SUMMARY
Arabidopsis seedlings display rhythmic growth when grown under diurnal conditions, with maximal elongation rates occurring at the end of the night under short-day photoperiods. Current evidence indicates that this behavior involves the action of the growth-promoting bHLH factors PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) and PHYTOCHROME-INTERACTING FACTOR 5 (PIF5) at the end of the night, through a coincidence mechanism that combines their transcriptional regulation by the circadian clock with control of protein accumulation by light. To assess the possible role of PIF3 in this process, we have analyzed hypocotyl responses and marker gene expression in pif single- and higher-order mutants. The data show that PIF3 plays a prominent role as a promoter of seedling growth under diurnal light/dark conditions, in conjunction with PIF4 and PIF5. In addition, we provide evidence that PIF3 functions in this process through its intrinsic transcriptional regulatory activity, at least in part by directly targeting growth-related genes, and independently of its ability to regulate phytochrome B (phyB) levels. Furthermore, in sharp contrast to PIF4 and PIF5, our data show that the PIF3 gene is not subject to transcriptional regulation by the clock, but that PIF3 protein abundance oscillates under diurnal conditions as a result of a progressive decline in PIF3 protein degradation mediated by photoactivated phyB, and consequent accumulation of the bHLH factor during the dark period. Collectively, the data suggest that phyB-mediated, post-translational regulation allows PIF3 accumulation to peak just before dawn, at which time it accelerates hypocotyl growth, together with PIF4 and PIF5, by directly regulating the induction of growth-related genes.
Keywords: PIF3, hypocotyl elongation, short day, phytochrome-mediated degradation, transcriptional regulation, Arabidopsis
INTRODUCTION
Light is fundamental for plants as a source of energy as well as an indicator of their living environment. Plants perceive and respond to ambient light signals through informational photoreceptors that include the phytochrome family (phyA–phyE in Arabidopsis) (Rockwell et al., 2006; Schafer and Nagy, 2006; Quail, 2010). The phytochromes perceive red (660 nm) and far red (720 nm) light of the solar spectrum, and monitor changes in light quality, quantity and duration to control developmental and growth responses such as germination, seedling de-etiolation, shade avoidance and flowering time (Franklin and Quail, 2010; Strasser et al., 2010). phyA is the only receptor for continuous far red light, but both phyA and phyB contribute to perception of continuous red light during early de-etiolation, with phyB being the dominant if not exclusive regulator of the hypocotyl-elongation response to continuous red light (Rockwell et al., 2006; Schafer and Nagy, 2006; Tepperman et al., 2006; Franklin and Quail, 2010). The phytochromes reversibly photoconvert between two conformers: the inactive red light-absorbing Pr form and the biologically active far red light-absorbing Pfr form. Pr to Pfr photoconversion takes place within seconds upon absorption of red light photons (Linschitz and Kasche, 1966), and reversion of Pfr to Pr occurs in far red light-enriched environments (Franklin, 2008), and also in the dark. In seedlings grown in the light, Pfr remains active upon initial transfer to the dark, but slowly reverts, at least partially, back to Pr with a half-life of approximately 60 min (Sweere et al., 2001; Rausenberger et al., 2010).
Phytochromes are synthesized in the cytoplasm in the inactive Pr form, and, upon photoactivation to Pfr, are translocated into the nucleus (Nagatani, 2004), where they associate with a subset of basic helix-loop-helix (bHLH) transcription factors called phytochrome-interacting factors (PIFs). The PIFs (PIF1, PIF3, PIF4, PIF5, PIF6 and PIF7 in Arabidopsis) accumulate in the dark and interact photoreversibly and conformer-specifically with the active Pfr phytochrome in the light (Leivar and Quail, 2011). This light-induced interaction between the Pfr phytochrome and PIF initiates a cascade of transcriptional changes that allows the seedling to adjust to the new light environment (Castillon et al., 2007; Jiao et al., 2007; Bae and Choi, 2008; Leivar and Quail, 2011). For a subset of these PIFs (PIF1, PIF3, PIF4 and PIF5), interaction with phyA and/or phyB triggers rapid phosphorylation and degradation of the PIF proteins within minutes (Bauer et al., 2004; Park et al., 2004; Shen et al., 2005; Oh et al., 2006; Nozue et al., 2007; Shen et al., 2007; Lorrain et al., 2008; Shen et al., 2008), establishing a new lower steady-state level of PIFs in continuous light (Monte et al., 2004). Concomitantly, exposure to light induces rapid phyA degradation (with a half-life of <2 h), and a slower and more modest degradation of phyB (Hennig et al., 1999; Khanna et al., 2007; Al-Sady et al., 2008), which remains relatively abundant in the light, together with phyC–phyE (Hirschfeld et al., 1998). During prolonged growth in continuous light, the PIFs induce phyB proteolytic degradation through the proteasome system using COP1 as an E3 ligase (Khanna et al., 2007; Al-Sady et al., 2008; Leivar et al., 2008a; Jang et al., 2010), suggesting the existence of a mutually negative feedback loop between the phyB and PIF proteins (Leivar and Quail, 2011). This light-induced phyB degradation is expected to contribute to the progressive decline in Pfr levels during the dark period under diurnal conditions (light/dark cycles). In addition, the PIFs re-accumulate in light-grown seedlings upon exposure to darkness (such as under diurnal conditions) (Monte et al., 2004; Shen et al., 2005; Nozue et al., 2007) or far red light-enriched environments (such as vegetational shade) (Lorrain et al., 2008) through a process that depends on the activation state (or Pfr/Pr ratio) of the phytochromes.
Hypocotyl elongation is a well-established light-regulated response that is maximal in seedlings grown in continuous dark. In post-germinative darkness, the PIF proteins promote hypocotyl elongation through their intrinsic transcription factor capacity, regulating a transcriptional network that sustains etiolated growth (Leivar et al., 2009; Shin et al., 2009). This conclusion is supported by the observation that a quadruple mutant deficient in PIF1, 3, 4 and 5 (pifq) exhibits a partial constitutively photomorphogenic phenotype in the dark, characterized by a short-hypocotyl phenotype (Leivar et al., 2008b). In continuous light, under which PIFs induce phyB degradation, PIF-deficient mutants display a hypersensitive short-hypocotyl phenotype that is interpreted to be, at least partially, the result of enhanced photosensitivity of the seedling due to elevated photoreceptor levels (Khanna et al., 2007; Al-Sady et al., 2008; Leivar et al., 2008a).
Under diurnal conditions, with an alternating light/dark cycle, the extent of hypocotyl elongation depends on the duration of the dark period (Niwa et al., 2009). During dark hours, the hypocotyl elongation rate is maximal at the end of the night in seedlings grown under short-day (SD) photoperiods (Nozue et al., 2007). Studies have indicated that PIF4 and PIF5 are positive regulators of this response (Nozue et al., 2007; Niwa et al., 2009). The precise regulation of their time of action at the end of the dark period has been proposed to involve a coincidence mechanism that combines regulation of PIF4 and PIF5 transcript levels by the circadian clock, superimposed on the control of PIF protein accumulation by light (Nozue et al., 2007; Nusinow et al., 2011). In addition to PIF4 and PIF5, the current model predicts that additional, yet to be identified, factors are involved in the regulation of seedling growth under SD conditions.
In this study, we have used single and multiple pif3, pif4 and pif5 mutants, combined with analyses of PIF3 protein accumulation and target gene expression, to define the role of PIF3 in the regulation of hypocotyl elongation in seedlings grown under diurnal conditions, and have examined the relative contributions of PIF3, PIF4 and PIF5 to this response. Our results suggest that phytochromes generate an oscillation of PIF3 abundance under SD conditions such that it peaks just before dawn, at which time PIF3 plays a prominent role in promoting elongation growth, in conjunction with PIF4 and PIF5, at least in part by directly regulating the expression of growth-related genes.
RESULTS
The pattern of PIF3 accumulation under SD conditions is regulated by phyA and phyB and is independent of transcriptional regulation by the clock
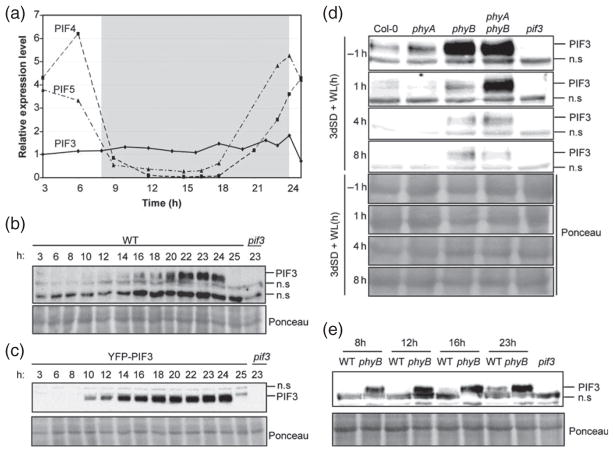
To establish the pattern of PIF3 expression under diurnal SD conditions [8 h white light + 16 h darkness], we analyzed PIF3 transcript levels over 24 h during the third day of seedling growth under SD conditions (Figure 1a), and compared them to the expression pattern of PIF4 and PIF5. PIF3 transcript levels remained fairly constant over the 24 h photoperiod (Figure 1a). In sharp contrast, PIF4 and PIF5 transcript levels decreased during the day, stayed low during most of the dark period, and increased again to peak at the end of the night (Figure 1a), consistent with the previously reported circadian clock regulation of PIF4 and PIF5 transcript levels (Yamashino et al., 2003; Nozue et al., 2007; Niwa et al., 2009). These results indicate that, in contrast to PIF4 and PIF5, PIF3 transcript levels do not oscillate under diurnal conditions, and suggest that the circadian clock does not regulate PIF3 transcription under SD conditions.
Figure 1. PIF3 protein accumulation under SD conditions.
Seedlings were grown under SD conditions for 2 days (a–c, e) or 3 days (d), and samples were taken during the following day at the specified time points.
(a) The expression of PIF3, PIF4 and PIF5 was analyzed by quantitative RT-PCR. Values were normalized to PP2A, and expression levels relative to PIF3 at 3 h are shown. Values are means of technical triplicates.
(b) Immunoblot of protein extracts from WT seedlings.
(c) Immunoblot of protein extracts from seedlings over-expressing YFP–PIF3.
(d) Immunoblot of protein extracts from WT, phyA-211, phyB-9 and phyA phyB seedlings.
(e) Immunoblot of protein extracts from WT and phyB-9 seedlings.
For (b–e), a PIF3-specific polyclonal antibody was used as the probe (top). As an antibody specificity control, a protein extract from pif3-3 harvested at time 23 h was included. Ponceau staining was used as a loading control (bottom). n.s., non-specific cross-reacting bands.
We next examined the pattern of accumulation of the endogenous PIF3 protein under diurnal light/dark cycles. To do this, we grew seedlings under SD conditions and tested the levels of endogenous PIF3 protein every 1–3 h over a period of 24 h. PIF3 protein started to accumulate at the start of the dark period, as early as 2 h after the transition from light to dark (10 h time point, Figures 1b and S1), and kept accumulating progressively to reach a maximum at the end of the night, after 14–16 h of darkness (22, 23 and 24 h time points, Figure 1b). PIF3 protein levels then dropped to below the detection limit after exposure of seedlings to white light for 1 h (25 h time point, Figure 1b). Transgenic plants overexpressing a YFP–PIF3 fusion (Al-Sady et al., 2006) showed a similar pattern of YFP–PIF3 accumulation under SD conditions, with low levels during the light period and a progressive increase during the dark to peak at the end of the night (Figure 1c). A similar pattern was also observed in transgenic lines over-expressing PIF4:HA and PIF5:HA, although in these experiments the seedlings were grown under SD/3 conditions (i.e. 8 h light/dark cycles comprising 160 min light + 320 min dark) (Nozue et al., 2007).
Together, the above experiments indicate that, under SD conditions, PIF3 protein levels are very low during the light period, but increase progressively during the night (Figure 1b) through post-transcriptional regulation (Figure 1a). In order to examine the role of phytochrome activity in regulation of this pattern of PIF3 accumulation, we measured PIF3 levels at the end of the night (−1 h) and after 1, 4 and 8 h of light exposure in phyA and phyB single and double mutants (Figure 1d). Wild-type (WT) seedlings accumulated PIF3 protein during the dark period, and light induced a rapid reduction in these levels within 1 h. Compared to WT, phyA phyB double mutants accumulated higher levels of PIF3 both at the end of the night and during the light period (Figure 1d), suggesting that phyA and/or phyB act to reduce PIF3 levels under SD conditions. Detailed single phyA and phyB mutant analysis at various time points suggests that the two photoreceptors contribute differentially to this activity. First, phyA mutants showed similar levels of PIF3 at the end of the night compared to WT (Figure 1d), but the levels in phyB and phyA phyB mutants were much higher (Figure 1d). Second, in contrast to the rapid light-induced degradation of PIF3 observed in WT, phyA and phyB seedlings, PIF3 levels remained relatively constant in phyA phyB double mutants after 1 h of illumination (Figure 1d). Finally, PIF3 levels further decreased and remained below the detection limit in phyA mutants during the day (4 and 8 h time points), similar to the WT (Figure 1d). In phyA phyB mutants, PIF3 levels also decreased between 1 and 4 h of illumination but PIF3 was still detectable after 4 and 8 h of light. In contrast, PIF3 levels in the phyB mutant did not further decrease after 1 h of illumination, and its levels were similar during the rest of the light period. Together, these results suggest that phyA and phyB act redundantly to rapidly reduce PIF3 levels within 1 h of illumination, and that at least one other photoreceptor is involved in the decrease in PIF3 levels at later time points (between 1 and 4 h). This scenario is similar to that reported during seedling de-etiolation, where phyD was shown to act together with phyA and phyB to induce PIF3 degradation in etiolated seedlings transferred to light (Bauer et al., 2004; Al-Sady et al., 2006). In addition, phyB activity is required to induce complete PIF3 degradation during the light period, and to prevent re-accumulation of PIF3 during the dark hours, in a process that requires little or no participation of phyA.
To obtain further insight into the role of phyB in preventing re-accumulation of PIF3 during the night in SD-grown seedlings, we performed a more detailed comparison of PIF3 levels in WT and the phyB mutant during the dark period. Figure 1(e) shows that PIF3 levels at the start of the night (8 h time point) were higher in phyB compared to WT seedlings, and rapidly increased in phyB during the first 4 h of darkness (8–12 h time points), reaching a new steady-state level that remained relatively constant until the end of the night (23 h). In contrast, PIF3 re-accumulation in the WT was slower during the first hours of darkness, and much lower levels were observed at the end of the night (Figure 1e). Together, our results suggest that the induction of PIF3 degradation by photoactive phyB Pfr during the light period extends into the first hours of the subsequent dark period. This possibility is in accordance with previous data showing that a far red light pulse given at the start of a 12 h dark period (removing the Pfr phytochrome pool from the cell) induced faster re-accumulation of GUS activity in GUS:PIF3 over-expressing seedlings grown under day-neutral conditions (Monte et al., 2004), and with the observation that the Pfr form of the photoreceptor continues to function in the dark to induce PIF3 degradation (Al-Sady et al., 2006).
PIF3 is necessary for hypocotyl growth under SD conditions
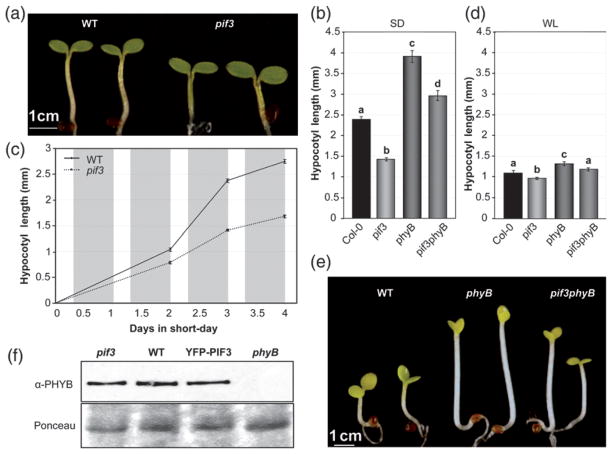
To examine the role of PIF3 during seedling growth under SD conditions, we measured hypocotyl elongation in seedlings lacking PIF3 (Monte et al., 2004). Hypocotyls of 3-day-old SD-grown pif3 mutants were approximately 40% shorter than the Col-0 control under these conditions (Figure 2a,b). In detailed time-course analyses, we found that WT hypocotyls elongated from 2 days onwards after germination under SD conditions, but the growth rate was severely reduced in the pif3 mutants (Figure 2c). The impact of PIF3 deficiency on growth was already apparent 48 h after initial exposure to SD, the first time point at which it was possible to measure seedling length (Figure 2c). In comparison to SD conditions, WT seedlings were shorter when grown under continuous white light (Figure 2b,d), and pif3 mutants grown under continuous white light were only slightly shorter than the WT (Figure 2d). These data indicate that the 16 h dark period in SD-grown seedlings accelerates hypocotyl elongation, consistent with previous reports (Niwa et al., 2009). Together with the PIF3 protein accumulation pattern (Figure 1), our data suggest that PIF3 is an important component of the cellular machinery that induces growth during the night hours.
Figure 2. PIF3 promotes hypocotyl growth under SD conditions (8 h light/16 h dark).
(a) Visual phenotype of 3-day-old SD-grown WT and pif3 mutant seedlings.
(b) Hypocotyl length in 3-day-old SD-grown WT and pif3 seedlings.
(c) Growth curves for hypocotyl length in WT and pif3 seedlings grown under SD for 4 days.
(d) Hypocotyl length in 3-day-old WT and pif3 seedlings grown under continuous white light (WL).
(e) Visual phenotype of 3-day-old SD-grown WT, pif3, phyB and pif3 phyB mutant seedlings.
(f) Immunoblots of protein extracts from 3-day-old WT and pif3 seedlings. Seedlings were grown under SD conditions for 2 days, and samples were harvested during the third day at the specified time points. Extracts were probed using phyB-specific monoclonal antibodies (top). Ponceau staining was used as a loading control (bottom).
For (b–d), data are means ± SE of at least 30 seedlings. For (b, d), different letters indicate significant differences among means (P < 0.05).
In contrast to the short phenotype of pif3 (Figure 2b), phyB mutant seedlings had more elongated hypocotyls than WT under SD conditions (Michael et al., 2008; Niwa et al., 2009), indicating an antagonistic functional relationship between phyB and PIF3 in regulating this response. Characterization of phyB and pif3 single and double mutants showed that phyB seedlings grown under SD conditions were approximately 1.5 mm taller than the corresponding WT (Figure 2b,e), and that genetic removal of PIF3 partially and significantly suppressed the phyB phenotype by 1 mm (Figure 2b, phyB versus pif3 phyB). These data suggest that the increased levels of PIF3 (Figure 1d) are at least partially responsible for the elongated hypocotyl phenotype of phyB mutant seedlings. In addition, compared to SD conditions, phyB mutant seedlings grown under continuous white light displayed a much reduced tall-hypocotyl phenotype (Figure 2d) and reduced suppression of this phenotype by the pif3 mutation (Figure 2d, phyB versus pif3 phyB). These data suggest that the dark period is necessary for full expression of the phyB mutant phenotype, probably by allowing higher accumulation of PIF3 protein under SD conditions compared to continuous white light (Figure 1d,e). Correlation between PIF3 levels and hypocotyl elongation was further observed in phyA phyB mutants (Figure S2). Compared to phyB, the double phyA phyB mutant had slightly longer hypocotyls under SD conditions, in agreement with the higher PIF3 levels detected at the start of the day in phyA phyB compared to phyB (Figure 1d).
Previously, PIF3-deficient mutants were shown to have increased phyB levels under continuous red light, contributing to their hypersensitive hypocotyl phenotype (Monte et al., 2004; Al-Sady et al., 2008; Leivar et al., 2008a). To examine whether the described negative feedback modulation of phyB photoreceptor levels by PIF3 under prolonged continuous red light and continuous white light conditions (Leivar et al., 2012a) operates under SD conditions, we measured phyB levels in the pif3 mutant and in YFP–PIF3 over-expressing lines at the end of the light period (8 h time point). Figure 2(f) shows that there were no significant differences in phyB levels between genotypes after 8 h of illumination, suggesting that PIF3-induced down-regulation of phyB requires more extended periods of light exposure. Together with our observation that PIF3 promotes growth under SD conditions in the absence of phyB (Figure 2b), these results indicate that PIF3 function under SD conditions is not exerted indirectly through regulation of phyB levels, and instead suggest that the PIF3 contribution to hypocotyl length under SD conditions is exerted through its intrinsic transcriptional activity, in accordance with previous data in etiolated seedlings (Al-Sady et al., 2008; Leivar et al., 2008b; Moon et al., 2008; Leivar et al., 2009; Shin et al., 2009; Sentandreu et al., 2011) and during shade avoidance (Hornitschek et al., 2009).
Expression of phytochrome-regulated, growth-related genes peaks at the end of the night under SD conditions and requires PIF3
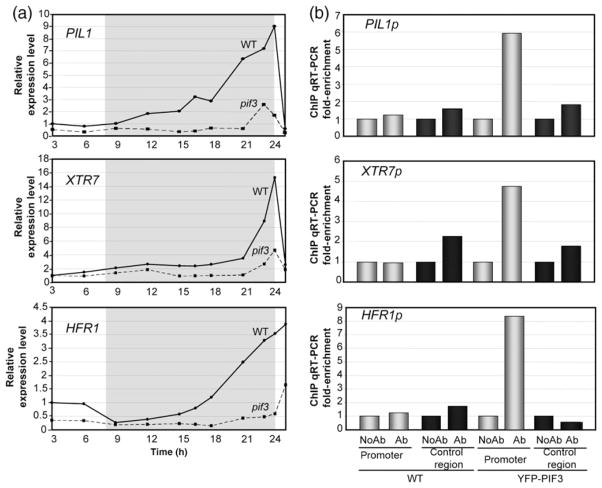
To test whether PIF3 regulates growth-related genes under SD conditions, we measured the expression of PIL1 (PHYTOCHROME-INTERACTING FACTOR 3-LIKE 1), HFR1 (LONG HYPOCOTYL IN FAR-RED 1) and XTR7 (XYLOGLUCAN ENDOTRANSGLYCOSYLASE 7) in WT and pif3 seedlings. These genes are repressed by the phytochromes and are up-regulated under conditions in which hypocotyl elongation is induced (Salter et al., 2003; Lorrain et al., 2008; Hornitschek et al., 2009; Leivar et al., 2009; Nozue et al., 2011), and have been proposed to be direct targets of transcriptional regulation by PIF4 in dark-adapted plants (de Lucas et al., 2008) and/or by PIF5 during shade avoidance (Hornitschek et al., 2009). PIL1 and HFR1 are PIF-related transcription factors (Leivar and Quail, 2011), and XTR7 encodes a xyloglucan endotransglycosylase-related protein that is potentially involved in cell-wall growth (Sasidharan et al., 2010).
Time-course expression analysis indicated that the expression levels of these three genes under SD conditions remain low during the light period in the WT, and start accumulating during the dark, peaking at the end of the night (Figure 3a). The expression levels at the end of the night were 4-, 10- and 16-fold greater than the levels during the light period for HFR1, PIL1 and XTR7, respectively (Figure 3a). Interestingly, the pattern of expression of these genes parallels the accumulation pattern of PIF3 protein (Figure 1b,c), rendering them good candidate genes for regulation by PIF3. Expression analysis by quantitative RT-PCR showed that their transcript levels are clearly reduced in the pif3 mutant, with the amplitude of the peak at the end of the night reduced by 80–90% for the three genes tested (Figure 3a). These data indicate that PIF3 induces expression of PIL1, HFR1 and XTR7 during the dark period under SD conditions, and suggest that PIF3 promotes growth under diurnal conditions by regulating expression of growth-related genes.
Figure 3. PIF3 regulates gene expression under SD conditions.
(a) Expression of PIL1, XTR7 and HFR1 was analyzed by quantitative RT-PCR in 2-day-old SD-grown seedlings. Samples were taken during the third day under SD conditions at the specified time points. Values were normalized to PP2A. Expression levels relative to WT at 3 h are shown. Data are the means of technical triplicates.
(b) Chromatin immunoprecipitation (ChIP) from 3-day-old SD-grown WT and YFP–PIF3 seedlings. Immunoprecipitated DNA was quantified by quantitative RT-PCR using primers in promoter regions containing G-boxes (promoter) or control regions without G-boxes (control region). Experiments include samples processed with anti-GFP antibody (Ab) and controls processed without antibody (NoAb). Data shown correspond to one representative ChIP experiment. The results of an additional ChIP experiment are shown in Figure S4. Data are the means of at least two technical replicates.
PIF3 directly binds to G-box-containing promoters of growth marker genes in vivo
HFR1, PIL1 and XTR7 genes harbor G-boxes in their promoters (Hornitschek et al., 2009) (Figure S3), suggesting that they are possible direct targets of PIF3 (Martínez-García et al., 2000; Shin et al., 2007). We analyzed the binding of PIF3 to the promoters of HFR1, PIL1 and XTR7 by chromatin imunoprecipitation (ChIP) using plants expressing PIF3 tagged with YFP (YFP–PIF3). ChIP was performed using an anti-GFP antibody, and immunoprecipitated G-box-containing and control DNA fragments were quantified by quantitative RT-PCR. Control DNA regions included non-G-box-containing regions of tested or unrelated genes. As controls, we used YFP–PIF3 plants processed without anti-GFP antibody, and Col-0 plants subjected to the same process with and without antibody. We performed these experiments in seedlings grown under SD conditions for 3 days and harvested at the end of the night (time point 23 h), when the maximum levels of PIF3 (Figure 1b) and the peak of expression of these genes coincide (Figure 3a). We observed significant enrichment of binding of PIF3 to the regions of HFR1, PIL1 and XTR7 promoters containing the G-box (Figures 3b and S4). These data indicate that PIF3 directly binds to the promoter regions of HFR1, PIL1 and XTR7, presumably through the G-box motif, and suggest that these genes are direct targets of transcriptional regulation by PIF3 under SD conditions.
PIF3 regulates hypocotyl growth and gene expression under SD conditions, together with PIF4 and PIF5
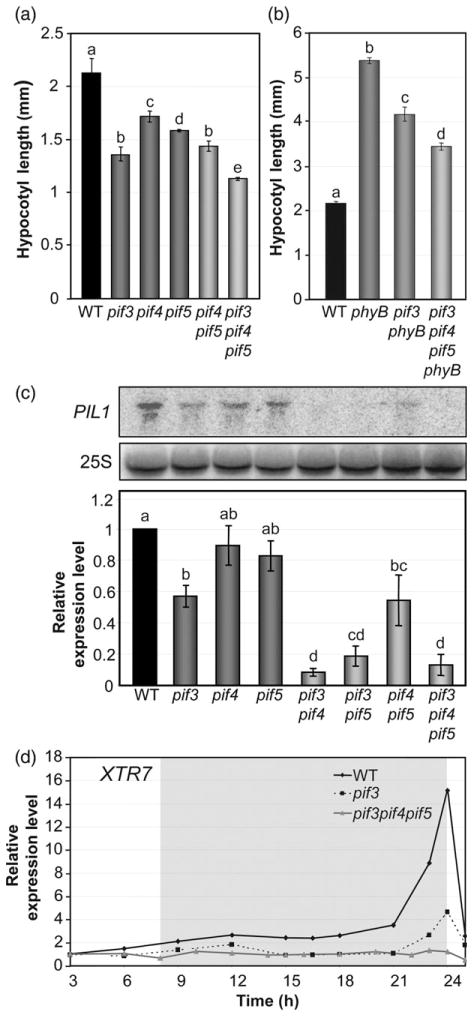
The observation that pif3 seedlings exhibit a reduced but still significant growth response to SD conditions compared to WT (Figure 2b,c) indicates that factors other than PIF3 are involved in the induction of hypocotyl elongation under SD conditions. Evidence obtained using pif4 and pif5 mutants indicates that these additional factors are probably PIF4 and PIF5 (Nozue et al., 2007; Niwa et al., 2009). To obtain insight into the contribution of PIF3 to the promotion of hypocotyl elongation under SD conditions relative to that of PIF4 and PIF5, we first analyzed the hypocotyl length of 3-day-old SD-grown pif3, pif4, pif5, pif4 pif5 and pif3 pif4 pif5 mutant seedlings. Under these conditions, pif4 and pif5 single mutants showed a quantitatively similar short-hypocotyl phenotype compared to the WT, an effect that was additive in the pif4 pif5 mutant, in accordance with previous reports (Figure 4a) (Nozue et al., 2007). In comparison, pif3 seedlings had more prominent short-hypocotyl phenotype than either pif4 or pif5, and this phenotype was similar in magnitude to that of the double pif4 pif5 mutant (Figure 4a). Moreover, the triple pif3 pif4 pif5 mutant had slightly shorter hypocotyls compared to pif3 (Figure 4a), confirming that PIF4 and PIF5 promote at least part of the residual growth of pif3 seedlings under SD conditions. We also compared the hypocotyl lengths of pif3 phyB and the pif3 pif4 pif5 phyB quadruple mutant. Our data indicate that removal of PIF4 and PIF5 in pif3 pif4 pif5 phyB had an additive effect over removal of PIF3 in pif3 phyB, and further suppressed the phyB tall phenotype (Figure 4b). Altogether, these results suggest that PIF3, PIF4 and PIF5 collectively function in the promotion of hypocotyl length under SD conditions, with the role of PIF3 probably being more prominent.
Figure 4. PIF3, PIF4 and PIF5 collectively regulate hypocotyl length and gene expression under SD conditions.
(a) Hypocotyl length in 3-day-old SD-grown WT, pif3, pif4, pif5, pif4 pif5 and pif3 pif4 pif5 seedlings.
(b) Hypocotyl length in 3-day-old SD-grown WT, phyB, pif3 phyB and pif3 pif4 pif5 phyB seedlings.
(c) The expression of PIL1was analyzed by RNA blots of 3-day-oldSD-grownWT, pif3, pif4, pif5, pif3 pif4, pif3 pif5, pif4 pif5 and pif3 pif4 pif5 seedlings. A representative blot is shown (top). Quantitative data (bottom) are means of three biological replicates; bars represent SE. Values were normalized to 25S rRNA.
(d) Expression of XTR7 was analyzed by quantitative RT-PCR in 2-day-old SD-grown WT, pif3 and pif3 pif4 pif5 seedlings during the third day of SD conditions. Values were normalized to PP2A. Expression levels relative to WT at 3 h are shown. Data are the means of technical triplicates.
For (a, b), data are means ± SE of at least 30 seedlings. For (a–c), different letters indicate significant differences among means (P < 0.05).
To examine the interactions between PIF3, PIF4 and PIF5 in regulating gene expression under SD conditions, we analyzed PIL1 expression in various pif3, pif4 and pif5 mutant combinations at the end of the night when expression of this gene peaks in the WT (23 h time point) (Figure 3a). The data show that individual deficiencies in PIF4 or PIF5 marginally reduced the level of PIL1 transcript (WT versus pif4 and pif5 single mutants), but a greater (and significant) reduction was observed in pif3 seedlings (Figure 4c). In addition, double mutant combinations including pif3 (pif3 pif4 and pif3 pif5) showed a further dramatic reduction in PIL1 expression (Figure 4c), indicating synergistic interactions between these factors for induction of PIL1 expression under SD conditions. Similar to the phenotypic analysis (Figure 4a), the magnitude of the reduction in PIL1 gene expression in pif3 mutants was similar to that of the double pif4 pif5 mutant (Figure 4c), and a further reduction in PIL1 expression was observed in pif3 pif4 pif5. These results suggest that PIF3 dominates the induction of PIL1 under SD conditions at the 23 h time point, and PIF4 and PIF5 are responsible for the residual PIL1 expression observed in pif3 single mutant seedlings at the end of the night (Figures 3a and 4c). Consistent with this observation, time-course analysis of XTR7 expression over the night showed that the peak of expression detected in pif3 (Figures 3a and 4d) is essentially absent in pif3 pif4 pif5 (Figure 4d), again suggesting that PIF4 and PIF5 are responsible for the residual XTR7 expression observed in pif3 single mutant seedlings at the end of the night. Together, the morphological (Figure 4a,b) and gene expression analyses (Figure 4c,d) suggest that PIF3, in conjunction with PIF4 and PIF5, plays a prominent role in induction of growth-related genes at the end of the night to promote growth under SD conditions.
PIF3 is required to promote growth at the end of the night under SD conditions
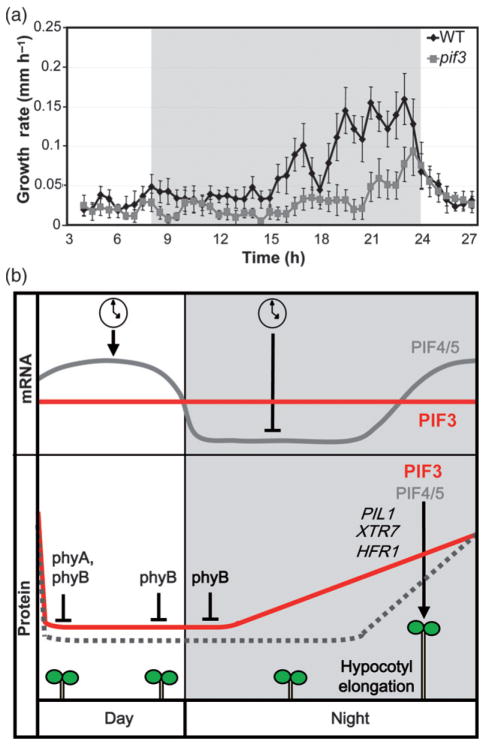
When grown under SD conditions, seedlings display rhythmic growth, with maximal elongation rates occurring at the end of the night (Nozue et al., 2007). To test whether the pif3 mutant shows an impaired growth pattern, we monitored seedling growth during a 24 h cycle, and calculated the growth rate of pif3 seedlings compared to WT. Our data show that WT seedlings, in accordance with previously published results (Nozue et al., 2007), maintain low growth rates during the day and the first half of the night, and the growth rate peaks at the end of the night (Figure 5a). In contrast, pif3 seedlings show a strong reduction in this growth peak (Figure 5a). These results are in accordance with the progressive pattern of PIF3 protein accumulation in the dark (Figure 1b,c) and the occurrence of PIF3-induced gene expression at the end of the night (Figure 3a), and suggest that the short hypocotyls in the pif3 mutant under SD conditions (Figure 2a,b) are mainly the result of a reduced growth rate at dawn. Together, these results indicate that PIF3 is required for hypocotyl elongation under diurnal conditions by promoting growth at the end of the night, as has been previously shown for PIF4 and PIF5 (Nozue et al., 2007).
Figure 5. PIF3 is required to promote growth at the end of the night under SD conditions.
(a) Hypocotyl elongation rate under SD conditions. Infrared imaging was used to monitor seedling growth from 2 days onwards. The growth rate is plotted as a function of time. Values are means ± SE of seven seedlings.
(b) Simplified model depicting PIF3- and PIF4/5-mediated hypocotyl growth under SD conditions. Top: the circadian clock regulates oscillation of PIF4 and PIF5 transcript abundance in SD-grown seedlings, whereas PIF3 remains constant throughout the day. Bottom: phyA and phyB activities induce degradation of PIF3 during the day (probably with an additional contribution from phyD), and phyB is active during the first part of the night to keep PIF3 levels low. As the night proceeds, phyB activity decreases and PIF3 progressively accumulates, peaking at the end of the night. For PIF4 and PIF5, coincidence of the circadian clock and light regulation ensures that protein accumulation peaks at the end of the night (Nozue et al., 2007). Endogenous PIF3 protein oscillation is indicated by a solid line, and the predicted endogenous PIF4/5 protein oscillation is indicated by a dashed line. PIF3 directly induces the expression of growth-related genes at the end of the night (exemplified by PIL1, HFR1 and XTR7), in conjunction with PIF4 and PIF5, to induce hypocotyl growth before dawn.
DISCUSSION
For Arabidopsis seedlings grown under SD conditions, the growth rate peaks at the end of the dark period. This rhythmic growth is implemented in part by the growth-promoting factors PIF4 and PIF5, and coincidence of both light and the circadian clock regulation determines their time of action just before dawn (Nozue et al., 2007; Niwa et al., 2009). The experiments presented here examine whether and through what mechanism PIF3 contributes to seedling growth under diurnal conditions. The data indicate that PIF3 protein accumulates progressively during the night under the control of phyB through a mechanism that does not involve transcriptional regulation by the clock, and provide evidence that PIF3, in conjunction with PIF4 and PIF5, is a major component of the cellular machinery that promotes hypocotyl elongation at dawn during growth under SD conditions, functioning at least in part through direct regulation of expression of growth-related genes (Figure 5b).
Our phenotypic and marker gene expression analyses of pif single and higher-order mutants provide evidence that PIF3 is necessary for seedling growth under SD conditions in conjunction with PIF4 and PIF5, and suggest that the PIF3 contribution is comparable to that of PIF4 and PIF5 combined. Various lines of evidence support this conclusion. First, the pif3 mutant displays a more prominent short-hypocotyl phenotype than either pif4 or pif5 under SD conditions, and this phenotype is similar in magnitude to that of the pif4 pif5 double mutant (Figure 4a). Second, the PIL1 expression level in pif3 shows a greater reduction with respect to WT than in either single pif4 or pif5 mutants, and a similar level of reduction to pif4 pif5 (Figure 4c). Finally, the pif3 pif4 and pif3 pif5 mutants show even more reduced PIL1 gene expression compared to pif3 or pif4 pif5, and this is similar to the triple mutant pif3 pif4 pif5 (Figure 4c). These data thus suggest that PIF3, PIF4 and PIF5 act together to promote hypocotyl elongation under diurnal conditions, and that PIF3 appears to play a more prominent role. Interestingly, the relative contributions of PIF3 and PIF4/PIF5 to the promotion of seedling growth under diurnal conditions appear to be different from the relative contribution of each PIF under other growth conditions. For example, the roles of PIF3, PIF4 or PIF5 in induction of hypocotyl growth in etiolated seedlings are mainly apparent in the absence of PIF1, the PIF with the strongest contribution to the hypocotyl response in post-germinative growth in the dark (Leivar et al., 2012b). In contrast, although no individual PIF appears to dominate the growth response to a continuous low red/far red ratio (Leivar et al., 2012b), PIF3 makes a greater contribution to afternoon shade events under diurnal conditions (Sellaro et al., 2012), and PIF4 is the strongest contributor to high-temperature effects (Koini et al., 2009; Stavang et al., 2009; Franklin et al., 2011). Together, these data suggest that the contribution of a given PIF to hypocotyl elongation varies between growth situations.
Previous results have shown that PIF3 demonstrates dual functioning during seedling de-etiolation: (i) as a transcriptional regulator during development in the dark and in the initial dark-to-light transition, and (ii) as a regulator of phyB homeostasis during sustained growth under prolonged light conditions. Evidence presented here suggests that, under SD conditions, a growth regime that alternates dark and light periods, PIF3 does not regulate phyB levels (Figure 2). Given the slow dynamics of PIF-induced phyB degradation in response to the initial light signal (Khanna et al., 2007; Al-Sady et al., 2008), it is likely that the short length of the light period (only 8 h) under SD conditions is not enough to promote a detectable effect. Instead, the role of PIF3 as promoter of hypocotyl growth appears to be mediated through its intrinsic transcriptional activity directly regulating the expression of growth-related genes (Figure 3). Our results show that, under SD conditions, PIF3 binds to the promoters and probably directly regulates expression of target genes that were previously reported to be growth-related during etiolation and shade avoidance, such as PIL1, HFR1 and XTR7 (Figure 3). These genes have been previously shown to be direct targets of PIF4 and/or PIF5 in dark-adapted plants (de Lucas et al., 2008) and under shade conditions (Hornitschek et al., 2009), respectively, and therefore it was not unexpected to find that PIF4 and PIF5 regulate their expression also under SD conditions (Figure 4), in accordance with recent data from Nozue et al. (2011) for HFR1 and XTR7. However, as indicated by the results for PIL1, the contribution of each of these PIFs to full induction appears to vary between growth conditions: whereas PIF3 is the strongest contributor under SD conditions (Figure 4b), PIF5 dominates in shade (Lorrain et al., 2008; Leivar et al., 2012b). These results suggest different target affinity and/or different relative levels of each PIF depending on the growth conditions.
The results presented here indicate that phyA and phyB are redundant in the rapid phytochrome-mediated degradation of PIF3 within 1 h after transition from darkness to light under SD conditions (Figure 1d), mirroring the phytochrome-mediated degradation of PIF3 during early stages of illumination of etiolated seedlings (Bauer et al., 2004; Al-Sady et al., 2006). These data suggest that PIF3 degradation under SD conditions may also require direct interaction with the phytochrome photoreceptor, leading to rapid phosphorylation of the transcription factor and degradation via the ubiquitin–proteasome system, as described for etiolated seedlings (Bauer et al., 2004; Al-Sady et al., 2006). In addition, our results show that the absence of phyB in phyB and phyA phyB mutants results in over-accumulation of PIF3 during the dark period, and that these elevated levels are reduced to a certain extent in response to prolonged light conditions (after 4 h), indicating that another photoreceptor is also involved in regulation of PIF3 degradation during the day. These results again mirror those observed in de-etiolation experiments, suggesting that this additional photoreceptor may be phyD (Bauer et al., 2004; Al-Sady et al., 2006). However, adding to previous data for dark-grown seedlings exposed to light (Bauer et al., 2004; Al-Sady et al., 2006), our evidence that the pool of PIF3 protein is not degraded in the absence of phyB indicates that phyB is necessary to mediate complete degradation of PIF3 during the light period under diurnal conditions (Figure 1e). This result provides evidence that phyB regulates degradation of PIF3 under SD conditions during the last part of the day. In addition, the observed re-accumulation of PIF3 in the absence of phyB during the first part of the night (Figure 1e) provides evidence that phyB also targets PIF3 for degradation at the start of the dark period. The extent of phyB action during the night is presumably determined by its dark reversion rate, which has been estimated to have a half-life of 1 h (Sweere et al., 2001; Rausenberger et al., 2010), as well as potentially via selective degradation of the Pfr form.
Our observation that phytochrome regulation keeps PIF3 protein levels low during the day and the first part of the night, with subsequent progressive accumulation, provides evidence for a phytochrome-mediated mechanism of PIF3 oscillation under SD conditions. Although phytochrome-imposed regulation of PIF3 protein accumulation may be sufficient to ensure timing of action of PIF3 at the end of the night, without additional transcriptional regulation by the circadian clock, a scenario in which the clock post-translationally regulates or fine-tunes PIF3 accumulation and/or activity indirectly cannot be completely discounted. DELLA proteins have been shown to interfere with PIF3 and PIF4 binding to DNA (de Lucas et al., 2008; Feng et al., 2008), and a recent report showed that DELLA proteins accumulate at the start of the night in seedlings grown under diurnal conditions (Arana et al., 2011). Therefore, DELLA proteins could represent a mechanism to prevent PIF3 from binding and inducing its target genes when its levels start to increase during the first part of the night. Further investigation is required to address this possibility.
Taken together, the data presented here indicate that PIF3 has a prominent role as a promoter of hypocotyl elongation under SD conditions, at least in part by directly regulating the expression of growth-related genes. Our work also reveals that phyA, phyB and possibly phyD induce degradation of PIF3 during the dark-to-light transition and the light period of diurnally grown seedlings, and residual photoactivated phyB prevents re-accumulation of PIF3 during the first part of the night. Our findings imply that PIFs regulating growth under diurnal conditions do not necessarily have to be transcriptionally regulated by the clock as previously shown for PIF4 and PIF5, and that phytochrome-mediated regulation may be sufficient. However, the existence of other more indirect layers of regulation of PIF3 by the clock and/or factors such as DELLA proteins (or other unknown mechanisms) cannot be excluded, and these may fine-tune the timing of PIF3 action under SD conditions.
EXPERIMENTAL PROCEDURES
Seedling growth and hypocotyl measurements
Wild-type and mutant Arabidopsis thaliana seeds used in these studies were all in the Columbia (Col-0) ecotype and have been described elsewhere, including pif3-3 (Monte et al., 2004), pif4-2 and pif3 pif4 (Leivar et al., 2008a), pif5-3 (Khanna et al., 2007), pif3 pif5, pif4 pif5 and pif3 pif4 pif5 phyB (Leivar et al., 2012a), pif3 pif4 pif5 (Leivar et al., 2008b), phyB-9 (Reed et al., 1993), pif3 phyB (Al-Sady et al., 2008), phyA-211 (Nagatani et al., 1993), phyA phyB (Cerdan and Chory, 2003) and pif3::YFP-PIF3 (Al-Sady et al., 2006).
Seeds were sterilized and plated on germination medium (GM) (Valvekens et al., 1988) without sucrose as previously described (Monte et al., 2003). Seedlings were then stratified for 4 days at 4°C in darkness, and then placed under short-day (SD) conditions [8 h white light (85 μmol m−2 sec−1) + 16 h dark] for the time indicated in each experiment. For hypocotyl measurements, seedlings were arranged horizontally on a plate, photographed using a digital camera (Nikon D80, http://www.nikon.com/) and measured as described previously (Monte et al., 2003). At least 30 seedlings for each line were measured to calculate the mean and standard error. For time-lapse photography, seedlings were grown on vertical plates, and, after 2 days of growth, photographs were taken at 30 min intervals for 24 h. To acquire images in the dark, 5 sec illumination was provided by an infrared light-emitting diode, and photographs were taken using an infrared-sensitive digital camera (Nikon D80). Hypocotyls of seven Col-0 and pif3 seedlings were measured, and the growth rate was calculated for each individual seedling.
Protein extraction and immunoblots
Protein extracts were prepared from 2- and 3-day-old seedlings grown under SD conditions as indicated. Tissue samples were collected and frozen in liquid nitrogen, and samples were manually ground under frozen conditions before resuspension in extraction buffer. The extraction buffer used and protein quantification were as previously described (Leivar et al., 2008a). Total protein extracts were subjected to SDS–PAGE (7.5%) for immunodetection of phyB and YFP–PIF3 protein (80 μg) or endogenous PIF3 (200 μg). Proteins were transferred to Hybond C membrane (Amersham Biosciences), and the membrane was stained with Ponceau S as a loading control. Immunodetection of PIF3 and YFP–PIF3 was performed using a rabbit anti-PIF3 polyclonal antibody (Al-Sady et al., 2006), incubated overnight with Hikari solution (Nacalai Tesque), and immunodetection of phyB was performed using mouse monoclonal anti-phyB (B1 and B7) antibodies (Somers et al., 1991). Peroxidase-linked anti-rabbit secondary antibody (Amersham Biosciences) for PIF3 and anti-mouse secondary antibody for phyB (Amersham Biosciences) and a SuperSignal West Femto chemiluminescence kit (Pierce) were used for detection of luminescence using a LAS-4000 Image imaging system (Fujifilm).
Gene expression analysis
For RNA blots, total RNA was extracted from 4-day-old SD-grown seedlings as described by Monte et al. (2003) (see Table S1 for primer sequences used to amplify the PIL1 probe). Hybridization signal was quantified using a Storm 860 PhosphorImager (Molecular Dynamics) and normalized to 25S rRNA levels.
For quantitative RT-PCR analysis, RNA extraction, cDNA synthesis and quantitative RT-PCR were performed as described previously (Sentandreu et al., 2011). Gene expression was measured in three technical replicates for each biological sample. PP2A (AT1G13320) was used as a normalization control as described previously (Shin et al., 2007). Table S1 lists primer sequences.
Chromatin immunoprecipitation (ChIP) assays
ChIP assays were performed using 3-day-old SD-grown pif3::YFP-PIF3 and Col-0 seedlings as described previously (Gendrel et al., 2002). After sonication, protein was quantified, and the inputs used in the subsequent immunoprecipitation step were equivalent for all samples. Antibody samples were immunoprecipitated by overnight incubation with GFP antibody-bound resin (GFP Agarose Beads, MBL). Mock ChIP reactions were performed without antibody to measure non-specific binding to target sequences. After immunoprecipitation, purified DNA was subjected to quantitative RT-PCR using promoter- and control-specific primers (Table S1) for each gene of interest. Quantitative RT-PCR results in the presence or absence of antibody for each genotype were first normalized to their input, and fold enrichment was then calculated for each antibody-containing sample relative to the corresponding sample lacking antibody.
Statistics
Data were analyzed by one-way ANOVA, and the differences between means were evaluated using Duncan’s post-hoc multiple comparison test (SPSS statistics software, IBM). Statistically significant differences were defined as those with a P value < 0.05.
Supplementary Material
Figure S1. Accumulation of PIF3 in SD-grown seedlings.
Figure S2. Hypocotyl phenotype of phyA, phyB and phyA phyB seedlings under SD conditions.
Figure S3. Relative position of ChIP primers.
Figure S4. Additional biological replicate for the ChIP experiment shown in Figure 3(b).
Table S1. List of primer sequences.
Acknowledgments
This work was supported by a JAE pre-doctoral fellowship (Jae-Pre_08_01049) and a JAE Estancia Breve grant (2010ESTCSIC-12125 for a short stay in S.P.’s laboratory) from CSIC to J.S., a ‘Comissionat per a Universitats i Recerca del Departament d’Innovació, Universitats i Empresa’ fellowship from the Generalitat de Catalunya (Beatriu de Pinós program) and Marie Curie International Reintegration Grant PIRG06-GA-2009-256420 to P.L., by National Institutes of Health Grant GM-47475, Department of Energy Grant DEFG03-87ER13742, and USDA Agricultural Research Service Current Research Information System Grant 5335-21000-027-00D to P.H.Q., and Marie Curie International Reintegration Grant 046568, grants from the Spanish Ministerio de Ciencia e Innovación (BIO2006-09254 and BIO2009-07675), and a grant from the Generalitat de Catalunya (2009-SGR-206) to E.M.
Footnotes
Additional Supporting Information may be found in the online version of this article
Please note: As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
References
- Al-Sady B, Ni W, Kircher S, Schafer E, Quail PH. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell. 2006;23:439–446. doi: 10.1016/j.molcel.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Al-Sady B, Kikis EA, Monte E, Quail PH. Mechanistic duality of transcription factor function in phytochrome signaling. Proc Natl Acad Sci USA. 2008;105:2232–2237. doi: 10.1073/pnas.0711675105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arana MV, Marin-de la Rosa N, Maloof JN, Blazquez MA, Alabadi D. Circadian oscillation of gibberellin signaling in Arabidopsis. Proc Natl Acad Sci USA. 2011;108:9292–9297. doi: 10.1073/pnas.1101050108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae G, Choi G. Decoding of light signals by plant phytochromes and their interacting proteins. Annu Rev Plant Biol. 2008;59:281–311. doi: 10.1146/annurev.arplant.59.032607.092859. [DOI] [PubMed] [Google Scholar]
- Bauer D, Viczian A, Kircher S, et al. Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell. 2004;16:1433–1445. doi: 10.1105/tpc.021568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon A, Shen H, Huq E. Phytochrome interacting factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 2007;12:514–521. doi: 10.1016/j.tplants.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Cerdan PD, Chory J. Regulation of flowering time by light quality. Nature. 2003;423:881–885. doi: 10.1038/nature01636. [DOI] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451:475–479. doi: 10.1038/nature06448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA. Shade avoidance. New Phytol. 2008;179:930–944. doi: 10.1111/j.1469-8137.2008.02507.x. [DOI] [PubMed] [Google Scholar]
- Franklin KA, Quail PH. Phytochrome functions in Arabidopsis development. J Exp Bot. 2010;61:11–24. doi: 10.1093/jxb/erp304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, et al. Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci USA. 2011;108:20231–20235. doi: 10.1073/pnas.1110682108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel AV, Lippman Z, Yordan C, Colot V, Martienssen RA. Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science. 2002;297:1871–1873. doi: 10.1126/science.1074950. [DOI] [PubMed] [Google Scholar]
- Hennig L, Buche C, Eichenberg K, Schafer E. Dynamic properties of endogenous phytochrome A in Arabidopsis seedlings. Plant Physiol. 1999;121:571–577. doi: 10.1104/pp.121.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschfeld M, Tepperman JM, Clack T, Quail PH, Sharrock RA. Coordination of phytochrome levels in phyB mutants of Arabidopsis as revealed by apoprotein-specific monoclonal antibodies. Genetics. 1998;149:523–535. doi: 10.1093/genetics/149.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P, Lorrain S, Zoete V, Michielin O, Fankhauser C. Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J. 2009;28:3893–3902. doi: 10.1038/emboj.2009.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang IC, Henriques R, Seo HS, Nagatani A, Chua NH. Arabidopsis Phytochrome interacting factor proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell. 2010;22:2370–2383. doi: 10.1105/tpc.109.072520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW. Light-regulated transcriptional networks in higher plants. Nat Rev Genet. 2007;8:217–230. doi: 10.1038/nrg2049. [DOI] [PubMed] [Google Scholar]
- Khanna R, Shen Y, Marion CM, Tsuchisaka A, Theologis A, Schafer E, Quail PH. The basic helix-loop-helix transcription factor PIF5 acts on ethylene biosynthesis and phytochrome signaling by distinct mechanisms. Plant Cell. 2007;19:3915–3929. doi: 10.1105/tpc.107.051508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koini MA, Alvey L, Allen T, Tilley CA, Harberd NP, Whitelam GC, Franklin KA. High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr Biol. 2009;19:408–413. doi: 10.1016/j.cub.2009.01.046. [DOI] [PubMed] [Google Scholar]
- Leivar P, Quail PH. PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 2011;16:19–28. doi: 10.1016/j.tplants.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Al-Sady B, Carle C, Storer A, Alonso JM, Ecker JR, Quail PH. The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell. 2008a;20:337–352. doi: 10.1105/tpc.107.052142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH. Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol. 2008b;18:1815–1823. doi: 10.1016/j.cub.2008.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Tepperman JM, Monte E, Calderon RH, Liu TL, Quail PH. Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell. 2009;21:3535–3553. doi: 10.1105/tpc.109.070672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Cohn MM, Quail PH. Phytochrome signaling in green Arabidopsis seedlings: impact assessment of a mutually-negative phyB–PIF feedback loop. Mol Plant. 2012a;5:208–223. doi: 10.1093/mp/sss031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Tepperman JM, Cohn MM, Monte E, Al-Sady B, Erickson E, Quail PH. Dynamic Antagonism Between Phytochromes and PIF-family bHLHs Induces Selective Reciprocal Responses to Light and Shade in a Rapidly Responsive Transcriptional Network in Arabidopsis. Plant Cell. 2012b;24:1398–1419. doi: 10.1105/tpc.112.095711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linschitz H, Kasche V. The kinetics of phytochrome conversion. J Biol Chem. 1966;241:3395–3403. [PubMed] [Google Scholar]
- Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 2008;53:312–323. doi: 10.1111/j.1365-313X.2007.03341.x. [DOI] [PubMed] [Google Scholar]
- de Lucas M, Daviere JM, Rodriguez-Falcon M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blazquez MA, Titarenko E, Prat S. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- Martínez-García JF, Huq E, Quail PH. Direct targeting of light signals to a promoter element-bound transcription factor. Science. 2000;288:859–863. doi: 10.1126/science.288.5467.859. [DOI] [PubMed] [Google Scholar]
- Michael TP, Breton G, Hazen SP, Priest H, Mockler TC, Kay SA, Chory J. A morning-specific phytohormone gene expression program underlying rhythmic plant growth. PLoS Biol. 2008;6:e225. doi: 10.1371/journal.pbio.0060225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte E, Alonso JM, Ecker JR, Zhang Y, Li X, Young J, Austin-Phillips S, Quail PH. Isolation and characterization of phyC mutants in Arabidopsis reveals complex crosstalk between phytochrome signaling pathways. Plant Cell. 2003;15:1962–1980. doi: 10.1105/tpc.012971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte E, Tepperman JM, Al-Sady B, Kaczorowski KA, Alonso JM, Ecker JR, Li X, Zhang Y, Quail PH. The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc Natl Acad Sci USA. 2004;101:16091–16098. doi: 10.1073/pnas.0407107101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J, Zhu L, Shen H, Huq E. PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc Natl Acad Sci USA. 2008;105:9433–9438. doi: 10.1073/pnas.0803611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani A. Light-regulated nuclear localization of phytochromes. Curr Opin Plant Biol. 2004;7:708–711. doi: 10.1016/j.pbi.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Nagatani A, Reed JW, Chory J. Isolation and initial characterization of Arabidopsis mutants that are deficient in phytochrome A. Plant Physiol. 1993;102:269–277. doi: 10.1104/pp.102.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa Y, Yamashino T, Mizuno T. The circadian clock regulates the photoperiodic response of hypocotyl elongation through a coincidence mechanism in Arabidopsis thaliana. Plant Cell Physiol. 2009;50:838–854. doi: 10.1093/pcp/pcp028. [DOI] [PubMed] [Google Scholar]
- Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JN. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448:358–361. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]
- Nozue K, Harmer SL, Maloof JN. Genomic analysis of circadian clock-, light-, and growth-correlated genes reveals Phytochrome-Interacting Factor5 as a modulator of auxin signaling in Arabidopsis. Plant Physiol. 2011;156:357–372. doi: 10.1104/pp.111.172684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farre EM, Kay SA. The ELF4–ELF3–LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011;475:398–402. doi: 10.1038/nature10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Yamaguchi S, Kamiya Y, Bae G, Chung WI, Choi G. Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 2006;47:124–139. doi: 10.1111/j.1365-313X.2006.02773.x. [DOI] [PubMed] [Google Scholar]
- Park E, Kim J, Lee Y, Shin J, Oh E, Chung WI, Liu JR, Choi G. Degradation of phytochrome interacting factor 3 in phytochrome-mediated light signaling. Plant Cell Physiol. 2004;45:968–975. doi: 10.1093/pcp/pch125. [DOI] [PubMed] [Google Scholar]
- Quail PH. Phytochromes. Curr Biol. 2010;20:R504–R507. doi: 10.1016/j.cub.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausenberger J, Hussong A, Kircher S, Kirchenbauer D, Timmer J, Nagy F, Schafer E, Fleck C. An integrative model for phytochrome B mediated photomorphogenesis: from protein dynamics to physiology. PLoS One. 2010;5:e10721. doi: 10.1371/journal.pone.0010721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J. Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell. 1993;5:147–157. doi: 10.1105/tpc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Su YS, Lagarias JC. Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol. 2006;57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter MG, Franklin KA, Whitelam GC. Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature. 2003;426:680–683. doi: 10.1038/nature02174. [DOI] [PubMed] [Google Scholar]
- Sasidharan R, Chinnappa CC, Staal M, Elzenga JT, Yokoyama R, Nishitani K, Voesenek LA, Pierik R. Light quality-mediated petiole elongation in Arabidopsis during shade avoidance involves cell wall modification by xyloglucan endotransglucosylase/hydrolases. Plant Physiol. 2010;154:978–990. doi: 10.1104/pp.110.162057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer E, Nagy F. Photomorphogenesis in Plants and Bacteria. Dordrecht, The Netherlands: Springer; 2006. [Google Scholar]
- Sellaro R, Pacín M, Casal JJ. Diurnal dependence of growth responses to shade in Arabidopsis: role of hormone, clock, and light signaling. Mol Plant. 2012;5:93–102. doi: 10.1093/mp/ssr122. [DOI] [PubMed] [Google Scholar]
- Sentandreu M, Martin G, Gonzalez-Schain N, Leivar P, Soy J, Tepperman JM, Quail PH, Monte E. Functional profiling identifies genes involved in organ-specific branches of the PIF3 regulatory network in Arabidopsis. Plant Cell. 2011;23:3974–3991. doi: 10.1105/tpc.111.088161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Moon J, Huq E. PIF1 is regulated by light-mediated degradation through the ubiquitin–26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J. 2005;44:1023–1035. doi: 10.1111/j.1365-313X.2005.02606.x. [DOI] [PubMed] [Google Scholar]
- Shen Y, Khanna R, Carle CM, Quail PH. Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol. 2007;145:1043–1051. doi: 10.1104/pp.107.105601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Zhu L, Castillon A, Majee M, Downie B, Huq E. Light-induced phosphorylation and degradation of the negative regulator Phytochrome-Interacting Factor1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell. 2008;20:1586–1602. doi: 10.1105/tpc.108.060020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Park E, Choi G. PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J. 2007;49:981–994. doi: 10.1111/j.1365-313X.2006.03021.x. [DOI] [PubMed] [Google Scholar]
- Shin J, Kim K, Kang H, Zulfugarov IS, Bae G, Lee CH, Lee D, Choi G. Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc Natl Acad Sci USA. 2009;106:7660–7665. doi: 10.1073/pnas.0812219106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Sharrock RA, Tepperman JM, Quail PH. The hy3 long hypocotyl mutant of Arabidopsis is deficient in phytochrome B. Plant Cell. 1991;3:1263–1274. doi: 10.1105/tpc.3.12.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavang JA, Gallego-Bartolome J, Gomez MD, Yoshida S, Asami T, Olsen JE, Garcia-Martinez JL, Alabadi D, Blazquez MA. Hormonal regulation of temperature-induced growth in Arabidopsis. Plant J. 2009;60:589–601. doi: 10.1111/j.1365-313X.2009.03983.x. [DOI] [PubMed] [Google Scholar]
- Strasser B, Sanchez-Lamas M, Yanovsky MJ, Casal JJ, Cerdan PD. Arabidopsis thaliana life without phytochromes. Proc Natl Acad Sci USA. 2010;107:4776–4781. doi: 10.1073/pnas.0910446107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweere U, Eichenberg K, Lohrmann J, Mira-Rodado V, Baurle I, Kudla J, Nagy F, Schafer E, Harter K. Interaction of the response regulator ARR4 with phytochrome B in modulating red light signaling. Science. 2001;294:1108–1111. doi: 10.1126/science.1065022. [DOI] [PubMed] [Google Scholar]
- Tepperman JM, Hwang YS, Quail PH. phyA dominates in transduction of red-light signals to rapidly responding genes at the initiation of Arabidopsis seedling de-etiolation. Plant J. 2006;48:728–742. doi: 10.1111/j.1365-313X.2006.02914.x. [DOI] [PubMed] [Google Scholar]
- Valvekens D, Montagu MV, Van Lijsebettens M. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashino T, Matsushika A, Fujimori T, Sato S, Kato T, Tabata S, Mizuno T. A link between circadian-controlled bHLH factors and the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol. 2003;44:619–629. doi: 10.1093/pcp/pcg078. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Accumulation of PIF3 in SD-grown seedlings.
Figure S2. Hypocotyl phenotype of phyA, phyB and phyA phyB seedlings under SD conditions.
Figure S3. Relative position of ChIP primers.
Figure S4. Additional biological replicate for the ChIP experiment shown in Figure 3(b).
Table S1. List of primer sequences.