Abstract
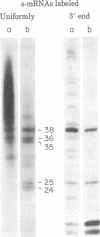
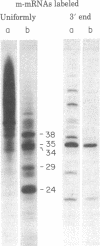
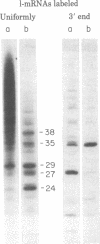
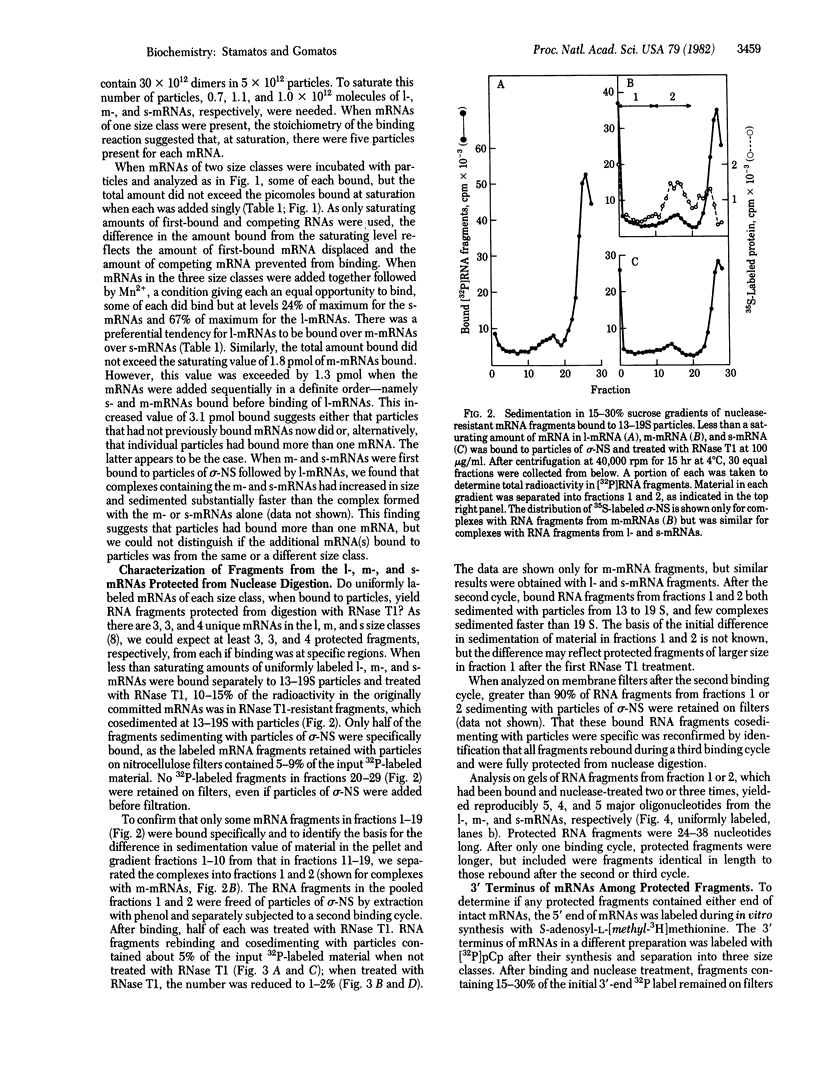
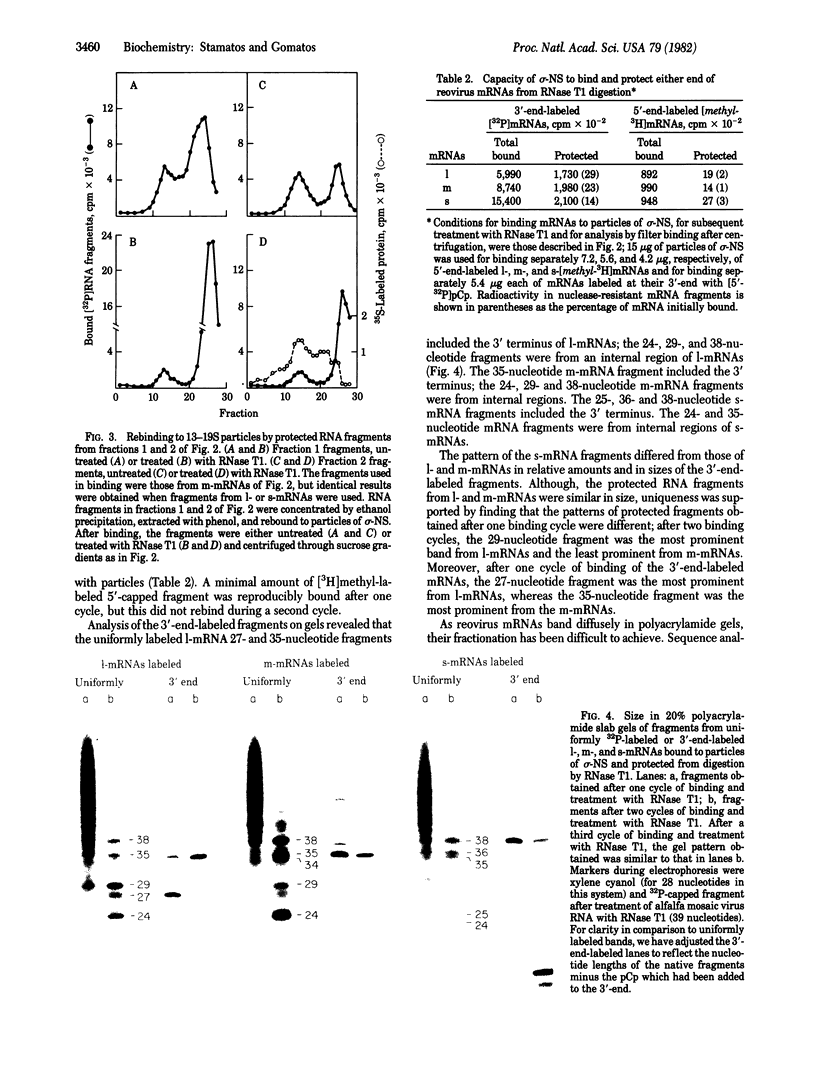
When assembled into 13--19S particles, the reovirus nonstructural protein sigma-NS selectively binds single-stranded RNAs. Sedimentation analyses combined with binding to nitrocellulose membrane filters showed that 1--2 pmol of reovirus mRNAs from the large, medium, or small size classes saturated in vitro the binding site(s) on 13--19S particles containing 100 pmol of sigma-NS. All mRNA segments in each size class bound to particles, and no mRNAs in one size class excluded the binding of mRNAs in any other class. In competition experiments, the maximal binding of all reovirus mRNAs to particles of sigma-NS was achieved when medium and small mRNAs were bound before the large mRNAs. This preferred order of addition of mRNAs to sigma-NS resulted in a marked increase in the size of some of the complexes. This finding suggests that the addition of large mRNAs last to particles promoted the formation of complexes with more than one RNA segment bound per particle. The 13--19S particles of sigma-NS protected 20- to 40-nucleotide RNA fragments from nuclease digestion. At least one of the protected fragments from mRNAs of each size class included the 3' terminus; the remaining were from internal regions of the mRNAs. The protected RNA fragments rebound to particles during a second or third cycle of binding in a configuration in which they were fully protected from nuclease digestion. We conclude that binding of particles of sigma-NS to reovirus mRNAs was not at random sites but was to specific regions unique for members of each size class.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Both G. W., Lavi S., Shatkin A. J. Synthesis of all the gene products of the reovirus genome in vivo and in vitro. Cell. 1975 Feb;4(2):173–180. doi: 10.1016/0092-8674(75)90124-5. [DOI] [PubMed] [Google Scholar]
- England T. E., Uhlenbeck O. C. 3'-terminal labelling of RNA with T4 RNA ligase. Nature. 1978 Oct 12;275(5680):560–561. doi: 10.1038/275560a0. [DOI] [PubMed] [Google Scholar]
- Franklin R. M. Purification and properties of the replicative intermediate of the RNA bacteriophage R17. Proc Natl Acad Sci U S A. 1966 Jun;55(6):1504–1511. doi: 10.1073/pnas.55.6.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomatos P. J., Prakash O., Stamatos N. M. Small reovirus particle composed solely of sigma NS with specificity for binding different nucleic acids. J Virol. 1981 Jul;39(1):115–124. doi: 10.1128/jvi.39.1.115-124.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomatos P. J., Stamatos N. M., Sarkar N. H. Small reovirus-specific particle with polycytidylate-dependent RNA polymerase activity. J Virol. 1980 Nov;36(2):556–565. doi: 10.1128/jvi.36.2.556-565.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huismans H., Joklik W. K. Reovirus-coded polypeptides in infected cells: isolation of two native monomeric polypeptides with affinity for single-stranded and double-stranded RNA, respectively. Virology. 1976 Apr;70(2):411–424. doi: 10.1016/0042-6822(76)90282-8. [DOI] [PubMed] [Google Scholar]
- Ito Y., Joklik W. K. Temperature-sensitive mutants of reovirus. I. Patterns of gene expression by mutants of groups C, D, and E. Virology. 1972 Oct;50(1):189–201. doi: 10.1016/0042-6822(72)90359-5. [DOI] [PubMed] [Google Scholar]
- Kozak M., Shatkin A. J. Characterization of ribosome-protected fragments from reovirus messenger RNA. J Biol Chem. 1976 Jul 25;251(14):4259–4266. [PubMed] [Google Scholar]
- Li J. K., Keene J. D., Scheible P. P., Joklik W. K. Nature of the 3'-terminal sequences of the plus and minus strands of the S1 gene of reovirus serotypes 1, 2 and 3. Virology. 1980 Aug;105(1):41–51. doi: 10.1016/0042-6822(80)90154-3. [DOI] [PubMed] [Google Scholar]
- Li J. K., Scheible P. P., Keene J. D., Joklik W. K. The plus strand of reovirus gene S2 is identical with its in vitro transcript. Virology. 1980 Aug;105(1):282–286. doi: 10.1016/0042-6822(80)90181-6. [DOI] [PubMed] [Google Scholar]
- McCrae M. A., Joklik W. K. The nature of the polypeptide encoded by each of the 10 double-stranded RNA segments of reovirus type 3. Virology. 1978 Sep;89(2):578–593. doi: 10.1016/0042-6822(78)90199-x. [DOI] [PubMed] [Google Scholar]
- McCrae M. A. Terminal structure of reovirus RNAs. J Gen Virol. 1981 Aug;55(Pt 2):393–403. doi: 10.1099/0022-1317-55-2-393. [DOI] [PubMed] [Google Scholar]
- Mustoe T. A., Ramig R. F., Sharpe A. H., Fields B. N. Genetics of reovirus: identification of the ds RNA segments encoding the polypeptides of the mu and sigma size classes. Virology. 1978 Sep;89(2):594–604. doi: 10.1016/0042-6822(78)90200-3. [DOI] [PubMed] [Google Scholar]
- Plotch S. J., Bouloy M., Ulmanen I., Krug R. M. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell. 1981 Mar;23(3):847–858. doi: 10.1016/0092-8674(81)90449-9. [DOI] [PubMed] [Google Scholar]
- Silverstein S. C., Christman J. K., Acs G. The reovirus replicative cycle. Annu Rev Biochem. 1976;45:375–408. doi: 10.1146/annurev.bi.45.070176.002111. [DOI] [PubMed] [Google Scholar]
- de Wachter R., Fiers W. Preparative two-dimensional polyacrylamide gel electrophoresis of 32 P-labeled RNA. Anal Biochem. 1972 Sep;49(1):184–197. doi: 10.1016/0003-2697(72)90257-6. [DOI] [PubMed] [Google Scholar]