Abstract
How fish larvae are protected from infection before the maturation of adaptive immunity, a process which may take up to several weeks in most species, has long been a matter of speculation. Using a germ-free model, we show that colonization by commensals in newly hatched zebrafish primes neutrophils and induces several genes encoding proinflammatory and antiviral mediators, increasing the resistance of larvae to viral infection. Commensal microbe recognition was found to be mediated mainly through a TLR/MyD88 signaling pathway, and professional phagocytes were identified as the source of these immune mediators. However, the induction of proinflammatory and antiviral genes, but not of antimicrobial effector genes, also required the covalent modification of histone H3 at gene promoters. Interestingly, chromatin modifications were not altered by commensal microbes or hatching. Taken together, our results demonstrate that gene-specific chromatin modifications are associated with the protection of zebrafish larvae against infectious agents before adaptive immunity has developed and prevent pathologies associated with excessive inflammation during development.
Keywords: epigenetic, cytokines, evolution, gene regulation, live imaging
Our understanding of the development of the vertebrate immune system has increased greatly during recent decades. Findings so far strongly suggest that events that occur in the early life stages of life have a profound impact on the organism’s later development (1), structure (2), and function (3). Traditionally, the study of animal development has focused on understanding how interactions among animal cells trigger developmental pathways. A frequent assumption is that all the steps involved in the development of a complex multicellular organism are genetically preordained (4). Only now, however, the field of ecological developmental biology begun to focus on the idea that some developmental triggers may come from the environment (5). Particularly interesting are the events that happen around the time of birth. In this early developmental stage, all vertebrates are subjected to an imminent colonization by a diverse microbiota inhabiting the surrounding environment. Among humans of developed countries, such microbial influence has tended to be avoided, or, at least diminished, through high standards of hygiene. In contrast, researchers into host–microbe relationships recently have established that the first contact between the two entities is essential for the maturation of immunity. This phenomenon has been called “developmental immunologic programming” (DIP) (6). DIP is a process whereby an environmental factor acting during a sensitive or vulnerable developmental period exerts effects that impact the structure and function of organs in ways that, in some cases, persist throughout life (7). This process is not exclusive to higher vertebrates; indeed, in lower taxa, such as fish, it has been known as “bacterial priming” (8). Therefore, DIP seems to be a conserved feature that has been preserved throughout the evolutionary process.
Immune-competent cells recognize microbial components that are not present in any of their structures but are conserved among pathogens. Recognition occurs mainly via receptors that are expressed in all cells of a given type (9). Receptors of the innate immune system, called “pattern-recognition receptors,” form two well-studied families with huge recognition capacities: the transmembrane Toll-like receptors (TLRs) and the intracytoplasmic Nod-like receptors (10). Their role in sensing is indispensable, and in mammals it is well recognized that all TLRs discovered to date, with the exception of TLR3, signal via their associated adaptor molecule, myeloid differentiation primary response protein 88 (MyD88) (11). Similarly, an almost complete set of TLRs has been described in the pufferfish Fugu rubripes and in the zebrafish Danio rerio (12), and, with the exception of TLR4 (13, 14), these TLRs seem to be functional orthologs of mammalian TLRs. Thus, they are able to sense the same ligands (15–17), use similar adaptor molecules for signaling (15), and activate the transcription factor NF-κB (13, 15, 18). In addition, it has been shown that in zebrafish MyD88 modulates innate immune responses to microbes (19–22).
Apart from the recognition mechanisms, vertebrates have developed means to support large societies of microbial partners during their life cycles. However, one puzzling issue is that the supported microbial partners have the same conformational, molecular, or locomotive structures as closely related pathogens, indicating that microbe-associated molecular patterns (MAMPs) are not limited to pathogens. Therefore, herein we use the term “MAMPs” instead of the term “pathogen-associated molecular patterns” (PAMPs).
This recognition feature is particularly conspicuous in teleost fish, because, although this phylogenetic group has developed a completely and fully functional immune system, it lives in one of the most aggressive habitats, the aquatic ecosystem. In this unique habitat an impressive number and diversity of microorganisms coexist (23–26). Therefore, fish might have specific functional mechanisms for discriminating between the threats of a pathogenic origin and the signals that come from commensals. The mechanisms controlling this fragile equilibrium are largely unknown but are thought to be mediated by specialized receptors, such as the TLRs. Although beneficial, this response could threaten the host integrity when uncontrolled and directed toward the host. In an effort to understand how microbial commensals are host-supported, powerful germ-free (GF) experimental approaches have been developed. For example, it has been noticed that intestines of GF mice can initiate but cannot complete their differentiation when particular associations between host and microbes are lacking (4). Using a similar model, Hrncir et al. (27) found that bacterial LPS, which is present as a contaminant in mouse food, is needed to develop a healthy phenotype. Unfortunately, available model species are scarce, and most findings under GF conditions are derived largely from a few mammalian species, mainly rodents (28) and swine (29). Only recently, another species, the zebrafish, has emerged as a key and powerful model organism to widen basic and biomedical animal research (30–35). To date, the study of host–microbe interactions in GF conditions in fish has proven effective for studying developmental processes, including immunity (36, 37). Growing evidence in GF zebrafish larvae throws light on the profound impact that bacteria have in the gut when it first is internally colonized. As in other vertebrate models, the main changes involved rapid epidermal degeneration and altered enterocyte morphology and proliferation (36, 38–40). In addition, a recent study showed that the presence not only of commensals but also of multiple bacterial secretion factors is essential for influencing intestinal epithelial cell proliferation, suggesting that normal gut microbiota exert a direct modulation of the β-cathenin pathway, independent of inflammation (22). In the present study, we used zebrafish larvae as a vertebrate model of innate immunity to test the hypothesis that the contact with commensal microbiota after hatching is an essential external environmental triggering factor to shape the development of immunity and disease resistance.
Results
Zebrafish Innate Immune Development Is Regulated by Microbial Presence.
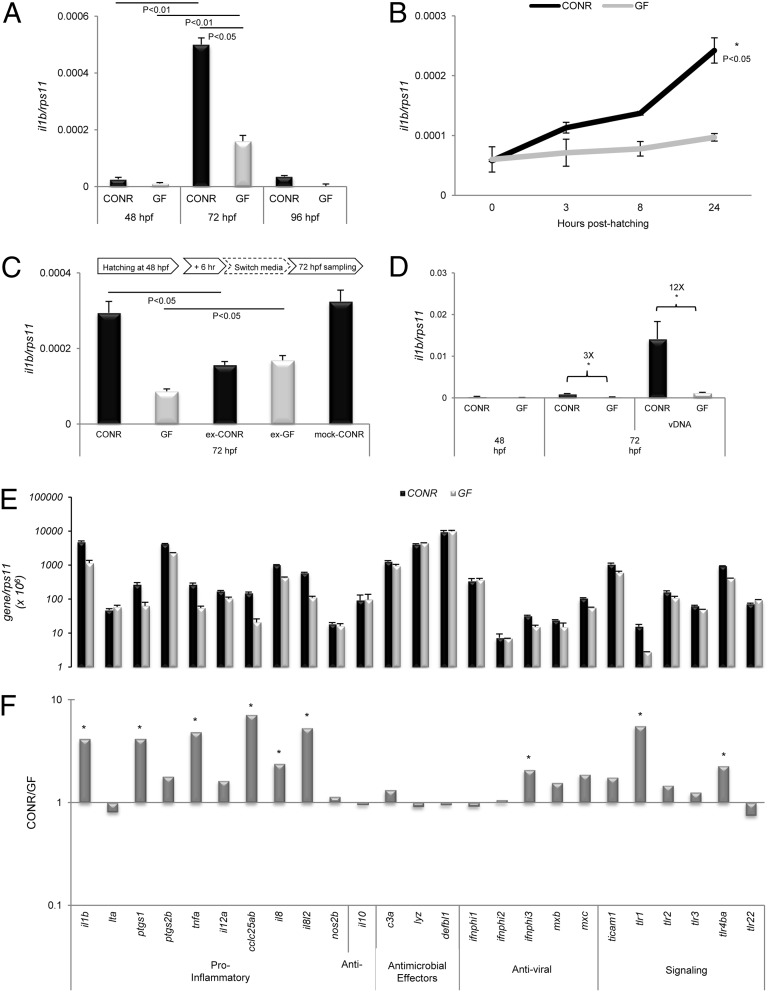
To characterize the host response of zebrafish embryos after their first exposure to commensal microorganisms in rearing water, wild-type zebrafish embryos were raised under GF or conventional (CONR) conditions until 96 h postfertilization (hpf). Under standard culture conditions, the animals started to hatch at 48 hpf. Thus, to study the effect of the contact of CONR animals with commensal microbes after hatching, analyses were conducted at 48, 72, and 96 hpf. At 48 hpf, animals in both groups presented similar levels of IL-1β transcripts. Differences between CONR and GF fish became more evident at 72 hpf, returning to basal levels at 96 hpf (Fig. 1A). To clarify whether this response increased gradually during the time of continuous exposure to commensal microbes, a time-series analysis was performed. Zebrafish embryos were allowed to hatch naturally from their eggs at 48 hpf, and 30 individual samples from each group were collected and pooled at 0, 3, 8, and 24 h posthatching (hph). Datasets revealed a time-dependant response of CONR hatched embryos compared with the GF embryos (Fig. 1B). The response of the CONR group was characterized by the gradually increasing expression of IL-1β gene transcripts, with a maximum at 72 hpf, i.e., 24 hph. However, no significant changes were observed in the GF group in the same time period.
Fig. 1.
Bacterial stimulation in the newly hatched zebrafish results in the activation of a mild inflammatory reaction followed by immunological tolerance. (A) qRT-PCR assay (IL-1β) of whole 48-, 72-, and 96-hpf naturally hatched larvae reared CONR or GF. (B) Starting 3 h after hatching (48 hpf), significantly higher IL-1β transcripts were found in CONR than in GF larvae. (C) Animals in both groups were raised following standard CONR or GF techniques and conventionalized at 48 hpf, 6 h after hatching. A mock switch (Mock-CONR) with CONR larvae handled identically but maintained in the same condition was included also. A switch in IL-1β transcript levels was observed in 72-hpf ex-CONR and ex-GF larvae. (D) Proinflammatory activity in zebrafish larvae raised in CONR or GF conditions revealed a threefold change between groups starting at 72 hpf. At this time, we tested the microbial priming effect using 50 larvae of each group that were incubated with 50 μg/mL vDNA for 4 h. (E) Commensals up-regulate genes intimately associated with innate immunity (identified in F) to significantly higher levels in CONR than in GF animals at the time of hatching. (F) Fold increases of statistical significance (denoted by asterisks) were recorded in several genes of the CONR group. Data are representative of two repeated trials in which all samples were run in triplicate (Student’s t test, P < 0.05). Error bars indicate SD.
Next we investigated whether the observed differences in IL-1β expression in CONR and GF zebrafish could be attributed only to the fish sensing of commensal microbes. This hypothesis was tested through a short reversal experiment in which fish raised as CONR or GF were switched to the opposite medium at 54 hph (Fig. 1C). As expected, a significant shift occurred in the IL-1β expression of ex-CONR and -GF groups as compared with the normal basal response at 72 hpf. However, IL-1β expression was unaffected in a mock swap control in which the CONR larvae were handled identically but maintained in the same condition (Fig. 1C). This reversal process suggests that the IL-1β induction previously recorded in CONR and GF conditions was triggered as a response to a major environmental pressure which could be exerted only by commensal microbes. Interestingly, these observations also confirmed that only a short period of contact with commensal microbes is needed to start the induction of the proinflammatory IL-1β gene. To analyze the innate immune response further, Vibrio anguillarum DNA (vDNA) was selected as immunogenic MAMP in addition to commensal microbes in the CONR or as a first signal in GF fish. The larvae of both groups were treated with a bath of vDNA for 4 h at 48 and 72 hpf. At 48 hpf, only a slight, nonsignificant increase was observed in the CONR group, indicating that zebrafish embryos in both groups respond similarly if they have not been exposed to microbes for prolonged periods of time. At 72 hpf, two major changes in the mRNA levels of the IL-1β gene could be distinguished. In the groups without vDNA pretreatment, a significant but moderate increase in the response was recorded in the CONR group as compared with the GF group (Fig. 1D). Nevertheless, both groups pretreated with vDNA for 4 h exhibited a strong induction of IL-1β expression. In the CONR group, particularly, IL-1β gene expression increased significantly, by more than 12-fold as compared with the treated GF group or more than 30-fold compared with the CONR group without vDNA pretreatment.
Quantitative real-time RT-PCR (qPCR) analysis of several gene markers of innate immunity in both CONR and GF zebrafish at 72 hpf revealed several shared features of the host response and reinforced the hypothesis that the presence of commensal microbes triggers the overall inflammatory immune response in CONR fish, although not all inflammatory genes responded to the microbial presence (Fig. 1E). The group of induced genes included the proinflammatory effectors IL-1β, prostaglandin-endoperoxidase synthase 1 (PTGS1, formerly COX1), and TNFα; the chemokines C-C motif chemokine C25ab (CCL-C25ab), IL-8, and IL-8–like 2; and the antiviral mediator IFNΦ3. It became particularly evident that the presence of commensal microbiota failed to trigger a significant induction of genes encoding antimicrobial effectors, such as lysozyme (LYZ), defensin β-like 1 (DEFBL1), complement component c3a (C3a), and anti-inflammatory cytokines, such as IL-10 (Fig. 1F). Intriguingly, a few genes encoding TLRs, such as TLR1 and TLR4ba, also were induced drastically in the CONR group.
Zebrafish Neutrophil Functions, but Not Myelopoiesis, Are Affected by the Microbiota.
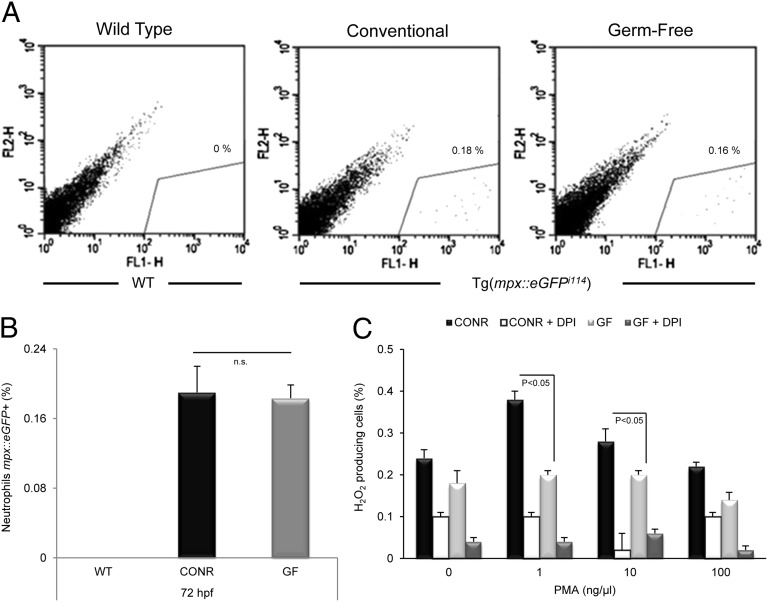
We next investigated the impact of the presence of commensal microbes on myelopoiesis and neutrophil activity using Tg(mpx::eGFPi114) zebrafish larvae, whose neutrophils express eGFP (32). The results showed that the numbers of eGFP-expressing neutrophils within total cell suspensions from CONR and GF zebrafish did not differ significantly (Fig. 2 A and B). However, a strong difference between the CONR and GF groups was observed in response to phorbol-myristate 13-acetate (PMA) (Fig. 2C). Thus, although PMA was able to trigger H2O2 production in cells from the CONR group, no response was observed in GF cells. The specificity of the reaction was confirmed using the NADPH oxidase inhibitor diphenyleneiodonium (DPI), which inhibited H2O2 production almost completely. These observations suggest that, even if microbial signals do not promote a higher rate of myelopoiesis, their presence primes professional phagocytes.
Fig. 2.
Microbial presence primes neutrophils but does not affect myelopoiesis. Single-cell suspensions were prepared from wild-type whole larva raised in CONR conditions and Tg(mpx::eGFPi114) larvae with positive eGFP neutrophils raised in CONR and GF conditions (n = 200 per group). (A) Representative dot-plots of FL1 vs. FL2 of the three groups showing as percentages the relative contribution of the gated compartment. (B) Average of three independent replicas of mpx::eGFP flow cytometric detection analysis demonstrating that the cell population labeled eGFP-positive was not significantly different in CONR and GF larvae. (C) Microbial presence primed the respiratory burst of total cell suspensions of 72-hpf zebrafish larvae (n = 3). To achieve this response, control cells were incubated for 1 h with the NADPH oxidase inhibitor DPI to avoid spontaneous respiratory burst activity. The respiratory burst was measured as the total DHR-fluorescence triggered by PMA. Data are presented as fold increase relative to cells incubated with medium alone. Statistically significant differences between the CONR and GF groups are shown (Student’s t test; P < 0.05). Error bars indicate SD. n.s., nonsignificant.
Commensal Microbes Modulate Neutrophil Recruitment and Activation.
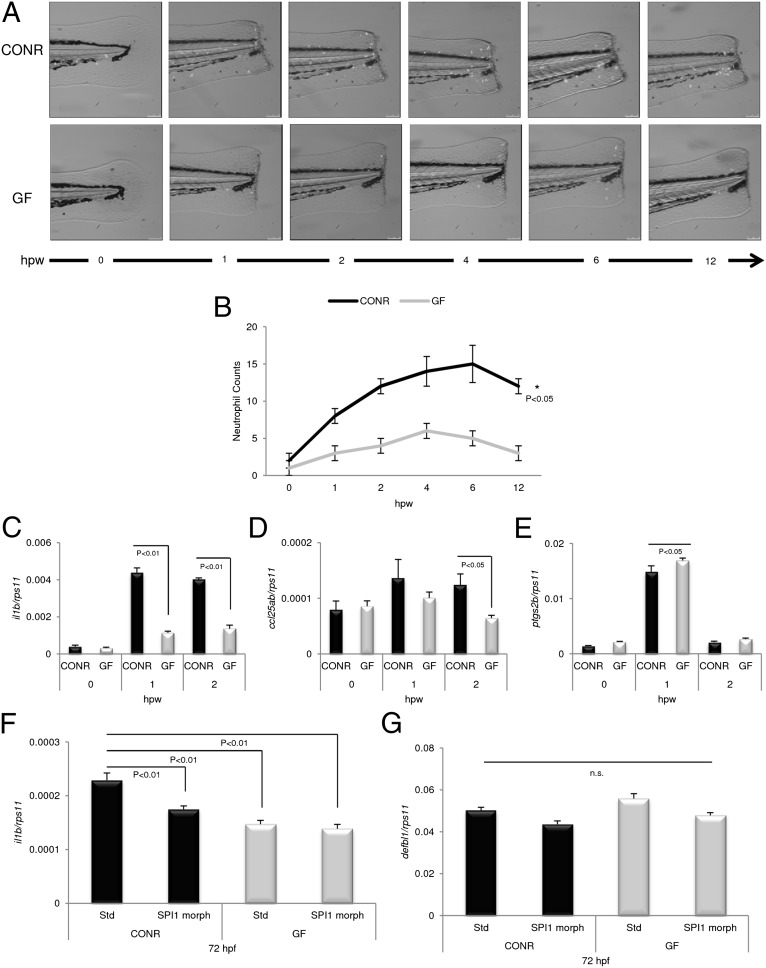
So far, we have shown that immediately after hatching CONR embryos recognize the presence of microbes and that such recognition is translated into microbial priming. Our next goal was to explore whether this priming affects neutrophil kinetics after the wounding of Tg(mpx::eGFPi114) zebrafish raised in CONR and GF conditions. Following sterile protocols, the tip of the tail fin in each larva was transected at 72 hpf. Wounding induced a faster and more robust recruitment of neutrophils to the injury site in the CONR group (Fig. 3A), although the injury took longer to resolve than in GF embryos (Fig. 3B). To explore the mechanisms related to this response, we collected the last segment of the fish, from anus to tail tip, and used qPCR to quantify the gene expression of the inflammatory mediators after wounding, namely two proinflammatory genes (IL-1β and PTGS2B) and the gene encoding chemokine CCL-C25ab. We found that exposure to microbes before and during wounding was sufficient to cause the rapid induction of both IL-1β (Fig. 3C) and CCL-C25ab (Fig. 3D) genes, beginning just 1 h after injury. In contrast, PTGS2b transcript levels increased similarly in both CONR and GF groups after wounding (Fig. 3E). The impaired induction of IL-1β and CCL-C25ab after wounding in the GF group may explain the reduced neutrophil recruitment observed in this group. In addition, the results also confirm that not all proinflammatory genes are affected by commensal priming.
Fig. 3.
Microbial presence increases neutrophil recruitment in vivo and induces the expression of proinflammatory mediators. Tail fins of Tg(mpx::eGFPi114) CONR and GF zebrafish were transected at 72 hpf, and the number of fluorescent neutrophils visible in the tail was assessed by fluorescence microscopy. (A) Representative images of positive neutrophils expressing eGFP recruited to wounding site in a CONR and a GF zebrafish larva. hpw, hours postwounding. (B) Positive cells in animals from both groups were quantified visually from high-quality pictures from a 12-h time series. Inflammatory events were significantly faster, stronger, and took more time to resolve in the CONR group than in GF embryos (*P < 0.05). n = 50 larvae per group and sampling point. (C–E) At the time points indicated, 80 individual fish were anesthetized and then a sterile scalpel was used to incise the body between the anus and the wounded tail tip. Individual samples of the same batch were pooled and were immersed immediately in TRIzol for quantification of IL-1β (C), CCL-C25ab (D), and PTGS2B (E) mRNA levels by qPCR. (F and G) Zebrafish eggs were microinjected at the one-cell stage with 8 ng STD or SPI morpholinos per egg and then were divided into two batches. One batch was raised as CONR, and the other was derived as GF. IL-1β (F) and DEFBL1 (G) transcript levels, assayed by qPCR, of 72-hpf SPI morphants and their STD morphant siblings, raised CONR or GF. Error bars indicate the SD of three independent experiments, each using 30 pooled larvae per treatment. n.s., nonsignificant.
These results, together with the recent observation that neutrophils and macrophages mediate the proinflammatory effects of IFN-γ in zebrafish larvae (41), led us to hypothesize that these cells also might be involved in the priming by commensal microbes in newly hatched larvae. To test this hypothesis, neutrophils and macrophages were depleted by silencing the master myeloid transcription factor SPI1 (also known as “PU.1”) with a translation-blocking morpholino (42). Strikingly, an impaired induction of IL-1β gene expression was observed in SPI1-deficient larvae of the CONR group (Fig. 3F), but the expression of the antimicrobial effector DEFBL1 was unaffected (Fig. 3G).
Microbial Priming by Commensal Microbes Is Essential for Protecting Zebrafish Larvae from Viral Infection.
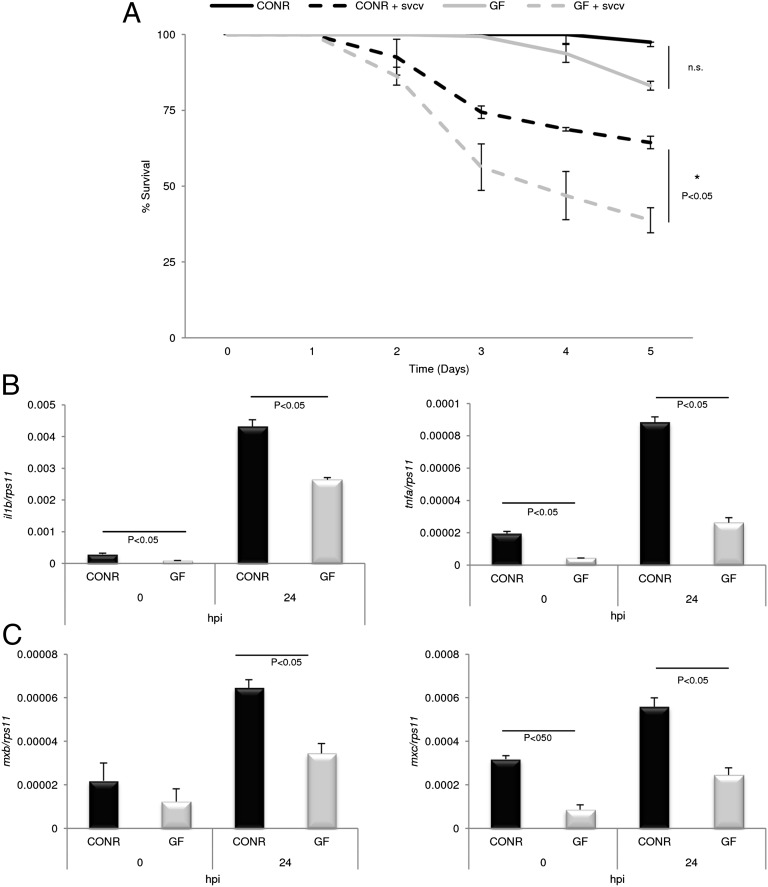
We next evaluated the disease resistance of CONR and GF zebrafish larvae at 72 hpf using a spring viremia of carp virus (SVCV) infection model (43) in which the virus is cleared by the IFN system (43, 44). Interestingly, the naturally primed CONR group showed increased resistance to SVCV as compared with GF larvae (Fig. 4A). Differences in mortality were not statistically significant under basal conditions. To look for direct evidence of enhanced transcriptional regulation as a result of the microbial priming, we analyzed the expression profile of several genes encoding proinflammatory and antiviral effectors in CONR and GF groups at 0 and 24 h postinfection (hpi) with SVCV. The results showed that, although the infection resulted in increased mRNA levels of proinflammatory (Fig. 4B) and antiviral (Fig. 4C) genes at 24 hpi in both groups, the induction of all genes was more robust in CONR than in GF larvae. These results strongly suggest that microbial priming at hatching is essential for further fortifying the innate immune response of fish and that the lack of such priming is detrimental for the resolution of infection.
Fig. 4.
Commensal priming after hatching potentiates zebrafish disease resistance. (A) Condition-dependent killing of CONR vs. GF 72-hpf zebrafish larvae exposed to 108 TCID50/mL SVCV by immersion. Survival curves differ among groups (log-rank test, P < 0.05). n = 30 larvae in duplicate experiments. (B and C) The transcript levels of proinflammatory IL-1β and TNFα (B) and antiviral MxB and MxC (C), measured by qRT-PCR, were significantly lower in GF larvae than in CONR at 24 hpi (P < 0.05). Data are representative of two separate trials in which all samples were run in triplicate. Error bars indicate SD. n.s., nonsignificant.
Commensal Microbes Are Recognized Through a TLR/MyD88 Signaling Pathway in Zebrafish Larvae.
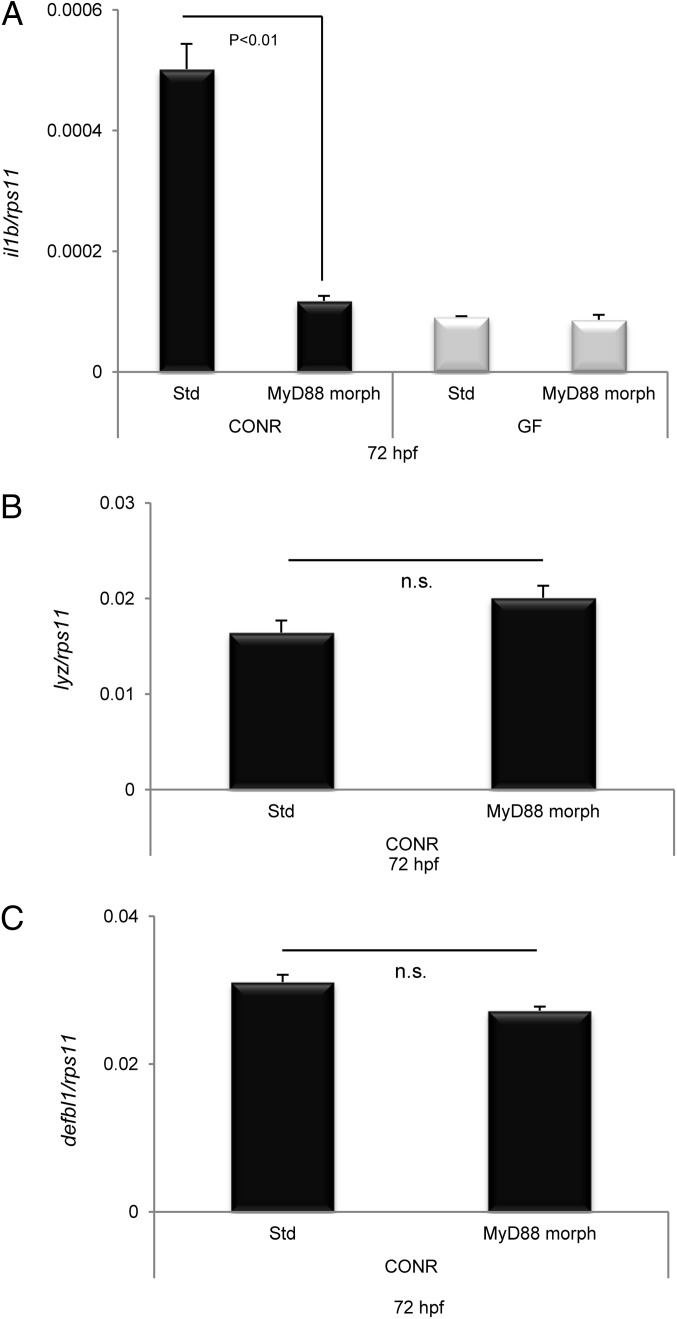
At this stage, we hypothesized that commensal microbes could influence the observed inflammatory response of newly hatched zebrafish embryos through direct transmembrane TLR signaling. To test this hypothesis, MyD88 was inhibited genetically using a translation-blocking morpholino (20). mRNA levels of IL-1β were significantly lower in MyD88-deficient fish than in their normal siblings (Fig. 5A). Strikingly, these levels correlated with those observed in their wild-type siblings raised under GF conditions. In sharp contrast, however, the mRNA levels of the LYZ (Fig. 5B) and DEFBL1 (Fig. 5C) genes in response to commensals was similar in MyD88-deficient fish and their wild-type siblings.
Fig. 5.
MAMPs signal through a TLR/MyD88 signaling pathway. Zebrafish eggs were microinjected at the one-cell stage with STD or MyD88 morpholinos, 4 ng per egg, and then were divided into two batches. One batch was raised as CONR, and the other was derived as GF. IL-1β (A), LYZ (B), and DEFBL1 (C) transcript levels, assayed by qPCR, of 72-hpf MyD88 morphant zebrafish and their STD morphant siblings raised CONR or GF. Error bars indicate the SD of two independent experiments, each using 30 pooled larvae per treatment. n.s., nonsignificant.
Histone Modifications Regulate Innate Immune Development.
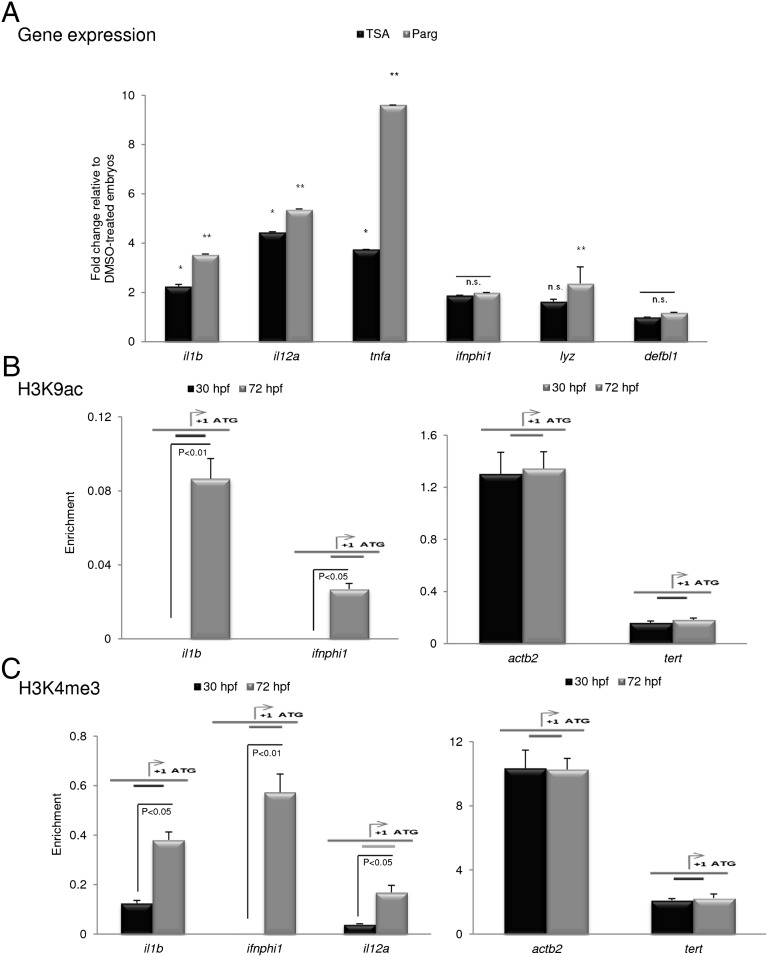
The induction of IL-1β expression in GF zebrafish after hatching, although at much lower levels than in CONR animals, suggests the existence of another microbe-independent mechanism that would activate a transient immune response when the embryos are devoid of their protective chorion. Furthermore, we previously observed the poor induction of genes encoding proinflammatory and antiviral molecules in larvae upon injection of MAMPs (13) or after viral infection (44), even though NF-κB was strongly induced. We then asked whether the expression of the immune genes was regulated at the level of chromatin remodeling via the covalent modification of histones, as has been observed in murine macrophages (45). Because acetylation of histone H3 at lysine 9 (H3K9ac) and trimethylation at lysine 4 (H3K4me3) mark transcriptionally active genes (46, 47), we treated zebrafish embryos microinjected with vDNA at the one-cell stage with trichostatin A (TSA), a histone deacetylase inhibitor, or pargyline, an inhibitor of the H3K4 demethylase LSD1 (48), and measured the induction of genes encoding IL-1β, IL-12α, TNFα, IFNφ1, LYZ, and DEFBL1. We confirmed that TSA and pargyline increased H3K9 acetylation and H3K4 trimethylation, respectively, at the IL-1β promoter (Fig. S1). In addition, vDNA injection significantly increased the mRNA levels of IL-1β and IFNφ1 (Fig. S2), as expected from its ability to activate NF-κB robustly in zebrafish embryos (13, 18). More interestingly, both TSA and pargyline facilitated the vDNA-mediated induction of IL-1β, IL-12α, and TNFα genes in embryos at 24 hpf but had a weak effect on the expression of IFNφ1, LYZ, and DEFBL1 (Fig. 6A). In addition, at 72 hpf zebrafish showed a marked increase in H3K9 acetylation (Fig. 6B) and H3K4 trimethylation (Fig. 6C) at the IL-1β and IFNφ1 promoters. Although the trimethylation of H3K4 at the IL-12α promoter showed a tendency similar to that of IL-1β and IFNφ1 (Fig. 6C), we did not observe H3K9 acetylation at this promoter, suggesting that other residues of this or other histones, most probably histone H4 (49), were acetylated. Importantly, these chromatin modifications were specific for the genes encoding these proinflammatory and antiviral factors, because the level of H3K9 acetylation (Fig. 6B) and H3K4 trimethylation (Fig. 6C) at the housekeeping genes β-actin2 and telomerase reverse transcriptase was similar in embryos (30 hpf) and larvae (72 hpf). In addition, neither of these histone modifications was observed at the promoters of LYZ and DEFBL1. Finally, we examined H3K4me3 at the IL-1β and β-actin2 promoters at a later time point, when inflammatory gene expression has dropped back down, to determine whether transient induction of inflammatory genes is accompanied by stable chromatin remodeling. The results showed that although H3K4me3 remained stable at the β-actin 2 promoter, it declined significantly at the IL-1β promoter (Fig. S3).
Fig. 6.
Histone modifications are differentially regulated in proinflammatory and antimicrobial effector genes. (A) Zebrafish eggs microinjected at the one-cell stage with 5–10 ng vDNA per egg were dechorionated manually at 20 hpf and were treated with 3 μM pargyline (Parg) or 100 nM TSA for 5 h. IL-1β, IL-12a, TNFα, IFNφ1, LYZ, and DEFBL1 transcript levels then were assayed by qPCR. (B and C) CONR embryos/larvae were analyzed by ChIP (H3K9ac and H3K4me3, respectively, at the indicated times. The amplicon used for each promoter is indicated by a diagram above the bars. Error bars indicate the SD of two independent experiments, each using 30 pooled larvae per treatment. *P < 0.05 vs. DMSO-treated embryos. **P < 0.01 vs. DMSO-treated embryos. n.s., nonsignificant.
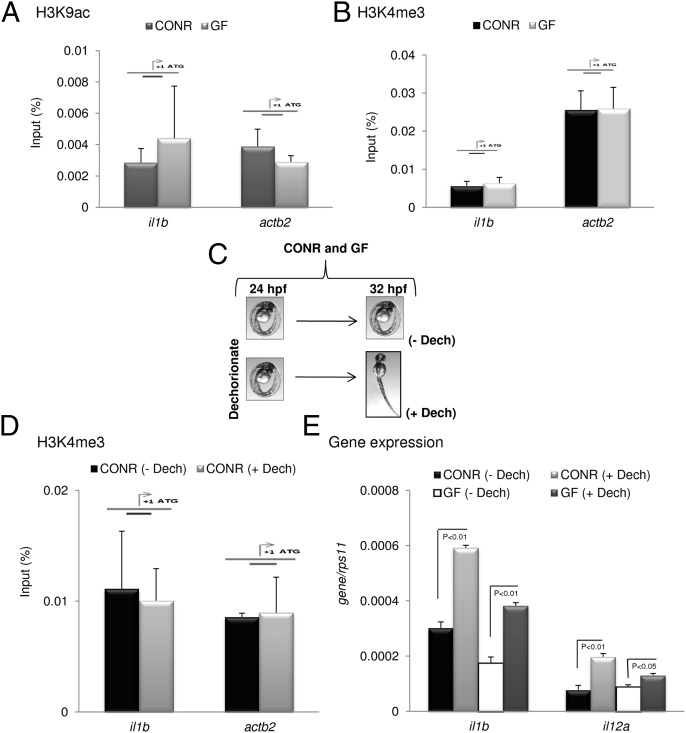
We next examined whether the level of H3K9ac and H3K4me3 at the promoter of IL-1β was regulated by the presence of microbes during development. The results pointed to a similar degree of acetylation and trimethylation of these residues in CONR and GF larvae (Fig. 7 A and B). However, injection of vDNA was able to increase H3K9ac and H3K4me3 robustly at the IL-1β promoter (Fig. S4), suggesting that a stronger inflammatory stimulus may alter these histone covalent modifications. Furthermore, we also found that the level of trimethylation of H3K4 was not triggered by hatching, because it was similar at IL-1β and β-actin2 promoters in manually dechorionated and nondechorionated CONR embryos at 32 hpf (Fig. 7 C and D). In sharp contrast, however, 8 h later manually dechorionated CONR and GF embryos showed higher IL-1β and IL-12α transcript levels than nondechorionated embryos (Fig. 7 C and E), suggesting that a third factor, in addition to the presence of commensal microbes and chromatin modifications, also orchestrates the induction of immune genes after hatching.
Fig. 7.
Histone modifications are not regulated by commensals or hatching. (A and B) Naturally hatched CONR and GF zebrafish larvae were analyzed by ChIP (H3K9ac and H3K4me3) at 72 hpf. (C) Scheme showing the generation of CONR and GF embryos after manual dechorionation. These embryos were analyzed by ChIP (H3K4me3) (D) or gene expression (IL-1β and IL-12a) (E) at 32 hpf. The amplicon used for each promoter is indicated by a diagram above the bars. Error bars indicate the SD of two independent experiments, each using 30 pooled larvae per treatment.
Discussion
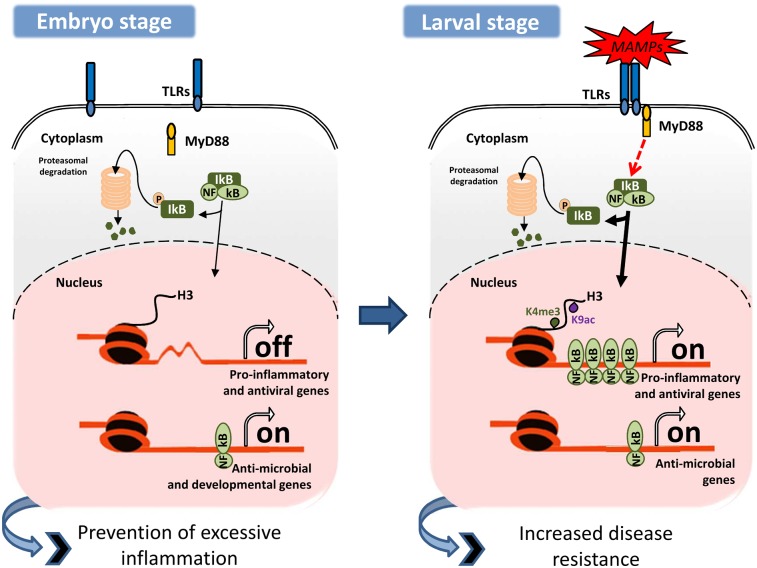
In the present study commensal microbes were found to have the capacity to prime the innate immunity of zebrafish larvae by inducing a transitory inflammatory response that, together with constitutively expressed antimicrobial effectors, acts to increase the resistance of zebrafish larvae to infectious disease soon after hatching. Commensals immediately surround cells, tissues, and indeed, the whole organism in vivo, colonizing the external surfaces of the vertebrate body and soon assembling into dense gastrointestinal communities (50). Vertebrate exposure to indigenous gut microbiota and their microbial products has been recognized widely to be a key element for the development and maturation of the mammalian immune system (4, 36, 51). Recently, Rawls’ team (40) found that the zebrafish digestive tract is robustly responsive to microbial colonization because of the impressive number and diversity of bacteria that assemble in the organ compared with the amount present in the microenvironment of the larvae. Thus, the majority of previous host–microbe studies targeted the gastrointestinal tract (40). However, our present data in CONR- and GF-raised zebrafish larvae revealed increased transcripts of major proinflammatory markers, chemokines, and some antiviral effectors and TLRs as early as 54 hpf, when definitive hematopoiesis already has begun (52); the levels of these transcripts increased until 72 hpf, when maximum activity was recorded. These differences, together with the priming of neutrophils observed in CONR larvae and the inability of larvae depleted of neutrophils and macrophages to induce proinflammatory genes after hatching, demonstrate that the CONR group is able to respond to several environmental pressures at hatching. Comparison with the GF group shows that commensal microbes are mainly responsible for this response. Intriguingly, several studies with zebrafish larvae have suggested that commensal microbiota colonize the larvae as soon as they hatch, but all assign great importance to the role played by gut microbiota in modifying innate immunity, nutrient metabolism, impaired intestinal differentiation, and function when microbes invade the internal organs (19, 22, 36–38, 40, 53). Also, in contrast to our work, it previously was proposed that host defense mechanisms, such as proinflammatory cytokines, are inducible mostly upon recognition of virulence factors of pathogenic bacteria (9, 52, 54–56). However, it should be noted that spontaneous infection was not recorded in the present study; thus the “inflammatory state” observed in CONR fish was transient, rapidly returning to basal levels, marked here by the levels of IL-1β expression similar to those observed in the GF fish 24 h later (i.e., at 96 hpf). It is possible that the immunological tolerance observed here could be a direct response to microbial burden on the skin. In vitro studies in mammals have suggested that apical contact of commensals with the epithelium attenuates NF-κB activation via the regulation of PPARδ and inhibition of the cellular proteosome activity required for IκB degradation (57, 58). Further studies are necessary to clarify these issues. In any case, these results again indicate that the ability of immune-competent cells is strikingly reinforced by the mere presence of commensal microbes at hatching. However, we observed no difference between the two groups in the number of neutrophils, suggesting that commensal microbes do not regulate neutrophil differentiation or apoptosis. In contrast, it has been reported recently that LPS is able to promote granulopoiesis in zebrafish embryos through the induction of granulocyte-colony stimulating factor (59). These differences might be explained by the nature, duration, and potency of the stimuli provided.
GF animals provide a healthy but susceptible host for experimental colonization with defined microbial populations (60). Here, we have established that the transient inflammatory response of young larvae could be reversed partially in both directions, as seen at 72 hpf, from the increased or decreased mRNA IL-1β levels in the respective experimental groups Previous studies in which the GF zebrafish phenotype was rescued by exposing animals to commensal microbiota 3 or 6 days postfertilization support these findings (36). However, studies with GF mice colonized with human or mouse fecal microflora observed only a partial protection to oral tolerance, suggesting that colonization of GF mice after birth is unlikely to recapitulate fully the colonization of newborn mice (61, 62) and that bacterial subproducts released during the egg stage may influence the development of the immune system. Indeed, the development of host immunity is a complex process that requires a series of coordinated events, including the functional differentiation of immune cells, expansion of immune populations, and the expression of other immune functions (52). In the present study, the administration of bacterial CpG motifs in zebrafish larvae at 72 hpf induced the expression of IL-1β in the CONR group, confirming that previous exposure to commensals increases immunocompetence in fish, similar to findings in mammals (63).
Consistent with our results, the presence of assembled commensal microbes in newborn mice has been identified as a potent stimulus for an increase in immune-related genes (61, 62). In addition, a transcriptomic analysis in zebrafish demonstrated that some immune genes were expressed specifically in response to certain microorganisms, indicating that the early colonization of the gastrointestinal tract by a particular microorganism may be responsible for the changed metabolism in the fish larvae (36). In a functional genomics analysis, the same authors observed induction of the complement factor component 3 (C3) after bacterial colonization of the gastrointestinal tract. Notably we observed that C3, like the other genes encoding antimicrobial effectors, showed similar expression levels in CONR and GF animals. This result was not unexpected, because fish surfaces comprise a large area of delicate epithelium, the major route of entry for pathogenic microorganisms; thus, healthy fish, as well as mammals, are capable of limiting infection effectively through the release of systemically expressed antimicrobial effectors that neutralize a broad range of microbes (64–67). Indeed we speculate that these mechanisms are shared features among several phyla and constitute a primitive immune defense mechanism among a wide range of eukaryotic organisms. Priming could be either an evolutionary selective pressure, in which the functionality of the neonate immune system is tested and members presenting any immunodeficiency are excluded from the group through pathogenic microorganisms, or, alternatively, priming simply may be a method of training the immune system to support the enormous number of bacteria that the gut will harbor throughout the life of the vertebrate.
Our results also present clear evidence that MAMPs are sensed via a TLR/MyD88 signaling pathway, which in turn leads to the transcriptional up-regulation of proinflammatory and antiviral genes, and suggest that TLRs are responsible for sensing MAMPs and the induction of inflammation in CONR animals. Strikingly, however, the expression of genes encoding antimicrobial effectors is independent of MyD88. Taken together, these results further suggest the importance of the TLR/MyD88 signaling pathway in the maturation of innate immunity triggered by commensal microbes and that the expression of gene-encoding antimicrobial effectors is independent of MyD88 or the presence of microbes, pathogenic or not. Similarly, it was found recently that the colonization of GF zebrafish by a commensal microbiota activated NF-κB via a MyD88 signaling pathway and led to the up-regulation of its target genes in intestinal and extraintestinal tissues of the digestive tract (40).
Our study also demostrates the involvement of chromatin modification in the gene-specific control of immunity during development. Although the genes encoding antimicrobial effectors, which are particularly essential for the defense of embryos/larvae in the absence of adaptive immunity and do not have the potential to cause tissue damage, are constitutively expressed at high levels during development, the genes encoding proinflammatory and antiviral mediators are activated transiently after hatching through the extensive methylation and acetylation of histone H3 at their promoters. Although these chromatin modifications were observed similarly in CONR and GF animals, our data cannot rule out an effect of commensals in their regulation at later developmental stages. In fact, injection of vDNA, which can induce NF-κB activation strongly (13, 18), robustly increased H3K9ac and H3K4me3 at the IL-1β promoter but not at the β-actin2 promoter, suggesting that a stronger inflammatory stimulus, which may be provided by pathogenic microbes, also may fine-tune the regulation of chromatin covalent modifications at proinflammatory gene promoters. In any case, because the major signaling molecules of TLRs, including NF-κB (68), MAPK (69), and PI3K (70), are required for the regulation of important developmental genes, we propose that nucleosome remodeling through covalent histone modification regulates the activation of proinflammatory and antiviral genes after hatching while preventing the pathology associated with excessive inflammation during early development. This mechanism is reminiscent of the one regulating the expression of proinflammatory (class T, tolerizeable) and antimicrobial (NT, nontolerizeable) genes in mouse macrophages (45), so we speculate that this mechanism is evolutionarily conserved in all vertebrate classes.
In summary, our results demonstrate that sensing of commensal microbes via both the TLR/MyD88/NF-κB signaling pathway and gene-specific chromatin modifications is associated with the protection of zebrafish larvae against infectious agents before adaptive immunity has developed and at the same time prevents pathologies associated with excessive inflammation during development.
Materials and Methods
Zebrafish Husbandry.
All experiments with live animals were performed using protocols approved by the European Union Council Guidelines (86/609/EU) and the Bioethical Committee of the University of Murcia (approval no, 333/2008). Zebrafish gametes were naturally expressed from wild-type (obtained from the Zebrafish International Resource Center), and the Tg(mpx::gfp)i114 (32) line held at our facilities following standard husbandry practices. Animals were constantly subjected to a 12/12-h light/dark cycle and maintained at 28.5 °C.
In each experiment, zebrafish embryos were produced through natural breeding conditions in clean breeding tanks with autoclaved system egg water containing 60 μg/mL sea salts in distilled water. As fish spawned, embryos were collected for 1 h and were transferred immediately to a sterile Petri dish. To generate and rear GF zebrafish embryos, one half of the total eggs collected was washed with antiseptics and derived as GF in a Telstar class II/B3 biological safety cabinet, following the protocol described in ref. 71 with slight modifications. The modifications were the use of glutaraldehyde (0.2 μg⋅mL−1) for 2 min to disinfect egg surfaces (72) and the addition of antibiotics [penicillin and streptomycin (Sigma-Aldrich) at a final concentration of 10 μ⋅ mL−1] to the autoclaved egg water before filtration with a 0.22-μm membrane filter (gnotobiotic zebrafish medium with antibiotics; AB-GZM). After derivation, GF animals were kept in sterile, vented tissue-culture flasks at an average density of 10 individuals per milliliter of autoclaved and filtered egg water without antibiotics (GZM). On alternate days, GF animals were monitored for sterility using standard microbial culture methods as described elsewhere (71), and 25% (vol/vol) of the total medium contained in each flask was replaced with fresh GZM. The remaining half of the total collected eggs, which did not receive any treatment, was raised conventionally (CONR) in culture vessels at the same densities as the GF group to standardize conditions. In both groups, dead eggs were removed aseptically twice each day.
Germ-Free Colonization Test.
Most larvae in GF experiments started to hatch 48 hpf, and 6 h later all had lost their protective chorions. At this time point every single larva in each GF or CONR batch was recovered using a soft sieve and was transferred immediately to a new cell-culture flask. Each flask with GF larvae was filled with medium collected from tanks housing CONR zebrafish (71); CONR zebrafish simply were transferred to GF medium.
In Vivo MAMP Tests.
Repeats of molecular CpG motifs obtained as phenol-extracted genomic vDNA (73) were used to stimulate whole zebrafish larvae by dip immersion at selected time points. Briefly, 50 μg⋅mL−1 vDNA was added to 25-cm2 ventilated cell-culture flasks containing 50 larvae in 15 mL medium. Depending on the experiment, incubation was carried out for 4 and/or 24 h at 28.5 °C. Once incubation was finished, larvae from each flask were collected in 1.5-mL Eppendorf tubes containing 500 μL TRIzol reagent and were frozen immediately at −80 °C for further analysis.
Flow Cytometry Analysis of Zebrafish Neutrophils.
Tg(mpx:eGFP)114i zebrafish were reared under CONR or GF conditions. At 72 hpf larvae from each batch were killed with an overdose of MS-222 (Sigma). In each experiment, 100 animals per condition were pooled after killing and were disaggregated as described elsewhere (41). Single-cell suspensions were analyzed by a FACS bench-top cytometer (BD Biosciences). Live cells (106) were analyzed by flow-cytometry using the CellQuest software (Becton Dickinson Immunocytometry Systems).
Gene-Expression Analysis.
Total RNA was isolated using TRIzol (Invitrogen) following the manufacturer’s specifications and treated with amplification grade DNase I (1 U/μg RNA; Invitrogen). The SuperScript III RNase H− reverse transcriptase (Invitrogen) was used to synthesize the first strand of cDNA with an oligo(dT) primer from 1 μg of total RNA for 50 min at 50 °C. Real-time PCR was performed with an ABI PRISM 7500 instrument (Applied Biosystems) using SYBR Green PCR core reagents (Applied Biosystems). Reaction mixtures were incubated for 10 min at 95 °C, followed by 40 cycles of 15 s at 95 °C, 1 min at 60 °C, and finally 15 s at 95 °C, 1 min at 60 °C, and 15 s at 95 °C. For each mRNA, the gene expression was normalized to the ribosomal protein S11 content in each sample using the Pfaffl method (74). The primers used are shown in Table S1. In all cases, each PCR was performed with triplicate samples and was repeated at least twice.
Respiratory Burst Activity Using Dyhydrorodamine 123 (DHR).
Total cells suspensions (5 × 106 cells⋅mL−1) of CONVR or GF 72-hpf zebrafish larvae diluted in RPMI-1640 culture medium (Life Technologies) containing 5% (vol/vol) fetal bovine serum (LIfe Technologies) were split into several aliquots for each treatment. Thereafter, one half of the aliquots was incubated with 100 μL of 10 μM DPI (an inhibitor of NADPH oxidase) for 1 h at room temperature. Then, all aliquots containing inhibited or naive cells were stained for 5 min at room temperature with 100 μL of 10 μM DHR solution in HBSS (75). The DHR-loaded cells were incubated further for 30 min at room temperature with 100 μL of PMA (Sigma-Aldrich) at 0, 1, 10, or 100 ng⋅ml−1 to trigger the respiratory burst. After incubation, samples were analyzed immediately by flow cytometry for total and mean fluorescence. Results were collected on the bright green fluorescence as FL1 and were compared with the FL2 signal.
Microinjection of Morpholino Nucleotides or RNA into Zebrafish Embryos.
Morpholino antisense oligomers targeting MyD88 (TAGCAAAACCTCTGTTATCCAGCGA) (20), the myeloid transcription factor SPI1 (GATATACTGATACTCCATTGGTGGT) (42), or a standard control (STD) CCTCTTACCTCAGTTACAATTTATA) were obtained from Gene Tools, LLC, solubilized in water to produce a stock solution (1 mM), and kept at −80 °C in small aliquots until use. Zebrafish embryos at the one-cell stage were microinjected with a mix of morpholino (4 ng per egg) in 0.5× Tango buffer and 0.05% phenol red as indicator using sterile egg supports consisting of 2% (wt/vol) low-melting-point agarose (Sigma-Aldrich) dissolved in sterile embryo medium (76) and a microinjector Narishige IM-300 with an attached glass capillary needle. After microinjection, eggs were removed aseptically from the agarose matrix and were left in egg water. CONR eggs were placed in tissue-culture flasks without any astringent solution, and GF eggs were derived immediately. After the derivation was verified, GF embryos were maintained in GZM until the end of each trial.
Imaging the Tail-Transection Wound.
Tg(mpx::eGFP)114i zebrafish were reared under CONR or GF conditions. At 72 hpf, larvae were anesthetized in tricaine and then were mounted in 1% (wt/vol) low-melting-point agarose dissolved in embryo medium. Complete transection of the tail was performed with a disposable sterile scalpel. The success of transection was confirmed immediately through a fluorescence stereomicroscope MZ16FA (Leica) equipped with green fluorescent filters. Each image was imaged at transection, which was established as time zero. Thereafter, images were captured at the selected times while animals were kept in their agar matrixes with the added medium at 28.5 °C. All images were acquired with the integrated camera on the stereomicroscope and were controlled under the Leica application suite using Windows operational system.
Infection Assays.
The SVCV isolate 56/70 was obtained as previously reported (43). Briefly, the virus stock was propagated in EPC cells and titrated in 96-well plates. Thirty 72-hpf zebrafish larvae were challenged at 25 °C in disposable Petri dishes by immersion in SVCV [108 50% tissue culture infective dose (TCID50)]. After challenge, the fish were monitored every 12 h over an 8-d period, and mortality was scored.
ChIP.
vDNA (5–10 ng per egg) was microinjected into the yolk sac of embryos at the one-cell state (13). At the indicated times, embryos were dechorionated manually and treated with 3 μM pargyline or 100 nM TSA for 5 h and, if required, were raised under CONR or GF conditions. Embryos/larvae were processed for RNA extraction as described above or for ChIP using the MAGnify Chromatin Immunoprecipitation System (Life Technologies). Briefly, for each immunoprecipitation 200 embryos (30 hpf) or 80 larvae (72 hpf) were cross-linked with 1% formaldehyde for 8 min, and the chromatin was sheared by sonication to an average fragment size of 500–1,000 bp (77). Lysates were cleared by centrifugation and diluted 1:10 in the dilution buffer of the ChIP kit, and immunoprecipitation was performed with 2–4 μg of antibody. ChIP and input DNA were amplified by qPCR using specific primers for the 5′ upstream sequence of the different genes (Table S2). qChIP values are given as percent input or DNA recovery normalized to total immunoprecipitated histone H3. ChIP with control mouse and rabbit anti-IgG showed no enrichment of any target promoters. ChIPs were performed with at least two independent chromatin preparations. The antibodies used in this assay were anti-H3 (ab1791; Abcam), anti-H3K4me3 (ab1012; Abcam), anti-H3K9ac (06–942; Millipore), and control mouse and rabbit anti-IgG provided by the MAGnify kit. All these antibodies have been validated in zebrafish previously (77), and the epitopes that they recognize are identical in zebrafish and mammals.
Statistical Analysis.
Statistics were generated with GraphPad Prism 5 software using Student’s t tests for paired groups and ANOVA and post hoc Tukey’s test for multiple comparisons. The log-rank test was used to calculate the differences in survival of the different experimental groups. Differences with P values <0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We thank Inma Fuentes and Pedro J. Martínez for expert technical assistance, Dr. S. A. Renshaw for Tg(mpx:eGFP114i), Dr. M. P. Somalo for the 56/70 strain of SVCV, and Prof. A. Estepa for the SVCV titration. This work was supported in part by National Council of Science and Technology (Mexico) Grant-in-Aid 8028 (to J.G.-V); Fundação para a Ciência e Tecnologia Fellowship SFRH/BD/62674/2009 (to S.d.O.); Spanish Ministry of Science and Innovation Grants BIO2008-01379, BIO2011-23400, and CSD2007-00002 (to V.M.); Fundación Séneca-Murcia Grant 04538/GERM/06 (to V.M.); and Fundación Marcelino Botín Grant (to V.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.V.C. is a guest editor invited by the Editorial Board.
See Author Summary on page 15545 (volume 109, number 39).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1209920109/-/DCSupplemental.
References
- 1.Herbst T, et al. Dysregulation of allergic airway inflammation in the absence of microbial colonization. Am J Respir Crit Care Med. 2011;184:198–205. doi: 10.1164/rccm.201010-1574OC. [DOI] [PubMed] [Google Scholar]
- 2.Jang HR, Gandolfo MT, Ko GJ, Racusen L, Rabb H. The effect of murine anti-thymocyte globulin on experimental kidney warm ischemia-reperfusion injury in mice. Transpl Immunol. 2009;22:44–54. doi: 10.1016/j.trim.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 3.Tlaskalová-Hogenová H, et al. The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: Contribution of germ-free and gnotobiotic animal models of human diseases. Cell Mol Immunol. 2011;8:110–120. doi: 10.1038/cmi.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hooper LV. Bacterial contributions to mammalian gut development. Trends Microbiol. 2004;12:129–134. doi: 10.1016/j.tim.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Dusheck J. It’s the ecology, stupid! Nature. 2002;418:578–579. doi: 10.1038/418578a. [DOI] [PubMed] [Google Scholar]
- 6.Kaplan JL, Shi HN, Walker WA. The role of microbes in developmental immunologic programming. Pediatr Res. 2011;69:465–472. doi: 10.1203/PDR.0b013e318217638a. [DOI] [PubMed] [Google Scholar]
- 7.Heijtz RD, et al. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci USA. 2011;108:3047–3052. doi: 10.1073/pnas.1010529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olafsen JA. Bacterial antigen priming of marine fish larvae. Adv Exp Med Biol. 1995;371A:349–352. doi: 10.1007/978-1-4615-1941-6_73. [DOI] [PubMed] [Google Scholar]
- 9.Medzhitov R, Janeway C., Jr Innate immune recognition: Mechanisms and pathways. Immunol Rev. 2000;173:89–97. doi: 10.1034/j.1600-065x.2000.917309.x. [DOI] [PubMed] [Google Scholar]
- 10.Akira S. TLR signaling. Curr Top Microbiol Immunol. 2006;311:1–16. doi: 10.1007/3-540-32636-7_1. [DOI] [PubMed] [Google Scholar]
- 11.Meijer AH, Spaink HP. Host-pathogen interactions made transparent with the zebrafish model. Curr Drug Targets. 2011;12:1000–1017. doi: 10.2174/138945011795677809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roach JC, et al. The evolution of vertebrate Toll-like receptors. Proc Natl Acad Sci USA. 2005;102:9577–9582. doi: 10.1073/pnas.0502272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sepulcre MP, et al. Evolution of lipopolysaccharide (LPS) recognition and signaling: Fish TLR4 does not recognize LPS and negatively regulates NF-kappaB activation. J Immunol. 2009;182:1836–1845. doi: 10.4049/jimmunol.0801755. [DOI] [PubMed] [Google Scholar]
- 14.Sullivan C, et al. The gene history of zebrafish tlr4a and tlr4b is predictive of their divergent functions. J Immunol. 2009;183:5896–5908. doi: 10.4049/jimmunol.0803285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phelan PE, Mellon MT, Kim CH. Functional characterization of full-length TLR3, IRAK-4, and TRAF6 in zebrafish (Danio rerio) Mol Immunol. 2005;42:1057–1071. doi: 10.1016/j.molimm.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 16.Takano T, et al. Molecular cloning and characterization of Toll-like receptor 9 in Japanese flounder, Paralichthys olivaceus. Mol Immunol. 2007;44:1845–1853. doi: 10.1016/j.molimm.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 17.Yoon SI, et al. Structural basis of TLR5-flagellin recognition and signaling. Science. 2012;335:859–864. doi: 10.1126/science.1215584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alcaraz-Pérez F, Mulero V, Cayuela ML. Application of the dual-luciferase reporter assay to the analysis of promoter activity in Zebrafish embryos. BMC Biotechnol. 2008;8:81. doi: 10.1186/1472-6750-8-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bates JM, Akerlund J, Mittge E, Guillemin K. Intestinal alkaline phosphatase detoxifies lipopolysaccharide and prevents inflammation in zebrafish in response to the gut microbiota. Cell Host Microbe. 2007;2:371–382. doi: 10.1016/j.chom.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van der Sar AM, et al. MyD88 innate immune function in a zebrafish embryo infection model. Infect Immun. 2006;74:2436–2441. doi: 10.1128/IAI.74.4.2436-2441.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hall C, et al. Transgenic zebrafish reporter lines reveal conserved Toll-like receptor signaling potential in embryonic myeloid leukocytes and adult immune cell lineages. J Leukoc Biol. 2009;85:751–765. doi: 10.1189/jlb.0708405. [DOI] [PubMed] [Google Scholar]
- 22.Cheesman SE, Neal JT, Mittge E, Seredick BM, Guillemin K. Epithelial cell proliferation in the developing zebrafish intestine is regulated by the Wnt pathway and microbial signaling via Myd88. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4570–4577. doi: 10.1073/pnas.1000072107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: The unseen majority. Proc Natl Acad Sci USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kemp PF, Aller JY. Bacterial diversity in aquatic and other environments: What 16S rDNA libraries can tell us. FEMS Microbiol Ecol. 2004;47:161–177. doi: 10.1016/S0168-6496(03)00257-5. [DOI] [PubMed] [Google Scholar]
- 25.Slapeta J, Moreira D, López-García P. The extent of protist diversity: Insights from molecular ecology of freshwater eukaryotes. Proc Biol Sci. 2005;272:2073–2081. doi: 10.1098/rspb.2005.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zinger L, Gobet A, Pommier T. Two decades of describing the unseen majority of aquatic microbial diversity. Mol Ecol. 2012;21:1878–1896. doi: 10.1111/j.1365-294X.2011.05362.x. [DOI] [PubMed] [Google Scholar]
- 27.Hrncir T, Stepankova R, Kozakova H, Hudcovic T, Tlaskalova-Hogenova H. Gut microbiota and lipopolysaccharide content of the diet influence development of regulatory T cells: Studies in germ-free mice. BMC Immunol. 2008;9:65. doi: 10.1186/1471-2172-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang ZL. Important aspects of Toll-like receptors, ligands and their signaling pathways. Inflamm Res. 2010;59:791–808. doi: 10.1007/s00011-010-0208-2. [DOI] [PubMed] [Google Scholar]
- 29.Butler JE, et al. The piglet as a model for B cell and immune system development. Vet Immunol Immunopathol. 2009;128:147–170. doi: 10.1016/j.vetimm.2008.10.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mione MC, Trede NS. The zebrafish as a model for cancer. Dis Model Mech. 2010;3:517–523. doi: 10.1242/dmm.004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin JS, Renshaw SA. Using in vivo zebrafish models to understand the biochemical basis of neutrophilic respiratory disease. Biochem Soc Trans. 2009;37:830–837. doi: 10.1042/BST0370830. [DOI] [PubMed] [Google Scholar]
- 32.Renshaw SA, et al. A transgenic zebrafish model of neutrophilic inflammation. Blood. 2006;108:3976–3978. doi: 10.1182/blood-2006-05-024075. [DOI] [PubMed] [Google Scholar]
- 33.Langenau DM, Zon LI. The zebrafish: A new model of T-cell and thymic development. Nat Rev Immunol. 2005;5:307–317. doi: 10.1038/nri1590. [DOI] [PubMed] [Google Scholar]
- 34.Trede NS, Langenau DM, Traver D, Look AT, Zon LI. The use of zebrafish to understand immunity. Immunity. 2004;20:367–379. doi: 10.1016/s1074-7613(04)00084-6. [DOI] [PubMed] [Google Scholar]
- 35.Lieschke GJ. Zebrafish—an emerging genetic model for the study of cytokines and hematopoiesis in the era of functional genomics. Int J Hematol. 2001;73:23–31. doi: 10.1007/BF02981899. [DOI] [PubMed] [Google Scholar]
- 36.Rawls JF, Samuel BS, Gordon JI. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc Natl Acad Sci USA. 2004;101:4596–4601. doi: 10.1073/pnas.0400706101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bates JM, et al. Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Dev Biol. 2006;297:374–386. doi: 10.1016/j.ydbio.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 38.Kanther M, Rawls JF. Host-microbe interactions in the developing zebrafish. Curr Opin Immunol. 2010;22:10–19. doi: 10.1016/j.coi.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheesman SE, Guillemin K. We know you are in there: Conversing with the indigenous gut microbiota. Res Microbiol. 2007;158:2–9. doi: 10.1016/j.resmic.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 40.Kanther M, et al. Microbial colonization induces dynamic temporal and spatial patterns of NF-κB activation in the zebrafish digestive tract. Gastroenterology. 2011;141:197–207. doi: 10.1053/j.gastro.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.López-Muñoz A, et al. Evolutionary conserved pro-inflammatory and antigen presentation functions of zebrafish IFNγ revealed by transcriptomic and functional analysis. Mol Immunol. 2011;48:1073–1083. doi: 10.1016/j.molimm.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 42.Rhodes J, et al. Interplay of pu.1 and gata1 determines myelo-erythroid progenitor cell fate in zebrafish. Dev Cell. 2005;8:97–108. doi: 10.1016/j.devcel.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 43.López-Muñoz A, Roca FJ, Meseguer J, Mulero V. New insights into the evolution of IFNs: Zebrafish group II IFNs induce a rapid and transient expression of IFN-dependent genes and display powerful antiviral activities. J Immunol. 2009;182:3440–3449. doi: 10.4049/jimmunol.0802528. [DOI] [PubMed] [Google Scholar]
- 44.López-Muñoz A, Roca FJ, Sepulcre MP, Meseguer J, Mulero V. Zebrafish larvae are unable to mount a protective antiviral response against waterborne infection by spring viremia of carp virus. Dev Comp Immunol. 2010;34:546–552. doi: 10.1016/j.dci.2009.12.015. [DOI] [PubMed] [Google Scholar]
- 45.Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- 46.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 47.Schneider R, et al. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- 48.Metzger E, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 49.Bernstein BE, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 50.Roeselers G, et al. Evidence for a core gut microbiota in the zebrafish. ISME J. 2011;5:1595–1608. doi: 10.1038/ismej.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lundin A, et al. Gut flora, Toll-like receptors and nuclear receptors: A tripartite communication that tunes innate immunity in large intestine. Cell Microbiol. 2008;10:1093–1103. doi: 10.1111/j.1462-5822.2007.01108.x. [DOI] [PubMed] [Google Scholar]
- 52.Clatworthy AE, et al. Pseudomonas aeruginosa infection of zebrafish involves both host and pathogen determinants. Infect Immun. 2009;77:1293–1303. doi: 10.1128/IAI.01181-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rawls JF, Mahowald MA, Ley RE, Gordon JI. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. 2006;127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stockhammer OW, Zakrzewska A, Hegedûs Z, Spaink HP, Meijer AH. Transcriptome profiling and functional analyses of the zebrafish embryonic innate immune response to Salmonella infection. J Immunol. 2009;182:5641–5653. doi: 10.4049/jimmunol.0900082. [DOI] [PubMed] [Google Scholar]
- 55.Prajsnar TK, Cunliffe VT, Foster SJ, Renshaw SA. A novel vertebrate model of Staphylococcus aureus infection reveals phagocyte-dependent resistance of zebrafish to non-host specialized pathogens. Cell Microbiol. 2008;10:2312–2325. doi: 10.1111/j.1462-5822.2008.01213.x. [DOI] [PubMed] [Google Scholar]
- 56.Brodsky IE, Medzhitov R. Targeting of immune signalling networks by bacterial pathogens. Nat Cell Biol. 2009;11:521–526. doi: 10.1038/ncb0509-521. [DOI] [PubMed] [Google Scholar]
- 57.Ribet D, Cossart P. Pathogen-mediated posttranslational modifications: A re-emerging field. Cell. 2010;143:694–702. doi: 10.1016/j.cell.2010.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wells JM, Loonen LM, Karczewski JM. The role of innate signaling in the homeostasis of tolerance and immunity in the intestine. Int J Med Microbiol. 2010;300:41–48. doi: 10.1016/j.ijmm.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 59.Liongue C, Hall CJ, O’Connell BA, Crosier P, Ward AC. Zebrafish granulocyte colony-stimulating factor receptor signaling promotes myelopoiesis and myeloid cell migration. Blood. 2009;113:2535–2546. doi: 10.1182/blood-2008-07-171967. [DOI] [PubMed] [Google Scholar]
- 60.Bloom SM, et al. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe. 2011;9:390–403. doi: 10.1016/j.chom.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gaboriau-Routhiau V, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 62.Gaboriau-Routhiau V, Raibaud P, Dubuquoy C, Moreau MC. Colonization of gnotobiotic mice with human gut microflora at birth protects against Escherichia coli heat-labile enterotoxin-mediated abrogation of oral tolerance. Pediatr Res. 2003;54:739–746. doi: 10.1203/01.PDR.0000086902.52137.C9. [DOI] [PubMed] [Google Scholar]
- 63.Jarchum I, Pamer EG. Regulation of innate and adaptive immunity by the commensal microbiota. Curr Opin Immunol. 2011;23:353–360. doi: 10.1016/j.coi.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Borkowski AW, Gallo RL. The coordinated response of the physical and antimicrobial peptide barriers of the skin. J Invest Dermatol. 2011;131:285–287. doi: 10.1038/jid.2010.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lai Y, et al. Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nat Med. 2009;15:1377–1382. doi: 10.1038/nm.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 67.Lai Y, et al. Activation of TLR2 by a small molecule produced by Staphylococcus epidermidis increases antimicrobial defense against bacterial skin infections. J Invest Dermatol. 2010;130:2211–2221. doi: 10.1038/jid.2010.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu X, et al. NF-kappaB and Snail1a coordinate the cell cycle with gastrulation. J Cell Biol. 2009;184:805–815. doi: 10.1083/jcb.200806074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Krens SF, Spaink HP, Snaar-Jagalska BE. Functions of the MAPK family in vertebrate-development. FEBS Lett. 2006;580:4984–4990. doi: 10.1016/j.febslet.2006.08.025. [DOI] [PubMed] [Google Scholar]
- 70.Hong CC, Kume T, Peterson RT. Role of crosstalk between phosphatidylinositol 3-kinase and extracellular signal-regulated kinase/mitogen-activated protein kinase pathways in artery-vein specification. Circ Res. 2008;103:573–579. doi: 10.1161/CIRCRESAHA.108.180745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pham LN, Kanther M, Semova I, Rawls JF. Methods for generating and colonizing gnotobiotic zebrafish. Nat Protoc. 2008;3:1862–1875. doi: 10.1038/nprot.2008.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dierckens K, et al. Development of a bacterial challenge test for gnotobiotic sea bass (Dicentrarchus labrax) larvae. Environ Microbiol. 2009;11:526–533. doi: 10.1111/j.1462-2920.2008.01794.x. [DOI] [PubMed] [Google Scholar]
- 73.Pelegrín P, Chaves-Pozo E, Mulero V, Meseguer J. Production and mechanism of secretion of interleukin-1beta from the marine fish gilthead seabream. Dev Comp Immunol. 2004;28:229–237. doi: 10.1016/j.dci.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 74.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stafford JL, McLauchlan PE, Secombes CJ, Ellis AE, Belosevic M. Generation of primary monocyte-like cultures from rainbow trout head kidney leukocytes. Dev Comp Immunol. 2001;25:447–459. doi: 10.1016/s0145-305x(01)00015-5. [DOI] [PubMed] [Google Scholar]
- 76.Westerfield M. 2007. The Zebrafish Book, A Guide for the Laboratory Use of Zebrafish (Danio rerio) (Univ of Oregon Press, Eugene, OR,) 5th Ed.
- 77.Lindeman LC, Vogt-Kielland LT, Aleström P, Collas P. Fish’n ChIPs: Chromatin immunoprecipitation in the zebrafish embryo. Methods Mol Biol. 2009;567:75–86. doi: 10.1007/978-1-60327-414-2_5. [DOI] [PubMed] [Google Scholar]