Abstract
There is controversy about whether traditional medicine can guide drug discovery, and investment in bioprospecting informed by ethnobotanical data has fluctuated. One view is that traditionally used medicinal plants are not necessarily efficacious and there are no robust methods for distinguishing those which are most likely to be bioactive when selecting species for further testing. Here, we reconstruct a genus-level molecular phylogenetic tree representing the 20,000 species found in the floras of three disparate biodiversity hotspots: Nepal, New Zealand, and the Cape of South Africa. Borrowing phylogenetic methods from community ecology, we reveal significant clustering of the 1,500 traditionally used species, and provide a direct measure of the relatedness of the three medicinal floras. We demonstrate shared phylogenetic patterns across the floras: related plants from these regions are used to treat medical conditions in the same therapeutic areas. This finding strongly indicates independent discovery of plant efficacy, an interpretation corroborated by the presence of a significantly greater proportion of known bioactive species in these plant groups than in random samples. We conclude that phylogenetic cross-cultural comparisons can focus screening efforts on a subset of traditionally used plants that are richer in bioactive compounds, and could revitalize the use of traditional knowledge in bioprospecting.
Keywords: ethnobotany, ethnopharmacology, herbal medicine, phylogeny, systematics
Many pharmaceutical drugs are derived from plants that were first used in traditional systems of medicine (1), and according to the World Health Organization ∼25% of medicines are plant-derived (http://www.who.int/mediacentre/factsheets/fs134). Discoveries of novel molecules and advances in production of plant-based products (2, 3) have revived interest in natural product research. Traditional knowledge has proven a useful tool in the search for new plant-based medicines (4–8). The number of traditionally used plant species worldwide is estimated to be between 10,000 and 53,000 (9, 10); however, only a small proportion have been screened for biological activity (11, 12) and the plants from some regions are less studied than others. For example, only 1% of tropical floras have been investigated (12). Moreover, there has been no systematic study to determine whether traditionally used species are significantly more likely to yield valuable bioactive compounds. This lack of data creates controversy about whether traditional medicine can guide drug discovery (1, 11, 13–15), and investment in ethnobotanically led bioprospecting has fluctuated (5, 14, 15). Methods put forward for distinguishing those plants most likely to be bioactive when selecting species for further testing have been criticized, and criteria proposed to prioritize traditionally used species have not been rigorously tested (16, 17). For example, use of the same or related plants by people from different regions and cultures provides indirect evidence for bioactivity, if the similarities in medicinal floras are the result of independent discoveries rather than cultural transmissions (18).
Despite studies that show traditional use is concentrated in certain taxonomic groups that are sometimes the same in different cultures (19), cross-cultural comparisons have had limited use in bioprospecting. The disparate regions that have experienced limited cultural contact are floristically disparate too, so different cultures will not be exposed to the same species, genera, or even families (19). Molecular phylogenetic trees can overcome the limitations of taxonomic approaches by measuring phylogenetic distance (20) between the plant species that compose medicinal floras. Thus, phylogenetic methods can overcome the limitations of taxon-based cross-cultural comparisons and provide powerful tools to synthesize and interpret cross-cultural patterns in medicinal floras. Phylogenetic tools have been used to demonstrate clustering of traditionally used (21) and bioactive species (22, 23), and to interpret cross-cultural trends in plant use (21, 24). Here, using methods initially developed for community ecology, we expand these phylogenetic methods further to provide a direct measure of the relatedness of medicinal floras from widely separated parts of the globe.
Careful selection of regional floras to minimize the likelihood of cultural transmission is the basis for demonstrating independent discovery of related plants. If plants from the same lineages are independently discovered, then these plant groups should be strong candidates for bioprospecting. We collated information on medicinal plant use from three geographically separated and botanically disparate regions (Nepal, New Zealand, and the Cape of South Africa). These floras comprise ∼7,000, 4,000, and 9,000 species in total, of which 982 (14%), 165 (4.1%), and 323 (3.6%), respectively, have documented medicinal use. Plant use was divided into 13 categories of medical conditions, and for each region we recorded which plant genera were used medicinally and in which category.
We generated sequence data from one exemplar species for each genus in the three regions, and reconstructed a phylogeny for each of the three floras, as well as a combined phylogeny to represent all three floras. We investigated the distribution of medicinal plants that these hyperdiverse floras encompass and identified nodes on the phylogeny that include significantly more plants traditionally used in medicine (referred to here as “hot nodes”). The identification of these nodes allowed us to determine whether lineages used across the regions are also richer in plants scientifically shown to be bioactive. Our study is unique in being a phylogenetic analysis of traditionally used plants spanning whole floras and different cultures. It shows that related plants are traditionally used as medicines in different regions, and these plant groups coincide with groups that are used to produce pharmaceutical drugs. Some commentators argue that species with perceived magical and cultural significance, which may be acting solely through the placebo effect, are frequently used, and that use in a traditional medical system may not indicate efficacy (25). That we recover a phylogenetic signal in our cross-cultural comparisons is strongly indicative of independent discovery of efficacy, and provides unique large-scale evidence that plant bioactivity underlies traditional medicine.
Results and Discussion
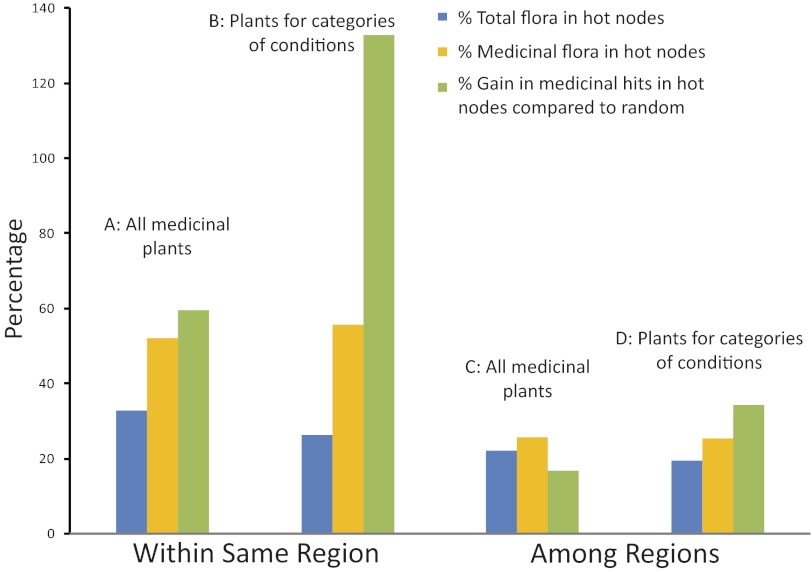
Our investigations of the three separate phylogenetic trees recovered strong phylogenetic signal in medicinal plant use in each of the three regions for the medicinal floras as a whole [using the “comstruct” command in PHYLOCOM v4.1 (26)] (Fig. 1). Phylogenetic clustering was also found for the 13 categories of conditions, and was significant in half of the cases (Table S1). Hot nodes were identified with the option “nodesig” in the same package. On average over the three floras, these nodes encompass 60% more traditionally used plants than expected in a random sample (P < 0.001) (Fig. 2A and Table S2). Medicinal plants for each category of conditions show even more node specificity: on average, hot nodes include 133% more medicinal plants compared with random samples of the floras (P < 0.001) (Fig. 2B, Fig S1A, and Table S2).
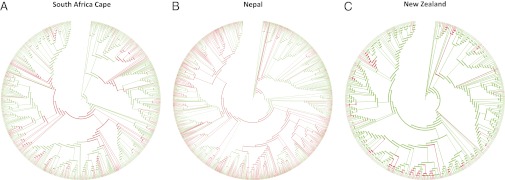
Fig. 1.
Phylogenetic clustering of medicinal floras. Maximum likelihood phylogenetic trees of the floras of the Cape of South Africa (A), Nepal (B), and New Zealand (C), including 80%, 86%, and 88% of the local flora at the genus level, respectively. Traditional medicinal plant use is not scattered randomly, but is concentrated in certain parts of the phylogenetic trees (red branches).
Fig. 2.
Concentration of medicinal plants in phylogenetic hot nodes. Blue and orange bars show percentage of total and medicinal flora included in hot nodes, respectively. Green bars show percentage gain in medicinal plants included in hot nodes. Gain is the difference between the number of medicinal plants in hot nodes, and the number of medicinal plants expected in a random sample of the same size. Within region data (A and B) show the average from the separate values for each of the three regions (Nepal, the Cape of South Africa, and New Zealand). Among regions data (C and D) show the average of pair-wise comparisons on a combined phylogenetic tree of the three floras. These comparisons assessed whether hot nodes from one region can predict medicinal plant use from the other two. Both within region (A and B) and among regions (C and D) hot nodes contain a higher percentage of the medicinal than of the total flora. In all cases (A–D) there is a gain in medicinal hits, compared with a random sample. We show medicinal plants are concentrated in few hot nodes, which overlap noticeably among regions.
Sequence data from the three regions were combined to produce a large phylogenetic tree for the three floras to investigate relationships among medicinal floras. The command “comdist” in PHYLOCOM v4.1 (26) was used to calculate phylogenetic distances between medicinal floras or subsets of them used in different therapeutic areas. We found that these phylogenetic distances were significantly smaller than expected by chance (Table 1), revealing strong agreement in lineages used in the three medicinal floras (Figs. S2–S4). The same is true for plants used for each category of conditions (Table 1). The strong degree of phylogenetic conservatism in plant use observed in this and other studies (21, 27) and the agreement in plant use shown here creates the expectation that hot nodes from one region can predict medicinal lineages in the other two regions. Using the combined phylogenetic tree of the three floras, we recorded the efficiency of the hot nodes from each region to predict medicinal plant use from the other two regions. On average, the hot nodes of one region contain 17% more medicinal plants than expected from the other regions (Fig. 2C). Similarly, hot nodes from one region include 38% more condition-specific medicinal plants than expected from the other regions, which in some cases is a significant increase (Fig. 2D, Fig. S1B, and Table S3).
Table 1.
Agreement in medicinal plant lineages used in Nepal, the Cape of South Africa, and New Zealand
| Floras and categories | Nepal/Cape | Nepal/New Zealand | Cape/New Zealand |
| Medicinal floras† | *** | *** | **(√) |
| Categories‡ | |||
| Cardiovascular/blood purity | *(√) | **(√) | **(√) |
| Dentistry/mouth | **(√) | n.s. | n.s. |
| Gastrointestinal | *** | *** | *** |
| General | **(√) | *** | **(√) |
| Gynecology/fertility | *** | *** | ** |
| Musculoskeletal | *** | *** | *(√) |
| Neurology | *** | **(√) | *(√) |
| Ophthalmology | *(√) | **(√) | n.s. |
| Other | *** | **(√) | **(√) |
| Otorhinolaryngology | *** | *(√) | ** |
| Respiratory/pulmonary | ** | ** | *** |
| Skin | *** | *** | **(√) |
| Urinary | n.s. | n.s. | n.s. |
†Whole medicinal floras.
‡Specific categories of conditions. Significance was assessed by comparing observed values of pairwise phylogenetic distances to those from 10,000 random comparisons per case and P values were the frequency of randomizations with smaller pair-wise distance. *** P < 0.001, **P < 0.01, *P < 0.05; n.s., nonsignificant agreement. Results followed by (√) are those that remain significant after applying a Bonferroni correction within columns to adjust the α-threshold. Note that in uncorrected data, 1/20 significant comparisons may be spurious (type I errors), but the Bonferroni correction adjusts the α-threshold so the study-wide error rate remains at 0.05.
These cross-cultural comparisons reveal that some lineages are more heavily used than others and that these plant lineages are not specific to cultures. Although common cross-cultural patterns in ethnobotany can be attributed to cultural exchange (28), we selected three floras to represent regions with markedly different floristic compositions (Table S4), making communication of plant use less likely. Documented cases of transmission of knowledge report cross-cultural use of the same of species, and more rarely congeners (29–32). However, even when excluding medicinal plant genera found in more than one region to minimize the effect of possible cultural transmission in our analysis, congruence in plant use remains significant (Table S5).
A more likely explanation for cross-cultural phylogenetic conservatism in the selection of species is that it can be attributed to phylogenetic conservatism in phytochemistry (33) and that these lineages have been selected independently because of their bioactivity. Recent studies have shown that demonstrable medicinal properties are shared between close relatives (21, 23), corroborating this view. To assess whether underlying bioactivity shapes the distribution of the lineages highlighted from these cross-cultural comparisons, we extracted all of the plant genera from which pharmaceuticals are either approved or under trial from a recent large-scale study (22) (Table S6) and evaluated the presence of bioactive species in these lineages. First, we found that there are statistically more drug genera in the three medicinal floras under study than expected by chance (P = 0.041), demonstrating drug development has concentrated around plants with known uses in traditional medicine. Second, a randomization sampling across the phylogenetic tree showed that the hot nodes encompass more drug genera than expected (P = 0.001). This finding suggests that plants included in hot nodes have high potential to deliver new medicines. Here, we list the genera found in the hot nodes of at least two of the three regions for the medicinal floras and the 13 categories of conditions (Tables S7–S20).
More than 80% of plant species have not been investigated for bioactivity (11) and methods to distinguish those plants most likely to be bioactive when selecting species for further testing are needed. The finding that medicinal plant use shows strong phylogenetic clustering indicates targeting close relatives of plants with known bioactivity (5, 23) or phylogenetic medicinal hotspots identified as hot nodes (21) is a good strategy for focused screening. However, we propose a more sophisticated framework of identifying plants with high medicinal potential based on traditional medicine, combining two criteria: phylogenetic signal and cross-cultural agreement. We have shown that lineages fulfilling these two criteria are significantly richer in plants with demonstrable bioactivity than a random sample. Several taxa from these plant clusters have no reported traditional uses, suggesting this method could lead to new discoveries not only from species with recorded uses, but also from previously overlooked plants. Our observations reveal the predictive power of traditional medicine in bioprospecting and promise to have implications for bioprospecting.
Random collection of plants to be evaluated for bioactivity has been the method of choice for pharmaceutical companies over the past few decades, because automated high-throughput screening has enabled the rapid testing of thousands of samples against specific targets and requires little or no previous knowledge of the medicinal uses of the plant. Ethnobotanically directed strategies, on the other hand, seemingly require more financial resources, because fieldwork needs to be carried out over long periods by trained ethnobotanists (5). Although there are numerous case studies highlighting a possible role for ethnobotanically directed plant collections in delivering new bioactive molecules (5, 7, 8, 34) and in research leading to the discovery of new medicines (35, 36), alone they cannot attest to the power of ethnobotanically directed approaches over those involving random screening. However, traditional knowledge goes beyond identifying plants that are likely to deliver new medicines or lead molecules for further development. Data on the medical conditions for which plants are used traditionally can suggest suitable targets to investigate in assays, information regarding which plant parts are normally used will avoid the unnecessary testing of inactive plant organs, and details of how the plant material is processed and the route of administration can indicate how best to prepare an extract for testing. Once plant groups that are most likely to deliver new products have been highlighted based on traditional knowledge, increasing the number of relevant assays on these plants can also contribute to improving hit rates. The more detailed the ethnobotanical knowledge available, especially on the nature of illnesses treated by a particular plant, the better the assays can be targeted. For example, the type of pain treated may indicate whether a plant has antinociceptive or anti-inflammatory (or both) effects, which will enable more appropriate assays to be selected. Application of the novel approaches tested in this study can contribute to the compilation of traditional knowledge “into a usable form” (1) to substantially increase outcomes of focused bioscreening efforts over random collections.
A decrease in investment in ethnobotanically directed natural products research schemes in the 1990s has been attributed to the Convention on Biological Diversity (CBD) (37). As more access and benefit-sharing partnerships and agreements are established, it appears that safeguarding the intellectual property rights of the people originating traditional knowledge need not be incompatible with the commercialization of natural products. Nevertheless, bioprospecting is still viewed with suspicion by many developing nations (38) and although issues have been addressed by the Nagoya Protocol adopted by the CBD, several concerns persist (39). Ethnobotanically directed natural products research could enable capacity building and enhance health in human study populations, outcomes which would be supported by contemporary ethnobotanists, whose primary objective is to provide benefit to local and indigenous communities and their associated ecosystems (40). From this perspective, there may be an ethical mandate to test the safety and efficacy of all traditionally used species, rather than just the ones that are potential drug leads. Conversely, in terms of compliance with the CBD, any method which can focus searches will not only improve success rates and reduce costs, but may also render drug discovery initiatives easier to negotiate. These approaches can only be realized when robust ethnobotanical data have been gathered. We have exploited relatively well-known pharmacopoeias, but further development of these methods depends on support for ethnobotanical fieldwork, which has never been strong by funding agencies. Particularly as traditional medicine practices and knowledge are eroded (41, 42), there is a pressing need for ethnobotanical fieldwork to complete inventories. Traditional knowledge enhances health worldwide and is considered crucial in preventing the deterioration of local healthcare (43). As new research initiatives demonstrate the revived interest in traditional knowledge (44–46), we show that disentangling the tree of life and novel methods of analysis can help better use the natural world in the future.
Materials and Methods
Ethnomedicinal Information.
For the three regions of study ethnomedicinal uses in the local ethnopharmacopoeias were recorded for each species and information was then organized at the genus level. Data for the medicinal uses were gathered from compilations of ethnobotanical uses for New Zealand (47–49), Nepal (50, 51), and the Cape of South Africa (52, 53), as well as the online databases of Ngā Tipu Whakaoranga database for New Zealand (http://maoriplantuse.landcareresearch.co.nz) and the Survey of Economic Plants for Arid and Semi-Arid Lands for the Cape of South Africa (http://www.kew.org/ceb/sepasal). We selected regions for which pharmacopoeias are well known. Pharmacopoeias and floras correspond geographically for Nepal and New Zealand. It is possible that some species distributed both outside and within the Cape find use outside the Cape but not within it, but are scored as medicinal. The high level of endemism of the Cape flora (70% the flora of the Cape of South Africa is endemic to that region) limits the numbers of species for which this may be true. That some medicinal plant use may not be recorded renders our analyses more conservative and our interpretations more robust. Medical conditions were grouped into 13 categories of conditions used in similar ethnobotanical studies (54, 55). These categories were: cardiovascular problems/blood purity, dentistry/mouth, gastrointestinal conditions, general, gynecology/fertility, neurological conditions, ophthalmology, musculoskeletal conditions, skin conditions, other, otorhinolaryngology, respiratory/pulmonary ailments, and urinary conditions. The use of each medicinal genus was scored for each of these categories.
DNA Sequencing and Phylogeny Reconstruction.
Phylogenetic relationships of the three floras were reconstructed at the genus level, sampling one exemplar species from each genus. Where possible, that species was present in the local flora, but in cases where a DNA sequence or plant material was not available, species of the same genus from other localities were used. The plastid DNA marker rbcL was selected for this study because of the availability of the marker, its successful amplification across plant lineages, and its capacity to resolve phylogenetic relationships at this level in large-scale studies (27, 56, 57). Publicly available sequences were compiled and previously unavailable sequences for 327 genera were generated. Sampling reached a total of 792 sequences for the Cape of South Africa (∼80% of the flora), 494 sequences for New Zealand (>88% of the flora), and 1,335 genera from Nepal (>85% of the total flora). Plant material was collected from herbarium specimens from the herbaria of the Royal Botanic Garden Edinburgh, the Royal Botanic Gardens Kew, and the University of Reading, and DNA material for certain genera was provided from the DNA Bank of the Royal Botanic Gardens Kew (http://apps.kew.org/dnabank/homepage.html). All accessions are detailed in Tables S21–S23. Total DNA was extracted from 0.2 to 0.3 g of leaf or flower tissue from herbarium or silica-gel dried material using a modification (58) of the CTAB method (59) or with the Qiagen DNeasy Plant mini kit (Qiagen) following the manufacturer’s protocol. DNA was purified using QIAquick columns (Qiagen) following the manufacturer’s protocol. Amplification and sequencing of the rbcL gene were performed for two overlapping fragments with combinations of primers and the protocols from Olmstead et al. (60) and Fay et al. (61) as used in Forest et al. (27). PCR purification and DNA sequencing from both strands were performed partly by Macrogen and partly at the Royal Botanic Gardens Kew using an Applied Biosystems 3730 capillary DNA automated sequencer (Applied Biosystems) using Big Dye terminator v3.1 chemistry, following the manufacturer’s protocols (Applied Biosystems). Complementary strands were assembled and edited with EditSeq (DNASTAR). Alignment of rbcL sequences was performed in BioEdit v. 7.0 using CLUSTAL W (62) and adjustments were made manually. Phylogenetic trees were constructed for individual floras and a large phylogenetic tree representing plant relationships in all three regions was also constructed by combining the sequence data from the three floras. For this analysis, when a genus was present in more than one flora, it was included only once in the matrix. Sequence data were analyzed under the maximum-likelihood (ML) criterion, under the GTR+I+Γ model as implemented in RAxML 7.2.8 (63).
Phylogenetic and Statistical Analyses.
To assess phylogenetic signal in medicinal plant use, the “comstruct” command in PHYLOCOM v4.1 (26) was used. This metric assesses the significance of phylogenetic signal for a set of taxa subsampled from a phylogeny by calculating the mean phylogenetic distance (MPD) for each sample (group of species on the phylogeny) and comparing it to MPD values for 1,000 randomly generated samples. For each run, the samples were randomized by shuffling species labels across the entire phylogeny. The one-tailed P value for the significance of phylogenetic signal was calculated by dividing the number of randomly generated samples that were more clustered on the phylogeny than the observed sample (rankLow) by the number of runs (1,000). Additionally, we calculated the net relatedness index (NRI), a standardized effect size measure of phylogenetic community structure that describes the difference between average phylogenetic distances in the observed and randomly generated samples, standardized by the SD of phylogenetic distances in the null communities. The sign of NRI informs whether the observed sample was more clustered (NRI > 0) or more dispersed (NRI < 0) than the null samples on the phylogeny (26).
To identify hot nodes (nodes that encompass significantly more medicinal plants than the rest of the tree), the “nodesig” command in PHYLOCOM v4.1 (26) was implemented for each of the three phylogenetic trees. This option was used to determine the position of phylogenetic clustering by testing each node of the phylogenetic tree for overabundance in medicinal terminal taxa distal to it. Observed patterns for each of the three medicinal floras were compared with those for random samples of the same size per case, drawn from the phylogeny. For these hot nodes in each of the three regions, we recorded the percentage of the total and medicinal flora included in them. We compared the observed number of traditionally used taxa encompassed in the hot nodes to the one expected to be found randomly in the percentage of the flora encompassed in the hot nodes; this was the gain in percentage of medicinal hits compared with random. Significance of gain in medicinal hits was assessed with binomial tests.
The above analyses were performed for the medicinal floras as a whole and for the plants in the different categories of conditions. For all statistical tests where multiple comparisons across therapeutic areas are made, both uncorrected and Bonferroni-corrected results are presented. The correction provides a conservative estimate of significance, minimizing type I errors but inflating type II errors; considering uncorrected results, 5% of significant findings may be type I errors. In all cases the correction was performed within table columns to adjust the α-threshold.
Cross-Cultural Congruence Analyses.
To test whether the same lineages are used medicinally in the three regions (cross-cultural agreement), we used the option “comdist” in PHYLOCOM v4.1 (26) that outputs the pair-wise distance matrix between samples, based on MPDs of all possible pairs of taxa in one sample to the taxa in the other. Pair-wise MPDs were calculated between the medicinal floras, as well as for the subsets of medicinal floras used for the 13 categories of conditions between Nepal, New Zealand, and the Cape of South Africa. These tests were performed on the combined phylogenetic tree of the three floras. To assess significance of observed MPDs between samples, 100 random samples of the same size as the observed one per case were generated, drawing only from the local total flora on the phylogenetic tree. MPDs were calculated between these random samples (10,000 permutations per pair-wise comparison) and the frequency of the cases with smaller MPD than the observed was the P value. These randomizations were performed for the medicinal floras and for the subsets used for each category of conditions. To assess whether cultural exchange could be one of the factors shaping cross-cultural patterns, we omitted all medicinal plant genera found in more than one region and performed the same analysis for the three medicinal floras without these genera. To test the extent to which phylogenetic patterns in one region (source region) predict traditional use in the other two regions (target regions), the following test was performed: on the combined phylogenetic tree of the three regions, hot nodes were identified for each of the three source regions, as described earlier. The percentage of the medicinal and total floras of the two target regions included within those nodes were recorded, as well as the gain in medicinal hits compared with random. This analysis was performed for the three medicinal floras and for the13 categories of conditions. In each case, the significance of gain in medicinal hits was assessed with binomial tests.
Traditional Use and Bioactivity.
To assess whether traditional use coincides with drug use, we extracted the list of genera producing drugs or under trials for drug production (drug genera) from a recent large-scale study (22) (Table S4) and recorded the number of these genera found in the three medicinal floras under study. To assess the significance in the number of drug genera in the medicinal floras, we generated 1,000 samples of genera of the same size as the three medicinal floras from The Plant List (http://www.theplantlist.org), a recently published database that aims to record all plant species. We calculated the number of drug genera in each of the samples. To estimate whether hot nodes are significantly rich in bioactive plants, we recorded the number of drug genera within these nodes and in 1,000 random sets of genera of the same size sampled from the phylogeny. In both cases, the frequency of the instances with more drug genera in the random sample than the observed was the one-tailed P value.
Supplementary Material
Acknowledgments
We thank Martyn Powell, Andrew Meade, Stephanie Bird, and the members of the Royal Botanic Gardens Kew DNA Bank Database for technical assistance; Campbell Webb for providing PHYLOCOM support; the University of Reading Statistical Advisory Service for advice on statistical analyses; and Ketan Patel and three anonymous reviewers for comments on initial drafts of the manuscript. This study was financially supported by a John Spedan Lewis Fellowship (to J.A.H, V.S., and E.M.W.) to support the doctoral studies of C.H.S.-L.; and V.S. was supported by the European Research Council, the Natural Environment Research Council, Leverhulme Trust, and the Royal Society.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. B.B.S. is a guest editor invited by the Editorial Board.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. JQ933200–JQ933526).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1202242109/-/DCSupplemental.
References
- 1.Fabricant DS, Farnsworth NR. The value of plants used in traditional medicine for drug discovery. Environ Health Perspect. 2001;109(Suppl 1):69–75. doi: 10.1289/ehp.01109s169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ro D-K, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- 3.Graham IA, et al. The genetic map of Artemisia annua L. identifies loci affecting yield of the antimalarial drug artemisinin. Science. 2010;327:328–331. doi: 10.1126/science.1182612. [DOI] [PubMed] [Google Scholar]
- 4.Cox PA. Will tribal knowledge survive the millennium? Science. 2000;287:44–45. doi: 10.1126/science.287.5450.44. [DOI] [PubMed] [Google Scholar]
- 5.Cox PA, Balick MJ. The ethnobotanical approach to drug discovery. Sci Am. 1994;270:82–87. [PubMed] [Google Scholar]
- 6.Taniguchi M, Kubo I. Ethnobotanical drug discovery based on medicine men’s trials in the African savanna: Screening of east African plants for antimicrobial activity II. J Nat Prod. 1993;56:1539–1546. doi: 10.1021/np50099a012. [DOI] [PubMed] [Google Scholar]
- 7.Lewis W. Pharmaceutical discoveries based on ethnomedicinal plants: 1985–2000 and beyond. Econ Bot. 2003;57(1):126–134. [Google Scholar]
- 8.Balick MJ. In: Bioactive Compounds from Plants. Chadwick DJ, Marsh J, editors. Chichester: Wiley & Sons; 1990. pp. 22–31. [Google Scholar]
- 9.Schippmann U, Leaman DJ, Cunningham AB. 2002. in Biodiversity and the Ecosystem Approach in Agriculture, Forestry and Fisheries. Satellite Event on the Occasion of the Ninth Regular Session of the Commission on Genetic Resources for Food and Agriculture Rome, 12–13 October. Inter-Departmental Working Group on Biological Diversity for Food and Agriculture, ed. (Food and Agriculture Organization, Rome)
- 10.McChesney JD, Venkataraman SK, Henri JT. Plant natural products: Back to the future or into extinction? Phytochemistry. 2007;68:2015–2022. doi: 10.1016/j.phytochem.2007.04.032. [DOI] [PubMed] [Google Scholar]
- 11.Soejarto DD, et al. Ethnobotany/ethnopharmacology and mass bioprospecting: issues on intellectual property and benefit-sharing. J Ethnopharmacol. 2005;100:15–22. doi: 10.1016/j.jep.2005.05.031. [DOI] [PubMed] [Google Scholar]
- 12.Gurib-Fakim A. Medicinal plants: Traditions of yesterday and drugs of tomorrow. Mol Aspects Med. 2006;27:1–93. doi: 10.1016/j.mam.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 13.Makhubu L. Bioprospecting in an African context. Science. 1998;282:41–42. doi: 10.1126/science.282.5386.41. [DOI] [PubMed] [Google Scholar]
- 14.Firn RD. Bioprospecting – Why is it so unrewarding? Biodivers Conserv. 2003;12:207–216. [Google Scholar]
- 15.Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod. 2012;75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gertsch J. How scientific is the science in ethnopharmacology? Historical perspectives and epistemological problems. J Ethnopharmacol. 2009;122:177–183. doi: 10.1016/j.jep.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 17.Heinrich M, Edwards S, Moerman DE, Leonti M. Ethnopharmacological field studies: A critical assessment of their conceptual basis and methods. J Ethnopharmacol. 2009;124:1–17. doi: 10.1016/j.jep.2009.03.043. [DOI] [PubMed] [Google Scholar]
- 18.Bletter N. A quantitative synthesis of the medicinal ethnobotany of the Malinké of Mali and the Asháninka of Peru, with a new theoretical framework. J Ethnobiol Ethnomed. 2007;3:36. doi: 10.1186/1746-4269-3-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saslis-Lagoudakis CH, Williamson EM, Savolainen V, Hawkins JA. Cross-cultural comparison of three medicinal floras and implications for bioprospecting strategies. J Ethnopharmacol. 2011;135:476–487. doi: 10.1016/j.jep.2011.03.044. [DOI] [PubMed] [Google Scholar]
- 20.Faith DP. Conservation evaluation and phylogenetic diversity. Biol Conserv. 1992;61(1):1–10. [Google Scholar]
- 21.Saslis-Lagoudakis CH, et al. The use of phylogeny to interpret cross-cultural patterns in plant use and guide medicinal plant discovery: An example from Pterocarpus (Leguminosae) PLoS ONE. 2011;6:e22275. doi: 10.1371/journal.pone.0022275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu F, et al. Clustered patterns of species origins of nature-derived drugs and clues for future bioprospecting. Proc Natl Acad Sci USA. 2011;108:12943–12948. doi: 10.1073/pnas.1107336108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rønsted N, Savolainen V, Mølgaard P, Jager AK. Phylogenetic selection of Narcissus species for drug discovery. Biochem Syst Ecol. 2008;36:417–422. [Google Scholar]
- 24.Hart KH, Cox PA. A cladistic approach to comparative ethnobotany: Dye plants of the the southwestern United States. Journal of Ethnobiology. 2000;20:303–325. [Google Scholar]
- 25.Firenzuoli F, Gori L. Herbal medicine today: Clinical and research issues. Evid Based Complement Alternat Med. 2007;4(Suppl 1):37–40. doi: 10.1093/ecam/nem096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Webb CO, Ackerly DD, Kembel SW. Phylocom: Software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics. 2008;24:2098–2100. doi: 10.1093/bioinformatics/btn358. [DOI] [PubMed] [Google Scholar]
- 27.Forest F, et al. Preserving the evolutionary potential of floras in biodiversity hotspots. Nature. 2007;445:757–760. doi: 10.1038/nature05587. [DOI] [PubMed] [Google Scholar]
- 28.Reyes-Garcia V, et al. Ethnobotanical knowledge shared widely among Tsimane’ Amerindians, Bolivia. Science. 2003;299:1707. doi: 10.1126/science.1080274. [DOI] [PubMed] [Google Scholar]
- 29.Leporatti ML, Ivancheva S. Preliminary comparative analysis of medicinal plants used in the traditional medicine of Bulgaria and Italy. J Ethnopharmacol. 2003;87:123–142. doi: 10.1016/s0378-8741(03)00047-3. [DOI] [PubMed] [Google Scholar]
- 30.Voeks R. Sacred leaves of Brazilian Candomble. Geogr Rev. 1990;80(2):118–131. [Google Scholar]
- 31.de Albuquerque UP, Andrade LHC. Etnobotánica del género Ocimum L. (Lamiaceae) en las comunidades afrobrasileñas [Ethnobotany of the genus Ocimum L. (Lamiaceae) in Afro-Brazilian communities] An Jardin Botanico Madr. 1998;56(1):107–118. Spanish. [Google Scholar]
- 32.Bennet BC, Gomez AP. In: Las Plantas y el Hombre [Plants and Man] Rios M, Borgtoff Pedersen H, editors. Quito, Ecuador: Abya-Yala; 1991. pp. 129–137. Spanish. [Google Scholar]
- 33.Agrawal AA, Salminen J-P, Fishbein M. Phylogenetic trends in phenolic metabolism of milkweeds (Asclepias): Evidence for escalation. Evolution. 2009;63:663–673. doi: 10.1111/j.1558-5646.2008.00573.x. [DOI] [PubMed] [Google Scholar]
- 34.Cox PA. In: Bioactive Compounds from Plants. Chadwick DJ, Marsh J, editors. Chichester: Wiley & Sons; 1990. pp. 40–47. [Google Scholar]
- 35.Sehgal N, et al. Withania somnifera reverses Alzheimer’s disease pathology by enhancing low-density lipoprotein receptor-related protein in liver. Proc Natl Acad Sci USA. 2012;109:3510–3515. doi: 10.1073/pnas.1112209109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harvey AL. Natural products in drug discovery. Drug Discov Today. 2008;13:894–901. doi: 10.1016/j.drudis.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 37.Wynberg R, Laird S. 2009. Indigenous Peoples, Consent and Benefit-Sharing. Learning from the San-Hoodia Case, eds Wynberg R, Schroeder D, Chennells R (Springer, Dordrecht, The Netherlands) pp. 69–86.
- 38.Dalton R. Natural resources: Bioprospects less than golden. Nature. 2004;429:598–600. doi: 10.1038/429598a. [DOI] [PubMed] [Google Scholar]
- 39.Watanabe KN, Teh GH. Wanted: Bioprospecting consultants. Nat Biotechnol. 2011;29:873–875. doi: 10.1038/nbt.2001. [DOI] [PubMed] [Google Scholar]
- 40.McClatchey WC, Mahady GB, Bennett BC, Shiels L, Savo V. Ethnobotany as a pharmacological research tool and recent developments in CNS-active natural products from ethnobotanical sources. Pharmacol Ther. 2009;123:239–254. doi: 10.1016/j.pharmthera.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Voeks R, Leony A. Forgetting the forest: Assessing medicinal plant erosion in Eastern Brazil. Econ Bot. 2004;58:S294–S306. [Google Scholar]
- 42.Srithi K, Balslev H, Wangpakapattanawong P, Srisanga P, Trisonthi C. Medicinal plant knowledge and its erosion among the Mien (Yao) in northern Thailand. J Ethnopharmacol. 2009;123:335–342. doi: 10.1016/j.jep.2009.02.035. [DOI] [PubMed] [Google Scholar]
- 43.McDade TW, et al. Ethnobotanical knowledge is associated with indices of child health in the Bolivian Amazon. Proc Natl Acad Sci USA. 2007;104:6134–6139. doi: 10.1073/pnas.0609123104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bagla P. Traditional medicine. Piercing the veil of Ayurveda. Science. 2011;334:1491. doi: 10.1126/science.334.6062.1491. [DOI] [PubMed] [Google Scholar]
- 45.Tian P. Convergence: Where West meets East. Nature. 2011;480:S84–S86. doi: 10.1038/480S84a. [DOI] [PubMed] [Google Scholar]
- 46.Xu Z. Modernization: One step at a time. Nature. 2011;480:S90–S92. doi: 10.1038/480S90a. [DOI] [PubMed] [Google Scholar]
- 47.Brooker SG, Cambie RC, Cooper RC. New Zealand Medicinal Plants. Auckland: Heinemann; 1981. [Google Scholar]
- 48.Brooker SG, Cambie RC, Cooper RC. Economic native plants of New Zealand. Econ Bot. 1989;43(1):79–106. [Google Scholar]
- 49.Riley M. Māori Healing and Herbal. Paraparaumu: Viking Sevenseas; 1997. [Google Scholar]
- 50.Manandhar NP. Plants and People of Nepal. Portland, Oregon: Timber Press; 2002. [Google Scholar]
- 51.Suwal PN. Medicinal Plants of Nepal. Thapathali: Department of Medicinal Plants, His Majesty's Government of Nepal, Ministry of Forest and Soil Conservation; 1993. [Google Scholar]
- 52.Neuwinger HD. African Traditional Medicine. Stuttgart: Medpharm Scientific; 2000. [Google Scholar]
- 53.Van Wyk B-E, van Oudtshoorn B, Gericke N. Medicinal Plants of South Africa. Pretoria: Briza; 1997. [Google Scholar]
- 54.Leonti M, Ramirez FR, Sticher O, Heinrich M. Medicinal flora of the Popoluca, Mexico: A botanical systematical perspective. Econ Bot. 2003;57:218–230. [Google Scholar]
- 55.Moerman DE. The medicinal flora of Native North America: An analysis. J Ethnopharmacol. 1991;31:1–42. doi: 10.1016/0378-8741(91)90141-y. [DOI] [PubMed] [Google Scholar]
- 56.Chase MW, et al. Phylogenetics of seed plants—An analysis of nucleotide-sequences from the plastid gene rbcL. Ann Mo Bot Gard. 1993;80:528–580. [Google Scholar]
- 57.Savolainen V, et al. Phylogenetics of flowering plants based on combined analysis of plastid atpB and rbcL gene sequences. Syst Biol. 2000;49:306–362. doi: 10.1093/sysbio/49.2.306. [DOI] [PubMed] [Google Scholar]
- 58.Csiba L, Powell MP. 2006. in DNA and Tissue Banking for Biodiversity and Conservation: Theory, Practice and Uses, eds. Savolainen V, Powell MP, Davis K, Reeves G, Corthals A (Royal Botanic Gardens Kew, Richmond, Surrey), pp. 114–117.
- 59.Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin. 1987;19(1):11–15. [Google Scholar]
- 60.Olmstead RG, Michaels HJ, Scott KM, Palmer JD. Monophyly of the Asteridae and identification of their major lineages inferred from DNA sequences of rbcL. Ann Mo Bot Gard. 1992;79:249–265. [Google Scholar]
- 61.Fay MF, et al. Plastid rbcL sequence data indicate a close affinity between Diegodendron and Bixa. Taxon. 1998;47(1):43–50. [Google Scholar]
- 62.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.