Abstract
Background
Amelogenesis imperfecta (AI) is an inherited disorder characterized by abnormal formation of tooth enamel. Although several genes responsible for AI have been reported, not all causative genes for human AI have been identified to date. AMI rat has been reported as an autosomal recessive mutant with hypoplastic AI isolated from a colony of stroke-prone spontaneously hypertensive rat strain, but the causative gene has not yet been clarified. Through a genetic screen, we identified the causative gene of autosomal recessive AI in AMI and analyzed its role in amelogenesis.
Methods
cDNA sequencing of possible AI-candidate genes so far identified using total RNA of day 6 AMI rat molars identified a novel responsible mutation in specificity protein 6 (Sp6). Genetic linkage analysis was performed between Sp6 and AI phenotype in AMI. To understand a role of SP6 in AI, we generated the transgenic rats harboring Sp6 transgene in AMI (Ami/Ami + Tg). Histological analyses were performed using the thin sections of control rats, AMI, and Ami/Ami + Tg incisors in maxillae, respectively.
Results
We found the novel genetic linkage between a 2-bp insertional mutation of Sp6 gene and the AI phenotype in AMI rats. The position of mutation was located in the coding region of Sp6, which caused frameshift mutation and disruption of the third zinc finger domain of SP6 with 11 cryptic amino acid residues and a stop codon. Transfection studies showed that the mutant protein can be translated and localized in the nucleus in the same manner as the wild-type SP6 protein. When we introduced the CMV promoter-driven wild-type Sp6 transgene into AMI rats, the SP6 protein was ectopically expressed in the maturation stage of ameloblasts associated with the extended maturation stage and the shortened reduced stage without any other phenotypical changes.
Conclusion
We propose the addition of Sp6 mutation as a new molecular diagnostic criterion for the autosomal recessive AI patients. Our findings expand the spectrum of genetic causes of autosomal recessive AI and sheds light on the molecular diagnosis for the classification of AI. Furthermore, tight regulation of the temporospatial expression of SP6 may have critical roles in completing amelogenesis.
Keywords: Amelogenesis imperfecta, AI, AMI, Animal disease model, Autosomal recessive, Sp6
Background
Amelogenesis imperfecta (AI) is a genetic disorder characterized by morphological and functional defects of tooth enamel formation. AI disease entity has been classified into four types: hypoplastic, hypocalcified, hypomaturation, and hypomaturation-hypoplastic with taurodontism based on the clinical symptoms and hereditary modes [1,2]. Several genes responsible for AI such as AMELXAMBNENAMMMP20KLK4DLX3WDR72FAM83H, and FAM20A have been so far reported [3-6]. Mutations of AMELX cause X chromosome-linked AI. Mutations of AMBNENAMDLX3, and FAM83H cause autosomal dominant AI. For autosomal recessive AI, mutations of ENAMMMP20KLK4, WDR72, and FAM20A have been demonstrated as shown in Table 1. However, not all causative genes for AI-related disease have been identified to date [5-8].
Table 1.
Causative genes of amelogenesis imperfecta
| Inheritance pattern | Gene | Human chromosome | Rat chromosome | Mutation in AMI | Reference in human |
|---|---|---|---|---|---|
| X-linked |
Amelx |
X |
X |
n.d.a |
[9-21] |
| Autosomal dominant |
Ambn |
4 |
14 |
n.d. |
n.d. |
| |
Enam |
4 |
14 |
no |
[14,22-27] |
| |
Dlx3 |
17 |
10 |
no |
[28] |
| |
Fam83h |
8 |
7 |
n.d. |
[29-34] |
| Autosomal recessive |
Enam |
4 |
14 |
no |
[35,36] |
| |
Mmp20 |
11 |
8 |
no |
[26,37-39] |
| |
Klk4 |
19 |
1 |
no |
[40] |
| |
Wdr72 |
15 |
8 |
no |
[41] |
| Fam20a | 17 | 10 | no | [3,4] |
an.d.: not determined.
Forward genetics using animal disease models is a powerful approach to identify novel pathogenic genes responsible for the disease, and to elucidate the molecular basis of gene functions. In this study, we focused on AI using a disease model, AMI, which has been reported as a spontaneous AI mutant rat with chalky white teeth isolated from a colony of stroke-prone spontaneously hypertensive rat (SHR-SP) strain [42] (Figure 1A). In general, the surface of incisor enamel (solely on the labial side) in rodent shows yellowish-brown color. The molecular mechanisms of pigmentation are not yet clear, however, it is observed to store the irons with ferritin in the ameloblasts at pigmentation stage [43]. The iron-ferritin complex is transferred to lysosomes for degradation. Then, metabolically processed iron is released from ameloblasts and deposited on the surface of enamel matrix as a yellowish-brown layer. The whitish appearance of incisors indicates these processes are not complete, in terms of the defects of ameloblast differentiation, and resulting the enamel defect. The original SHR-SP strain, hereafter referred to as WT strain, does not have any AI phenotype. The AI phenotype in AMI is hypoplastic, and inherited as an autosomal single recessive trait; however, the causative gene has not been determined yet [42]. In this study, we clarified the genetic causes of autosomal recessive AI in AMI, and dissected the possible function of SP6 in amelogenesis.
Figure 1.
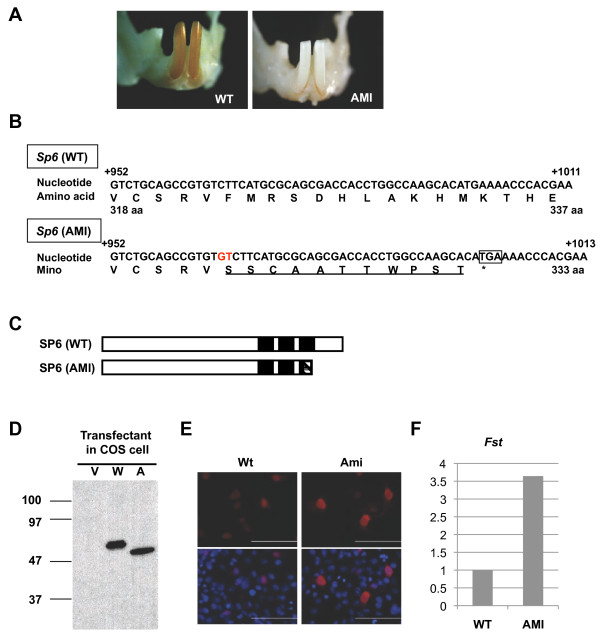
Identification of a 2-bp insertion inSp6 in AMI rats. A. Representative phenotype of WT and AMI rats. Photographs show mandibular incisors from each of the rats. B. The nucleotide sequence of Sp6 in wild-type (WT) and AMI rats. The 2-bp insertion is highlighted (red). The square represents the premature stop codon. The amino acid sequence deduced from the codons is shown below the nucleotide sequence. The altered amino acid sequence in Ami-SP6 is underlined. C. Schematic representation of SP6. The N-terminal region is located to the left. Black boxes represent the position of the zinc finger motifs. The box with diagonal lines shows the altered amino acid sequence in Ami-SP6. D. Western blot analyses. COS7 cells were transfected with an expression vector only (V) or with FLAG-tagged Wt- (W) or Ami- (A) cDNA. Samples were blotted with FLAG antibodies. E. Subcellular localization of FLAG-tagged SP6 by immunocytochemical analysis. Red, FLAG-tagged protein; Blue, Nuclei. F. Relative expression levels of Fst in WT and AMI molars.
Methods
Animals
Animal experiments were approved by the Ethics Committee for Animal Experiments of the University of Tokushima (No.06105). Both SHR-SP and AMI rats were supplied from Daiichi Seiyaku, Co., Ltd. and maintained at the animal facility in the University of Tokushima. To investigate whether the Sp6 transgene can rescue the AI phenotype in AMI rats, we crossed Sp6 transgenic (Tg) rats, which we generated previously [43], with AMI rats to generate a Sp6 transgene homozygous mutant (Ami/Ami + Tg), and used for further analyses.
cDNA sequencing of AI candidate genes andSp6
The coding regions for Sp6 and other candidate genes were amplified from molar cDNA samples from WT and AMI rats by PCR using the primers shown in Table 2 with KOD plus DNA polymerase (TOYOBO, Osaka, Japan) or PrimeSTAR HS DNA polymerase (Takara, Shiga, Japan). PCR products were cloned into the pGEM-T easy vector (Promega, Madison, WI). The sequences of the products were analyzed with an ABI PRISM 3100-Avant Genetic Analyzer (Applied Biosystems, Foster City, CA).
Table 2.
Gene-specific primers for coding regions
| Gene | Primer sequence |
|---|---|
|
Dlx3 |
5′-CCAGCATGAGCGGCTCCTTCGATCGCAAGC-3′ |
| |
5′-GGTACTCAGTACACAGCCCCAGGGTTAGGC-3′ |
|
Dlx4 |
5′-CCGCAATGACCTCTTTACCCTGTCCCCTTCC-3′ |
| |
5′-CAGACTCACATCCTCTGAGGCAGCGCCAGC-3′ |
|
Enam |
5′-AATAAATGTGTCTTGCTCCTTGGCTCTCTG-3′ |
| |
5′- AATACCTAAGCCTGAAGCAGTAAACAGCCG-3′ |
|
Fam20a |
5′-GGGCCATGCCCGGACTGCGCAGGGACCGCC-3′ |
| |
5′-TTGCTGCCGTTAGCTTGTCAGATTAGCCTG-3′ |
|
Klk4 |
5′-CCAATATGATGGTCACTGCACGAACCCCC-3′ |
| |
5′-CAGAGCTATCTTGTCTGAATGGTGGTCCAG-3′ |
|
Mmp20 |
5′-AGGAGATGAAGGTGCTACCTGCCTCTGGCC-3′ |
| |
5′-ACGATTCAGCAACCAACCCAGGAGCTGG-3′ |
|
Ngfr |
5′-GTGCAATGAGGAGGGCAGGTGCTGCCTGCAG-3′ |
| |
5′-TGAGTTCACACTGGGGATGTGGCAGTGGAC-3′ |
|
Sp2 |
5′-GCAACATGACGCTCACCCTGCCACTCAACAACC-3′ |
| |
5′-TGGCCTTACAAGCCCTTCGTGCCTAGGTGGGTC-3′ |
|
Sp6 |
5′-CCGGCAATGCTAACCGCTGTCTGTG-3′ |
| |
5′-GGCTCAGTTGGAGGACGCCGAGCTG-3′ |
|
aWdr72(a) |
5′-CTAACATGAGGAGTGCTCTGCAGGCTGTGG-3′ |
| |
5′-TGCGCATCATCGCCGCAGTT-3′ |
|
aWdr72(b) |
5′-GCGTGCCCGGAAGCACCTTT-3′ |
| 5′-TCGGGACTCCATCCATCTAGCAC-3′ |
aWdr72: We divided the coding region into two fragments (a and b) for cloning.
Genotyping
Genomic DNA was isolated from the tail tips of the rats and subjected to PCR analysis using the following four primers in a single tube. For WT Sp6 amplification, WT-Sp6.F-CTT (5′-GTC TGC AGC CGT GTC TT-3′), rSp6.genomeR (5′-CTG GCA GCC TAA ATA ATA TTC AAG CAG-3′), GAPDH-S (5′-CAT TGA CCT CAA CTA CAT GG-3′), and GAPDH-AS (5′-CTC AGT GTA GCC CAG GAT GC-3′) were used, and for AMI Sp6, AMI-Sp6.F (5′-GTC TGC AGC CGT GTG TC-3′) was used instead of WT-Sp6.F-CTT. Primer concentrations were as follows: Sp6, 0.75 mM; Gapdh, 0.25 mM. Sp6 (344 and 346 bp for WT and AMI, respectively) and Gapdh (722 bp) were amplified using a thermal cycler (94 °C for 4 min, 33 cycles of 94 °C for 30 s, 63 °C for 30 s, 72 °C for 30 s, and an extension step at 72 °C for 7 min) using Taq DNA polymerase (Promega).
Western blot analysis and immunocytochemistry
COS7 cells were transfected with a pCI-neo mammalian expression vector (Promega) carrying FLAG-tagged rat Sp6 (WT or AMI) or a mock vector control. Immunodetection and immunocytochemical analysis were performed as described previously [44].
Immunohistochemistry
Immunohistochemical analysis of postnatal day 1 incisors was performed as described previously [44]. For longitudinal sections of adult incisors, craniofacial regions were isolated from adult rats after intracardiac perfusion with phosphate-buffered saline containing 5 U/ml heparin followed by fixation with 0.1 M phosphate buffer (pH 7.4) containing 4 % paraformaldehyde. Postfixation was performed by immersion of the dissected specimen into the same fixative for 2 h. Postfixed samples were decalcified in 10 % formic acid–4 % citric acid decalcification solution for 2 weeks or more at 4 °C. Decalcified maxillae were trimmed and divided into three pieces vertically to the longitudinal axis of the incisors, as described previously [43,45]. Each incisor segment was embedded in paraffin, and a series of longitudinal sections (6-μm thick) were prepared. The sections were treated with 3 % hydrogen peroxide in methanol for 20 min and boiled in antigen-unmasking solution (Vector, Burlingame, CA, USA) using a microwave. For immunostaining, samples were blocked with 3 % horse serum in phosphate-buffered saline (−), incubated with rabbit anti-rat SP6 antiserum [44] or normal rabbit serum as primary antibodies (1:400), and incubated with Histofine Simple Stain Rat MAX-PO (R) (Nichirei, Tokyo, Japan). DAB-buffer tablets (Merck, Darmstadt, Germany) were used to visualize signals. Sections were slightly cross-stained with hematoxylin and mounted in Entellan New (Merck).
Histological analysis
Deparaffinized longitudinal sections of the maxillary incisors (12 μm) were stained with hematoxylin for histological analysis. The ameloblast differentiation stages were determined from the apical to the incisal end of the incisors using a modification of Warshawsky and Smith’s classification [46]. Presecretory ameloblasts face pulp or dentin, and the secretory stage starts from the position where the enamel matrix appears. The maturation stage starts from the point where the ameloblasts rapidly begin to decrease in size. In the reduced stage, ameloblasts become cuboidal in shape without a cytoplasmic region above the nucleus. Images of the longitudinal sections of hematoxylin-stained incisors were captured under a light microscope connected to a CCD camera. The length of the ameloblast layer at each stage was measured using the ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Macroscopic observation
Incisors (7- to 9-week-old), digits, and whiskers (5- to 8-month-old) were recorded with a digital camera. The whiskers of newborn pups were observed under a MZ16 stereomicroscope (Leica Microsystems, Wetzlar, Germany).
Linkage analysis
Sixty-two heterozygous (Wt/Ami) and 56 homozygous (Ami/Ami) mutant rats were examined to determine the correlation between AI and Ami. The JoinMap 4 software (Kyazma, Wageningen, Netherlands) was used for genetic linkage analysis.
X-ray analysis
Radiographs of rat teeth were obtained using MCT-CB100MF (Hitachi Medico, Tokyo, Japan). The X-ray analysis system was operated at a 50 kV accelerating voltage with a 100 mA probe current.
Results and discussion
To explore the causative gene of AMI, we first examined the sequences of the coding region of all reported causative genes for autosomal recessive AI, such as Enam, Mmp20, Klk4, Wdr72, and Fam20a using total RNA of day 6 AMI rat molars and compared these with those of WT rats. We did not find any mutations among them (summarized in Table 1), suggesting that a novel gene may cause AI.
We next examined the Sp6 gene, which has been identified as a key player in tooth development [47,48]. SP6 is a member of the SP/KLF family of transcription factors, which have three tandem zinc finger domains of the classical C2H2 type at the C-terminal region [48,49]. Loss-of-function analyses demonstrated that deficiency of Sp6 in mice leads to malformation of teeth, hair, digits, and lungs [50,51]. In particular, tooth anomalies are observed including supernumerary teeth (hyperdontia), abnormal dentin crystal structure, and enamel agenesis [50,51]. In addition, our in vitro gain-of-function studies of SP6 identified several downstream target genes in the dental epithelial cell line [44,52]. These findings indicate that SP6 may play important roles in ameloblast differentiation. When we performed the sequence analysis of Sp6, we found a 2-bp insertion between the nucleotide positions 965 and 966 in the coding region of Sp6 in AMI rats (Figure 1B). This insertion caused a frameshift mutation with the addition of 11 cryptic amino acid residues from amino acid position 323 of SP6, disrupting the third zinc finger domain of SP6 (Figure 1C). To confirm whether the inserted Sp6 (Ami) could be translated into a protein in cells, we transduced a CMV promoter-driven FLAG-tagged Wt (wild type-Sp6) or FLAG-tagged Ami expression vector into COS7 cells. The anti-FLAG antibody could detect both the Wt and Ami-type SP6 protein on western blot analysis (Figure 1D). Furthermore, subcellular localization of Ami-SP6 was detected mainly in the nuclei and, to some extent, in the cytosol, similar to Wt-SP6 (Figure 1E). Furthermore, we examined the expression level of follistatin (Fst) mRNA in postnatal day 6 (d6) molars from WT and AMI rats, because we previously demonstrated that Fst is one of the SP6 downstream target genes [44]. Fst expression was higher in AMI molars than in WT molars (Figure 1F). These results demonstrated that Ami-SP6 can be translated and located in the same manner as Wt-SP6, but it has different effects on downstream target gene expression.
To determine whether the 2-bp insertion in Sp6 correlates with the AI phenotype in AMI rats, we performed a linkage analysis. F1 rats were backcrossed with AMI to obtain descendants carrying Ami either heterozygously or homozygously. Although the color of the incisors in the first backcross generation was yellowish brown when the rats were heterozygotes with both Wt and Ami, Ami homozygote rats had chalky white incisors (Figure 2B). The Sp6 genotypes in WT, AMI, and their F1 hybrids rats were analyzed by genomic PCR (Figure 2A), and we confirmed that all AMI rats were homozygous for Ami. Linkage analysis with 62 heterozygous (Wt/Ami) and 56 homozygous (Ami/Ami) mutant rats revealed that the AI phenotype and Ami homozygosity were strongly linked with a LOD score of 35.46.
Figure 2.
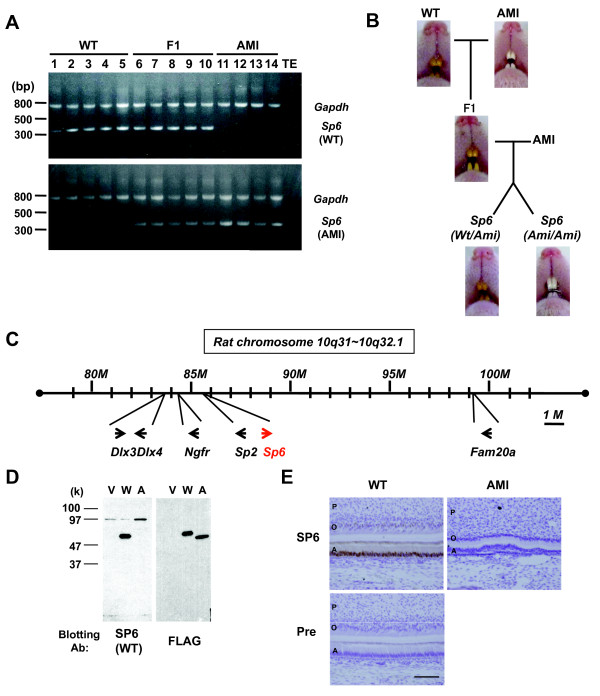
Linkage analysis betweenSp6 and AI in AMI rats. A. Genomic PCR products amplified by WT- (upper panel) or AMI- (lower panel) Sp6-specific primers. Sample numbers and the characteristics of the animals are denoted above the gel image. B. The color of the incisors was examined. Pups derived from the cross between F1 and AMI rats were sorted by the Sp6 genotype to determine the correlation between Sp6 and AI in AMI rats (Wt/Ami, n = 62; Ami/Ami, n = 56). C. Schematic diagram of focal gene localization in rat chromosome 10q31–10q32.1. D. Western blot analyses. COS7 cells were transfected with expression vector only (V) or with FLAG-tagged Wt- (W) or Ami- (A) cDNA. Samples were blotted with the indicated antibodies. E. Immunohistochemical analysis of incisors from newborn pups. Sections from secretory stage incisors of WT rats and those of the corresponding region from AMI rats were stained with the anti-Wt-SP6 antiserum (SP6) or preimmune rabbit serum (Pre). Signals were obtained from DAB (brown). Sections were counterstained with Hematoxylin. Scale bar: 100 mm. A, ameloblasts; O, odontoblasts; P, pulp.
Sp6 is located in rat chromosome 10q31, and this locus is well conserved in human chromosome 17q21. There are several tooth-related genes and family member genes within 2 Mb of Sp6 (Figure 2C). Sp2 is a member of the same family as Sp6[49]. Dlx3 has been reported as a causative gene for autosomal dominant AI [6], and Dlx4 is located next to Dlx3[53]. Ngfr, also known as p75NTR, has been reported to be important for early ameloblast differentiation [54]. Since we found a strong link between this locus and the AI phenotype associated with Ami, we further analyzed these genes by cDNA cloning and sequencing in order to eliminate candidate AI causative genes. We could not find any mutations in Sp2Dlx3Dlx4, or Ngfr genes using d6 AMI molar RNA as a result (data not shown).
In order to distinguish Wt-SP6 from Ami-SP6, we prepared rabbit polyclonal antibodies against specific peptides for Wt-SP6 (amino acid residues 362–376) [44]. The specificity of the antibodies was confirmed using cell lysates of COS7 cells transfected with FLAG-tagged Wt or FLAG-tagged Ami by western blot analysis (Figure 2D). Next, we examined endogenous SP6 localization in the mandibular incisors of WT and AMI newborn pups by immunohistochemical analysis. In WT incisors, SP6 was detected in the nuclei of early secretory ameloblasts and contralateral odontoblasts, whereas no signal was observed in AMI incisors (Figure 2E).
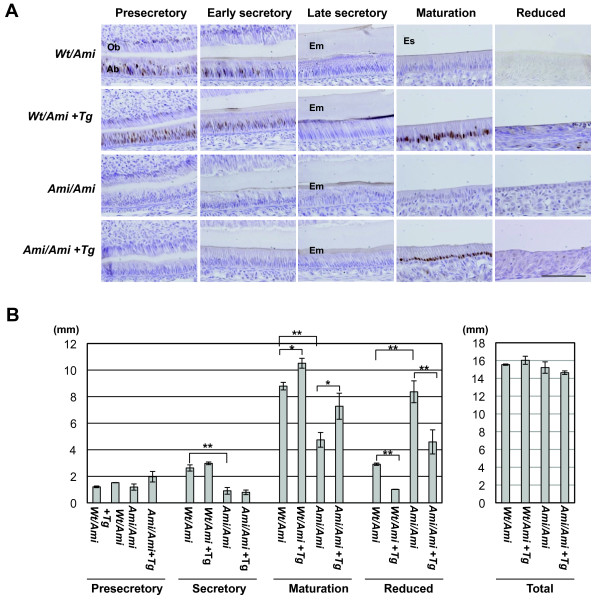
Ameloblast differentiation is morphologically classified into presecretory, secretory, and maturation stages [46,55]. In this classification, the reduced stage is treated as a substage of the maturation stage. In order to verify the functional relationship between AI and SP6 protein in detail, we divided ameloblast differentiation into five stages: presecretory; early secretory; late secretory; maturation; and reduced. We then analyzed the localization of the SP6 protein in the ameloblasts of maxillary incisors from 6-week-old rats (Figure 3A). In Wt/Ami rats, SP6 signals were strong in the presecretory and early secretory ameloblasts and weak in the contralateral odontoblasts. SP6 signals were not detected in late secretory, maturation, and reduced stage ameloblasts (Figure 3A, top panels). On the other hand, homozygotes with the mutant Sp6 (Ami/Ami) did not express the WT-SP6 protein at all, and defective ameloblast morphology was observed, such as shortened cells with immature polarization compared to the corresponding regions in Wt/Ami rats (Figure 3A, third panel).
Figure 3.
Analyses of longitudinal sections of the maxillary incisors of 6-week-old rats. A. Immunohistochemical analysis. Incisor sections were immunostained with antiserum against rat Wt-SP6. The sections were prepared from rats heterozygous (Wt/Ami) or homozygous (Ami/Ami) for mutant Sp6. Animals were sorted based on the transgenic Sp6 (Tg) genotype. Scale bar, 100 mm. Ab, ameloblasts; Em, enamel matrix; Es, enamel space; Ob, odontoblasts. B. The length of the ameloblast layer for the indicated differentiation stages. Columns represent the average of two to three independent samples; bars indicate standard deviation. Statistical significance was evaluated by unpaired t tests for an indicated set of data. *p < 0.05, **p < 0.01.
To investigate whether the Sp6 transgene can rescue the AI phenotype in AMI rats, we then crossed Sp6 transgenic (Tg) rats with AMI rats to generate a Sp6 transgene homozygous mutant (Ami/Ami + Tg). The Sp6 Tg rat model was recently established by the introduction of a human CMV immediate early enhancer/promoter-driven Sp6 coding sequence into Sprague–Dawley rats [43] Since tooth development is also driven by epithelial-mesenchymal interaction, we have used CMV enhancer/promoter to expect SP6 expression in both ameloblasts and odontoblasts. In Ami/Ami + Tg rats, the morphology of ameloblasts was similar to that observed in Wt/Ami rats from secretory to maturation stages (Figure 3A, fourth panel). The ameloblasts in Ami/Ami + Tg rats were aligned, well polarized, and their cellular height was restored to the levels similar to those in Wt/Ami rats (Figure 3A, top panels). However, tooth color, enamel calcification, and enamel matrix expression could not be rescued (data not shown). Subsequently, we examined SP6 expression in Ami/Ami + Tg rats. We could not detect any Wt-SP6 protein in the regions corresponding to the presecretory, secretory, and reduced stages in Ami/Ami + Tg rats, except for exclusive expression in maturation stage ameloblasts (Figure 3A, fourth panel). Consistent with these results, Wt-SP6 signals were detected in maturation stage ameloblasts, in addition to both presecretory and secretory ameloblasts, in the Sp6 transgene heterozygous mutants (Wt/Ami + Tg) (Figure 3A, second panel). One of the possible reasons for the failure to rescue the AI phenotype is the silencing of Sp6 transgene expression at the correct time and region because we recently found that SP6 is a short-lived protein (t1/2 ; 40 min) under the control of a proteasome pathway [52] and epigenetic mechanisms [56].
Next, we measured the length of the ameloblast layer at each differentiation stage using longitudinal sections of the maxillary incisors to determine whether there was a correlation between ameloblast differentiation and SP6 function (Figure 3B). In Ami/Ami incisors, the length of the ameloblast layer at the secretory and maturation stages was shorter, and the reduced stage was elongated compared with the Wt/Ami incisors (Figure 3B). In Wt/Ami + Tg and Ami/Ami + Tg rat incisors, we found that the length of the maturation stage ameloblast layer was longer than that of the non-Tg controls Wt/Ami and Ami/Ami, but the length of their reduced stage was much shorter compared with that of the same non-Tg controls. These results suggested that SP6 expression regulates cell morphology by an unknown mechanism. However, further analyses are necessary to confirm the molecular mechanism.
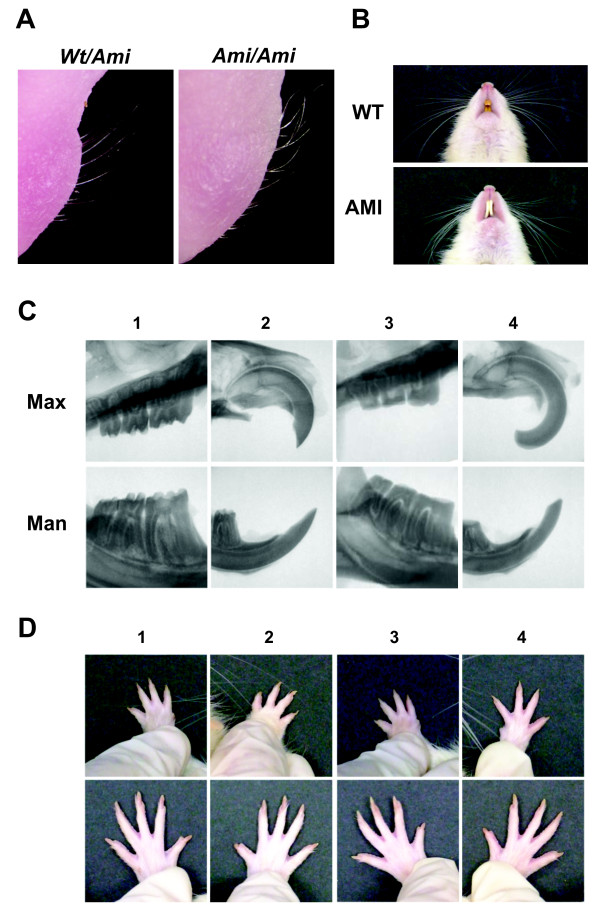
Sp6-deficient mice have several developmental abnormalities such as hyperdontia (supernumerary), enamel agenesis, oligodactyly, syndactyly, and hairless phenotypes [50,51,57]. Therefore, we carefully examined other organogenesis processes in AMI rats that required SP6. The whiskers were present, but curly, in newborn AMI rats, whereas they were straight in WT rats (Figure 4A). In adulthood, the whiskers were feeble in AMI rats, but not in WT rats (Figure 4B). The number of teeth and their structure in both WT and AMI rats were normal as judged by X-ray analyses, except for enamel formation in AMI rats (Figure 4C). Because of this enamel defect, the incisors of AMI showed less-sharp ends of incisor tips and elongation of incisor length due to impossible grinding and continuous growing.
Figure 4.
Organogenesis requiringSp6 activity. A. Whiskers of rats at postnatal day 1. Each Sp6 genotype in the 1st backcrossed generation was examined. (Wt/Ami, n = 13; Ami/Ami, n = 21) B. Whiskers of adult rats. WT and AMI rats were examined when they were 6–8 months old (WT, n = 3; AMI, n = 14) C. X-ray analysis of the maxillae (Max) and mandibles (Man) of WT (1, 2) and AMI (3, 4) rats. Molars (1, 3) and incisors (2, 4) are highlighted (WT, n = 2; AMI, n = 2) D. Normal number of digits in AMI rats. Digits of the left (1, 3) and right (2, 4) limbs of WT (1, 2) and AMI (3, 4) rats are shown. Upper panels, forelimb; lower panels, hindlimb (WT, n = 3; AMI, n = 14).
Furthermore, we observed normal numbers and morphology of the digits of both the forelimbs and hindlimbs of AMI and WT rats (Figure 4D). Basically, the AMI model shows only the AI phenotype. We summarized the phenotypic comparison between AMI rats and Sp6-deficient mice in Table 3.
Table 3.
Abnormalities in AMI rats andSp6-deficient mice
| Organ | Category |
AMI rats this study, [42,58] |
Sp6-deficient mice [50,51,57] |
|---|---|---|---|
| Teeth |
Number |
Normal |
Excess (hyperdontia) |
| |
Enamel |
Thin, calcified |
Absence or thin |
| |
Ameloblasts |
|
|
| |
- Nuclear |
Polarized |
Not well-polarized |
| |
-Tome’s process |
No |
Atypical |
| |
-Morphological differentiation |
Early regression |
Defect |
| Hair |
|
Normal |
Short, alopecia, no fur |
| Whisker |
Pups (at birth) |
Curly |
Rare, short, curly, alopecia |
| |
Adult |
Feeble |
Delayed appearance |
| Digits | Normal | Syndactyly, oligodactyly |
Recently, there was a notable report of a frameshift mutation of Sp7/Osterix in a patient that caused a similar disruption of the third zinc finger domain with additional amino acids and resulted in autosomal recessive osteogenesis imperfecta (OI) [59]. The known OI causative genes are restricted to type I collagen metabolic pathway, while Sp7/Osterix is a member of the SP/KLF family, which plays an essential role in osteoblast differentiation as a downstream target of RUNX2 [60]; therefore, it was an unexpected finding that a Sp7/Osterix mutation could cause OI. Whereas Sp7/Osterix-deficient mice could not survive [60], the OI phenotype of the frameshift mutant is far milder. Our study revealed a similar relationship between the Sp6 frameshift mutation and AI as that between the Sp7 frameshift mutation and OI. So far, there have been no other reports of the association between an Sp6 mutation and AI patients, but our findings could encourage additional surveys of patients with AI of an unknown cause, particularly an autosomal recessive type of AI.
SP6 has three tandem zinc finger domains of the classical C2H2 type at the C-terminal region, which is common among SP/KLF family members [48,49]. Each finger recognizes the specific nucleotide sequences and contributes to the binding strength to DNA. In addition, the finger plays a role of the interface with the regulatory protein(s) [61]. SP1 is a well-characterized member of the SP/KLF family, and all three zinc fingers are essential for binding to importin alpha and contribute to nuclear localization [62]. Here, we identified a novel frameshift mutation caused by a 2-bp insertion in the Sp6 coding sequence, resulting in disruption of the third zinc finger domain with an addition of an unrelated 11 amino acids (Figure 1B,C). This SP6 mutant form could enter the nuclei (Figure 1E). Our results indicate that the SP6 mutant form may have the following effects: 1) decrease in the affinity of DNA binding, and either loss or reduce of SP6 function as a transcription factor (Figure 1F); 2) decrease in the recruitment of interacting molecule(s), and a change in the gene expression pattern; 3) a novel recruitment or disappearance of protein–protein interactions through the cryptically added 11 amino acids; 4) a dominant-negative effect by chelating the regulatory domain interacting protein(s) at the N-terminal region of SP6; and 5) differential effects induced by the Sp6 antisense transcript. Further investigations are necessary to confirm the possible effects induced by the SP6 mutant and understand the molecular basis for amelogenesis.
Amelogenesis imperfecta is an inherited disorder of enamel formation with both clinical and genetic heterogeneity [1,2]. In many cases, the nomenclature of the inherited diseases has been developed based on the phenotypes and inheritance patterns. Therefore, it is difficult to distinguish the highly variable clinical phenotypes. Recently, molecular basis of these inherited diseases are reorganized based on the phenotype-genotype relationship. Online Mendelian Inheritance in Man (http://www.ncbi.nlm.nih.gov/omim) is a well-organized and updated genetic disease database. However, there is very limited description about the autosomal recessive form of hypoplastic AI in the section of amelogenesis imperfect. Here, we demonstrated the novel genetic linkage between SP6 frameshift mutation and autosomal recessive type of AI using a spontaneous AI animal model. We presents the evidence for a novel genotype–phenotype correlation of hereditary enamel defect, and it is encouraged to screen Sp6 gene in the of AI patients with unknown cause. Further studies on molecular basis of SP6 in amelogenesis could promote the development of the therapeutic application, such as gene therapy.
Conclusion
We propose the addition of Sp6 mutation in the AI classification as a new molecular criterion to properly diagnose and to apply it to development of therapeutic tools. Further analyses for the regulation of the temporospatial expression of SP6 and its function may clarify the mechanisms of AI disease and amelogenesis.
Abbreviations
AI: Amelogenesis Imperfecta; Fst: Follistatin; SHR-SP: Stroke-prone Spontaneously Hypertensive Rat; Sp6: Specificity protein 6; TG: Transgenic.
Competing interests
The authors declare no competing interests.
Authors’ contributions
TM maintained rats, contributed to the experimental design, performed genotyping, western blot analysis, immunohistochemistry, histological analyses, macroscopic analysis, linkage analysis, X-ray analysis, and the manuscript preparation. KM maintained rats, contributed to the coordination of study, performed cDNA sequencing, western blot analysis, immunocytochemistry, and the manuscript preparation. TH maintained rats, and TH and HH performed cDNA sequencing. TN contributed to the project design and data interpretation, coordination of study, and the manuscript preparation. All authors read and approved the final manuscript.
Contributor Information
Taro Muto, Email: muto@cdb.riken.jp.
Keiko Miyoshi, Email: miyoshi@dent.tokushima-u.ac.jp.
Taigo Horiguchi, Email: thoriguc@dent.tokushima-u.ac.jp.
Hiroko Hagita, Email: hagita-hiroko@dent.tokushima-u.ac.jp.
Takafumi Noma, Email: ntaka@tokushima-u.ac.jp.
Acknowledgments
This work was partly supported by grants-in-aid for scientific research (Nos. 18791368 and 21791789 to T.M.; 17689051 and 21791805 to K.M., 23592735 to T.N.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan and by a Research Grant from KAO Health Science Research (to T.N.).
References
- Crawford PJ, Aldred M, Bloch-Zupan A. Amelogenesis imperfecta. Orphanet J Rare Dis. 2007;2:17. doi: 10.1186/1750-1172-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkop CJ Jr. Amelogenesis imperfecta, dentinogenesis imperfecta and dentin dysplasia revisited: problems in classification. J Oral Pathol. 1988;17:547–553. doi: 10.1111/j.1600-0714.1988.tb01332.x. [DOI] [PubMed] [Google Scholar]
- Cho SH, Seymen F, Lee KE, Lee SK, Kweon YS, Kim KJ, Jung SE, Song SJ, Yildirim M, Bayram M, Tuna EB, Gencay K, Kim JW. Novel FAM20A mutations in hypoplastic amelogenesis imperfecta. Hum Mutat. 2011;33:91–94. doi: 10.1002/humu.21621. [DOI] [PubMed] [Google Scholar]
- O'Sullivan J, Bitu CC, Daly SB, Urquhart JE, Barron MJ, Bhaskar SS, Martelli-Junior H, dos Santos Neto PE, Mansilla MA, Murray JC, Coletta RD, Black GC, Dixon MJ. Whole-Exome sequencing identifies FAM20A mutations as a cause of amelogenesis imperfecta and gingival hyperplasia syndrome. Am J Hum Genet. 2011;88:616–620. doi: 10.1016/j.ajhg.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos MC, Hart PS, Ramaswami M, Kanno CM, Hart TC, Line SR. Exclusion of known gene for enamel development in two Brazilian families with amelogenesis imperfecta. Head Face Med. 2007;3:8. doi: 10.1186/1746-160X-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urzua B, Ortega-Pinto A, Morales-Bozo I, Rojas-Alcayaga G, Cifuentes V. Defining a new candidate gene for amelogenesis imperfecta: from molecular genetics to biochemistry. Biochem Genet. 2011;49:104–121. doi: 10.1007/s10528-010-9392-6. [DOI] [PubMed] [Google Scholar]
- Kim JW, Simmer JP, Lin BP, Seymen F, Bartlett JD, Hu JC. Mutational analysis of candidate genes in 24 amelogenesis imperfecta families. Eur J Oral Sci. 2006;114(Suppl 1):3–12. doi: 10.1111/j.1600-0722.2006.00278.x. [DOI] [PubMed] [Google Scholar]
- Wright JT, Torain M, Long K, Seow K, Crawford P, Aldred MJ, Hart PS, Hart TC. Amelogenesis imperfecta: genotype-phenotype studies in 71 families. Cells Tissues Organs. 2011;194:279–283. doi: 10.1159/000324339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldred MJ, Crawford PJ, Roberts E, Thomas NS. Identification of a nonsense mutation in the amelogenin gene (AMELX) in a family with X-linked amelogenesis imperfecta (AIH1) Hum Genet. 1992;90:413–416. doi: 10.1007/BF00220469. [DOI] [PubMed] [Google Scholar]
- Collier PM, Sauk JJ, Rosenbloom SJ, Yuan ZA, Gibson CW. An amelogenin gene defect associated with human X-linked amelogenesis imperfecta. Arch Oral Biol. 1997;42:235–242. doi: 10.1016/s0003-9969(96)00099-4. [DOI] [PubMed] [Google Scholar]
- Greene SR, Yuan ZA, Wright JT, Amjad H, Abrams WR, Buchanan JA, Trachtenberg DI, Gibson CW. A new frameshift mutation encoding a truncated amelogenin leads to X-linked amelogenesis imperfecta. Arch Oral Biol. 2002;47:211–217. doi: 10.1016/s0003-9969(01)00111-x. [DOI] [PubMed] [Google Scholar]
- Hart PS, Aldred MJ, Crawford PJ, Wright NJ, Hart TC, Wright JT. Amelogenesis imperfecta phenotype-genotype correlations with two amelogenin gene mutations. Arch Oral Biol. 2002;47:261–265. doi: 10.1016/s0003-9969(02)00003-1. [DOI] [PubMed] [Google Scholar]
- Hart S, Hart T, Gibson C, Wright JT. Mutational analysis of X-linked amelogenesis imperfecta in multiple families. Arch Oral Biol. 2000;45:79–86. doi: 10.1016/s0003-9969(99)00106-5. [DOI] [PubMed] [Google Scholar]
- Kida M, Ariga T, Shirakawa T, Oguchi H, Sakiyama Y. Autosomal-dominant hypoplastic form of amelogenesis imperfecta caused by an enamelin gene mutation at the exon-intron boundary. J Dent Res. 2002;81:738–742. doi: 10.1177/0810738. [DOI] [PubMed] [Google Scholar]
- Kim JW, Simmer JP, Hu YY, Lin BP, Boyd C, Wright JT, Yamada CJ, Rayes SK, Feigal RJ, Hu JC. Amelogenin p.M1T and p.W4S mutations underlying hypoplastic X-linked amelogenesis imperfecta. J Dent Res. 2004;83:78–383. doi: 10.1177/154405910408300505. [DOI] [PubMed] [Google Scholar]
- Kindelan SA, Brook AH, Gangemi L, Lench N, Wong FS, Fearne J, Jackson Z, Foster G, Stringer BM. Detection of a novel mutation in X-linked amelogenesis imperfecta. J Dent Res. 2000;79:1978–1982. doi: 10.1177/00220345000790120901. [DOI] [PubMed] [Google Scholar]
- Lagerstrom M, Dahl N, Nakahori Y, Nakagome Y, Backman B, Landegren U, Pettersson U. A deletion in the amelogenin gene (AMG) causes X-linked amelogenesis imperfecta (AIH1) Genomics. 1991;10:971–975. doi: 10.1016/0888-7543(91)90187-j. [DOI] [PubMed] [Google Scholar]
- Lagerstrom-Fermer M, Nilsson M, Backman B, Salido E, Shapiro L, Pettersson U, Landegren U. Amelogenin signal peptide mutation: correlation between mutations in the amelogenin gene (AMGX) and manifestations of X-linked amelogenesis imperfecta. Genomics. 1995;26:159–162. doi: 10.1016/0888-7543(95)80097-6. [DOI] [PubMed] [Google Scholar]
- Lench NJ, Brook AH, Winter GB. SSCP detection of a nonsense mutation in exon 5 of the amelogenin gene (AMGX) causing X-linked amelogenesis imperfecta (AIH1) Hum Mol Genet. 1994;3:827–828. doi: 10.1093/hmg/3.5.827. [DOI] [PubMed] [Google Scholar]
- Lench NJ, Winter GB. Characterisation of molecular defects in X-linked amelogenesis imperfecta (AIH1) Hum Mutat. 1995;5:251–259. doi: 10.1002/humu.1380050310. [DOI] [PubMed] [Google Scholar]
- Sekiguchi H, Tanakamaru H, Minaguchi K, Machida Y, Yakushiji M. A case of amelogenesis imperfecta of deciduous and all permanent teeth. Bull Tokyo Dent Coll. 2001;42:45–50. doi: 10.2209/tdcpublication.42.45. [DOI] [PubMed] [Google Scholar]
- Kang HY, Seymen F, Lee SK, Yildirim M, Tuna EB, Patir A, Lee KE, Kim JW. Candidate gene strategy reveals ENAM mutations. J Dent Res. 2009;88:266–269. doi: 10.1177/0022034509333180. [DOI] [PubMed] [Google Scholar]
- Gutierrez SJ, Chaves M, Torres DM, Briceno I. Identification of a novel mutation in the enamalin gene in a family with autosomal-dominant amelogenesis imperfecta. Arch Oral Biol. 2007;52:503–506. doi: 10.1016/j.archoralbio.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Kim JW, Seymen F, Lin BP, Kiziltan B, Gencay K, Simmer JP, Hu JC. ENAM mutations in autosomal-dominant amelogenesis imperfecta. J Dent Res. 2005;84:278–282. doi: 10.1177/154405910508400314. [DOI] [PubMed] [Google Scholar]
- Mardh CK, Backman B, Holmgren G, Hu JC, Simmer JP, Forsman-Semb K. A nonsense mutation in the enamelin gene causes local hypoplastic autosomal dominant amelogenesis imperfecta (AIH2) Hum Mol Genet. 2002;11:1069–1074. doi: 10.1093/hmg/11.9.1069. [DOI] [PubMed] [Google Scholar]
- Ozdemir D, Hart PS, Ryu OH, Choi SJ, Ozdemir-Karatas M, Firatli E, Piesco N, Hart TC. MMP20 active-site mutation in hypomaturation amelogenesis imperfecta. J Dent Res. 2005;84:1031–1035. doi: 10.1177/154405910508401112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajpar MH, Harley K, Laing C, Davies RM, Dixon MJ. Mutation of the gene encoding the enamel-specific protein, enamelin, causes autosomal-dominant amelogenesis imperfecta. Hum Mol Genet. 2001;10:1673–1677. doi: 10.1093/hmg/10.16.1673. [DOI] [PubMed] [Google Scholar]
- Dong J, Amor D, Aldred MJ, Gu T, Escamilla M, MacDougall M. DLX3 mutation associated with autosomal dominant amelogenesis imperfecta with taurodontism. Am J Med Genet A. 2005;133A:138–141. doi: 10.1002/ajmg.a.30521. [DOI] [PubMed] [Google Scholar]
- El-Sayed W, Shore RC, Parry DA, Inglehearn CF, Mighell AJ. Ultrastructural analyses of deciduous teeth affected by hypocalcified amelogenesis imperfecta from a family with a novel Y458X FAM83H nonsense mutation. Cells Tissues Organs. 2010;191:235–239. doi: 10.1159/000252801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart PS, Becerik S, Cogulu D, Emingil G, Ozdemir-Ozenen D, Han ST, Sulima PP, Firatli E, Hart TC. Novel FAM83H mutations in Turkish families with autosomal dominant hypocalcified amelogenesis imperfecta. Clin Genet. 2009;75:401–404. doi: 10.1111/j.1399-0004.2008.01112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun HK, Lee SK, Lee KE, Kang HY, Kim EJ, Choung PH, Kim JW. Identification of a novel FAM83H mutation and microhardness of an affected molar in autosomal dominant hypocalcified amelogenesis imperfecta. Int Endod J. 2009;42:1039–1043. doi: 10.1111/j.1365-2591.2009.01617.x. [DOI] [PubMed] [Google Scholar]
- Kim JW, Lee SK, Lee ZH, Park JC, Lee KE, Lee MH, Park JT, Seo BM, Hu JC, Simmer JP. FAM83H mutations in families with autosomal-dominant hypocalcified amelogenesis imperfecta. Am J Hum Genet. 2008;82:489–494. doi: 10.1016/j.ajhg.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Hu JC, Bartlett JD, Lee KE, Lin BP, Simmer JP, Kim JW. Mutational spectrum of FAM83H: the C-terminal portion is required for tooth enamel calcification. Hum Mutat. 2008;29:E95–99. doi: 10.1002/humu.20789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JT, Frazier-Bowers S, Simmons D, Alexander K, Crawford P, Han ST, Hart PS, Hart TC. Phenotypic variation in FAM83H-associated amelogenesis imperfecta. J Dent Res. 2009;88:356–360. doi: 10.1177/0022034509333822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart TC, Hart PS, Gorry MC, Michalec MD, Ryu OH, Uygur C, Ozdemir D, Firatli S, Aren G, Firatli E. Novel ENAM mutation responsible for autosomal recessive amelogenesis imperfecta and localised enamel defects. J Med Genet. 2003;40:900–906. doi: 10.1136/jmg.40.12.900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdemir D, Hart PS, Firatli E, Aren G, Ryu OH, Hart TC. Phenotype of ENAM mutations is dosage-dependent. J Dent Res. 2005;84:1036–1041. doi: 10.1177/154405910508401113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Simmer JP, Hart TC, Hart PS, Ramaswami MD, Bartlett JD, Hu JC. MMP-20 mutation in autosomal recessive pigmented hypomaturation amelogenesis imperfecta. J Med Genet. 2005;42:271–275. doi: 10.1136/jmg.2004.024505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Seymen F, Kang HY, Lee KE, Gencay K, Tuna B, Kim JW. MMP20 hemopexin domain mutation in amelogenesis imperfecta. J Dent Res. 2010;89:46–50. doi: 10.1177/0022034509352844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papagerakis P, Lin HK, Lee KY, Hu Y, Simmer JP, Bartlett JD, Hu JC. Premature stop codon in MMP20 causing amelogenesis imperfecta. J Dent Res. 2008;87:56–59. doi: 10.1177/154405910808700109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart PS, Hart TC, Michalec MD, Ryu OH, Simmons D, Hong S, Wright JT. Mutation in kallikrein 4 causes autosomal recessive hypomaturation amelogenesis imperfecta. J Med Genet. 2004;41:545–549. doi: 10.1136/jmg.2003.017657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sayed W, Parry DA, Shore RC, Ahmed M, Jafri H, Rashid Y, Al-Bahlani S, Al Harasi S, Kirkham J, Inglehearn CF, Mighell AJ. Mutations in the beta propeller WDR72 cause autosomal-recessive hypomaturation amelogenesis imperfecta. Am J Hum Genet. 2009;85:699–705. doi: 10.1016/j.ajhg.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi K, Iino T, Sekiguchi F. Amelogenesis imperfecta, a new dental mutation in rats. Lab Anim Sci. 1990;40:16–20. [PubMed] [Google Scholar]
- Muto T, Miyoshi K, Horiguchi T, Noma T. Dissection of morphological and metabolic differentiation of ameloblasts via ectopic SP6 expression. J Med Invest. 2012;59:59–68. doi: 10.2152/jmi.59.59. [DOI] [PubMed] [Google Scholar]
- Ruspita I, Miyoshi K, Muto T, Abe K, Horiguchi T, Noma T. Sp6 downregulation of follistatin gene expression in ameloblasts. J Med Invest. 2008;55:87–98. doi: 10.2152/jmi.55.87. [DOI] [PubMed] [Google Scholar]
- Smith CE. A method for preparing longitudinal semi-thin Epon sections of entire rat incisors. Arch Oral Biol. 1974;19:1045–1048. doi: 10.1016/0003-9969(74)90093-4. [DOI] [PubMed] [Google Scholar]
- Warshawsky H, Smith CE. Morphological classification of rat incisor ameloblasts. Anat Rec. 1974;179:423–446. doi: 10.1002/ar.1091790403. [DOI] [PubMed] [Google Scholar]
- Jimenez-Rojo L, Ibarretxe G, Aurrekoetxea M, de Vega S, Nakamura T, Yamada Y, Unda F. Epiprofin/Sp6: a new player in the regulation of tooth development. Histol Histopathol. 2010;25:1621–1630. doi: 10.14670/HH-25.1621. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Unda F, de-Vega S, Vilaxa A, Fukumoto S, Yamada KM Yamada Y. The Kruppel-like factor epiprofin is expressed by epithelium of developing teeth, hair follicles, and limb buds and promotes cell proliferation. J Biol Chem. 2004;279:626–634. doi: 10.1074/jbc.M307502200. [DOI] [PubMed] [Google Scholar]
- Suske G, Bruford E, Philipsen S. Mammalian SP/KLF transcription factors: bring in the family. Genomics. 2005;85:551–556. doi: 10.1016/j.ygeno.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Hertveldt V, Louryan S, van Reeth T, Dreze P, van Vooren P, Szpirer J, Szpirer C. The development of several organs and appendages is impaired in mice lacking Sp6. Dev Dyn. 2008;237:883–892. doi: 10.1002/dvdy.21355. [DOI] [PubMed] [Google Scholar]
- Nakamura T, de Vega S, Fukumoto S, Jimenez L, Unda F, Yamada Y. Transcription factor epiprofin is essential for tooth morphogenesis by regulating epithelial cell fate and tooth number. J Biol Chem. 2008;283:4825–4833. doi: 10.1074/jbc.M708388200. [DOI] [PubMed] [Google Scholar]
- Utami TW, Miyoshi K, Hagita H, Yanuaryska RD, Horiguchi T, Noma T. Possible linkage of SP6 transcriptional activity with amelogenesis by protein stabilization. J Biomed Biotechnol. 2011, Article ID 320987. [DOI] [PMC free article] [PubMed]
- Quinn LM, Johnson BV, Nicholl J, Sutherland GR, Kalionis B. Isolation and identification of homeobox genes from the human placenta including a novel member of the Distal-less family, DLX4. Gene. 1997;187:55–61. doi: 10.1016/s0378-1119(96)00706-8. [DOI] [PubMed] [Google Scholar]
- Mitsiadis TA, Couble P, Dicou E, Rudkin BB, Magloire H. Patterns of nerve growth factor (NGF), proNGF, and p75 NGF receptor expression in the rat incisor: comparison with expression in the molar. Differentiation. 1993;54:161–175. doi: 10.1111/j.1432-0436.1993.tb01599.x. [DOI] [PubMed] [Google Scholar]
- Kallenbach E. Fine structure of rat incisor enamel organ during late pigmentation and regression stages. J Ultrastruct Res. 1970;30:38–63. doi: 10.1016/s0022-5320(70)90063-8. [DOI] [PubMed] [Google Scholar]
- Utami TW, Miyoshi K, Hagita H, Yanuaryska RD, Horiguchi T, Noma T. Dynamic Changes of Sp6 Transgene Expression in Dental Epithelial Cells during Long-term Culture. Indones J Dent Res, . in press.
- Talamillo A, Delgado I, Nakamura T, de-Vega S, Yoshitomi Y, Unda F, Birchmeier W, Yamada Y, Ros MA. Role of Epiprofin, a zinc-finger transcription factor, in limb development. Dev Biol. 2009;337:363–374. doi: 10.1016/j.ydbio.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima T, Baba O, Takano Y, Ishibashi K, Oida S. Analysis of enamel formation in mutant rats with hypoplastic type of amelogenesis imperfeta. Dent Jpn. 2003;39:8–14. [Google Scholar]
- Lapunzina P, Aglan M, Temtamy S, Caparros-Martin JA, Valencia M, Leton R, Martinez-Glez V, Elhossini R, Amr K, Vilaboa N, Ruiz-Perez VL. Identification of a frameshift mutation in Osterix in a patient with recessive osteogenesis imperfecta. Am J Hum Genet. 2010;87:110–114. doi: 10.1016/j.ajhg.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, de Behringer RR, Crombrugghe B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- Ito T, Kitamura H, Uwatoko C, Azumano M, Itoh K, Kuwahara J. Interaction of Sp1 zinc finger with transport factor in the nuclear localization of transcription factor Sp1. Biochem Biophys Res Commun. 2010;403:161–166. doi: 10.1016/j.bbrc.2010.10.036. [DOI] [PubMed] [Google Scholar]