Abstract
Purpose
A major factor limiting the effective clinical management of colorectal cancer (CRC) is resistance to chemotherapy. Therefore, the identification of novel, therapeutically targetable mediators of resistance is vital.
Experimental design
We used a CRC disease-focused microarray platform to transcriptionally profile chemotherapy-responsive and non-responsive pre-treatment metastatic CRC liver biopsies and in vitro samples, both sensitive and resistant to clinically relevant chemotherapeutic drugs (5-FU and oxaliplatin). Pathway and gene set enrichment analyses (GSEA) identified candidate genes within key pathways mediating drug resistance. Functional RNAi screening identified regulators of drug resistance.
Results
MAPK signalling, focal adhesion, cell cycle, insulin signalling and apoptosis were identified as key pathways involved in mediating drug resistance. The G-protein coupled receptor galanin receptor 1 (GalR1) was identified as a novel regulator of drug resistance. Notably, silencing either GalR1 or its ligand galanin, induced apoptosis in drug-sensitive and resistant cell lines and synergistically enhanced the effects of chemotherapy. Mechanistically, GalR1/galanin silencing resulted in down-regulation of the endogenous caspase 8 inhibitor FLIPL, resulting in induction of caspase 8-dependent apoptosis. Galanin mRNA was found to be overexpressed in colorectal tumours, and importantly, high galanin expression correlated with poor disease-free survival of early stage CRC patients.
Conclusion
This study demonstrates the power of systems biology approaches to identify key pathways and genes that are functionally involved in mediating chemotherapy resistance. Moreover, we have identified a novel role for the GalR1/galanin receptor-ligand axis in chemo-resistance, providing evidence to support its further evaluation as a potential therapeutic target and biomarker in CRC.
INTRODUCTION
Colorectal cancer (CRC) is the second leading cause of cancer-related deaths in Europe and North America. 5-Fluorouracil (5-FU)-based chemotherapy regimens remain the standard treatment for CRC in both the adjuvant and advanced disease settings. However, response rates to 5-FU therapy are between 10-20% in the metastatic setting (1). Combining 5-FU with the DNA-damaging agent oxaliplatin has significantly improved response rates for advanced colorectal cancer to 40-50%, however 5 year overall survival remains less than 5% (2). Resistance to chemotherapy, either intrinsic or acquired, ultimately results in treatment failure for the vast majority of patients with metastatic disease and also is a major problem in the locally advanced disease setting, where 40-50% patients relapse (3).Therefore, novel therapeutic strategies are urgently required.
The anti-cancer activities of 5-FU include thymidylate synthase (TS) inhibition and incorporation of its metabolites into RNA and DNA (4). Known determinants of 5-FU response include expression levels of TS and the 5-FU catabolic enzyme dihydropyrimidine dehydrogenase (DPD) which mediates the degradation of 5-FU (4). Oxaliplatin is a third generation platinum compound which forms platinum-DNA adducts leading to cytotoxicity. Known determinants of response include expression of excision repair cross-complementing 1 protein (ERCC1) and xeroderma pigmentosum group A (XPA). These are central components of the nucleotide excision repair (NER) pathway, which is the major repair pathway of platinum drug-induced DNA damage (5). Despite our understanding of many of the pathways that regulate 5-FU- and oxaliplatin-induced cell death and the mechanisms by which resistance to these chemotherapeutic agents arises, this knowledge has failed to translate to clinical practice. There is, therefore, an urgency to identify novel mediators of 5-FU/oxaliplatin resistance that are “druggable” and/or useful predictive biomarkers of clinical response.
In the present study, we used a systems biology approach incorporating microarray profiling of clinical and in vitro samples, bioinformatic analyses and functional RNAi screens to identify novel mediators of resistance to 5-FU and oxaliplatin. Our group has previously used gene expression profiles derived from cell line models of known drug sensitivity to generate classifiers that predict clinical response to 5-FU/SN-38 therapy (6). This type of approach also provides novel mechanistic insights into how drug resistance arises. RNAi has proved to be a powerful tool in the identification of genes that are synthetically lethal with oncogenes such as Kras (7), and genes that impact on drug sensitivity in vitro (8). In the present study, we identified the mitogen-activated protein kinase (MAPK) signaling, focal adhesion, cell cycle, insulin signaling and apoptosis pathways as important regulators of chemo-resistance in CRC. In particular, for the first time we report the stem cell marker and regulator, galanin (9-14), and its receptor the G-protein coupled receptor galanin receptor 1 (GalR1) as key determinants of drug resistance and potential therapeutic targets for combating drug resistance. Mechanistically, we identify a novel role for the GalR1/galanin receptor-ligand axis as an important upstream regulator of expression of the anti-apoptotic protein FLIPL. Clinically, galanin mRNA was found to be overexpressed in colorectal tumours, and notably, high galanin mRNA expression correlated with poor disease-free survival in early stage disease.
MATERIALS AND METHODS
Materials
5-FU and Oxaliplatin were purchased from Sigma Chemical Co. (St. Louis, MO) and Abatra Technology Co, LTD (China) respectively. zVAD (OMe)-FMK was purchased from Calbiochem (Darmstadt, Germany). All siRNAs were purchased from Qiagen (Crawley, UK).
Cell culture
Authentication and culture of CRC cell lines HCT116, LS174T, RKO, HT29, SW620 and LoVo, has been described previously (15). LS174T (2008), SW620 (2008) and RKO (2001) cells were obtained from the American Type Culture Collection (ATCC). The p53 wild-type HCT116 human colon cancer cell line, kindly provided by Prof. Bert Vogelstein (Johns Hopkins University, Baltimore, MD) in 2003. The 5-FU-, SN-38 and oxaliplatin-resistant HCT116 sub-lines and the oxaliplatin-resistant p53 null HCT116 sub-line were generated in our laboratory as previously described (16). The LoVo (2004) cells were obtained from the European Collection of Cell Cultures (ECACC). RKO and HT-29 human cell lines were provided by the National Cancer Institute. The 5-FU-and SN-38-resistant LoVo and HT29 sub-lines were generated in our laboratory by continuous exposure of increasing doses of chemotherapy over an ~10 month period. The c-FLIPL overexpressing cell lines were previously described (17).
Patient Samples
Eight patients with metastatic colorectal cancer were included in the present study. All patients provided written fully informed consent as per IRB guidelines in the University of Southern California and approval was granted from this body. Patients underwent biopsy of colorectal liver metastases prior to commencing oxaliplatin/5-FU chemotherapy. CT imaging for response evaluation using WHO criteria was performed every 6 weeks and patients were subsequently designated as responders, stable disease or progressive disease. Of these 8 patients, 1 had a partial response to treatment, 3 had stable disease and 4 had progressive disease on treatment. For the purpose of this study we have further defined ‘responders’ as those patients with either partial response or stable disease and ‘non-responders’ as those patients with progressive disease.
Microarray analysis
Total RNA was extracted from 8 pre-treatment metastatic tumour biopsies from patients with advanced colorectal cancer and profiled on the Colorectal Disease Specific Array (DSA™) Research Tool (Almac Diagnostics, Craigavon, UK). In addition, in vitro analyses were also carried out using the Colorectal DSA™. The in vitro analyses have been previously described for the 5-FU experiment (18) and the same experimental design was used for the oxaliplatin experiment. Detailed experimental protocols and raw expression data are available at ArrayExpress repository (19) (Accession number E-MEXP-3368 (Clinical analysis) and E-MEXP-1691 (in vitro analysis)).
Bioinformatic analysis
Generation of genelists
All data analysis was completed within Genespring v7.3.1 (Agilent Technologies, UK). All genelists were generated as previously described (6). The filtering criteria used was (i) all data passing Affymetrix flag calls (P/M) in all samples were retained, (ii) all data passing the cross gene error model with genes displaying control values greater than the average base/proportional value being retained (this was required in all samples for any gene), (iii) the list was filtered using a 1.5-fold cut-off for each gene relative to the control array and (iv) all data passing P<0.05 were retained. All data are displayed as log2 transformed. For the in vitro analysis, the untreated parental samples were compared to the drug-treated parental samples (inducible parental), the untreated drug-resistant samples were compared to the drug treated drug-resistant samples (inducible resistant) and the untreated parental cells were compared to the untreated drug-resistant cells (basally deregulated), for both the 5-FU experiment and the oxaliplatin experiment. For the clinical analysis a genelist was created of those genes that were altered between 5-FU/oxaliplatin responding (PR+SD) and non-responding patients (PD). No patients achieved a complete response (CR).
Pathway analysis
All pathway analysis was carried out using Genespring v7.3.1 (Agilent Technologies, UK) using both KEGG and GenMAPP pathways. Briefly, the final working genelists for both the in vitro experiments and the clinical experiments were used as the starting genelists and pathways were selected that contained greater than 5 genes per pathway. Statistical analysis for each pathway was carried out using hypergeometric statistics.
Gene Set Enrichment Analysis (GSEA)
GSEA is a computational method for determining whether a rank-ordered list of genes for a particular comparison of interest is enriched in genes derived from an independently generated gene set (20). GSEA was performed with the expression dataset derived from the in vitro samples (inducible parental, inducible resistant and constitutively altered) and then the clinical samples (responders v non-responders). Gene sets from the Molecular Signatures Database (http://www.broad.mit.edu/gsea/msigdb/index.jsp) were used for the enrichment study including the curated gene sets from online pathway databases, publications in PubMed and knowledge of domain experts (C2) and the gene ontology (GO) gene sets (C5). A false discovery rate (FDR) below 25% was considered for significant enrichment.
RNAi Screening
This was performed using siRNAs targeting pre-selected genes identified from microarray analysis. siRNA transfection conditions have been previously described (15). The drugs used were 5-FU (IC30(48h)) and oxaliplatin (IC30(48h)). Cell viability was determined by using a 3-(4, 5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT, Sigma) assay as described previously (17). Combination index (CI) values were calculated as previously described (21). The ToxiLight® BioAssay Kit was purchased from Lonza Rockland, Inc. (Rockland, ME, USA) and was used according to the manufacturer’s instructions.
The RNAi screen comprised primary, secondary and tertiary screens. The primary screen used single siRNAs targeting 84 candidate genes. Two-way ANOVA was used to classify positive hits as targets which induced a significant increase (P<0.05) in relative toxicity (cell death/cell viability), with either siRNA alone or siRNA combined with drug. This identified 30 positive hits which were carried forward to a secondary RNAi screen incorporating an additional 3 siRNA sequences per target. Positive hits were classified by a significant increase in relative toxicity with either siRNA alone or siRNA combined with drug for ≥2 of the additional siRNA sequences. This identified 11 positive hits which were carried forward to a tertiary RNAi screen. For each target, the optimum chemo-sensitizing siRNA from the primary and secondary RNAi screens was used in a panel of 6 independent CRC cell lines. siFGFR4_4 displayed particularly high levels of relative toxicity alone and was not considered. A positive hit was classified as a significant increase in relative toxicity with either siRNA alone or siRNA combined with drug in ≥3 CRC cell lines; this identified 6 positive hits.
Q-PCR analysis
siRNA silencing of the candidate genes was confirmed by Q-PCR, as described previously (15). Final expression values were normalised to GAPDH expression and presented relative to the untreated time-matched siRNA control (SC).
Western blot analysis
Western blots were performed as previously described (22). Caspase 8 (12F5; Alexis) and Poly-ADP-Ribose-Polymerase-1 (PARP; eBioscience, San Diego, CA) mouse monoclonal antibodies were used in conjunction with a horseradish peroxidase-conjugated sheep anti-mouse secondary antibody (Amersham, UK). Caspase 3 rabbit polyclonal antibody (Cell Signaling Technology, Inc.) was used in conjunction with anti-rabbit secondary antibody (Amersham, UK). GALR1 (Sigma) goat polyclonal antibody was used in conjunction with donkey anti-goat secondary antibody (Santa Cruz). Equal loading was assessed using GAPDH or actin (AbD Serotec, UK).
Annexin V/Propidium Iodide analysis
Cells were harvested and analysed according to the manufacturer’s instructions (BD Biosciences, Oxford, UK). Annexin V/Propidium iodide (PI) analysis was carried out using the EPICS XL Flow Cytometer (Coulter, Miami, FL, USA). Briefly, levels of apoptosis were calculated as the sum of FITC-Annexin V positive/PI negative (early apoptosis) and FITC-Annexin V positive/PI positive (late apoptosis) cell population.
Caspase Activation Assays
Caspase activity in cell protein isolates was measured using Caspase Glo 3/7, 8 and 9 assays (Promega).
Analysis of public CRC datasets
Association of target gene expression with CRC tumour
Public CRC microarray datasets were accessed (23) and CRC tumour expression data for Galanin mRNA was downloaded (24, 25).
Association of target gene expression with clinical response
Public CRC microarray datasets were accessed (26) and Galanin expression data sets GSE17536 (27) and GSE14333 (28) were downloaded. GraphPad Prism 5 software was used to generate a Kaplan-Meier survival curves for high and low expression groups. These were dichotomized at the optimal cutpoint, as calculated by log-rank test (26).
RESULTS
Generation of in vitro- and clinically-derived genelists
An overview of our experimental approach is depicted in figure 1. Bioinformatic analysis of the 5- FU in vitro data identified 1329 genes that were constitutively altered between the parental and resistant cell lines (Table S1a), 1389 genes that were 5-FU inducible in parental cells (Table S1b), and 922 genes that were 5-FU-inducible in 5-FU-resistant cells (Table S1c). For oxaliplatin, 1164 genes were identified that were constitutively altered between the parental and resistant cell lines (Table S1d), 643 genes that were oxaliplatin-inducible in parental cells (Table S1e) and 373 genes that were oxaliplatin-inducible in oxaliplatin-resistant cells (Table S1f). In the clinical samples, 939 genes were differentially expressed when responding (PR+SD) and non-responding (PD) tumours were compared (Table S1g).
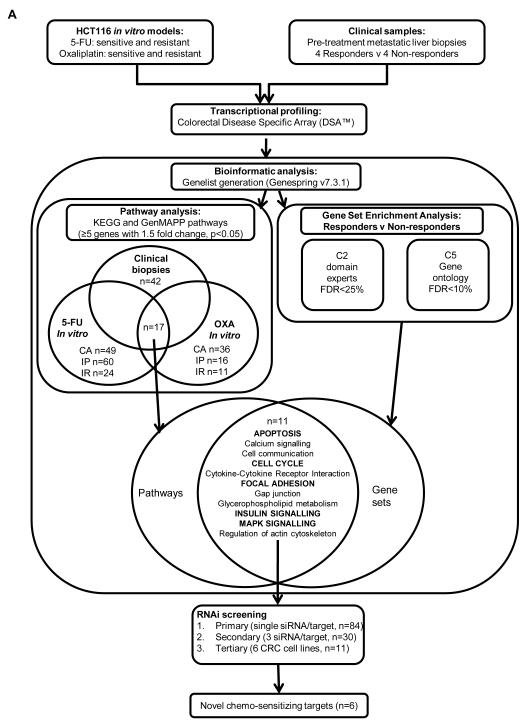
Fig. 1. Experimental overview.
Schematic of the systems biology approach used to identify novel determinants of resistance to 5-FU and oxaliplatin in CRC. Microarray profiling was carried out on sensitive and resistant in vitro models and clinical samples from responding and non responding patients. Genelists generated from the microarray data were analysed by pathway analysis and gene set enrichment analysis to identify key pathways mediating resistance to 5-FU and oxaliplatin. Candidate genes from the selected pathways were functionally assessed by RNAi screening. CA=constitutively altered, IP=inducible parental, IR=inducible resistant, FDR=false discovery rate.
Pathway Analysis
An overview of our bioinformatic analyses is outlined in figure 1. Pathways were selected that contained >5 differentially expressed genes. This was applied to the clinical genelist, and 3 in vitro gene lists: (i) genes inducible in the parental cell line (IP); (ii) genes constitutively altered between parental and resistant cell lines (CA) and (iii) genes inducible in the resistant cell line (IR), and this was carried out for both the 5-FU and oxaliplatin in vitro data sets. Pathways that were found to be present in ≥4 genelists are listed in Table 1. In the 5-FU experiment, 49 pathways were constitutively altered (Table S2a), 60 pathways were 5-FU-inducible in parental cells (Table S2b) and 24 pathways were 5-FU-inducible in 5-FU-resistant cells (Table S2c). In the oxaliplatin in vitro experiment, 36 pathways were constitutively altered (Table S2d), 16 pathways were oxaliplatin-inducible in parental cells (Table S2e), and 11 pathways were oxaliplatin-inducible in oxaliplatin-resistant cells (Table S2f). When pathway analysis was carried out on those genes altered between responding and non-responding patients, a large number (n=42) of differentially regulated pathways were identified (Table S2g). The most significant pathways identified in these analyses are presented in Table 1; notably, these pathways overlap significantly with those identified in the in vitro analyses.
Table 1.
Pathways common to Gene set enrichment analysis (GSEA) of the clinical genelist and pathway analysis of in vitro and clinical genelists. Pathways identified by pathway analysis contained ≥5 genes with a 1.5 fold-change and P<0.05. Only pathways identified from ≥4 genelists are listed.
| PATHWAY | PATHWAY ANALYSIS | GSEA | ||||||
|---|---|---|---|---|---|---|---|---|
| 5-FU in vitro |
Oxaliplatin in vitro |
Clinical 5FU/OXA |
Clinical 5FU/OXA |
|||||
|
Inducible
Parental |
Constitutively
Altered |
Inducible
Resistant |
Inducible
Parental |
Constitutively
Altered |
Inducible
Resistant |
R v NR | R v NR | |
| Adherens Junction | 6 | 13 | 8 | 12 | ||||
| Apoptosis | 7 | 5 | 7 | 8 | 5 |
|
||
| Axon guidance | 14 | 11 | 8 | 13 | ||||
| Calcium signalling | 8 | 9 | 7 | 9 | 8 | 10 |
|
|
| Cell communication | 5 | 6 | 5 | 14 |
|
|||
| Cell cycle | 21 | 11 | 13 | 8 | 14 | 14 |
|
|
| Cytokine-cytokine receptor interaction |
5 | 9 | 6 | 6 | 17 |
|
||
| Focal adhesion | 16 | 10 | 13 | 12 | 11 | 19 |
|
|
| Gap junction | 5 | 7 | 6 | 6 |
|
|||
| Glycerophospholipid metabolism |
8 | 6 | 6 | 7 |
|
|||
| Insulin signalling | 20 | 8 | 12 | 7 | 5 | 5 | 9 |
|
| MAPK signalling | 19 | 12 | 14 | 18 | 6 | 17 |
|
|
| Purine metabolism | 23 | 17 | 12 | 10 | 20 | 17 | ||
| Pyrimidine metabolism |
19 | 10 | 12 | 5 | 21 | 15 | ||
| Regulation of Actin cytoskeleton |
15 | 8 | 16 | 14 | 5 | 22 |
|
|
| Tight Junction | 6 | 7 | 14 | 12 | 14 | |||
| Wnt signalling | 12 | 8 | 8 | 5 | 8 | 10 | ||
Gene Set Enrichment Analysis (GSEA)
GSEA was carried out using the clinical responder versus non-responder genelist as the molecular profile data with the C2 and C5 geneset databases. GSEA using the C2 geneset database, which contains the functional sets, identified 48 genesets were significantly enriched (false discovery rate (FDR) of <25%) in responding patients, while 2 genesets were significantly enriched (FDR<25%) in non-responding patients (Table S3a). When the analysis was carried out using the C5 geneset database, which contains the GO genesets, 171 genesets were significantly enriched (FDR<10%) in responding patients, while 10 genesets were significantly enriched (FDR<10%) in non-responding patients (Table S3b). GSEA was also carried out using the clinical responder versus non-responder genelist as the molecular profile data with the oxaliplatin basally deregulated in vitro gene list as a user-defined geneset. This identified a list of genes (n=508) that were enriched and up-regulated in both oxaliplatin-resistant cells and non-responding patients (Table S3c).
Overlap between pathway analysis and GSEA
When the C2 and C5 GSEA results were compared to the pathway analysis results, apoptosis, calcium signalling, cell cycle, cell communication, cytokine-cytokine receptor interaction, focal adhesion/MAPK signalling, Gap junction, glycerophospholipid metabolism, insulin signalling and regulation of actin cytoskeleton were found to significantly correlate with the GSEA (Figure 1 and Table 1). From this list, we selected apoptosis, cell cycle, focal adhesion, MAPK signalling and insulin signalling to study further.
Candidate gene selection
Candidate genes (n=84) were chosen that passed a 1.5-fold change in expression with a P value <0.05 within the identified pathways, and nodal points within these pathways were prioritized (Table S4). Ligands such as the fibroblast growth factors were identified within the MAPK signaling pathway, however, we opted to silence their receptors, to reduce the number of candidate genes to be screened.
Primary RNAi screen
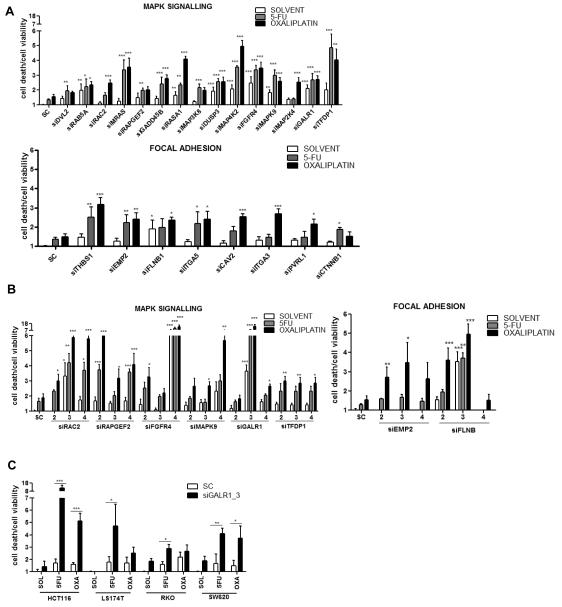
To identify genes which when down-regulated enhanced sensitivity to 5-FU/oxaliplatin, we used the HCT116 cell line for the primary RNAi screen and single siRNAs against 84 candidate genes. A positive hit was defined as a significant increase (P<0.05) in relative toxicity (cell death/cell viability) with the targeting siRNA either alone or combined with chemotherapy, and this was determined by 2-way-ANOVA. Thirty positive hits were identified in the primary RNAi screen: 15 MAPK targets, 8 focal adhesion targets (Fig. 2A), 3 cell cycle targets, 2 insulin signalling targets, and 2 apoptosis targets (Fig. S1A). The positive hits were carried forward to the secondary RNAi screen. A significant interaction (P<0.05) between targeting siRNA and drug was observed for 11 of these genes (Table S5a).
Fig. 2. Positive hits from RNAi screening.
(A) The primary RNAi screen was carried out in the HCT116 cell line with 1 siRNA sequence per target to functionally assess 84 candidate genes from key pathways. A positive ‘hit’ was defined by 2-way ANOVA as a significant (P<0.05) increase in relative toxicity (cell death/cell viability) with siRNA alone or siRNA combined with drug. (B) A secondary RNAi screen used an additional 3 siRNA sequences per target. A positive hit was classified as a significant increase in relative toxicity alone or combined with drug with ≥2 additional siRNA sequences. (C) A tertiary RNAi screen used the optimum chemo-sensitizing siRNA from the previous screens and incorporated a panel of 6 independent CRC cell lines. A positive hit was classified as a significant increase in relative toxicity alone or combined with drug in ≥3 cell lines. Results are given as mean values ± SEM of triplicate measurements. *P<0.05; **P<0.01; ***P<0.001. SC=siRNA control; si=targeted siRNA; SOL=solvent control.
Secondary RNAi screen
To validate the primary screen ‘hits’, 3 additional siRNAs per target were used, and sensitizing effects were determined as before. Multiple siRNAs reproduced a sensitizing phenotype for 11 of these targets: 6 MAPK targets (RAC2, RAPGEF2, FGFR4, MAPK9, GALR1, and TFDP1), 2 focal adhesion targets (EMP2 and FLNB) (Fig. 2B), and 3 cell cycle targets (CENPE, CDC20 and SMC4) (Fig. S1B). A significant chemotherapy interaction was observed for siRNAs targeting RAC2, RAPGEF2, MAPK9 or GalR1 (Table S5b). Gene silencing was confirmed by Q-PCR (Fig. S2A and B).
Tertiary RNAi screen
The focus of further validation was based on the MAPK/focal adhesion targets identified in Fig. 2B. To rule out cell line-specific effects, a panel of 6 independent colorectal cancer cell lines (HCT116, LS174T, RKO, SW620, LoVo and HT29) were incorporated into a tertiary RNAi screen. The optimum chemo-sensitizing siRNA for each target from the previous screens was used. Significant 5-FU/oxaliplatin sensitizing effects (P<0.05) were determined as before. A sensitizing phenotype was observed across the cell line panel for 6 targets: GalR1 (Fig. 2C), MAPK9, RAC2, FGFR4, EMP2 and TFDP1 (Fig. S1C). As a highly novel and potentially druggable target, the GalR1/galanin receptor/ligand axis, was selected for further functional analyses.
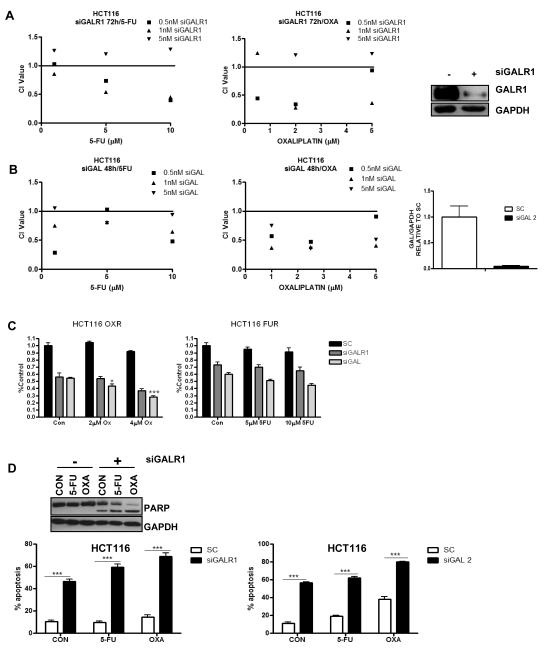
GalR1/galanin silencing synergistically enhances the effects of chemotherapy
HCT116 cells were transfected for 24h with GalR1 siRNA (siGALR1) or galanin siRNA (siGAL) prior to a 24h/48h co-treatment with IC30(48h) doses of 5-FU/oxaliplatin. GalR1 silencing was confirmed by Western blot (Fig. 3A, inset) and galanin silencing was confirmed by Q-PCR (Fig. 3B, inset). Cell viability was determined by MTT assay, and combination index (CI) values were calculated (Fig. 3A). These analyses demonstrated that siGALR1 was synergistic with 5-FU and oxaliplatin treatment, particularly at lower concentrations of siRNA. Furthermore, synergy was also observed between siGAL and chemotherapy treatment (Fig. 3B). Synergy was also demonstrated in the RKO and LS174T cell lines following GalR1 silencing or galanin silencing (Fig. S3A and data not shown) and the H630 cell line following galanin silencing (data not shown).
Fig. 3. Galanin/GalR1 silencing is synergistic with chemotherapy treatment.
Cell viability assays were conducted in the HCT116 parental cell line and the method of Chou and Talalay was used to evaluate the interaction between (A) GalR1 siRNA (siGALR1) or (B) galanin siRNA (siGAL) and increasing concentrations of either 5-FU or oxaliplatin at 48/72h. CI values <1, =1, and >1 indicating synergism, additivity, and antagonism, respectively. GALR1 silencing was confirmed by Western blot (A, inset) and galanin silencing was confirmed by Q-PCR (B, inset). (C) Cell viability of HCT116 oxaliplatin-resistant (OXR) and 5-FU-resistant (FUR) cell lines following galanin/GalR1 silencing (10nM siRNA) for 24h prior to 48h co-treatment with parental IC30(48h) and IC50(48h) doses of chemotherapy. A synergistic interaction was determined by 2-way ANOVA. *P<0.05; ***P<0.001 (D) Annexin V/PI flow cytometric analysis was used to measure apoptosis levels in HCT116 parental cells following galanin/GalR1 silencing (5nM siRNA) for 24h prior to a 24h co-treatment with IC30(48h) doses of either 5-FU or oxaliplatin. (D, inset) Western blot showing PARP cleavage in HCT116 cells transfected with 5nM siGALR1 for 24h prior to a 24h co-treatment with ~IC30(48h) doses of either 5-FU or oxaliplatin. Results are given as mean values ± SEM of triplicate measurements. *P<0.05; **P<0.01; ***P<0.001. SC=siRNA control.
Drug resistant HCT116 models were also transfected for 24h with siGALR1 or siGAL prior to 48h co-treatment with parental IC30(48h) and IC50(48h) doses of 5-FU/oxaliplatin. A significant reduction in cell viability was observed following GalR1 or galanin silencing alone in both the oxaliplatin-and 5-FU resistant HCT116 models, and this was further reduced when combined with chemotherapy treatment (Fig. 3C). This reduction in viability was synergistic for siGAL and oxaliplatin treatment. A similar interaction was observed in the oxaliplatin-resistant HCT116 p53 null model (Fig. S3B). Silencing of galanin alone but not GalR1, significantly reduced cell viability in other 5-FU-resistant models (LoVo and HT29) (Fig. S3C). Of note, galanin/GalR1 silencing also significantly reduced cell viability in models (HCT116, LoVo and HT29) resistant to the active metabolite of irinotecan, the other chemotherapeutic agent currently approved for use in advanced CRC (Fig. S3D). These results indicate that drug resistant models maintain the dependency on galanin signalling observed in the parental drug-sensitive cell lines.
GalR1/galanin silencing induces apoptosis in CRC cell lines
To determine the mechanism of chemo-sensitization following silencing of GalR1 and galanin, HCT116 cells were transfected with 5nM siGALR1 or siGAL for 24h prior to a 24h co-treatment with ~IC30(48h) doses of 5-FU or oxaliplatin; apoptosis levels were determined by flow cytometry. GalR1 silencing for 48h resulted in significantly higher levels of apoptosis (~47%) compared to ~10% in the control siRNA (SC) transfected cells (P<0.001) (Fig. 3D). This was further enhanced to ~59% when combined with 5-FU and ~69% when combined with oxaliplatin; 2-way ANOVA confirmed a significant interaction between siGALR1 and 5-FU/oxaliplatin (P<0.001). Similar results were observed following galanin silencing (Fig. 3D) and Western blot analysis demonstrated PARP cleavage (a hallmark of apoptosis) following GalR1 silencing (Fig. 3D, inset), supporting the flow cytometry results. This chemo-sensitizing phenotype was reproduced using a second siRNA sequence for both GalR1 (siGALR1_1) and galanin (siGAL_5) (Fig. S4A). To assess whether the effects were cell line-specific, LS174T and RKO CRC cell lines were analysed (Fig. S4B). In these models, the effect of GalR1/galanin silencing on induction of apoptosis was less than in the HCT116 cells. However, the sensitization to chemotherapy-induced apoptosis was still observed in both these models and was again particularly marked for oxaliplatin. A significant induction of apoptosis was observed following GalR1 or galanin silencing alone in drug resistant models (Fig. S4C) and Western blot analysis demonstrated PARP cleavage and activation of the executioner caspase, caspase 3 following GalR1/galanin silencing, further supporting the flow cytometry results (Fig. S4D).
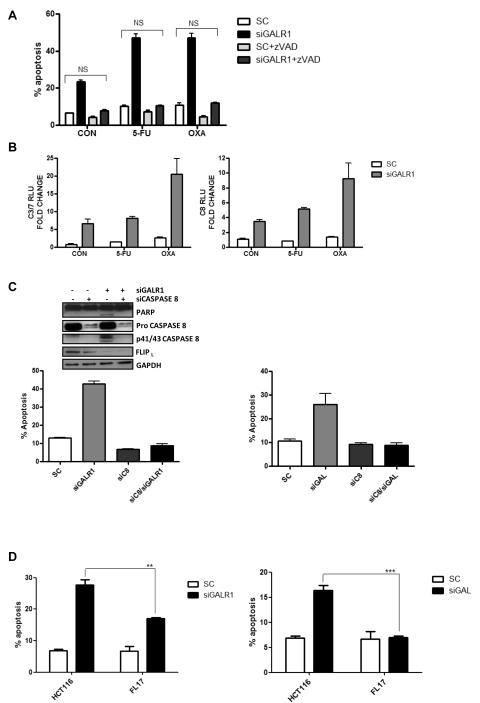
GalR1/galanin silencing down-regulates FLIPL and activates caspase 8-dependent apoptosis
Caspase-dependent apoptosis following GALR1 silencing was confirmed using the pan-caspase inhibitor, zVAD, which completely attenuated apoptosis (Fig. 4A). Analysis of caspase activity following GalR1 silencing indicated that there was a close correlation between the activity of the executioner caspases 3 and 7 and the initiator caspase, caspase 8 (Pearson’s correlation coefficient of r=0.99), suggesting a role for caspase 8 in initiation of the apoptotic phenotype (Fig. 4B). To investigate this further, HCT116 cells were co-transfected with siGALR1 and siCaspase 8 for 48h. Notably, co-silencing of caspase 8 resulted in inhibition of siGALR1- induced apoptosis (Fig. 4C). Western blot analysis demonstrated that GalR1 silencing induced PARP cleavage and processing of procaspase 8 and that PARP cleavage was blocked by co-silencing of caspase 8 (Fig. 4C, inset). Similarly, the apoptosis induced by galanin silencing (Fig. 4C and S5A) was blocked by co-transfection with siCaspase 8 (Fig. 4C).
Fig. 4. Galanin/GalR1 silencing induces caspase 8-dependent apoptotic cell death.
Annexin V/PI flow cytometric analysis was used to measure apoptosis levels. (A) HCT116 cells were transfected with 5nM siGALR1 ± 10μM zVAD for 24h prior to a 24h co-treatment with IC30(48h) doses of either 5-FU or oxaliplatin. (B) Caspase 3/7 and 8 activity levels in HCT116 cells following 24h GalR1 silencing (5nM siRNA) prior to a 24h co-treatment with IC30(48h) doses of either 5-FU or oxaliplatin. (C) HCT116 cells following 48h GalR1/galanin (5nM siRNA) and Caspase 8 (10nM siRNA) co-silencing. (C, inset) Western blotting was used to measure expression of PARP, caspase 8 and FLIPL following 24h GalR1 and Caspase 8 co-silencing. (D) HCT116 parental cells and the FLIPL overexpressing cell line, FL17, following 24h GalR1/galanin silencing (5nM siRNA). Results are mean values ± SEM of triplicate measurements. *P<0.05; **P<0.01; ***P<0.001. SC=siRNA control.
Given the caspase 8-dependent manner of siGALR1/siGAL-induced apoptosis, the expression of FLIPL, an endogenous inhibitor of caspase 8 activation, was investigated. FLIPL levels were found to be down-regulated following transfection with siGALR1 (Fig. 4C, inset and S5B) or siGAL (Fig. S5A). FLIPL down-regulation was confirmed with 2 additional siRNA sequences (29) (siGALR1_1 and siGALR1_4) (Fig. S5C) and an additional CRC cell line (RKO) (Fig. S5D). Moreover, a FLIPL overexpressing cell line FL17 (17) was significantly resistant to apoptosis induction following GalR1/galanin silencing compared to the parental HCT116 cell line (P<0.01, Fig. 4D). These results indicate for the first time that FLIPL is a key downstream effector of GalR1/galanin-mediated anti-apoptotic signalling.
Clinical Relevance of Galanin expression in CRC
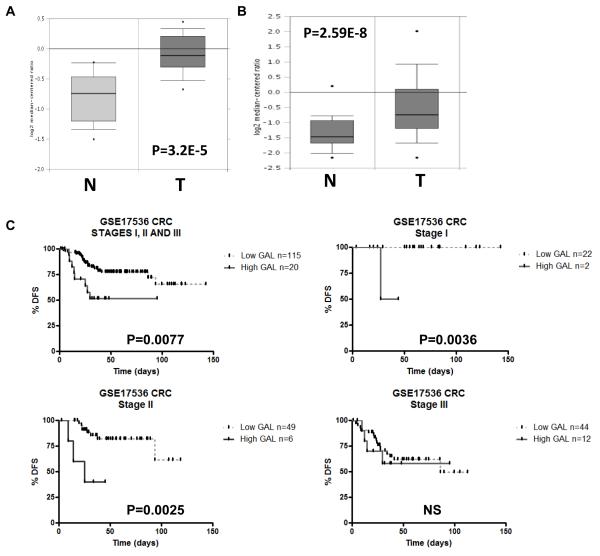
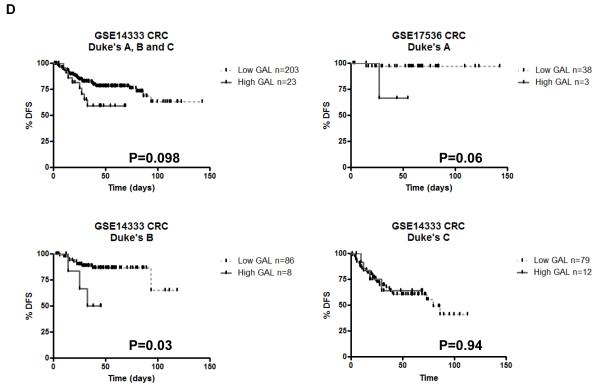
Using publicly available data sets, galanin mRNA expression was found to be significantly up-regulated in CRC tumours compared to normal colon from 2 independent studies: a 1.7-fold increase; P=3.2E-5 (Fig. 5A;(23, 24)) and a 1.9-fold increase; P=2.59E-8 (Fig. 5B, (23, 25)). Having identified galanin and its receptor as determinants of chemotherapy resistance, we examined whether galanin mRNA expression was a potentially useful clinical biomarker in CRC. Using the oxaliplatin constitutively altered (CA) genelist (Table S1d), GSEA identified genes that were enriched and up-regulated in non-responding patients (FDR<25%). Galanin was identified in the top 24 genes that were significantly up-regulated (P<0.05) and enriched in both the oxaliplatin-resistant cells and non-responding patients (Table S3C). We then examined the prognostic value of galanin mRNA expression, since predictive biomarkers may also have prognostic value. Importantly, analysis of galanin mRNA expression in the early stage patients within the GSE17536 CRC dataset (27) revealed a significant correlation between high galanin expression and decreased disease-free survival (DFS) (Fig. 5C; P=0.0077; n=135). Notably, when the early stage patient cohort was broken down into individual stages, this correlation was lost in the stage III cohort, but remained highly significant in the stage I and stage II cohorts (Figure 5C, P=0.0036 and P=0.0025 respectively). In a separate CRC dataset (GSE14333) (28), a significant correlation between high galanin mRNA expression and poorer disease-free survival was also observed in the Dukes stage B (stage II) patients (Fig. 5D, P=0.03). Non-significant trends were observed between high galanin mRNA expression and poorer DFS in Dukes A disease (stage I, P=0.06) and when stages A, B and C were combined (P=0.098). This data suggests a potential association between high galanin mRNA expression and poor patient outcome in early stage CRC. There were insufficient numbers of clinical samples within these public data sets to explore the potential of galanin as a predictive biomarker; ongoing studies aim to further examine the clinical relevance of galanin mRNA expression as both a predictive and prognostic factor.
Fig. 5. Clinical Galanin expression data.
(A and B) Public data sets of galanin mRNA expression in CRC tumour (T) compared to Normal (N). Kaplan-Meier analysis, using the Log-rank (Mantel-Cox) statistical test, of (C) CRC GSE17536 dataset (probe ID: 214240_at) revealed that high galanin expression significantly correlated with decreased disease-free survival in stages I, II and III patients combined (HR=0.2143; 95% CI of ratio=0.06918 to 0.6641; P=0.0077; n=135), stage I only patients (P=0.0036) and stage II only patients (P=0.0025) but not stage III patients (NS) and (D) CRC GSE14333 dataset (probe ID: 214240_at) revealed a trend for high galanin and decreased disease-free survival in combined stages (P=0.098; n=246) and Duke’s A patients (P=0.06). There was a significant correlation between high galanin expression and decreased disease free survival in the Duke’s B patients (P=0.03).
DISCUSSION
Resistance to chemotherapy remains a major problem in CRC, both in the adjuvant and advanced disease settings. In this study, we used a systems biology approach comprising both in vitro and clinical microarray data to identify novel genes and pathways that regulate 5- FU/oxaliplatin resistance together with a functional genetic approach to identify chemo-sensitizing interactions between specific proteins and chemotherapy in CRC cells. Pathway analysis and GSEA identified MAPK signalling, insulin signalling, cell cycle, focal adhesion and apoptosis as key determinants of drug sensitivity. Using an siRNA screening approach, we examined whether down-regulating specific nodal points within these pathways enhanced 5- FU/oxaliplatin sensitivity. From the chemo-sensitizing targets identified in our study, GalR1 was considered to be highly novel and was prioritized for further functional analysis.
GalR1 is one of 3 galanin receptors and is the predominantly expressed galanin receptor in the human colon (30). The galanin receptor ligands are neuropeptides and include galanin (GAL), galanin-message-associated peptide (GMAP), a derivative from the same peptide precursor as galanin, galanin-like peptide (GALP), which is encoded by a different gene, and alarin, which is encoded by a splice variant of the GALP gene. Galanin’s functions in the GI tract include inhibition of gastric acid secretion and inhibition of the release of pancreatic peptides such as insulin, amylase, glucagon and somatostatin. This study focused on GalR1 and the ligand, galanin, as to date, no specific GI-tract functions have been delineated for GalR2, GalR3 or the ligands GMAP, GALP or alarin (30).
There are many reports of cancer-promoting properties for GalR1. It is known to signal through the MAPK pathway, and mitogenic effects of galanin have been reported in pancreatic cancer cells (31), small cell lung cancer cells (32) and rat pituitary tumour cells in vitro (33). Galanin has also been found to be up-regulated in the serum of CRC patients compared to healthy donors (34), and CGH analyses showed gains at the galanin locus, 11q13, in CRC metastases (35), which has been associated with poor outcome (36). In contrast, GalR1 has been implicated as a tumour suppressor since loss of the GALR1 locus, 18q23, have been reported in HNSCC (37) and metastatic CRC tumours (30, 38). In addition, galanin stimulation of GalR1 overexpressing oral SCCs was shown to induce ERK activation, and this was associated with suppressed cell proliferation, inhibition of colony formation and suppressed tumour growth in vivo (39). It has been suggested that the different signal transduction pathways associated with each galanin receptor may account for distinct biological activities of galanin in different types and possibly different stages of cancer (30). Therefore the impact of galanin signalling may depend on the expression level of each receptor, which in turn may vary in a tissue-and tumour-specific manner.
Galanin is considered to be a marker of pluripotent stem cells. Four of the most significantly over-expressed genes in undifferentiated embryonic tissue are galanin, POU5F1 (Oct3), NANOG and DPPA4 (10, 12) with galanin highlighted as the most abundantly expressed in human and rodent embryonic stem cells (ESCs) (10, 14). Furthermore, a 92 gene molecular signature of “stemness” in 6 human ESC lines found that galanin mRNA was the most abundant transcript, along with those of Pou5F1, Nanog, Sox2 and FoxD3 (9, 10). Up-regulation of galanin mRNA and protein expression has been reported in undifferentiated embryonal carcinoma, suggesting it may be a diagnostic marker for undifferentiated tumour cells (40). Cancer cells possessing stem-cell properties have been described in solid tumours, and it is hypothesised that tumour cells with stem-cell attributes are chemo-refractory (41, 42). It is possible therefore that galanin’s potential role in maintaining stem cell-like properties is linked to the novel role we have identified for this ligand in mediating chemotherapy resistance in CRC. Further work is ongoing to explore this possibility.
Analysis of our microarray data sets using a customised GSEA found that high galanin mRNA expression was enriched in the oxaliplatin-resistant setting in vitro and in non-responding patients. Further investigation of galanin mRNA expression in public data-sets revealed that galanin was over-expressed in colorectal tumours compared to normal colon. Importantly, high galanin expression was also significantly associated with poorer DFS of CRC patients, specifically in early stage disease. The clinical management of stage II CRC is a matter of debate as the majority of patients are cured by surgery alone (43). The identification of the ~15-20% of stage II CRC patients who are most likely to relapse is therefore highly clinically relevant as these patients could be selected for treatment with adjuvant chemotherapy, whereas the patients who are unlikely to relapse could be spared the toxic side-effects of chemotherapy treatment. A number of studies, including one involving our group, have attempted to identify prognostic biomarkers for stage II CRC (44-46). The preliminary data presented in this study suggest that galanin mRNA expression may be a useful prognostic biomarker for stage II disease; this is currently being examined in larger patient cohorts.
Previous findings by our group have identified FLIP, in particular FLIPL as a key determinant of drug resistance in CRC (3). The mechanism by which GalR1 silencing enhanced chemo-sensitivity in CRC was shown to involve caspase 8-dependent apoptosis following down-regulation of FLIPL. This is the first demonstration of a link between FLIP and galanin/GalR1; the mechanistic basis by which GalR1/galanin regulates the expression of FLIP and other downstream apoptosis-regulating proteins is the subject of ongoing studies. We previously found that high FLIP expression was an independent prognostic factor in stage II/III CRC (47). Given the prognostic significance of galanin expression and its role as an upstream regulator of FLIP expression, it is possible that high galanin levels promote high FLIP expression in CRC tissues resulting in a more aggressive disease phenotype. This would implicate the galanin signalling pathway as a potentially important therapeutic target in CRC.
In summary, using a systems biology approach, we have identified several pathways as potential mediators of 5-FU/oxaliplatin response and resistance. From these analyses, we identified GalR1/galanin as a novel receptor-ligand system that regulates CRC cell survival and drug resistance. The mechanism by which GalR1/galanin mediates 5-FU/oxaliplatin resistance in CRC was found to be due (at least in part) to its upstream regulation of FLIPL expression. Notably, galanin mRNA expression was found to be up-regulated in colorectal tumour versus normal tissue and overexpressed and enriched in the non-responding patients of this study. Moreover, high galanin mRNA levels also correlated with a poorer prognosis in CRC patient cohorts, particularly in stage II disease. Together, these findings provide the foundation for further research into GalR1/galanin as a novel therapeutic target and/or prognostic biomarker in CRC.
Supplementary Material
STATEMENT OF CLINICAL RELEVANCE.
Chemotherapy resistance is a major factor limiting effective clinical management of colorectal cancer (CRC). Therefore, the identification of therapeutically targetable mediators of resistance is vital. The aim of this study was to identify potentially druggable targets using a sytems biology approach. This resulted in the identification of the receptor-ligand axis, G-protein coupled receptor galanin receptor 1 (GalR1) and galanin as a novel regulator of drug resistance. Silencing GalR1 or its ligand galanin induced apoptosis in drug-sensitive and drug-resistant cells and synergistically enhanced chemotherapy. Galanin was overexpressed in colorectal tumours and high galanin expression correlated with poor disease-free survival of early stage CRC patients. Collectively, these results demonstrate a novel role for GalR1/galanin in chemo-resistance, providing evidence to support its further evaluation as a potential therapeutic target and biomarker in CRC.
Acknowledgements
We would like to thank Cathy Fenning for technical assistance.
Financial Support: This work was supported by Cancer Research UK (C212/A7402) (P. G. Johnston); Cancer Research UK Bobby Moore Fellowship and Research and Development Office Northern Ireland, Department of Health, Social Services and Public Safety (RRG/3261/05, RRG 6.42).
Footnotes
Conflicts of Interest: P.G. Johnston is employed by and has an ownership interest in Almac Diagnostics and Fusion Antibodies. He has received a research grant from Invest NI/McClay Foundation/QUB. He is a consultant/advisor for Chugai pharmaceuticals, Sanofi-Aventis and on the board of the Society for Translational Oncology. He has honoraria from Chugai Pharmaceuticals, Sanofi-Aventis and Precision therapeutics.
References
- 1.Johnston PG, Kaye S. Capecitabine: a novel agent for the treatment of solid tumors. Anticancer Drugs. 2001;12:639–646. doi: 10.1097/00001813-200109000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Giacchetti S, Perpoint B, Zidani R, Le Bail N, Faggiuolo R, Focan C, et al. Phase III multicenter randomized trial of oxaliplatin added to chronomodulated fluorouracil-leucovorin as first-line treatment of metastatic colorectal cancer. J Clin Oncol. 2000;18:136–147. doi: 10.1200/JCO.2000.18.1.136. [DOI] [PubMed] [Google Scholar]
- 3.Longley DB, Johnston PG. Molecular mechanisms of drug resistance. J Pathol. 2005;205:275–292. doi: 10.1002/path.1706. [DOI] [PubMed] [Google Scholar]
- 4.Longley DB, Harkin DP. Johnston PG 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer. 2003;3:330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 5.Longley DB, Allen WL, Johnston PG. Drug resistance, predictive markers and pharmacogenomics in colorectal cancer. Biochim Biophys Acta. 2006;1766:184–196. doi: 10.1016/j.bbcan.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 6.Allen WL, Coyle VM, Jithesh PV, Proutski I, Stevenson L, Fenning C, et al. Clinical determinants of response to irinotecan-based therapy derived from cell line models. Clin Cancer Res. 2008;14:6647–6655. doi: 10.1158/1078-0432.CCR-08-0452. [DOI] [PubMed] [Google Scholar]
- 7.Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swanton C, Marani M, Pardo O, Warne PH, Kelly G, Sahai E, et al. Regulators of mitotic arrest and ceramide metabolism are determinants of sensitivity to paclitaxel and other chemotherapeutic drugs. Cancer Cell. 2007;11:498–512. doi: 10.1016/j.ccr.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 9.Bhattacharya B, Miura T, Brandenberger R, Mejido J, Luo Y, Yang AX, et al. Gene expression in human embryonic stem cell lines: unique molecular signature. Blood. 2004;103:2956–2964. doi: 10.1182/blood-2003-09-3314. [DOI] [PubMed] [Google Scholar]
- 10.Zeng X, Miura T, Luo Y, Bhattacharya B, Condie B, Chen J, et al. Properties of pluripotent human embryonic stem cells BG01 and BG02. Stem Cells. 2004;22:292–312. doi: 10.1634/stemcells.22-3-292. [DOI] [PubMed] [Google Scholar]
- 11.Tarasov KV, Tarasova YS, Crider DG, Anisimov SV, Wobus AM, Boheler KR. Galanin and galanin receptors in embryonic stem cells: accidental or essential? Neuropeptides. 2002;36:239–245. doi: 10.1016/s0143-4179(02)00050-1. [DOI] [PubMed] [Google Scholar]
- 12.Kimber SJ, Sneddon SF, Bloor DJ, El-Bareg AM, Hawkhead JA, Metcalfe AD, et al. Expression of genes involved in early cell fate decisions in human embryos and their regulation by growth factors. Reproduction. 2008;135:635–647. doi: 10.1530/REP-07-0359. [DOI] [PubMed] [Google Scholar]
- 13.Louridas M, Letourneau S, Lautatzis ME, Vrontakis M. Galanin is highly expressed in bone marrow mesenchymal stem cells and facilitates migration of cells both in vitro and in vivo. Biochem Biophys Res Commun. 2009;390:867–871. doi: 10.1016/j.bbrc.2009.10.064. [DOI] [PubMed] [Google Scholar]
- 14.Anisimov SV, Tarasov KV, Tweedie D, Stern MD, Wobus AM, Boheler KR. SAGE identification of gene transcripts with profiles unique to pluripotent mouse R1 embryonic stem cells. Genomics. 2002;79:169–176. doi: 10.1006/geno.2002.6687. [DOI] [PubMed] [Google Scholar]
- 15.Allen WL, Stevenson L, Coyle VM, Jithesh PV, Proutski I, Carson G, et al. A systems biology approach identifies SART1 as a novel determinant of both 5-fluorouracil and SN38 drug resistance in colorectal cancer. Mol Cancer Ther. 2012;11:119–131. doi: 10.1158/1535-7163.MCT-11-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyer J, McLean E, Aroori S, Wilson P, McCulla A, Carey P, et al. Characterization of p53 wild-type and null isogenic colorectal cancer cell lines resistant to 5-fluorouracil, oxaliplatin, and irinotecan. Clin Cancer Res. 2004;10:2158–2167. doi: 10.1158/1078-0432.ccr-03-0362. [DOI] [PubMed] [Google Scholar]
- 17.Longley DB, Wilson TR, McEwan M, Allen WL, McDermott U, Galligan L, et al. c-FLIP inhibits chemotherapy-induced colorectal cancer cell death. Oncogene. 2006;25:838–848. doi: 10.1038/sj.onc.1209122. [DOI] [PubMed] [Google Scholar]
- 18.Stevenson L, Allen WL, Proutski I, Stewart G, Johnston L, McCloskey K, et al. Calbindin 2 (CALB2) regulates 5-fluorouracil sensitivity in colorectal cancer by modulating the intrinsic apoptotic pathway. PLoS One. 6:e20276. doi: 10.1371/journal.pone.0020276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. http://www.ebi.ac.uk/arrayexpress/
- 20.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 22.Wilson TR, McLaughlin KM, McEwan M, Sakai H, Rogers KM, Redmond KM, et al. c-FLIP: a key regulator of colorectal cancer cell death. Cancer Res. 2007;67:5754–5762. doi: 10.1158/0008-5472.CAN-06-3585. [DOI] [PubMed] [Google Scholar]
- 23. http://www.oncomine.org.
- 24.Graudens E, Boulanger V, Mollard C, Mariage-Samson R, Barlet X, Gremy G, et al. Deciphering cellular states of innate tumor drug responses. Genome Biol. 2006;7:R19. doi: 10.1186/gb-2006-7-3-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ki DH, Jeung HC, Park CH, Kang SH, Lee GY, Lee WS, et al. Whole genome analysis for liver metastasis gene signatures in colorectal cancer. Int J Cancer. 2007;121:2005–2012. doi: 10.1002/ijc.22975. [DOI] [PubMed] [Google Scholar]
- 26. http://gibk21.bio.kyutech.ac.jp/PrognoScan/index.html.
- 27.Smith JJ, Deane NG, Wu F, Merchant NB, Zhang B, Jiang A, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 138:958–968. doi: 10.1053/j.gastro.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jorissen RN, Gibbs P, Christie M, Prakash S, Lipton L, Desai J, et al. Metastasis-Associated Gene Expression Changes Predict Poor Outcomes in Patients with Dukes Stage B and C Colorectal Cancer. Clin Cancer Res. 2009;15:7642–7651. doi: 10.1158/1078-0432.CCR-09-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The ENCODE (ENCyclopedia Of DNA Elements) Project Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 30.Lang R, Gundlach AL, Kofler B. The galanin peptide family: receptor pharmacology, pleiotropic biological actions, and implications in health and disease. Pharmacol Ther. 2007;115:177–207. doi: 10.1016/j.pharmthera.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 31.Tjomsland V, El-Salhy M. Effects of single, double or triple combinations of octreotide, galanin and serotonin on a human pancreatic cancer cell line. Histol Histopathol. 2005;20:537–541. doi: 10.14670/HH-20.537. [DOI] [PubMed] [Google Scholar]
- 32.Seufferlein T, Rozengurt E. Galanin, neurotensin, and phorbol esters rapidly stimulate activation of mitogen-activated protein kinase in small cell lung cancer cells. Cancer Res. 1996;56:5758–5764. [PubMed] [Google Scholar]
- 33.Hammond PJ, Smith DM, Akinsanya KO, Mufti WA, Wynick D, Bloom SR. Signalling pathways mediating secretory and mitogenic responses to galanin and pituitary adenylate cyclase-activating polypeptide in the 235-1 clonal rat lactotroph cell line. J Neuroendocrinol. 1996;8:457–464. doi: 10.1046/j.1365-2826.1996.04747.x. [DOI] [PubMed] [Google Scholar]
- 34.Kim KY, Kee MK, Chong SA, Nam MJ. Galanin is up-regulated in colon adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 2007;16:2373–2378. doi: 10.1158/1055-9965.EPI-06-0740. [DOI] [PubMed] [Google Scholar]
- 35.Nakao M, Kawauchi S, Furuya T, Uchiyama T, Adachi J, Okada T, et al. Identification of DNA copy number aberrations associated with metastases of colorectal cancer using array CGH profiles. Cancer Genet Cytogenet. 2009;188:70–76. doi: 10.1016/j.cancergencyto.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 36.Vendrell E, Ribas M, Valls J, Sole X, Grau M, Moreno V, et al. Genomic and transcriptomic prognostic factors in R0 Dukes B and C colorectal cancer patients. Int J Oncol. 2007;30:1099–1107. [PubMed] [Google Scholar]
- 37.Misawa K, Ueda Y, Kanazawa T, Misawa Y, Jang I, Brenner JC, et al. Epigenetic inactivation of galanin receptor 1 in head and neck cancer. Clin Cancer Res. 2008;14:7604–7613. doi: 10.1158/1078-0432.CCR-07-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knosel T, Petersen S, Schwabe H, Schluns K, Stein U, Schlag PM, et al. Incidence of chromosomal imbalances in advanced colorectal carcinomas and their metastases. Virchows Arch. 2002;440:187–194. doi: 10.1007/s004280100493. [DOI] [PubMed] [Google Scholar]
- 39.Kanazawa T, Iwashita T, Kommareddi P, Nair T, Misawa K, Misawa Y, et al. Galanin and galanin receptor type 1 suppress proliferation in squamous carcinoma cells: activation of the extracellular signal regulated kinase pathway and induction of cyclin-dependent kinase inhibitors. Oncogene. 2007;26:5762–5771. doi: 10.1038/sj.onc.1210384. [DOI] [PubMed] [Google Scholar]
- 40.Skotheim RI, Lind GE, Monni O, Nesland JM, Abeler VM, Fossa SD, et al. Differentiation of human embryonal carcinomas in vitro and in vivo reveals expression profiles relevant to normal development. Cancer Res. 2005;65:5588–5598. doi: 10.1158/0008-5472.CAN-05-0153. [DOI] [PubMed] [Google Scholar]
- 41.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 42.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 43.Johnston PG. Stage II colorectal cancer: to treat or not to treat. Oncologist. 2005;10:332–334. doi: 10.1634/theoncologist.10-5-332. [DOI] [PubMed] [Google Scholar]
- 44.Buhmeida A, Bendardaf R, Hilska M, Collan Y, Laato M, Syrjanen S, et al. Prognostic significance of matrix metalloproteinase-9 (MMP-9) in stage II colorectal carcinoma. J Gastrointest Cancer. 2009;40:91–97. doi: 10.1007/s12029-009-9091-x. [DOI] [PubMed] [Google Scholar]
- 45.Bertagnolli MM, Redston M, Compton CC, Niedzwiecki D, Mayer RJ, Goldberg RM, et al. Microsatellite instability and loss of heterozygosity at chromosomal location 18q: prospective evaluation of biomarkers for stages II and III colon cancer--a study of CALGB 9581 and 89803. J Clin Oncol. 2011;29:3153–3162. doi: 10.1200/JCO.2010.33.0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kennedy RD, Bylesjo M, Kerr P, Davison T, Black JM, Kay EW, et al. Development and independent validation of a prognostic assay for stage II colon cancer using formalin-fixed paraffin-embedded tissue. J Clin Oncol. 2011;29:4620–4626. doi: 10.1200/JCO.2011.35.4498. [DOI] [PubMed] [Google Scholar]
- 47.McLornan DP, Barrett HL, Cummins R, McDermott U, McDowell C, Conlon SJ, et al. Prognostic significance of TRAIL signaling molecules in stage II and III colorectal cancer. Clin Cancer Res. 2010;16:3442–3451. doi: 10.1158/1078-0432.CCR-10-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.