Abstract
Transcriptional antiterminator proteins of the BglG family control the expression of enzyme II (EII) carbohydrate transporters of the bacterial phosphotransferase system (PTS). In the PTS, phosphoryl groups are transferred from phosphoenolpyruvate (PEP) via the phosphotransferases enzyme I (EI) and HPr to the EIIs, which phosphorylate their substrates during transport. Activity of the antiterminators is negatively controlled by reversible phosphorylation catalyzed by the cognate EIIs in response to substrate availability and positively controlled by the PTS. For the Escherichia coli BglG antiterminator, two different mechanisms for activation by the PTS were proposed. According to the first model, BglG is activated by HPr-catalyzed phosphorylation at a site distinct from the EII-dependent phosphorylation site. According to the second model, BglG is not activated by phosphorylation, but solely through interaction with EI and HPr, which are localized at the cell pole. Subsequently BglG is released from the cell pole to the cytoplasm as an active dimer. Here we addressed this discrepancy and found that activation of BglG requires phosphorylatable HPr or the HPr homolog FruB in vivo. Further, we uniquely demonstrate that purified BglG protein becomes phosphorylated by FruB as well as by HPr in vitro. Histidine residue 208 in BglG is essential for this phosphorylation. These data suggest that BglG is in fact activated by phosphorylation and that there is no principal difference between the PTS-exerted mechanisms controlling the activities of BglG family proteins in Gram-positive and Gram-negative bacteria.
Keywords: histidine protein, carbon catabolite repression, RNA binding protein, β-glucoside operon, antagonistically acting phosphorylations
The Escherichia coli BglG antiterminator protein represents the prototype of a family of regulators that are widespread in bacteria. BglG regulates expression of the bglGFBH operon coding for BglG itself, the aryl-β-glucoside transporter BglF (EIIBgl) and additional proteins required for utilization of aryl-β-glucoside sugars (1). BglG controls the bgl operon by inactivating two transcriptional terminators that frame the bglG gene thereby allowing transcription elongation (2, 3). Activity of BglG is negatively controlled by BglF. BglF is a transporter belonging to the phosphoenolpyruvate (PEP)-dependent phosphotransferase system (PTS). In the PTS, phosphoryl groups are transferred from PEP via the two general phosphotransferases enzyme I (EI) and HPr to the sugar-specific transport proteins (enzymes II, EIIs), which finally phosphorylate their substrates during uptake (4). In the absence of β-glucosides, BglG is recruited to the cell membrane by BglF, which transfers the phosphoryl groups to BglG rather than to the sugar, whereas the reverse process takes place in the presence of substrate (5–7). The BglF-catalyzed phosphorylation/dephosphorylation triggers the monomer/dimer transition and thereby activity of BglG. Exclusively dimeric BglG is active in antitermination (8, 9). Antiterminator proteins of the BglG family are present in Gram-negative as well as Gram-positive bacteria, such as Bacillus subtilis. Where known, these BglG homologs control synthesis of a specific EII and associated functions for utilization of a particular sugar via a transcription antitermination mechanism reminiscent of the bgl system. The EIIs in turn negatively control the activities of their cognate antiterminator proteins by reversible phosphorylation similar to the BglG/BglF interplay (4, 10, 11).
Several of the BglG homologs in Gram-positive bacteria are subject to an additional control. They require the general PTS proteins EI and HPr to gain antitermination activity. The data indicate that HPr phosphorylates and thereby activates these antiterminators at site(s) that differ from the EII-dependent phosphorylation site (11–14). Antiterminators of the BglG family exhibit a modular structure consisting of three domains. The N-terminal RNA binding domain is followed by two PTS regulatory domains (PRD1 and PRD2), which are homologous to each other and presumably evolved by duplication (11). Each PRD contains two conserved histidine residues, which are the targets of the PTS-catalyzed phosphorylation events. Collectively, the results obtained with various BglG homologs showed that the first conserved histidine in PRD1 is phosphorylated by the cognate EII, whereas HPr phosphorylates the histidines in PRD2 (13–15). The E. coli BglG protein, like its homologs in Gram-positive bacteria, requires the general PTS enzymes EI and HPr to gain antiterminator activity (16, 17). This activation is independent of the negative control of BglG by BglF, i.e., it also operates in the absence of BglF (16, 17). In vivo phosphorylation assays indicated that BglG becomes phosphorylated in EI- and HPr-dependent reactions also in the absence of BglF. Consequently, we suggested that BglG is activated by phosphorylation catalyzed by HPr (16). Analysis of BglG mutants in vivo indicated that His101 located in PRD1 is phosphorylated by BglF. In contrast, the HPr-dependent phosphorylation site was mapped to His208 in PRD2. BglG proteins bearing mutations in this site lost activity and concomitantly phosphorylation by HPr and EI in vivo (9). Taken together, we proposed that BglG is regulated by the PTS through antagonistically acting phosphorylations similar to its Gram-positive homologs. Thus, activity of these proteins requires dephosphorylation by the cognate EII in PRD1 and simultaneously phosphorylation by HPr in PRD2.
However, more recently it was proposed that activation of BglG occurs through formation of a ternary complex with EI and HPr and does not involve the physical transfer of phosphoryl groups from HPr to BglG. This conclusion was drawn from two observations (17). First, activation of BglG also occurred in a strain bearing a mutation in the His15 phosphorylation site of HPr, suggesting that phosphorylatable HPr is not required for this process. Second, phosphorylation of a maltose binding protein (MBP)–BglG fusion protein by HPr in vitro could never be observed (e.g., ref. 6). BglG forms insoluble inclusion bodies upon overexpression and so far its purification has only been possible by fusing it to the large MBP (6). Further, elegant analyses of the cellular localization of BglG and the PTS enzymes EI and HPr revealed that in the absence of the specific sugar, BglG is sequestered to the membrane by BglF. Upon addition of substrate, BglG is released from BglF and recruited to the cell pole by interaction with HPr and EI. This recruitment is independent of phosphorylation of EI, HPr, and BglG, respectively. Subsequently, BglG is released from the cell pole to the cytoplasm. As mechanism of BglG activation by the PTS enzymes, stabilization of the BglG dimer conformation by HPr and EI at the cell pole was discussed (7).
To clarify the discrepancies, we reinvestigated the mechanism of activation of BglG by the PTS. We confirm that BglG still exhibits activity in a strain carrying a nonphosphorylatable HPr variant, in agreement with the work of Raveh et al. (17). However, we found that the requirement of BglG activity for phosphorylatable HPr is masked by the presence of the FruB protein, which carries a domain homologous to HPr. In a strain lacking FruB, BglG requires phosphorylatable HPr and EI for activity. Genetic and in vivo phosphorylation analyses indicate that HPr as well as FruB can both transfer phosphoryl groups from EI to BglG leading to its activation. Most importantly, we succeeded in purification of soluble BglG carrying the Strep epitope at its N terminus. PEP-dependent in vitro phosphorylation assays unequivocally show that FruB and HPr are both capable of transferring phosphoryl groups from EI directly to BglG. Mutation of His208 in BglG abolishes its phosphorylation by HPr or FruB, supporting the conclusion that phosphorylation at this site is required for BglG activity.
Results
BglG Requires EI and Either Phosphorylatable HPr or FruB for Activity.
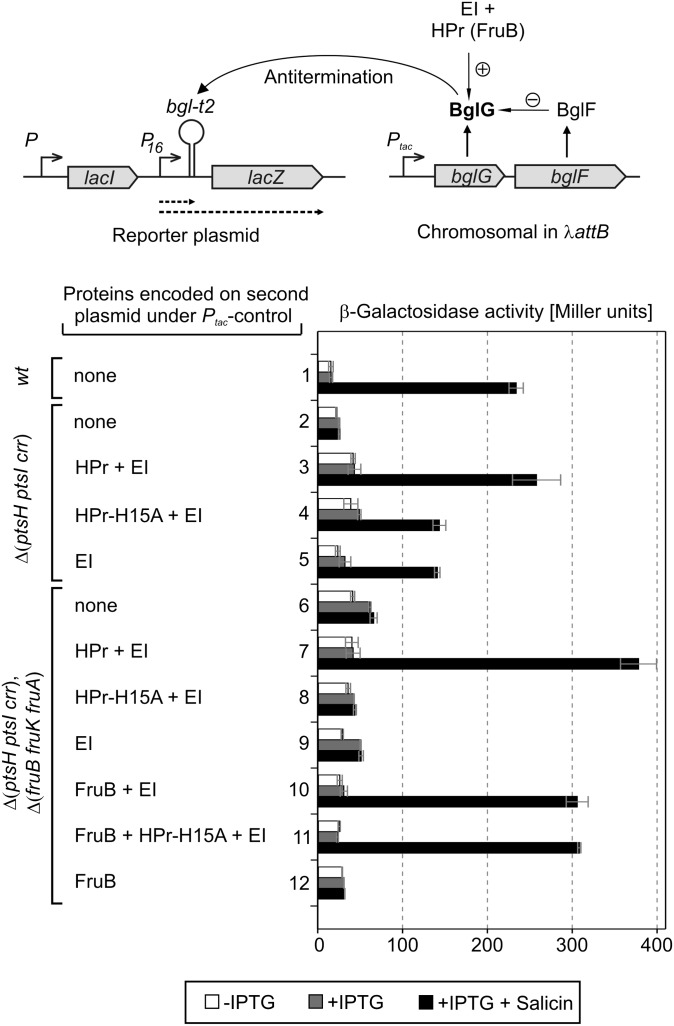
The bgl operon is subject to multiple levels of regulation at the transcriptional as well as posttranscriptional levels (18). To reevaluate the genetic requirements for activity of BglG, we therefore used a reporter system that allows determination of BglG activity independent of the context of the bgl operon (Fig. 1, Top). The examined strains lack the natural bgl operon, but carry an artificial bglG–bglF operon under control of the isopropyl-β-d-1-thiogalactopyranoside (IPTG)-inducible tac promoter in the chromosomal λattB site. BglG activity was measured using antitermination reporter plasmid pFDX3158. This plasmid carries a fusion of the bglt2 terminator to lacZ, which is transcribed from the constitutive promoter P16 (19). In the absence of IPTG and thus bglG expression, most transcripts terminate at the bglt2 terminator resulting in only low β-galactosidase activities (Fig. 1, white bars). Induction of bglG–bglF expression by IPTG did not increase these activities, because BglG is inhibited by BglF-mediated phosphorylation. In the additional presence of salicin, which is a substrate for BglF, β-galactosidase activities increased 15-fold (Fig. 1, black bar 1). This increase reflects the release of BglG from inhibition by BglF, resulting in antitermination of the bglt2–lacZ transcript. BglG activity was completely abolished in an isogenic strain lacking the ptsHIcrr operon, which encodes HPr, EI, and EIIAGlc (Fig. 1, black bar 2). Complementation of this strain with a compatible plasmid carrying genes ptsH and ptsI under Ptac control restored high BglG activity (Fig. 1, black bar 3), confirming previous reports that BglG requires the general PTS proteins for activity (9, 16, 17). Interestingly, activity was also restored to some extent (i.e., 56% of the activity obtained with the ptsH–ptsI wild-type construct) when the phosphorylation site His15 in HPr was mutated to an alanine (Fig. 1, black bar 4), in agreement with data by Raveh et al. (17). At first glance, this result might indicate that EI and HPr jointly activate BglG in a phosphorylation-independent manner. However, previously we provided initial evidence that the product of the fruB gene can substitute for HPr in activation of BglG, thereby masking the requirement for phosphorylated HPr (16). FruB, which is encoded in the fruBKA operon, carries a C-terminal domain with homology to HPr. To clarify the involvement of FruB, we investigated BglG activity in a strain lacking the ptsHIcrr and the fruBKA operon. In this double mutant, activity of BglG was similarly low as in the Δpts single mutant (Fig. 1, black bar 6). Significantly, high BglG activity was restored when the ΔptsHIcrr ΔfruBKA mutant was complemented with the plasmid carrying ptsH–ptsI, but not when the isogenic construct carrying the ptsH–H15A allele or a construct carrying only ptsI was introduced (Fig. 1, black bars 7, 8, and 9). Strikingly, BglG gained also high activity, when plasmids were introduced that carried the fruB gene in addition to ptsI or ptsI and ptsH–H15A (Fig. 1, black bars 10 and 11). A plasmid carrying fruB alone was not able to restore BglG activity (Fig. 1, black bar 12). These data show that BglG requires EI and either HPr or FruB for activity. Furthermore, HPr is only able to activate BglG, if its His15 phosphorylation site is intact. However, this requirement becomes visible only in a strain lacking FruB.
Fig. 1.
Activation of BglG requires intact HPr that can be phosphorylated and this requirement can partially be complemented by FruB. Strains were tested that carried deletions of the bgl and lac operons (WT) and additional deletions as indicated at Left. All strains also carried an artificial bglG–bglF operon under control of the IPTG-inducible Ptac promoter ectopically integrated into the λattB site on the chromosome (Upper Right). Strains were cotransformed with plasmid pFDX3158 as reporter for BglG antitermination activity (Upper Left) and an additional plasmid carrying the genes as indicated under Ptac control. Bacteria were grown in M9 glycerol medium. IPTG (1 mM) for induction of Ptac-controlled genes and salicin (7 mM) as substrate for BglF were added as indicated. β-Galactosidase activities were determined from exponentially grown cells. The following strains carrying plasmid pFDX3158, and in addition the plasmids given in parentheses, were used: Bars 1, R1752; bars 2, R2013; bars 3, R2013 (pFDX3851); bars 4, R2013 (pFDX3852); bars 5, R2013 (pFDX3161); bars 6, R1977; bars 7, R1977 (pFDX3851); bars 8, R1977 (pFDX3852); bars 9, R1977 (pFDX3161); bars 10, R1977 (pFDX3221); bars 11, R1977 (pFDX4732); and bars 12, R1977 (pFDX3214).
BglG Becomes Phosphorylated in ΔbglF Cells in a Reaction That Requires EI and Either HPr or FruB.
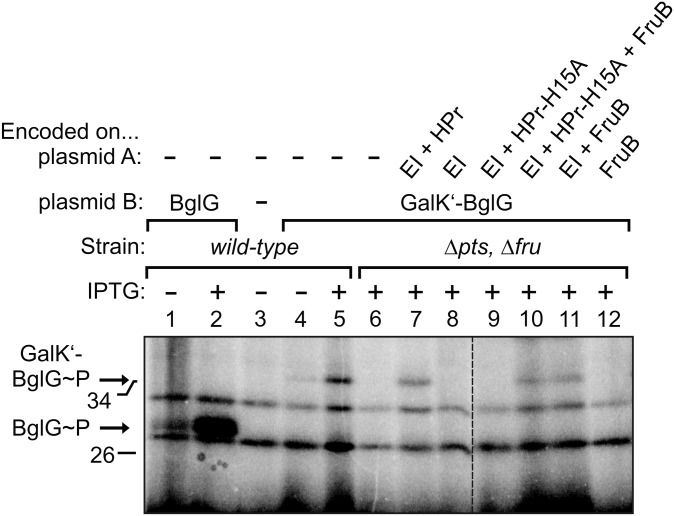
The genetic requirements for activation of BglG are most easily explained by a mechanism that involves the physical transfer of phosphoryl groups from EI to BglG via HPr or FruB. To assess whether BglG can be phosphorylated by these proteins, we used metabolic H3[32P]PO4 labeling of cells, which allows detection of phosphorylated proteins in vivo. In agreement with previous results (16), phosphorylated BglG (molecular weight, MW = 32.1 kDa) became detectable when its gene was expressed from a plasmid under control of the tac promoter, and signal intensity strongly increased upon induction of the promoter with IPTG (Fig. 2, lanes 1 and 2). Because there was no BglF present in the cells, this phosphorylation must be catalyzed by different protein(s). To avoid misinterpretation of the data by a nearby migrating band of another phosphorylated protein, we analyzed a GalK′–BglG fusion protein with increased molecular weight (MW = 35.6 kDa). The phosphorylation signal of the GalK′–BglG fusion protein was decreased in comparison with wild-type BglG (Fig. 2, lanes 4 and 5), which can be attributed to its lower amount in the cell (9). To determine whether EI, HPr, or FruB are required for the BglF-independent phosphorylation of GalK′–BglG, we analyzed its phosphorylation in the mutant lacking the ptsHIcrr and fruBKA operons. Phosphorylation of GalK′–BglG was not detectable in this strain, but it could be restored by introduction of a low copy plasmid coexpressing ptsI and ptsH (Fig. 2, lanes 6 and 7). Significantly, introduction of isogenic plasmids carrying ptsI either alone or together with the ptsH–H15A allele did not restore phosphorylation of GalK′–BglG (Fig. 2, lanes 8 and 9). However, when the fruB gene was additionally present on these constructs, phosphorylated GalK′–BglG became detectable again (Fig. 2, lanes 10 and 11). This phosphorylation did not occur when a plasmid expressing solely fruB was used for complementation, demonstrating that it requires EI (Fig. 2, lane 12). Collectively, the data indicate that BglG is phosphorylated in vivo also in the absence of BglF, by a pathway that requires EI and either phosphorylatable HPr or its paralog FruB.
Fig. 2.
BglG becomes phosphorylated in vivo also in the absence of BglF, and this phosphorylation requires EI and either HPr or its paralog FruB, but not EIIAGlc. Phosphorylation of BglG or the GalK′–BglG fusion protein was addressed in various genetic backgrounds by metabolic labeling of proteins with H3[32P]PO4. The wild type (strain R1279; lanes 1–5) and the double mutant lacking the chromosomal ptsHIcrr and fruBKA operons (strain R1969; lanes 6–12) were tested. Both strains lacked the natural bgl operon. With exception of lane 3, the strains harbored a p15A plasmid (plasmid B), which carried either bglG (lanes 1 and 2; plasmid pFDX2942) or the galK′–bglG fusion gene (lanes 4–12; plasmid pFDX3225) under control of the IPTG-inducible Ptac promoter. In lanes 7–12, the ΔptsHIcrr ΔfruBKA mutant was complemented with additional compatible pSC101-type plasmids (plasmid A), which expressed the PTS proteins as indicated from a constitutive promoter. These plasmids were pFDX4735 (lane 7), pFDX4733 (lane 8), pFDX4736 (lane 9), pFDX4738 (lane 10), pFDX4737 (lane 11), and pFDX4739 (lane 12). Lac repressor was delivered from the ColEI-type plasmid pFDY226. The transformants were grown in minimal medium containing glycerol and 100 μM IPTG for induction of Ptac-controlled genes as indicated. The dashed line indicates cropping of one lane from the original autoradiograph.
BglG Becomes Phosphorylated in Vitro in a Reaction Depending on PEP, EI, and HPr.
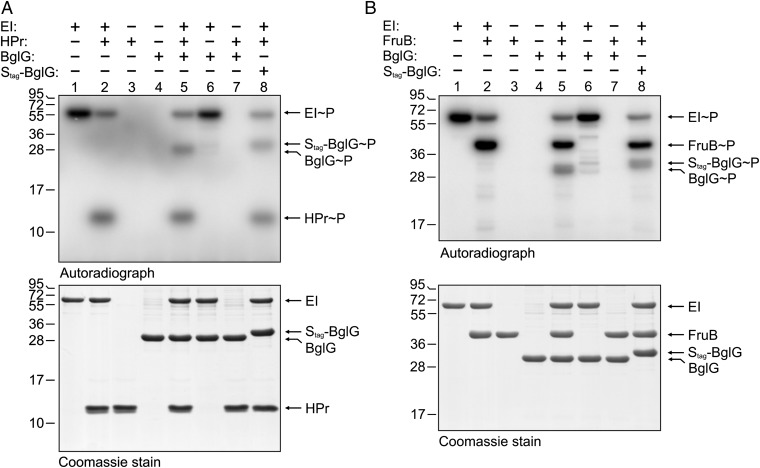
The genetic analyses and the results from the in vivo phosphorylation assays were in support of our hypothesis that BglG requires phosphorylation by HPr or FruB to gain activity. However, the in vivo analyses so far cannot exclude the possibility that the BglF-independent phosphorylation of BglG (Fig. 2) is indirect and catalyzed by an unknown protein that in turn receives phosphoryl groups from HPr∼P or FruB∼P. In vitro experiments are required to decide about this issue. Fortuitously, we recently observed that BglG fused to the C terminus of the Strep-tag epitope remains soluble, allowing purification of recombinant Strep–BglG. This provided the opportunity to study phosphorylation of Strep–BglG in vitro. To this end, recombinant His10–EI and His10–HPr proteins were purified and [32P]-PEP–dependent phosphorylation assays were carried out testing the purified proteins in all possible combinations (Fig. 3A). As expected, autophosphorylation of EI could be easily detected upon incubation with [32P]-PEP, whereas no signal was obtained when HPr was tested alone (Fig. 3A, lanes 1 and 3). When both EI and HPr were present in the assay, phosphorylated HPr also became detectable, i.e., phosphoryl groups were transferred from [32P]-PEP to HPr via EI (Fig. 3A, lane 2). When BglG was incubated with [32P]-PEP either alone or together with either EI or HPr, a signal corresponding to phosphorylated BglG could not be detected (Fig. 3A, lanes 4, 6, and 7). However, when EI and HPr were simultaneously present in the assay, a 32P-labeled signal appeared migrating at the position of BglG (Fig. 3A, lane 5). To verify that this signal corresponds to BglG∼P and not to a phosphorylated degradation product of EI, we used a purified BglG variant with increased molecular weight. This variant additionally carries the S tag between the N-terminal Strep tag and the BglG sequence, which increases the molecular weight by 1.75 kDa. This resulted in a slightly slower migration behavior in SDS gels compared with Strep–BglG (Fig. 3A Lower, compare lanes 7 and 8). Indeed, when incubated together with [32P]-PEP, EI, and HPr a signal for phosphorylated Strep–Stag–BglG became visible at the expected position (Fig. 3A, lane 8). Phosphorylation of the Strep–Stag–BglG protein required the simultaneous presence of EI and HPr in the assay. If one or both of these proteins were absent, phosphorylated Strep–Stag–BglG could not be observed (Fig. S1). Collectively, these results indicate that phosphoryl groups are transferred from PEP to BglG via EI and HPr. Thus, HPr is capable of directly phosphorylating BglG.
Fig. 3.
BglG becomes phosphorylated in vitro by HPr (A) as well as by FruB (B). PEP-dependent phosphorylation assays were carried out containing purified His10–EI, His10–HPr, Strep–BglG, or Strep–Stag–BglG as indicated. In the assays shown in B, His10–HPr was replaced by His10–FruB. Assays were carried out using either [32P]-PEP (Upper) or 1 μM cold PEP (Lower). Proteins were subsequently separated by denaturing gel electrophoresis using 15% (A) or 12% polyacrylamide gels (B). Gels were analyzed by phosphoimaging (Upper) or staining with Coomassie brilliant blue (Lower). Positions of the molecular weight marker are given at Left.
FruB Can Substitute for HPr in in Vitro Phosphorylation of BglG.
Our in vivo analyses indicated that BglG can be phosphorylated in the absence of HPr, leading to its concomitant activation. This reaction requires the FruB protein (Figs. 1 and 2). To test whether FruB phosphorylates BglG directly or indirectly, we repeated in vitro phosphorylation assays, but replaced His10–HPr by purified His10–FruB. In agreement with previous studies (20), a strong phosphorylation signal was obtained for FruB, when incubated together with EI in the presence of [32P]-PEP, whereas this signal was missing when EI was omitted from the assay (Fig. 3B, lanes 2 and 3). FruB is phosphorylated by EI in the C-terminal HPr-like domain, from where the phosphoryl group is subsequently transferred to the N-terminal IIA domain. Hence FruB is doubly phosphorylated in the presence of EI, which explains its stronger phosphorylation signal compared with HPr (compare Fig. 3 B and A). Noteworthy, a radio-labeled signal corresponding to phosphorylated Strep–BglG became visible, when this protein was additionally added to the assay (Fig. 3B, lane 5). The same result was obtained when using Strep–Stag–BglG (Fig. 3B, lane 8). Phosphorylation of Strep–BglG did not occur, when either EI or FruB or both proteins were absent from the assay (Fig. 3B, lanes 4, 6, and 7). In conclusion, FruB phosphorylates Strep–BglG directly and with a similar efficiency as HPr (compare lanes 5 in Fig. 3 A and B).
Histidine Residue 208 in BglG Is Essential for Its Phosphorylation by HPr or FruB.
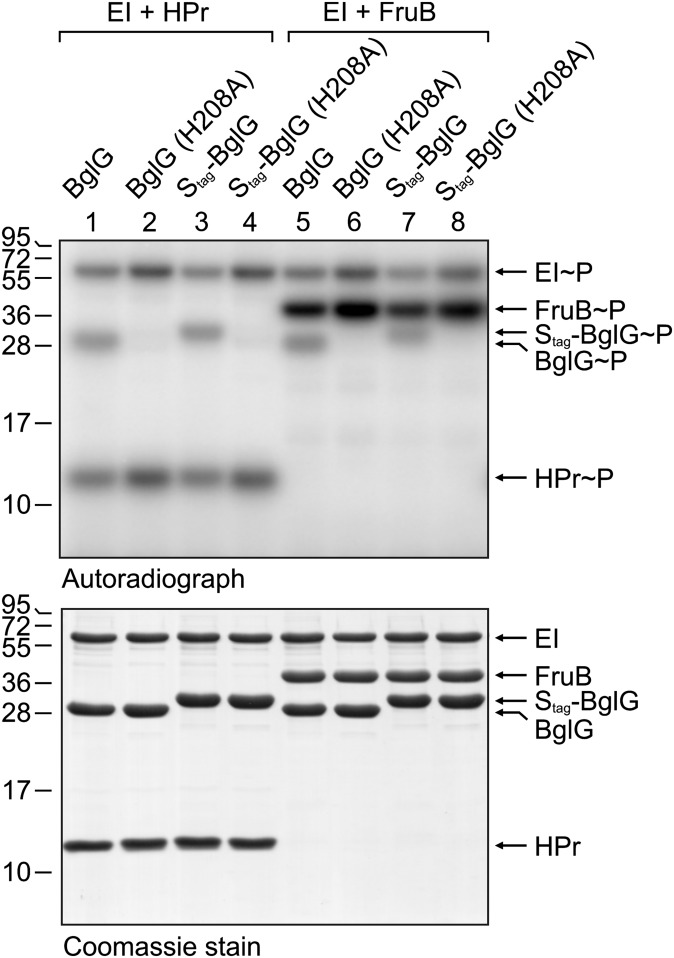
Previous in vivo studies indicated that BglG is phosphorylated by BglF at the conserved histidine 101 in PRD1, whereas the HPr-dependent phosphorylation site was mapped to histidine 208 in PRD2 (9). To see whether this also holds true in vitro, we constructed plasmids coding for Strep-tagged BglG–H101A and BglG–H208A variants. The Strep–BglG–H101A protein was entirely insoluble making its purification impossible. However, we succeeded in purification of the Strep–BglG–H208A protein (Fig. 4 Lower, lanes 2 and 6) and therefore we tested whether this mutant is still amenable to phosphorylation by HPr or FruB in the presence of EI and [32P]-PEP. Intriguingly, no phosphorylation signals were obtained for the Strep–BglG–H208A variant regardless of whether HPr or FruB was present in the assay (Fig. 4, lanes 2 and 6). In contrast, wild-type Strep–BglG, which was tested in parallel, became readily phosphorylated, confirming the results obtained before (Fig. 4, lanes 1 and 5). To verify these results unambiguously, we repeated the experiments using the BglG derivatives with an additional S tag and thus increased molecular weight. Once again, phosphorylation of the H208A variant was completely abolished, whereas the corresponding wild-type protein became phosphorylated by HPr as well as FruB (Fig. 4, lanes 3, 4, 7, and 8). Taken together, these data establish that the histidine 208 in BglG is essential for its phosphorylation by HPr or FruB.
Fig. 4.
The conserved His208 residue in BglG is essential for its phosphorylation by HPr or FruB in vitro. Shown are PEP-dependent phosphorylation assays containing EI and HPr (lanes 1–4) or EI and FruB (lanes 5–8). In addition, the following BglG variants were present: Strep–BglG (lanes 1 and 5), Strep–BglG–H208A (lanes 2 and 6), Strep–Stag–BglG (lanes 3 and 7), or Strep–Stag–BglG–H208A (lanes 4 and 8). Assays contained either [32P]-PEP (Upper) or 1 μM cold PEP (Lower). Proteins were subsequently separated on 15% SDS-polyacrylamide gels and gels were analyzed by phosphoimaging (Upper) or Coomassie brilliant blue staining (Lower).
Discussion
The aim of the current work was to clarify the mechanism underlying activation of the antiterminator protein BglG by the PTS enzymes EI and HPr. Our data suggest that BglG is activated by phosphorylation at histidine residue 208 located in the PRD2 domain. This conclusion is based on in vivo analyses, which demonstrate that EI and either HPr or its homolog FruB are required for BglG phosphorylation and activity. Further, phosphorylation of BglG was reconstituted in vitro using purified BglG, EI, and HPr or FruB proteins, whereas a BglG–H208A mutant was not phosphorylated in vitro. Activation of BglG by phosphorylation within the PRD2 domain in E. coli resembles activation of BglG family proteins in Gram-positive bacteria. This mechanism provides a means to tightly control BglG activity, as the availability of phosphorylated HPr (and FruB) for phosphorylation and thus activation of BglG depends on the general carbohydrate supply.
Previously we proposed that HPr phosphorylates BglG leading to its concomitant activation (9, 16), whereas more recently it was suggested that BglG becomes activated by a phosphorylation-independent mechanism through interaction with HPr and EI, suggesting that activation of BglG is possibly mediated by an EI and HPr induced conformational change stabilizing the BglG dimer (7, 17). In fact, the latter model appears to be supported by the result that BglG is active in a strain carrying a HPr–H15A mutant (our data and ref. 17). However, additional data suggested that the FruB protein, which carries a HPr-like domain, can substitute for HPr in activation of BglG (16), and here we demonstrated that the nonphosphorylatable HPr–H15A mutant together with EI is unable to activate BglG in a strain lacking FruB (Fig. 1, bars 8). Thus, FruB complements HPr in activation of BglG. Involvement of an additional HPr homolog such as NPr of the PTSNtr (20) in activation of BglG is unlikely, because BglG remains unphosphorylated and concomitantly inactive in a strain lacking both HPr and FruB (Fig. 1, bars 9 and Fig. 2, lane 8). Interestingly, FruB can also substitute for HPr in BglF-dependent β-glucoside transport, i.e., it can phosphorylate BglF (Fig. S2). FruB-mediated phosphorylation of BglF in the HPr–H15A mutant explains that addition of the BglF substrate salicin is required to release BglG from negative regulation by BglF (Fig. 1, bars 4). Thus, a phosphorylation-independent mechanism of regulation of BglG activity by BglF in HPr mutants, as proposed in ref. 17, also appears unlikely, as FruB complements for phosphorylation of both BglG and BglF.
Taken together, most of the data by Raveh et al. (17) are in agreement with our results. However, as one discrepancy we observed significant activity of BglG in a strain lacking HPr but possessing EI (Fig. 1, bars 5) (16), in agreement with complementation of HPr mutants by FruB. In contrast, in the study of Raveh et al., BglG gained almost no activity in a comparable genetic background (17). Different EI expression levels generated from the complementing plasmid constructs or differences in the strain backgrounds or the reporter systems used to measure BglG activity could account for this remaining discrepancy. In vitro phosphorylation experiments were previously performed with BglG protein fused to the large MBP domain (43.4 kDa), because native BglG is insoluble upon overexpression. However, in these experiments no phosphorylation of MBP–BglG by HPr and EI from various sources (e.g., E. coli and B. subtilis) was detected (6, 17). Here, we succeeded in purification of a BglG variant carrying the short Strep-tag epitope at its N terminus. [32P]-PEP–dependent phosphorylation assays carried out with Strep–BglG in vitro (Fig. 3) demonstrate that BglG becomes phosphorylated in the sequential reaction PEP→EI→HPr (FruB)→BglG (Fig. 5).
Fig. 5.
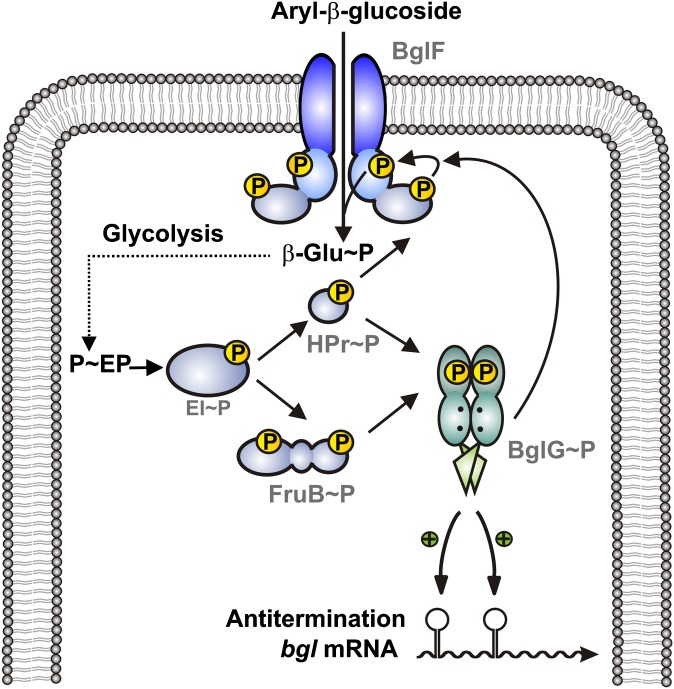
Model for activation of the BglG antiterminator protein by the PTS. BglG requires two distinct (de)phosphorylation events to gain activity. On the one hand, BglG must be dephosphorylated by the β-glucoside transporter BglF, which only occurs when BglF is engaged in transport (5, 6). In addition to this dephosphorylation, which according to previous data occurs in domain PRD1 of BglG (9), BglG requires phosphorylation at a different site to become active (ref. 16) and this work). This phosphorylation is directly catalyzed by HPr, and its paralog FruB can substitute HPr in this reaction (this work). In vivo (9) and biochemical evidence (this work) suggests that this activating phosphorylation takes place at residue His208 in PRD2. According to previous analysis (16), it serves as a carbon catabolite mechanism down-regulating BglG activity, when other PTS sugars become available in addition to β-glucosides. Transport of these additional PTS sugars drains phosphoryl groups from HPr, leading to a smaller population of active BglG molecules.
Which are the sites in BglG that are phosphorylated by HPr and BglF, respectively? For LicT, the closest relative of BglG in Gram-positive bacteria (42% amino acid sequence identity), genetic and biochemical evidence demonstrated that inhibition by its cognate enzyme II, BglP, occurs through phosphorylation of the first histidine in PRD1 and that the second histidine in this PRD contributes to this regulation. In contrast, the conserved histidines in PRD2 were identified as targets of the positively acting phosphorylations catalyzed by HPr (12, 13). We previously proposed a similar model for BglG with the exception that the second conserved histidine in PRD2 is lacking in BglG (9). The analyses suggested His101 as primary target of BglF-mediated phosphorylation and an auxiliary role for His160, similar to LicT. In contrast, BglG mutants bearing mutations in the conserved His208 in PRD2 lost activity and concomitantly phosphorylation by HPr in vivo. Consequently, His208 was proposed to be the positive site of regulation phosphorylated by HPr (9). This proposal is supported by the current study, in which we show that His208 is essential for phosphorylation of BglG by HPr (or FruB) in vitro (Fig. 4). Taken together, data presented in this work, cellular localization studies (7), and previous data (5, 6, 9, 16) suggest the following mechanism of BglG regulation by the PTS. In the absence of the specific substrate, BglG is sequestered to the membrane by BglF and inactivated by phosphorylation at His101 in the PRD1 domain. Upon addition of a specific β-glucoside, phosphorylation and tethering to the membrane by BglF is released. Then BglG localizes to the cell pole by interaction with EI and HPr in a phosphorylation-independent manner. There, phosphorylation of BglG by HPr (or FruB) at His208 within the PRD2 domain activates BglG, and active BglG relocalizes to the cytoplasm. Therefore, one might speculate that phosphorylation of BglG at the cell pole is the trigger for release of the active dimeric BglG–H208∼P to the cytoplasm, which remains to be tested.
In conclusion, the data suggest that the mechanism of how HPr activates BglG-type antiterminators is similar in Gram-negative and Gram-positive bacteria. This is further supported by our observation that HPr from B. subtilis is also able to phosphorylate and activate BglG (Fig. S3 and ref. 21). Such a similar mechanism of activation (through phosphorylation) of antiterminator proteins of the BglG family in both groups of bacteria appears reasonable because the enterobacterial bgl operon was most likely acquired late in evolution by horizontal gene transfer from a low-GC content Gram-positive bacterium (22).
Materials and Methods
Strains, plasmids, and growth conditions are described in SI Materials and Methods.
Determination of β-Galactosidase Activity.
β-Galactosidase activity assays were performed as described previously (16).
Labeling of Phosphorylated Proteins in Vivo.
Metabolic labeling of phosphorylated proteins using H3[32P]PO4 was carried out as described previously (16). Phosphorylated proteins were separated on 12.5% SDS gels and dried gels were analyzed by phosphoimaging.
Protein Purification.
E. coli strain FT1/pLysS was used for overproduction of the recombinant proteins, which were encoded on plasmids (Table S1). Strep-tagged and His-tagged proteins were purified as described previously (23). However, for Strep-tagged BglG proteins the buffers additionally contained 1 M NaCl to increase the stringency of purification.
In Vitro Phosphorylation Assays.
[32P]-PEP was prepared by isotope exchange reaction using pyruvate kinase as described previously (24). Briefly, the reaction mixture containing 100 mM triethylamine/HCl pH 7.6, 15 mM KCl, 3 mM MgCl2, 165 μM PEP, 1 mM pyruvate, 5 μM ATP, 60 μCi [γ-32P]-ATP (5,000 Ci/mmol) and 40 units pyruvate kinase (Sigma) was set up in a volume of 250 μL and incubated at 30 °C for 90 min. Phosphorylation assays were carried out as described previously (12) and contained in 20 μL volume 25 mM Tris/HCl pH 7.5, 10 mM MgCl2, 1 mM DTT, 1 μL [32P]-PEP solution and the following proteins as indicated: 80 pmol EI, 300 pmol HPr, 100 pmol FruB, and 250 pmol BglG. The reactions were incubated at 37 °C for 30 min and then stopped by adding SDS sample buffer. The samples were heated at 40 °C for 5 min and subsequently separated by 12% or 15% SDS-polyacrylamide gel electrophoresis as indicated in the figure legends. Gels were dried and analyzed by phosphoimaging.
Supplementary Material
Acknowledgments
We thank Sabine Lentes and Elge Koalick for excellent technical assistance and Sebastian Hübner for the gift of plasmid pGP438. This work was supported by Grants from the Deutsche Forschungsgemeinschaft (DFG) (GO1355/7-1 to B.G. and SFB860 to J.S.) and by the DFG Graduiertenkolleg “Biochemie der Enzyme” (to T.B and B.R.). T.B. and F.M.R. were supported by stipends from the “Konrad Adenauer Stiftung” and the “Studienstiftung des Deutschen Volkes,” respectively.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1210443109/-/DCSupplemental.
References
- 1.Schnetz K, Toloczyki C, Rak B. Beta-glucoside (bgl) operon of Escherichia coli K-12: Nucleotide sequence, genetic organization, and possible evolutionary relationship to regulatory components of two Bacillus subtilis genes. J Bacteriol. 1987;169:2579–2590. doi: 10.1128/jb.169.6.2579-2590.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schnetz K, Rak B. Regulation of the bgl operon of Escherichia coli by transcriptional antitermination. EMBO J. 1988;7:3271–3277. doi: 10.1002/j.1460-2075.1988.tb03194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mahadevan S, Wright A. A bacterial gene involved in transcription antitermination: Regulation at a rho-independent terminator in the bgl operon of E. coli. Cell. 1987;50:485–494. doi: 10.1016/0092-8674(87)90502-2. [DOI] [PubMed] [Google Scholar]
- 4.Deutscher J, Francke C, Postma PW. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev. 2006;70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schnetz K, Rak B. Beta-glucoside permease represses the bgl operon of Escherichia coli by phosphorylation of the antiterminator protein and also interacts with glucose-specific enzyme III, the key element in catabolite control. Proc Natl Acad Sci USA. 1990;87:5074–5078. doi: 10.1073/pnas.87.13.5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Q, Arents JC, Bader R, Postma PW, Amster-Choder O. BglF, the sensor of the E. coli bgl system, uses the same site to phosphorylate both a sugar and a regulatory protein. EMBO J. 1997;16:4617–4627. doi: 10.1093/emboj/16.15.4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lopian L, Elisha Y, Nussbaum-Shochat A, Amster-Choder O. Spatial and temporal organization of the E. coli PTS components. EMBO J. 2010;29:3630–3645. doi: 10.1038/emboj.2010.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amster-Choder O, Wright A. Modulation of the dimerization of a transcriptional antiterminator protein by phosphorylation. Science. 1992;257:1395–1398. doi: 10.1126/science.1382312. [DOI] [PubMed] [Google Scholar]
- 9.Görke B. Regulation of the Escherichia coli antiterminator protein BglG by phosphorylation at multiple sites and evidence for transfer of phosphoryl groups between monomers. J Biol Chem. 2003;278:46219–46229. doi: 10.1074/jbc.M308002200. [DOI] [PubMed] [Google Scholar]
- 10.van Tilbeurgh H, Declerck N. Structural insights into the regulation of bacterial signalling proteins containing PRDs. Curr Opin Struct Biol. 2001;11:685–693. doi: 10.1016/s0959-440x(01)00267-6. [DOI] [PubMed] [Google Scholar]
- 11.Stülke J, Arnaud M, Rapoport G, Martin-Verstraete I. PRD—a protein domain involved in PTS-dependent induction and carbon catabolite repression of catabolic operons in bacteria. Mol Microbiol. 1998;28:865–874. doi: 10.1046/j.1365-2958.1998.00839.x. [DOI] [PubMed] [Google Scholar]
- 12.Lindner C, Galinier A, Hecker M, Deutscher J. Regulation of the activity of the Bacillus subtilis antiterminator LicT by multiple PEP-dependent, enzyme I- and HPr-catalysed phosphorylation. Mol Microbiol. 1999;31:995–1006. doi: 10.1046/j.1365-2958.1999.01262.x. [DOI] [PubMed] [Google Scholar]
- 13.Tortosa P, et al. Sites of positive and negative regulation in the Bacillus subtilis antiterminators LicT and SacY. Mol Microbiol. 2001;41:1381–1393. doi: 10.1046/j.1365-2958.2001.02608.x. [DOI] [PubMed] [Google Scholar]
- 14.Gosalbes MJ, Esteban CD, Pérez-Martínez G. In vivo effect of mutations in the antiterminator LacT in Lactobacillus casei. Microbiology. 2002;148:695–702. doi: 10.1099/00221287-148-3-695. [DOI] [PubMed] [Google Scholar]
- 15.Schmalisch MH, Bachem S, Stülke J. Control of the Bacillus subtilis antiterminator protein GlcT by phosphorylation. Elucidation of the phosphorylation chain leading to inactivation of GlcT. J Biol Chem. 2003;278:51108–51115. doi: 10.1074/jbc.M309972200. [DOI] [PubMed] [Google Scholar]
- 16.Görke B, Rak B. Catabolite control of Escherichia coli regulatory protein BglG activity by antagonistically acting phosphorylations. EMBO J. 1999;18:3370–3379. doi: 10.1093/emboj/18.12.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raveh H, Lopian L, Nussbaum-Shochat A, Wright A, Amster-Choder O. Modulation of transcription antitermination in the bgl operon of Escherichia coli by the PTS. Proc Natl Acad Sci USA. 2009;106:13523–13528. doi: 10.1073/pnas.0902559106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Venkatesh GR, et al. BglJ-RcsB heterodimers relieve repression of the Escherichia coli bgl operon by H-NS. J Bacteriol. 2010;192:6456–6464. doi: 10.1128/JB.00807-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwartz E, Herberger C, Rak B. Second-element turn-on of gene expression in an IS1 insertion mutant. Mol Gen Genet. 1988;211:282–289. doi: 10.1007/BF00330605. [DOI] [PubMed] [Google Scholar]
- 20.Powell BS, et al. Novel proteins of the phosphotransferase system encoded within the rpoN operon of Escherichia coli. Enzyme IIANtr affects growth on organic nitrogen and the conditional lethality of an erats mutant. J Biol Chem. 1995;270:4822–4839. doi: 10.1074/jbc.270.9.4822. [DOI] [PubMed] [Google Scholar]
- 21.Reichenbach B, Breustedt DA, Stülke J, Rak B, Görke B. Genetic dissection of specificity determinants in the interaction of HPr with enzymes II of the bacterial phosphoenolpyruvate:sugar phosphotransferase system in Escherichia coli. J Bacteriol. 2007;189:4603–4613. doi: 10.1128/JB.00236-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sankar TS, et al. Fate of the H-NS-repressed bgl operon in evolution of Escherichia coli. PLoS Genet. 2009;5:e1000405. doi: 10.1371/journal.pgen.1000405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landmann JJ, et al. Crh, the paralogue of the phosphocarrier protein HPr, controls the methylglyoxal bypass of glycolysis in Bacillus subtilis. Mol Microbiol. 2011;82:770–787. doi: 10.1111/j.1365-2958.2011.07857.x. [DOI] [PubMed] [Google Scholar]
- 24.Roossien FF, Brink J, Robillard GT. A simple procedure for the synthesis of [32P]phosphoenolpyruvate via the pyruvate kinase exchange reaction at equilibrium. Biochim Biophys Acta. 1983;760:185–187. doi: 10.1016/0304-4165(83)90141-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.