Abstract
Computational analyses of systematic measurements on the states and activities of signaling proteins (as captured by phosphoproteomic data, for example) have the potential to uncover uncharacterized protein-protein interactions and to identify the subset that are important for cellular response to specific biological stimuli. However, inferring mechanistically plausible protein signaling networks (PSNs) from phosphoproteomics data is a difficult task, owing in part to the lack of sufficiently comprehensive experimental measurements, the inherent limitations of network inference algorithms, and a lack of standards for assessing the accuracy of inferred PSNs. A case study in which 12 research groups inferred PSNs from a phosphoproteomics data set demonstrates an assessment of inferred PSNs on the basis of the accuracy of their predictions. The concurrent prediction of the same previously unreported signaling interactions by different participating teams suggests relevant validation experiments and establishes a framework for combining PSNs inferred by multiple research groups into a composite PSN. We conclude that crowdsourcing the construction of PSNs—that is, outsourcing the task to the interested community—may be an effective strategy for network inference.
Obstacles and Opportunities in Signaling Network Inference
The availability of antibodies that recognize phosphorylated residues on specific signaling proteins are the basis of an expanding number of quantitative phosphoproteomics assays (1–4). To use these quantitative protein phosphorylation data sets, approaches that infer the structure of signaling networks from protein state data have been developed (5–9). However, a number of technical difficulties conspire to derail network inference. Experimental studies that support current network inference procedures typically involve stimuli and perturbations to the system that are large in number from a practical perspective but that are barely sufficient from the perspective of network inference. Furthermore, many proteins (or phosphorylation or activity states) important for signaling networks remain unmeasured either because appropriate reagents (typically antibodies) are not available for their detection or because the proteins and modifications are not yet annotated. Existing inference algorithms also face the problem that they do not necessarily yield directly testable and mechanistically plausible inferences—for example, by confusing correlation with causation. Last, even if a protein signaling network (PSN) can be successfully reconstructed, it is not obvious how to quantify its accuracy. In most cases, the “most likely” network given the observed data is selected, but there is a degree of ambiguity in the definition of most likely. For example, we may say most likely with respect to the currently accepted network models, most likely given the data at hand, or most likely on the basis of an arbitrary cost function.
We describe some of the difficulties and successes encountered in assessing the accuracy of signaling-network inference and the specific strategies used in the DREAM4 Predictive Signaling Network Challenge. This challenge, which took place in the summer of 2009, was organized under the umbrella of the DREAM (Dialogue on Reverse Engineering Assessment and Methods) project (http://www.the-dream-project.org). Twelve groups participated in the task of predicting the response of human liver carcinoma cells (the HepG2 cell line) to different extracellular ligands (“stimuli”) in the presence or absence of several smallmolecule kinase inhibitors (“perturbations”). The data consisted of measuring the abundance of seven phosphoproteins by using sandwich immunoassays with the xMAP platform (Luminex, Austin, Texas).
Network Assessment by Accuracy of Predicted Interactions
One possible metric for assessing the accuracy of an inferred PSN is the “recall,” defined as the fraction of previously known biological interactions (or edges in network jargon) recovered by an algorithm. It is not uncommon to see claims that hundreds of predicted interactions are justified based on showing a few previously reported inter actions. This metric of inference accuracy is misleading because a completely connected network (one in which every node—or protein—is linked to every other protein) has perfect recall, but it is clearly inaccurate. A metric complementary to recall is “precision,” defined as the fraction of inferred edges that are correct (with respect to the current knowledge base). Assuming a complete and error-free prior knowledge base, a combination of precision and recall may be a good measure to quantify the accuracy of inferred networks (10). Unfortunately, the current knowledge base for signaling interactions is vastly incomplete (missing many nodes and edges) and contains an unknown number of incorrect edges. Moreover, current knowledge often lacks biological specificity, because the data from many different cellular contexts and organisms are frequently combined into single-network diagrams that are intended to represent the current state of information. Thus, the safest recourse for deciding whether an inferred edge that is absent from the knowledge base is a true positive or a false positive is to experimentally test each prediction. The number of validation experiments required to achieve this laudable goal is overwhelming, both in effort and in cost. Moreover, in many cases, network inference lacks the mechanistic detail required for unambiguous experimental validation. To take maximum advantage of the benefits of network inference from high-throughput data, we require metrics other than recall and precision to score an inferred PSN. Those networks with the highest scores should provide the basis for the most promising follow-up experiments for the discovery of new interactions or for the validation of known interactions.
Network Assessment by Accuracy of Quantitative Predictions
The metric adopted in the DREAM4 Predictive Signaling Network Challenge for assessing the accuracy of inferred PSNs was based on a comparison between the predicted and actual phosphoproteomics measurements.
Each team created a “model network” using a training data set (phosphoproteomics measurements observed under a specific set of conditions). Each team’s model networks related stimuli and measurement values using different mathematical formalisms, such as differential equations (7, 8) or Boolean logical functions (11). From the model network, each team predicted changes in the phosphoproteome that would occur in response to a different combination of stimuli and perturbations (the test data set). Last, the effectiveness of each model network was assessed by comparing the accuracy of the predicted measurements with those of the actual test data set.
This framework in which data from some experimental conditions are set aside for testing how well a fitted model generalizes to new data are common in the machine-learning community. The framework rewards predictive accuracy without regard to plausible biological mechanisms or interpretability of the predictive models as an interaction network. If mechanistic understanding is the ultimate goal, it is up to the researcher to work with a type of model that is interpretable in the form of an interaction network. For example, a weight matrix of interaction strengths between variables representing protein phosphorylation values can easily be interpreted as a network, whereas models that process data with kernel methods—which makes predictions after mapping the data into a higher, maybe infinite, dimensional feature space— or principal components analysis—which projects the data into a reduced dimensional feature space—may not necessarily be interpretable as a network.
We explored several metrics for assessing the accuracy of predicted measurements, including the sum of square errors both in linear and logarithmic scales, mismatches in temporal trends, and correlation measures. The actual assessment was made by using the residual sum of squares (RSS) normalized by the sum of the technical and biological variance (NRSS) as defined in Eq. 1:
| (1) |
The NRSS is calculated over the set of all the predicted phosphoproteomics values for each phosphoprotein. We estimated technical variance from the lower detection limit of the measurement apparatus, which is 300 arbitrary fluorescence units. We estimated the biological variance from the coefficient of variation, which was 0.08 in independent assays using the same xMAP measurement platform.
The NRSS makes no assumption about the validity of the computational model or the distribution of the differences between measured and predicted protein abundances. To estimate the significance of the NRSS achieved by the predictions for a given phosphoprotein, we simulated the empirical distribution of the NRSS under the null hypothesis that the predicted values are randomly sampled from the values in the training data set for that phosphoprotein. From the resulting empirical null distribution, a P value can be readily obtained for any realization of the NRSS.
Specifics of the DREAM4 Predictive Signaling Network Challenge
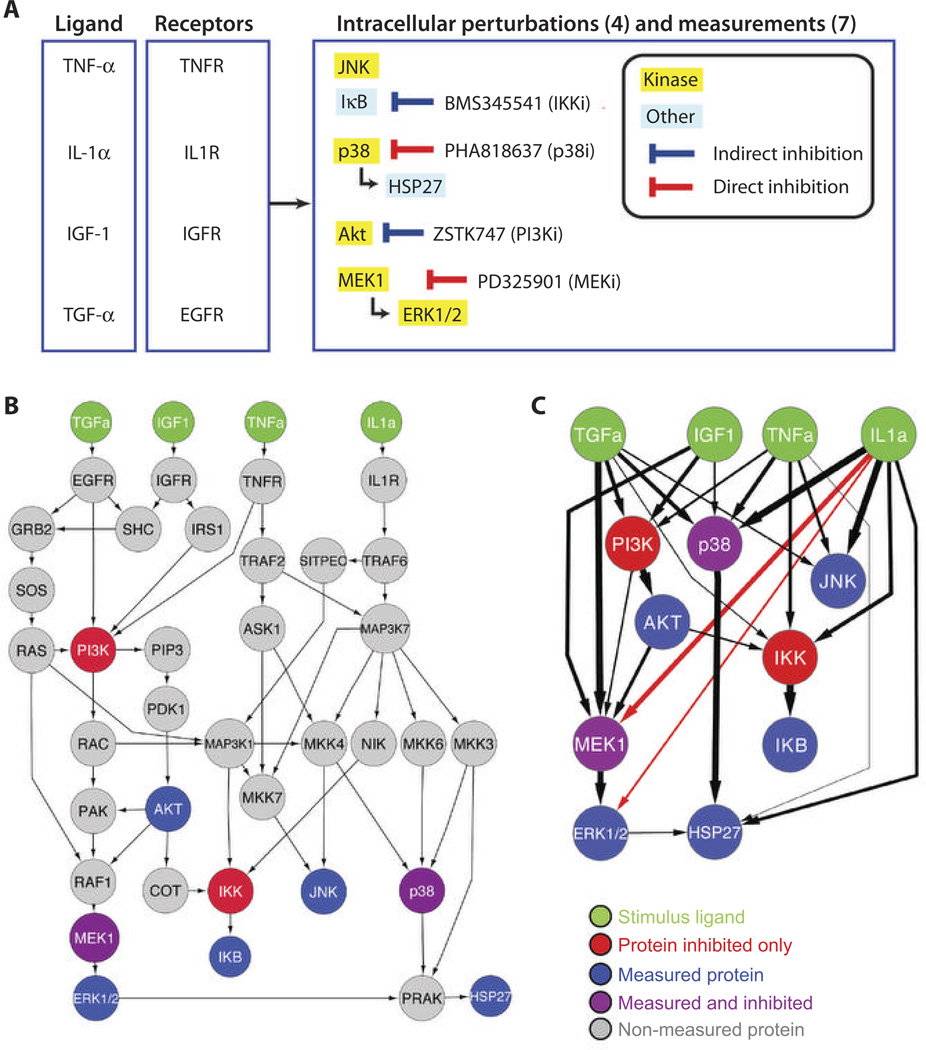
The training set was composed of measurements of activating phosphorylation events on seven measured phosphorylated proteins or groups of protein isoforms [the kinase Akt, the mitogen-activated protein kinase (MAPK) family members ERK1 and -2, which are detected with the same antibody and denoted as ERK1/2; JNK1, -2, and -3, which are detected with the same antibody and denoted as JNK; p38; the MAPK kinase MEK1; inhibitor of nuclear factor κB (NF-κB) (denoted as IKB); and heat shock protein HSP27] observed at three time points (0, 30, and 180 min) after stimulation by one of four ligands [transforming growth factor–α (denoted as TGFa), insulin-like growth factor 1 (IGF1), tumor necrosis factor–α (denoted as TNFa), or interleukin-1α (denoted as IL1a) in human hepatocellular liver carcinoma HepG2 cells (Fig. 1A). Measurements were obtained with and without pretreatment of cells with potent and relatively specific small-molecule inhibitors of cytosolic kinases (p38i, MEKi, PI3Ki, and IKKi, where “i” denotes inhibitor) that inhibited p38, MEK1, phosphatidylinositol 3-kinase (PI3K), or inhibitor of nuclear factor κB (IκB) kinase (IKK) as described (6). Participants attempted to predict phosphorylation measurements of the same seven proteins at 30 min after stimulation by various individual and pair-wise combinations of the ligands, in the presence of pair-wise combinations of the inhibitors (12). The experimental conditions comprising the training set were mutually exclusive with the experimental conditions comprising the test set. The complete challenge description and the data can be obtained from the DREAM project Web site (http://www.the-dream-project.org).
Fig. 1. The DREAM4 Challenge and resulting network.
(A) Experimental approach: HepG2 cells were pretreated with four different inhibitors and subsequently stimulated with four different ligands. For each combination of inhibitor and ligand, the phosphorylation activity of seven proteins or groups of protein isoforms was measured. (B) PKN pathway map (derived from the Ingenuity database) summarizing the signaling pathway relevant to the DREAM4 phosphoproteomics challenge. Green, ligands used as stimuli; red, proteins that were inhibited with small-molecule inhibitors but the phosphorylation status of which was not measured; blue, proteins that were measured in all experiments but were not targeted by small molecule inhibitors; purple, proteins targeted by small molecule inhibitors and also for which phosphorylation status was measured; gray, proteins known to be part of the network but were neither measured nor targeted by small-molecule inhibitors in the data sets supplied for the challenge. (C) Summary of the compressed model networks with edges weighted according to the frequency of their appearance in the networks submitted by each team. Only ligands, measured phosphoproteins, and proteins targeted by small-molecule inhibitors are included. Black arrows correspond to compressed interactions from the figure in (B), in which an edge exists between two nodes if there is a path from the source to the target nodes in the PKN. The thicknesses of the black arrows correspond to the number of teams that used an interaction to account for the training set (the thickest edge was used by 12 teams, whereas the thinnest was used by just one team). The red arrows are the most frequently predicted interactions that were not present in the PKN. Both the left and right networks were created with Cytoscape (http://www.cytoscape.org/). The protein ERK1/2 represents both ERK1 and -2, and JNK represents JNK1, -2, and -3.
In addition to the training set, participants received a prior knowledge network (PKN; a directed graph with edges specified as activating or inhibitory) compiled from the scientific literature as based on the Ingenuity Systems (Redwood, California) database encompassing the pathways known to be responsive to the ligands used for the challenge (Fig. 1B). In addition to the prediction task, the challenge entailed adding and removing edges to the PKN to capture those interactions that were essential to explain the training data. This task encouraged participants to go beyond “black box” prediction algorithms to enable some mechanistic interpretation of the quantitative models used to predict the test data set. Although some participants applied models that were interpretable as a network, others focused on the prediction task only and did not attempt to interpret their model in terms of a network. Anecdotally, the team with the highest prediction score used a model that was not readily interpretable as a network, suggesting that maximizing mechanistic interpretability of a model might compromise predictive accuracy.
The NRSS was evaluated separately for each of the seven proteins because measurements of phosphorylation status (Table 1) between proteins are not directly comparable because of different affinities of the antibodies for their targets and variation in protein abundances. The seven P values for each of the measured phosphoproteins represent the probability that the prediction accuracy on the test set is better than a naïve prediction assembled by randomly sampling from the phosphorylation status in the training set. The “Prediction Score” for a team summarizes the team’s overall predictive performance and was defined as the negative of the log10 of the geometric mean of the P values obtained by that team across all the predicted proteins (Eq. 2):
| (2) |
Table 1.
The performance scores for each of the phoshoproteins predicted by the teams and assessed in the DREAM4 phosphoproteomics challenge. The blue entries are the most significant predictions, with P values (PVAL) smaller than 10−10. The red entries are the least significant, with P values larger than 0.01. The overall score is computed as the prediction score (defined as minus the log to the base 10 of the geometric mean of the phosphoprotein P values), minus a penalty that scales with the number of edges in the predicted HepG2 networks (Eq. 3).
| Team | Overall score |
Edge count |
Prediction score |
Akt PVAL | ERK1/2 PVAL |
IκB PVAL | JNK PVAL | p38 PVAL | HSP27 PVAL |
MEK1 PVAL |
|---|---|---|---|---|---|---|---|---|---|---|
| Team 1 | 6.68 | 18 | 8.17 | 4.70E–5 | 1.60E–09 | 3.80E–10 | 1.70E–10 | 1.10E–08 | 8.30E–11 | 1.60E–07 |
| Team 2 | 6.32 | 17 | 7.73 | 9.40E–4 | 3.50E–14 | 6.00E–09 | 1.30E–10 | 1.40E–10 | 5.40E–06 | 4.00E–05 |
| Team 3 | 6.28 | 26 | 8.43 | 2.10E–5 | 1.90E–16 | 3.80E–10 | 3.80E–10 | 1.90E–06 | 1.90E–06 | 4.90E–09 |
| Team 4 | 5.02 | 18 | 6.51 | 7.80E–4 | 1.10E–08 | 1.30E–08 | 5.30E–11 | 4.80E–05 | 9.30E–07 | 1.10E–06 |
| Team 5 | 4.69 | 17 | 6.09 | 3.30E–5 | 5.90E–11 | 1.00E–07 | 2.30E–04 | 1.10E–05 | 1.20E–08 | 3.70E–05 |
| Team 6 | 4.58 | 22 | 6.4 | 6.80E–5 | 2.80E–08 | 1.10E–09 | 7.30E–11 | 1.70E–04 | 2.60E–04 | 2.40E–07 |
| Team 7 | 3.72 | 15 | 4.96 | 2.50E–6 | 8.20E–10 | 2.30E–04 | 4.80E–05 | 2.60E–02 | 6.60E–03 | 4.90E–09 |
| Team 8 | 3.1 | 27 | 5.33 | 1.80E–4 | 1.50E–05 | 2.60E–07 | 6.90E–10 | 1.30E–04 | 4.40E–04 | 1.70E–06 |
| Team 9 | 2.21 | 18 | 3.7 | 9.40E–4 | 2.10E–07 | 4.10E–05 | 1.20E–04 | 2.00E–02 | 2.20E–04 | 3.10E–03 |
| Team 10 | 1.55 | 10 | 2.37 | 1.10E–3 | 1.80E–05 | 1.50E–02 | 1.30E–03 | 5.00E–03 | 4.90E–02 | 2.60E–01 |
| Team 11 | 0.4 | 19 | 1.97 | 6.80E–4 | 8.50E–01 | 9.80E–05 | 1.00E+00 | 2.30E–03 | 1.30E–03 | 8.90E–02 |
| Team 12 | 0 | 54 | 4.47 | 1.00E–3 | 6.10E–08 | 3.60E–04 | 3.80E–11 | 1.30E–03 | 4.10E–03 | 1.20E–02 |
A high prediction score corresponds to high statistical significance for the accuracy of the prediction (a low average P value).
In prediction problems, a model with a number of fitted parameters that is smaller than the number of constraints in the problem (for example, the number of experiments) is generally preferred to a model with more parameters than constraints because the former is more parsimonious, less prone to overfitting, and typically more interpretable (7, 12). Also, empirical evidence suggests that biological networks are sparse (13), that is, the number of edges are of the order N (the number of nodes) rather than of order N2. We imposed a sparseness criterion on the selection of the best-performer using a cost function that rewards prediction accuracy and penalizes densely connected model networks to calculate the “Overall Score” for each team (Eq. 3):
| (3) |
Cost per edge was calibrated to the actual prediction scores and networks of the teams by taking the minimum (Prediction Score/Number of Edges) over all teams. The most accurate team was third by this criterion, whereas the second most accurate participant was first. [For the methodology used by the best performing team, Team 1, see (14).] This Overall Score cost function is ad hoc, and other formulations could rank the teams differently. One take-home message is that predictive accuracy, as measured by the Prediction Score and without regard to model complexity, model interpretability, or mechanistic plausibility, may be valuable in some tasks but not necessarily in the task of network inference. Indeed, the correlation between edge count and prediction score was low (0.03), indicating that increasing the number of edges does not automatically improve the predictability of the model. For a Boolean model, we previously showed that removing edges with no empirical support improved predictive accuracy (12). Networks with sparse connectivity, therefore, might be expected to score better than highly connected networks. However, it remains an open problem to design a cost function that rewards desirable attributes and penalizes undesirable attributes in a model network.
Crowdsourcing as a Strategy for Signaling Network Reconstruction
Networks inferred by different research groups can be combined into a composite network in different ways. For example, edges can be aggregated by using a majority vote (an edge is included only if it is predicted by more than a minimum number of groups) or using a scheme that weighs edges predicted by each team according to the team’s Prediction Score or Overall Score. Other methods of aggregation have also been proposed (15, 16). Because only some of the nodes in the provided PKN were measured or manipulated in the HepG2 cell line data, we asked participants to submit HepG1 networks containing only the observable nodes (ligands, measured phosphoproteins, and molecules targeted by the inhibitors) and the edges linking them. Thus, the submitted networks were representations of a “compressed” network (Fig. 1C) comprising only the observed nodes in which an edge is included if a path (direct or indirect) exists from the source node to the target node in the original PKN.
Not all teams used the compressed-network edges similarly. Some edges were used by all 12 teams, some were used only by 2 of the 12 teams, and one was invoked by just one team (Table 2). This analysis suggests that for HepG2 cells and these ligands, much of the “signal transduction” occurs through a subset of the edges present the original PKN (which is not specific to hepatocytes). Nine of the 12 teams added an edge between IL1α and MEK1, and of the three teams that did not add this edge (Teams 5, 10, and 11), two of them (Team 10 and 11) had the lowest Prediction Scores. Four teams added an interaction between IL1α and ERK1/2, which conveys a similar signal transduction as that conveyed by the added interaction between IL1α and MEK1. Most of the other added interactions were invoked by individual teams. The consensus among the ensemble of predictions that a currently uncharacterized pathway connects the stimulus IL1α and MEK1 in HepG2 cells should prove fruitful in designing testable hypothesis for follow-up experiments and is consistent with an independent study (12). Because literature-derived networks are an amalgam of signaling interactions observed in many cell types and environments, the actual cell-specific and condition-specific signaling networks may be quite different from the canonical one derived from the literature. Additionally, the importance of specific pathways may be different depending on the cellular context, as suggested by the addition to or subtraction from the original PKN of specific edges in the final network resulting from the team’s predictions.
Table 2.
Frequency of use of original and additional direct interactions used by the teams to create the model networks. The left side of each set represents the interaction and the right side represents the number of teams using that interaction.
| ORIGINAL INTERACTIONS | ADDED INTERACTIONS | ||
|---|---|---|---|
| TGFα→MEK1 | 12 | IL1α→MEK1 | 9 |
| IKK→IKB | 12 | IL1α→ERK1 | 4 |
| IL1α→JNK | 12 | P38→MEK1 | 2 |
| IL1α→ p38 | 12 | AKT→ JNK | 2 |
| P13K→AKT | 11 | IKK→ERK1/2 | 2 |
| p38→HSP27 | 11 | MEK1→AKT | 1 |
| MEK1→ERK1/2 | 10 | JNK→AKT | 1 |
| TGFα→PI3K | 7 | JNK→IKB | 1 |
| IGF1→MEK1 | 7 | p38→AKT | 1 |
| TNFα→p38 | 7 | IKB→HSP27 | 1 |
| IL1A→IKK | 7 | JNK→MEK1 | 1 |
| TGFα→p38 | 7 | ERK1/2→MEK1 | 1 |
| IGF1→PI3K | 7 | IKK→PI3K | 1 |
| TNFα→IKK | 6 | IKB→ERK1/2 | 1 |
| IL1α→HSP27 | 6 | IKK→HSP27 | 1 |
| AKT→MEK1 | 6 | HSP27→ERK1/2 | 1 |
| TNFα→JNK | 5 | MEK1→JNK | 1 |
| TNFα→PI3K | 4 | IKB→AKT | 1 |
| AKT→IKK | 3 | ||
| TGFα→JNK | 3 | ||
| PI3K→MEK1 | 3 | ||
| IGF1→p38 | 3 | ||
| ERK1/2→HSP27 | 2 | ||
| TGFα→IKK | 2 | ||
| TNFα→HSP27 | 1 | ||
Conclusions
Our network assessment strategy embraces the viewpoint that predictions are meaningful only when their direct consequences can have an experimental counterpart. In such cases, the plausibility of the predictions can be measured by cost functions that quantify the deviation between prediction and observation. The framework of setting aside some experiments to evaluate how well a computational model generalizes to previously unseen experimental perturbations is well suited to the data-driven network inference methods studied in the DREAM4 challenge. However, more research is needed on formulating cost functions for network inference that balance predictive accuracy with model complexity. For example, the sparseness criterion used in our Overall Score can be complemented with additional constraints on the abundance of different network motifs (17, 18). Although all network inference methods can make errors, we suggest that a way to improve network predictions is to blend an ensemble of networks, based on the same data, generated by a diversity of independent mathematical approaches into a composite network (16). The probability of finding one edge by chance may be high for any single technique, but the probability of finding that same edge by chance in many independent predictions decreases as the number of aggregate predictions increases (assuming the methods are independent). In this sense, the suggestion from the DREAM4 teams that a connection exists between IL1α and MEK1 is statistically significant.
Community experiments, such as the DREAM challenges, can become a powerful tool for network prediction: By aggregating the intelligence of the “crowds” (researchers), comprehensive and accurate inference of PSNs could become a reachable goal. Moreover, the development of increasingly high-throughput methods for measuring thousands of proteins and their posttranslational modifications in hundreds of samples promises to make collecting some of the data necessary for sophisticated network inference increasingly possible. The technical challenge of combining the intelligence of crowds with the latest instrumentation as a means to tackle problems of outstanding biomedical importance remains.
Algorithm development by the crowd is labor intensive, as is the process of evaluating and combining its conclusions. Because effective data generation requires a conceptual basis or underlying hypothesis (however general), the researchers collecting the data will likely be involved in an initial round of analysis. We are skeptical of the idea that data generation should be separated from analysis modeling. However, we think that the true value of the research will only be realized upon subsequent or concurrent crowdsourcing of data analysis. We believe the crowd will likely develop more sophisticated and important conclusions than those of the initial data generators. Critically, the computational and managerial machinery needed to enable crowdsourcing needs to be supported, and the likelihood (or past history) of data reuse needs to be part of the justification for data collecting in the first place. Many of these ideas are familiar to the DNA sequencing community, but they are not part of the current ethos for “functional,” perturbation-rich experimentation on cells and tissues. Moreover, in the case of modeling networks (but not genome sequencing), data release and computational predictions need to run in a closed loop, with one round of predictions informing the next round of data generation. Over time, the data available for training computational models will grow, leading to more refined predictions. New structures for assigning credit are needed in a scientific environment where data producers and (competing) data analysts might never collaborate in the traditional sense of the word, although the work is clearly collaborative in a fundamental sense.
Acknowledgments
We thank I. N. Melas for help with the analysis of submissions. We acknowledge the efforts of all the teams for their participation in the DREAM4 predictive signaling network challenge.
Funding sources: We acknowledge support for the DREAM project by the National Institutes of Health (NIH) Roadmap Initiative, through the Columbia University Center for Multiscale Analysis Genomic and Cellular Networks (MAGNet), and grant GM68762 for data generation; the LINCS program is currently funding data collection for future DREAM challenges (U54HG006097).
Footnotes
Meeting Information: The DREAM4 Predictive Signaling Network Challenge took place in the summer of 2009. Results were presented at the DREAM4 conference, December 2009, The Broad Institute of MIT and Harvard, Cambridge, Massachusetts.
Competing interests: P.K.S. is the cofounder and chair of the scientific advisory board of Merrimack Pharmaceuticals; on the board of directors of Applied Precision and Rarecye; and a consultant for Pfizer and Boehringer Ingelheim.
References and Notes
- 1.Bandura DR, Baranov VI, Ornatsky OI, Antonov A, Kinach R, Lou X, Pavlov S, Vorobiev S, Dick JE, Tanner SD. Mass cytometry: Technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal. Chem. 2009;81:6813–6822. doi: 10.1021/ac901049w. [DOI] [PubMed] [Google Scholar]
- 2.Bendall SC, Simonds EF, Qiu P, Amir AD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, Balderas RS, Plevritis SK, Sachs K, Pe’er D, Tanner SD, Nolan GP. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolf-Yadlin A, Sevecka M, MacBeath G. Dissecting protein function and signaling using protein microarrays. Curr. Opin. Chem. Biol. 2009;13:398–405. doi: 10.1016/j.cbpa.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciaccio MF, Wagner JP, Chuu CP, Lauffenburger DA, Jones RB. Systems analysis of EGF receptor signaling dynamics with micro-western arrays. Nat. Methods. 2010;7:148–155. doi: 10.1038/nmeth.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sachs K, Perez O, Pe’er D, Lauffenburger DA, Nolan GP. Causal protein-signaling networks derived from multiparameter single-cell data. Science. 2005;308:523–529. doi: 10.1126/science.1105809. [DOI] [PubMed] [Google Scholar]
- 6.Alexopoulos LG, Saez-Rodriguez J, Cosgrove BD, Lauffenburger DA, Sorger PK. Networks inferred from biochemical data reveal profound differences in toll-like receptor and inflammatory signaling between normal and transformed hepatocytes. Mol. Cell. Proteomics. 2010;9:1849–1865. doi: 10.1074/mcp.M110.000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelander S, Wang W, Nilsson B, She QB, Pratilas C, Rosen N, Gennemark P, Sander C. Models from experiments: Combinatorial drug perturbations of cancer cells. Mol. Syst. Biol. 2008;4:216. doi: 10.1038/msb.2008.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bansal M, Belcastro V, Ambesi-Impiombato A, di Bernardo D. How to infer gene networks from expression profiles. Mol. Syst. Biol. 2007;3:78. doi: 10.1038/msb4100120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitsos A, Melas IN, Siminelakis P, Chairakaki AD, Saez-Rodriguez J, Alexopoulos LG. Identifying drug effects via pathway alterations using an integer linear programming optimization formulation on phosphoproteomic data. PLOS Comput. Biol. 2009;5 doi: 10.1371/journal.pcbi.1000591. e1000591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stolovitzky G, Prill RJ, Califano A. Lessons from the DREAM2 Challenges. Ann. N.Y. Acad. Sci. 2009;1158:159–195. doi: 10.1111/j.1749-6632.2009.04497.x. [DOI] [PubMed] [Google Scholar]
- 11.Morris MK, Saez-Rodriguez J, Sorger PK, Lauffenburger DA. Logic-based models for the analysis of cell signaling networks. Biochemistry. 2010;49:3216–3224. doi: 10.1021/bi902202q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saez-Rodriguez J, Alexopoulos LG, Epperlein J, Samaga R, Lauffenburger DA, Klamt S, Sorger PK. Discrete logic modelling as a means to link protein signalling networks with functional analysis of mammalian signal transduction. Mol. Syst. Biol. 2009;5:331. doi: 10.1038/msb.2009.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leclerc RD. Survival of the sparsest: Robust gene networks are parsimonious. Mol. Syst. Biol. 2008;4:213. doi: 10.1038/msb.2008.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eduati F, Corradin A, Di Camillo B, Toffolo G. A Boolean approach to linear prediction for signaling network modeling. PLoS ONE. 2010;5:e12789. doi: 10.1371/journal.pone.0012789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marbach D, Mattiussi C, Floreano D. Combining multiple results of a reverse-engineering algorithm: Application to the DREAM five-gene network challenge. Ann. N.Y. Acad. Sci. 2009;1158:102–113. doi: 10.1111/j.1749-6632.2008.03945.x. [DOI] [PubMed] [Google Scholar]
- 16.Marbach D, Prill RJ, Schaffter T, Mattiussi C, Floreano D, Stolovitzky G. Revealing strengths and weaknesses of methods for gene network inference. Proc. Natl. Acad. Sci. U.S.A. 2010;107:6286–6291. doi: 10.1073/pnas.0913357107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma’ayan A, Cecchi GA, Wagner J, Rao AR, Iyengar R, Stolovitzky G. Ordered cyclic motifs contribute to dynamic stability in biological and engineered networks. Proc. Natl. Acad. Sci. U.S.A. 2008;105:19235–19240. doi: 10.1073/pnas.0805344105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma’ayan A, Jenkins SL, Neves S, Hasseldine A, Grace E, Dubin-Thaler B, Eungdamrong NJ, Weng G, Ram PT, Rice JJ, Kershenbaum A, Stolovitzky GA, Blitzer RD, Iyengar R. Formation of regulatory patterns during signal propagation in a mammalian cellular network. Science. 2005;309:1078–1083. doi: 10.1126/science.1108876. [DOI] [PMC free article] [PubMed] [Google Scholar]