Abstract
LUBAC (linear ubiquitin chain assembly complex) activates the canonical NF-κB pathway through linear polyubiquitination of NEMO (NF-κB essential modulator, also known as IKKγ) and RIP1. However, the regulatory mechanism of LUBAC-mediated NF-κB activation remains elusive. Here, we show that A20 suppresses LUBAC-mediated NF-κB activation by binding linear polyubiquitin via the C-terminal seventh zinc finger (ZF7), whereas CYLD suppresses it through deubiquitinase (DUB) activity. We determined the crystal structures of A20 ZF7 in complex with linear diubiquitin at 1.70–1.98 Å resolutions. The crystal structures revealed that A20 ZF7 simultaneously recognizes the Met1-linked proximal and distal ubiquitins, and that genetic mutations associated with B cell lymphomas map to the ubiquitin-binding sites. Our functional analysis indicated that the binding of A20 ZF7 to linear polyubiquitin contributes to the recruitment of A20 into a TNF receptor (TNFR) signalling complex containing LUBAC and IκB kinase (IKK), which results in NF-κB suppression. These findings provide new insight into the regulation of immune and inflammatory responses.
Keywords: A20, deubiquitinase, lymphoma, NF-κB, ubiquitin
Introduction
Nuclear factor-κB (NF-κB) is a pivotal transcription factor involved in many cellular processes, including innate and adaptive immune responses, inflammation, cell adhesion and cell survival (Vallabhapurapu and Karin, 2009; Hayden and Ghosh, 2012). NF-κB is activated by a vast array of stimuli, including viral and bacterial pathogens, proinflammatory cytokines, genotoxic agents, ultraviolet radiation and oxidative stress (Vallabhapurapu and Karin, 2009; Hayden and Ghosh, 2012). Thus, aberrant NF-κB signalling is implicated in multiple disorders, such as cancer and autoimmune, chronic inflammatory and metabolic diseases (Pasparakis, 2009; Ben-Neriah and Karin, 2011; Perkins, 2012). Typically, NF-κB activation is mediated by both canonical and non-canonical pathways, which have distinct biological roles. In the canonical NF-κB pathway, proinflammatory cytokines and pathogen-associated molecular patterns trigger the rapid activation of the canonical IKK complex, composed of two kinase subunits, IKKα and IKKβ, and a regulatory subunit, NEMO, resulting in the nuclear translocation of an NF-κB heterodimer, predominantly composed of p65 and p50 (Vallabhapurapu and Karin, 2009; Hayden and Ghosh, 2012).
The activation of the canonical NF-κB pathway is controlled by various types of polyubiquitin chains, such as Lys (K)11-, K48-, and K63-linked chains, and Met(M)1-linked linear chains (Iwai, 2012). We previously reported that the LUBAC ubiquitin ligase, composed of SHARPIN, HOIL-1L (also known as RBCK1) and HOIP (also known as RNF31), generates a novel type of M1-linked linear polyubiquitin, in which the C-terminal Gly76 of a distal ubiquitin moiety is conjugated to the α-amino group of the N-terminal Met1 of a proximal ubiquitin moiety (Kirisako et al, 2006; Tokunaga and Iwai, 2012). Upon proinflammatory cytokine stimulation, LUBAC linearly ubiquitinates NEMO and RIP1, and induces canonical NF-κB activation (Tokunaga et al, 2009; Gerlach et al, 2011; Ikeda et al, 2011; Tokunaga et al, 2011). Recent studies showed that LUBAC regulates the interferon production pathway (Inn et al, 2011), the genotoxic stress response (Niu et al, 2011), tumour metastasis (Tomonaga et al, 2012), osteogenesis (Xia et al, 2011) and innate immunity (Damgaard et al, 2012), highlighting the physiological significance of LUBAC-catalysed linear ubiquitination in a wide variety of NF-κB-related cellular responses.
NF-κB signalling is generally attenuated by deubiquitinating enzymes (DUBs), such as A20 (also known as TNFAIP3), Cezanne and CYLD (Wertz and Dixit, 2010; Harhaj and Dixit, 2011). A20 and Cezanne belong to the ovarian tumour (OTU) DUB family. A20 is composed of an OTU domain and seven Cys2/Cys2 zinc fingers (Figure 1A), while Cezanne is composed of an OTU domain and a single Cys2/Cys2 zinc finger. CYLD, a cylindromatosis tumour suppressor gene product, belongs to the ubiquitin-specific protease (USP) DUB family (Brummelkamp et al, 2003). However, it remains elusive how DUBs physiologically downregulate LUBAC-mediated NF-κB activation.
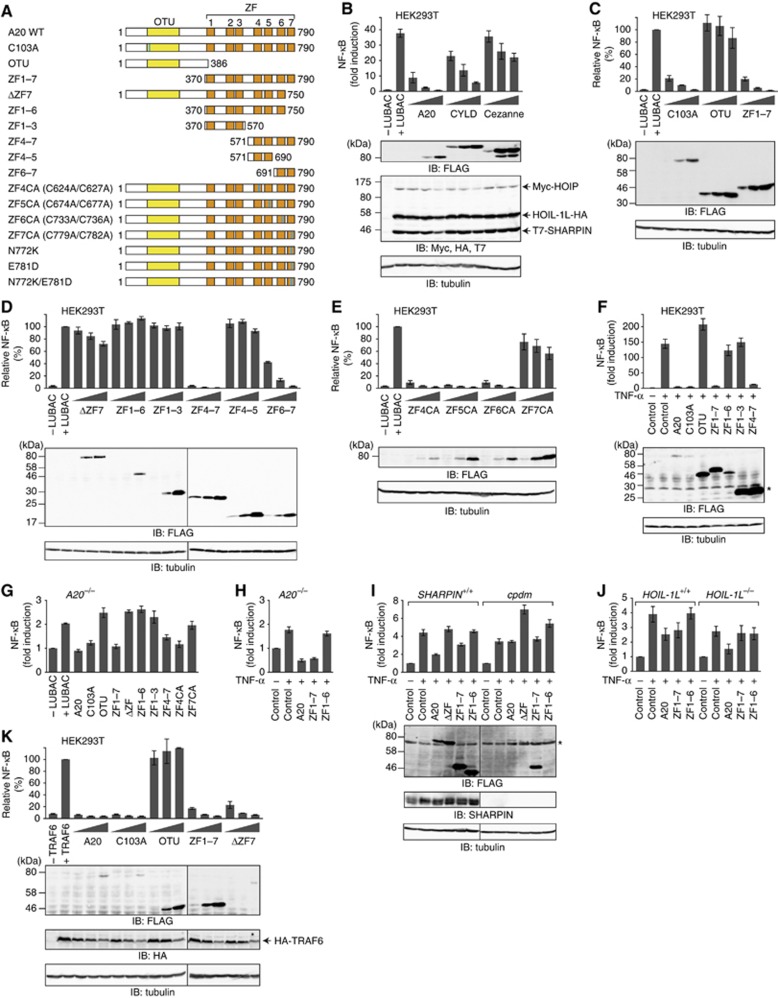
Figure 1.
A20 ZF7 is crucial for the suppression of LUBAC-mediated NF-κB activation. (A) Schematic representation of A20 WT and mutants. Mutation sites are indicated by blue lines. (B) Expression of A20 and CYLD, but not that of Cezanne, suppresses LUBAC-induced NF-κB activation. Luciferase activities are shown as mean±s.d. (n=3), and the DUB and LUBAC expression levels are shown in the lower panels. (C) A20 ZFs, but not A20 OTU, participate in the NF-κB suppression. (D) A20 ZF7 participates in NF-κB suppression. (E) Intact A20 ZF7 is required for NF-κB suppression. (F) A20 ZF7 participates in NF-κB suppression in TNF-α-stimulated HEK293T cells. (G, H) A20 ZF7 participates in NF-κB suppression in LUBAC-overexpressing A20−/− MEFs (G) and TNF-α-stimulated A20−/− MEFs (H). (I, J) Effects of A20 WT and mutants on TNF-α-induced NF-κB activation in SHARPIN-ablated cpdm MEFs (I) and HOIL-1L−/− MEFs (J). (K) A20 ZF7 is dispensable for TRAF6-induced NF-κB activation. Relative luciferase activities (C–E, K) and induction folds of NF-κB activity (B, F, G–J) are shown as mean±s.d. (n=3). Expression of FLAG-tagged A20 proteins and the loading control tubulin are shown in the lower panels (C–F, I, K). *, Non-specific signal (F, I).
Here, we report that A20 and CYLD suppress LUBAC-mediated NF-κB activation through distinct molecular mechanisms. We showed that the DUB activity of CYLD, but not that of A20, is important for NF-κB suppression, whereas the binding of A20 to linear polyubiquitin via the C-terminal ZF7 is essential for the suppression of LUBAC-mediated NF-κB activation. We determined the crystal structures of A20 ZF7 in complex with linear diubiquitin and tetraubiquitin, which revealed the molecular basis for linear polyubiquitin recognition by A20 ZF7. We found that the mutations associated with B cell lymphomas map to the ubiquitin-binding sites in A20 ZF7, and that the binding of A20 ZF7 to linear polyubiquitin plays a crucial role in the recruitment of A20 to a TNFR signalling complex, resulting in the NF-κB suppression. Our present findings highlight the physiological significance of linear polyubiquitination in NF-κB signalling.
Results
A20 and CYLD suppress LUBAC-induced NF-κB activation
To address the regulatory mechanism of LUBAC-mediated NF-κB activation, we first focused on A20, CYLD and Cezanne, the well-characterized DUBs in the NF-κB pathway (Wertz and Dixit, 2010). We overexpressed these DUBs in HEK293T cells and examined their inhibitory effects on LUBAC-induced NF-κB activation, using luciferase reporter assays. Transient co-expression of LUBAC components (SHARPIN, HOIL-1L and HOIP) induced ∼40-fold enhancement of NF-κB activity (Figure 1B). Transient co-expression of A20 and CYLD, but not that of Cezanne, together with the LUBAC components, resulted in strong suppression of LUBAC-induced NF-κB activation (Figure 1B). CYLD efficiently hydrolysed linear and K63-linked polyubiquitin chains, as described (Komander et al, 2008; Supplementary Figure S1A and B), and the catalytically inactive CYLD C601A mutant showed no inhibitory effect on LUBAC-induced NF-κB activation (Supplementary Figure S2). These results revealed that the CYLD DUB activity is indispensable for the suppression of LUBAC-mediated NF-κB activation.
Previous studies indicated that A20 attenuates NF-κB signalling by removing K63-linked ubiquitin chains from RIP1 through the DUB activity of the OTU domain, followed by conjugating K48-linked ubiquitin chains to RIP1 through the E3 ubiquitin ligase activity of the ZF4 domain, resulting in the proteasome degradation of RIP1 (Wertz et al, 2004). However, the co-expression of the catalytically inactive A20 C103A mutant still resulted in the suppression of LUBAC-induced NF-κB activation, and the co-expression of the A20 OTU domain alone had almost no inhibitory effect (Figure 1C), indicating that the A20 DUB activity is dispensable for the suppression of LUBAC-induced NF-κB activation. A20 and Cezanne hydrolyse K48- and K11-linked ubiquitin chains, respectively, but not linear ubiquitin chains (Komander and Barford, 2008; Bremm et al, 2010). Consistent with these previous reports, our biochemical analysis showed that A20 hydrolyses K48- and K63-linked chains (Supplementary Figure S1B), but neither unanchored linear polyubiquitin nor NEMO-conjugated linear polyubiquitin (Supplementary Figure S1C).
A20 ZF7 participates in the suppression of LUBAC-mediated NF-κB activation
A20 is a multidomain protein composed of an OTU domain and seven zinc fingers (Figure 1A). To identify the region required for the suppression of LUBAC-mediated NF-κB activation, we overexpressed deletion and point mutants of A20 in HEK293T cells, and examined their inhibitory effects on LUBAC-induced NF-κB activation. The A20 mutants containing ZF7 (ZF1–7, ZF4–7 and ZF6–7) suppressed LUBAC-induced NF-κB activation as effectively as wild-type (WT) A20 (Figure 1C and D), whereas the A20 mutants lacking ZF7 (ΔZF7, ZF1–6, ZF1–3 and ZF4–5) and the C779A/C782A mutant (ZF7CA) failed to suppress it (Figure 1D and E), suggesting the involvement of A20 ZF7 in NF-κB suppression. To determine the physiological significance of A20, we examined the inhibitory effects of A20 WT and mutants in TNF-α-stimulated HEK293T cells. As in LUBAC-overexpressing HEK293T cells, the A20 mutants containing ZF7, but not the A20 mutants lacking ZF7, showed inhibitory effects on TNF-α-induced NF-κB activation (Figure 1F). To exclude the potential effects of endogenous A20, we examined the inhibitory effects of A20 WT and mutants in A20-ablated mouse embryonic fibroblasts (MEFs). We found that A20 ZF7 is indispensable for NF-κB suppression in LUBAC-overexpressing A20−/− MEFs (Figure 1G) and TNF-α-stimulated A20−/− MEFs (Figure 1H).
To further explore the physiological significance of A20 ZF7, we examined the inhibitory effects of A20 WT and mutants in LUBAC-deficient cells, such as SHARPIN-ablated MEFs (cpdm MEFs) and HOIL-1L−/− MEFs. In control MEFs, A20 WT and ZF1–7, but not the A20 mutants lacking ZF7 (ΔZF7 and ZF1–6), suppressed TNF-α-induced NF-κB activation (Figure 1I and J). In contrast, the inhibitory effects of A20 WT and ZF1–7 were attenuated in both cpdm and HOIL-1L−/− MEFs, suggesting that A20 ZF7 plays an inhibitory role downstream of LUBAC in the TNF-α-induced NF-κB activation pathway.
TRAF6 is an E3 ligase that predominantly generates K63-linked polyubiquitin chains and participates in IL-1β-induced NF-κB activation (Wang et al, 2001). To determine whether A20 ZF7 specifically participates in the suppression of the LUBAC-mediated NF-κB pathway, we examined the inhibitory effects of A20 WT and mutants in TRAF6-overexpressing HEK293T cells. A20 WT, C103A and ZF1–7, but not A20 OTU, suppressed TRAF6-induced NF-κB activation (Figure 1K), as in LUBAC-induced NF-κB activation (Figure 1C and D). However, unlike the LUBAC-induced NF-κB activation, A20 ΔZF7 also suppressed TRAF6-induced NF-κB activation, suggesting that A20 ZF7 is dispensable for the suppression of TRAF6-mediated NF-κB activation. Taken together, these results suggested that A20 ZF7 is critical for the suppression of the LUBAC-mediated, but not the TRAF6-mediated, NF-κB activation pathway.
A20 ZF7 specifically binds linear diubiquitin
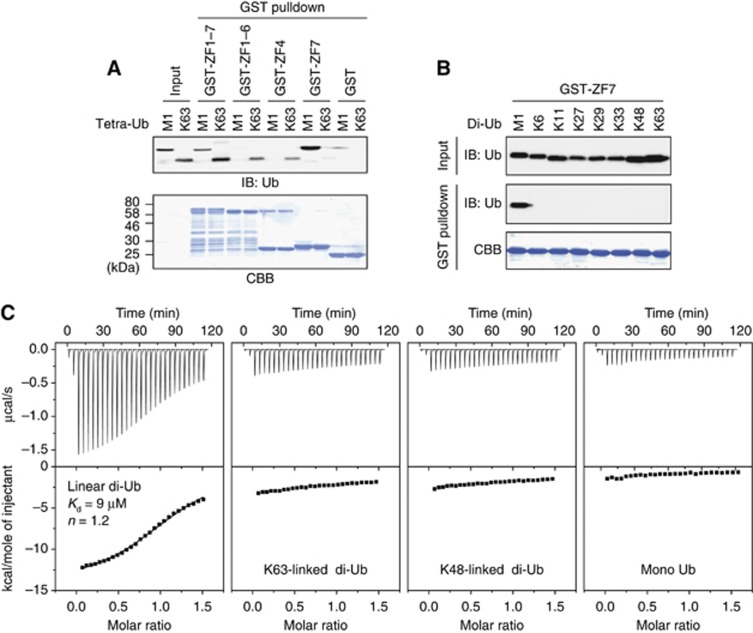
To determine the mechanism by which A20 ZF7 suppresses the LUBAC-mediated NF-κB activation, we examined whether A20 ZF7 inhibits the E3 activity of LUBAC. Neither A20 ZF7 nor A20 ZF4 inhibited the LUBAC-catalysed generation of either free linear polyubiquitin or NEMO-conjugated linear polyubiquitin (Supplementary Figure S3). Among the seven zinc fingers of A20, A20 ZF7 shares 36% sequence identity with A20 ZF4, which binds monoubiquitin and K63-linked polyubiquitin (Bosanac et al, 2010). We thus re-examined the ability of the A20 ZFs to bind linear and K63-linked tetraubiquitin by glutathione S-transferase (GST) pulldown assays. GST-ZF1–7 bound both the linear and K63-linked chains, whereas GST-ZF1–6 and GST-ZF4 bound only the K63-linked chains, but not the linear chains (Figure 2A). Consistently, GST-ZF7 specifically bound M1-linked linear chains (Figure 2A). We next examined the ability of A20 ZF7 to bind diubiquitins with different linkages, and found that GST-ZF7 specifically binds M1-linked linear diubiquitin (Figure 2B). Furthermore, we examined the binding of A20 ZF7 to diubiquitin by isothermal titration calorimetry (ITC), and found that A20 ZF7 binds linear diubiquitin with a Kd value of 9 μM, but not monoubiquitin, K48-linked diubiquitin and K63-linked diubiquitin (Figure 2C). Altogether, these results indicated that A20 ZF7 specifically recognizes linear diubiquitin, but not Lys-linked diubiquitins.
Figure 2.
A20 ZF7 specifically binds linear ubiquitin chains. (A) Binding of A20 ZFs to linear and K63-linked tetraubiquitins. (B, C) Binding of A20 ZF7 to diubiquitin with different linkages in GST pulldown assays (B) and ITC experiments (C).
Crystal structure of A20 ZF7 in complex with linear diubiquitin
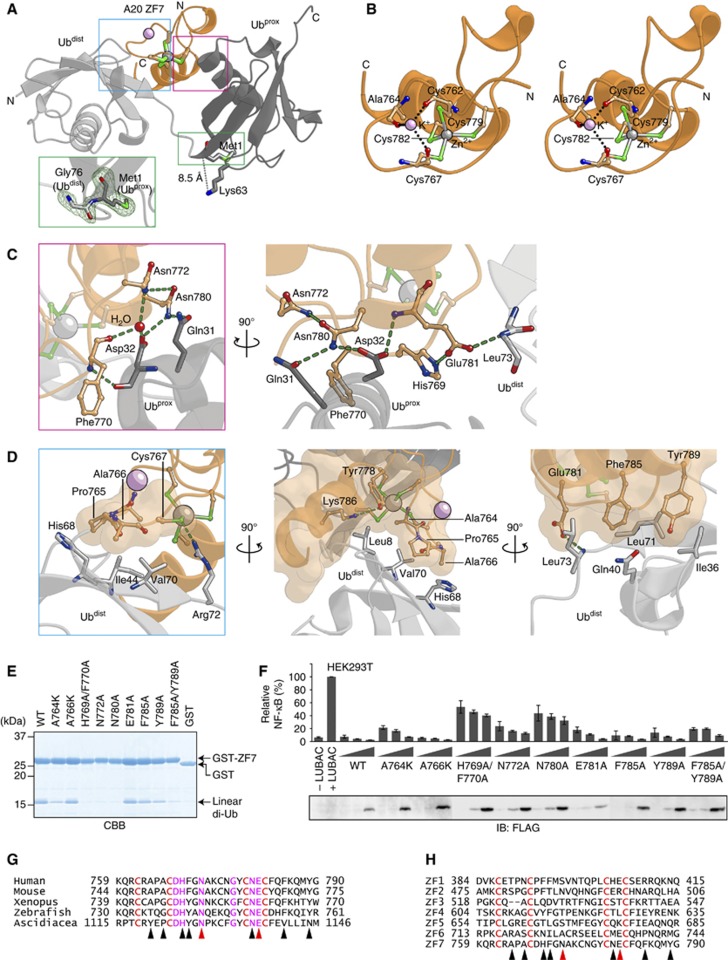
To elucidate the mechanism of linear diubiquitin recognition by A20 ZF7, we crystallized A20 ZF7 in the presence of linear diubiquitin or linear tetraubiquitin, and determined two crystal structures of A20 ZF7 in complex with linear diubiquitin (Form I, 1.95 Å resolution; and Form II, 1.70 Å resolution) and one in complex with linear tetraubiquitin (Form II, 1.98 Å resolution) (Supplementary Figure S4; Table I). A20 ZF7 interacts with three and four adjacent ubiquitin molecules in Forms I and II, respectively, and it interacts with Met1-linked, adjacent ubiquitin molecules in a similar manner in both Forms I and II (Supplementary Figure S4), indicating the physiological significance of this A20 ZF7–linear diubiquitin complex. The A20 ZF7–linear diubiquitin complexes in Forms I and II have a virtually identical conformation (r.m.s. deviations are <1.04 Å for aligned Cα atoms), while the linkage between the ubiquitin moieties is the most clearly defined in the electron density map of the tetraubiquitin complex (Figure 3A, inset). Thus, we will hereafter describe the A20 ZF7–linear diubiquitin complex structure in the tetraubiquitin complex. A20 ZF7 consists of a loop region (residues 758–779) and an α-helix (residues 780–789), with a bound zinc ion stabilizing the overall architecture (Figure 3A). The zinc ion is coordinated by Cys762, Cys767, Cys779 and Cys782 (Figure 3B). A potassium ion derived from the crystallization buffer is coordinated by the main-chain carbonyl groups of Cys762, Ala764 and Cys767 in the loop region, and helps maintain the loop conformation (Figure 3B), although the physiological relevance of the bound potassium ion is currently unknown.
Table 1. Data collection and refinement statistics.
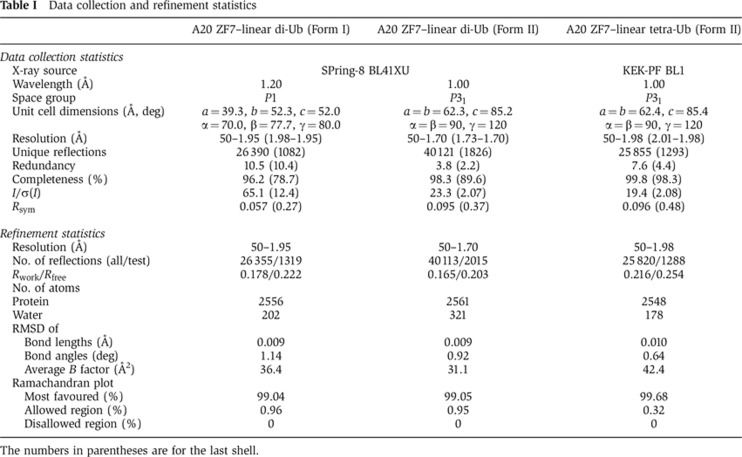
The numbers in parentheses are for the last shell.
Figure 3.
A20 ZF7 specifically recognizes linear diubiquitin. (A) Crystal structure of A20 ZF7 in complex with linear diubiquitin (crystallized in the presence of linear tetraubiquitin). A20 ZF7 and ubiquitin are coloured orange and grey, respectively. Bound zinc and potassium ions are shown as grey and pink spheres, respectively. Binding sites of the proximal and distal ubiquitin moieties are indicated by magenta and cyan boxes, respectively. A close-up view of the linkage between the ubiquitin moieties is shown in the inset. Fo−Fc omit maps for Met1 of the proximal ubiquitin and Gly76 of the distal ubiquitin are shown as a green mesh (contoured at 2.5σ). (B) Crystal structure of A20 ZF7 (stereoview). (C) Recognition of the proximal ubiquitin. (D) Recognition of the distal ubiquitin. Hydrogen bonds are shown as green dashed lines (C, D). (E) Binding of A20 ZF7 mutants to linear diubiquitin in GST pulldown assays. (F) Suppression of LUBAC-induced NF-κB activation by full-length A20 mutants. Luciferase assays and protein expression were analysed as in Figure 1B. (G) Multiple sequence alignment of A20 ZF7 proteins from different organisms. (H) Multiple sequence alignment of human A20 ZF1–7. Zinc-coordinating cysteine residues are indicated by red letters, and residues involved in linear-diubiquitin binding are indicated by triangles, with Asn772 and Glu781 highlighted by red triangles (G, H).
A20 ZF7 simultaneously interacts with both the proximal and distal ubiquitin moieties. A20 ZF7 mainly recognizes an α-helix region of the proximal ubiquitin through a hydrogen bonding network (Figure 3C). Asp32 on the α-helix in the proximal ubiquitin is recognized by Phe770, Asn772, Asn780 and Glu781 of ZF7, through direct and water-mediated hydrogen bonds. Asn772 of ZF7 forms a water-mediated hydrogen bond with Asp32 of the proximal ubiquitin, and a hydrogen bond with Asn780 of ZF7, which in turn hydrogen bonds with Gln31 and Asp32 of the proximal ubiquitin. In addition, His769 and Phe770 of ZF7 form a surface complementary to the C-terminal edge of the α-helix of the proximal ubiquitin. In contrast, A20 ZF7 mainly recognizes a hydrophobic patch of the distal ubiquitin by shape complementarity (Figure 3D). Ala764, Pro765 and Ala766 on the zinc-coordinating loop interact with the hydrophobic patch formed by Leu8, Ile44, His68 and Val70 of the distal ubiquitin. Tyr778, Cys782, Phe785, Lys786 and Tyr789 of ZF7 form a hydrophobic surface that interacts with Leu8, Ile36, Gln40, Leu71 and Leu73 of the distal ubiquitin. Cys767 forms a salt bridge with Arg72 of the distal ubiquitin. In addition, Glu781 forms hydrogen bonds with His769 in ZF7 and the main-chain amide group of Leu73 in the distal ubiquitin (Figure 3C). Taken together, our crystallographic analysis showed that A20 ZF7 specifically recognizes linear diubiquitin through simultaneous interactions with the proximal and distal ubiquitin moieties. In the present structure, the Met1 main-chain nitrogen atom and the Lys63 side-chain Nε atom of the proximal ubiquitin are 8.5 Å apart (Figure 3A). These observations indicated that, unlike the linear chains, the K63-linked chains cannot adopt a configuration that allows A20 ZF7 to simultaneously interact with the proximal and distal ubiquitin moieties, explaining why A20 ZF7 specifically binds linear diubiquitin, but not the structurally similar K63-linked diubiquitin.
Mutational analysis of the linear diubiquitin-binding sites of A20 ZF7
To confirm the functional significance of the interactions between A20 ZF7 and linear diubiquitin observed in the crystal structure, we examined the ability of A20 ZF7 mutants to bind linear diubiquitin, using GST pulldown assays. The H769A/F770A, N772A, N780A and F785A/Y789A mutants of GST-A20 ZF7 showed markedly decreased ubiquitin binding (Figure 3E). The A764K, F785A and Y789A mutants of GST-A20 ZF7 exhibited moderate decreases in ubiquitin binding, and the A766K and E781A mutants of GST-A20 ZF7 showed slightly reduced ubiquitin binding activities (Figure 3E). These results indicated that the integrity of both binding sites for the proximal and distal ubiquitin moieties is required for linear diubiquitin recognition by A20 ZF7. To exclude the possibility that these mutants were unfolded and thus exhibited defects in ubiquitin binding, we analysed these A20 ZF7 mutants by size-exclusion chromatography. Like A20 WT, all of the mutants eluted as a single peak from the size-exclusion column (Supplementary Figure S5), suggesting that the mutants are correctly folded.
To further confirm the functional significance of the interactions between A20 ZF7 and linear ubiquitin chains for NF-κB suppression, we overexpressed full-length A20 proteins bearing mutations within ZF7 in HEK293T cells, and examined their inhibitory effects on LUBAC-induced NF-κB activation. Notably, the full-length A20 proteins bearing the H769A/F770A, N772A, N780A and F785A/Y789A mutations, which abolished the linear diubiquitin binding activity, displayed considerable defects in the suppression of LUBAC-induced NF-κB activation (Figure 3F). Other A20 mutants also exhibited defects in NF-κB suppression, depending on their ability to bind linear diubiquitin (Figure 3F). These results indicated that the binding of A20 ZF7 to linear ubiquitin chains is critical for the suppression of LUBAC-mediated NF-κB activation.
In addition to the zinc-coordinating cysteine residues, the ubiquitin-binding residues of A20 ZF7 are conserved among the A20 ZF7 proteins from different organisms, including Ascidiacea (Saccoglossus kowalevskii), a primitive species that possesses the TNF-α signalling pathway (Figure 3G), highlighting the functional significance of these ubiquitin binding residues. In contrast, these residues are not conserved in A20 ZF1–ZF6 (Figure 3H), which may reflect functional differences between the A20 ZFs.
Mutations in the linear ubiquitin-binding site of A20 ZF7 are associated with B cell lymphomas
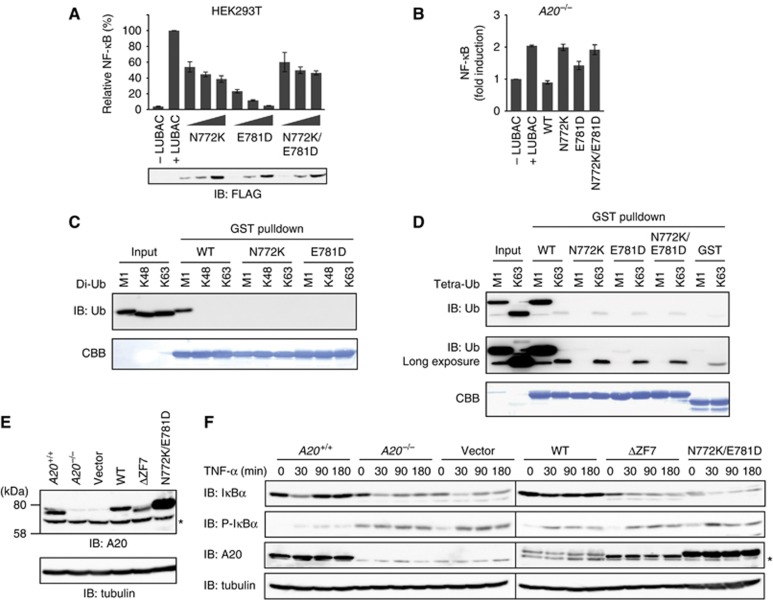
Genetic mutations of A20 result in Hodgkin’s and non-Hodgkin’s B cell lymphomas (Compagno et al, 2009; Kato et al, 2009; Schmitz et al, 2009; Hymowitz and Wertz, 2010), and polymorphisms of A20 are associated with multiple autoimmune and inflammatory diseases, including Sjögren syndrome, rheumatoid arthritis, systemic lupus erythematosus, psoriasis and type I diabetes (Vereecke et al, 2009). In addition, the A20 mutant lacking ZF7 failed to suppress NF-κB activation (Kato et al, 2009), and missense mutations in A20 ZF7, such as N772K and E781D, cause primary mediastinal B cell lymphoma (Schmitz et al, 2009).
To address the mechanism by which A20 mutations cause B cell lymphomas, we overexpressed full-length A20 proteins bearing N772K, E781D and N772K/E781D in HEK293T cells and A20−/− MEFs, and examined their inhibitory effects on LUBAC-induced NF-κB activation. The N772K, E781D and N772K/E781D mutants all showed impaired inhibitory effects on LUBAC-induced NF-κB activation in HEK293T cells (Figure 4A) and A20−/− MEFs (Figure 4B), suggesting that these mutations compromise the suppression of NF-κB activation, which results in pathological conditions. The present structure revealed that Asn772 and Glu781 participate in linear diubiquitin recognition (Figure 3C). We next examined the effects of these mutations on the ubiquitin-binding ability of A20 ZF7, using GST pulldown assays. GST-ZF7 N772K and GST-ZF7 E781D failed to bind linear di- and tetra-ubiquitins (Figure 4C and D). GST-ZF7 N772K/E781D also failed to bind linear tetraubiquitin (Figure 4D). A long exposure revealed the faint binding of linear tetraubiquitin with GST-ZF7 E781D, but not GST-ZF7 N772K, suggesting that the N772K mutation has a more serious impact on the linear-ubiquitin binding ability of A20 ZF7, as compared with the E781D mutation. The N772K mutation also exerted a more serious impact on NF-κB suppression, as compared with the E781D mutation (Figure 4A and B). These observations support the notion that the binding of A20 ZF7 to linear ubiquitin chains is important for the suppression of LUBAC-mediated NF-κB activation.
Figure 4.
Mutations in A20 ZF7 are associated with B cell lymphomas. (A, B) The full-length A20 protein bearing the N772K and E781D mutations, found in non-Hodgkin’s lymphoma, failed to suppress LUBAC-induced NF-κB activation in HEK293T cells (A) and A20−/− MEFs (B). (C, D) The N772K and E781D mutants of A20 ZF7 failed to bind linear diubiquitin (C) and linear tetraubiquitin (D) in GST pulldown assays. (E) Retroviral expression of A20 WT, ΔZF7 and N772K/E781D in A20−/− MEFs. (F) A20 ΔZF7 and N772K/E781D failed to rescue the defects in TNF-α-induced NF-κB activation in A20−/− MEFs. IκBα phosphorylation and degradation were analysed after TNF-α stimulation (10 ng/ml) in A20+/+ MEFs and A20−/− MEFs infected with retroviruses expressing A20 WT, ΔZF7 or N772K/E781D. *, Non-specific signal (E, F).
To explore the physiological significance of A20 ZF7, we expressed A20 WT, A20 ΔZF7 and A20 N772K/E781D in A20−/− MEFs, and then examined the TNF-α-induced phosphorylation and degradation of IκBα. Western blotting confirmed that A20 WT, A20 ΔZF7 and A20 N772K/E781D were expressed in A20−/− MEFs at levels similar to that of endogenous A20 in A20+/+ MEFs (Figure 4E). In A20+/+ MEFs, IκBα degradation was observed 30 min after TNF-α stimulation, followed by IκBα re-accumulation (Figure 4F). In contrast, re-accumulation of IκBα was not observed in A20−/− MEFs, due to the enhanced phosphorylation and degradation of IκBα upon TNF-α-stimulation (Figure 4F), as previously described (Lee et al, 2000). The restoration of A20 WT resulted in the regulated degradation and re-accumulation of IκBα in A20−/− MEFs (Figure 4F). In contrast, the restoration of A20 ΔZF7 and A20 N772K/E781D failed to rescue the defects in degradation and re-accumulation of IκBα in A20−/− MEFs (Figure 4F). These results indicated that the ability of A20 ZF7 to bind linear ubiquitin chains is crucial for the regulation of TNF-α-induced NF-κB activation.
The ability of A20 ZF7 to bind linear ubiquitin chains is sufficient for the suppression of LUBAC-mediated NF-κB activation
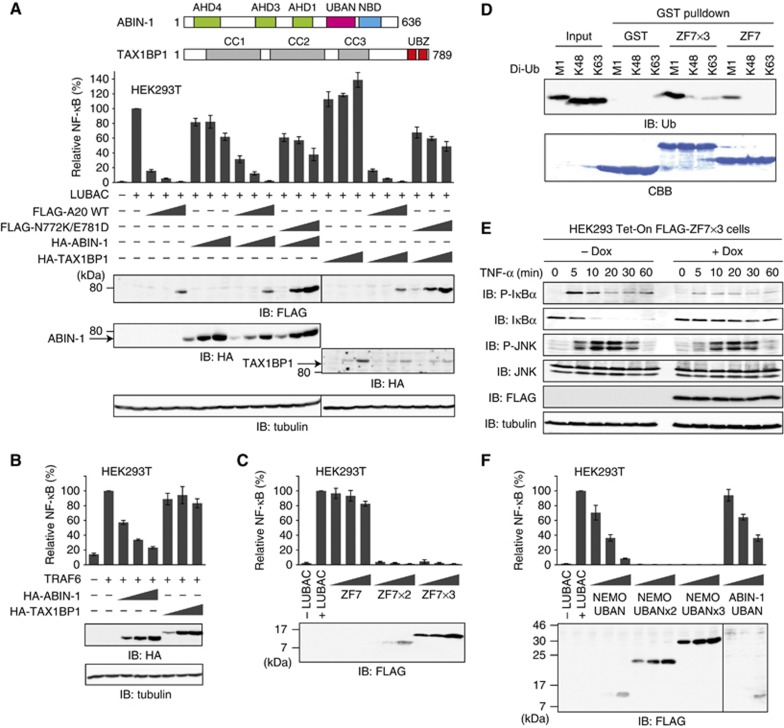
A20 forms a multi-molecular complex with ubiquitin-binding adaptor proteins, such as ABIN-1 (Heyninck et al, 1999) and TAX1BP1 (De Valck et al, 1999). We thus examined the effects of ABIN-1 and TAX1BP1 on LUBAC-induced NF-κB activation in HEK293T cells. The expression of ABIN-1 and TAX1BP1 did not exert any remarkable inhibitory effects on LUBAC-induced NF-κB activation (Figure 5A). In addition, the co-expression of A20 WT or A20 N772K/E781D, together with ABIN-1 or TAX1BP1, showed similar effects on LUBAC-induced NF-κB activation, as compared with the expression of A20 WT and A20 N772K/E781D alone. In contrast, ABIN-1 showed higher inhibitory effects on TRAF6-induced NF-κB activation, as compared with those on LUBAC-induced NF-κB activation (Figure 5B), consistent with previous reports (Iha et al, 2008; Verstrepen et al, 2009). These results indicated that A20 ZF7, but not ABIN-1 and TAX1BP1, plays a primary regulatory role in the suppression of LUBAC-mediated NF-κB activation.
Figure 5.
The binding of A20 ZF7 to linear ubiquitin chains is sufficient for the suppression of LUBAC-mediated NF-κB activation. (A) ABIN-1 and TAX1BP1 are not involved in the suppression of LUBAC-induced NF-κB activation. Increasing amounts of ABIN-1 or TAX1BP1 were expressed alone or together with A20 WT or A20 N772K/E781D, and their effects on LUBAC-induced NF-κB activation were examined by luciferase reporter assays (mean±s.d., n=3). (B) ABIN-1 and TAX1BP1 affect TRAF6-induced NF-κB activation. Luciferase reporter assays were performed as in Figure 5A. (C) Tandem conjugates of the multiple ZF7s suppress LUBAC-induced NF-κB activation. Relative luciferase activities are shown as mean±s.d. (n=3), and protein expression is shown in the lower panel. (D) A20 ZF7 × 3 binds larger amounts of linear diubiquitin, as compared with A20 ZF7. GST pulldown assays were performed as in Figure 2B, and the bound diubiquitin was detected by western blotting, using an anti-ubiquitin antibody. (E) A20 ZF7 is specifically involved in the NF-κB pathway. TNF-α-induced phosphorylation and degradation of IκBα and JNK were examined in HEK293 Tet-On cells expressing A20 ZF7 × 3 in the presence of doxycycline (Dox). (F) The UBAN domains of NEMO and ABIN-1 also suppress LUBAC-induced NF-κB activation. Luciferase reporter assays were performed as in Figure 5C.
To address the mechanism of A20 ZF7-mediated NF-κB suppression, we examined the inhibitory effects of A20 ZF7 on LUBAC-induced NF-κB activation in HEK293T cells. A20 ZF7 exhibited a minimal inhibitory effect, probably due to its poor expression in HEK293T cells (Figure 5C). A tandemly repeated ubiquitin-binding domain (UBD) recognizes ubiquitin with higher affinity, as compared with the single domain (Hjerpe et al, 2009). We examined the inhibitory effects of tandem conjugates of two and three ZF7s (ZF7 × 2 and ZF7 × 3) on LUBAC-induced NF-κB activation. Unlike A20 ZF7 alone, ZF7 × 2 and ZF7 × 3 strongly suppressed LUBAC-induced NF-κB activation (Figure 5C). Pulldown assays revealed that A20 ZF7 × 3 binds larger amounts of linear diubiquitin, as compared with A20 ZF7 alone (Figure 5D). To determine whether A20 ZF7 specifically suppresses LUBAC-induced NF-κB activation, we examined the phosphorylation and degradation of IκBα and JNK upon TNF-α stimulation in HEK293 Tet-On cells expressing FLAG-ZF7 × 3 in the presence of doxycycline (Dox). Upon TNF-α stimulation, the phosphorylation and degradation of IκBα, but not those of JNK, were inhibited in the presence of Dox (Figure 5E), indicating that A20 ZF7 × 3 specifically attenuates the NF-κB pathway. These results revealed that the binding of A20 ZF7 to linear ubiquitin chains is sufficient for the suppression of LUBAC-mediated NF-κB activation.
The UBAN domains in NEMO and ABINs preferentially bind linear ubiquitin chains (Rahighi et al, 2009; Kensche et al, 2012). To determine whether the binding of UBDs to linear ubiquitin chains is sufficient for the suppression of LUBAC-mediated NF-κB activation, we examined the inhibitory effects of the UBAN domains of NEMO and ABIN-1 on LUBAC-induced NF-κB activation. NEMO UBAN suppressed LUBAC-induced NF-κB activation, and tandemly repeated NEMO UBANs (NEMO UBAN × 2 and NEMO UBAN × 3) suppressed it more effectively (Figure 5F). ABIN-1 UBAN also showed an inhibitory effect on LUBAC-induced NF-κB activation (Figure 5F). These results indicated that the binding of UBDs to linear ubiquitin chains is sufficient for the suppression of LUBAC-mediated NF-κB activation. The present crystal structure suggested that full-length A20 can bind linear polyubiquitin in a manner similar to that of A20 ZF7 alone (Supplementary Figure S4A). Unlike A20, full-length ABIN-1 showed a lower inhibitory effect on LUBAC-induced NF-κB activation than ABIN-1 UBAN alone (Figure 5A), suggesting that full-length ABIN-1 binds linear polyubiquitin less effectively than ABIN-1 UBAN alone, due to steric hindrances.
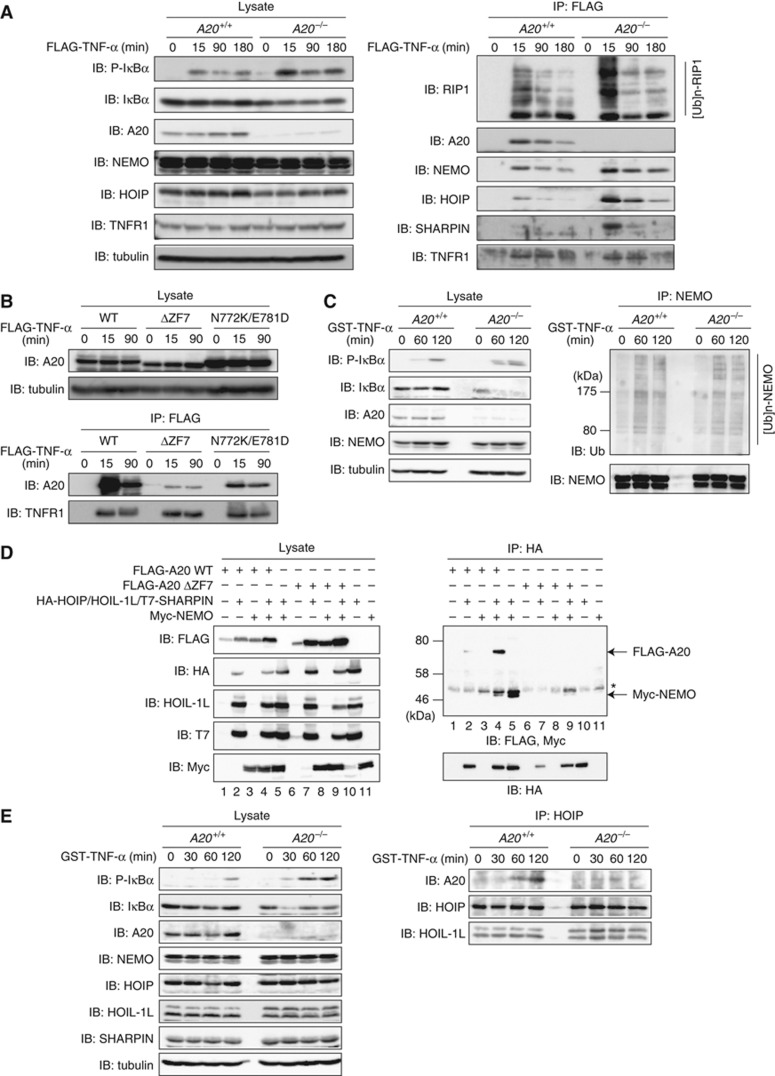
A20 recruitment to the TNFR signalling complex via the interaction between A20 ZF7 and linear polyubiquitin is critical for NF-κB suppression
Upon TNF-α stimulation, specific proteins, such as RIP1, TRADD, TRAF2/TRAF5, c-IAP-1/2 and LUBAC, are transiently recruited to TNFR to form signalling complex I, which plays a crucial role in canonical IKK activation (Micheau and Tschopp, 2003; Walczak et al, 2012). To explore the mechanism of NF-κB suppression by A20 ZF7, we stimulated A20+/+ and A20−/− MEFs with FLAG-TNF-α, and then examined the TNFR signalling complex formation. The phosphorylation and degradation of IκBα were enhanced in A20−/− MEFs, as compared with those in A20+/+ MEFs (Figure 6A). Together with RIP1, NEMO, HOIP and SHARPIN, A20 co-immunoprecipitated with FLAG-TNF-α in TNF-α-stimulated A20+/+ MEFs (Figure 6A). As compared with the levels in A20+/+ MEFs, larger amounts of RIP1, NEMO, HOIP and SHARPIN co-immunoprecipitated with FLAG-TNF-α in TNF-α-stimulated A20−/− MEFs (Figure 6A), indicating that A20 recruitment to TNFR facilitates the dissociation of the TNFR signalling complex, which results in NF-κB suppression. In addition, the polyubiquitination of RIP1 was enhanced in A20−/− MEFs, as compared with that in A20+/+ MEFs (Figure 6A). To clarify the role of A20 ZF7 in the recruitment of A20 to the TNFR signalling complex, we stimulated A20−/− MEFs expressing A20 WT, A20 ΔZF7 or A20 N772K/E781D with FLAG-TNF-α, and then examined the TNFR signalling complex formation. As compared with A20 WT, reduced amounts of A20 ΔZF7 and A20 N772K/E781D were recruited to the TNFR signalling complex (Figure 6B), suggesting that the binding of A20 ZF7 to linear polyubiquitin is important for the recruitment of A20 to the TNFR signalling complex. NEMO undergoes various ubiquitination modifications, including linear and K63-linked polyubiquitination during NF-κB activation (Chen et al, 2006; Tokunaga et al, 2009, 2011). Therefore, we stimulated A20+/+ and A20−/− MEFs with FLAG-TNF-α, and then examined the polyubiquitination of NEMO. The polyubiquitination levels of NEMO were similar in A20+/+ and A20−/− MEFs (Figure 6C), consistent with our data showing that A20 neither cleaves linear polyubiquitin (Supplementary Figure S1) nor inhibits LUBAC E3 activity (Supplementary Figure S3).
Figure 6.
The binding of A20 ZF7 to linear polyubiquitin is critical for the recruitment of A20 to the TNFR signalling complex. (A) A20 attenuates the recruitment of LUBAC and NEMO to TNFR complex I. A20+/+ and A20−/− MEFs were stimulated with FLAG-TNF-α (2 μg/ml) for the indicated times, and then TNFR complex I was immunoprecipitated using an anti-FLAG antibody, followed by western blotting using the indicated antibodies. (B) A20 ZF7 plays a critical role in the recruitment of A20 to the TNFR signalling complex. A20 WT, ΔZF7 and N772K/E781D were expressed in A20−/− MEFs, and then analysed as in Figure 6A. (C) A20 does not affect the TNF-α-induced polyubiquitination of NEMO. A20+/+ and A20−/− MEFs were stimulated with GST-TNF-α (1 μg/ml) for the indicated times, and then NEMO was immunoprecipitated using an anti-NEMO antibody, followed by western blotting with an anti-ubiquitin antibody. (D) A20 ZF7 participates in complex formation with LUBAC and NEMO. Cell lysates and immunoprecipitates from HEK293T cells expressing the indicated proteins were analysed by western blotting, using the indicated antibodies. (E) Endogenous association of A20 with LUBAC. A20+/+ and A20−/− MEFs were stimulated with GST-TNF-α (1 μg/ml), and then HOIP was immunoprecipitated using an anti-HOIP antibody, followed by western blotting with the indicated antibodies.
To further address the mechanism of the signalling complex formation, we overexpressed A20, LUBAC and NEMO in HEK293T cells, and then examined their interactions by co-immunoprecipitation. A20 WT, but not A20 ΔZF7, co-immunoprecipitated with LUBAC and NEMO (Figure 6D, lanes 4 and 9). In addition, the A20 mutants containing ZF7 (ZF1–7, ZF4–7, C103A and ZF4CA), but not those lacking intact ZF7 (OTU, ZF1–3 and ZF7CA), co-immunoprecipitated with LUBAC (Supplementary Figure S6), indicating that A20 ZF7 is crucial for the complex formation with LUBAC. Moreover, endogenous A20 co-immunoprecipitated with endogenous LUBAC upon TNF-α stimulation (Figure 6E). Taken together, these results suggested that the binding of A20 ZF7 to linear polyubiquitin contributes to the recruitment of A20 to the TNFR signalling complex upon TNF-α stimulation, resulting in the inhibition of IKK activity, which ultimately leads to the suppression of LUBAC-mediated NF-κB activation.
Discussion
A20 is a pleiotropic regulatory protein. In addition to the OTU DUB domain, A20 contains seven ZFs, which are involved in E3 activity (ZF4) (Wertz et al, 2004), RIP1 binding (ZF1) (Bosanac et al, 2010), ubiquitin binding (ZF4) (Bosanac et al, 2010), E2 binding (ZF5–7) (Bosanac et al, 2010), E2 degradation (ZF4) (Shembade et al, 2010), and complex formation with ABINs, TAX1BP1 (Klinkenberg et al, 2001; Verstrepen et al, 2011), and the E3 ubiquitin ligases RNF11 and Itch (Shembade et al, 2009; Verstrepen et al, 2010). A20 ZF7 also regulates IRF3 activation (Lin et al, 2006) and TNF-α-induced NF-κB activation (Natoli et al, 1998). In this study, we showed that A20 and CYLD downregulate LUBAC-mediated NF-κB activation through distinct molecular mechanisms. A20 suppressed NF-κB activation by binding linear ubiquitin chains through ZF7. In contrast, CYLD suppressed NF-κB activation by degrading linear ubiquitin chains, consistent with previous studies showing that CYLD degrades linear ubiquitin chains (Komander et al, 2008) and inhibits the linear ubiquitination of NEMO (Niu et al, 2011).
Both the linear polyubiquitination of NEMO by LUBAC and the specific recognition of linear polyubiquitin by the NEMO UBAN domain are critical for NF-κB activation (Lo et al, 2009; Rahighi et al, 2009). In addition, the specific recognition of linear polyubiquitin by the HOIL-1L NZF domain is also important for NF-κB activation (Sato et al, 2011). These observations suggested a model for LUBAC-mediated NF-κB activation, in which the trans-phosphorylation of IKKβ is facilitated by the recruitment of the IKK complexes via the interaction between the NEMO UBAN domain and linear polyubiquitin conjugated to NEMO, although the precise mechanism remains elusive (Tokunaga et al, 2011; Kensche et al, 2012). Our functional analysis indicated that (1) the binding of A20 ZF7 to linear polyubiquitin is necessary and sufficient for the suppression of LUBAC-mediated NF-κB activation, (2) the recruitment of A20 to the TNFR signalling complex upon TNF-α stimulation is important for NF-κB suppression, (3) the binding of A20 ZF7 to linear polyubiquitin is essential for the recruitment of A20 to the TNFR signalling complex, and (4) A20 ZF7 participates in the complex formation of A20 with LUBAC and NEMO. Based on these observations, we propose a model for the suppression of LUBAC-mediated NF-κB activation, in which A20 is recruited to the TNFR signalling complex upon TNF-α stimulation via the interaction between A20 ZF7 and linear polyubiquitin on NEMO and RIP1, facilitating the dissociation of LUBAC and NEMO from TNFR. Moreover, our structural and functional analyses revealed that Asn772 and Glu781 of A20 ZF7, with mutations associated with B cell lymphomas (Schmitz et al, 2009), participate in linear diubiquitin recognition (Figure 3C), and the N772K and E781D mutations abolished the abilities of A20 ZF7 to bind linear ubiquitin chains (Figure 4C and D) and to suppress LUBAC-induced NF-κB activation (Figure 4A and B). These observations highlight the physiological significance of A20 ZF7 binding to linear polyubiquitin in NF-κB regulation. The conjugate of multiple A20 ZF7s potently suppressed NF-κB activation (Figure 5C), suggesting that it could be a therapeutic agent for B cell lymphomas.
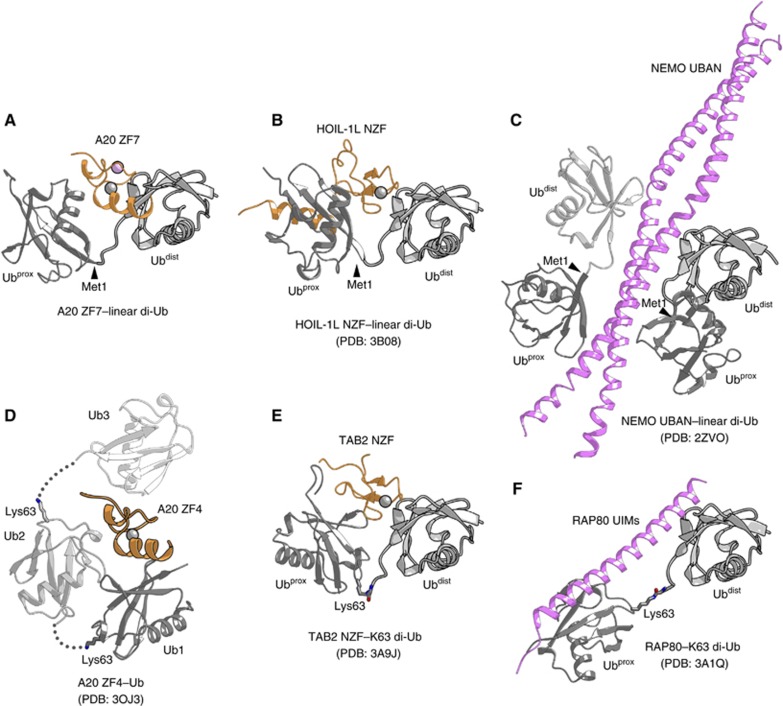
A structural comparison of the A20 ZF7–diubiquitin complex with the A20 ZF4–ubiquitin complex (Bosanac et al, 2010) revealed a distinct ubiquitin-binding mode (Figure 7), consistent with their functional differences. Moreover, the crystal structures of the NEMO UBAN–linear diubiquitin (Lo et al, 2009; Rahighi et al, 2009), HOIL-1L NZF–linear diubiquitin (Sato et al, 2011), TAB2/3 NZF–K63-linked diubiquitin (Kulathu et al, 2009; Sato et al, 2009b) and RAP80 UIMs–K63-linked diubiquitin (Sims and Cohen, 2009; Sato et al, 2009a) complexes showed that, despite their distinct ubiquitin-binding modes, these UBDs selectively recognize diubiquitin with a specific linkage, through simultaneous interactions with the proximal and distal ubiquitin moieties (Figure 7). A structural comparison of the present A20 ZF7–linear diubiquitin complex with the NEMO UBAN–linear diubiquitin (Rahighi et al, 2009) and HOIL-1L NZF–linear diubiquitin (Sato et al, 2011) complexes revealed that A20 ZF7 recognizes linear diubiquitin in a novel manner (Figure 7), thus providing new insight into the linkage-specific recognition mechanism by UBDs. In addition, this structural comparison uncovered notable differences in the overall conformations of the polyubiquitin chains, highlighting the flexible nature of the linear and K63-linked chains. The present structure revealed that A20 ZF7 does not directly contact the linkage between the proximal and distal ubiquitin moieties, but recognizes the specific configuration of linear diubiquitin through simultaneous interactions with the proximal and distal ubiquitin moieties. Thus, the present structure reinforces the notion that UBDs recognize a specific configuration of diubiquitin that is unique to the particular linkage, thereby achieving linkage-specific binding to polyubiquitin.
Figure 7.
Structural comparison of UBD–diubiquitin complexes. (A) The A20 ZF7–linear diubiquitin complex. (B) The HOIL-1L NZF–linear diubiquitin complex (Sato et al, 2011) (PDB code 3B08). (C) The NEMO UBAN–linear diubiquitin complex (Rahighi et al, 2009) (PDB code 2ZVO). (D) The A20 ZF4–monoubiquitin complex (Bosanac et al, 2010) (PDB code 3OJ3). (E) The TAB2 NZF–K63-linked diubiquitin complex (Kulathu et al, 2009; Sato et al, 2009b) (PDB code 3A9J). (F) The RAP80 UIMs–K63-linked diubiquitin complex (Sims and Cohen, 2009; Sato et al, 2009a) (PDB code 3A1Q). These structures were aligned based on the distal ubiquitin moieties, except for the A20 ZF4–monoubiquitin complex, which was aligned with the A20 ZF7–linear diubiquitin complex based on the ZFs.
K63-linked ubiquitination is required for IL-1β-induced NF-κB activation, whereas it is dispensable for TNF-α-induced NF-κB activation (Xu et al, 2009). In the IL-1β-induced NF-κB pathway, the TAK1–TAB1–TAB2/3 complex and the IKK complex bind K63-linked polyubiquitin through the TAB2/3 NZF domain and the NEMO UBAN domain, respectively, resulting in IKK activation by TAK1-mediated phosphorylation of IKKβ (Chen, 2012). A recent study proposed a model, in which A20 is recruited to the IKK complex through bipartite binding to the N-terminal region of NEMO and unanchored K63-linked polyubiquitin chains, thus inhibiting IKK activation by TAK1 (Skaug et al, 2011). In this model, multiple A20 ZFs, especially A20 ZF7, participate in K63-linked polyubiquitin binding, whereas A20 ZF7 alone is not sufficient for K63-linked polyubiquitin binding. It seems likely that, although A20 ZF7 specifically binds linear ubiquitin chains (Figure 2), it can also weakly interact with K63-linked long polyubiquitin chains, due to the avidity provided by the multiple ubiquitin moieties present in polyubiquitin. NEMO UBAN shows about 100-fold higher affinity to linear diubiquitin than K63-linked diubiquitin (Lo et al, 2009; Rahighi et al, 2009). These differences were explained by the crystal structures of NEMO UBAN in complex with linear diubiquitin (Lo et al, 2009; Rahighi et al, 2009) and K63-linked diubiquitin (Yoshikawa et al, 2009), which revealed that NEMO UBAN simultaneously binds the proximal and distal ubiquitin moieties of linear diubiquitin, but not K63-linked diubiquitin. In addition, our recent study showed that NEMO functions as a high affinity receptor for linear ubiquitin chains and a low affinity receptor for long Lys-linked ubiquitin chains (Kensche et al, 2012). Taken together, these observations indicated that A20 ZF7 specifically binds linear polyubiquitin and plays a crucial role in the linear polyubiquitin-mediated pathways, whereas it also plays an auxiliary role in the K63-linked polyubiquitin-mediated pathways. Further studies will be needed to understand the mechanisms underlying the activation and inhibition of LUBAC-mediated NF-κB activation.
Materials and methods
Plasmids, antibodies and reagents
The open reading frames of human A20, CYLD and Cezanne were amplified by RT–PCR, and the other cDNAs were prepared as described previously (Tokunaga et al, 2009, 2011). The cDNAs were ligated to the appropriate epitope sequences and cloned into the pcDNA3.1 (Invitrogen), pGEX-6P-1 (GE Healthcare) or pTRE (Clontech) vector. HA-tagged TAX1BP1 in the pCAGGS plasmid was kindly provided by Dr Hidekatsu Iha (Oita University, Japan). GST-tagged proteins were expressed in E. coli, and purified using glutathione Sepharose (GE Healthcare). Site-directed mutagenesis was performed using PrimeSTAR MAX DNA polymerase (Takara Bio). The following antibodies were used: FLAG (Stratagene), A20 (A-12), ubiquitin (P4D1), TNFR1 (H271) and NEMO (FL-419) (Santa Cruz Biotechnology), tubulin (Cedarlane), RIP1 (BD Transduction Laboratories), A20 (AM63) (Calbiochem), NEMO (EA2-6) (MBL), and A20 (#5630), P-IκBα (#9246), IκBα (#4812), P-JNK (#4688), and JNK (#9258) (Cell Signalling). K6-, K11-, K27-, K29- and K33-linked diubiquitins were purchased from Boston Biochem. Linear di- and tetra-ubiquitins were expressed in E. coli, and purified. K48- and K63-linked di- and tetra-ubiquitins were prepared as described (Komander et al, 2008). Other reagents were obtained as described previously (Tokunaga et al, 2009, 2011).
Cell culture, transfection and luciferase assay
HEK293T, HOIL-1L+/+ and HOIL-1L−/− MEFs (Tokunaga et al, 2009), SHARPIN+/+ and SHARPIN-ablated cpdm MEFs (Tokunaga et al, 2011), A20+/+ and A20−/− MEFs (Lee et al, 2000), and HEK293 Tet-On cells were cultured in DMEM, containing 10% fetal bovine serum, 100 IU/ml penicillin G and 100 μg/ml streptomycin, at 37°C under a 7.5% CO2 atmosphere. For the luciferase assay, HEK293T cells or various MEFs were transfected with the luciferase reporter plasmids, pGL4-NF-κB-Luc and pGL4-Renilla-Luc/TK (Promega), the LUBAC component plasmids, pcDNA3.1-Myc-HOIP, pcDNA3.1-HOIL-1L-HA and pcDNA3.1-T7-SHARPIN, and appropriate plasmids, using Lipofectamine 2000 (Invitrogen), ExGen500 (Thermo), or Turbofect (Thermo). At 24 h after transfection, the cells were lysed and the luciferase activity was measured in a Lumat luminometer (Berthold), using the Bright-Glo luciferase assay system (Promega), as previously described (Tokunaga et al, 2009, 2011). In some experiments, the cells were treated with TNF-α (10 ng/ml) for 6 h before harvest.
Retroviral expression
HA-tagged WT and mutant human A20 cDNAs were ligated into the pMXs-IP vector, and the plasmids were transfected into Plat E packaging cells, as described (Tokunaga et al, 2009, 2011). The retrovirus thus produced was used to infect A20−/− MEFs, and puromycin-resistant cells were selected.
TNFR1 complex analysis
TNFR complex formation upon TNF-α stimulation was analysed using FLAG-tagged TNF-α, as described (Schneider, 2000). Briefly, MEFs (5 × 107 cells/ml) were stimulated for the indicated times with 2 μg/ml FLAG-tagged TNF-α, and then lysed in 1 ml lysis buffer (20 mM Tris–HCl, pH 7.4, 150 mM NaCl, 0.2% NP-40, 10% glycerol, and complete protease inhibitor cocktail) for 15 min on ice. Lysates were precleared with 20 μl Sepharose-6B (Sigma) for 2 h at 4°C and then immunoprecipitated with 20 μl anti-FLAG M2 beads (Sigma) overnight at 4°C. The beads were recovered by centrifugation, washed five times with 1 ml of lysis buffer, and then analysed by SDS–PAGE and western blotting, as described (Micheau and Tschopp, 2003).
Construction of tandem conjugates of A20 ZF7 and UBAN and its inducible cells
Tandem conjugates of human A20 ZF7 (residues 758–790) with a Gly linker were constructed by PCR, using following primers: BamHI-5 CGGGATCCTCTGGTGGAGGTCCCAAGCAGCGTTGCCGG, EcoRI-3 GGAATTCTCCACCGCCGCCGCCATACATCTGCTTGAA, EcoRI-5 GGAATTCTCTGGTGGAGGTCCCAAGCAGCGTTGCCGG, SalI-3 ACGAGTCGACCACCGCCGCCGCCATACATCTGCTTGAA, SalI-5 ACGCGTCGACCCTCTGGTGGAGGTCCCAAGCAGCGTTGCCGG, and NotI-3 TAAAGCGGCCGCTAACCACCGCCGCCGCCATACATCTGCTTGAA. PCR fragments were digested with the designated restriction enzymes and inserted into the pGEX-6P-1, pcDNA3.1 or pTRE vector. To prepare cells expressing FLAG-tagged tandem conjugates of three ZF7s (FLAG-ZF7 × 3), HEK293 Tet-On cells were co-transfected with pTRE-FLAG-ZF7 × 3 and pXS-Puro, and puromycin-resistant clones were selected. FLAG-ZF7 × 3 was expressed in the stable cells with 1 μg/ml doxycycline for 24 h, followed by stimulation with TNF-α (10 ng/ml). SDS–PAGE and western blotting were performed as described previously (Tokunaga et al, 2009, 2011).
Similarly, single and tandem conjugates of human NEMO UBAN (residues 257–346) and human ABIN-1 UBAN (residues 452–512) were constructed by PCR, using following primers: NEMO UBAN-BamHI-5 CGGGATCCGGAATGCAGCTGGAAGAT, NEMO UBAN-EcoRI-3 GGAATTCGCTGGCCTTCAGTTTGCT, NEMO UBAN-EcoRI-5 GGAATTCGGAATGCAGCTGGAAGAT, NEMO UBAN-SalI-3 ACGCGTCGACCGCTGGCCTTCAGTTTGCT, NEMO UBAN-SalI-5 ACGCGTCGACTCGGAATGCAGCTGGAAGAT, NEMO UBAN-XhoI-3 CCGCTCGAGGCTGGCCTTCAGTTTGCT, ABIN-1 UBAN-BamHI-5 CGGGATCCAAACAGGAGCTGGTCACG, and ABIN-1 UBAN-EcoRI-3 GGAATTCTCATTTGAATGCTTTTAGCTG.
GST pulldown assay
GST-tagged A20 ZFs were expressed in E. coli Rosetta2(DE3) (Novagen) or BL21-CodonPlus(DE3)-RIPL (Agilent), and were purified using glutathione Sepharose. To examine the ubiquitin-binding ability, GST-ZF proteins (4 μM) and diubiquitin (2 μM) or tetraubiquitin (0.5 μM) were incubated in reaction buffer (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1 mM DTT, 0.1% NP-40 and 250 μg/ml BSA) at 4°C for 2 h, followed by the addition of glutathione Sepharose beads. The samples were further incubated at 4°C for 1 h with gentle rotation, and then the beads were washed three times with the buffer without BSA, and analysed by SDS–PAGE. The bound ubiquitin was detected by western blotting using an anti-ubiquitin antibody, and GST-ZF proteins were stained with Coomassie Brilliant Blue.
To investigate the linear-diubiquitin binding of A20 ZF7 mutants, GST-ZF7 proteins (4 μM) were incubated with linear diubiquitin (4 μM) and MagneGST beads (Promega) at 4°C for 2 h in reaction buffer (50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 5 mM DTT and 0.1% NP-40). The beads were washed three times with the buffer, and the bound proteins were eluted with the buffer supplemented with 10 mM glutathione. The eluted proteins were analysed by 10–20% SDS–PAGE, and stained with SimplyBlue Safestain (Invitrogen).
Crystallography
GST-tagged human A20 ZF7 (residues 758–790) was expressed in E. coli Rosetta2(DE3), and purified by glutathione Sepharose chromatography. The protein was treated with Turbo3C Protease (Nacalai Tesque) to remove the GST tag, and was further purified by HiLoad Superdex 75 16/60 gel filtration chromatography (GE Healthcare). Monoubiquitin, linear diubiquitin, and linear tetraubiquitin were expressed in E. coli Rosetta2(DE3), and were purified by heat treatment, followed by chromatography on HiTrap SP (GE Healthcare) and HiLoad Superdex 75 16/60 gel filtration columns.
Crystallization was performed at 20°C. Crystals of the A20 ZF7–diubiquitin complex (Form I) were obtained using the sitting drop vapour diffusion method, by mixing 1 μl of protein solution (2 mg/ml A20 ZF7, 7 mg/ml diubiquitin, 10 mM Tris–HCl, pH 8.0, 150 mM NaCl and 1 mM DTT) and 1 μl of reservoir solution (0.2 M Na/K tartrate and 20% PEG 3350). Crystals of the A20 ZF7–diubiquitin complex (Form II) were obtained using the hanging drop vapour diffusion method, by mixing 1 μl of protein solution (1 mg/ml A20 ZF7, 2 mg/ml diubiquitin, 10 mM Tris–HCl, pH 8.0, 150 mM NaCl and 1 mM DTT) and 1 μl of reservoir solution (0.2 M Na/K tartrate and 20% PEG 3350). Crystals of the A20 ZF7–tetraubiquitin complex (Form II) were obtained using the hanging drop vapour diffusion method, by mixing 2 μl of protein solution (1 mg/ml A20 ZF7, 2 mg/ml tetraubiquitin, 10 mM Tris–HCl, pH 8.0, 150 mM NaCl and 1 mM DTT) and 2 μl of reservoir solution (0.2 M Na/K tartrate and 8% PEG 3350).
X-ray diffraction data were collected at 100 K on beamlines BL41XU at SPring-8 (Hyogo, Japan) and BL1 at PF-AR (Tsukuba, Japan). Crystals were cryoprotected in the reservoir solution supplemented with 20% ethylene glycol. Diffraction data were processed using HKL2000 (HKL Research Inc.). The structure of the A20 ZF7–diubiquitin complex (Form I) was determined by molecular replacement with MOLREP (Vagin and Teplyakov, 2010), using the monoubiquitin molecule in the TAB2 NZF–K63-linked diubiquitin complex (PDB code 3A9J) as a search model. The structures of A20 ZF7 in complex with diubiquitin and tetraubiquitin (Form II) were determined by molecular replacement. Manual model rebuilding and refinement were performed using Coot (Emsley and Cowtan, 2004) and PHENIX (Adams et al, 2002). As the crystals of the A20 ZF7–diubiquitin complex (Form II) were merohedrally twinned, the structure was refined using the twin target function implemented in the program PHENIX (Adams et al, 2002). The resulting twinning fraction was 0.28 for the twinning operator (h, −h−k, −l). Data collection and refinement statistics are provided in Table I. Molecular graphics were prepared using CueMol (http://www.cuemol.org).
Isothermal titration calorimetry
ITC measurements were performed using a VP-ITC (GE Healthcare). Monoubiquitin, linear diubiquitin, and K48- and K63-linked diubiquitin (50–58 μM) were each loaded into the cell (1.4 ml), and A20 ZF7 (0.37 mM) was loaded into a syringe (0.3 ml). The samples were dialysed against PBS buffer supplemented with 1 mM TCEP. Titrations were performed at 25°C, using 29 injections (2 μl for injections 1–2 and 10 μl for injections 3–29), with 4-min intervals between injections. The stirring rate was 307 r.p.m. Data from injections into the buffer were subtracted from the sample data before data analysis, using the Origin7 software (MicroCal).
Accession codes
The atomic coordinates have been deposited in the Protein Data Bank with accession codes 3VUW (A20 ZF7–linear diubiquitin, Form I), 3VUX (A20 ZF7–linear diubiquitin, Form II) and 3VUY (A20 ZF7–linear tetraubiquitin).
Supplementary Material
Acknowledgments
We thank to Dr Hidekatsu Iha (Department of Infectious Diseases, Faculty of Medicine, Oita University) for the TAX1BP1 plasmid. This work was supported by a grant from the Japan Society for the Promotion of Science (JSPS), through its ‘Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST program)’, to ON, by the Core Research for Evolutional Science and Technology (CREST) Program ‘The Creation of Basic Medical Technologies to Clarify and Control the Mechanisms Underlying Chronic Inflammation’ of Japan Science and Technology Agency (JST) to FT and ON, by a grant for the National Project on Protein Structural and Functional Analyses, from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), to ON and KI, by a Grant-in-Aid for Scientific Research from MEXT to FT, RI, KI and ON, and by a grant from The Uehara Memorial Foundation to FT.
Author contributions: FT, EG, TN, and KK performed biochemical and cell biological experiments and HN performed biochemical experiments. HN and RI performed the crystallographic analysis. KM performed the biophysical analysis of ubiquitin. AM provided materials and participated in discussions. FT, RI and ON conceived the project. FT, HN, RI, KI and ON wrote the manuscript. All authors commented on the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954 [DOI] [PubMed] [Google Scholar]
- Ben-Neriah Y, Karin M (2011) Inflammation meets cancer, with NF-κB as the matchmaker. Nat Immunol 12: 715–723 [DOI] [PubMed] [Google Scholar]
- Bosanac I, Wertz IE, Pan B, Yu C, Kusam S, Lam C, Phu L, Phung Q, Maurer B, Arnott D, Kirkpatrick DS, Dixit VM, Hymowitz SG (2010) Ubiquitin binding to A20 ZnF4 is required for modulation of NF-κB signaling. Mol Cell 40: 548–557 [DOI] [PubMed] [Google Scholar]
- Bremm A, Freund SM, Komander D (2010) Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nat Struct Mol Biol 17: 939–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Nijman SM, Dirac AM, Bernards R (2003) Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-κB. Nature 424: 797–801 [DOI] [PubMed] [Google Scholar]
- Chen F, Bhatia D, Chang Q, Castranova V (2006) Finding NEMO by K63-linked polyubiquitin chain. Cell Death Differ 13: 1835–1838 [DOI] [PubMed] [Google Scholar]
- Chen ZJ (2012) Ubiquitination in signaling to and activation of IKK. Immunol Rev 246: 95–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compagno M, Lim WK, Grunn A, Nandula SV, Brahmachary M, Shen Q, Bertoni F, Ponzoni M, Scandurra M, Califano A, Bhagat G, Chadburn A, Dalla-Favera R, Pasqualucci L (2009) Mutations of multiple genes cause deregulation of NF-κB in diffuse large B-cell lymphoma. Nature 459: 717–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damgaard RB, Nachbur U, Yabal M, Wong WW, Fiil BK, Kastirr M, Rieser E, Rickard JA, Bankovacki A, Peschel C, Ruland J, Bekker-Jensen S, Mailand N, Kaufmann T, Strasser A, Walczak H, Silke J, Jost PJ, Gyrd-Hansen M (2012) The ubiquitin ligase XIAP recruits LUBAC for NOD2 signaling in inflammation and innate immunity. Mol Cell 46: 746–758 [DOI] [PubMed] [Google Scholar]
- De Valck D, Jin DY, Heyninck K, Van de Craen M, Contreras R, Fiers W, Jeang KT, Beyaert R (1999) The zinc finger protein A20 interacts with a novel anti-apoptotic protein which is cleaved by specific caspases. Oncogene 18: 4182–4190 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Gerlach B, Cordier SM, Schmukle AC, Emmerich CH, Rieser E, Haas TL, Webb AI, Rickard JA, Anderton H, Wong WW, Nachbur U, Gangoda L, Warnken U, Purcell AW, Silke J, Walczak H (2011) Linear ubiquitination prevents inflammation and regulates immune signalling. Nature 471: 591–596 [DOI] [PubMed] [Google Scholar]
- Harhaj EW, Dixit VM (2011) Deubiquitinases in the regulation of NF-κB signaling. Cell Res 21: 22–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S (2012) NF-κB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev 26: 203–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyninck K, De Valck D, Vanden Berghe W, Van Criekinge W, Contreras R, Fiers W, Haegeman G, Beyaert R (1999) The zinc finger protein A20 inhibits TNF-induced NF-κB-dependent gene expression by interfering with an RIP- or TRAF2-mediated transactivation signal and directly binds to a novel NF-κB-inhibiting protein ABIN. J Cell Biol 145: 1471–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjerpe R, Aillet F, Lopitz-Otsoa F, Lang V, England P, Rodriguez MS (2009) Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep 10: 1250–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hymowitz SG, Wertz IE (2010) A20: from ubiquitin editing to tumour suppression. Nat Rev Cancer 10: 332–341 [DOI] [PubMed] [Google Scholar]
- Iha H, Peloponese JM, Verstrepen L, Zapart G, Ikeda F, Smith CD, Starost MF, Yedavalli V, Heyninck K, Dikic I, Beyaert R, Jeang KT (2008) Inflammatory cardiac valvulitis in TAX1BP1-deficient mice through selective NF-κB activation. EMBO J 27: 629–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda F, Deribe YL, Skanland SS, Stieglitz B, Grabbe C, Franz-Wachtel M, van Wijk SJ, Goswami P, Nagy V, Terzic J, Tokunaga F, Androulidaki A, Nakagawa T, Pasparakis M, Iwai K, Sundberg JP, Schaefer L, Rittinger K, Macek B, Dikic I (2011) SHARPIN forms a linear ubiquitin ligase complex regulating NF-κB activity and apoptosis. Nature 471: 637–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inn KS, Gack MU, Tokunaga F, Shi M, Wong LY, Iwai K, Jung JU (2011) Linear ubiquitin assembly complex negatively regulates RIG-I- and TRIM25-mediated type I interferon induction. Mol Cell 41: 354–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai K (2012) Diverse ubiquitin signaling in NF-κB activation. Trends Cell Biol 22: 355–364 [DOI] [PubMed] [Google Scholar]
- Kato M, Sanada M, Kato I, Sato Y, Takita J, Takeuchi K, Niwa A, Chen Y, Nakazaki K, Nomoto J, Asakura Y, Muto S, Tamura A, Iio M, Akatsuka Y, Hayashi Y, Mori H, Igarashi T, Kurokawa M, Chiba S et al. (2009) Frequent inactivation of A20 in B-cell lymphomas. Nature 459: 712–716 [DOI] [PubMed] [Google Scholar]
- Kensche T, Tokunaga F, Ikeda F, Goto E, Iwai K, Dikic I (2012) Analysis of nuclear factor-κB (NF-κB) essential modulator (NEMO) binding to linear and lysine-linked ubiquitin chains and its role in the activation of NF-κB. J Biol Chem 287: 23626–23634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, Sano S, Tokunaga F, Tanaka K, Iwai K (2006) A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J 25: 4877–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinkenberg M, Van Huffel S, Heyninck K, Beyaert R (2001) Functional redundancy of the zinc fingers of A20 for inhibition of NF-κB activation and protein-protein interactions. FEBS Lett 498: 93–97 [DOI] [PubMed] [Google Scholar]
- Komander D, Barford D (2008) Structure of the A20 OTU domain and mechanistic insights into deubiquitination. Biochem J 409: 77–85 [DOI] [PubMed] [Google Scholar]
- Komander D, Lord CJ, Scheel H, Swift S, Hofmann K, Ashworth A, Barford D (2008) The structure of the CYLD USP domain explains its specificity for Lys63-linked polyubiquitin and reveals a B box module. Mol Cell 29: 451–464 [DOI] [PubMed] [Google Scholar]
- Kulathu Y, Akutsu M, Bremm A, Hofmann K, Komander D (2009) Two-sided ubiquitin binding explains specificity of the TAB2 NZF domain. Nat Struct Mol Biol 16: 1328–1330 [DOI] [PubMed] [Google Scholar]
- Lee EG, Boone DL, Chai S, Libby SL, Chien M, Lodolce JP, Ma A (2000) Failure to regulate TNF-induced NF-κB and cell death responses in A20-deficient mice. Science 289: 2350–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Yang L, Nakhaei P, Sun Q, Sharif-Askari E, Julkunen I, Hiscott J (2006) Negative regulation of the retinoic acid-inducible gene I-induced antiviral state by the ubiquitin-editing protein A20. J Biol Chem 281: 2095–2103 [DOI] [PubMed] [Google Scholar]
- Lo YC, Lin SC, Rospigliosi CC, Conze DB, Wu CJ, Ashwell JD, Eliezer D, Wu H (2009) Structural basis for recognition of diubiquitins by NEMO. Mol Cell 33: 602–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheau O, Tschopp J (2003) Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell 114: 181–190 [DOI] [PubMed] [Google Scholar]
- Natoli G, Costanzo A, Guido F, Moretti F, Bernardo A, Burgio VL, Agresti C, Levrero M (1998) Nuclear factor κB-independent cytoprotective pathways originating at tumor necrosis factor receptor-associated factor 2. J Biol Chem 273: 31262–31272 [DOI] [PubMed] [Google Scholar]
- Niu J, Shi Y, Iwai K, Wu ZH (2011) LUBAC regulates NF-κB activation upon genotoxic stress by promoting linear ubiquitination of NEMO. EMBO J 30: 3741–3753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasparakis M (2009) Regulation of tissue homeostasis by NF-κB signalling: implications for inflammatory diseases. Nat Rev Immunol 9: 778–788 [DOI] [PubMed] [Google Scholar]
- Perkins ND (2012) The diverse and complex roles of NF-κB subunits in cancer. Nat Rev Cancer 12: 121–132 [DOI] [PubMed] [Google Scholar]
- Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D, Randow F, Wakatsuki S, Dikic I (2009) Specific recognition of linear ubiquitin chains by NEMO is important for NF-κB activation. Cell 136: 1098–1109 [DOI] [PubMed] [Google Scholar]
- Sato Y, Fujita H, Yoshikawa A, Yamashita M, Yamagata A, Kaiser SE, Iwai K, Fukai S (2011) Specific recognition of linear ubiquitin chains by the Npl4 zinc finger (NZF) domain of the HOIL-1L subunit of the linear ubiquitin chain assembly complex. Proc Natl Acad Sci USA 108: 20520–20525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Yoshikawa A, Mimura H, Yamashita M, Yamagata A, Fukai S (2009a) Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80. EMBO J 28: 2461–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Yoshikawa A, Yamashita M, Yamagata A, Fukai S (2009b) Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by NZF domains of TAB2 and TAB3. EMBO J 28: 3903–3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R, Hansmann ML, Bohle V, Martin-Subero JI, Hartmann S, Mechtersheimer G, Klapper W, Vater I, Giefing M, Gesk S, Stanelle J, Siebert R, Kuppers R (2009) TNFAIP3 (A20) is a tumor suppressor gene in Hodgkin lymphoma and primary mediastinal B cell lymphoma. J Exp Med 206: 981–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider P (2000) Production of recombinant TRAIL and TRAIL receptor: Fc chimeric proteins. Methods Enzymol 322: 325–345 [DOI] [PubMed] [Google Scholar]
- Shembade N, Ma A, Harhaj EW (2010) Inhibition of NF-κB signaling by A20 through disruption of ubiquitin enzyme complexes. Science 327: 1135–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shembade N, Parvatiyar K, Harhaj NS, Harhaj EW (2009) The ubiquitin-editing enzyme A20 requires RNF11 to downregulate NF-κB signalling. EMBO J 28: 513–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims JJ, Cohen RE (2009) Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of rap80. Mol Cell 33: 775–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaug B, Chen J, Du F, He J, Ma A, Chen ZJ (2011) Direct, noncatalytic mechanism of IKK inhibition by A20. Mol Cell 44: 559–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga F, Iwai K (2012) LUBAC, a novel ubiquitin ligase for linear ubiquitination, is crucial for inflammation and immune responses. Microbes Infect 14: 563–572 [DOI] [PubMed] [Google Scholar]
- Tokunaga F, Nakagawa T, Nakahara M, Saeki Y, Taniguchi M, Sakata S, Tanaka K, Nakano H, Iwai K (2011) SHARPIN is a component of the NF-κB-activating linear ubiquitin chain assembly complex. Nature 471: 633–636 [DOI] [PubMed] [Google Scholar]
- Tokunaga F, Sakata S, Saeki Y, Satomi Y, Kirisako T, Kamei K, Nakagawa T, Kato M, Murata S, Yamaoka S, Yamamoto M, Akira S, Takao T, Tanaka K, Iwai K (2009) Involvement of linear polyubiquitylation of NEMO in NF-κB activation. Nat Cell Biol 11: 123–132 [DOI] [PubMed] [Google Scholar]
- Tomonaga M, Hashimoto N, Tokunaga F, Onishi M, Myoui A, Yoshikawa H, Iwai K (2012) Activation of nuclear factor-kappa B by linear ubiquitin chain assembly complex contributes to lung metastasis of osteosarcoma cells. Int J Oncol 40: 409–417 [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A (2010) Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr 66: 22–25 [DOI] [PubMed] [Google Scholar]
- Vallabhapurapu S, Karin M (2009) Regulation and function of NF-κB transcription factors in the immune system. Annu Rev Immunol 27: 693–733 [DOI] [PubMed] [Google Scholar]
- Vereecke L, Beyaert R, van Loo G (2009) The ubiquitin-editing enzyme A20 (TNFAIP3) is a central regulator of immunopathology. Trends Immunol 30: 383–391 [DOI] [PubMed] [Google Scholar]
- Verstrepen L, Carpentier I, Verhelst K, Beyaert R (2009) ABINs: A20 binding inhibitors of NF-κB and apoptosis signaling. Biochem Pharmacol 78: 105–114 [DOI] [PubMed] [Google Scholar]
- Verstrepen L, Verhelst K, Carpentier I, Beyaert R (2011) TAX1BP1, a ubiquitin-binding adaptor protein in innate immunity and beyond. Trends Biochem Sci 36: 347–354 [DOI] [PubMed] [Google Scholar]
- Verstrepen L, Verhelst K, van Loo G, Carpentier I, Ley SC, Beyaert R (2010) Expression, biological activities and mechanisms of action of A20 (TNFAIP3). Biochem Pharmacol 80: 2009–2020 [DOI] [PubMed] [Google Scholar]
- Walczak H, Iwai K, Dikic I (2012) Generation and physiological roles of linear ubiquitin chains. BMC Biol 10: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ (2001) TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412: 346–351 [DOI] [PubMed] [Google Scholar]
- Wertz IE, Dixit VM (2010) Signaling to NF-κB: regulation by ubiquitination. Cold Spring Harb Perspect Biol 2: a003350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz IE, O’Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, Ma A, Koonin EV, Dixit VM (2004) De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature 430: 694–699 [DOI] [PubMed] [Google Scholar]
- Xia T, Liang Y, Ma J, Li M, Gong M, Yu X (2011) Loss-of-function of SHARPIN causes an osteopenic phenotype in mice. Endocrine 39: 104–112 [DOI] [PubMed] [Google Scholar]
- Xu M, Skaug B, Zeng W, Chen ZJ (2009) A ubiquitin replacement strategy in human cells reveals distinct mechanisms of IKK activation by TNFα and IL-1β. Mol Cell 36: 302–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa A, Sato Y, Yamashita M, Mimura H, Yamagata A, Fukai S (2009) Crystal structure of the NEMO ubiquitin-binding domain in complex with Lys 63-linked di-ubiquitin. FEBS Lett 583: 3317–3322 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.