Background: Low molecular weight protein-tyrosine phosphatase A, MptpA, is a key virulence factor of Mycobacterium tuberculosis.
Results: We determined the apo-MptpA NMR structure and identified the binding site of kinase PtkA and of inorganic phosphate.
Conclusion: There is a major rearrangement in the D-loop in the apo-state of MptpA.
Significance: Detailed understanding of the intramolecular architecture and intermolecular interactions of bacterial apo-state phosphatases is crucial for design of novel anti-infectives.
Keywords: Bacterial Protein Kinases, Bacterial Protein Phosphatases, Drug Resistance, Mycobacterium tuberculosis, NMR, MptpA, PtkA, Low Molecular Weight PTP
Abstract
Protein-tyrosine phosphatases (PTPs) and protein-tyrosine kinases co-regulate cellular processes. In pathogenic bacteria, they are frequently exploited to act as key virulence factors for human diseases. Mycobacterium tuberculosis, the causative organism of tuberculosis, secretes a low molecular weight PTP (LMW-PTP), MptpA, which is required for its survival upon infection of host macrophages. Although there is otherwise no sequence similarity of LMW-PTPs to other classes of PTPs, the phosphate binding loop (P-loop) CX5R and the loop containing a critical aspartic acid residue (D-loop), required for the catalytic activity, are well conserved. In most high molecular weight PTPs, ligand binding to the P-loop triggers a large conformational reorientation of the D-loop, in which it moves ∼10 Å, from an “open” to a “closed” conformation. Until now, there have been no ligand-free structures of LMW-PTPs described, and hence the dynamics of the D-loop have remained largely unknown for these PTPs. Here, we present a high resolution solution NMR structure of the free form of the MptpA LMW-PTP. In the absence of ligand and phosphate ions, the D-loop adopts an open conformation. Furthermore, we characterized the binding site of phosphate, a competitive inhibitor of LMW-PTPs, on MptpA and elucidated the involvement of both the P- and D-loop in phosphate binding. Notably, in LMW-PTPs, the phosphorylation status of two well conserved tyrosine residues, typically located in the D-loop, regulates the enzyme activity. PtkA, the kinase complementary to MptpA, phosphorylates these two tyrosine residues in MptpA. We characterized the MptpA-PtkA interaction by NMR spectroscopy to show that both the P- and D-loop form part of the binding interface.
Introduction
Tuberculosis is a chronic infectious disease caused by the facultative pathogen Mycobacterium tuberculosis, which is one of the oldest known pathogens, and has remained a major global health problem, with an estimated total of up to 8.8 million new cases and 1.4 million tuberculosis-related deaths annually (1). Although there are several treatment options available, the emergence of multidrug-resistant and extensive drug-resistant M. tuberculosis strains prioritizes the need for the development of new strategies to treat this widespread disease (2, 3). One of the emerging strategies in the drug development process is the approach to target bacterial virulence factors, which play a key role in bacterial entry and survival in the host cell (4). Protein-tyrosine phosphatases (PTPs)3 and protein-tyrosine kinases act as major virulence determinants by modifying host-pathogen signaling pathways (5–8) and are considered to be potential drug targets for anti-tuberculosis therapeutics (9). The genome of M. tuberculosis encodes two PTPs, MptpA and MptpB, which are secreted during infection and act as key virulence factors important for bacterial survival during macrophage infection (6, 10). Protein phosphorylation and dephosphorylation regulate various cellular functions, including pathogenicity (9). The dephosphorylation is catalyzed by different enzymes, which are categorized due to their specificity and structural properties into (i) serine/threonine and tyrosine phosphatases, (ii) dual specific phosphatases, and (iii) polymerase-histidinol phosphatases. The tyrosine phosphatases are represented by two major subclasses, the HMW-PTPs (∼30 kDa) and LMW-PTPs (∼20 kDa). MptpA (17.5 kDa) is a member of the LMW-PTP family. The MptpA deletion mutant of M. tuberculosis reveals that MptpA is essential for infection of host macrophages (11). The phagosome-lysosome fusion is important for macrophage microbicidal activity, and some organisms, including M. tuberculosis, evade this process for survival (12). MptpA dephosphorylates the host macrophage protein VPS33B (vacuolar protein sorting 33B), a protein that is responsible for the regulation of phagosome-lysosome fusion and trafficking (11). Dephosphorylation of VPS33B by MptpA is mediated by subunit H of the macrophage vacuolar H+-ATPase machinery, a multisubunit protein complex in the phagosome membrane-driven luminal acidification (13). Inactivation of VPS33B by dephosphorylation leads to arrest of the phagosome-lysosome fusion, thereby inhibiting the cellular response to infection. Recently, it has been shown that protein-tyrosine kinase A (PtkA) located immediately upstream from MptpA in the same operon, phosphorylates MptpA on two adjacent conserved tyrosine residues (14). Although the exact role of this phosphorylation is not yet determined, it was speculated that phosphorylation might increase the activity of MptpA or regulate its secretion (14). The important role of MptpA as mediator of M. tuberculosis virulence is evident, which makes it a potential drug target.
The HMW- and LMW-PTPs share a common cysteine-catalyzed reaction mechanism. However, except for the catalytically important phosphate binding CX5R motif called the P-loop and the critical aspartic acid residue in the D-loop, they exhibit no other sequence similarity (15, 16). The engagement of the phosphotyrosine substrate promotes a major conformational change in the PTP catalytic motifs (17), and notably, it has been speculated that modifications in the D-loop influence the catalytic activity of the protein (15, 16, 18). In most HMW-PTPs, the binding of ligand or substrate at the P-loop triggers a large structural change in the D-loop and swings its “open” conformation by ∼10 Å to a “closed” conformation (18, 19). In contrast, the role of the D-loop in LMW-PTPs is poorly understood (20). This is not surprising because all of the previously reported structures of LMW-PTPs (21–27), including the x-ray structure of MptpA (28), are obtained in the presence of ligands (phosphate/anions or substrate mimetics), and hence, the conformation (and dynamics) of the D-loop in the absence of ligand or substrate is unknown. In addition, a hallmark of the LMW-PTPs is the presence of two adjacent and well conserved tyrosine residues in the D-loop, whose phosphorylation status regulates its functional activity (29, 30). Upon phosphorylation, two effects are observed for several LMW-PTPs: (i) increase in activity (29, 30) and (ii) negative regulation (31). The regulation of the human LMW-PTP HCPTP-A, which has 37% sequence similarity to MptpA, is carried out by the phosphorylation on these two adjacent tyrosine residues (Tyr131 and Tyr132) (30). The in vitro phosphorylation of HCPTP-A Tyr131/Tyr132 by pp60v-Src has been reported and shows different effects on the enzyme activity. Whereas phosphorylation of Tyr131 increases the enzyme activity 25-fold, phosphorylation of Tyr132 does not affect the enzyme activity but leads to the recruitment of an adaptor protein Grb2 Src homology 2 domain, important for signal transduction. These findings indicate that both tyrosine residues are of crucial importance for the structural and functional regulation of HCPTP-A. Biochemical assays showed that the residues Tyr128 and Tyr129 in MptpA are phosphorylated by the kinase PtkA (14). Both the phosphorylation of these tyrosines and the structural conformation of the D-loop are supposed to play an important role in the structure-function relationship of MptpA.
Here, we present a high resolution solution nuclear magnetic resonance (NMR) structure of ligand-free MptpA, which reveals that the D-loop adopts an open conformation compared with the closed conformation observed in ligand-bound structures. Based on NMR chemical shift perturbations (CSPs), we were able to map the MptpA binding site for inorganic phosphate (Pi), a competitive inhibitor of LMW-PTPs, and to clarify the involvement of both the P- and D-loop in the catalytic mechanism. Furthermore, we show by mass spectrometry that PtkA can phosphorylate MptpA and show by NMR that the P- and D-loop of MptpA form part of the PtkA binding interface.
Our results report a previously unknown conformation of the D-loop in LMW-PTPs and refine our view of how the D-loop shuttles between the open and closed conformation upon ligand binding. The substantially different conformation of D-loop amino acids in the apo-structure, presenting the hydrophobic residues Tyr128 and Tyr129 in other conformations, allows for more specific structure-based drug design strategies to the apo-state of MptpA in the future.
EXPERIMENTAL PROCEDURES
MptpA Cloning, Expression, and Purification
The plasmid pET16bTEV vector containing the MptpA sequence according to RV2234 was obtained from Dr. Anil Koul. From the pET16bTEV-MptpA vector, the base pairs corresponding to full-length MptpA (amino acid residues 1–163; see Fig. 1A) were amplified by PCR using 5′-AAA CCA TGG GGA TGT CTG ATC CG-3′ and 5′-TTA TTG CTC AGC GGT GGC AGC A-3′ as primers. The PCR product was double-digested with NcoI/BlpId and cloned into the modified pKM263 (His6-tagged ProtGB1-TEV between NdeI and XhoI) vector, which contains the ProtGB1 for enhancement of solubility of the expressed fusion partner. The authenticity of the clone was validated via nucleotide sequencing. Protein expression was performed in Escherichia coli strain BL21(DE3)pLysS (Novagen) containing the desired plasmid using Lysogeny broth (LB) medium at 37 °C supplemented with 100 μg/ml ampicillin and 34 μg/ml chloramphenicol. For induction of protein expression, 1 mm isopropyl-1-thio-β-d-galactopyranoside at A600 of 0.6–0.7 was added. The culture was incubated in 5-liter Erlenmeyer flasks for 12 h with aeration (150 rpm) at 16 °C before harvesting the cells by centrifugation (6,000 × g, 30 min, 4 °C). The cell pellet was resuspended in lysis buffer (300 mm NaCl, 50 mm Tris-HCl (pH 8.0), 10 mm β-mercaptoethanol) with the addition of complete protease inhibitor (1 tablet/100 ml of EDTA-free, Roche Applied Science). After cell disruption (Microfluidizer, 15,000 p.s.i., 2 cycles) the soluble fraction was isolated from cell debris by centrifugation (18,000 × g, 45 min, 4 °C). The supernatant was applied to a Ni-NTA FastFlow column (GE Healthcare) following the manufacturer's recommendations. His6-tagged ProtGB1 was removed by tobacco etch virus (TEV)-protease cleavage via dialysis overnight at 4 °C (300 mm NaCl, 50 mm Tris-HCl (pH 8.0), 10 mm β-mercaptoethanol) and excluded by a Ni-NTA column. Further purification of the protein was performed by gel filtration on a HiLoad 26/60 Superdex 75 preparative grade column (GE Healthcare) in 25 mm HEPES (pH 7.0), 150 mm NaCl, and 1 mm DTT. The protein-containing fractions were pooled, concentrated, and stored at −80 °C or immediately used for experimental procedures. Doubly (13C,15N) labeled protein was expressed in M9 (32) minimal medium containing 15NH4Cl (1 g/liter) as nitrogen and [13C]glucose (2 g/liter) as sole carbon source.
FIGURE 1.
A, full-length MptpA amino acid sequence construct (Rv2234, Met1–Ser163). Additional residues G(−3)–G(0) originate from subcloning. B, 1H,15N TROSY-HSQC spectrum of MptpA at 900 MHz. The inset shows the enlargement of the region between 7.4 and 9.0 ppm in 1H and between 117.6 and 124.2 ppm in the 15N dimension. Amide protons of side chain resonances are indicated by horizontal lines. The spectrum was recorded at T = 303 K in the following buffer: 1.2 mm MptpA, 50 mm arginine/glutamate, 25 mm HEPES (pH 7.0), 10 mm DTT, 10% D2O, and 150 μm 2,2-dimethyl-2-silapentane-5-sulfonic acid. Additional peaks seen in the TROSY-HSQC spectrum with low intensities possibly arose from sample heterogeneity.
PtkA Cloning, Expression, and Purification
The plasmid vector (pET151/D-TOPO, Invitrogen) containing His6-tagged TEV PtkA sequence was a kind gift from the laboratory of Prof. Yossef Av-Gay (University of British Columbia). Using site-directed mutagenesis, the PtkA sequence according to RV2232 (supplemental Fig. S1) was generated. The following primers were used: 5′-GAT CCC TTC ACC CTT AAG ATG TCT TCG CCT CGT GAA CG-3′, 5′-CGT TCA CGA GGC GAA GAC ATC TTA AGG GTG AAG GGA TC-3′, 5′-CAC GGT GGT GGT CGG CTG GGG CTA CGG GCG CG-3′, and 5′-CGC GCC CGT AGC CCC AGC CGA CCA CCA CCG TG-3′. For the expression of unlabeled protein, ZYM-5052 autoinduction medium (33) was used. The culture was incubated in 5-liter Erlenmeyer flasks for 12 h with aeration (160 rpm) at 25 °C before harvesting the cells by centrifugation (6,000 × g, 30 min, 4 °C). The cell pellet was resuspended in lysis buffer (300 mm NaCl, 50 mm Tris-HCl (pH 8.0), 10 mm 2-mercaptoethanol) with the addition of complete protease inhibitor (1 tablet/100 ml of EDTA-free, Roche Applied Science). After cell disruption (Microfluidizer, 15,000 p.s.i., 2 cycles), the soluble fraction was isolated from cell debris by centrifugation (18,000 × g, 45 min, 4 °C). The supernatant was applied to a Ni-NTA FastFlow column (GE Healthcare) following the manufacturer's recommendations. The His6 tag was removed by TEV-protease cleavage via dialysis overnight at 4 °C (300 mm NaCl, 50 mm Tris-HCl (pH 8.0), 10 mm β-mercaptoethanol) and excluded by a Ni-NTA column. For further purification of the protein, gel filtration chromatography on a HiLoad 26/60 Superdex 75 preparative grade column (GE Healthcare) in 25 mm HEPES (pH 7.0), 300 mm NaCl, and 10 mm DTT was used. The protein-containing fractions were pooled, concentrated, and stored at −80 °C or immediately used for experimental procedures.
Phosphatase Assay
Phosphatase activity was determined by adding 20 μm MptpA to the buffer (25 mm Tris-HCl (pH 7.0), 50 mm NaCl, 2 mm EDTA) containing 25 mm p-nitrophenyl phosphate as substrate. The absorbance at 410 nm in a time-dependent manner was followed using a UV spectrometer (Varian, Cary50Bio) (supplemental Fig. S2). In parallel, assays with samples lacking MptpA were performed.
Luciferase Assay
Autophosphorylation activity was determined by using the Kinase-Glo® Plus luminescent kinase assay (Promega). Different enzyme concentrations (500 nm and 1, 10, and 50 μm) were added to the assay buffer (300 mm NaCl, 50 mm Tris-HCl (pH 8.0), 10 mm DTT, 10 mm MgCl2) containing 1, 5, 10, and 100 μm ATP. After 60 min of incubation, the luciferase mixture was added and incubated for 30 min before measuring the relative light units with a luminometer (96-well plate; Promega).
Phosphorylation Reaction
For the phosphorylation of 200 μm MptpA by 50 μm PtkA in 300 mm NaCl, 50 mm Tris-HCl (pH 8.0), 10 mm DTT, the proteins were incubated with 10 mm MgCl2 and 10 mm ATP for 12 h at 4 °C. Subsequently, MALDI-MS as well as 31P NMR spectra were recorded to verify the phosphorylation reaction.
NMR Spectroscopy
NMR experiments were conducted at T = 303 K on a Bruker 500-MHz room temperature TXI-HCN probe as well as Bruker 600-, 700-, 800-, 900-, and 950-MHz spectrometers equipped with TXI-HCN cryogenic probes. The spectrometers were locked on D2O. As an internal standard for spectral calibration, 2,2-dimethyl-2-silapentane-5-sulfonic acid (150 μm) was used. The backbone resonance assignment at T = 303 K (1.2 mm 15N,13C-labeled MptpA in 50 mm arginine/glutamate, 25 mm HEPES (pH 7.0), 10 mm DTT) was obtained using TROSY versions of HNCO (34), HNCA (34), HNCACB (35), and HN(CO)CACB (35) triple resonance experiments and was verified utilizing the NMR backbone resonances of MptpA at T = 298 K as reported previously (36). The aliphatic side chain resonance assignment was obtained from (H)CC(CO)NH-TOCSY (37, 38) (mixing time, 21.1 ms) and H(C)CH-TOCSY (39) (mixing time, 18 ms) experiments. For the aromatic side chain resonance assignment, (H)CB(CGCC-TOCSY)Har (40) (Phe/Tyr mixing time, 8.6 ms; Trp mixing time, 5.75 and 11.5 ms), three-dimensional Har(CC-TOCSY-CGCBCACO)NH (40), and (H)C(NC)H and HCD(CG)CB-TROSY (41, 42) experiments were performed. NOE-based distance restraints were obtained from three-dimensional 1H,1H-NOESY-15N TROSY (43) (900 MHz; mixing time, 70 ms) and three-dimensional 1H,1H-NOESY-13C-HSQC (aliphatic) and constant time 13C,1H-TROSY (44) based three-dimensional 13C-separated 1H,1H-NOESY (aromatic) (950 MHz; mixing time, 70 ms) experiments. Heteronuclear 15N relaxation experiments ((1H)-15N hetNOE, T1 and T2) (45–47) were performed with a 15N-labeled MptpA (1.0 mm) sample (150 mm NaCl, 50 mm HEPES (pH 7.0), 10 mm DTT) at T = 298 K using a 600-MHz spectrometer with cryogenic probe. The T1 longitudinal 15N relaxation rates were determined from spectra with different delays of 10, 100, 400, 600, 800, 1000, 1200, and 1500 ms. T2 transverse 15N relaxation rates were determined using the following delays: 35.2, 70.4, 105.6, 140.8, 176.0, 211.2, 246.4, and 281.6 ms. The (1H)-15N hetNOEs were obtained from the ratio of peak intensities (INOE/Ino NOE) in spectra recorded with and without the saturation of amide protons. The 3J(HN,Hα) coupling constants were obtained via a three-dimensional HNHα-HMQC experiment (48, 49). Residual dipolar couplings were measured in pf1 bacteriophages as alignment medium (5 g/liter; Profos) at 600 MHz. The 1D (HN,N) couplings were extracted from IPAP-(15N,1H,)-HSQC (50) spectra. As the end point of the titration of MptpA with PtkA (with steps of 0.5), a ratio of 1:2 (MptpA/PtkA) was used. The spectra were processed using Topspin version 2.1 (Bruker Biospin) and analyzed using either SPARKY version 3.114 or CARA version 1.8.4.2. The calculation of dynamics (S2) by using hetNOE and T1/T2 ratios was executed with TENSOR2 (51) and ModelFree (52, 53).
Structure Calculation
Structure calculations were performed using the software packages CYANA (54–56) and ARIA/CNS (57, 58). Initial structure calculations and optimizations were done independently with CYANA version 3.9 and ARIA version 1.2. The final structure calculation was performed with CYANA version 3.9 and refined using ARIA version 1.2 protocols. The MptpA resonances were manually assigned and used as input together with unassigned NOESY peak lists for fully automated NOESY resonance assignment and calibration. The three-dimensional 15N and 13C separated NOESY spectra were peak-picked using the restricted peak-picking routine of SPARKY version 3.114 (59). The NOESY peak lists were refined by manual inspection of the resonances around crowded regions and the diagonal. Furthermore, all of the resonances between 4.72 and 4.76 ppm were excluded because of overlap with the water signal. The chemical shift tolerances were set to 0.10 ppm for the heavy atoms and 0.02 and 0.03 ppm for the bound protons and other protons, respectively. In addition to NOE data, hydrogen bond distances and amply defined dihedral angle restraints (based on TALOS+ predictions (60, 61) and confirmed with NOESY resonance assignments and initial structure calculations) as well as 1D (H,N) residual dipolar coupling and 3J(HN,Hα) coupling constant (Karplus relation) restraints were included in the standard structure calculation with CYANA version 3.9 (100 structures per iteration, 15,000 refinement steps). The final bundle of the 20 best structures was used as input for optimization with CNS version 1.1 (62) using the ARIA version 1.2 setup and protocols for refinement in explicit water (63). The NOE distance restraints were converted to ARIA/CNS format. Next to the 1D(H,N) residual dipolar couplings (anisotropy, −18.72, rhombicity 0.17), 3J(HN,Hα) coupling constants, hydrogen bonds, and dihedral angle restraints, T1/T2 relaxation values were also included as diffusion anisotropy restraints (anisotropy, 1.25; rhombicity, 0.39; τc = 12.12) in this refinement stage. The standard protocols and allhdg5.3 force field were used with optimized potentials for liquid simulations non-bonded parameters. The final structure bundle was analyzed with PROCHECK version 3.5.4 (64, 65) and CING (66).
RESULTS
NMR Resonance and NOE Cross-peak Assignment of MptpA
The 1H, 15N, and 13C resonances of MptpA were assigned at T = 303 K and verified with the previously reported backbone amide resonances at T = 298 K (36) (Biological Magnetic Resonance Data Bank entry 6722). We carried out additional NMR experiments to assign the aromatic side chain resonances ((H)CB(CGCC-TOCSY)Har (40), three-dimensional Har(CC-TOCSY-CGCBCACO)NH, (H)C(NC)H, HCD(CG)CB-TROSY (41, 42)) and completed the assignment of the aliphatic side chain resonances using (H)CC(CO)NH-TOCSY (37, 38) and H(C)CH-TOCSY (39). These experiments were all performed at T = 303 K, which allowed us also to extend the backbone amide resonance assignment of MptpA with a nearly complete side chain assignment. We were able to assign 96.8% of all proton resonances at pH 7.0 and T = 303 K (Fig. 1B). In the two-dimensional 1H,15N TROSY experiment, 151 backbone amide protons of 156 expected resonances were resolved, and 15 side chain resonances were detected. The resonances for a stretch of amino acids, including Thr12, Gly13, and Arg17 (interestingly being part of the active site), as well as the single amino acids Thr119 and His132 were missing, although for all of those except Arg17 the side chain assignment could be achieved. For the automated NOESY cross-peak assignment, peak lists containing intensities were generated from the three-dimensional 15N and 13C separated NOESY spectra by using the restricted peak-picking routine of SPARKY. In total, 11,735 peaks were selected as input data for CYANA, whereas peaks close to the H2O signal (4.72–4.76 ppm) were excluded from further analysis. The automated NOESY assignment routine assigned 7,764 peaks, resulting in 4,821 NOE-based distance restraints (on average about 29 per residue). After the structure calculation with CYANA and refinement in explicit water with CNS using ARIA protocols, a bundle of 20 structures (Fig. 2A) with an average backbone RMSD of 0.22 ± 0.04 Å and all heavy atom RMSD of 0.62 ± 0.04 Å, for the core of the protein (residues 3–114 and 132–158) excluding the flexible termini and part of the D-loop, was calculated (Table 1).
FIGURE 2.
Solution structure of apo-MptpA. A, superposition of the backbone traces showing the bundle of the 20 lowest energy structures and rotated by 180°. B, ribbon representation of the lowest energy structure. The loop regions involved in substrate binding are highlighted. Blue, P-loop (Cys11–Arg17) connecting β1-α1; green, W-loop (Ser44–Glu55) connecting β2-α2; magenta, D-loop (Arg111–Asp131). The side chain residues crucial for the catalytic activity (Cys11 (P-loop) and Asp126 (D-loop)) and substrate specificity (Trp48 (W-loop)) are shown as sticks. The overall structure of MptpA consists of a four-stranded parallel folded β-sheet (β1(Leu5–Cys11), β2(Ala37–Ala43), β3(Leu85–Leu89), β4(Arg106–Leu110)) connected via five α-helices (α1(Cys16-Arg32), α2(Asp55–His65), α3(Gly77–Ala82), α4(Asp90–Leu100), α5(His132-Asn160)) also known as the Rossmann fold. The P-loop is flanked by the W- and D-loop. C, backbone dynamics of MptpA measured at 600 MHz and T = 303 K. Mapping the S2 values on the backbone bundle of 20 lowest energy structures. Colors range from blue (S2 = 1) to red (S2 = 0) to gray (no data/no reliable fit). The very low order parameters for the N and C terminus (white, Met1, Ser2, Gly161, and Ser163) are not shown. The figure was generated by PyMOL.
TABLE 1.
Structural statistics for the ensemble of the 20 best NMR structures of MptpA
| Parameter | Value |
|---|---|
| NOESY assignmenta | |
| Selected cross-peaks | 11,735 |
| Assigned cross-peaks | 7,764 |
| Experimental restraints | |
| NOE-based distance restraints | |
| Total | 4,821 |
| Intraresidual (|i − j| = 0) | 815 |
| Sequential (|i − j| = 1) | 1,292 |
| Medium range (2 ≤|i − j| ≤ 4) | 1,103 |
| Long range (|i − j| ≥ 5) | 1,611 |
| Restrained hydrogen bonds | 69 |
| Dihedral angle restraints | |
| Torsion angle (φ/ψ) | 301 |
| Diffusion anisotropy restraints | |
| T1/T2 values for HN | 108 |
| Residual dipolar couplings | |
| 1D(H,N) coupling restraints | 119 |
| Scalar couplings | |
| 3J(HN,Hα) | 83 |
| RMSD from average for residue (Å)b | |
| Backbone N, CA, C′ | 0.22 ± 0.04 |
| Heavy atoms | 0.62 ± 0.04 |
| Ramachandran plot (%)c | |
| Most favorable | 81.8 |
| Additional allowed | 17.9 |
| Generously allowed | 0.3 |
| Disallowed | 0.0 |
a Automated NOESY peak picking performed by SPARKY. Values were obtained by using CYANA.
b Values were calculated for the core of the protein comprising residues 3–114 and 132–158 (thus excluding the flexible termini and part of the D-loop) by using MOLMOL.
c Values were calculated for all residues excluding glycines and prolines by using PROCHECK.
The Overall Structure of Apo-MptpA
MptpA adopts the Rossmann fold typical for LMW-PTPs consisting of a four-stranded parallel β-sheet (β1(Leu5–Cys11), β2(Ala37–Ala43), β3(Leu85–Leu89), and β4(Arg106–Leu110)) connected via five α-helices (α1(Cys16–Arg32), α2(Asp55–His65), α3(Gly77–Ala82), α4(Asp90–Leu100), and α5(His132-Asn160)). The active site motif 11CTGNICRS18 is highly conserved among all known LMW-PTPs containing the catalytic active Cys11, which is buried in a crevice flanked by three loops (β2–α2, α2–α3, and β4–α5) containing hydrophobic residues (Trp48, Tyr128, and Tyr129). The β2–α2 loop containing residue Trp48 (W-loop, Gly44–Asp55) and the β4–α5 loop containing residue Asp126 (D-loop, Arg111–Asp131) are both in spatial proximity to the P-loop and contain crucial residues for substrate specificity (Trp48 and His49) as well as residues involved in the catalytic mechanism (Asp126) (Fig. 2B). The amide protons of most of the active site residues (Thr12, Gly13, Asn14, Ile15, Cys16, Arg17, and Ser18) are pointing toward the inside of the P-loop, in agreement with structures of the LMW-PTP of Bos taurus (23).
Comparison of Apo- and Ligand-bound MptpA
The available x-ray structures of MptpA are in complex with either chloride (PDB entry 1U2P) or glycerol (PDB entry 1U2Q) (28). Comparison of MptpA x-ray structure (PDB entries 1U2P and 1U2Q) and our solution structure bundle (PDB entry 2LUO) reveals an RMSD of 1.31 ± 0.05 and 1.34 ± 0.05 Å (all heavy atoms and backbone residues 4–158), respectively. We performed a structural alignment, based on our structure, of only the P- and D-loop motifs, which confirmed the conserved P-loop structure (0.31 ± 0.06 and 0.27 ± 0.03 Å) but revealed major deviations in the D-loop orientation represented by an RMSD of 2.14 ± 0.21 and 1.92 ± 0.19 Å (Table 2). Detailed analysis of the x-ray structures of MptpA (PDB entries 1U2P and 1U2Q) (28) revealed crystal packing artifacts concerning the D-loop region. The crystal packing restricts the orientation of the D-loop residues Glu125 and Asp126. The structural comparison of the D-loop conformation in solution and in the x-ray structures of MptpA (PDB entries 1U2P and 1U2Q) (28) reveals major deviations for the residues Glu125, Asp126, Pro127, Tyr128, Tyr129, and Gly130. Notably, the side chain orientation of residues Tyr128 and Tyr129 in the D-loop is different as well as for Trp48 in the W-loop. Specific distance measurements between the residues in the D-loop and residues in the P-loop and W-loop clearly indicate this and show the more open confirmation for the binding pocket (Fig. 3, A and B). In particular, the distance between Trp48 Nϵ1 and Tyr128 Oη is nearly 13 Å displaced. Consequently, the surface surrounding the active site is influenced because the hydrophobic residues Tyr128/Tyr129 are pointing in the opposite direction of the P-loop (Fig. 4, E and F). Furthermore, the key residue Asp126 involved in catalysis is about 3 Å further apart from the active site as observed in the x-ray structures (Fig. 3, A and B).
TABLE 2.
Comparison of 20 best NMR structures of MptpA P-loop and D-loop motifs and of different LMW-PTPs
Average pairwise RMSD values of P-loop and D-loop motif. RMSD values are given in Å. All 20 best structures from MptpA were taken into account. For the alignment, only the backbone atoms in question (P-loop, Cys11–Arg17; as part of the D-loop, Lys122–Asp131), respectively, were superimposed before the RMSD was calculated using MOLMOL 2K.2. For the comparison with other NMR structures (BPTP, TPTP, YxiE, and Cj1258), every possible combination between the clusters of available structures was superimposed, before calculating the RMSD. The coordinate files of MptpA (PDB entries 1U2P and 1U2Q, x-ray), YxiE (PDB entry 1ZGG, NMR), HCPTP-A (PDB entry 5PNT, x-ray), LTP1 (PDB entry 1D1P, x-ray), TPTP (PDB entry 1P8A, NMR), BPTP (PDB entry 1BVH, NMR), and Cj1258 (PDB entry 2GI4, NMR) were obtained from the Protein Data Bank.
| Protein | Species | P-loop | D-loop |
|---|---|---|---|
| MptpA 2LUO | M. tuberculosis | 0.23 ± 0.17 | 0.87 ± 0.23 |
| MptpA 1U2P | 0.31 ± 0.06 | 2.14 ± 0.21 | |
| MptpA 1U2Q | 0.27 ± 0.03 | 1.92 ± 0.19 | |
| YxiE | B. subtilis | 0.77 ± 0.09 | 1.74 ± 0.14 |
| HCPTP-A | Human | 0.26 ± 0.04 | 2.36 ± 0.22 |
| LTP1 | Yeast | 0.38 ± 0.04 | 2.62 ± 0.21 |
| TPTP | T. foetus | 0.92 ± 0.33 | 3.22 ± 0.49 |
| BPTP | Bovine | 1.07 ± 0.25 | 3.25 ± 0.40 |
| Cj1258 | C. jejuni | 0.48 ± 0.11 | 5.51 ± 0.16 |
FIGURE 3.
D-loop movement reveals open and closed conformation. A, D-loop distance measurements reveal closed conformation of MptpA x-ray structure (PDB entry 1U2P) in the chloride-bound state. The distance between Trp48 Nϵ1 and Tyr128 Oη of 7.2 Å reveals a more closed binding pocket compared with the apo-state of MptpA, as well as the Cα distances between Arg17 and Tyr128/Tyr129 of 8.9 and 12.5 Å, respectively. B, open conformation of binding pocket observed in apo-MptpA (PDB entry 2LUO) due to a distance of 19.8 ± 3.5 Å between Trp48 Nϵ1 and Tyr128 Oη. The dashed lines indicate the Cα distances between Arg17 and Tyr128/Tyr129 of 14.0 ± 1.3 and 15.9 ± 0.8 Å, respectively. C and D, the following PDB entries are used: MptpA NMR structure (PDB entry 2LUO) (cyan), MptpA x-ray structure (PDB entry 1U2P) (magenta), and bovine BPTP (PDB entry 1PNT) (green). C, superposition of the P-loop surrounding area observed in solution (cyan) and x-ray structure (magenta) of MptpA in complex with chloride. Representation of the P-loop electrostatic surface with charge distribution. Secondary structure elements are depicted in white. D, same scheme as in C, replacing the x-ray structure of MptpA with the x-ray structure of bovine BPTP (green) in complex with phosphate. All images were generated in PyMOL. The solvent-accessible electrostatic surface was calculated using APBS Tools version 2.1.
FIGURE 4.
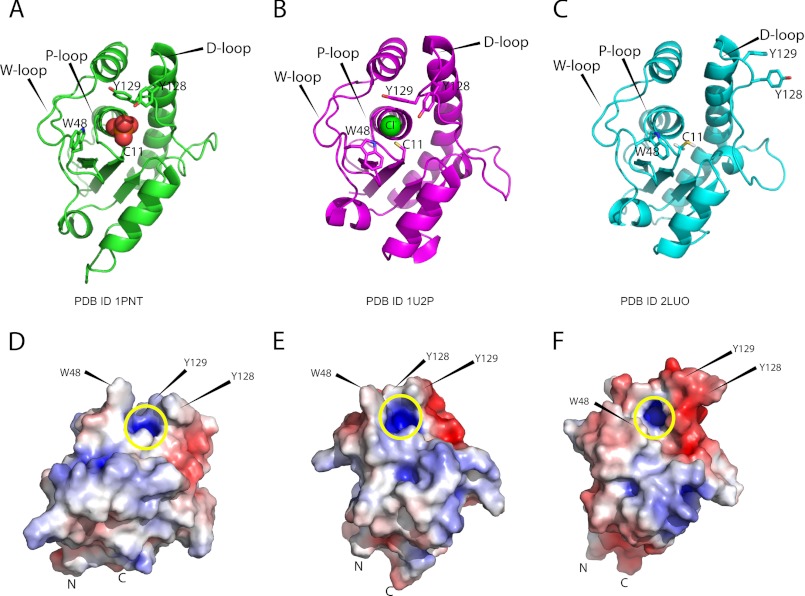
Structural changes observed upon ligand binding in LMW-PTPs. The following PDB entries are used: bovine BPTP (PDB entry 1PNT, x-ray) (A and D), MptpA x-ray structure (PDB entry 1U2P) (B and E), and MptpA NMR structure (PDB entry 2LUO) (C and F). A–C, comparison of secondary structure elements of bovine BPTP (A), MptpA x-ray structure (B), and MptpA solution structure (C). D–F, solvent-accessible surface mapped on bovine BPTP (D), MptpA x-ray structure (E), and MptpA solution structure (F). Yellow circle, positively charged binding pocket. The locations of hydrophobic residues (Trp48, Tyr128, and Tyr129) flanking the active site are highlighted. The solvent-accessible electrostatic surface was calculated using APBS Tools version 2.1.
Structure Comparisons with Other LMW-PTPs
Our solution structure of M. tuberculosis MptpA in its apo-state was compared with the x-ray crystal and NMR solution structures of LMW-PTPs of other organisms: yeast (LTP1 from Saccharomyces cerevisiae), human (HCPTP-A), bovine (BPTP), eukaryotic parasite (TPTP from Tritrichomonas foetus), and bacterial (YwiE from Bacillus subtilis, cj1258 from Campylobacter jejuni and Wzb from Escherichia coli). The LMW-PTP sequences from these different organisms have identities between 26 and 38% (Fig. 5). The active site motif (H/V)CX5R(S/T) (P-loop) is well conserved in all species, as well as Ser42 and the D-loop motif DP (Asp126 and Pro127 in MptpA), which is followed by hydrophobic residues (Tyr128 and Tyr129 in MptpA). Furthermore, Ser42, which is part of the hydrogen bonding network surrounding the P-loop, is conserved among all species. Additionally, Glu22 is highly conserved and is also found to be in close spatial proximity to His71, which is involved in formation of the P-loop conformation. Although there is hardly any sequence similarity for the W-loop, around residue Trp48 (only residues Gly51 and Gly55 are conserved to some extent), it shows a high structural resemblance to the other LMW-PTPs (Fig. 5, green). The overall fold of MptpA secondary structure elements is highly consistent with structures found in other eukaryotic and prokaryotic LMW-PTPs.
FIGURE 5.
Multiple sequence alignment. Shown is a sequence alignment of LMW-PTPs, with the secondary structure elements of MptpA shown on top. The figure was prepared using ESPript2.2 (67) by applying the BLOSUM62 scoring matrix. The P-, W-, and D-loop are colored in blue, green, and magenta, respectively.
Comparison of P- and D-loop Motifs
Superimposing only P-loop (Cys11–Arg17) and a part of the D-loop (Leu122–Asp131) residues of all compared LMW-PTPs confirmed the conserved conformation of the P-loop and revealed a reorientation of the D-loop (Table 2). The side chain orientation of residues Tyr128/Tyr129 of MptpA in the x-ray structure is consistent with bovine BPTP (Fig. 4, A and B). Furthermore, similar deviations concerning the conformation of the D-loop are observed. Fig. 3, C and D, shows a detailed view of the P-loop electrostatic surface after superposition of the x-ray structures from bovine BPTP and MptpA with the here elucidated solution structure of MptpA. Both x-ray structures, in complex with chloride/phosphate, show the same orientation of the two adjacent tyrosine residues in the D-loop, pointing toward the active site motif (Fig. 3, C and D, magenta and green). However, the side chains of Tyr128/Tyr129 represented by our NMR structure in the apo-state are not oriented toward the active site cavity (Fig. 3, C and D, cyan). We should also mention that although in bovine LMW-PTP (BPTP), in the presence of phosphate, a protein concentration-dependent dimerization induced by the interaction of W- and D-loop residues is observed (20), we do not observe a similar concentration-dependent dimerization in MptpA.
Hydrogen Bond Analysis
The hydrogen-bonding network observed in the bundle of the 20 best structures of apo-MptpA revealed a stabilization of the Cys11 amide by interacting with the carbonyl of Ala43; thus, the catalytic active thiolate group of Cys11 is pointing inside the substrate binding pocket. The hydrogen bond network interconnecting the residues Asn14, Ser18, Ser42, and His71, found in all LMW-PTPs, is reported to stabilize the observed strained, left-handed (φ = 60°; ψ = 45°) conformation of residue Asn14. The conformation of Asn14 in x-ray structures (of bovine BPTP, human HCPTP-A, yeast LPT1, and M. tuberculosis MptpA) is described as a strained, left-handed helix with a positive φ angle. The solution structures solved by NMR spectroscopy (bovine BPTP, human HCPTP-A, yeast LPT1, T. foetus TPTP, and C. jejuni cy1258) in the presence of at least 20 mm phosphate lead to the assumption of a transient hydrogen bonding network, including Asn14, Ser42, and His71. Due to the proposed transient hydrogen bond network, Asn14 was reported to exist in two possible conformations. In solution and in the presence of phosphate, Asn14 was found in the α-helical as well as in the αL-helical conformation (68). Our apo-state solution structure of MptpA solved here reveals a predominantly left-handed conformation of Asn14. The α-helical conformation of Asn14 in the solution structure of MptpA was observed in only two of the 20 best structures. This 2-foldness could be a feature representing the flexibility and dynamics of this region in solution, but it should be noted that this could also be influenced to some extent by the missing amide resonances for the nearby residues Thr12 and Gly13. In the apo-state solution structure of MptpA, the left-handed conformation of Asn14 (φ = 59.0 ± 6.2°; ψ = 21.4 ± 5.5°) is stabilized by the potential hydrogen network of Asn14, Ser18, Ser42, and His71. The distance measured between Asn14 Nδ2 and Ser18 Oγ, Ser42 Oγ, and His71 Hϵ2 (5.4 ± 1.5, 4.2 ± 1.9, and 4.0 ± 1.5 Å) support the presence of a hydrogen bond network in this region. In 13 of the 20 best structures, distances from Asn14 Nδ2 to Ser18 Oγ of 4.6 ± 1.0 Å, of 3.1 ± 0.4 Å to Ser42 Oγ, and of 3.4 ± 0.2 Å to His71 Hϵ2 are observed. Spatial proximity between Asn14 and Ser18, Ser42, and His71 is observed frequently, indicating the presence of a hydrogen bond network.
Analysis of the Active Site-stabilizing Network
In addition to the aromatic residues Trp48, Tyr128, and Tyr129 surrounding the active site, the amino acids His49, Asp123, and Arg91 place a unique charge distribution around the active site (28). The interaction between Thr12 Oγ1 and His93 Nϵ2 further stabilizes the P-loop. This interaction is a unique feature of bacterial LMW-PTPs, whereas His93 is found to be replaced by aspartic acid in all known mammalian LMW-PTPs. Distance analysis of Thr12 Oγ1 and His93 Nϵ2 revealed a close spatial proximity (3.0 ± 0.5 Å) of these atoms, confirming a potential hydrogen bond interaction. In turn, the backbone amide proton of His93 interacts with Asp90 Oδ2, which is found to be conserved among bacterial LMW-PTPs. Hence, the interaction between His93 and the side chain of Asp90 is of crucial importance for the substrate specificity and confirmed in our structure. Distance measurements based on the apo-state solution structure show that Glu22 is in spatial proximity to the imidazole ring of His71. Multiple sequence alignment identifies Glu22 and His71 as highly conserved among LMW-PTPs (Fig. 5). Therefore, the electrostatic interaction between Glu22 and His71 must be of pivotal importance for the conformation of the active site motif. The distance between Glu22 Oϵ1 and His71 Nδ1 of 4.15 ± 0.35 Å and of 2.98 ± 0.19 Å between Glu22 Oϵ2 and His71 Nδ1 of apo-MptpA is in good agreement to the observed distances of 3.71 ± 0.68 and 3.72 ± 0.80 Å in YxiE, the LMW-PTP of B. subtilis (27).
Dynamic Properties of MptpA
Relaxation analysis of MptpA revealed order parameters S2 of 0.7–0.9 for the rigid core residues of the protein, with the exception of the flexible loop regions and the termini. Mapping of the order parameter S2 onto the backbone structure reveals a few distinct regions and residues with different dynamical properties (Fig. 2C). The N- and C-terminal residues Met1, Ser2, Gly161, and Ser163 have low order parameters of S2 ≤ 0.3, indicating an unrestricted motion that is commonly observed at the termini of a protein (supplemental Table S1 and Fig. S3). Residue Asn14, located in the P-loop, has a high order parameter of S2 = 0.861 ± 0.005, exhibiting Asn14 as relatively rigid. Due to exchange broadening the P-loop, residues Cys11–Gly13 and Ile15–Ser18 were not detectable. The D-loop motif (Arg111–Gly131) contains two residues with a low order parameter, indicating that those are more flexible, Gly118 S2 = 0.662 ± 0.046 and Val124 S2 = 0.656 ± 0.004. Notably, the (1H)-15N hetNOE relaxation experiment (supplemental Table S1 and Fig. S3) reveals a high flexibility for the D-loop residues Arg116–Asp123. These residues are located in the D-loop region, which undergoes structural reorientations during ligand binding. Furthermore, residues Thr119–Ala121 as well as Glu125 and Pro127 located in the D-loop are not observable due to exchange broadening. Interestingly, residue Asp126, which is critical during dephosphorylation, has a high order parameter of ∼0.8, which indicates that this residue has a strict orientation.
Interaction with Phosphate Ions
Titration experiments of MptpA with Pi, as competitive inhibitor, resulted in a decreasing number of amide backbone resonances in the two-dimensional 1H,15N TROSY experiment. In the presence of 20 mm NaH2PO3, the resonances of six residues (Asn14, Cys16, Trp48, Asp90, Tyr128, and Tyr129) located in the P-loop and in spatial proximity to the phosphate binding motif (D- and W-loop) are no longer observable due to line broadening. Plotting CSPs as a function of residue number reveals five regions that are considerably affected by phosphate binding to the active site. These regions are located in the loop regions of the protein and coincide with the following residues: Asn14–Met24 (P-loop), Ala43–Glu56 (W-loop), Arg72–Gln75, Lys89 and Asp90, and Asp123–His150 (D-loop and hinge region in α5-helix). CSPs around Arg72–Gln75 can be explained by the proximity to the phosphate binding moiety of the P-loop. The occurrence of the changes in the chemical shift of Lys89 and Asp90 can be an indirect effect caused by changes in the chemical environment of Thr12, which is in close proximity. The phosphate titration experiments thus indicate the specific binding of phosphate to MptpA involving the functionally important P-, W-, and D-loop as well as other spatially related regions. Experiments in the presence of sulfate ions do not show comparable CSPs in the two-dimensional 1H,15N TROSY spectrum, further revealing the localized binding of phosphate ions to MptpA. The residues and regions involved in MptpA phosphate binding are comparable with the results of a phosphate titration reported for the TPTP LMW-PTP from T. foetus (68). However, in contrast to our observations for MptpA, phosphate binding to TPTP at pH 5.2 was reported to lead to the detection of amide backbone signals that were invisible in the absence of phosphate (68), indicating a slow exchange rate. Based on our experiments, which show broadening of signals, we propose that the binding of phosphate to MptpA at pH 7.0 can be described by an intermediate exchange rate.
Phosphorylation of MptpA by PtkA
Phosphorylation of MptpA Tyr128 and Tyr129 is catalyzed by the kinase PtkA, which was previously determined via TLC (14). We performed luciferase assays (Fig. 6) that confirmed the autophosphorylation activity expected for PtkA as well as the lack of autophosphorylation activity proposed for MptpA. Phosphorylation of MptpA Tyr128/Tyr129 was confirmed by 31P NMR spectroscopy as well as MALDI-MS (supplemental Figs. S4 and S5). The phosphorylation reaction did not, however, yield a sufficient amount of monophosphorylated MptpA for further investigations by multidimensional NMR spectroscopy. We therefore can only speculate that an intermolecular dephosphorylation of the monophosphorylated MptpA species occurs because the phosphorylation of one of the two adjacent tyrosine residues is proposed to increase the activity of the enzyme 25-fold (30), at least in other LMW-PTPs. Previous studies concerning the phosphorylation of human LMW-PTP by pp60v-src demonstrated that in the presence of phenylarsine oxide, a selective PTP inhibitor, the phosphorylation level is notably increased (70). This assumption suggests that the autodephosphorylation activity of MptpA is in accordance with the low yield of phosphorylated MptpA.
FIGURE 6.

Autophosphorylation activity monitored by Luciferase assay. Different enzyme concentrations (500 nm and 1 and 50 μm) were added to the assay buffer (300 mm NaCl, 50 mm Tris-HCl (pH 8.0), 10 mm DTT, and 10 mm MgCl2) containing 1, 5, 10, and 100 μm ATP. A, in the autophosphorylation assay for MptpA, no decrease in the amount of emitted light (relative light units; RLU) is observed. The comparison of the negative control (−MptpA; red line) with varying enzyme concentrations does not show the deviation associated with autophosphorylation. B, in the autophosphorylation assay for PtkA, the decrease in the amount of emitted light is associated with autophosphorylation activity of PtkA.
Mapping the Binding Interface of the MptpA-PtkA Complex by NMR Spectroscopy
The CSPs observed during NMR titration experiments of 15N-labeled MptpA with unlabeled PtkA (up to a molar ratio of 1:2) at T = 298 K and pH 7.0 were mapped on the solution structure of the apo-state MptpA. The overlay of the two-dimensional 1H,15N TROSY spectra of the apo-state MptpA and the MptpA-PtkA complex is shown in (Fig. 7A), showing a significant number of small CSPs. The assignment of the shifted resonances was obtained by tracing back the shifted resonances to the origin in the free form. All resonances could be assigned unambiguously. The backbone amides of MptpA with the largest changes (CSP ≥ 0.015 ppm) obtained upon binding of PtkA are Asn14, Cys16, Met20, Ala21, Asn47, Trp48, His49, Gly77, Thr78, Leu89, Asp90, Val124, Glu125, Asp126, Tyr129, Gly148, and His150. Furthermore, the amide resonances of Cys16 and Trp48 disappeared in the two-dimensional 1H,15N TROSY spectrum at a complex ratio of 1:2, whereas at a ratio of 1:1, the signals of Cys16 and Trp48 were still detectable. The corresponding one-dimensional 1H slices extracted from the two-dimensional 1H,15N TROSY spectrum of complex ratio 1:1 and 1:2 show the chemical shift changes as well as the loss of intensity. Mapping of the CSPs onto the structure reveals a distinct interface of MptpA (Fig. 7, B and C) comprising the P-, W-, and D-loop motifs. Four additional regions, including residues Met20, Gly77, and Asp90 as well as Ala145, can be detected. Contrary to all other residues, Met20 and Ala21 are not solvent-accessible in the apo-structure. The CSPs observed for residues Met20 and Ala21 are most likely caused by the interaction of the adjacent P-loop residues (Asn14 and Cys16) with PtkA. Hydrogen bond analysis indicates the interactions of His93 HN and Asp90 Oδ1, whereas structural analysis revealed the close contact between His93 Hϵ2 and Thr12 Oγ1. Thus, the observed CSPs in region Asp90 can be explained due to the close contact of Asp90, His93, and Thr12 and are therefore not directly caused by the interaction with PtkA. Because the structure and resonance assignment of PtkA are not yet available, a reciprocal experiment for mapping the binding site on the structure of PtkA could not be conducted.
FIGURE 7.
MptpA-PtkA interaction studied by NMR spectroscopy. A, NMR spectroscopy of complex titration. Overlay of two-dimensional 1H,15N TROSY spectrum of MptpA with and without PtkA. Blue, apo-MptpA; orange, MptpA-PtkA (end point of the titration experiment with ratio 1:2). The arrows highlight the trajectory of chemical shift changes. B, combined CSPs of backbone amide residues in ppm as a function of the MptpA residue number obtained via titration of MptpA with PtkA (1:2). Due to a loss of intensities, values of Cys16 and Trp48 are missing, as indicated by vertical dashed lines. C, mapping of CSPs on the MptpA solution structure. CSPs ≥ 0.015 ppm (orange) are taken into account for the mapping of the binding site of PtkA on the NMR solution structure of MptpA. Important motifs (P-, W-, and D-loop) are labeled.
DISCUSSION
All known classical PTPs, vaccinia virus H1-like dual specific phosphatases, and the LMW-PTPs share a common active site motif that is located in a crevice on the molecular surface. The signature motif (H/V)CX5R(S/T) (P-loop) binds not only phosphorylated protein substrates but also oxyanions, including phosphate, tungstate, or sulfate. The active site motif is flanked by the D-loop, containing the catalytic aspartate residue, which is opposite to the nucleophilic cysteine residue. The superposition of the ligand-free and ligand-bound structures of HMW-PTPs has provided insight into the movement of the highly conserved WPD (Trp-Pro-Asp)-loop toward the catalytic center covering the active site like a “flap.” In the ligand-bound state, the aspartate residue is pointing toward the bound oxyanion and donates a proton to the tyrosine leaving group. The tungstate-bound state of the Yersinia HMW-PTP describes a movement of the aspartate (Asp356) by 6 Å toward the active site, thus positioning the carboxylate in spatial proximity to the oxyanion oxygen (19). This conformational change has also been described in the human PTP1B C215S mutant during the binding of phosphotyrosine, bringing the aspartate (Asp181) into the catalytic active site (71). The ligand-induced loop closure observed for HMW-PTPs has thus far been assumed to be operative for LMW-PTPs as well.
However, up to now, the structural characterization of apo-LMW-PTPs has not been reported, and a conclusive statement about the movement of the loop containing the catalytic aspartate residue therefore could not be made. In previous studies, the structural characteristics of LMW-PTPs in solution were exclusively described in complex with inorganic phosphate (27, 68, 72, 73), which serves as a competitive inhibitor and stabilizes the conformational plasticity of the phosphate binding loop (P-loop). Here, we present the solution structure of a LMW-PTP in its apo-state. Fig. 3, A and B, demonstrates the spatial differences of the D-loop orientation between our apo-MptpA solution structure (PDB entry 2LUO) and the x-ray structure (PDB entry 1U2P) of MptpA in complex with chloride. The substrate-binding pocket (P-loop) is flanked by the D-loop containing Asp126 and Tyr128/Tyr129. The difference in distance between Asp126 Cα and Arg17 Cα in the chloride-bound and apo-MptpA is about 3 Å. Therefore, a movement of the D-loop similar to the one observed in HMW-PTPs is also observed in LMW-PTPs. Furthermore, the distances from the active site residue Arg17 Cα to the Tyr128 and Tyr129 Cα backbone carbons are about 3 and 5 Å larger, respectively, in apo-MptpA than in holo-MptpA. Besides the D-loop, the W-loop containing Trp48, which is important for substrate specificity, also flanks the active site and is supposed to interact with substrates. The indole side chain of Trp48 reorients upon ligand binding and therefore modulates the surface of the binding pocket between the apo- and the holo-state. The distance between Trp48 Nϵ1 (W-loop) and Tyr128 Oη (D-loop) in apo-MptpA is significantly increased as compared to the holo-structure (a displacement of nearly 13 Å). The concerted movement of the D-loop and residue Trp48 results in a more open conformation in the absence of ligands. Examination of LMW-PTPs from other species in complex with phosphate also reveals this deviation in the D-loop conformation. On the basis of our first apo-structure of a LMW-PTP, we propose comparable D-loop dynamics during ligand binding as observed for the Yersinia PTP.
The conformational dynamics of kinases are very pronounced in solution (74–77), and therefore it is interesting to compare the structural dynamics of our LMW-PTP with those dynamics observed for the catalytic domains of other kinases. In particular, the dynamics of the so-called DFG (Asp-Phe-Gly)-loop have been exploited to generate very potent new classes of inhibitors that are non-ATP-competitive. In the apo-state of the protein, the DFG-loop is in equilibrium between the DFG-in and -out state (74). Ligands known to bind the DFG-in conformation do not influence this equilibrium, whereas DFG-out ligands shift the equilibrium to the DFG-out conformation. The observation of similar dynamics and large conformational rearrangements upon substrate binding for phosphatases might open a strategy where the unique situation of the two activity-related adjacent tyrosine residues pointing away from the binding pocket could be utilized for drug targeting. Thanks to the structure solved here, the approach of designing inhibitors for different binding modes can now also be used for LMW-PTPs. Because the development of tight binding inhibitors for LMW-PTPs based on the ligand-bound structures has not yet resulted in potential drug candidates (69), the ligand-free structure of MptpA might now lead to a more successful design of potent inhibitors.
Furthermore, in order to design potential inhibitors, the structure-function relationship has to be taken into account, by employing strategies that influence the activation status of a potential drug target. LMW-PTPs are regulated by different mechanisms: (i) phosphate binding, (ii) oxidation of catalytic active cysteine residues, and (iii) phosphorylation. While binding of phosphate to the phosphatase leads to a non-covalent inactivation, it at the same time protects the enzyme against oxidation of the nucleophilic cysteine by reactive oxygen species. The phosphorylation of tyrosine residues located in the D-loop is of high importance for the regulation as observed for the human LMW-PTP HCPTP-A. As shown in vitro, phosphorylation of HCPTP-A Tyr131 by pp60v-src increases the enzyme activity 25-fold. However, the direct phosphorylation of active HCPTP-A did not lead to detectable yields of phosphorylated Tyr131. In order to induce such phosphorylation, the inhibitor phenylarsine oxide was added to the reaction mixture and resulted in inactivated HCPTP-A. In these experiments, autodephosphorylation was inhibited, and Tyr131-phosphorylated HCPTP-A could be detected (70). Previous studies have shown that the residues Tyr128 and Tyr129 of MptpA are phosphorylated by the kinase PtkA (14). Based on evidence for the HCPTP-A LMW-PTP, it can be assumed that phosphorylation of MptpA Tyr128/Tyr129 increases the enzyme activity considerably. NMR titration experiments, provided here, support the previously assumed (14) protein-protein complex and resolve the binding surface with atomic resolution. The observed binding sites in the apo-state solution structure of MptpA involve the P-, W-, and D-loop as well as two additional adjacent regions. Because the mechanism of MptpA secretion into the host macrophages remains unclear, it might be of crucial interest to elucidate the consequences of phosphorylation driven by PtkA. Furthermore, the inhibition of the interaction between MptpA and PtkA might serve as a new strategy for the design of potential drug candidates by developing inhibitors of this protein-protein complex vital for pathogen survival.
In summary, our findings reveal the first apo-structure of an LMW-PTP elucidating the rearrangement of the D-loop upon ligand binding. The apo-structure can be described as more open compared with the structures in complex with Pi or ligand mimetic, which seem to have a more closed conformation. We were able to map the binding site of PtkA on MptpA, elucidating the protein-protein interaction. Knowledge of the binding site for PtkA and the apo-structure of MptpA leads to further understanding of the regulation and ligand binding behavior of MptpA, which might serve as a basis for the successful design of new anti-tuberculosis therapeutics.
Acknowledgments
We thank Prof. Av-Gay for providing the PtkA plasmid vector. In addition, we thank Prof. Peter Güntert for passing on the latest software package of CYANA version 3.9 to us. We thank Dr. Denis Kudlinzki, Robert Silvers, and Jochen Stehle for inspiring discussions.
This work was supported by the European Union Commission in the program WeNMR (to H. J. and T. S.). The Center for Biomolecular Magnetic Resonance (BMRZ) is supported by the state of Hesse.

This article contains supplemental Table S1 and Figs. S1–S5.
The atomic coordinates and structure factors (code 2LUO) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The resonance-specific assignment of MptpA was updated and deposited in the Biological Magnetic Resonance Data Bank with the accession code 18533.
- PTP
- protein-tyrosine phosphatase
- HMW- and LMW-PTP
- high and low molecular weight PTP, respectively
- PtkA
- protein-tyrosine kinase A
- MptpA and -B
- M. tuberculosis protein-tyrosine phosphatase A and B, respectively
- hetNOE
- heteronuclear NOE
- TEV
- tobacco etch virus
- TOCSY
- total correlation spectroscopy
- RMSD
- root mean square deviation
- TROSY
- transverse relaxation-optimized spectroscopy
- CSP
- chemical shift perturbation
- Ni-NTA
- nickel-nitrilotriacetic acid
- PDB
- Protein Data Bank.
REFERENCES
- 1. World Health Organization (2012) 2011/2012 Tuberculosis Global Facts, World Health Organization, Geneva [Google Scholar]
- 2. De Cock K. M., Chaisson R. E. (1999) Will DOTS do it? A reappraisal of tuberculosis control in countries with high rates of HIV infection. Int. J. Tuberc. Lung Dis. 3, 457–465 [PubMed] [Google Scholar]
- 3. Koul A., Arnoult E., Lounis N., Guillemont J., Andries K. (2011) The challenge of new drug discovery for tuberculosis. Nature 469, 483–490 [DOI] [PubMed] [Google Scholar]
- 4. Clatworthy A. E., Pierson E., Hung D. T. (2007) Targeting virulence. A new paradigm for antimicrobial therapy. Nat. Chem. Biol. 3, 541–548 [DOI] [PubMed] [Google Scholar]
- 5. Cozzone A. J., Grangeasse C., Doublet P., Duclos B. (2004) Protein phosphorylation on tyrosine in bacteria. Arch. Microbiol. 181, 171–181 [DOI] [PubMed] [Google Scholar]
- 6. Koul A., Choidas A., Treder M., Tyagi A. K., Drlica K., Singh Y., Ullrich A. (2000) Cloning and characterization of secretory tyrosine phosphatases of Mycobacterium tuberculosis. J. Bacteriol. 182, 5425–5432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chao J., Wong D., Zheng X., Poirier V., Bach H., Hmama Z., Av-Gay Y. (2010) Protein kinase and phosphatase signaling in Mycobacterium tuberculosis physiology and pathogenesis. Biochim. Biophys. Acta 1804, 620–627 [DOI] [PubMed] [Google Scholar]
- 8. DeVinney R., Steele-Mortimer O., Finlay B. B. (2000) Phosphatases and kinases delivered to the host cell by bacterial pathogens. Trends Microbiol. 8, 29–33 [DOI] [PubMed] [Google Scholar]
- 9. Whitmore S. E., Lamont R. J. (2012) Tyrosine phosphorylation and bacterial virulence. Int. J. Oral Sci. 4, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cowley S. C., Babakaiff R., Av-Gay Y. (2002) Expression and localization of the Mycobacterium tuberculosis protein tyrosine phosphatase PtpA. Res. Microbiol. 153, 233–241 [DOI] [PubMed] [Google Scholar]
- 11. Bach H., Papavinasasundaram K. G., Wong D., Hmama Z., Av-Gay Y. (2008) Mycobacterium tuberculosis virulence is mediated by PtpA dephosphorylation of human vacuolar protein sorting 33B. Cell Host Microbe 3, 316–322 [DOI] [PubMed] [Google Scholar]
- 12. Armstrong J. A., Hart P. D. (1971) Response of cultured macrophages to Mycobacterium tuberculosis, with observations on fusion of lysosomes with phagosomes. J. Exp. Med. 134, 713–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wong D., Bach H., Sun J., Hmama Z., Av-Gay Y. (2011) Mycobacterium tuberculosis protein-tyrosine phosphatase (PtpA) excludes host vacuolar H+-ATPase to inhibit phagosome acidification. Proc. Natl. Acad. Sci. U.S.A. 108, 19371–19376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bach H., Wong D., Av-Gay Y. (2009) Mycobacterium tuberculosis PtkA is a novel protein tyrosine kinase whose substrate is PtpA. Biochem. J. 420, 155–160 [DOI] [PubMed] [Google Scholar]
- 15. Barford D., Jia Z., Tonks N. K. (1995) Protein tyrosine phosphatases take off. Nat. Struct. Biol. 2, 1043–1053 [DOI] [PubMed] [Google Scholar]
- 16. Zhang Z. Y. (1998) Protein-tyrosine phosphatases. Biological function, structural characteristics, and mechanism of catalysis. Crit. Rev. Biochem. Mol. Biol. 33, 1–52 [DOI] [PubMed] [Google Scholar]
- 17. Zhang Z. Y. (1997) Structure, mechanism, and specificity of protein-tyrosine phosphatases. Curr. Top. Cell Regul. 35, 21–68 [DOI] [PubMed] [Google Scholar]
- 18. Schubert H. L., Fauman E. B., Stuckey J. A., Dixon J. E., Saper M. A. (1995) A ligand-induced conformational change in the Yersinia protein-tyrosine phosphatase. Protein Sci. 4, 1904–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stuckey J. A., Schubert H. L., Fauman E. B., Zhang Z. Y., Dixon J. E., Saper M. A. (1994) Crystal structure of Yersinia protein-tyrosine phosphatase at 2.5 Å and the complex with tungstate. Nature 370, 571–575 [DOI] [PubMed] [Google Scholar]
- 20. Akerud T., Thulin E., Van Etten R. L., Akke M. (2002) Intramolecular dynamics of low molecular weight protein-tyrosine phosphatase in monomer-dimer equilibrium studied by NMR. A model for changes in dynamics upon target binding. J. Mol. Biol. 322, 137–152 [DOI] [PubMed] [Google Scholar]
- 21. Wang S., Tabernero L., Zhang M., Harms E., Van Etten R. L., Stauffacher C. V. (2000) Crystal structures of a low molecular weight protein-tyrosine phosphatase from Saccharomyces cerevisiae and its complex with the substrate p-nitrophenyl phosphate. Biochemistry 39, 1903–1914 [DOI] [PubMed] [Google Scholar]
- 22. Zhang M., Van Etten R. L., Stauffacher C. V. (1994) Crystal structure of bovine heart phosphotyrosyl phosphatase at 2.2 Å resolution. Biochemistry 33, 11097–11105 [DOI] [PubMed] [Google Scholar]
- 23. Zhang M., Zhou M., Van Etten R. L., Stauffacher C. V. (1997) Crystal structure of bovine low molecular weight phosphotyrosyl phosphatase complexed with the transition state analog vanadate. Biochemistry 36, 15–23 [DOI] [PubMed] [Google Scholar]
- 24. Zhang M., Stauffacher C. V., Lin D., Van Etten R. L. (1998) Crystal structure of a human low molecular weight phosphotyrosyl phosphatase. Implications for substrate specificity. J. Biol. Chem. 273, 21714–21720 [DOI] [PubMed] [Google Scholar]
- 25. Zabell A. P., Schroff A. D., Jr., Bain B. E., Van Etten R. L., Wiest O., Stauffacher C. V. (2006) Crystal structure of the human B-form low molecular weight phosphotyrosyl phosphatase at 1.6 Å resolution. J. Biol. Chem. 281, 6520–6527 [DOI] [PubMed] [Google Scholar]
- 26. Su X. D., Taddei N., Stefani M., Ramponi G., Nordlund P. (1994) The crystal structure of a low molecular weight phosphotyrosine protein phosphatase. Nature 370, 575–578 [DOI] [PubMed] [Google Scholar]
- 27. Xu H., Xia B., Jin C. (2006) Solution structure of a low molecular weight protein tyrosine phosphatase from Bacillus subtilis. J. Bacteriol. 188, 1509–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Madhurantakam C., Rajakumara E., Mazumdar P. A., Saha B., Mitra D., Wiker H. G., Sankaranarayanan R., Das A. K. (2005) Crystal structure of low molecular weight protein-tyrosine phosphatase from Mycobacterium tuberculosis at 1.9 Å resolution. J. Bacteriol. 187, 2175–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tailor P., Gilman J., Williams S., Couture C., Mustelin T. (1997) Regulation of the low molecular weight phosphotyrosine phosphatase by phosphorylation at tyrosines 131 and 132. J. Biol. Chem. 272, 5371–5374 [DOI] [PubMed] [Google Scholar]
- 30. Bucciantini M., Chiarugi P., Cirri P., Taddei L., Stefani M., Raugei G., Nordlund P., Ramponi G. (1999) The low Mr phosphotyrosine protein phosphatase behaves differently when phosphorylated at Tyr131 or Tyr132 by Src kinase. FEBS Lett. 456, 73–78 [DOI] [PubMed] [Google Scholar]
- 31. Schwarzer D., Zhang Z., Zheng W., Cole P. A. (2006) Negative regulation of a protein-tyrosine phosphatase by tyrosine phosphorylation. J. Am. Chem. Soc. 128, 4192–4193 [DOI] [PubMed] [Google Scholar]
- 32. Sambrook J., Russel D. W. (2006) Molecular Cloning: A Laboratory Manual, 2nd Ed., p. 718, Cold spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 33. Studier F. W. (2005) Protein production by autoinduction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 34. Salzmann M., Pervushin K., Wider G., Senn H., Wüthrich K. (1998) TROSY in triple-resonance experiments. New perspectives for sequential NMR assignment of large proteins. Proc. Natl. Acad. Sci. U.S.A. 95, 13585–13590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Salzmann M., Wider G., Pervushin K., Senn H., Wüthrich K. (1999) TROSY-type triple-resonance experiments for sequential NMR assignments of large proteins. J. Am. Chem. Soc. 121, 844–848 [Google Scholar]
- 36. Saxena K., Elshorst B., Berk H., Betz M., Grimme S., Langer T., Pescatore B., Schieborr U., Vogtherr M., Schwalbe H. (2005) Backbone NMR assignment of the low molecular weight protein-tyrosine phosphatase (MPtpA) from Mycobacterium tuberculosis. J. Biomol. NMR 33, 136. [DOI] [PubMed] [Google Scholar]
- 37. Grzesiek S., Anglister J., Bax A. (1993) Correlation of backbone amide and aliphatic side-chain resonances in 13C/15N-enriched proteins by isotropic mixing of 13C magnetization. J. Magn. Reson. B 101, 114–119 [Google Scholar]
- 38. Logan T. M., Olejniczak E. T., Xu R. X., Fesik S. W. (1993) A general method for assigning NMR spectra of denatured proteins using 3D HC(CO)NH-TOCSY triple resonance experiments. J. Biomol. NMR 3, 225–231 [DOI] [PubMed] [Google Scholar]
- 39. Kay L. E., Xu G. Y., Singer A. U., Muhandiram D. R., Formankay J. D. (1993) A gradient-enhanced HCCH-TOCSY experiment for recording side-chain 1H and 13C correlations in H2O samples of proteins. J. Magn. Reson. B 101, 333–337 [Google Scholar]
- 40. Löhr F., Hänsel R., Rogov V. V., Dötsch V. (2007) Improved pulse sequences for sequence-specific assignment of aromatic proton resonances in proteins. J. Biomol. NMR 37, 205–224 [DOI] [PubMed] [Google Scholar]
- 41. Löhr F., Katsemi V., Betz M., Hartleib J., Rüterjans H. (2002) Sequence-specific assignment of histidine and tryptophan ring 1H, 13C and 15N resonances in 13C/15N- and 2H/13C/15N-labeled proteins. J. Biomol. NMR 22, 153–164 [DOI] [PubMed] [Google Scholar]
- 42. Löhr F., Rogov V. V., Shi M., Bernhard F., Dötsch V. (2005) Triple-resonance methods for complete resonance assignment of aromatic protons and directly bound heteronuclei in histidine and tryptophan residues. J. Biomol. NMR 32, 309–328 [DOI] [PubMed] [Google Scholar]
- 43. Pervushin K. V., Wider G., Riek R., Wüthrich K. (1999) The 3D NOESY-[1H,15N,1H]-ZQ-TROSY NMR experiment with diagonal peak suppression. Proc. Natl. Acad. Sci. U.S.A. 96, 9607–9612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pervushin K., Riek R., Wider G., Wüthrich K. (1998) Transverse relaxation-optimized spectroscopy (TROSY) for NMR studies of aromatic spin systems in 13C-labeled proteins. J. Am. Chem. Soc. 120, 6394–6400 [Google Scholar]
- 45. Dayie K. T., Wagner G. (1994) Relaxation-rate measurements for 15N-1H groups with pulsed-field gradients and preservation of coherence pathways. J. Magn. Reson. A 111, 121–126 [Google Scholar]
- 46. Farrow N. A., Muhandiram R., Singer A. U., Pascal S. M., Kay C. M., Gish G., Shoelson S. E., Pawson T., Forman-Kay J. D., Kay L. E. (1994) Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33, 5984–6003 [DOI] [PubMed] [Google Scholar]
- 47. Kay L. E., Nicholson L. K., Delaglio F., Bax A., Torchia D. A. (1992) Pulse sequences for removal of the effects of cross-correlation between dipolar and chemical shift anisotropy relaxation mechanisms on the measurement of heteronuclear T1 and T2 values in proteins. J. Magn. Reson. 97, 359–375 [Google Scholar]
- 48. Vuister G. W., Bax A. (1994) Measurement of four-bond HN-Hα J-couplings in staphylococcal nuclease. J. Biomol. NMR 4, 193–200 [DOI] [PubMed] [Google Scholar]
- 49. Vuister G. W., Bax A. (1993) Quantitative J correlation. A new approach for measuring homonuclear three-bond J(HNHα) coupling constants in 15N-enriched proteins. J. Am. Chem. Soc. 115, 7772–7777 [Google Scholar]
- 50. Ottiger M., Delaglio F., Bax A. (1998) Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J. Magn. Reson. 131, 373–378 [DOI] [PubMed] [Google Scholar]
- 51. Dosset P., Hus J. C., Blackledge M., Marion D. (2000) Efficient analysis of macromolecular rotational diffusion from heteronuclear relaxation data. J. Biomol. NMR 16, 23–28 [DOI] [PubMed] [Google Scholar]
- 52. Mandel A. M., Akke M., Palmer A. G. (1995) Backbone dynamics of Escherichia Coli ribonuclease HI. Correlations with structure and function in an active enzyme. J. Mol. Biol. 246, 144–163 [DOI] [PubMed] [Google Scholar]
- 53. Palmer A. G., Rance M., Wright P. E. (1991) Intramolecular motions of a zinc finger DNA-binding domain from Xfin characterized by proton-detected natural abundance C-12 heteronuclear NMR spectroscopy. J. Am. Chem. Soc. 113, 4371–4380 [Google Scholar]
- 54. López-Méndez B., Güntert P. (2006) Automated protein structure determination from NMR spectra. J. Am. Chem. Soc. 128, 13112–13122 [DOI] [PubMed] [Google Scholar]
- 55. Güntert P. (2003) Automated NMR protein structure calculation. Prog. Nucl. Magn. Reson. Spectrosc. 43, 105–125 [Google Scholar]
- 56. Güntert P. (2004) Automated NMR structure calculation with CYANA. Methods Mol. Biol. 278, 353–378 [DOI] [PubMed] [Google Scholar]
- 57. Linge J. P., O'Donoghue S. I., Nilges M. (2001) Automated assignment of ambiguous nuclear Overhauser effects with ARIA. Methods Enzymol. 339, 71–90 [DOI] [PubMed] [Google Scholar]
- 58. Linge J. P., Habeck M., Rieping W., Nilges M. (2003) ARIA. Automated NOE assignment and NMR structure calculation. Bioinformatics 19, 315–316 [DOI] [PubMed] [Google Scholar]
- 59. Goddard T. D., Kneller D. G. (2002) SPARKY, version 3.105, University of California, San Francisco [Google Scholar]
- 60. Cornilescu G., Delaglio F., Bax A. (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13, 289–302 [DOI] [PubMed] [Google Scholar]
- 61. Shen Y., Delaglio F., Cornilescu G., Bax A. (2009) TALOS+. A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography and NMR system. A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 63. Linge J. P., Williams M. A., Spronk C. A., Bonvin A. M., Nilges M. (2003) Refinement of protein structures in explicit solvent. Proteins 50, 496–506 [DOI] [PubMed] [Google Scholar]
- 64. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) PROCHECK. A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 65. Laskowski R. A., Rullmann J. A. C., MacArthur M. W., Kaptein R., Thornton J. M. (1996) AQUA and PROCHECK-NMR. Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 66. Doreleijers J. F., Vranken W. F., Schulte C., Markley J. L., Ulrich E. L., Vriend G., Vuister G. W. (2012) NRG-CING. Integrated validation reports of remediated experimental biomolecular NMR data and coordinates in wwPDB. Nucleic Acids Res. 40, D519–D524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gouet P., Courcelle E., Stuart D. I., Métoz F. (1999) ESPript. Multiple sequence alignments in PostScript. Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]
- 68. Gustafson C. L., Stauffacher C. V., Hallenga K., Van Etten R. L. (2005) Solution structure of the low molecular weight protein tyrosine phosphatase from Tritrichomonas foetus reveals a flexible phosphate binding loop. Protein Sci. 14, 2515–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Silva A. P., Tabernero L. (2010) New strategies in fighting TB. Targeting Mycobacterium tuberculosis-secreted phosphatases MptpA and MptpB. Future Med. Chem. 2, 1325–1337 [DOI] [PubMed] [Google Scholar]
- 70. Rigacci S., Degl'Innocenti D., Bucciantini M., Cirri P., Berti A., Ramponi G. (1996) pp60vsrc phosphorylates and activates low molecular weight phosphotyrosine-protein phosphatase. J. Biol. Chem. 271, 1278–1281 [DOI] [PubMed] [Google Scholar]
- 71. Barford D., Flint A. J., Tonks N. K. (1994) Crystal structure of human protein tyrosine phosphatase 1B. Science 263, 1397–1404 [PubMed] [Google Scholar]
- 72. Lescop E., Hu Y., Xu H., Hu W., Chen J., Xia B., Jin C. (2006) The solution structure of Escherichia coli Wzb reveals a novel substrate recognition mechanism of prokaryotic low molecular weight protein-tyrosine phosphatases. J. Biol. Chem. 281, 19570–19577 [DOI] [PubMed] [Google Scholar]
- 73. Tolkatchev D., Shaykhutdinov R., Xu P., Plamondon J., Watson D. C., Young N. M., Ni F. (2006) Three-dimensional structure and ligand interactions of the low molecular weight protein-tyrosine phosphatase from Campylobacter jejuni. Protein Sci. 15, 2381–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Vogtherr M., Saxena K., Hoelder S., Grimme S., Betz M., Schieborr U., Pescatore B., Robin M., Delarbre L., Langer T., Wendt K. U., Schwalbe H. (2006) NMR characterization of kinase p38 dynamics in free and ligand-bound forms. Angew. Chem. Int. Ed. Engl. 45, 993–997 [DOI] [PubMed] [Google Scholar]
- 75. Levinson N. M., Kuchment O., Shen K., Young M. A., Koldobskiy M., Karplus M., Cole P. A., Kuriyan J. (2006) A Src-like inactive conformation in the Abl tyrosine kinase domain. PLoS Biol. 4, e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Morgan D. O., De Bondt H. L. (1994) Protein kinase regulation. Insights from crystal structure analysis. Curr. Opin. Cell Biol. 6, 239–246 [DOI] [PubMed] [Google Scholar]
- 77. Berteotti A., Cavalli A., Branduardi D., Gervasio F. L., Recanatini M., Parrinello M. (2009) Protein conformational transitions. The closure mechanism of a kinase explored by atomistic simulations. J. Am. Chem. Soc. 131, 244–250 [DOI] [PubMed] [Google Scholar]