Abstract
The acetyltransferase Esa1 is essential in the yeast Saccharomyces cerevisiae and plays a critical role in multiple cellular processes. The most well-defined targets for Esa1 are lysine residues on histones. However, an increasing number of nonhistone proteins have recently been identified as substrates of Esa1. In this study, four genes (LYS20, LEU2, VAP1, and NAB3) were identified in a genetic screen as high-copy suppressors of the conditional temperature-sensitive lethality of an esa1 mutant. When expressed from a high-copy plasmid, each of these suppressors rescued the temperature-sensitivity of an esa1 mutant. Only NAB3 overexpression also rescued the rDNA-silencing defects of an esa1 mutant. Strengthening the connections between NAB3 and ESA1, mutants of nab3 displayed several phenotypes similar to those of esa1 mutants, including increased sensitivity to the topoisomerase I inhibitor camptothecin and defects in rDNA silencing and cell-cycle progression. In addition, nuclear localization of Nab3 was altered in the esa1 mutant. Finally, posttranslational acetylation of Nab3 was detected in vivo and found to be influenced by ESA1.
Keywords: Nrd1, Sen1, chromatin, nonhistone acetylation, KAT
Nucleosomes containing the core histones (H2A, H2B, H3, and H4) form the basic packaging unit of DNA that organizes chromatin into higher-order structures. The N-terminal tails of histones are subject to multiple covalent modifications that can influence gene expression locally at specific promoters or within large regions of chromatin. Increased histone acetylation is associated with both transcriptional activation and repression. Lysine acetyltransferases (KAT), the enzymes that catalyze the acetylation reaction on histones, have been ascribed multiple cellular functions. Recently, nonhistone targets have also been identified for many KATs, including Esa1 (Lin et al. 2009) [reviewed in Yang and Seto (2008)].
The Esa1 KAT of Saccharomyces cerevisiae is a member of the deeply conserved MYST family of acetyltransferases and is essential in yeast (Smith et al. 1998a; Clarke et al. 1999). Esa1 is the catalytic component of the NuA4 and piccolo complexes that acetylate histone H4, H2A, and its variant H2A.Z (Allard et al. 1999; Babiarz et al. 2006; Keogh et al. 2006; Millar et al. 2006). Many of the NuA4 subunits, including Esa1, are essential (Galarneau et al. 2000; Loewith et al. 2000; Eisen et al. 2001), indicating that this complex has critical cellular roles.
Esa1 has a role in regulating expression of ribosomal protein genes (Reid et al. 2000). Further, genome-wide expression analysis reveals widespread transcriptional changes in esa1 mutants (Durant and Pugh 2006), and genome-wide binding profiles show Esa1 bound to the promoters of actively transcribed genes (Robert et al. 2004). Esa1 also functions in transcriptional silencing of the rDNA and at telomeres (Clarke et al. 2006). The variety of genomic targets identified thus far suggests Esa1 activity regulates transcription at many loci, indicative of its function in multiple cellular processes.
Genetic analysis further defines Esa1’s role in diverse cellular functions. Temperature-sensitive mutants of esa1 display a G2/M cell-cycle arrest at the restrictive temperature that is dependent upon the RAD9 DNA damage checkpoint (Clarke et al. 1999) and are hypersensitive to the topoisomerase I inhibitor camptothecin (Bird et al. 2002). Esa1 localizes to double-strand breaks where it functions in repair of DNA damage (Downs et al. 2004). Together, these results suggest Esa1 activity is required for cell-cycle regulation and genomic integrity, although Esa1’s catalytic activity may not be its only essential role (Decker et al. 2008).
Suppression analyses have linked ESA1 to the deacetylase Sir2, a key silencing protein. Overexpression of Sir2 was found to suppress esa1 rDNA-silencing defects, thereby suggesting that Sir2 and Esa1 may function coordinately to silence the rDNA array (Clarke et al. 2006). Several other studies have identified additional suppressors of conditional alleles of esa1 (Biswas et al. 2008; Lin et al. 2008; Chang and Pillus 2009; Scott and Pillus 2010).
To pursue genetic interactors of ESA1, a dosage suppression screen was performed on an esa1 mutant. Of the four high-copy suppressors identified, NAB3 became a focus for two primary reasons. First, only NAB3 overexpression rescued both the temperature-sensitivity and the silencing defects of esa1 mutants. Second, NAB3 has known roles in RNA processing, and this functional connection to Esa1 may establish a novel link between two nuclear processes. Numerous studies have characterized roles for Nab3 and its binding partner Nrd1 in 3′-end processing of several classes of small noncoding RNAs [reviewed in Lykke-Andersen and Jensen (2007)]. These classes of RNAs include small-nuclear (sn) RNAs, small-nucleolar (sno) RNAs, and cryptic unstable transcripts (CUT). Nab3 and Nrd1 each recognize specific RNA sequences for 3′-end formation and transcription termination (Carroll et al. 2004).
This study reports new mutant phenotypes of nab3, revealing roles for Nab3 in rDNA silencing, the DNA damage response, and cell-cycle progression. Further, Nab3 was found to be posttranslationally modified by acetylation. This acetylation was reduced in an esa1 conditional mutant that displays reduced Esa1 acetyltransferase activity, providing evidence that Nab3 is a nonhistone substrate of Esa1 whose function may be influenced by acetylation.
Materials and Methods
Dosage suppressor screen
A URA3-marked 2μ genomic library (generously provided by P. Hieter) was transformed into two isolates of the esa1-414 strain LPY3291 in six independent experiments, yielding a total of 130,000 transformants with an approximate 70-fold coverage of the genome. Transformants were grown under permissive conditions on SC-Trp-Ura plates, and then replica-plated and incubated at 28°, 35°, and 37°. Two hundred colonies were able to grow at 35° but not 37° (this was a secondary screen used to avoid recovering wild-type ESA1). These candidates were tested for plasmid dependence by growing original transformants on 5-fluoroorotic acid (5-FOA) plates. The resulting resistant strains, which had lost the URA3-marked plasmid, were tested for temperature sensitivity at 35°. This resulted in 34 suppressor strains being classified as plasmid-dependent. Suppressing plasmids were rescued from yeast, and inserts were sequenced using T3 and T7 primers. Of the 34 plasmids, 22 were WT ESA1, 3 were unidentified, and the remaining 9 comprised six independent clones containing one of the four following genes: LYS20, NAB3, VAP1, or LEU2. Library fragments that contained multiple ORFs were dissected by subcloning to identify the gene responsible for suppression. Strategy for identification of the four suppressors is described in detail (Clarke 2001). The plasmid subclones were retransformed into LPY3291 to confirm the suppressing phenotype.
Yeast methods and strain and plasmid construction
All yeast strains and plasmids used in this study are listed in Tables 1 and 2. The silencing markers rDNA::ADE2-CAN1 (Fritze et al. 1997) and TELVR::URA3 (Renauld et al. 1993) were introduced through standard genetic crosses. All nab3-10 strains originate from YPN100 (provided by M. Swanson) (Conrad et al. 2000). Nab3 Flag-tagging was carried out by amplification of pFA6a-2FLAG-kanMX6 and transformation into LPY5 (W303-1a) using the method described (Longtine et al. 1998) to make LPY15000. All library plasmids are in the pRS202 (pLP1402) backbone. pLP1238 (NAB3 in pRS202) and pLP2018 (NAB3 in pRS426) were subcloned from pLP1419 (NAB3 library construct) using EcoRI and XhoI. pLP1310 (NAB3 in pLP271) was subcloned from pLP1419 using EcoRI. Dilution assays for growth, silencing, and drug sensitivity were performed as described (Chang and Pillus 2009) and represent 5-fold serial dilutions starting from an A600 of 1.0. Images were captured after 2–4 days of growth at the indicated temperatures.
Table 1. Yeast strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| LPY5 (W303-1a) | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | Thomas and Rothstein 1989 |
| LPY3291 | MATa his3Δ200 leu2-3,112 trp1Δ1 ura3-52 esa1Δ::HIS3 + pLP863 (esa1-414) | Clarke et al. 1999 |
| LPY4774 | W303 MATa esa1-414 | |
| LPY4909 | W303 MATα rDNA::ADE2-CAN1 | Clarke et al. 2006 |
| LPY4911 | W303 MATα esa1-414 rDNA::ADE2-CAN1 | Clarke et al. 2006 |
| LPY4917 | W303 MATα TELVR::URA3 | Clarke et al. 2006 |
| LPY4919 | W303 MATα esa1-414 TELVR::URA3 | Clarke et al. 2006 |
| LPY4979 | W303 MATα sir2Δ::HIS3 TELVR::URA3 | |
| LPY5406 | W303 MATa nab3-10 rDNA::ADE2-CAN1 | |
| LPY5407 | W303 MATa nab3-10 TELVR::URA3 | |
| LPY10622 | W303 MATa nab3-10 | |
| LPY11286 | W303 MATa nab3-10 adh4::ADE2 TELVIIL | |
| LPY11300 | W303 MATa adh4::ADE2 TELVIIL | |
| LPY12154 | W303 MATa rpd3::kanMX | Chang and Pillus 2009 |
| LPY15000 | W303 MATa NAB3-2Flag::kanMX | |
| LPY15004 | W303 MATa esa1-414 NAB3-2Flag::kanMX |
Except where noted, strains were constructed during the course of this study or are part of the standard lab collection.
Table 2. Plasmids used in this study.
| Plasmid (Alias) | Description | Source/Reference |
|---|---|---|
| pLP362 (pRS426) | Vector URA3 2µ | Sikorski and Hieter 1989 |
| pLP1402 (pRS202) | Library vector URA3 2μ | P. Hieter |
| pLP37 | SIR2 URA3 2μ | |
| pLP271 | Vector TRP1 2µ | |
| pLP796 | ESA1 URA3 2μ | Clarke et al. 2006 |
| pLP798 | ESA1 TRP1 2µ | |
| pLP863 | esa1-414 TRP1 CEN | Clarke et al. 1999 |
| pLP1238 | NAB3 URA3 2μ | |
| pLP1259 | VAP1 URA3 2μ | |
| pLP1310 | NAB3 TRP1 2µ | |
| pLP1412 | LYS20 URA3 2μ | |
| pLP1405 | LYS20-library clone URA3 2μ | |
| pLP1406 | VAP1-library clone URA3 2μ | |
| pLP1417 | LEU2-library clone URA3 2μ | |
| pLP1419 | NAB3-library clone URA3 2μ | |
| pLP2018 | NAB3 URA3 2µ | |
| pLP2054 | NRD1 URA3 2µ |
Except where noted, plasmids were constructed during the course of this study or are part of the standard lab collection. “Library clone” represents a clone obtained directly in the suppressor screen, whereas others are subclones as detailed in Clarke (2001).
Northern analysis, protein immunoblots, and immunoprecipitations
RNA was isolated using the hot acid phenol protocol as described (Collart and Oliviero 2001). Northern blotting was performed as described (Cox and Walter 1996), and results were obtained by phosphorimager (Storm, GE Healthcare). Yeast extracts were prepared by bead beating as described previously (Clarke et al. 1999), separated on SDS-polyacrylamide gels (18% for detection of histones, 8% for Sir2 and Rpd3), and transferred to nitrocellulose (0.2 μm). Primary antisera used were anti-H4K5Ac (Serotec), anti-H4K8Ac (Serotec), anti-H4K12Ac (Serotec), anti-H4K16Ac (Upstate), anti-Sir2 (Garcia and Pillus 2002), anti-Rpd3 (Rundlett et al. 1996), anti-PGK (Baum et al. 1978), anti-FLAG (Sigma-Aldrich, F3165), and anti-acetyl-lysine (Cell Signaling, #9681). Secondary antibodies conjugated to horseradish peroxidase in combination with chemiluminescence reagents were used for detection on film. FLAG-Nab3 was immunoprecipitated with anti-FLAG M2 Affinity Gel (Sigma-Aldrich, A2220), eluted in SDS sample buffer, separated on a SDS-polyacrylamide gel, and immunoblotted with either anti-FLAG or anti-acetyl lysine. All experiments were performed in triplicate or more and a representative blot was chosen for quantification. Quantification of all immunoblots was performed with ImageQuant software.
Nab3 and Sir2 immunofluorescence
Immunofluorescence was performed as described (Gotta et al. 1997; Stone et al. 2000). WT and esa1 strains were grown in YPD for four hours at either 28° or 37°. Cells were fixed by adding paraformaldehyde to the cultures at a final concentration of 3.3% at 30° for 10 min. Samples were washed twice in YPD, resuspended at 1 ml per 0.1 g of cells in 0.1 M EDTA, KOH pH 8.0, and 19 mM DTT, and then incubated at 30° for 10 min with gentle agitation. The primary antibodies used were anti-Nab3 (mouse monoclonal 2F12) (Wilson et al. 1994) and anti-Sir2 (Garcia and Pillus 2002). Texas Red-conjugated goat anti-mouse and FITC-conjugated goat anti-rabbit were used as secondary antibodies. Staining was visualized with an Applied Precision Deltavision optical sectioning deconvolution microscope.
Flow cytometry
Cell-cycle profiles were obtained by flow cytometry of propidium iodide stained cells on a FACSCalibur machine (Becton Dickinson) and analyzed with CellQuest software (Becton Dickinson). Cells were grown to an A600 of between 0.6 and 1.0, fixed in ethanol overnight, and stained with propidium iodide. Stained cells were sonicated and then analyzed by flow cytometer. For each sample, 100,000 cells were counted and analyzed.
Results
Four suppressors of the esa1 temperature-sensitive phenotype
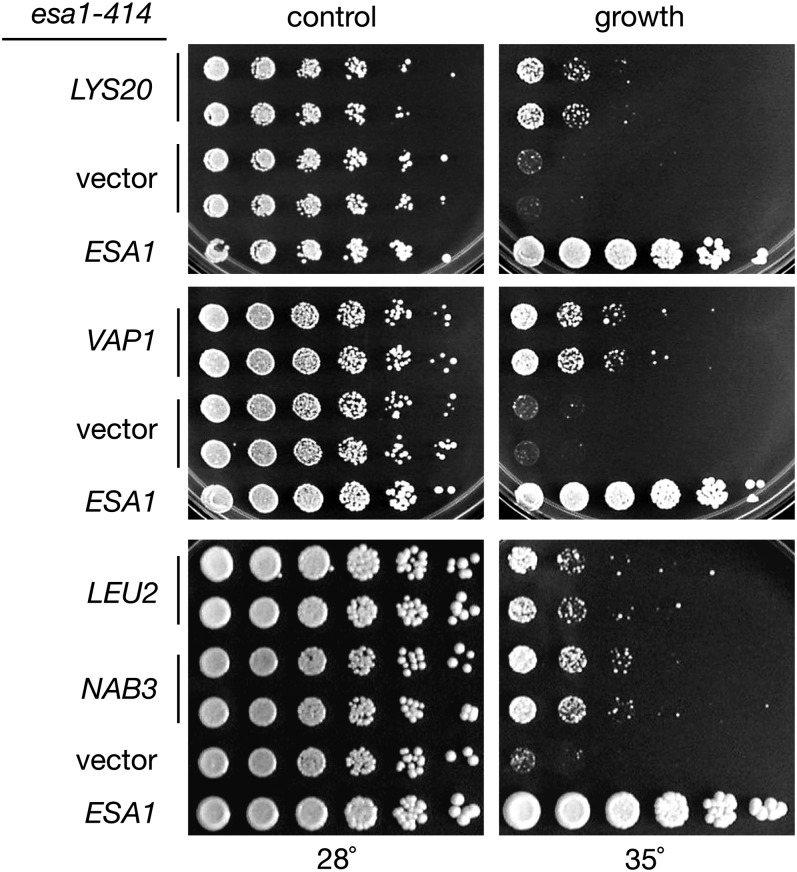
To identify genes that interact functionally with ESA1, a dosage-suppressor screen was performed utilizing a genomic 2μ plasmid library. The esa1-414 temperature-sensitive strain was transformed with the library, and transformants were tested for growth at both permissive and restrictive temperatures. Plasmids were rescued from transformants that grew at the restrictive temperature to determine the identity of suppressing genomic fragments. The results of this analysis revealed four esa1 dosage suppressors: LEU2, LYS20, NAB3, and VAP1 (Figure 1). None of these suppressors bypassed the inviable esa1Δ. When tested with other previously characterized esa1 alleles (Clarke et al. 1999), some allele-specificity was observed (supporting information, Table S1). The series of alleles was also tested for suppression of other esa1 phenotypes (see below).
Figure 1 .
The esa1 temperature-sensitive growth defect is partially suppressed by four genes expressed from 2μ plasmids. Increased gene dosage of LYS20 (pLP1405), VAP1 (pLP1406), LEU2 (pLP1417), or NAB3 (pLP1419) from a 2μ plasmid moderately suppresses the esa1-414 (LPY3291) growth defect at 35°. The esa1 growth defect at 35°, demonstrated by vector (pLP1402) transformants, is completely restored in cells transformed with an ESA1 plasmid (pLP796). All strains were plated on SC-Ura-Trp media. Suppression was not observed at higher temperatures. Multiple independent transformants were tested to examine any variability between transformants.
LEU2 and LYS20 are nonessential genes required for the biosynthesis of leucine and lysine [reviewed in Kohlhaw (2003) and Xu et al. (2006), respectively]. VAP1 is also involved in amino acid metabolism, encoding a transporter of several amino acids, including tyrosine, tryptophan, valine, and leucine (Schmidt et al. 1994). Characterization of LYS20 as a suppressor of esa1 revealed additional roles for this metabolic gene in DNA damage repair (Scott and Pillus 2010). NAB3, as noted, is an essential gene critical for 3′-end processing of nonpolyadenylated transcripts [reviewed in Lykke-Andersen and Jensen (2007)].
Increased dosage of NAB3 suppresses multiple esa1 mutant phenotypes
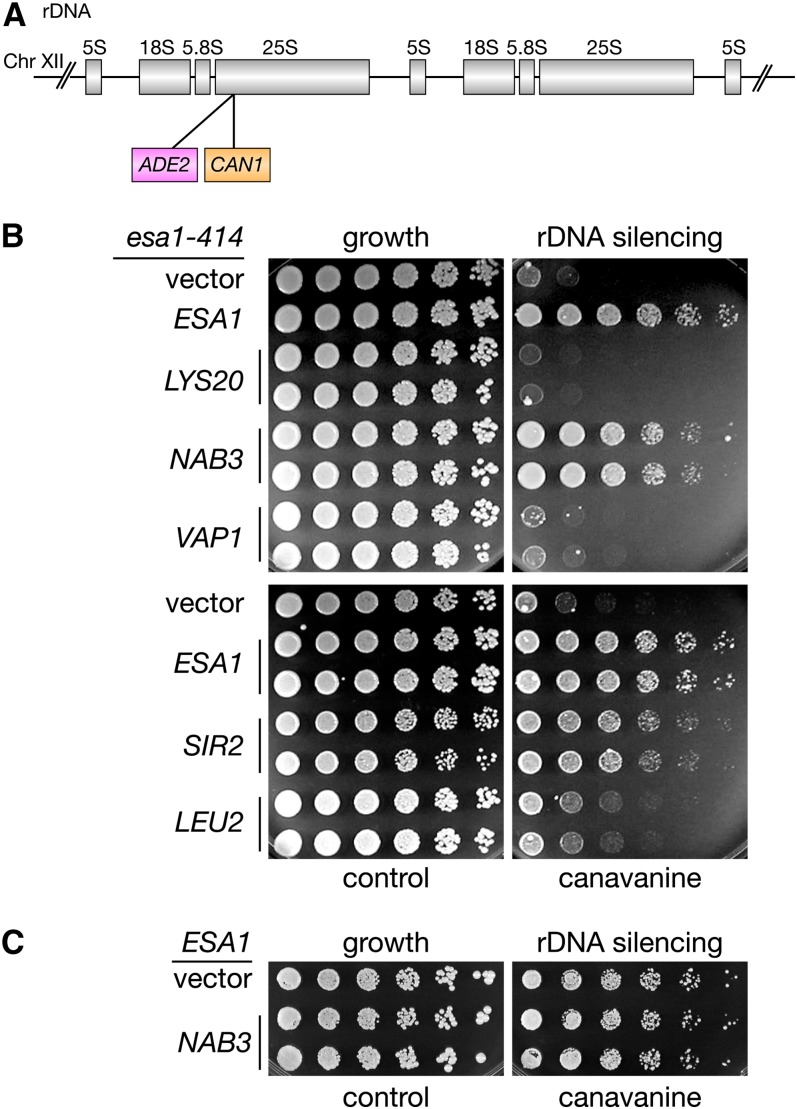
To understand the connection between the suppressors and Esa1 function, overexpression of the four genes was tested for suppression of esa1 mutant defects other than temperature sensitivity. One phenotype of esa1 mutants is a strong rDNA-silencing defect and a slight increase in mitotic rDNA recombination (Clarke et al. 2006). Previously, it was shown that increased gene dosage of SIR2 suppresses the esa1 rDNA-silencing defect (Clarke et al. 2006). Using an esa1 strain with the ADE2-CAN1 dual reporter integrated at a single 25S rDNA repeat (Fritze et al. 1997) (Figure 2A), the suppressors were tested for their effect on silencing of the rDNA locus. Only increased dosage of NAB3 robustly suppressed the esa1 rDNA-silencing defect, restoring silencing to near wild-type levels (Figure 2B). By contrast, LYS20 slightly exacerbated esa1’s silencing defect, whereas LEU2 and VAP1 had little to no effect (Figure 2B). Unlike increased gene dosage of SIR2 in a wild-type strain (Smith et al. 1998b), NAB3 did not enhance wild-type rDNA silencing (Figure 2C). None of the suppressors had significant effects on rDNA recombination. As there appeared to be a link between NAB3 and ESA1 for both silencing and growth, we chose to characterize NAB3 in greater detail.
Figure 2 .
The esa1 rDNA silencing defect is suppressed by increased gene dosage of NAB3. (A) Diagram and location of rDNA::ADE2-CAN1 silencing marker within the rDNA array on chromosome XII. (B) The esa1 rDNA silencing defect (vector) is restored in cells transformed with an ESA1 plasmid, and by NAB3. Increased dosage of LYS20, LEU2, or VAP1 does not rescue esa1’s rDNA silencing defect. An esa1 strain with the 25S rDNA::ADE2-CAN1 reporter (LPY4911) was transformed with vector (pLP1402), ESA1 (pLP796), LYS20 (pLP1412), NAB3 (pLP1238), VAP1 (pLP1259), SIR2 (pLP37), or LEU2 (pLP1417). To test for rDNA silencing defects, strains were plated on SC-Ade-Arg-Ura (growth) with and without 32 µg/ml canavanine (rDNA silencing) at 33°. (C) NAB3 overexpression (pLP1238) has no effect on rDNA silencing of a WT strain (LPY4909).
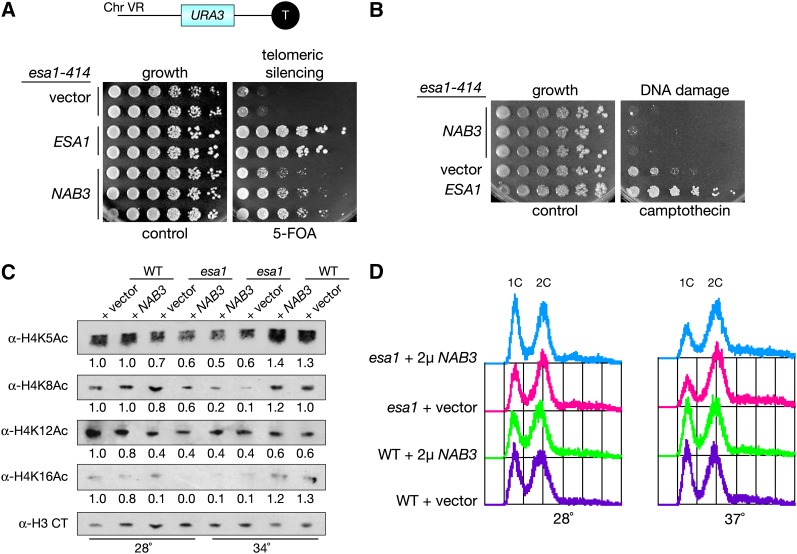
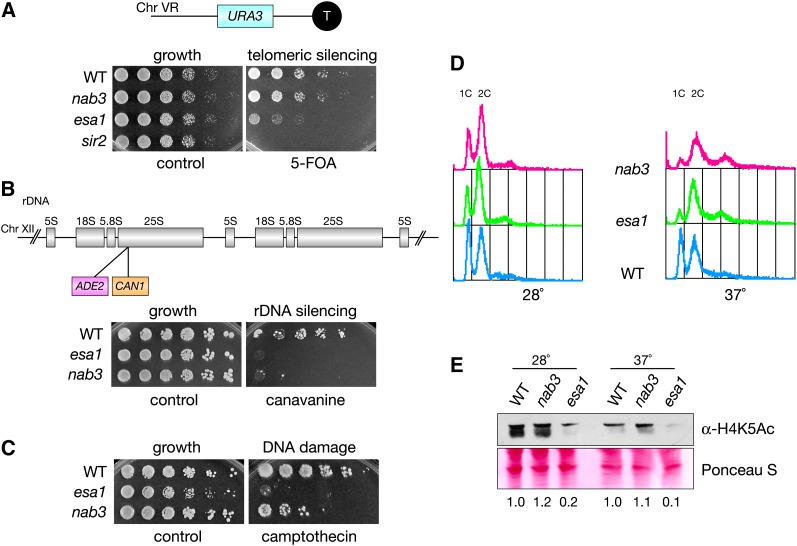
In addition to their rDNA-silencing defects, esa1 mutants are defective in telomeric silencing (Clarke et al. 2006) (Figure 3A), as shown by diminished growth on 5-FOA when using a URA3 reporter gene on the right arm of chromosome V (TELVR) (Renauld et al. 1993). Increased dosage of NAB3 in esa1 mutants allowed for increased growth on 5-FOA, thereby rescuing the sensitivity shown in the esa1 mutant (Figure 3A). Recent studies have shown that readout of this reporter-based assay for some genes may reflect changes in nucleotide metabolism instead of telomeric-silencing defects (Rossmann et al. 2011; Takahashi et al. 2011). Thus, based on these new studies, rescue of esa1’s 5-FOA sensitivity by NAB3 in strains carrying the URA3 telomeric reporter gene can be interpreted as the ability of NAB3 overexpression to suppress telomeric-silencing defects or nucleotide metabolism changes in an esa1 mutant. Because ESA1 has no known defects in HM silencing or mating efficiency (Clarke et al. 2006), NAB3 dosage was not tested for effects on mating efficiency.
Figure 3 .
Overexpression of NAB3 affects multiple esa1 mutant phenotypes. (A) Top: Diagram of TELVR::URA3 telomeric silencing marker on the right arm of chromosome V. Bottom: Increased gene dosage of NAB3 suppresses the esa1 5-FOA sensitivity in this assay. An esa1 strain with the TELVR::URA3 reporter (LPY4919) was transformed with vector (pLP271), ESA1 (pLP798), or NAB3 (pLP1310), and plated on SC-Trp (growth) with and without 5-FOA (telomeric silencing) at 33°. (B) Increased gene dosage of NAB3 exacerbates esa1’s sensitivity to the DNA damaging agent camptothecin. An esa1 strain (LPY4774) was transformed with vector (pLP326), ESA1 (pLP796), or NAB3 (pLP2018), and plated on SC-Ura with DMSO (growth) and 20 µg/ml camptothecin (DNA damage). (C) Overexpression of NAB3 does not increase global acetylation levels of H4K5, H4K8, H4K12, or H4K16 in esa1 mutants. Whole-cell extracts were made from wild-type (LPY5) and esa1 (LPY4774) strains with vector (pLP362) or 2μ NAB3 (pLP2018) grown in SC-Ura media at both permissive (28°) and elevated (34°) temperatures. These were immunoblotted for amounts of isoform-specific H4 acetylation and total H3. An H3 reprobe was performed for each individual H4 acetylation blot. Quantification data shown are normalized for H3 loading. (D) Overexpression of NAB3 does not influence esa1’s G2/M cell-cycle block. The same strains as in (C) were grown at 28° and shifted to 37° for 4 hr before fixing and staining with propidium iodide. Cell-cycle profiles were analyzed by flow cytometry.
Another phenotype of esa1 mutants is sensitivity to DNA damage induced by camptothecin, a topoisomerase I inhibitor that triggers double-strand breaks (Bird et al. 2002). NAB3 overexpression was tested for its ability to suppress this esa1 mutant defect in the DNA damage response and was found to exacerbate esa1’s camptothecin sensitivity (Figure 3B). This result is in contrast to NAB3-mediated suppression of esa1’s silencing defects, highlighting a difference between Nab3 and Esa1’s functions in transcriptional silencing and DNA damage repair.
At a molecular level, global H4 acetylation is dramatically reduced in esa1 mutants when grown at restrictive temperatures (Clarke et al. 1999). To determine whether increased dosage of NAB3 restores wild-type levels of histone acetylation to esa1 mutants, a series of protein immunoblots with isoform-specific antibodies was performed to define the global acetylation state in esa1 strains overexpressing NAB3. All the histone H4 lysine residues that Esa1 is known to acetylate (K5, K8, K12, and K16) (Clarke et al. 1999) were tested in these experiments (Figure 3C). Total histone levels were determined by probing with a control antibody specific to the C-terminus of histone H3. This series of immunoblots shows that increased dosage of NAB3 in esa1 strains did not restore H4 acetylation. Therefore, NAB3 overexpression does not rescue esa1 mutants by restoring global acetylation defects at substrate residues in the H4 N-terminal tail.
A distinct potential mechanism for NAB3 suppression is through Esa1’s role in the cell cycle. Since Esa1 is required for cell-cycle progression through G2/M, cell-cycle profiles of esa1 mutant strains with increased dosage of NAB3 were examined by flow cytometry to distinguish cellular DNA content before (1C) and after (2C) replication. The esa1 mutants at restrictive temperature have a well-defined G2/M cell-cycle block, visualized as a decrease in the 1C peak and an accumulation of the 2C peak (Clarke et al. 1999). With NAB3 overexpression, no change in the esa1 cell-cycle profile was observed (Figure 3D), indicating that NAB3 overexpression does not bypass the G2/M cell-cycle block of esa1 mutants. Thus, increased dosage of NAB3 suppresses a defined subset of esa1 mutant phenotypes, which includes silencing defects and temperature sensitivity.
Nab3 does not affect protein or transcript levels of histone-modifying enzymes
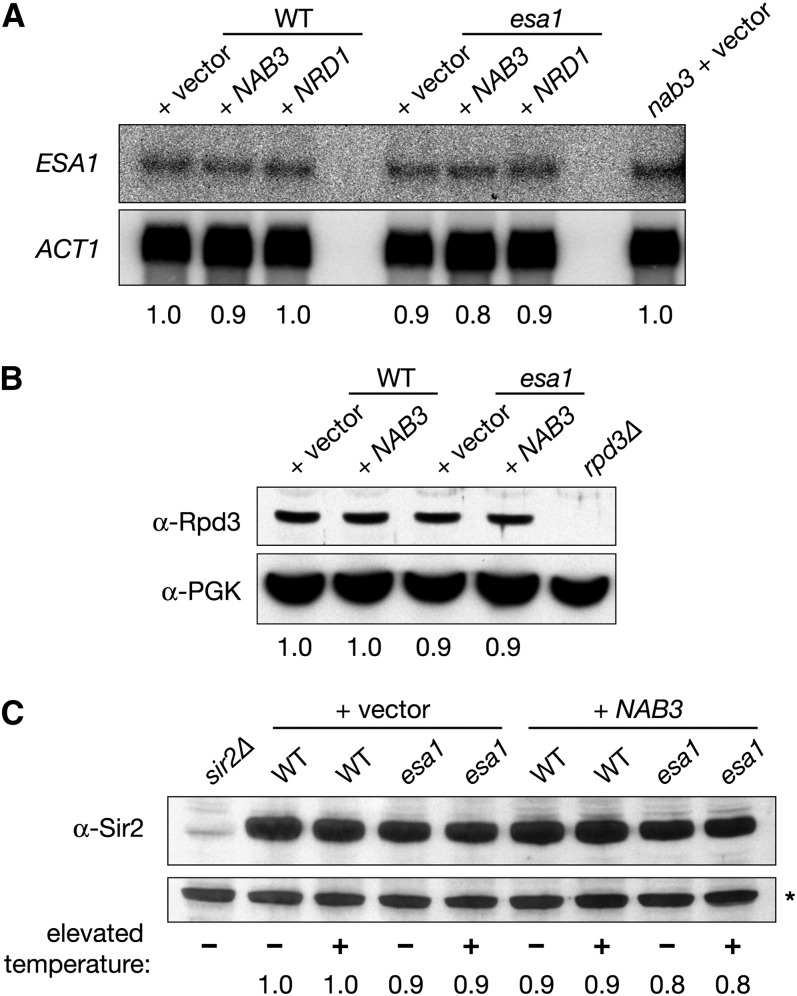
In addition to their function for termination of noncoding RNAs, there is evidence that Nab3 and its partner Nrd1 participate in 3′-end formation of protein-coding transcripts (Sugimoto et al. 1995; Arigo et al. 2006a; Darby et al. 2012). We considered the possibility that Nab3 might bind to ESA1 mRNA to direct its 3′-end formation. Nab3 binding sites have the simple UCUU consensus sequence (Carroll et al. 2004) that is found at several positions within the ESA1 transcript. Northern blotting was performed to determine whether there were any NAB3-dependent changes in the ESA1 transcript. NAB3 is an essential gene (Wilson et al. 1994) and, thus, the temperature-sensitive nab3-10 mutant was used in this study. The nab3-10 allele was described previously and specifies a single F371L amino acid substitution in its RNA-recognition motif (RRM) domain (Conrad et al. 2000). When ESA1 transcripts were examined in the nab3-10 mutant (Figure 4A), there were no detectable changes in either transcript levels or migration. Transcript levels of ESA1 were also found to be constant with or without increased dosage of NAB3 (Figure 4A). Increased dosage of NRD1, which encodes a binding partner of Nab3, also failed to influence ESA1 mRNA. In conclusion, NAB3 does not affect the transcriptional regulation of ESA1 itself.
Figure 4 .
NAB3 overexpression does not change transcript levels of ESA1 or protein levels of Rpd3 and Sir2. (A) NAB3 overexpression does not alter ESA1 mRNA levels. Total RNA was isolated from both WT (LPY5) and esa1 (LPY4774) mutant strains grown at an elevated temperature (35°) with vector control (pLP362), NAB3 overexpression (pLP2018), or NRD1 overexpression (pLP2054). Northern analysis was performed with an ESA1-specific probe, and results were obtained by phosphorimager scan. (B) Overexpression of NAB3 does not influence Rpd3 protein levels. Whole-cell lysates from WT and esa1 strains grown at an elevated temperature (35°) with vector control or NAB3 overexpression [same strains as in (A)] were examined by immunoblot with anti-Rpd3. An rpd3Δ strain (LPY12154) transformed with vector (pLP362) was used as a negative control, and anti-PGK1 (phosphoglycerate kinase) was used to determine equal loading between samples. (C) Overexpression of NAB3 does not influence Sir2 protein levels. Whole-cell lysates were made from WT (LPY5) and esa1 (LPY4774) strains grown at an elevated temperature (37°) with vector control (pLP1402) or NAB3 overexpression (pLP1238) and immunoblotted with anti-Sir2. Extract from a sir2Δ strain was used as a negative control. A nonspecific band (*) detected by anti-Sir2 was used to determine equal loading between samples.
We considered the possibility that NAB3 affects transcription of a histone deacetylase (HDAC) that acts in opposition to Esa1. Transcriptional downregulation of an HDAC could compensate for the lack of functional Esa1 and restore the imbalance of acetylation in the cell. For example, deletion of the histone deacetylase gene RPD3 suppresses the temperature-sensitivity and silencing defects of an esa1 mutant (Chang and Pillus 2009). To test whether NAB3 suppression of esa1 is mediated through changes in Rpd3 levels, its protein levels were examined by immunoblot. Comparing Rpd3 levels between wild type and esa1 strains with and without increased dosage of NAB3 revealed no NAB3-dependent changes (Figure 4B). Another HDAC candidate of interest is Sir2, an HDAC critical for establishment and maintenance of silent chromatin [reviewed in Rusche et al. (2003)]. Similar to NAB3 overexpression, SIR2 overexpression has been shown to rescue rDNA silencing in an esa1 mutant (Clarke et al. 2006). Sir2 levels were determined in wild-type and esa1 mutant strains overexpressing NAB3 by immunoblot, and no NAB3-dependent differences were observed (Figure 4C). Therefore, NAB3 overexpression does not alter expression of either Rpd3 or Sir2, demonstrating that suppression is not mediated through transcriptional regulation of either HDAC.
nab3 mutants share phenotypes with esa1 mutants
To characterize further the role of NAB3 in relation to ESA1, nab3 mutants were examined for established phenotypes of esa1 mutants. Since NAB3 overexpression rescued the esa1 telomeric- and rDNA-silencing defects (Figures 2B and 3A), it was possible that nab3 mutants might be defective in silencing. Telomeric silencing was tested using the same URA3 reporter assay as before (Figure 3A), and it revealed that nab3 mutants display growth on 5-FOA comparable to wild-type strains (Figure 5A). Use of an independent ADE2 color-based telomeric-silencing assay also showed no defects for nab3 mutants (Figure S1). Combined with the lack of defects observed for nab3 mutants in both our assays (Figure 5A), the earlier observation that NAB3 overexpression rescued esa1’s 5-FOA sensitivity (Figure 3A) likely results through an indirect mechanism. In contrast, when assayed for rDNA-silencing defects, nab3 mutants displayed a strong defect, similar to that observed in esa1 (Figure 5B). Together, these data suggest that Nab3 functions directly in rDNA silencing but not telomeric silencing.
Figure 5 .
nab3 mutants display defects similar to esa1 mutants. (A) nab3 mutants display no defect in the telomeric 5-FOAS assay. WT (LPY4917), nab3-10 (LPY5407), esa1 (LPY4919), and sir2 (LPY4979) strains with a TELVR::URA3 reporter were plated on SC and 5-FOA. (B) nab3 mutants have an rDNA silencing defect. WT (LPY4909), esa1 (LPY4911), and nab3 (LPY5406) strains with the 25S rDNA::ADE2-CAN1 reporter were assayed for rDNA silencing defects on SC-Ade-Arg with and without 16 μg/ml canavanine. (C) The nab3 mutant is sensitive to the DNA-damaging agent camptothecin. WT (LPY5), esa1 (LPY4774), and nab3 (LPY10622) were plated on DMSO (control) and camptothecin (40 μg/ml) to test for drug sensitivity. (D) nab3 mutants display a G2/M block when grown at an elevated temperature. The same strains as in (C) were fixed and stained with propidium iodide to analyze cell-cycle profiles after being grown at 28° and shifted to 37° for 4 hr. (E) nab3 mutants have wild-type levels of global H4K5 acetylation. The same strains as in (C) and (D) were grown in YPD at 28° and shifted to 37° for 2 hr before whole-cell extract preparation. Samples were immunoblotted to detect global H4K5 acetylation levels. Compared with the H4K5 acetylation levels in esa1 mutants shown in Figure 3C, a more severe temperature challenge is shown here, accounting for the greater magnitude in decreased acetylation.
NAB3 overexpression did not suppress the DNA damage and cell-cycle phenotypes of esa1 mutants (Figure 3, B and D). However, when nab3 mutants were examined for defects in DNA damage repair and cell-cycle progression, the results revealed a role for NAB3 in these processes. As seen in Figure 5C, nab3 mutants are sensitive to the topoisomerase I inhibitor camptothecin, although less so than esa1. Cell-cycle profiles of nab3 mutants also showed a G2/M block resembling that of esa1 mutants (Figure 5D). In addition to the defective rDNA silencing of nab3, the identification of these phenotypes for nab3 mutants reveals a more extensive functional overlap with esa1 mutants.
We earlier considered the possibility that a molecular link for Nab3 and Esa1 functions would be that Nab3 influences histone acetylation (Figure 3C). When tested for changes in acetylation of H4K5, the primary in vivo target of Esa1 (Clarke et al. 1999), global acetylation in nab3 mutants was maintained at wild-type levels (Figure 5E). Therefore, NAB3 does not directly influence the global histone acetylation activity of Esa1’s primary target.
Localization and posttranslational acetylation of Nab3 are altered in esa1 mutants
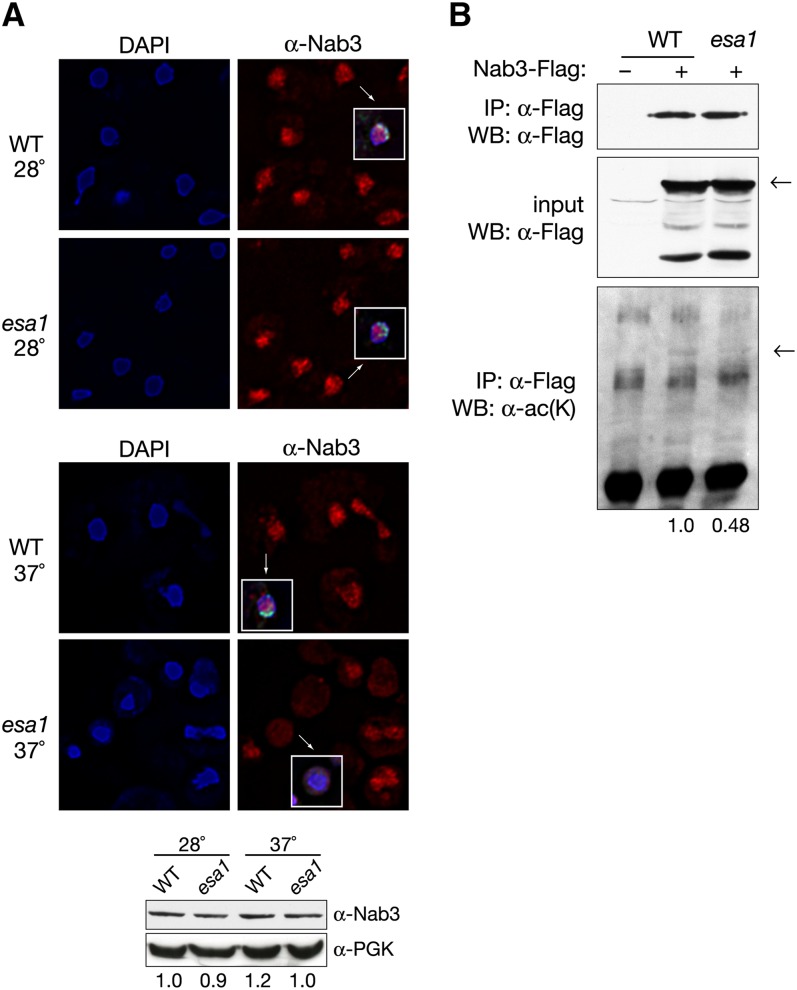
The nucleolus is a key compartment for RNA processing in the nucleus. Ultrastructural analysis has shown esa1 mutants to have aberrant nucleoli (Clarke et al. 1999), and esa1 mutants display strong rDNA-silencing defects and rDNA chromatin structure defects (Clarke et al. 2006). Because of these connections of Esa1 to nucleolar function and Nab3′s influence on rDNA silencing (Figure 5B), Nab3 localization was visualized in esa1 mutants. Immunofluorescence was performed using an antibody directed against Nab3 in wild-type and esa1 strains. In addition, Sir2 staining was used to demarcate the nucleolus.
Nab3 localization has been previously described as dispersed throughout the nucleus but distinct from nucleolar structure proteins (Wilson et al. 1994) (Figure 6A, top). At permissive temperatures, Nab3 localization appeared normal in both wild-type and esa1 cells. However, at restrictive temperature, Nab3 localization in esa1 became diffuse and no longer confined to the nucleus as defined by DAPI staining (Figure 6A, middle), indicating that Nab3 localization is altered in the esa1 mutant. Sir2 staining was also affected in the esa1 mutant and no longer found in discrete nucleolar and telomeric foci, although Sir2 protein expression appeared essentially normal at elevated temperature (Figure 4C). Nab3 protein levels were also found to be equal by immunoblot between wild-type and esa1 cells at both permissive and restrictive temperatures (Figure 6A, bottom).
Figure 6 .
Nab3 localization and acetylation is altered in esa1 cells. (A) Nab3 localization is aberrant in the esa1 mutant. Top: At a permissive temperature (28°), Nab3 staining in wild type (LPY4909) and esa1 (LPY4911) cells appears as punctate nuclear foci interspersed with diffuse nuclear staining. Sir2 localization demarcates the nucleolus in a crescent shape (inset, green) and is normal. At a restrictive temperature (37°), Nab3 staining is diffuse in the esa1 mutant but appears normal in the wild-type strain. No Sir2 foci are observed in the esa1 mutant. Bottom: WT and esa1 strains used above were grown at permissive and elevated temperatures and used for immunoblots to detect total Nab3 levels using anti-Nab3. Anti-PGK1 (phosphoglycerate kinase) was used as a loading control. (B) Nab3 is acetylated in vivo in an ESA1-dependent manner. To detect posttranslational acetylation of Nab3, a WT (LPY15000) and esa1 (LPY15004) strain containing a chromosomal FLAG-tagged version of Nab3 were grown at an elevated temperature (37°) and used in an anti-FLAG immunoprecipitation followed by an immunoblot with anti-acetyl lysine. Decreased levels of Nab3 acetylation are observed in the esa1 mutant. Quantification of films from independent experiments shows a 48% decrease in Nab3 acetylation in esa1 compared with wild-type. An untagged WT strain (LPY5) is used as a negative control. Nab3-FLAG levels are not themselves altered in the esa1 mutant, as demonstrated by control immunoblotting of immunoprecipitations and inputs with anti-FLAG.
Because WT levels of Nab3 were observed in esa1 mutants, whereas simple overexpression suppressed esa1 phenotypes, we considered the possibility that in the mutants, Nab3 protein differs not quantitatively but qualitatively. One such qualitative difference could be at the level of its posttranslational modification. We tested the idea that Nab3 might itself be an in vivo substrate for Esa1, a possibility first raised by a proteomics survey suggesting that Esa1 could acetylate Nab3 in vitro (Lin et al. 2009). To examine whether this modification occurs in vivo, an antibody that recognizes proteins with acetylated lysines was utilized. Immunoprecipitation of Nab3 followed by immunoblot detection with anti-acetyl-lysine revealed Nab3 to be acetylated in vivo (Figure 6B). To test whether Nab3 is a substrate for Esa1 acetylation, Nab3 acetylation levels were evaluated in an esa1 mutant. Since ESA1 is essential, the temperature-sensitive esa1 mutant was grown at nonpermissive temperatures and samples were prepared. As expected if Esa1 acetylates Nab3 in vivo, a decrease in acetylated-Nab3 was observed in the esa1 mutant. Quantification of the anti-acetyl-lysine immunoblot shows that Nab3 acetylation was reduced in the esa1 mutant to 48% of the level observed in the wild-type strain. Thus, it appears that a fraction of the acetylation of Nab3 was ESA1-dependent, although our data did not distinguish whether this acetylation was by Esa1 on Nab3 directly as a target or indirectly through another acetyltransferase influenced by Esa1.
Discussion
Genetic suppression has provided a valuable tool for expanding the understanding of Esa1’s nuclear functions (Biswas et al. 2008; Lin et al. 2008; Chang and Pillus 2009; Scott and Pillus 2010) and, in this case, its interactions with the RNA binding protein Nab3. Increased dosage of NAB3 was found to suppress a subset of esa1 mutant phenotypes, including temperature sensitivity and silencing defects. In addition, nab3 mutants shared overlapping phenotypes with esa1 mutants, displaying defects in rDNA silencing, cell-cycle progression, and the DNA damage response. Further strengthening these genetic interactions, nuclear localization and posttranslational acetylation of Nab3 were both altered in the esa1 mutant.
Nab3 is found in a complex with the RNA binding protein Nrd1 and the Sen1 helicase. This Nab3 complex ensures proper termination and 3′-end formation of many nonpolyadenylated transcripts, including snRNAs, snoRNAs, and CUTs (Steinmetz et al. 2001; Arigo et al. 2006b; Thiebaut et al. 2006). In addition, Nab3 physically associates with the nuclear exosome for processing and degradation of these transcripts (Vasiljeva and Buratowski 2006). Nab3 and Nrd1 form a heterodimer (Carroll et al. 2007), and each protein has a different consensus RNA recognition sequence (Carroll et al. 2004). Domain analysis suggests that both proteins bind RNA transcripts, whereas Nrd1 also physically associates with the C-terminal domain of Pol II (Conrad et al. 2000). In accordance with these tightly linked functions of Nab3 and Nrd1, we found that overexpression of NRD1 also suppresses some esa1 mutant phenotypes (Figure S2). Because genetic suppression by NRD1 was less dramatic than that by NAB3, our focus in this study was on NAB3’s genetic interaction with ESA1, but our observations with NRD1 support the idea that suppression is mediated by Nab3 in the context of the Nab3-Nrd1-Sen1 complex, and not via an independent role of Nab3 alone.
Because the ESA1 transcript was unchanged in the nab3-10 mutant (Figure 4A), this implies that the Nab3-Nrd1-Sen1 complex does not direct 3′-end termination of the ESA1 transcript. It should be noted that our study was restricted to this loss-of-function nab3-10 mutation. Thus, considering the genetic limitations of studying essential genes such as NAB3, we cannot fully eliminate the possibility that the Nab3 complex processes the ESA1 transcript, as we have not studied multiple mutant alleles of NAB3. However, we consider our in vivo data showing that Nab3 acetylation is influenced by Esa1 either directly or indirectly (Figure 6B) to provide a more likely explanation for the dosage suppression observed between ESA1 and NAB3. Consistent with these data, one potential model for the suppression is that Esa1 acetylation of Nab3 influences its function such that the reduced Nab3 acetylation in esa1 mutants results in its reduced cell viability and defects in rDNA silencing. Thus, suppression of these defects is obtained in the esa1 mutant by overexpressing NAB3 to compensate for the decreased pools of acetylated Nab3.
In S. cerevisiae, the rDNA is a repetitive array in the genome that is mainly transcribed by Pol I and Pol III. Reporter genes that are transcribed by Pol II and inserted in the array are known to undergo Sir2-mediated transcriptional silencing. An endogenous Pol II transcript has been detected in the “nontranscribed” spacer region (NTS1) of the rDNA. This transcript is a CUT that is processed by the Nab3 complex and degraded by the exosome (Houseley et al. 2007; Vasiljeva et al. 2008). In addition to uncovering an rDNA-silencing defect for nab3 mutants (Figure 5B), we observed that overexpression of NAB3 rescued the rDNA-silencing defects of esa1 mutants (Figure 2). Esa1 binding is enriched at the rDNA, and histone acetylation at the rDNA is reduced in the esa1 mutant (Clarke et al. 2006). Although Nab3 does not appear nucleolar by immunofluorescence (Wilson et al. 1994) (Figure 6A), a recent study found that Nab3 localizes to the rDNA via chromatin immunoprecipitation (Lepore and Lafontaine 2011). Thus, one possibility is that Nab3 recruitment to the CUTs within the rDNA is regulated by its acetylation status through Esa1 activity. Future studies will establish how Esa1 functions with the Nab3-Nrd1 complex in contributing to transcriptional silencing at the rDNA.
The number of nonhistone proteins known to be acetylated by Esa1 and the MYST family of KATs has expanded in recent years. Several schools of thought exist about the function of this posttranslational modification. In parallel with the models for histone acetylation, acetylation of nonhistone proteins may change the activity of these proteins or may serve as a recruitment platform for physical binding of other proteins [reviewed in Sapountzi and Côté (2011)]. Our finding that Nab3 is acetylated in vivo raises several possibilities regarding the function of this posttranslational modification. Whereas Nab3 acetylation is reduced in an esa1 mutant, overall levels of Nab3 remain constant (Figure 6). Therefore, it is unlikely that acetylation affects Nab3 stability but, rather, that it influences its activity or function. Knowing that Nab3 is aberrantly localized in the esa1 mutant, one possible scenario is that acetylation of Nab3 by Esa1 promotes proper Nab3 nuclear localization.
In contrast to NAB3, the other three suppressors identified in our dosage-suppression screen (LEU2, LYS20, and VAP1) are all involved in amino acid metabolism. A separate study defined the connections between LYS20 and ESA1 through DNA repair that could be distinguished from Lys20’s role in amino acid biosynthesis, potentially through a noncanonical role in acetylation (Scott and Pillus 2010). Recent findings report the prevalence of lysine acetylation as a posttranslational modification in the regulation of metabolic proteins in mammals (Zhao et al. 2010). In light of these studies and ours, it is possible that Esa1 acetylates the protein products of the genes we identified as dosage suppressors. Only Nab3, and not the other suppressors, was identified as a substrate in the in vitro proteomics study (Lin et al. 2009). However, a number of other metabolic enzymes were found, including the gluconeogenic enzyme Pck1 that is reciprocally deacetylated by Sir2, providing a link to our earlier suppression studies between ESA1 and SIR2 (Clarke et al. 2006). One potential explanation for our current findings of dosage suppression of esa1 by LEU2 and VAP1 is that Leu2 and Vap1 are acetylated by Esa1 in vivo. Future studies to determine in vivo Esa1 targets of nonhistone proteins will shed light on additional substrates and their functions.
Although it has been assumed that Esa1’s catalytic activity is its essential activity, it is unclear exactly why esa1Δ strains are inviable. One recent study found that an ESA1 strain bearing a mutation in a residue important for catalysis retained viability, proposing that there may be more to the essential nature of Esa1 than its histone acetyltransferase activity (Decker et al. 2008). Given that our screen highlights a strong genetic interaction between ESA1 and the essential gene NAB3, along with several genes encoding metabolic proteins (LEU2, VAP1, LYS20), one of Esa1’s essential functions may be the recognition and acetylation of important nonhistone substrates.
Suppressor analysis is a widely used strategy that facilitates the identification of functional relationships between different proteins. A recent investigation of hundreds of dosage suppressors in yeast revealed that dosage suppression provides functional links between two genes (Magtanong et al. 2011). In addition, dosage suppression can identify unique interactions that are not discovered through other types of genome-wide studies, such as protein-protein and synthetic sickness interactions. In our study, genetic suppression has provided an effective platform for identifying and characterizing potential new substrates for an enzyme primarily studied as an acetyltransferase targeting histones.
Supplementary Material
Acknowledgments
We thank M. Swanson for the nab3-10 strain and the Nab3 antibody, and E. Steinmetz, D. Brow, and J. Corden for nrd1 strains and iNRD1 plasmids. M. Grunstein provided the Rpd3 antibody, and T. Johnson, the anti-acetyl lysine antibody. We thank P. Hieter for the 2μ genomic library; J. DuRose and M. Niwa for assistance with northern analysis; M. Busse for help with Sir2 westerns; J. Feramisco and S. McMullen for assistance with deconvolution microscopy; A. Castillo and M. Winey for help with the dosage suppressor screen; the David Lab for access to its FACS instrument; and L. Clark, R. Garza, R. Otsuka, and A. L. Torres Machorro for technical assistance. We thank members of the Pillus lab for helpful advice throughout the course of this study, and A. L. Torres Machorro for critical reading of the manuscript. This work was supported by National Institutes of Health grants GM-56469, GM-90177, and T32-GM-007240.
Footnotes
Communicating editor: J. Rine
Literature Cited
- Allard S., Utley R. T., Savard J., Clarke A., Grant P., et al. , 1999. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 18: 5108–5119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arigo J. T., Carroll K. L., Ames J. M., Corden J. L., 2006a Regulation of yeast NRD1 expression by premature transcription termination. Mol. Cell 21: 641–651 [DOI] [PubMed] [Google Scholar]
- Arigo J. T., Eyler D. E., Carroll K. L., Corden J. L., 2006b Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol. Cell 23: 841–851 [DOI] [PubMed] [Google Scholar]
- Babiarz J. E., Halley J. E., Rine J., 2006. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes Dev. 20: 700–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum P., Thorner J., Honig L., 1978. Identification of tubulin from the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 75: 4962–4966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. W., Yu D. Y., Pray-Grant M. G., Qiu Q., Harmon K. E., et al. , 2002. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419: 411–415 [DOI] [PubMed] [Google Scholar]
- Biswas D., Takahata S., Stillman D. J., 2008. Different genetic functions for the Rpd3(L) and Rpd3(S) complexes suggest competition between NuA4 and Rpd3(S). Mol. Cell. Biol. 28: 4445–4458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll K. L., Pradhan D. A., Granek J. A., Clarke N. D., Corden J. L., 2004. Identification of cis elements directing termination of yeast nonpolyadenylated snoRNA transcripts. Mol. Cell. Biol. 24: 6241–6252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll K. L., Ghirlando R., Ames J. M., Corden J. L., 2007. Interaction of yeast RNA-binding proteins Nrd1 and Nab3 with RNA polymerase II terminator elements. RNA 13: 361–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. S., Pillus L., 2009. Collaboration between the essential Esa1 acetyltransferase and the Rpd3 deacetylase is mediated by H4K12 histone acetylation in Saccharomyces cerevisiae. Genetics 183: 149–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke A., 2001. Genetic and biochemical characterization of ESA1: an essential histone acetyltransferase involved in cell-cycle progression and transcriptional silencing, pp. 402 in Molecular, Cellular and Developmental Biology. University of Colorado, Boulder, CO [Google Scholar]
- Clarke A. S., Lowell J. E., Jacobson S. J., Pillus L., 1999. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol. Cell. Biol. 19: 2515–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke A. S., Samal E., Pillus L., 2006. Distinct roles for the essential MYST family HAT Esa1p in transcriptional silencing. Mol. Biol. Cell 17: 1744–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart M. A., Oliviero S., 2001. Preparation of yeast RNA. Curr. Protoc. Mol. Biol. Chapter 13: Unit13.12. [DOI] [PubMed] [Google Scholar]
- Conrad N. K., Wilson S. M., Steinmetz E. J., Patturajan M., Brow D. A., et al. , 2000. A yeast heterogeneous nuclear ribonucleoprotein complex associated with RNA polymerase II. Genetics 154: 557–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J. S., Walter P., 1996. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87: 391–404 [DOI] [PubMed] [Google Scholar]
- Darby M. M., Serebreni L., Pan X., Boeke J. D., Corden J. L., 2012. The Saccharomyces cerevisiae Nrd1-Nab3 transcription termination pathway acts in opposition to Ras signaling and mediates response to nutrient depletion. Mol. Cell. Biol. 32: 1762–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker P. V., Yu D. Y., Iizuka M., Qiu Q., Smith M. M., 2008. Catalytic-site mutations in the MYST family histone acetyltransferase Esa1. Genetics 178: 1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs J. A., Allard S., Jobin-Robitaille O., Javaheri A., Auger A., et al. , 2004. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol. Cell 16: 979–990 [DOI] [PubMed] [Google Scholar]
- Durant M., Pugh B. F., 2006. Genome-wide relationships between TAF1 and histone acetyltransferases in Saccharomyces cerevisiae. Mol. Cell. Biol. 26: 2791–2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen A., Utley R. T., Nourani A., Allard S., Schmidt P., et al. , 2001. The yeast NuA4 and Drosophila MSL complexes contain homologous subunits important for transcription regulation. J. Biol. Chem. 276: 3484–3491 [DOI] [PubMed] [Google Scholar]
- Fritze C. E., Verschueren K., Strich R., Easton Esposito R., 1997. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 16: 6495–6509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galarneau L., Nourani A., Boudreault A. A., Zhang Y., Heliot L., et al. , 2000. Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol. Cell 5: 927–937 [DOI] [PubMed] [Google Scholar]
- Garcia S. N., Pillus L., 2002. A unique class of conditional sir2 mutants displays distinct silencing defects in Saccharomyces cerevisiae. Genetics 162: 721–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotta M., Strahl-Bolsinger S., Renauld H., Laroche T., Kennedy B. K., et al. , 1997. Localization of Sir2p: the nucleolus as a compartment for silent information regulators. EMBO J. 16: 3243–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J., Kotovic K., El Hage A., Tollervey D., 2007. Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. EMBO J. 26: 4996–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh M. C., Mennella T. A., Sawa C., Berthelet S., Krogan N. J., et al. , 2006. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 20: 660–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlhaw G. B., 2003. Leucine biosynthesis in fungi: entering metabolism through the back door. Microbiol. Mol. Biol. Rev. 67: 1–15 table of contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepore N., Lafontaine D. L., 2011. A functional interface at the rDNA connects rRNA synthesis, pre-rRNA processing and nucleolar surveillance in budding yeast. PLoS ONE 6: e24962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. Y., Qi Y., Lu J. Y., Pan X., Yuan D. S., et al. , 2008. A comprehensive synthetic genetic interaction network governing yeast histone acetylation and deacetylation. Genes Dev. 22: 2062–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. Y., Lu J. Y., Zhang J., Walter W., Dang W., et al. , 2009. Protein acetylation microarray reveals that NuA4 controls key metabolic target regulating gluconeogenesis. Cell 136: 1073–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R., Meijer M., Lees-Miller S. P., Riabowol K., Young D., 2000. Three yeast proteins related to the human candidate tumor suppressor p33(ING1) are associated with histone acetyltransferase activities. Mol. Cell. Biol. 20: 3807–3816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., et al. , 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen S., Jensen T. H., 2007. Overlapping pathways dictate termination of RNA polymerase II transcription. Biochimie 89: 1177–1182 [DOI] [PubMed] [Google Scholar]
- Magtanong L., Ho C. H., Barker S. L., Jiao W., Baryshnikova A., et al. , 2011. Dosage suppression genetic interaction networks enhance functional wiring diagrams of the cell. Nat. Biotechnol. 29: 505–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar C. B., Xu F., Zhang K., Grunstein M., 2006. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 20: 711–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid J. L., Iyer V. R., Brown P. O., Struhl K., 2000. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell 6: 1297–1307 [DOI] [PubMed] [Google Scholar]
- Renauld H., Aparicio O. M., Zierath P. D., Billington B. L., Chhablani S. K., et al. , 1993. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 7: 1133–1145 [DOI] [PubMed] [Google Scholar]
- Robert F., Pokholok D. K., Hannett N. M., Rinaldi N. J., Chandy M., et al. , 2004. Global position and recruitment of HATs and HDACs in the yeast genome. Mol. Cell 16: 199–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann M. P., Luo W., Tsaponina O., Chabes A., Stillman B., 2011. A common telomeric gene silencing assay is affected by nucleotide metabolism. Mol. Cell 42: 127–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundlett S. E., Carmen A. A., Kobayashi R., Bavykin S., Turner B. M., et al. , 1996. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. USA 93: 14503–14508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche L. N., Kirchmaier A. L., Rine J., 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72: 481–516 [DOI] [PubMed] [Google Scholar]
- Sapountzi V., Côté J., 2011. MYST-family histone acetyltransferases: beyond chromatin. Cell. Mol. Life Sci. 68: 1147–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A., Hall M. N., Koller A., 1994. Two FK506 resistance-conferring genes in Saccharomyces cerevisiae, TAT1 and TAT2, encode amino acid permeases mediating tyrosine and tryptophan uptake. Mol. Cell. Biol. 14: 6597–6606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott E. M., Pillus L., 2010. Homocitrate synthase connects amino acid metabolism to chromatin functions through Esa1 and DNA damage. Genes Dev. 24: 1903–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P., 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E. R., Eisen A., Gu W., Sattah M., Pannuti A., et al. , 1998a ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc. Natl. Acad. Sci. USA 95: 3561–3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. S., Brachmann C. B., Pillus L., Boeke J. D., 1998b Distribution of a limited Sir2 protein pool regulates the strength of yeast rDNA silencing and is modulated by Sir4p. Genetics 149: 1205–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz E. J., Conrad N. K., Brow D. A., Corden J. L., 2001. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature 413: 327–331 [DOI] [PubMed] [Google Scholar]
- Stone E. M., Heun P., Laroche T., Pillus L., Gasser S. M., 2000. MAP kinase signaling induces nuclear reorganization in budding yeast. Curr. Biol. 10: 373–382 [DOI] [PubMed] [Google Scholar]
- Sugimoto K., Matsumoto K., Kornberg R. D., Reed S. I., Wittenberg C., 1995. Dosage suppressors of the dominant G1 cyclin mutant CLN3–2: identification of a yeast gene encoding a putative RNA/ssDNA binding protein. Mol. Gen. Genet. 248: 712–718 [DOI] [PubMed] [Google Scholar]
- Takahashi Y. H., Schulze J. M., Jackson J., Hentrich T., Seidel C., et al. , 2011. Dot1 and histone H3K79 methylation in natural telomeric and HM silencing. Mol. Cell 42: 118–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiebaut M., Kisseleva-Romanova E., Rougemaille M., Boulay J., Libri D., 2006. Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the nrd1-nab3 pathway in genome surveillance. Mol. Cell 23: 853–864 [DOI] [PubMed] [Google Scholar]
- Thomas B. J., Rothstein R., 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56: 619–630 [DOI] [PubMed] [Google Scholar]
- Vasiljeva L., Buratowski S., 2006. Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol. Cell 21: 239–248 [DOI] [PubMed] [Google Scholar]
- Vasiljeva L., Kim M., Terzi N., Soares L. M., Buratowski S., 2008. Transcription termination and RNA degradation contribute to silencing of RNA polymerase II transcription within heterochromatin. Mol. Cell 29: 313–323 [DOI] [PubMed] [Google Scholar]
- Wilson S. M., Datar K. V., Paddy M. R., Swedlow J. R., Swanson M. S., 1994. Characterization of nuclear polyadenylated RNA-binding proteins in Saccharomyces cerevisiae. J. Cell Biol. 127: 1173–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Andi B., Qian J., West A. H., Cook P. F., 2006. The alpha-aminoadipate pathway for lysine biosynthesis in fungi. Cell Biochem. Biophys. 46: 43–64 [DOI] [PubMed] [Google Scholar]
- Yang X. J., Seto E., 2008. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol. Cell 31: 449–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Xu W., Jiang W., Yu W., Lin Y., et al. , 2010. Regulation of cellular metabolism by protein lysine acetylation. Science 327: 1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.