Abstract
Biological membranes contain an extraordinary diversity of lipids. Phospholipids function as major structural elements of cellular membranes, and analysis of changes in the highly heterogeneous mixtures of lipids found in eukaryotic cells is central to understanding the complex functions in which lipids participate. Phospholipase-catalyzed hydrolysis of phospholipids often follows cell surface receptor activation. Recently, we demonstrated that granule fusion is initiated by addition of exogenous, nonmammalian phospholipases to permeabilized mast cells. To pursue this finding, we use positive and negative mode Fourier-transform ion cyclotron resonance mass spectrometry (FTICR-MS) to measure changes in the glycerophospholipid composition of total lipid extracts of intact and permeabilized RBL-2H3 (mucosal mast cell line) cells. The low energy of the electrospray ionization results in efficient production of molecular ions of phospholipids uncomplicated by further fragmentation, and changes were observed that eluded conventional detection methods. From these analyses we have spectrally resolved more than 130 glycerophospholipids and determined changes initiated by introduction of exogenous phospholipase C, phospholipase D, or phospholipase A2. These exogenous phospholipases have a preference for phosphatidylcholine with long polyunsaturated alkyl chains as substrates and, when added to permeabilized mast cells, produce multiple species of mono- and polyunsaturated diacylglycerols, phosphatidic acids, and lysophosphatidylcholines, respectively. The patterns of changes of these lipids provide an extraordinarily rich source of data for evaluating the effects of specific lipid species generated during cellular processes, such as exocytosis.
Keywords: mast cells, phospholipases, exocytosis
Phospholipids play important roles in transmembrane signaling processes as well as in dynamic aspects of cell membrane structure. The major phospholipids found in the membranes of mammalian cells include phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylinositol (PI), phosphatidylserine (PS), and phosphatidylglycerol (PG). Some exist as both sphingolipids and glycerophospholipids, as plasmalogens and glycerol diesters, as polyphosphorylated species (e.g., PIs), and with a diverse and specialized array of alkyl chains. Phospholipid metabolism is regulated by distinct types of extracellular receptor-regulated pathways in a variety of ways, including generation of first and second messengers [inositol trisphosphate, phosphatidylinositol 4,5-bisphosphate, platelet-activating factor, diacylglycerol (DAG), sphingosine, etc.]; modifications associated with membrane fusion, secretion, trafficking, and plasma membrane shape change; and to date vaguely described changes in bilayer structure needed to regulate the activities of enzymes, channels, and transport proteins. The chemical and physical properties of membranes are largely dependent on the phospholipid composition (1–4). Although the biological significance of lipid heterogeneity is not understood, several groups have shown that a cell membrane may be composed of more than 100 phospholipids. Such diversity suggests that lipid species play essential roles in cell survival.
Mast cells and basophils contribute to immune and allergic responses by release of secretory granule contents, which contain histamine and other bioactive substances. The antigen-mediated aggregation of high-affinity IgE on the surface of mast cells initiates a biochemical cascade that leads to activation of nonreceptor tyrosine kinases as early steps in the signal transduction process. These cascades culminate in alteration of the cytoskeleton, stimulated exocytosis, production of bioactive lipids by phospholipases, and transcriptional activation. Because activation of cell surface receptors initiates a complex cascade of events that alters both cytoskeletal proteins and production of lipid second messengers, we sought to study the effects of altering lipid composition alone. Despite extensive study, definition of the roles of phospholipids in secretion and membrane transport processes has been elusive. Recent studies in our laboratory (5) show that changes in membrane phospholipid composition as a result of phospholipase activity are associated with degranulation in mast cells. Because several of the activators of phospholipase D (PLD) activity in mammalian cells (e.g., Arf, Rho, protein kinase C) have themselves been implicated in exocytosis, we decided to explore the effects of changes in lipid composition by using phospholipase enzymes from lower organisms that do not require signal transduction proteins for catalytic regulation. Surprisingly, we found that three distinct classes of exogenous phospholipases, all of which were reported to hydrolyze PC preferentially, trigger degranulation in permeabilized RBL-2H3 cells, a mucosal mast cell line. As detected by TLC, production of phosphatidic acid (PA) by PLD, DAG by phospholipase C (PLC), and free fatty acids and lysophosphatidylcholine (LPC) by PLA2 initiate degranulation independent of endogenous cytosolic factors (5). Production of these bioactive lipids promoted release of granule contents through the plasma membrane. The exogenous phospholipase-induced degranulation pathways circumvent specific factors, such as Rho subfamily GTPases and classical isoforms of protein kinase C, activated after stimulation of the IgE receptor, as well as GTP- and ATP-dependent intracellular pathways. These findings highlight the importance of determining precise changes in membrane phospholipid composition during exocytosis. Conventional lipid analysis by radioisotopic labeling and TLC shows only a limited pool of labeled lipids. For more detailed elucidation of phospholipid function during regulated secretion in mast cells, it is important to determine the precise changes in molecular species composition of membrane lipids that occur during degranulation.
Comprehensive determination of the molecular species within a phospholipid class has been a challenging task because of the diversity of fatty acid chains, rapid metabolism, and limited abundance. Although HPLC allows the separation and quantification of many phospholipid classes, the information concerning the individual molecular species within each class still requires more elaborate techniques and separations (6–9). HPLC with UV detection of some important classes of lipids such as DAGs and lysolipids is impaired because of low UV absorbance. Many of these limitations can be overcome by the use of tandem electrospray ionization mass spectrometry (ESI-MS). ESI-mass spectrometry has become a powerful qualitative tool in analysis of biomolecules (10–13). It requires no derivatization, possesses high sensitivity, and has moderate experimental complexity, which affords highly reproducible results. The method has been successfully used for quantification of phospholipids from biological samples (14–17), but we have adapted this method to analyze changes in phospholipid composition of RBL-2H3 cells during degranulation induced by exogenous phospholipases. It was done by use of positive and negative mode electrospray ionization Fourier transform ion cyclotron mass spectrometry (ESI-FTMS). We have identified over 130 molecular species in nine classes of phospholipids by using both detection modes, and we have monitored relative changes in these species during secretory granule release. A major accomplishment of this procedure is that it measures changes in the cellular lipids in both spectra (positive and negative modes), which allows us to analyze relative changes in lipid species in the background of the total lipid pool. This methodology permits us to investigate changes in lipid composition that may be essential to mast cell secretory granule release. The ways in which the concentrations of these lipids change in response to cellular signaling potentially provides an extraordinarily rich source of data for classifying and evaluating the effects and interactions of incoming information.
Materials and Methods
Materials.
RBL-2H3 cells were grown and permeabilized, and degranulation was stimulated as described (5). Bacillus cereus PC-PLC (bcPLC) and Trimeresurus flavoviridis PLA2 (tfPLA2) were obtained from Calbiochem. Streptolysin O was from Abbott Diagnostics (Abbott Park, IL). Streptomyces chromofuscus PLD (scPLD) was obtained from Boehringer Mannheim. All solvents were HPLC grade and used without further purification. Chloroform and methanol were purchased from Mallinckrodt Baker; 1-butanol and diisopropyl ether were obtained from Fisher Scientific. Phospholipid standards were obtained from Avanti Polar Lipids and included phosphatidic acid (PA), PC, PE, PG, phosphatidylserine (PS), PI, DAG, LPC, and sphingomyelin (SM). PI was purified from bovine liver and SM from bovine brain. The other lipids were of synthetic origin: palmitoyl oleoyl PA, PE, and DAG; dioleoyl PS and PG; palmitoyl arachidonoyl PC; and arachidoyl LPC.
Cell Permeabilization and Lipase Treatment.
Intact cells were washed twice in ice-cold permeabilization buffer (PBuf) containing 10 mM Hepes, 115 mM KCl, 1 mM MgCl2, 1 mM DTT, 3 mM EGTA, and 5 mM glucose. Streptolysin O (SLO) in the same buffer was added to the cells at 4°C to a final concentration of 2 units/ml and incubated for 15 min. Cells were washed with ice-cold buffer to remove residual, unbound SLO. Permeabilized cells were stimulated with exogenous phospholipases, and degranulation was determined by a modified colorimetric assay to measure the release of β-hexosaminidase (5). While intact RBL-2H3 cells typically respond to antigen rapidly (over 20 min), we previously found that exogenous phospholipases required an extended incubation to observe production of lipid products and degranulation. Therefore phospholipases were incubated in the permeabilized cells for 60 min before extraction.
Preparation of Total Lipid Extracts.
RBL-2H3 cells (1 × 106) were extracted according to a modification of the Bligh and Dyer (19) procedure. Chloroform extracts were probe sonicated and lyophilized. The residue was resuspended in 300 μl of diisopropyl ether/1-butanol (60:40), mixed in a Vortex mixer, and bath sonicated for 3 min, followed by addition of 150 μl of 50 mM NaCl and Vortex mixing. After separation of the layers by centrifugation at 400 × g for 10 min the upper (organic) layer was collected and lyophilized for 7–10 h based on a published procedure (17). The modifications were shown to be important to obtaining reproducible spectra on the ESI-MS instrument. After lyophilization the lipids were dissolved in 100 μl of chloroform/methanol (1:1) and analyzed by mass spectrometry. Usually 10 μl of this solution was used in a single analysis. All phospholipid analyses performed in this study used the same extraction procedure.
ESI-MS.
ESI-mass spectra were acquired on a modified 6T Finnigan (Madison, WI) FTMS instrument, which is described in detail elsewhere (20). All samples were nanosprayed in positive and negative ion mode as described (21, 22). The effective mass range was m/z 500 to m/z 1000. Typically, less than 2 × 105 cell equivalents of lipid extract were needed to obtain a complete spectrum. On the basis of quantitation in CHO cells (14) we estimate there will be ≈9 nmol of total lipid in the analyzed cell samples. Absolute sensitivity of detection is 5–10 pmol of lipid. The relative responses for phospholipids are more dependent on the chemistry of the head group than on the fatty acid composition within a phospholipid class (23), which allows quantitation by using standard mixture sensitivities. The relative sensitivities of detection of different phospholipid classes were determined by using a mixture of PC/PE/SM/LPC/DAG (1:1:1:1:1 molar ratio) standards for the positive ion mode and a mixture of PI/PE/PS/PA/PG (1:1:1:1:1 molar ratio) standards for the negative ion mode. The standards were used in amounts of 30–50 pmol. PE intensities observed in both ion modes were used to normalize the spectra with respect to each other. As previously recognized (15), no multiply charged ions or aggregations occur in concentrations lower than 100 μmol, and the relationship between ion counts and lipid amount is linear in these limits.
Results and Discussion
The exceptional sensitivity and selectivity of FTMS for phospholipid analysis in both positive and negative ion modes were used to directly identify the structure and mole fraction of individual phospholipid components from chloroform-phase extracts of RBL-2H3 cells. All zwitterionic phospholipids (such as PC, LPC, PE, and SM), as well as DAG, can ionize in both positive and negative ion modes. However, they are more efficiently analyzed in positive ion mode (15). Anionic phospholipids maintain negative charge at neutral pH and thus negative ion ESI-mass spectra are obtained with only [M − H]− as the molecular ion peak.
The assignment of each of the spectrum peaks was possible because of the high sensitivity of FTMS, which produces two peaks for each of the molecular ions. One peak is at the exact molecular mass and the second peak is at 1 Da higher, reflecting the 1.1% natural abundance of 13C and also showing that these ions are singly charged (z = 1). All m/z values presented in this study refer to the monoisotopic molecular weight because the MS analyses were obtained with about a unit mass resolution over the complete mass range. Each phospholipid type was identified in the mixtures by subjecting the positive and negative ions generated in ESI to collisionally activated dissociation. PC, PE, SM, and LPC lose their polar head groups, and PI and PG lose their alkyl chains (12, 14, 17).
Because of their common head group PC, LPC, and SM show an abundant fragment ion at m/z 184. The [M + H]+ ions for phospholipids with nitrogen-containing polar head groups (PE, PC, and LPC) appeared at even masses, whereas SM (containing two nitrogen atoms) is detected at odd m/z values. (We take advantage of the fact that nitrogen is the only common element whose mass and valence are not both even-numbered or odd-numbered). The molecular species of LPC have significantly lower m/z values than PCs, and thus the two can be distinguished despite the same product ion formation. The characteristic product ion for PE class is [M + H − 141]+, reflecting the loss of neutral polar head group.
The principal fragment ions from all classes detected in negative mode (PA, PS, PG, and PI) result from cleavage of the acyl chains with charge retained on the free fatty acids (R1CH2COO− and R2CH2COO−). Less prominent ions are also detected that result from elimination of one or both acids from [M − H]− ions. The product ion mass spectra of PGs also contain an ion formed by the loss of the sn-2 substituent R2CH2COOH from PA ion [M − 76]−, which itself cannot be detected in the product spectrum. In PS, the formation of PA ion [M − 87]− by elimination of a neutral fragment serine − H2O is observed. A prominent characteristic ion at m/z 241 (inositol phosphate − H2O) is detected in the product ion spectrum of PIs. The major fragmentation pathways under low-energy collisionally activated dissociation for PI arise from neutral loss of free fatty acid substitution and neutral loss of ketenes (R2CH⩵C⩵O), followed by consecutive loss of the inositol head group (m/z 162, inositol − H2O). Because of their low concentrations, PA and DAG were not amenable to MS/MS analysis. However, because of the lower m/z values of their molecular ions they are directly identified from the initial negative or positive spectrum, respectively.
Each class of phospholipids is detected with different efficiency in a given mode. For quantitative distribution of phospholipid species we used PE, which can be detected in both positive and negative mode to normalize negative and positive spectra with respect to each other. We obtained relative sensitivities of detection for each class of phospholipids by using a mixture of known amounts of standards described in Materials and Methods. For PC/PE/SM/LPC/DAG they were 1:0.75:0.46:0.75:0.95. For anionic phospholipids PI/PG/PA/PS/PE the sensitivities were 1:1.1:0.5:0.46:0.12. The relative intensities for each class were calculated with a standard error of ± 10% within day-to-day experiments.
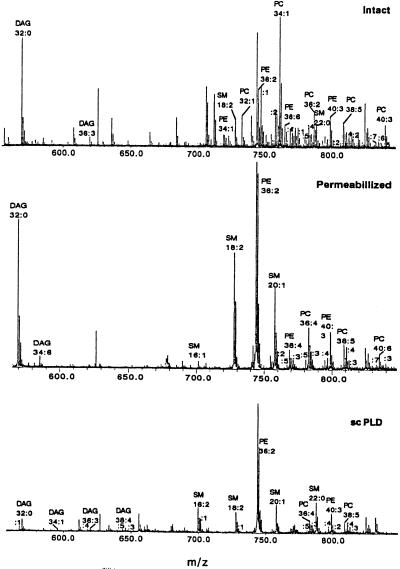
In this study we compared the phospholipid composition of RBL-2H3 cell extracts before and after treatment with exogenous phospholipases. The resulting degranulation was measured as a function of β-hexosaminidase release (5) in the same samples. Figs. 1 and 2 show representative positive and negative mass spectra from intact, permeabilized, and scPLD treated RBL-2H3 cell lipid extracts. In the lower mass range of the positive ion spectrum (m/z 500–700) we detect DAG and LPC (not shown) species. The presence of DAG in intact and permeabilized RBL-2H3 cell extracts is limited to two or three species predominantly with short saturated or short polyunsaturated alkyl chains. After scPLD treatment we detected DAG species consisting of mostly longer (18 or more C atoms) polyunsaturated alkyl chains. The major detected phospholipids in positive mode are PC, PE, and SM. By comparing the three panels of Fig. 1, we can conclude that there is an abundance of PE and PC molecules with different alkyl chain lengths and numbers of double bonds. After scPLD treatment the composition is different and the long (20 C atoms) alkyl chain and multiple double bond PC species are not present. There appears to be synthesis of new SM lipids after treatment by the exogenous phospholipases. Each of the exogenous phospholipases is demonstrated to metabolize the PC in the RBL-2H3 membranes. The breakdown of PC may lead to increases in available phosphocholine (directly or indirectly) and thereby increase production of SM. The significance of this observation is unclear, as sphingomyelin incorporation has been reported to decrease the rates of lipid mixing in large unilamellar vesicles in contrast to the facilitation of fusion observed when PA levels were increased (18).
Figure 1.
Representative positive ion spectra of intact, permeabilized, and scPLD (0.4 unit/ml)-treated RBL-2H3 cells, extracted and analyzed under identical conditions. Labels on the peaks indicate phospholipid species headgroups (xx:y, where xx = total number of carbon atoms in the fatty acid chains, and y = number of double bonds).
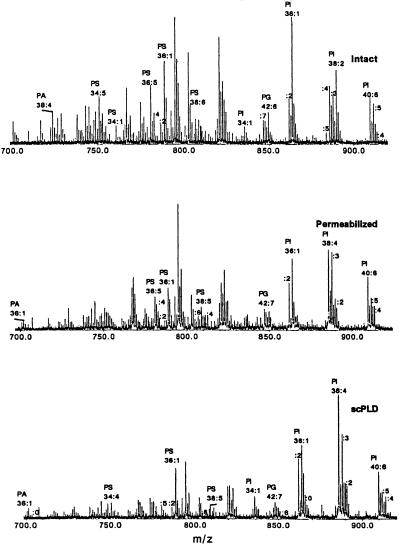
Figure 2.
Representative negative ion spectra of intact, permeabilized, and scPLD (0.4 unit/ml)-treated RBL-2H3 cells, extracted and analyzed under identical conditions.
Fig. 2 shows negative ion spectra of the same intact, permeabilized, and scPLD-treated RBL-2H3 cell lipid extracts. The major components of the extracts are PI species that conserve their composition (total carbon number and number of double bonds). The other detected phospholipids, PG and PS, are without a significant change in alkyl chain length or double bond distribution after treatment with the phospholipases. In the lower mass range (m/z 600–750) we also detect PA molecules.
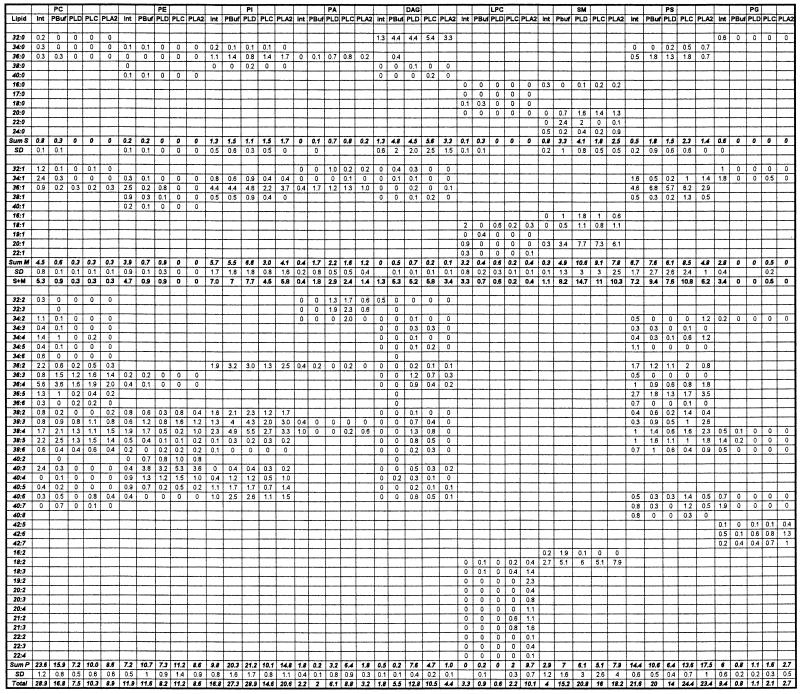
The quantitative analysis of the phospholipids from intact, permeabilized, and exogenous lipase-treated RBL-2H3 cell extracts is summarized in Table 1. All phospholipids are presented as (xx:y), where xx indicates the number of carbon atoms in the fatty acid chain and y is the total number of double bonds in the alkyl groups. The distribution of PC in the intact cell lipid extracts includes 2.8% not containing double bonds, 15.6% with one double bond in the fatty acid chains, and 81.7% containing more than two double bonds. Phospholipases hydrolyze all of the saturated PC species, 50% of monounsaturated species, and an average of 46% of polyunsaturated species, with the total percentage of hydrolysis being 55%, 39%, and 47% for scPLD, bcPLC, and tfPLA2, respectively. Because we are using PC-preferring exogenous phospholipases there is no significant PE hydrolysis. After permeabilization and phospholipase treatment we observed changes in SM species. The most dramatic difference lies in the quantities of monounsaturated SM species, which increased more than 2-fold after lipase treatment. Simultaneously, saturated and monounsaturated PG species are not present, and their polyunsaturated counterparts are 2- to 3-fold increased after lipase treatment (compared with permeabilized).
Table 1.
Normalized distribution (mol %) of phospholipid species detected in intact (Int), permeabilized (PBuf), and bcPLC-, scPLD-, or tfPLA2-treated RBL-2H3 cells
xx = total number of carbon atoms in the fatty acid chains, y = number of double bonds. Standard deviations (SD) are shown only for the sums within each group. The data are average of four experiments. Sum S, Sum M, and Sum P are, respectively, the sums of the saturated, monounsaturated, and polyunsaturated molecular species.
The accumulation of the three major phospholipid second messenger products after phospholipase treatment is summarized in Table 2. More precise analysis of the data from Table 1 reveals the distribution of the molecular species according to their saturation. Mostly saturated and polyunsaturated PA species have been synthesized as a result of lipase activity. Saturated PA species have increased 7- to 8-fold for scPLD- and bcPLC-treated RBL-2H3 cells, whereas polyunsaturated species have increased 16-, 32-, and 9-fold for scPLD-, bcPLC-, and tfPLA2-treated samples, respectively. We also must consider the subsequent metabolism of the primary products after treatment with the exogenous enzymes. The apparent accumulation of DAG after scPLD treatment (Table 1) likely occurs after conversion of PA into DAG by a lipid phosphate phosphatase. Similarly, the production of PA after bcPLC addition may be the result of DAG-kinase activity in the RBL-2H3 permeabilized cells. The subsequent metabolism of the primary products must be analyzed as a function of the onset of regulated degranulation in the preparations. A future goal is to be able to block synthesis of specific lipid products, such that we will be able to establish whether formation of specific lipid species is necessary to achieve exocytosis. Our initial goal has been to determine whether changes in lipid composition alone are able to facilitate release of secretory granule contents without apparent activation of parallel signal transduction pathways, such as through binding of antigen to the high-affinity IgE receptor. Therefore we sought to determine broadly the changes in lipid composition associated with fusion of granules with the plasma membrane during release of bioactive compounds from mast cells. The capability of ESI-MS to resolve and identify such a broad range of phospholipids simultaneously has made this achievable. The presence of close to as many as 30 different species of any given lipid class makes the absolute quantitation very difficult. The changes that we observed are mostly in saturated (≈3% of total) and long-chain polyunsaturated PCs (≈10% of total). Because absolute quantitation of specific lipid species requires inclusion of that same chemically defined species in the analysis (14), it is difficult to determine whether lipid substrate consumed is completely accounted for by lipid products generated. However, in most experiments the very low percentage of total mole fractions changing suggests that overall conservation of total lipids was maintained.
Table 2.
Accumulation of major phospholipid products after lipase treatment of RBL-2H3 cells
| Lipase | Fold increase
|
% degranulation | ||
|---|---|---|---|---|
| PA | LPC | DAG | ||
| scPLD | 3.0 | None | 2.3 | 41.0 ± 10.0 |
| bcPLC | 4.4 | 2.4 | 2.0 | 57.5 ± 7.3 |
| tfPLA2 | 1.6 | 11.2 | None | 36.0 ± 3.0 |
Each data point represents the mean of four experiments. Reproducible data were obtained with typical standard errors of ±13%. Standard deviations are shown for percent degranulation.
The distribution of DAG molecular species as a result of phospholipase stimulation shows almost no changes for species with either no or one double bond in the fatty acid part of the molecule. There is a significant increase of 38- and 23-fold in the amount of polyunsaturated DAG species for scPLD- and bcPLC-treated samples, respectively. Synthesis and accumulation of LPC occurs as a result of tfPLA2 treatment of the permeabilized cells, although there is also a modest increase observed after bcPLC treatment. While its total quantities increased more than 10-fold after tfPLA2 treatment (see Table 2), its molecular species distribution shows a 48-fold increase in the species containing more than two double bonds in the fatty acid chain and almost no changes in the saturated and monounsaturated species. This type of change undoubtedly has profound effects on the physical-chemical state of the membrane, particularly if changes are in highly localized regions.
There is a simple relationship between the amount of total PA produced and the magnitude of secretory granule release (see Table 2). The increasing of total PA content by 1.6- (tfPLA2), 3.0- (scPLD), and 4.4- (bcPLC) fold corresponds to degranulation responses of 36%, 41%, and 57.5%, respectively. Yet, no obvious correlation between the amount of any specific lipid species generated and the degree of degranulation was found. This result suggests that the involvement of phospholipids in degranulation is a function of multiple changes in membrane phospholipid content. Although only scPLD produces PA as a primary lipid product, all three exogenous phospholipases were found to increase PA formation by ESI-MS analysis. Interestingly, this increase in PA formation was not apparent from the TLC analysis (see ref. 5), probably because the [3H]myristate-labeled pools were not used in the metabolic formation of PA. It is also possible that tfPLA2 and bcPLC generate novel substrates that generate PA by secondary metabolism even in these broken cell systems. The exquisite sensitivity and reproducibility of ESI-MS allows us to observe these underlying changes in lipid composition that were not apparent when conventional types of analysis were used. These findings highlight that analysis of a broad spectrum of lipids during secretory granule release may be important in identifying patterns of changes that must proceed sequentially.
We have been able to resolve and quantify relative changes in distinct species of phospholipids during mast cell degranulation at a level not previously reported. The ESI-MS/MS technique used for quantification of phospholipids in permeabilized RBL-2H3 cell extracts presents a powerful tool for investigation of biological samples where sensitivity and speed are the most important considerations. ESI-MS/MS allows precise information about changes in membrane phospholipid composition to be obtained. The approach is analogous in concept to nucleic acid microarray technology in its ability to examine broad changes in biological molecules simultaneously and will eventually permit better definition of the mechanistic contributions of specific lipids in cell functions, such as regulated exocytosis.
Acknowledgments
We thank James Kerwin (Cornell Biotechnology Center) and Barbara Baird for numerous insightful conversations. This work was supported by funding from the U.S. Department of Agriculture Hatch Act Consolidated Research Program and National Institutes of Health Grant GM58516 (to H.A.B.). H.A.B. is a Sidney Kimmel Foundation for Cancer Research Scholar.
Abbreviations
- PLC
phospholipase C
- bcPLC
PLC from Bacillus cereus
- PLD
phospholipase D
- scPLD
PLD from Streptomyces chromofuscus
- PLA2
phospholipase A2
- tfPLA2
PLA2 from Trimeresurus flavoviridis
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PI
phosphatidylinositol
- PS
phosphatidylserine
- PG
phosphatidylglycerol
- PA
phosphatidic acid
- DAG
diacylglycerol
- LPC
lysophosphatidylcholine
- SM
sphingomyelin
- ESI-MS
electrospray ionization mass spectrometry
- FTICR-MS (or FTMS)
Fourier-transform ion cyclotron resonance mass spectrometry
References
- 1.Stubs C C, Smith A D. Biochim Biophys Acta. 1984;779:89–137. doi: 10.1016/0304-4157(84)90005-4. [DOI] [PubMed] [Google Scholar]
- 2.Muderhwa J M, Brockman H L. J Biol Chem. 1992;267:24184–24192. [PubMed] [Google Scholar]
- 3.Chernomordik L V, Leikina E, Frolov V, Bronk P, Zimmerberg J. J Cell Biol. 1997;136:81–93. doi: 10.1083/jcb.136.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barr F A, Shorter J. Curr Biol. 2000;10:R141–R144. doi: 10.1016/s0960-9822(00)00326-2. [DOI] [PubMed] [Google Scholar]
- 5.Cohen J S, Brown H A. Biochemistry. 2001;40:6589–6597. doi: 10.1021/bi0103011. [DOI] [PubMed] [Google Scholar]
- 6.Olsson N U, Salem N., Jr J Chromatogr B. 1997;692:245–256. doi: 10.1016/s0378-4347(96)00507-5. [DOI] [PubMed] [Google Scholar]
- 7.Mollova N N, Moore I M, Hutter J, Shram K H. J Mass Spectrom. 1995;30:1405–1420. [Google Scholar]
- 8.Mawatari S, Murakami K. Anal Biochem. 1998;264:118–123. doi: 10.1006/abio.1998.2830. [DOI] [PubMed] [Google Scholar]
- 9.Bernhard W, Linck M, Creutzburg H, Postle A D, Arning A, Martin-Carrera I, Sewing K-F. Anal Biochem. 1994;220:172–180. doi: 10.1006/abio.1994.1315. [DOI] [PubMed] [Google Scholar]
- 10.Kerwin J L, Tuininga A R, Ericsson L H. J Lipid Res. 1994;35:1102–1114. [PubMed] [Google Scholar]
- 11.Smith P B W, Snyder A P, Harden C S. Anal Chem. 1995;67:1824–1830. doi: 10.1021/ac00107a011. [DOI] [PubMed] [Google Scholar]
- 12.Han X, Gross R W. J Am Soc Mass Spectrom. 1995;6:1202–1210. doi: 10.1016/1044-0305(95)00568-4. [DOI] [PubMed] [Google Scholar]
- 13.Schneiter R, Brügger B, Sandhoff R, Zelling G, Leber A, Lampl M, Athenstaedt K, Hrastnik C, Eder S, Daum G, et al. J Cell Biol. 1999;146:741–754. doi: 10.1083/jcb.146.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brügger B, Erben G, Sandhoff R, Wieland F T, Lehmann W D. Proc Natl Acad Sci USA. 1997;94:2339–2344. doi: 10.1073/pnas.94.6.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han X, Gross R W. Proc Natl Acad Sci USA. 1994;91:10635–10639. doi: 10.1073/pnas.91.22.10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sweetman G, Trinei M, Modha J, Kusel J, Freestone P, Fishov I, Joseleau-Petit D, Redman C, Farmer P, Norris V. Mol Microbiol. 1996;20:233–234. doi: 10.1111/j.1365-2958.1996.tb02504.x. [DOI] [PubMed] [Google Scholar]
- 17.Fridricsson E K, Shipkova P A, Sheets E D, Holowka D A, Baird B, McLafferty F W. Biochemistry. 1999;38:8056–8063. doi: 10.1021/bi9828324. [DOI] [PubMed] [Google Scholar]
- 18.Brock T G, Nagaprakash K, Margolis D I, Smolen J E. J Membr Biol. 1994;141:139–148. doi: 10.1007/BF00238247. [DOI] [PubMed] [Google Scholar]
- 19.Bligh E G, Dyer W J. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 20.Beu S C, Senko M W, Quinn J P, Wampler F M, III, McLafferty F W. J Am Soc Mass Spectrom. 1993;4:557–565. doi: 10.1016/1044-0305(93)85017-R. [DOI] [PubMed] [Google Scholar]
- 21.Wilm M S, Mann M. Int J Mass Spectrom Ion Processes. 1994;136:167–180. [Google Scholar]
- 22.Wilm M S, Mann M. Anal Chem. 1996;68:1–8. doi: 10.1021/ac9509519. [DOI] [PubMed] [Google Scholar]
- 23.Kim H-Y, Wang T C L, Ma Y C. Anal Chem. 1994;66:3977–3982. doi: 10.1021/ac00094a020. [DOI] [PubMed] [Google Scholar]