-score 3.5, root mean square deviation of 3.0 over 72 residues; Fig. 2C), proteins that function as sensory components in signaling pathways (45, 46), especially for oxygen (47). The Rosetta-generated structural model is thus attractive in the context of ZirS extracellular localization and oxidative stress (OxyR) regulated expression (13).
ZirS Ig-Domain Binding Partners
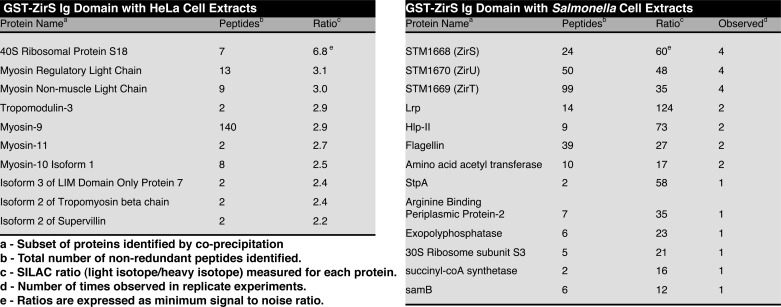
To search for potential glycan and protein interaction partners, ZirS(136–276) was screened using carbohydrate arrays within the Consortium for Functional Glycomics, and via SILAC experiments (48) with both mammalian and Salmonella protein extracts. The results of the glycan array with common mammalian sugars did not show significant binding and can be viewed in the public repository at the Consortium for Functional Glycomics (printed array version 4.1, H:1960). Likewise, SILAC experiments using HeLa extracts with GST-tagged ZirS(136–276) yielded weak hits, many of which were myosin (Table 1). Myosin is a common contaminant with SILAC (37), and additional experiments showed little binding (supplemental Fig. S2).
TABLE 1.
Proteins identified by MS from SILAC experiments
In contrast, SILAC experiments of GST-ZirS(136–276) with Salmonella extracts yielded significant heavy to light isotope ratio hits that were observed in all four replicates of the experiment (Table 1). These hits included both ZirS and ZirT, and the protein STM1670, encoded by an adjacent open reading frame (ORF) to ZirT (13). In line with the current nomenclature, STM1670 will be herein renamed ZirU.
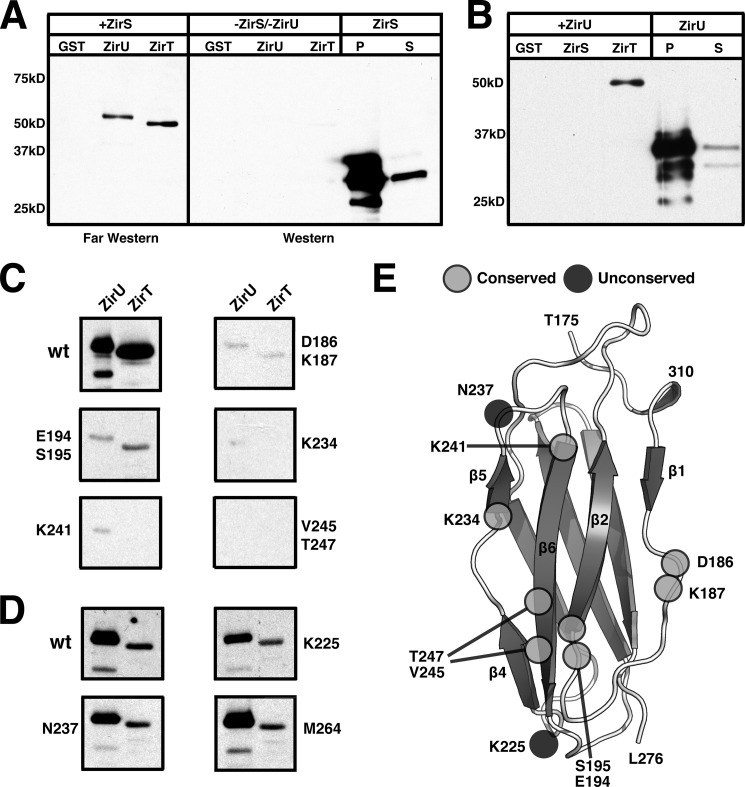
ZirU Is Secreted by ZirT
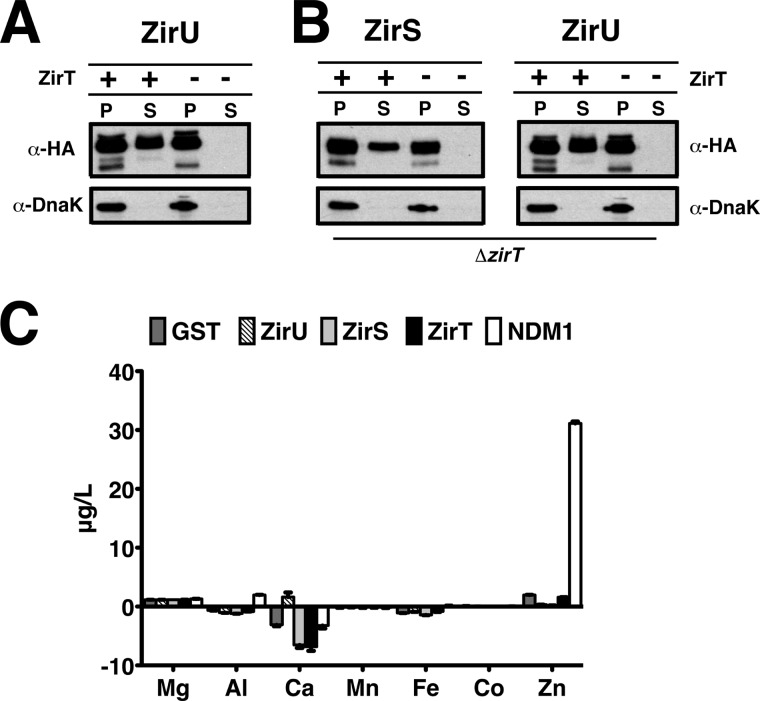
Similar to both ZirS and ZirT, ZirU is predicted to be expressed under an OxyR promoter region by Virtual Footprint (49), and to contain an N-terminal sec-cleavable sequence by the server SignalP (50) (supplemental Fig. S3A). To probe secretion of ZirU by ZirT, we cloned ZirU into the vector pWSK29 with a C-terminal 2HA tag, added ZirT in trans on vector pBR322 in Salmonella, and monitored the media for the presence of ZirU. In the presence of recombinant ZirT, ZirU was readily exported into the media, but was not exported by Salmonella containing only endogenous ZirT (Fig. 3A). This is similar to what was observed previously for ZirS, and is thought to be due to the requirement of a particular stoichiometry (13) and/or a specific set of unmet environmental conditions. For further verification, we also compared the ZirT-dependent secretion of ZirU to that of ZirS. Both ZirU and ZirS were secreted into the extracellular media when complemented with ZirT in a ΔzirT background (Fig. 3B). As our results indicate that ZirU is a substrate for ZirT and a binding partner for ZirS, we will refer to the zirTS antivirulence pathway as the zir antivirulence pathway for simplicity.
FIGURE 3.
ZirU is part of the zir pathway. A, secretion of ZirU by ZirT in cultured Salmonella as visualized by Western blot. P represents the cell fraction and S the filtered media. Lanes with ZirT+ indicate that ZirT has been added in trans. DnaK is shown as a negative secretion control. B, as in A with the secretion of both ZirS and ZirU by ZirT in a Salmonella zirT deletion strain, SL1344ΔzirT. C, metal binding analysis of the zir pathway by ICP-MS. The data show the average metal observed for each protein from three measurements. GST is the negative control and NDM-1 the positive control for zinc binding. The data are graphed by shaded box.
ZirU Is Homologous to Immunoglobulin Superfamily Members
The structure-based Robetta server (30) predicts ZirU to contain Ig-like domains (38–152 and/or 153–272) with a greater than 85% confidence (HHsearch score 2.2) (30, 51). This is supported by a sequence alignment between ZirS and ZirU showing that many of the residues in the ZirS IgSF hydrophobic core are conserved in ZirU (supplemental Fig. S3, B and D). In contrast, sequence analysis of ZirU using PSI-Blast (41) shows homology to the putative zinc finger protein 236 from Homo sapiens (e-value 4e−58, NP_0313713), although ZirU lacks a clear metal binding site (supplemental Fig. S3C) (Metaldetector (52)). To resolve this, we performed ICP-MS to probe for potential metal binding with purified ZirU(38–281) and ZirS(136–276) (Fig. 3C). As shown by the analysis, neither ZirU nor the ZirS IgSF domain co-purify with common biological metals, including zinc.
ZirT Domain Delineation
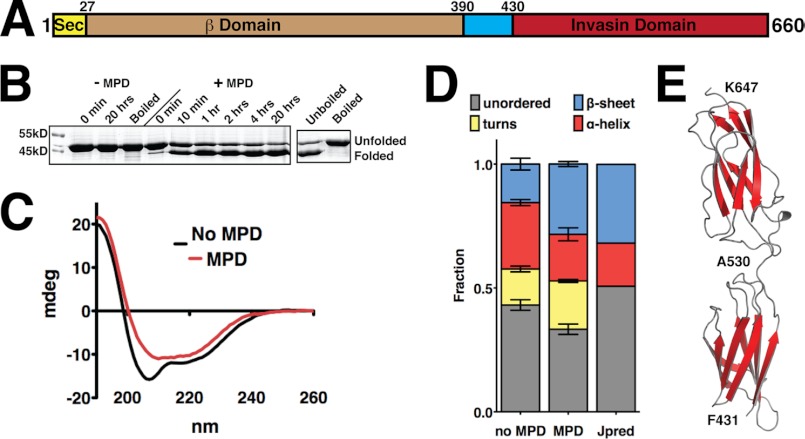
ZirT residues 88–368 are homologous to the β-barrel region of Invasin, or β domain (13, 19), and ZirT localizes to the outer membrane where it facilitates the secretion of ZirS (13) and ZirU (Fig. 3B). Furthermore, it is an interaction partner for the ZirS IgSF domain (Table 1), making ZirT a central element of the zir pathway. To explore these interactions we first characterized the domains of ZirT using biochemical and computational analysis (Fig. 4 and supplemental Fig. S4). To further delineate the exact ZirT β domain, various constructs were purified as inclusion bodies and solubilized in SDS. Refolding of porins and other β-barrel membrane proteins can be induced by the addition of MPD to SDS and visualized on SDS-PAGE gel for the appearance of a faster migrating band (53) (Fig. 4A and supplemental Fig. S4A). A construct encompassing only the predicted minimal β domain region (80–368) or extension of the C terminus (80–390) failed to properly refold (supplemental Fig. S4A), whereas a construct spanning residues 1–390 showed a similar in-gel migration change as full-length ZirT (13) (Fig. 4B). Correct folding of ZirT(1–390) was monitored by circular dichroism spectroscopy (CD), which showed formation of an expected β-sheet structure and similar secondary structure partitions as predicted by Jpred (∼42% β-strand and turns corresponding to ∼12–16 β-strands) (Fig. 4, C and D). As a final control, ZirT(1–390) maintained its fold when exchanged into other detergents such as C8E4 (supplemental Fig. S4B).
FIGURE 4.
Domain structure of ZirT shows a β domain linked to a soluble IgSF fold. A, schematic representation of the ZirT domains. B, refolding of the ZirT β domain by SDS and MPD as visualized by a Coomassie-stained SDS-PAGE gel. Bands corresponding to folded and unfolded species are indicated. C, MPD-induced ZirT β domain refolding monitored by circular dichroism spectroscopy. D, deconvolution of the recorded CD data with Dichroweb. The secondary structure fraction for ZirT(1–390) as predicted by Jpred was calculated by dividing the number of residues predicted to have secondary structure elements by the total residues. Unordered denotes no secondary structure prediction. Error bars show the standard deviation from three independent experiments. E, homology model of ZirT residues 430–660 as calculated by Phyre and Modeler. A530 is in the middle of the linker region between the two Ig folds. Residues C-terminal to K647 are predicted as disordered.
The C-terminal region of ZirT(430–660) was analyzed using the homology modeling server Phyre (34) and Modeller (35), then verified by biochemical characterization of constructs spanning the predicted structural regions. Both tools yielded a model of ZirT spanning residues 431 to 647 with 99% confidence based on the segments D1 and D2 of Invasin from Y. pseudotuberculosis (39) (Fig. 4E). Constructs spanning residues 430–660 and 430–647 were soluble, could be purified, and displayed a dispersed 15N-HSQC spectra characteristic of high β-sheet content (supplemental Fig. S4, A and D). For completeness, ZirT(430–660) was also tested by ICP-MS but showed no evidence of zinc binding (Fig. 3C).
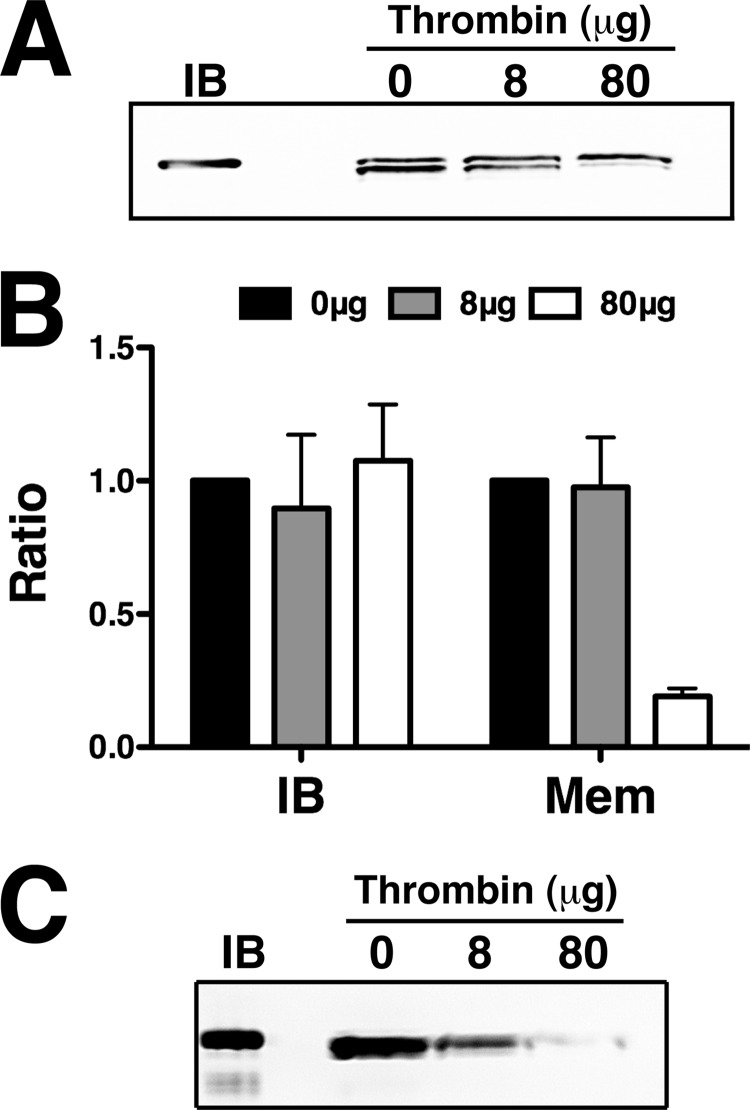
ZirT Topology
As ZirT residues 1–390 constitute an integral membrane β domain (Fig. 4), we asked if the C-terminal domains of ZirT(430–660) are periplasmic or extend into the extracellular medium. To test this, ZirT(1–660) was cloned into pBAD with a thrombin cleavable C-terminal His tag, and introduced into Salmonella (ΔinvG). ZirT expression was induced, followed by incubation of unlysed cells with thrombin and subsequent addition of PMSF to stop the proteolytic reaction. Membrane-enriched fractions were solubilized, boiled, and ZirT was visualized by a anti-His tag Western blot (Fig. 5). The untreated ZirT (0 thrombin) ran as two bands, one similar to purified inclusion bodies containing the sec-signal (verified by N-terminal sequencing), and a lower band that reflects N-terminal cleavage by the sec-system with the C-terminal His tag still present (Fig. 5A). ZirT that has yet to be acted upon by the sec-system (IB) retains its His tag, whereas the His tag of mature ZirT (Mem) is removed (Fig. 5, A and B), suggesting that the C terminus is extracellular. For further validation, ZirT induction was increased to 2 h to allow the amount of mature ZirT to overtake the full-length material, which clearly showed a dose-dependent loss of the C-terminal His tag (Fig. 5C). Finally, as the IgSF domains of ZirT are extracellular, we probed for a mammalian interaction partner, but found no significant hits (supplemental Table S2).
FIGURE 5.
The ZirT IgSF domains are extracellular. A, Western blot of the membrane fractions of cells with ZirT expression induced for 1 h after whole cells were treated with thrombin. IB represents purified inclusion bodies and the amount of thrombin used in each experiment is indicated above the panel. Each lane consists of an upper band corresponding to unprocessed protein and a lower band corresponding to processed protein inserted into Salmonella membranes. B, quantification of panel A performed in triplicate. IB represents the upper band and Mem the lower band. C, repeat experiment of panel A but at 2 h post-induction. Only the processed lower band is observed.
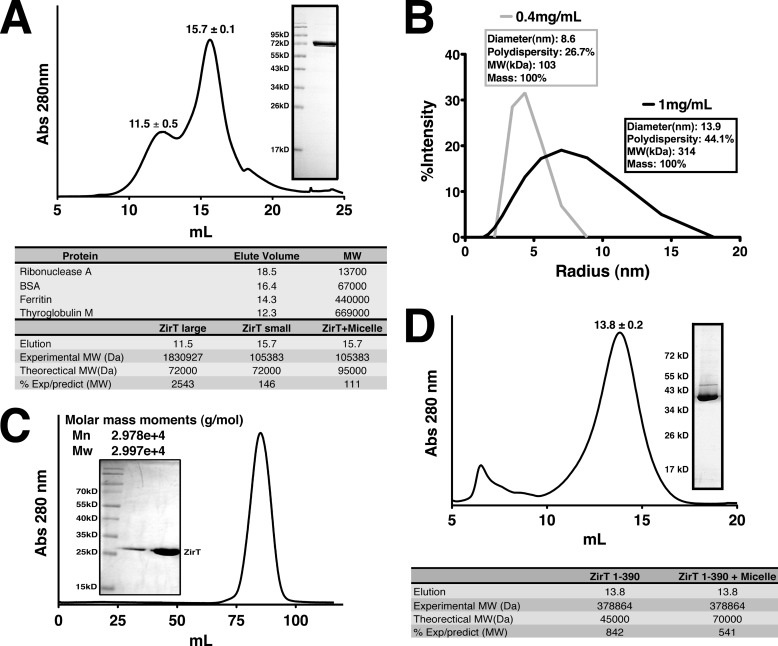
ZirT Self-association Requires the β Domain
Outer membrane proteins such as Invasin and Intimin are believed to oligomerize on the cell surface to mediate multivalent adhesion (54, 55). We tested if ZirT could also self-associate by examining the behavior of mature ZirT purified from the Salmonella outer membrane by gel-filtration. ZirT was injected at a concentration of 3 mg/ml, elutes as a peak close to the predicted size of a monomer as well as a second peak of large molecular size (Fig. 6A). Immediately following gel filtration, the ZirT monomer fraction was observed by dynamic light scattering at two concentrations (Fig. 6B). At a lower concentration (0.4 mg/ml) ZirT behaved as a single species with comparatively low polydispersity (26.7%) and had a predicted mass in agreement with the monomer seen in gel filtration. In contrast, a 2.5-fold more concentrated ZirT sample (1 mg/ml) displays higher polydispersity (44.1%) showing the presence of both the monomer and larger species. This suggests that ZirT can self-associate in a concentration-dependent manner. Interestingly, the mean size of the larger species was ∼300 kDa, implying a possible trimer. For further verification, chemical cross-linking was also performed on ZirT within the membrane fraction (supplemental Fig. S4C), which shows higher molecular weight ZirT bands that appear with increasing cross-linking agent.
FIGURE 6.
ZirT self-associates. A, chromatogram representing the gel-filtration profile of purified ZirT from Salmonella membranes. The elution volumes are the average with the S.D. of three separate purifications. The inset shows a Coomassie-stained SDS-PAGE gel of pure ZirT. The bottom panel lists the standard elution volumes of known proteins and the calculated size of ZirT. B, dynamic light scattering of ZirT at two different concentrations, 0.4 mg/ml in gray and 1 mg/ml in black. The calculated diameter, polydispersity, predicted molecular mass (MW) in kilodaltons, and percentage of mass is listed in the box next to the curve. C, gel filtration profile of the ZirT Invasin domain (430–660) with the calculated static light scattering size indicated on the figure. The inset shows the purified ZirT Invasin domain. D, gel filtration profile of the ZirT β domain (1–390). The elution volume is the average with the S.D. of three separate purifications. The inset shows a Coomassie-stained SDS-PAGE gel of pure ZirT(1–390). The bottom panel lists the calculated size of ZirT(1–390).
We further explored the nature of ZirT self-association by examining if the IgSF-like domain (430–660) or the β domain (1–390) is the point of contact. ZirT(430–660) was examined by gel filtration-coupled static light scattering at 3 mg/ml but showed only a single well defined monomeric species (Fig. 6C). The refolded β domain was injected at 3 mg/ml and also eluted at a single peak, but had a predicted size of ∼380 kDa (Fig. 6D) showing that it self-associates. As shown by the inset gel, the β domain remained folded (Fig. 4) indicating that the result was not due to simple nonspecific aggregation of unfolded protein.
ZirS and ZirU Interact with the C-terminal Domain of ZirT
The ZirS IgSF domain (Fig. 1) is an interaction partner for both ZirU (Fig. 3) and ZirT (Fig. 4) as shown by SILAC (Table 1). To further explore these protein-protein interactions, purified GST-tagged constructs of the ZirS-IgSF domain (136–276), the Ig-like ZirU(38–281), and the extracellular ZirT IgSF domains (430–660) were used as bait in a far Western experiment with the biological mature forms of ZirS and ZirU secreted from Salmonella (Fig. 7). In agreement with the SILAC results (Table 1), wild-type ZirS interacts with both ZirU and the extracellular domain of ZirT (Fig. 7A). Additionally, secreted ZirU also interacts with the Invasin domain of ZirT, but contrary to the SILAC results, does not interact with the ZirS IgSF domain (Fig. 7B). Affinity tagged protein experiments are often directional, and unlike SILAC, a far Western experiment first denatures the bait that can disrupt a biological interaction. Complementing these results, ZirU(38–281) and the ZirS IgSF domain were tested by both NMR titration and ITC showing a weak but identifiable interaction (supplemental Fig. S5). Both techniques showed similar results, suggesting a dissociation constant in the mid-micromolar range (50–120 μm). An accurate number could not be obtained due to a concentration limit of the ZirS IgSF of ∼1 mm.
FIGURE 7.
ZirS and ZirU bind the C-terminal domains of ZirT. A, far Western experiment with ZirS-HA. GST was used as a control and GST-ZirU or GST-ZirT(430–660) used as bait. A Western blot without treatment with ZirS-HA is also shown next to ZirS-HA isolated from Salmonella as a control. B, the equivalent experiment as described in panel A but with ZirU-HA and GST-ZirS and GST-ZirT(430–660) used as bait. P indicates cell pellet and S the media. Bait concentrations were equivalent. C, far Western binding experiments with mutants of the conserved surface of the ZirS IgSF domain. D, far Western binding experiments with mutants of unconserved residues within the ZirS IgSF domain. E, structure of the ZirS IgSF domain with the positions of the mutants from panels C and D shown. M264 is on the opposite side of the displayed surface.
The ZirS IgSF Domain Targets ZirS to ZirU and ZirT
To test if the conserved surface of the ZirS IgSF (Fig. 1C) is the contact point between ZirS and its binding partners, both conserved and unconserved surface residues were selected in full-length ZirS (Fig. 7E), substituted with alanine, and probed for binding (Fig. 7, C and D). Every mutant assayed was readily expressed at levels similar to wild-type (supplemental Fig. S6, C and D) but only the mutations of conserved residues seemed to significantly block the interaction with both ZirU and ZirT (Fig. 7C). This data, in combination with the SILAC data (Table 1), show that ZirS and ZirU not only interact with each other after secretion but also bind to the extracellular domains of ZirT, with these interactions mediated in part by the conserved ZirS IgSF surface.
DISCUSSION
In this work, we structurally and biochemically characterize the known components of the zir antivirulence pathway, the secreted ZirS and transporter ZirT, and identify a new protein component that interacts with the system, ZirU. We have solved the NMR solution structure of ZirS(136–276) (Fig. 1) and found that the ZirS IgSF domain interacts with both ZirT and ZirU, by using SILAC mass spectrometry methods supported by additional biochemical analysis (Table 1, Figs. 3, 4, and 7). Similar to ZirS, ZirU is secreted by ZirT and is likely also an immunoglobulin superfamily fold. As ZirT appears central to this pathway, biochemical and biophysical characterization of this integral membrane protein were performed, allowing us to delineate the span and topology of the β domain and extracellular tandem Ig-like adhesin domains of ZirT (Figs. 4 and 5), and show that ZirT can self-associate in a concentration-dependent manner through contacts in its integral membrane β domain (Fig. 6). Additionally, we observe that ZirT acts as a binding platform for ZirS and ZirU (Fig. 7).
The NMR solution structure of ZirS(136–276) revealed that this secreted protein is a member of the IgSF, with significant structural homology to a large number of known protein-binding modules. Closer inspection showed that one face of this domain exhibited high sequence conservation, suggestive of a potential interaction surface. Follow up experimentation with SILAC and site-specific mutagenesis methods verified that this was indeed the case, showing that the ZirS-IgSF domain facilitates an interaction with ZirT and ZirU. To date, the bacterial immunoglobulin-like domains (BID or Big) (56) that have been studied are expressed in response to a host environment and are virulence factors. These BIDs enhance virulence by mediating cell host adhesion through a direct contact with host proteins or other targets on the host cell (39, 55, 57). This includes the Ig-like domains that form the pilus secreted by the chaperone-usher system, which self-associate by donor strand complementation on the surface of enteric bacteria (58). Thus it was interesting to find that Salmonella also secrete BIDs, ZirS, and ZirU, in response to the host environment as part of an antivirulence mechanism. It was known that regions of the ZirT transporter are homologous to Invasin/Intimin (13) and therefore may function in adhesion, but it was surprising to find that ZirT serves as the central binding platform for its own secreted IgSF substrates. This instead evokes a potential similarity to the chaperone-usher system in that the Zir proteins may self-associate into a large multiple Ig-domain adhesive structure, but unlike the pilus the zir pathway does not seem to use donor-strand complementation (59, 60). Instead, based on our observations, the zir pathway appears to use multiple weak interactions between several multidomain Ig-like proteins, possibly acting in a synergistic effect to construct a strong scaffold. Our data thus reveal a new function for the IgSF and BIDs, that of self-adhesion for antivirulence.
The zir pathway had been shown to be homologous but distinctive to type V secretion (13). Specifically, it was thought to mirror two-partner secretion but have a separate mechanism for substrate recognition by the transporter. Here our SILAC results with the ZirS IgSF domain further separate the connection between this antivirulence secretion system and type V secretion. As we have found that an additional Salmonella protein ZirU not only interacts with ZirS but is also secreted by ZirT, this secretion system must either be classified as its own subfamily of type V or given its own secretion classification. We prefer the latter, as current bioinformatic results show a vast number of BIDs and associated operons of unknown function (56) that may secrete substrates in a manner similar to the zir pathway. As these systems are studied, a new secretion class that includes zir will likely be established. This likelihood is also exemplified by observations that ZirS and ZirU share no readily identifiable secretion signal for ZirT (supplemental Fig. S3), suggesting a mode of transporter recognition unique from currently known mechanisms.
The newly identified ZirU is predicted to also contain at least one IgSF domain, which is supported by both computational modeling with Robetta (30) and sequence comparison to ZirS. Similar to ZirS, these domains are likely protein-binding modules for interaction with ZirS and ZirT. However, more experimentation will be needed to search for any additional ZirU extracellular binding partners, and to understand the role of ZirU in antivirulence. For example, a multimeric assembly of the Zir proteins into an adhesive surface may be required to recognize other partners. Likewise, although the ab initio model of ZirS(50–146) predicts a potential environmental sensor, the role of this domain also remains to be explored.
The discovery of ZirU as a component of the zir antivirulence pathway prompts the question if other genes encoded near ZirS and ZirT also participate in this system. The answer is likely yes, as STM1667, which we name here ZirR, is a thioredoxin (13). This is especially convincing because ZirR is predicted to be a glutaredoxin (34), a known negative regulator of OxyR, which therefore may act together in regulation of the zir pathway. Another protein, STM1671 (encoded adjacent to ZirU), is a Rob-like transcription factor (as predicted by Phyre with 100% confidence) related to the activator SoxS involved in defense against oxidative damage in Enterobacteria (61). STM1666 (encoded adjacent to ZirR) is a predicted ferredoxin-like fold (Phyre 100% confidence), similar to heme-degrading proteins such as IsdG (62). STM1665 is a putative phenylalanyl-tRNA synthetase (Phyre 100% confidence), proteins whose activity has been shown to be influenced by zinc (63). In concert, these surrounding genes may provide intracellular signaling cues coordinated to the self-adhesive functions and/or stimulation provided extracellularly by ZirUTS.
In our refolding experiments with the ZirT β domain, we found it interesting that the sequence predicted a minimal domain (88–368 or 88–390) that was insufficient for proper refolding. Inclusion of the N-terminal elements (27–87) was required for folding and by inference structural integrity of the domain. From secondary structure prediction this N-terminal region is completely α-helical. A common feature of structurally related outer membrane domains is an α-helical plug in the central pore that functions to facilitate secretion of a substrate (64). As our topology results indicate that the ZirT Ig-like domains are extracellular, the N-terminal region of ZirT is likely the periplasmic point of interaction with ZirS and ZirU for secretion. Given that helices of ∼20 amino acids are required to span the length of the membrane, one can reason that a third of the N-terminal helical region may cover the central lumen of the ZirT porin, with the remaining 40 amino acids extending into the periplasm to recognize secretion partners.
Given the previously observed inhibitory effects of zinc on the zir pathway (13), binding of the cation was an obvious first possibility to test. However, our ICP-MS data show that none of the available extracellular zir pathway components tightly associate with zinc. Although, the N-terminal domain was not included in this analysis due to poor solubility, one must also allow for the possibility that the zir pathway does not interact directly with zinc, but is rather regulated by the ion at the level of protein expression. During bacterial infection zinc and other metals are rapidly sequestered, a response known as nutritional immunity (65). Changing concentrations of zinc in the environment may thus allow Salmonella to regulate expression of the zir pathway and to cue antivirulence in response to encountering a host environment.
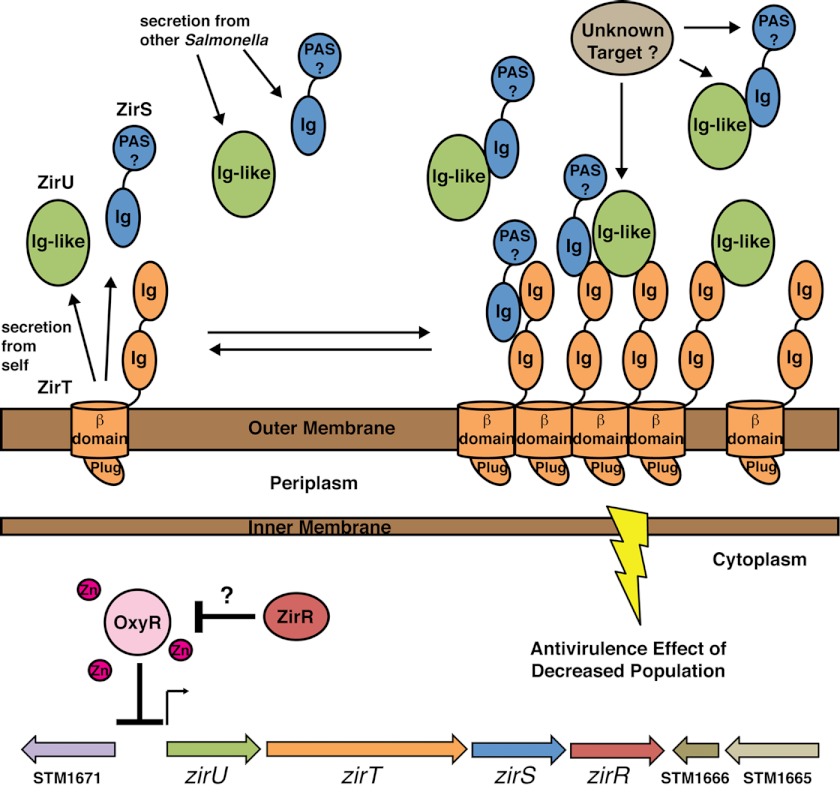
In light of the known biological regulation of the zir pathway combined with our structural and biochemical results, we can construct a first molecular model of this novel antivirulence mechanism (Fig. 8). In our model, the acute phase response (sequestering of zinc) and anaerobic respiration by Salmonella in the small intestine (17) sustain the observed high zir pathway levels (13). ZirT secretes both ZirS and ZirU into the extracellular space, or possibly the Salmonella containing vacuole, where they interact with themselves and with the extracellular Ig-like domains of ZirT that has self-associated through the β domain due to high concentrations from sustained expression. This patch of adhesive ZirT would form a binding platform for the building concentration of Zir proteins secreted from an increasing Salmonella population. We predict that a specific concentration of Zir complexes may provide a regulatory signal, the nature of which is still unknown, but could involve adhesion to other Salmonella in a manner similar to a biofilm or interaction with environmental solutes or host proteins through the ZirS N-terminal domain or perhaps ZirU. The build up of ZirS, and ZirU, would thus serve as a sensor of population or a change in the host environment to limit bacterial population growth and migration. Upon migration to systemic sites and exposure to reactive oxygen species from macrophages, OxyR activity is engaged and the zir pathway silenced to allow for increased survival and replication of Salmonella. In this model, the observed population differences between wild-type and zir deletions at the systemic sites (13) would stem from the zir antivirulence regulation in the intestine. We suspect that ZirR may also function in the intestine to prevent shut down of the system by low levels of OxyR, with the genes surrounding the zir pathway relaying intracellular signaling cues timed to the events occurring at the cell surface.
FIGURE 8.
Mechanistic model for the zir antivirulence pathway. Schematic representation of the structural and biochemical data presented in Figs. 1–7. Proteins are represented by colored ellipse and labeled by structural domain. The zir operon and surrounding genes are represented by a block arrow at the bottom of the figure. ZirRTSU are expressed during shedding and infection of the host gastrointestinal tract to regulate Salmonella growth. ZirT (orange) secretes both ZirS (blue) and ZirU (green), which are released into the extracellular space and can self-associate. ZirT maintains an equlibrium in the outer membrane, forming an Ig-rich adhesion surface for ZirS and ZirU. Upon the build up of enough ZirT, ZirS, and ZirU from infecting Salmonella a regulatory signal is engaged. This event may involve unknown protein targets (light brown) from Salmonella or the host that interact with the ZirS N-terminal domain or ZirU. Expression of the zir genes is negatively regulated by OxyR and zinc and may involve the thioredoxin protein ZirR (red). The adjacent genes STM1666, STM1665, and STM1671 encode predicted redox pathway proteins (see text) making them candidates for the cytoplasmic signaling events of the zir pathway.
Current research shows that the interaction between a pathogen and its host is an increasingly complex dynamic process that has been fine-turned by co-evolution. The zir pathway studied here is no exception, and based on our research, exerts its regulatory effects through an intricate mechanism involving the interactions between several proteins. In the context of the structural and biochemical data presented here, the zir antivirulence pathway is a multicomponent immunoglobulin adhesion system keyed to the environmental conditions of the host.
Supplementary Material
Acknowledgments
We thank Dr. Harold Jerome Coyne III for NMR tutelage, Dr. Emilie Lameignere for help with light scattering, Matthew Solomonson for help with ICP-MS, Dr. Fred Rosell of the LMB spectroscopy and kinetics hub for assistance with the circular dichroism measurements, Wanyin Deng for assistance with far Western experiments, and Dustin King for help with cross-linking.
This work was supported by the Human Frontier Science Program (HFSP), the Michael Smith Foundation for Health Research, the Canadian Institute of Health Research (CIHR), the Howard Hughes Medical Institute and Canada Research Chair Program. Instrument support was provided by the CIHR, the Canada Foundation for Innovation, the British Columbia Knowledge Development Fund, the UBC Blusson Fund, and the Michael Smith Foundation for Health Research.

This article contains supplemental Figs. S1–S6 and Tables S1 and S2.
The atomic coordinates and structure factors (code 2lv4) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The chemical shift assignments of ZirS have been deposited in the BioMagResBank under accession code 18553.
T. D. Goddard and D. G. Kneller, Sparky 3, University of San Francisco.
- MPD
- 2-methyl-2,4-pentanediol
- IgSF
- immunoglobulin superfamily
- SILAC
- stable isotope labeling of amino acids in cell culture
- ICP-MS
- inductively coupled plasma-mass spectrometry
- BID and Big
- bacterial immunoglobulin-like domains
- HSQC
- heteronuclear single quantum coherence.
REFERENCES
- 1. Coburn B., Sekirov I., Finlay B. B. (2007) Type III secretion systems and disease. Clin. Microbiol. Rev. 20, 535–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Galán J. E. (2009) Common themes in the design and function of bacterial effectors. Cell Host Microbe 5, 571–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Worrall L. J., Lameignere E., Strynadka N. C. (2011) Structural overview of the bacterial injectisome. Curr. Opin. Microbiol. 14, 3–8 [DOI] [PubMed] [Google Scholar]
- 4. Tegtmeyer N., Wessler S., Backert S. (2011) Role of the cag-pathogenicity island encoded type IV secretion system in Helicobacter pylori pathogenesis. FEBS J. 278, 1190–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marlovits T. C., Stebbins C. E. (2010) Type III secretion systems shape up as they ship out. Curr. Opin. Microbiol. 13, 47–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stebbins C. E., Galán J. E. (2001) Structural mimicry in bacterial virulence. Nature 412, 701–705 [DOI] [PubMed] [Google Scholar]
- 7. Matsumoto H., Young G. M. (2009) Translocated effectors of Yersinia. Curr. Opin. Microbiol. 12, 94–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Foreman-Wykert A. K., Miller J. F. (2003) Hypervirulence and pathogen fitness. Trends Microbiol. 11, 105–108 [DOI] [PubMed] [Google Scholar]
- 9. Lipsitch M., Moxon E. R. (1997) Virulence and transmissibility of pathogens. What is the relationship? Trends Microbiol. 5, 31–37 [DOI] [PubMed] [Google Scholar]
- 10. Cunningham M. L., Titus R. G., Turco S. J., Beverley S. M. (2001) Regulation of differentiation to the infective stage of the protozoan parasite Leishmania major by tetrahydrobiopterin. Science 292, 285–287 [DOI] [PubMed] [Google Scholar]
- 11. Mouslim C., Hilbert F., Huang H., Groisman E. A. (2002) Conflicting needs for a Salmonella hypervirulence gene in host and non-host environments. Mol. Microbiol. 45, 1019–1027 [DOI] [PubMed] [Google Scholar]
- 12. Parsons D. A., Heffron F. (2005) sciS, an icmF homolog in Salmonella enterica serovar Typhimurium, limits intracellular replication and decreases virulence. Infect. Immun. 73, 4338–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gal-Mor O., Gibson D. L., Baluta D., Vallance B. A., Finlay B. B. (2008) A novel secretion pathway of Salmonella enterica acts as an antivirulence modulator during salmonellosis PLoS Pathog. 4, e1000036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kingsley R. A., Bäumler A. J. (2002) Pathogenicity islands and host adaptation of Salmonella serovars. Curr. Top. Microbiol. Immunol. 264, 67–87 [PubMed] [Google Scholar]
- 15. Cano D. A., Martínez-Moya M., Pucciarelli M. G., Groisman E. A., Casadesús J., García-Del Portillo F. (2001) Salmonella enterica serovar Typhimurium response involved in attenuation of pathogen intracellular proliferation. Infect. Immun. 69, 6463–6474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grassl G. A., Finlay B. B. (2008) Pathogenesis of enteric Salmonella infections. Curr. Opin. Gastroenterol. 24, 22–26 [DOI] [PubMed] [Google Scholar]
- 17. Storz G., Imlay J. A. (1999) Oxidative stress. Curr. Opin Microbiol. 2, 188–194 [DOI] [PubMed] [Google Scholar]
- 18. McClelland M., Sanderson K. E., Spieth J., Clifton S. W., Latreille P., Courtney L., Porwollik S., Ali J., Dante M., Du F., Hou S., Layman D., Leonard S., Nguyen C., Scott K., Holmes A., Grewal N., Mulvaney E., Ryan E., Sun H., Florea L., Miller W., Stoneking T., Nhan M., Waterston R., Wilson R. K. (2001) Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413, 852–856 [DOI] [PubMed] [Google Scholar]
- 19. Fairman J. W., Dautin N., Wojtowicz D., Liu W., Noinaj N., Barnard T. J., Udho E., Przytycka T. M., Cherezov V., Buchanan S. K. (2012) Crystal structures of the outer membrane domain of intimin and invasin from enterohemorrhagic E. coli and enteropathogenic Y. pseudotuberculosis. Structure 20, 1233–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mazar J., Cotter P. A. (2007) New insight into the molecular mechanisms of two-partner secretion. Trends Microbiol. 15, 508–515 [DOI] [PubMed] [Google Scholar]
- 21. Sattler M., Schleucher J., Griesinger C. (1999) Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog. Nucl. Magn. Reson. Spectrosc. 34, 93–158 [Google Scholar]
- 22. Farrow N. A., Muhandiram R., Singer A. U., Pascal S. M., Kay C. M., Gish G., Shoelson S. E., Pawson T., Forman-Kay J. D., Kay L. E. (1994) Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33, 5984–6003 [DOI] [PubMed] [Google Scholar]
- 23. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) Nmrpipe, a multidimensional spectral processing system based on Unix Pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 24. Güntert P., Mumenthaler C., Wüthrich K. (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273, 283–298 [DOI] [PubMed] [Google Scholar]
- 25. Herrmann T., Güntert P., Wüthrich K. (2002) Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 319, 209–227 [DOI] [PubMed] [Google Scholar]
- 26. Shen Y., Delaglio F., Cornilescu G., Bax A. (2009) TALOS+. A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zwahlen C., Legault P., Vincent S. J. F., Greenblatt J., Konrat R., Kay L. E. (1997) Methods for measurement of intermolecular NOEs by multinuclear NMR spectroscopy. Application to a bacteriophage λ N-peptide/boxB RNA complex. J. Am. Chem. Soc. 119, 6711–6721 [Google Scholar]
- 28. Linge J. P., Williams M. A., Spronk C. A., Bonvin A. M., Nilges M. (2003) Refinement of protein structures in explicit solvent. Proteins 50, 496–506 [DOI] [PubMed] [Google Scholar]
- 29. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography & NMR system. A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 30. Chivian D., Kim D. E., Malmström L., Bradley P., Robertson T., Murphy P., Strauss C. E., Bonneau R., Rohl C. A., Baker D. (2003) Automated prediction of CASP-5 structures using the Robetta server. Proteins 53, 524–533 [DOI] [PubMed] [Google Scholar]
- 31. Raman S., Vernon R., Thompson J., Tyka M., Sadreyev R., Pei J., Kim D., Kellogg E., DiMaio F., Lange O., Kinch L., Sheffler W., Kim B. H., Das R., Grishin N. V., Baker D. (2009) Structure prediction for CASP8 with all-atom refinement using Rosetta. Proteins 77, 89–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cole C., Barber J. D., Barton G. J. (2008) The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36, W197–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Holm L., Rosenström P. (2010) Dali server. Conservation mapping in three-dimension. Nucleic Acids Res. 38, W545–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web. A case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 35. Eswar N., Eramian D., Webb B., Shen M. Y., Sali A. (2008) Protein structure modeling with MODELLER. Methods Mol. Biol. 426, 145–159 [DOI] [PubMed] [Google Scholar]
- 36. Whitmore L., Wallace B. A. (2008) Protein secondary structure analyses from circular dichroism spectroscopy. Methods and reference databases. Biopolymers 89, 392–400 [DOI] [PubMed] [Google Scholar]
- 37. Trinkle-Mulcahy L., Boulon S., Lam Y. W., Urcia R., Boisvert F. M., Vandermoere F., Morrice N. A., Swift S., Rothbauer U., Leonhardt H., Lamond A. (2008) Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J. Cell Biol. 183, 223–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Harpaz Y., Chothia C. (1994) Many of the immunoglobulin superfamily domains in cell adhesion molecules and surface receptors belong to a new structural set which is close to that containing variable domains. J. Mol. Biol. 238, 528–539 [DOI] [PubMed] [Google Scholar]
- 39. Hamburger Z. A., Brown M. S., Isberg R. R., Bjorkman P. J. (1999) Crystal structure of invasin. A bacterial integrin-binding protein. Science 286, 291–295 [DOI] [PubMed] [Google Scholar]
- 40. Ashkenazy H., Erez E., Martz E., Pupko T., Ben-Tal N. (2010) ConSurf 2010. Calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38, W529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Gapped BLAST and PSI-BLAST. A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dolinsky T. J., Czodrowski P., Li H., Nielsen J. E., Jensen J. H., Klebe G., Baker N. A. (2007) PDB2PQR. Expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 35, W522–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ward J. J., McGuffin L. J., Bryson K., Buxton B. F., Jones D. T. (2004) The DISOPRED server for the prediction of protein disorder. Bioinformatics 20, 2138–2139 [DOI] [PubMed] [Google Scholar]
- 44. Dyson H. J., Wright P. E. (2005) Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6, 197–208 [DOI] [PubMed] [Google Scholar]
- 45. Galperin M. Y. (2004) Bacterial signal transduction network in a genomic perspective. Environ. Microbiol. 6, 552–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Galperin M. Y. (2006) Structural classification of bacterial response regulators. Diversity of output domains and domain combinations. J. Bacteriol. 188, 4169–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hefti M. H., Françoijs K. J., de Vries S. C., Dixon R., Vervoort J. (2004) The PAS fold. A redefinition of the PAS domain based upon structural prediction. Eur. J. Biochem. 271, 1198–1208 [DOI] [PubMed] [Google Scholar]
- 48. Ong S. E., Kratchmarova I., Mann M. (2003) Properties of 13C-substituted arginine in stable isotope labeling by amino acids in cell culture (SILAC) J. Proteome Res. 2, 173–181 [DOI] [PubMed] [Google Scholar]
- 49. Münch R., Hiller K., Grote A., Scheer M., Klein J., Schobert M., Jahn D. (2005) Virtual footprint and PRODORIC. An integrative framework for regulon prediction in prokaryotes. Bioinformatics 21, 4187–4189 [DOI] [PubMed] [Google Scholar]
- 50. Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007) Locating proteins in the cell using TargetP, SignalP, and related tools. Nat. Protoc. 2, 953–971 [DOI] [PubMed] [Google Scholar]
- 51. Söding J. (2005) Protein homology detection by HMM-HMM comparison. Bioinformatics 21, 951–960 [DOI] [PubMed] [Google Scholar]
- 52. Lippi M., Passerini A., Punta M., Rost B., Frasconi P. (2008) MetalDetector. A web server for predicting metal-binding sites and disulfide bridges in proteins from sequence. Bioinformatics 24, 2094–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Michaux C., Pomroy N. C., Privé G. G. (2008) Refolding SDS-denatured proteins by the addition of amphipathic cosolvents. J. Mol. Biol. 375, 1477–1488 [DOI] [PubMed] [Google Scholar]
- 54. Dersch P., Isberg R. R. (1999) A region of the Yersinia pseudotuberculosis Invasin protein enhances integrin-mediated uptake into mammalian cells and promotes self-association. EMBO J. 18, 1199–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Luo Y., Frey E. A., Pfuetzner R. A., Creagh A. L., Knoechel D. G., Haynes C. A., Finlay B. B., Strynadka N. C. (2000) Crystal structure of enteropathogenic Escherichia coli intimin-receptor complex. Nature 405, 1073–1077 [DOI] [PubMed] [Google Scholar]
- 56. Tsai J. C., Yen M. R., Castillo R., Leyton D. L., Henderson I. R., Saier M. H., Jr. (2010) The bacterial intimins and invasins. A large and novel family of secreted proteins. PLoS One 5, e14403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Raman R., Rajanikanth V., Palaniappan R. U., Lin Y. P., He H., McDonough S. P., Sharma Y., Chang Y. F. (2010) Big domains are novel Ca2+-binding modules. Evidences from big domains of Leptospira immunoglobulin-like (Lig) proteins. PLoS One 5, e14377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Allen W. J., Phan G., Waksman G. (2012) Pilus biogenesis at the outer membrane of Gram-negative bacterial pathogens. Curr. Opin Struct. Biol., in press [DOI] [PubMed] [Google Scholar]
- 59. Le Trong I., Aprikian P., Kidd B. A., Forero-Shelton M., Tchesnokova V., Rajagopal P., Rodriguez V., Interlandi G., Klevit R., Vogel V., Stenkamp R. E., Sokurenko E. V., Thomas W. E. (2010) Structural basis for mechanical force regulation of the adhesin FimH via finger trap-like β sheet twisting. Cell 141, 645–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sauer F. G., Pinkner J. S., Waksman G., Hultgren S. J. (2002) Chaperone priming of pilus subunits facilitates a topological transition that drives fiber formation. Cell 111, 543–551 [DOI] [PubMed] [Google Scholar]
- 61. Kwon H. J., Bennik M. H., Demple B., Ellenberger T. (2000) Crystal structure of the Escherichia coli Rob transcription factor in complex with DNA. Nat. Struct. Biol. 7, 424–430 [DOI] [PubMed] [Google Scholar]
- 62. Lee W. C., Reniere M. L., Skaar E. P., Murphy M. E. (2008) Ruffling of metalloporphyrins bound to IsdG and IsdI, two heme-degrading enzymes in Staphylococcus aureus. J. Biol. Chem. 283, 30957–30963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mayaux J. F., Blanquet S. (1981) Binding of zinc to Escherichia coli phenylalanyl transfer ribonucleic acid synthetase. Comparison with other aminoacyl transfer ribonucleic acid synthetases. Biochemistry 20, 4647–4654 [DOI] [PubMed] [Google Scholar]
- 64. Clantin B., Delattre A. S., Rucktooa P., Saint N., Méli A. C., Locht C., Jacob-Dubuisson F., Villeret V. (2007) Structure of the membrane protein FhaC. A member of the Omp85-TpsB transporter superfamily. Science 317, 957–961 [DOI] [PubMed] [Google Scholar]
- 65. Kehl-Fie T. E., Skaar E. P. (2010) Nutritional immunity beyond iron. A role for manganese and zinc. Curr. Opin. Chem. Biol. 14, 218–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.