Abstract
In the central nervous system, three enzymes belonging to the serine hydrolase family are thought to regulate the life time of the endocannabinoid 2-arachidonoylglycerol (C20:4) (2-AG). From these, monoacylglycerol lipase (MAGL) is well characterized and, on a quantitative basis, is the main 2-AG hydrolase. The postgenomic proteins α/β-hydrolase domain containing (ABHD)6 and ABHD12 remain poorly characterized. By applying a sensitive fluorescent glycerol assay, we delineate the substrate preferences of human ABHD6 and ABHD12 in comparison with MAGL. We show that the three hydrolases are genuine MAG lipases; medium-chain saturated MAGs were the best substrates for hABHD6 and hMAGL, whereas hABHD12 preferred the 1 (3)- and 2-isomers of arachidonoylglycerol. Site-directed mutagenesis of the amino acid residues forming the postulated catalytic triad (ABHD6: S148-D278-H306, ABHD12: S246-D333-H372) abolished enzymatic activity as well as labeling with the active site serine-directed fluorophosphonate probe TAMRA-FP. However, the role of D278 and H306 as residues of the catalytic core of ABHD6 could not be verified because none of the mutants showed detectable expression. Inhibitor profiling revealed striking potency differences between hABHD6 and hABHD12, a finding that, when combined with the substrate profiling data, should facilitate further efforts toward the design of potent and selective inhibitors, especially those targeting hABHD12, which currently lacks such inhibitors.
Keywords: ABPP, monoacylglycerol lipase, monoglyceride lipase, TAMRA-FP
The human serine hydrolases comprises a large family of enzymes with a predicted number of ∼240 that fall into two subfamilies: the serine proteases (∼125 members) and the metabolic serine hydrolases (∼115 members) (1, 2). The metabolic serine hydrolases include lipases and amidases and use a conserved serine nucleophile to hydrolyze amide, ester, and thioester bonds. The majority of serine hydrolases contain an α/β-hydrolase domain (ABHD) fold and use a Ser-His-Asp (SHD) triad for catalysis. Although several members of the metabolic serine hydrolase family are relatively well defined, the majority remain poorly characterized with respect to their physiological substrates and functions.
Members of the metabolic serine family are also intimately involved in the generation and degradation of the endocannabinoid 2-arachidonoylyglycerol (C20:4) (2-AG). In brain regions endowed with 2-AG signaling, “on demand” biosynthesis of 2-AG occurs through phospholipase Cβ-catalyzed cleavage of the membrane phospholipid phoshatidylinositol bisphosphate to generate sn-2-arachidonoyl-containing diacylglycerol (DAG) species, which are subsequently hydrolyzed by sn-1-specific lipases (DAGLα and DAGLβ) to generate 2-AG (3). The endocannabinoids are involved in a broad range of (patho)physiological processes, including neurotransmission, appetite, nociception, addiction, inflammation, peripheral metabolism, and reproduction (4–6). The biological actions of 2-AG are mediated via two G protein-coupled receptors (CB1R and CB2R) that show unique and tissue-specific distribution. CB1R is highly enriched in the brain.
The major enzymatic route for 2-AG inactivation is via hydrolysis, generating arachidonic acid (AA) and glycerol. In the CNS, three serine hydrolases, namely monoacylglycerol lipase (MAGL) and the α/β-hydrolase domain (ABHD)-containing proteins ABHD6 and ABHD12, account for ∼99% of 2-AG hydrolysis (7, 8). From these, MAGL is relatively well characterized and on a quantitative basis appears to be the main 2-AG hydrolase. MAGL is a ∼33 kDa protein that was originally purified and cloned from adipose tissue (9), where it catalyzes the final step in lipolysis (9, 10). MAGL shows a wide tissue distribution (9) and is therefore generally thought to serve “house-keeping” functions in lipid metabolism (11). In addition, recent studies have illuminated pathophysiological roles for MAGL (12, 13). Methylarachidonoylfluorophosphonate (MAFP) is among the most potent MAGL inhibitors identified to date (14–16). MAFP inhibits MAGL irreversibly but lacks selectivity because it inhibits most members of the metabolic serine hydrolase family.
The post-genomic proteins ABHD6 and ABHD12 remain poorly characterized regarding their physiological substrates and functions. ABHD6 is a ∼30 kDa integral membrane protein with high expression reported in certain forms of tumors (17, 18). Based on hydropathy analysis and biochemical studies, ABHD6 appears to be an integral membrane protein whose active site is predicted to face the cell interior (7). Such an orientation suggests that ABHD6 might be well suited to guard the intracellular pool of 2-AG. Recent evidence suggests that ABHD6 can indeed control the levels and signaling efficacy of 2-AG in neurons (19, 20), but it is not known whether this enzyme uses other substrates as well. Few inhibitors have been identified that selectively target ABHD6.
ABHD12 is a ∼45 kDa glycoprotein whose potential role as a brain 2-AG hydrolase was disclosed using activity-based protein profiling (ABPP) with mouse brain proteome, and it was estimated that at the bulk brain level ABHD12 accounts for ∼9% of total 2-AG hydrolase activity (7). 2-AG is the only recognized substrate for ABHD12, and 2-AG hydrolase activity is the only feature potentially linking ABHD12 to the endocannabinoid system. Based on hydropathy analysis and biochemical data, ABHD12 is an integral membrane protein whose active site is predicted to face the lumen/extracellular space (7). Inactivating mutations in the ABHD12 gene have been causally linked to the neurodegenerative disease called PHARC (polyneuropathy, hearing loss, ataxia, retinitis pigmentosa, and cataract) (21). ABHD12 transcripts are abundant in various brain regions as well as in microglia and related peripheral cell types, such as macrophages and osteoclasts (21). Further characterization of ABHD12 is hampered by the lack of selective inhibitors.
Further research on the novel 2-AG hydrolases would greatly benefit from a versatile activity assay allowing profiling of a wide repertoire of substrates by a simple readout such as monitoring the end product glycerol. Although glycerol assays have been in routine use for decades in lipolysis research, as far as we are aware, the potential of this methodology has not been realized in the field of endocannabinoid hydrolases. By applying a sensitive fluorescent assay to kinetically monitor glycerol production “on line,” we delineate here the substrate profiles of human ABHD6 and ABHD12 in comparison with MAGL. After transient transfections in HEK293 cells, the three hydrolases degraded a variety of MAGs, each with distinct substrate and isomer preferences. The enzymes exhibited no detectable fatty acid amide hydrolase or lysophospholipase activity; nor did they hydrolyze di- or triacylglycerols. By site-directed mutagenesis, we identified the catalytic triad of ABHD12 as S246-D333-H372 and verified S148 as the catalytic nucleophile of ABHD6. Preliminary inhibitor profiling revealed striking potency differences between hABHD6 and hABHD12.
MATERIALS AND METHODS
Drugs, chemicals, and reagents
All reagents for the glycerol assay mix (Fig. 1b), and the following substrates were obtained from Sigma (St. Louis, MO): MAG-C8:0 (1-capryloyl-rac-glycerol [C8:0]); MAG-C10:0 (1-decanoyl-rac-glycerol [C10:0]); MAG-C12:0 (1-lauroyl-rac-glycerol [C12:0]); MAG-C14:0 (1-myristoyl-rac-glycerol [C14:0]); MAG-C16:0 (2-palmitoyl-rac-glycerol [C16:0]); MAG-C18:0 (1-stearoyl-rac-glycerol [C18:0]); the 1 (3)- and 2-isomers of MAG-C18:1; 1,2,3-trioleoyl-rac-glycerol; and 1-oleoyl(C18:1)-sn-glycero-3-phosphate (LPA). 1,2-dioleoyl-rac-glycerol (DAG), 1-palmitoyl-2-oleoyl-3-linoleoyl-rac-glycerol, and the 1 (3)- and 2-isomers of MAG-C18:2 and MAG-C20:4 were from Cayman Chemicals (Ann Arbor, MI). Hydrolase inhibitors were from the following sources: methyl arachidonoylfluorophosphonate (MAFP), isopropyl dodecylfluorophosphonate (IDFP), and WWL70 from Cayman Chemicals; THL (orlistat) and pristimerin from Sigma; RHC-80267 from Biomol (Enzo Life Sciences); and hexadecane-1-sulfonyl fluoride (HDSF) from Calbiochem.
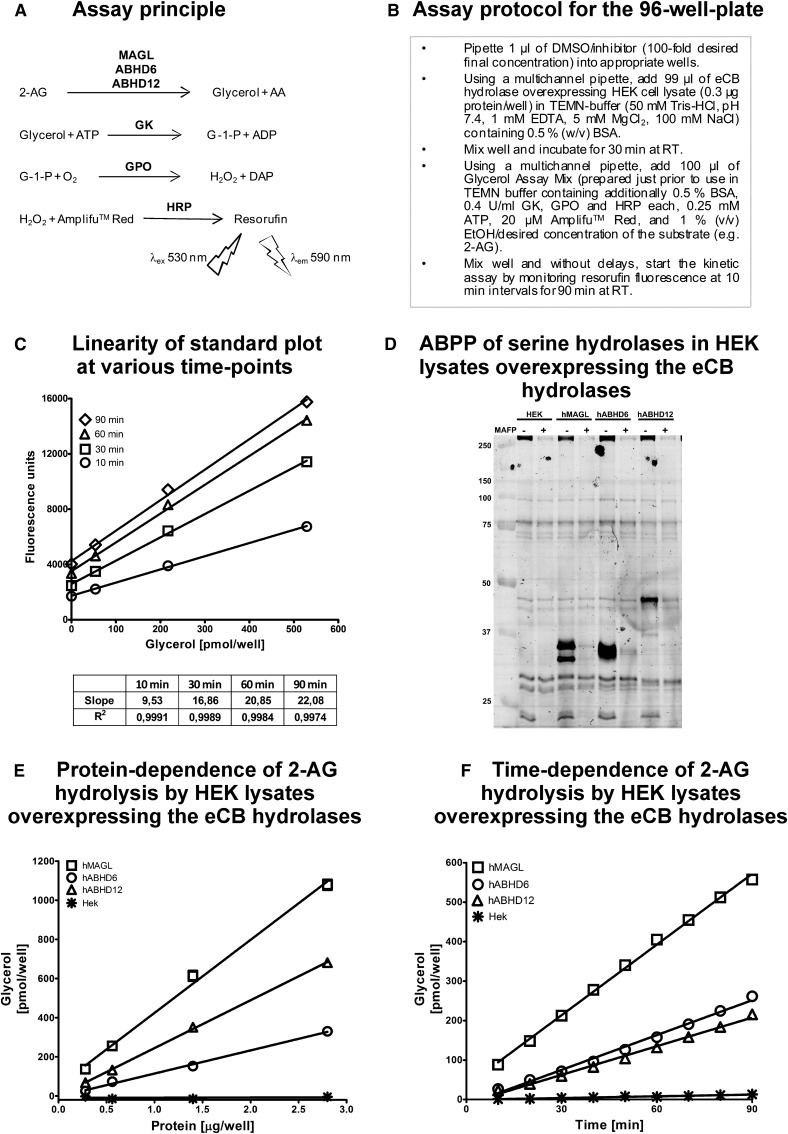
Fig. 1.
A sensitive fluorometric glycerol assay for endocannabinoid hydrolases. a: The assay is based on the following principle: The endocannanabinoid 2-AG is degraded to AA and glycerol by the endocannabinoid hydrolases MAGL, ABHD6, and ABHD12. Glycerol is converted to glycerol-1-phosphate (G-1-P) by glycerol kinase (GK). Glycerol phosphate oxidase (GPO)-catalyzed oxidation of G-1-P generates H2O2, which in the presence of HRP converts Amplifu™ Red to the fluorescent product resorufin. Resorufin fluorescence is monitored kinetically using excitation and emission wavelengths of 530 and 590 nm, respectively. b: Our assay protocol for the 96-well plate. c: Linearity of glycerol standard plot (0–520 pmol/well) at various time points. Although the coupled enzyme reactions generate maximal fluorescence only after prolonged (60 and 90 min) incubation time, the assay linearly detects glycerol at every time-point tested. d: Competitive ABPP of HEK cell lysates after transient transfections of the endocannabinoid hydrolases using the active site serine targeting fluorescent probe TAMRA-FP. After separation in SDS-electrophoresis gel (10%), serine hydrolases were visualized by in-gel fluorescent gel scanning as detailed in Materials and Methods. Molecular weight markers (kDa) are indicated by the numbers at left. Lysates were pretreated with DMSO or the serine-nucleophile targeting irreversible inhibitor MAFP (10−6 M) to demonstrate that TAMRA-FP labels the active-site serine residues of the endocannabinoid hydrolases. hMAGL and hABHD6 migrate as doublets (MAGL ∼33 and ∼35 kDa; ABHD6 ∼36 kDa), whereas hABHD12 migrates as single band (∼46 kDa). In addition to the endocannabinoid hydrolases, MAFP-sensitive TAMRA-FP labeling of endogenous serine hydrolases (<25 kDa and >250 kDa) is also evident. Data are from one typical transfection; transfections were repeated independently four times (hABHD6 or hABHD12) or twice (hMAGL). e and f: Linear protein-dependent (e) and time-dependent (f) glycerol response in lysates of HEK293 cells (0.3 µg protein/well) overexpressing the three human endocannabinoid hydrolases in assay mix containing 25 µM final 2-AG concentration. In panel e, glycerol was determined at time-point 30 min. In panels c, e, and f, values are means ± SD of duplicate wells from a typical experiment.
Generation of t ransient endocannabinoid hydrolase-overexpressing HEK293 cells
HEK293-cells were cultured as monolayers in DMEM (Euroclone, Milan, Italy) containing 10% FBS (Euroclone) under antibiotics (penicillin/streptomycin, Euroclone) at 37°C in a humidified atmosphere of 5% CO2/95% air. Plasmids containing wild-type (WT) or mutant cDNA were introduced to cells by a standard transient transfection procedure using X-tremeGENE Hp DNA Transfection reagent (Roche, Mannheim, Germany). HEK293 and/or Mock cells (cells transfected with an empty vector) were cultured in parallel for controlling expression and activity in later experiments. Cell lysates were prepared by washing cells two times with ice-cold PBS. Cells were scraped and pelleted at 250 g for 10 min at 4°C. Cell pellets were freeze-thawed three times, resuspended in ice-cold PBS, briefly sonicated, and aliquoted for storage at −80°C. Membranes were prepared by resuspending the cell pellet in PBS (pH 7.40), followed by brief sonication and centrifugation at 100,000 g for 45 min at 4°C. The pellet was resuspended in PBS by brief sonication and aliquoted for storage at −80°C. Protein concentrations were measured by using Pierce BCA Protein Assay Kit (Pierce, Rockford, IL) using BSA as a standard.
The fluorescent glycerol assay for endocannabinoid hydrolase activity
The set-up and validation of the fluorometric 96-well-plate glycerol assay is detailed in Fig. 1. Briefly, glycerol production was coupled via a three-step enzymatic cascade to a hydrogen peroxide (H2O2)-dependent generation of resorufin whose fluorescence (λex 530; λem 590 nm) was monitored using a Tecan Infinite M200 plate reader (Tecan Group Ltd., Männedorf, Switzerland). The assay protocol is detailed in Fig. 1b.
Activity-based protein profiling of serine hydrolases
Activity-based protein profiling (ABPP) was conducted using the fluorophosphonate probe TAMRA-FP (ActivX Fluorophosphonate Probes, Thermo Fisher Scientific Inc., Rockford, IL) following outlines of previously published methodology (7). Briefly, 25 µl of cellular lysates (5–10 µg protein) diluted in PBS were preincubated with 0.5 µl of the indicated inhibitors or a vehicle (DMSO) for 1 h at room temperature. Then, serine hydrolases were labeled with 0.5 µl of 100 µM TAMRA-FP for 1 h at RT. The reaction was stopped by adding 2× SDS-loading buffer, and proteins were separated in SDS-electrophoresis gel (10%) with molecular weight standards and compared with the pattern of labeled serine hydrolases in HEK293 or mock-transfected cells by following in-gel fluorescent gel scanning with Fujifilm FLA-3000 laser fluorescence scanner (Fujifilm, Tokyo, Japan) (Fluor. 532 nm; Filter: O580 nm).
Determination of substrate preferences
The substrate preferences of the endocannabinoid hydrolases were determined by monitoring glycerol production in lysates of HEK293 cells after transient expressing of each of the enzymes in assay mixes containing mono-, di-, or triglycerides or LPA (25 µM final concentration) with varying acyl chain length and saturation, as detailed in Fig. 2. To facilitate comparison between the three hydrolases under identical assay conditions, each substrate was tested in parallel using the three hydrolase preparations in the same experiment. For each tested substrate, assay blanks without enzyme, cellular background (HEK293/Mock cell lysates), as well as a glycerol quality control sample was included to monitor assay performance. Fluorescence of the assay blank was subtracted before calculation of the final results.
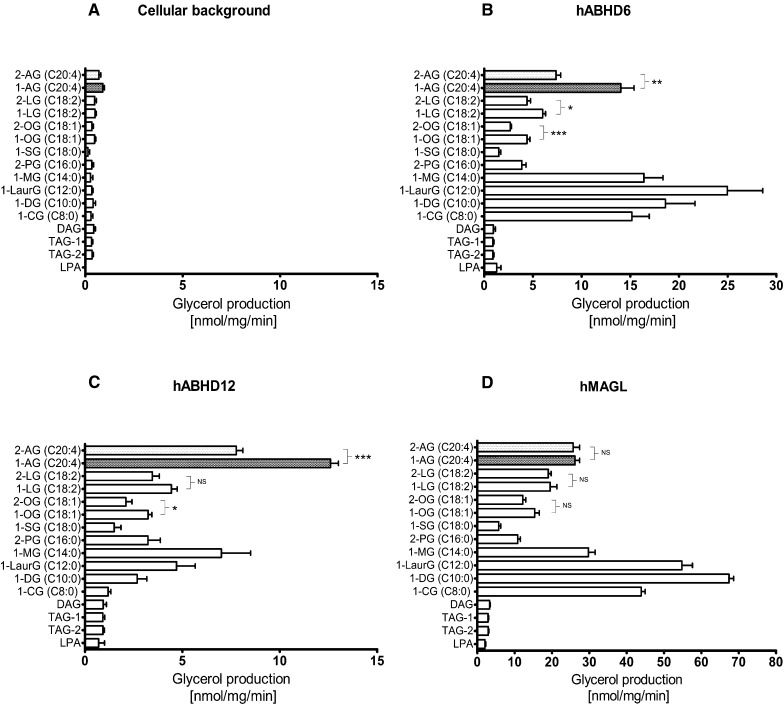
Fig. 2.
Substrate profiles of the human endocannabinoid hydrolases. HEK293 cells were transiently transfected with the cDNAs encoding hMAGL, hABHD6, or hABHD12 as detailed in Materials and Methods. After 48 h, cells were harvested, and lysates were prepared for hydrolase activity measurements using a sensitive fluorescent glycerol assay as described in Fig. 1. The substrate panel included MAGs with the indicated acyl chain length, isomer and degree of saturation, one DAG 1,2-dioleoyl(C18:1)-rac-glycerol, two triacylglycerols (TAG1, 1,2,3-trioleoyl(C18:1)glycerol; TAG2, 1-palmitoyl(C16:0)-2-oleoyl(C18:1)-3-linoleoyl(C18:2)-rac-glycerol, and LPA. Cellular lysates (0.3 µg/well) were incubated together with the indicated substrates (25 µM final concentration, added from 10 mM stock solutions in ethanol into the glycerol assay mix containing 0.5% [w/v] BSA and 1% [v/v ethanol]). Glycerol production was determined at 90 min. a: Background activity for the tested substrates. Cellular background was similar between HEK and mock-transfected cells (data not shown). b: Substrate profile of hABHD6. c: Substrate profile of hABHD12. d: Substrate profile of hMAGL. Data are mean + SEM from three to seven independent experiments using lysates of each enzyme from one transfection. Transfections were repeated independently four times (hABHD6 or hABHD12) or twice (hMAGL), and the relative profile for selected substrates (1-AG, 2-AG, 1-lauroyl-rac-glycerol [C14:0], and 1-myristoyl-rac-glycerol [C16:0]) was similar between different transfections (supplementary Fig. I). Statistical comparison between the MAG 1 (3)- and 2-isomers was done using a paired t-test (*P < 0.05; **P < 0.01; ***P < 0.001). The 1 (3)- and 2-isomers of MAG(C20:4) are highlighted in each panel.
Site-directed mutagenesis of the postulated catalytic triads
Plasmids (pCMV6-AC-hABHD6, pCMV6-XL4-hABHD12, and pCMV6-XL5-hMAGL transcript variant 1 [313 amino acid residues]) were purchased from Origene Technologies Inc. (Rockville, MD). Site-targeted mutagenesis to generate hABHD6 mutants S148A, D278A/E/N, and H306A/S/Y and hABHD12 mutants S246A, D333N, and H372A was performed by using a QuikChange® Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) following the manufacturer's instructions. Primers for the mutagenesis were purchased from Oligomer (www.oligomer.fi). After transformation of mutated material into competent bacterial cells, DNA was isolated, purified, and fully sequenced for confirmation of correct constructs.
Expression analysis of ABHD6 and ABHD12 SHD-mutants by Western blot
The expression level of WT and mutant hABHD6/12 in cells was analyzed by Western blot as follows. Samples of lysates from each HEK293 population carrying WT or mutant enzymes (20 µg protein of hABHD6 lysates and 50 µg protein of hABHD12 lysates), together with molecular weight markers, were fractionated by SDS-PAGE (10%) and transferred to a nitrocellulose membrane (Protran; Schleicher and Schell, Dassel, Germany). To block nonspecific binding, membranes were incubated with 0.5% (w/v) BSA and Tris-buffered saline containing 0.1% Tween (TBS-T) solution for 1 h at room temperature. Next, membranes were incubated overnight at 4°C with a rabbit anti-hABHD6 antibody (HPA017283, 1:1,000; Sigma) or with a mouse anti-hABHD12 antibody (ab68949, 1:200; Abcam) diluted in 0.5% BSA-TBS-T. After washing for 4 × 10 min in TBS-T, hABHD6 membranes were incubated with a goat anti-rabbit IgG secondary antibody ([H+L] 800, 1:30,000; Thermo Scientific) and hABHD12 membranes with a goat anti-mouse IgG secondary antibody (35521, Lot# MJ164339, [H+L] 800, 1:10,000; Thermo Scientific) diluted in 0.5% BSA-TBS-T for 1 h at 20°C, followed by washing 4 × 10 min with TBS-T. For the normalization of enzyme expression between separate WT and mutant cell transfections, expression of β-actin was determined from hABHD6 lysates by incubating membranes with a mouse anti-β-actin primary antibody (A5441, 1:2,000; Sigma) overnight at 4°C. After washing for 4 × 10 min in TBS-T, membranes were incubated with a secondary goat anti-mouse IgG antibody (cat#35521, DyLight 800 Conjugated, 1:10,000; Thermo Scientific). In the case of hABHD12, relative expression was calculated against expression of β-tubulin by using a mouse anti-β-tubulin primary antibody (1:500, cat#T4026; Sigma,) and goat anti-Mouse IgG secondary antibody (cat#35521, DyLight 800 Conjugated, 1:10,000; Thermo Scientific) with otherwise equal incubation conditions used with hABHD6 membranes. Immunoblots were visualized by Odyssey (Li-Cor Biosciences Inc., Lincoln, NB) and quantified by using ImageJ, a freely available Java-based image analysis software system developed in the National Institutes of Health (http://rsb.info.nih.gov/ij/).
Data reproducibility and statistical analyses
For hABHD6 and hABHD12, transient transfections were repeated independently four times (twice for hMAGL). For each enzyme batch, the relative hydrolysis rate of selected substrates was assessed to monitor the behavior and reproducibility of enzyme preparations between different transfections (supplementary Fig. I). A more detailed substrate profiling (Fig. 2) was performed using one particular batch of each enzyme and always so that that the three hydrolases were evaluated in parallel in the same experiment. With the exception of data presented in Fig. 1 and supplementary Figs. II and III, all numerical data are mean ± SEM from at least three independent experiments. The SHD mutants were analyzed from two to three separate transfections with similar outcome, and the numerical data presented in Fig. 3 are pooled from these experiment. The Km and Vmax values, inhibitor dose-response curves, and IC50 values derived thereof were calculated from nonlinear regressions using GraphPad Prism 5.0 for Windows. Statistical comparison between the MAG 1 (3)- and 2-isomers (Fig. 2) was done using paired t-test (* P < 0.05, **P < 0.01, and ***P < 0.001).
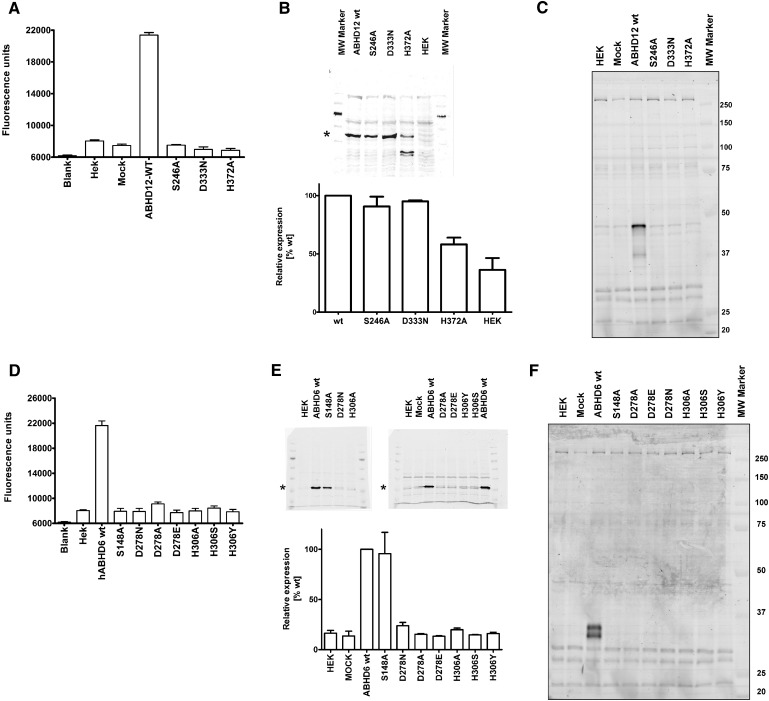
Fig. 3.
Analysis of hABHD6 and hABHD12 active site mutants. a: Raw fluorescence data of the hABHD12 SDH-mutants, analyzed using 2.8 µg cellular lysate per well. b: Western blots of the hABHD12 SDH-mutants (top) and the relative expression level of the mutants as compared with the wild-type enzyme (bottom), based on quantification of the protein bands indicated with an asterisk. The quantitative data are mean ± SEM from three separate transfections and were normalized against β-tubulin. c: TAMRA-FP labeling of the hABHD12 SDH-mutants. Molecular weight markers (kDa) are shown in the column at right. Data are from one set of transient transfections; transfections were repeated independently three times with identical outcome. d: Raw fluorescence data of the hABHD6 SDH-mutants analyzed using 2.8 µg cellular lysate per well. e: Western blots of the hABHD6 SDHmutants (top) and the relative expression level of the mutants as compared with the WT enzyme (bottom) based on quantification of the protein bands indicated with an asterisk. The quantitative data are mean ± SEM from three (WT, S148A, D278N, H306A) or two (all others) separate transfections and were normalized against β-actin. f: TAMRA-FP labeling of the hABHD6 SDH mutants. Molecular weight markers (kDa) are shown in the column at right. Data are from two sets of transient transfections; transfections were repeated independently three times (WT, S148A, D278N, H306A) or twice (all others) with identical outcome.
RESULTS
HEK293 cells as a convenient host for transient expression of human 2-AG hydrolases
The set-up and validation of the fluorometric endocannabinoid hydrolase assay is presented in Fig. 1a–c. Lysates were prepared from HEK293 cells 48 h after transient transfections with the cDNAs encoding hABHD6 and hABHD12. For comparative purposes, HEK293 cells were transfected also with the cDNA encoding hMAGL (313 amino acid residues). ABPP with the active site serine-targeting TAMRA-FP probe indicated that the three hydrolases were successfully expressed after transient transfections (Fig. 1d). TAMRA-FP recognized hMAGL migrating in SDS-PAGE gels as two protein bands (∼33 and ∼35 kDa). Similarly, hABHD6 migrated as a doublet with a molecular weight of ∼36 kDa. In contrast, hABHD12 migrated as a single band with a molecular weight of ∼46 kDa. In lysates pretreated with MAFP (10−6 M), TAMRA-FP labeling was fully prevented indicating that the fluorophosphonates shared a common target (i.e., the catalytic serine residues in each of the three hydrolases) (Fig. 1d).
The ABPP also indicated that HEK293 cells express negligible levels of endogenous MAGL, ABHD6, or ABHD12 because there was low-to-undetectable TAMRA-FP labeling of endogenous proteins migrating at the position of these enzymes. Moreover, TAMRA-FP labeling of the weak endogenous band with a size approximating that of ABHD12 was not prevented by MAFP, indicating that this band did not correspond to endogenous ABHD12. Transient expression of the three hydrolases resulted in robust time- and protein-dependent increase in 2-AG hydrolase activity as compared with the cellular background (Fig. 1e, f). With the tested protein concentrations, enzymatic activity increased linearly for the three hydrolases. Comparison of activities between HEK cell lysates and membranes indicated modest enrichment of hABHD6 and hABHD12 activity in membrane preparations (supplementary Fig. II), consistent with previous evidence implicating that the two hydrolases are integral membrane proteins (7). However, because cellular background was generally lower in lysates than in membranes and, on the other hand, hABHD6 and hABHD12 activities were readily detectable also in lysates, we used lysates instead of membranes in these studies. This was justified also from the economical point of view that only a fraction of cellular material was required to produce equal amounts of protein from lysates than from membranes. The high sensitivity of the fluorescent method allowed us to routinely perform activity assays in a 96-well format using only 0.3 µg cellular lysate per well; at this protein concentration hydrolase activity due to cellular background was negligible. The pH dependence of 2-AG hydrolysis was tested at pH values ranging from 5.3 to 9.1 (supplementary Fig. III). These experiments indicated that hABHD6 and hABHD12 exhibited a broad pH optimum between 7.2 and 9.1. Because the TEMN-BSA buffer (pH 7.4) (Fig. 1b) has been used in our previous studies exploring the signaling capacity and degradation of 2-AG in native cellular membranes (15, 22), we used this buffer also in the present study. Collectively, these experiments indicate that HEK293 cells serve a convenient host to express the three endocannabinoid hydrolases for further characterization.
Substrate preferences of hABHD6, hABHD12, and hMAGL
We delineated the substrate preferences of the three hydrolases by monitoring glycerol production in HEK293 cell lysates individually overexpressing each of the enzymes in assay mixes containing mono-, di-, and triglycerols (25 µM final concentration) with varying acyl chain length and saturation (Fig. 2). To facilitate comparison between the three hydrolases under identical assay conditions, each substrate was tested using the three hydrolase preparations in the same experiment. Cellular background activity was similar in HEK293 and mock-transfected cells (Fig. 3a, d and data not shown). Fig. 2a illustrates background activity with the tested substrates. This represented in most cases <10% of the glycerol produced by endocannabinoid hydrolase overexpressing lysates.
hABHD6 hydrolyzed a variety of MAGs with a distinct substrate preference (Fig. 2b). MAGs with saturated acyl chain length varying from C8:0 to C16:0 were readily hydrolyzed, with the relative activity order being C12:0 > C8:0 ≈ C10:0 ≈ C14:0 >> C16:0 > C18:0 (marginal). The preference for 1 (3)- vs. 2-acylglycerols was tested using oleoyl (C18:1), linoleoyl (C18:2), and arachidonoyl (C20:4) glycerols; in each case, hABHD6 preferred the 1 (3)-isomer with the relative activity order C20:4 >> C18:2 ≈ C18:1. hABHD6 exhibited negligible activity toward di- and triglycerides; nor did it hydrolyze 1-oleoyl-lysophosphatidic acid (LPA).
hABHD12 had a clearly different substrate profile (Fig. 1c). The preferred MAG substrate was 1 (3)-AG, followed by 2-AG and MAG(C14:0), which were hydrolyzed to the same extent. hABHD12 clearly preferred 1 (3)-AG over 2-AG, and a similar, although weaker, trend was observed for the 1 (3)-isomers of oleoyl (C18:1) and linoleoyl (C18:2) glycerol. For the unsaturated MAGs, the relative hydrolysis rate was C20:4 >> C18:2 ≈ C18:1. Although hABHD6 efficiently hydrolyzed MAGs with saturated acyl chain length varying from C8:0 to C16:0, hABHD12 used these substrates less efficiently and with a different preference (C14:0 > C12:0 ≈ C16:0 ≈ C10:0 > C18:0 ≈ C8:0). Like hABHD6, hABHD12 did not hydrolyze di- or triglycerides or LPA.
The overall substrate profile of hMAGL resembled that of hABHD6 with some notable exceptions (Fig. 1d). Like hABHD6, hMAGL hydrolyzed various MAGs with the following substrate preference: 1-decanoyl-rac-glycerol (C10:0) was the best substrate, and MAGs with saturated acyl chain were readily hydrolyzed, the relative activity being C10:0 > C12:0 > C8:0 > C14:0 >> C16:0 > C18:0 (marginal). In contrast to hABHD6 and hABHD12, which preferred the 1 (3)-isomers of unsaturated MAGs, hMAGL showed no such preference for the 1 (3)-isomers of linoleoyl (C18:2) and arachidonoyl (C20:4) glycerol. However, hMAGL slightly preferred the 1 (3)-isomer of C18:1, although this effect was not statistically significant. The relative activity toward unsaturated MAGs was C20:4 > C18:2 ≈ C18:1. Like hABHD6 and hABHD12, hMAGL did not hydrolyze di- or triglycerides or LPA.
Neither hABHD6 nor hABHD12 possessed fatty acid amide hydrolase activity; anandamide or its fluorogenic substrate analog N-decanoyl 7-amino-4-methyl coumarin were not used above cellular background by HEK293 cell preparations overexpressing these hydrolases (supplementary Table I).
Site-directed mutagenesis of the predicted catalytic triads
The MAGL catalytic triad (S122-D239-H269) was previously identified based on mutagenesis studies (9). To our knowledge, similar experimental verification is lacking concerning the predicted catalytic cores of ABHD6 and ABHD12. Amino acid sequence comparisons of the human, mouse, and rat orthologs indicated that the primary structures are well conserved between the species (supplementary Fig. IV). Homology for ABHD6 was 94.1% between human and mouse, 93.8% between human and rat, and 97.0% between mouse and rat. For ABHD12, the figures were 93.2% (human vs. mouse), 93.5% (human vs. rat), and 99.7% (mouse vs. rat). Typical α/β-hydrolase fingerprints, including the lipase motif and the predicted catalytic triads, are fully conserved.
We individually mutated the amino acids of the predicted catalytic triads of hABHD6 (S148-D278-H306) and hABHD12 (S246-D333-H372) and, after transient expression in HEK293 cells, assessed the ability of the mutants to catalyze 2-AG hydrolysis. As expected, the WT enzymes efficiently hydrolyzed 2-AG, whereas all mutants were devoid of enzymatic activity (Fig. 3a, d). Transfections were repeated two or three times, and each time the outcome was the same. Western blotting (Fig. 3b, e) was used to verify that the mutant proteins were properly expressed and in amounts comparable to those of their WT counterparts. This proved to be the case for all hABHD12 mutants, with the exception that the H372A mutant migrated as three immunoreactive bands (possibly due to increased sensitivity of this particular mutant toward enzymatic degradation), one corresponding to the size of the WT enzyme and two others with clearly smaller size (Fig. 3b). For hABHD6, the S148A mutant was properly expressed, whereas none of the mutants D278A/E/N or H306A/Y/S showed detectable expression (Fig. 3e). It is possible that these mutations disturbed proper protein folding, leading to total degradation in cells. Alternatively, the mutations may have altered protein conformation so that antibody recognition was severely compromised. Using ABPP, we assessed the capacity of the mutant hydrolases to bind TAMRA-FP (labeling of which requires the intact catalytic triad), with the outcome that none of the mutants was labeled by this probe (Fig. 3c, f).
Km and Vmax values for 2-AG
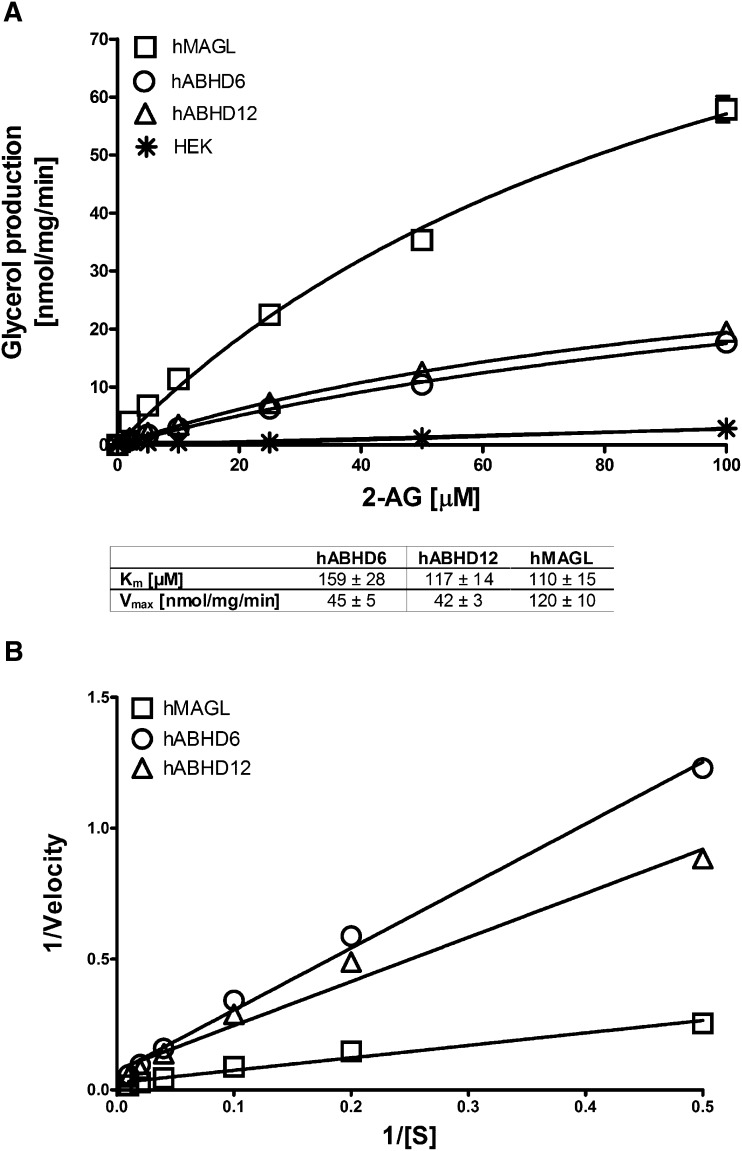
We compared the three hydrolases further by determining the Km and Vmax values for 2-AG as the substrate (Fig. 4). The apparent Km values were 159 ± 28 µM for hABHD6, 117 ± 14 µM for hABHD12, and 110 ± 15 µM for hMAGL. For these hydrolase batches, the Vmax values were 45 ± 5, 42 ± 3, and 120 ± 10 nmol/mg/min, respectively.
Fig. 4.
Km and Vmax values for 2-AG. a: HEK293 cells were transiently transfected with the cDNAs encoding hMAGL, hABHD6, or hABHD12 as detailed in Materials and Methods. After 48 h, cells were harvested, and lysates were prepared for hydrolase activity measurements as described in Fig. 1. Cellular lysates (0.3 µg/well) were incubated with the indicated concentrations of 2-AG, and the Km and Vmax values were determined at 60 min; substrate consumption was <10%. The Km and Vmax values are shown in the box and were calculated as nonlinear regressions using GraphPad Prism 5.0 for Windows. b: The Lineweaver-Burk plots of the data. Values are mean ± SEM from three independent experiments.
Inhibitor profiles of hABHD6 and hABHD12
We assessed the potency of a panel of inhibitors toward hABHD6 and hABHD12 using 2-AG as the substrate. The inhibitors included the fluorophosphonates MAFP (C20:4) and IDFP (C12:0), the sulfonylfluoride HDSF (C16:0), the lipase inhibitors THL (orlistat) and RHC-80267, the ABHD6-selective inhibitor WWL70, and the triterpene pristimerin. Dose-response curves were constructed for each inhibitor using five or six concentrations, and the IC50 values were calculated after nonlinear fitting of these curves (Table 1). We found that MAFP was the most potent inhibitor of both enzymes, followed by THL. IDFP and HDSF were ∼23-fold more potent toward hABHD6 than hABHD12, whereas for MAFP and THL, this difference was ∼5-fold. For hABHD6, the relative inhibitor potency order was MAFP > THL > WWL70 ≈ IDFP ≈ HDSF > RHC-80267 > pristimerin. For hABHD12, the relative potency order was MAFP > THL > IDFP ≈ HDSF >>> WWL70, RHC-80267, pristimerin (all inactive).
TABLE 1.
Inhibitor profiles of hABHD6 and hABHD12
| hABHD6 | hABHD12 | |
| Inhibitor | log[IC50] ± SEM | log[IC50] ± SEM |
| MAFP | −7.77 ± 0.04 | −7.06 ± 0.04 |
| IDFP | −6.97 ± 0.05 | −5.60 ± 0.06 |
| HDSF | −6.84 ± 0.10 | −5.67 ± 0.11 |
| THL | −7.32 ± 0.06 | −6.72 ± 0.07 |
| RHC-80267 | −6.18 ± 0.08 | Remaining activity, 96.7 ± 0.5% at 10 M |
| WWL70 | −7.07 ± 0.05 | Remaining activity, 101.0 ± 0.8% at 10 M |
| Pristimerin | −5.86 ± 0.07 | Remaining activity, 101.4 ± 3.4% at 10 M |
HEK293 cells were transiently transfected with the cDNAs encoding hABHD6 or hABHD12 as detailed in Materials and Methods. After 48 h, cells were harvested, and lysates were prepared for hydrolase activity measurements as described in Fig. 1. Cellular lysates (0.3 µg/well) were preincubated for 30 min at RT with DMSO (control) or with increasing concentrations of the inhibitors (MAFP, IDFP, HDSF, THL, RHC-8027, WWL70, and pristimerin). Thereafter, glycerol assay mix containing 2-AG (12.5 µM final concentration) was added, and glycerol production was monitored kinetically for 90 min at RT. Inhibitor dose-response curves were determined, and the IC50 values were calculated as nonlinear regressions using GraphPad Prism 5.0 for Windows. Data are mean ± SEM from three independent experiments.
DISCUSSION
Endocannabinoid hydrolysis is commonly studied using radiolabeled “natural” MAG substrates (9, 11, 14, 23–25) or colorigenic/fluorogenic artificial substrates (9, 16). In addition, “natural” MAG substrates with mass-spectrometry-based (7) and HPLC-based (15) detection have been successfully used. Although feasible, these methods have limitations, such as handling of radioactive material, restricted applicability to HTS format, and promiscuity of the artificial substrates toward nontarget hydrolases/esterases. Therefore, an initial goal of the present studies was to advance this methodology toward the direction where various natural substrates could be studied in a multiwell-plate format. Although glycerol assays coupled to colorimetric detection have been used in lipid research for decades (26, 27), we have here for the first time adapted and properly validated the sensitive fluorometric HTS glycerol assay for kinetic measurement of endocannabinoid hydrolase activity.
The versatile activity assay allowed us to delineate the substrate profiles of the endocannabinoid hydrolases by testing a panel of natural substrates. These studies reveal for the first time the substrate preferences of hABHD6 and hABHD12. We are not aware of any published study that has addressed this issue to a similar extent, even in the case of MAGL. Our studies indicated that the three hydrolases are genuine MAG lipases exhibiting negligible activity toward di- or triacylglycerols or fatty acid amides. None of the hydrolases catalyzed the LPA → MAG + Pi → glycerol + free fatty acid conversion, indicating that, in contrast to native brain membrane preparations that readily degrade LPA to generate Pi and glycerol by the concerted action of lipid phosphate phosphatases and MAG lipases (28), the three endocannabinoid hydrolases are devoid of lipid phosphate phosphatase activity that is required for the first step of this cascade.
We demonstrated that hABHD6 and hABHD12 clearly prefer the 1 (3)-acyl isomers of the unsaturated MAGs over their respective 2-acyl isomers, whereas hMAGL showed no such preference. A more detailed kinetic analysis (supplementary Fig. V) revealed that when glycerol output from the 1 (3)- and 2-isomers of MAG(C20:4) was determined at earlier time points, hABHD6 and hABHD12 hydrolyzed 1 (3)-AG at ∼2 to 3-fold faster rates, suggesting that nonenzymatic 2 → 1 (3) isomerization may be needed before enzymatic hydrolysis. In parallel incubations, hMAGL used both isomers equally well at every time point tested, consistent with previous modeling (29) and experimental data (15), showing that MAGL hydrolyzes 1 (3)-AG and 2-AG at similar rates. It was previously shown that when overexpressed in HeLa cells, rat MAGL hydrolyzes 2-OG at a 5-fold higher rate than 2-AG (11). In contrast, porcine brain MAGL clearly preferred 1 (3)- and 2-AG over 1 (3)-OG (14). In line with the latter observation, we found that hMAGL clearly preferred 2-AG over 2-OG. The observed differences likely reflect species and/or methodological differences. Our data are also in good agreement with earlier findings showing that mammalian MAGLs from various sources hydrolyze C20:4 from the sn-2-MAGs faster than C18:1 or C16:0 (30–32).
The in vitro substrate profile of hMAGL also closely matches that observed in vivo when analyzing MAG contents in brain and peripheral tissues of transgenic MAGL mice. For example, global MAGL knockout dramatically elevated 2-AG levels in brain and peripheral tissues (27, 33, 34). In line with our data, the largest elevations in brain MAG levels were observed for the unsaturated MAGs (C20:4 > C18:2 > C18:1), whereas levels of C16:0 or C18:0 (both were poor substrates in our study) remained also unchanged in MAGL-KO brains (27). Analysis of brain regional MAG levels in knock-in mice engineered to selectively overexpress MAGL in forebrain neurons indicated that levels of C20:4 (and in some cases C18:1) were lower in the transgenic mice, whereas levels of C16:0 and C18:0 were not altered (35).
Using a colorimetric glycerol assay analogous to the presently used fluorescent assay, Imamura and Kitaura (26) profiled a panel of MAGs toward purified bacterial MAGL. The substrate preference of bacterial MAGL in that study closely parallels that of hMAGL described here. MAGLs from both species showed the highest activity for MAGs(C8:0-C14:0), minimal activity for MAG(C18:0), and clearly improved activity with increasing C18 chain unsaturation (C18:1 and C18:2). Thus, MAGLs from species as distant as human and bacteria are strikingly similar in their substrate preference. It is generally thought that the substrate specificity of α/β hydrolases is largely determined by the so-called “cap region.” Recent elucidation of the crystal structure of bacterial MAGL revealed an unexpected conservation of the cap architecture between bacterial and human MAGL (36), providing structural support to the experimental data. Thus, regardless of species, MAGL is structurally conserved to carry out its principal role (i.e., MAG hydrolysis). It follows that tissue- and cell-type-dependent availability of MAG species determines to what extent each substrate is used in a given tissue or cell type. In adipose tissue MAGL is needed to complete the final step of lipolysis (10), whereas in the brain MAGL plays a key role in terminating the signaling function of 2-AG (16, 33, 34). Moreover, although phospholipase A2 activity is generally considered as the main source of AA for cyclooxygenase (COX)-mediated prostaglandin biosynthesis in peripheral tissues, MAGL-dependent 2-AG hydrolysis provides the principal source of AA for COX-mediated synthesis of neuroinflammatory prostaglandins in the CNS (13). In cancer cells, MAGL overexpression was proposed to act as the key metabolic switch orchestrating cancer cell malignancy by redirecting lipids from storage sites toward biosynthesis of cancer promoting signaling lipids (12). Collectively, these findings suggest that, despite the promiscuous substrate profile, MAGL serves highly specialized, tissue-specific (patho)physiological roles.
Less can be said regarding ABHD6. Our data revealed that hABHD6 and hMAGL share similar substrate preferences, suggesting that they could serve related functions. Recent evidence suggests that, like MAGL, ABHD6 can control the levels and signaling efficacy of 2-AG in neurons (19, 20). Overexpression of the two hydrolases has been found in certain types of cancer cells (12, 18). However, in contrast to MAGL inhibition (12), knockdown of ABHD6 did not inhibit tumor cell growth (18). Further studies with pharmacological or genetic ABHD6 inactivation should shed more light on this issue.
From the tested substrates, hABHD12 best hydrolyzed the isomers of the endocannabinoid 2-AG showing clear preference for 1 (3)-AG, although it moderately hydrolyzed also MAG(14:0). To achieve its retrograde signaling function in neurons, 2-AG is produced on demand postsynaptically and then activates CB1Rs on the presynaptic terminals (4–6). It is well established that in the aqueous milieu 2-AG undergoes rapid, nonenzymatic isomerization to 1 (3)-AG (15). This could represent an initial deactivation pathway for 2-AG, although 1 (3)-AG is not devoid of biological activity; it is only ∼3-fold less potent than 2-AG in activating the CB1Rs (22). Might ABHD12 provide further deactivation of the endocannabinoid by hydrolyzing 1 (3)-AG in particular? Hydropathy analysis and biochemical data suggest that ABHD12 is an integral membrane protein whose active site is predicted to face the lumen and/or extracellular space (7). Combined with the present findings showing that ABHD12 efficiently hydrolyses 1 (3)-AG, these observations support the notion (8) that ABHD12 is well suited to function as an ectohydrolase that is capable of guarding the extracellular signaling pool of 2-AG. However, there are no experimental data to support this. ABHD12 is highly expressed in various brain regions and in microglia (21). This suggests that ABHD12 could guard 2-AG-CB1R signaling in neurons. In addition, there is growing appreciation for the potential role of the endocannabinoids, and microglial CB2Rs in particular, as regulators of immune function in the CNS (37). It is therefore possible that ABHD12 might regulate 2-AG-CB2R signaling in glial cells.
With 2-AG as the substrate, we determined the apparent Km values of ∼160 µM and ∼120 µM for hABHD6 and hABHD12, respectively. In parallel incubations, the apparent Km for hMAGL was ∼110 µM. To our knowledge, these are the first Km estimates concerning hABHD6 and hABHD12. For MAGL, we are not aware of studies that have determined Km for 2-AG as the substrate. Instead, apparent Km values ranging from 0.2 mM to 0.5 mM were reported for purified MAGL preparations using 1 (3)-OG and/or 2-OG as the substrate (23–25). In one study, however, a Km value of 17 ± 3 µM was reported for hMAGL (38). The Km value we obtained here for hMAGL falls within the reported range. However, the Km values for lipophilic substrates should be considered only as approximates because of several factors that may affect the accuracy of Km determinations. Instead of purified enzymes, we used lysates of endocannabinoid hydrolase overexpressing cells, and, although the sensitive assay allowed us to minimize the signal due to cellular background, it was evident that the background slightly contributed to the total activity, especially in the low-substrate-concentration range. On the other hand, availability of the lipophilic substrates due to solubility and/or critical micellar concentration likely also compromises accurate Km determinations. Despite these limitations, our study offers a comparative view to the Km values of the endocannabinoid hydrolases, which were tested under identical conditions.
Our inhibitor profiling revealed that MAFP, IDFP, HDSF, and THL inhibited hABHD6 and hABHD12, whereas RHC-80126, WWL70, and pristimerin selectively targeted hABHD6. MAFP, which is the fluorophosphonate analog of 2-AG, was the most potent compound with IC50 values of ∼20 and ∼90 nM for hABHD6 and hABHD12, respectively. Compared with MAFP, the potency of IDFP (a fluorophosphonate with C12:0 acyl chain) was ∼6-fold lower for hABHD6 but almost 30-fold lower for hABHD12. The latter finding is consistent with the substrate profile of hABHD12 showing that MAG(C20:4) was the preferred substrate. We anticipate that this information could be useful in future design of potent and ABHD12-selective inhibitors. The potency of IDFP toward hABHD6 (IC50 ∼100 nM) is rather modest as compared with the ∼125-fold higher potency (IC50, 0.8 nM) of this compound toward MAGL (39). We can confirm the inferior potency of IDFP toward hMAGL also with the present methodology (our unpublished observations). The potency data are consistent with the hMAGL substrate profile showing that MAGs(C8:0-C12:0) were clearly the best substrates. The broad-spectrum lipase inhibitor THL also relatively potently inhibited hABHD6 and hABHD12 (IC50 ∼50 nM and ∼190 nM, respectively), whereas only hABHD6 was sensitive to RHC-80267. Previously, SDS-PAGE gels from competitive ABPP of mouse brain membrane proteome indicated that mouse ABHD12 was inhibited by THL but not by RHC-80267 (40). Our findings concerning hABHD12 are in line with the mouse data. However, the ABPP data also indicated that mouse ABHD6 was resistant to THL and RHC-80267 (40), whereas our data clearly show that hABHD6 was inhibited by both compounds. This discrepant outcome likely reflects methodological rather that species differences because we have observed that rat ABHD6 (which is 97% homologous to mouse ABHD6) (supplementary Fig. IV) is readily inhibited by THL and with a potency closely matching that for hABHD6 (unpublished observations). WWL70 was previously described as a selective and potent (IC50, 70 nM) ABHD6 inhibitor (41). In our experiments, WWL70 inhibited hABHD6 with almost identical potency (IC50, 85 nM) and showed no activity toward hABHD12. Finally, the triterpene pristimerin, which was previously shown to reversibly inhibit MAGL (IC50, 93 nM) (42), also inhibited hABHD6 (IC50, 1.4 µM), whereas hABHD12 was resistant to this compound. These findings, together with the recent discovery of similar inhibitory action of other triterpenoids (43), should encourage further studies to explore more thoroughly this family of naturally occurring compounds as reversible inhibitors of enzymatic 2-AG degradation.
In conclusion, we have delineated the substrate preferences, active cores, and inhibitor profiles of the human endocannabinoid hydrolases ABHD6 and ABHD12 in comparison with MAGL. We showed that all three hydrolases are genuine MAG lipases and that hMAGL and hABHD6 exhibited a broad and rather similar substrate profile showing highest activity toward medium-to-long-chain saturated MAGs. Thus, depending on substrate availability, the two lipases can handle various MAGs, as might be expected for enzymes with the active site facing the cell interior. In contrast, the 1 (3)- and 2-isomers of the endocannabinoid C20:4 were the preferred substrates for hABHD12 whose active site is predicted to face cellular lumen/extracellular space, collectively suggesting that hABHD12 is well suited to guard the extracellular signaling pool of 2-AG. Inhibitor profiling provided initial structure-activity data that together with the substrate profiling data should facilitate future efforts toward the design of potent and selective inhibitors, especially those targeting hABHD12.
Supplementary Material
Acknowledgments
The authors thank Susanna Saario, Ph.D., for participation in the early stages of this study and for help in setting up the ABPP assays; Ms. Satu Marttila and Ms. Taija Hukkanen for highly competent technical assistance in the laboratory; and Tapio Nevalainen, Ph.D., and Jayendra Z. Patel, M.Sc., for synthesizing N-decanoyl 7-amino-4-methyl coumarin for the fatty acid amide hydrolase assays.
Footnotes
Abbreviations:
- 2-AG
- 2-arachidonoyl glycerol (C20:4)
- AA
- arachidonic acid
- ABHD
- α/β-hydrolase domain containing
- ABPP
- activity-based protein profiling
- DAG
- 1,2-dioleoyl-rac-glycerol (diacylglycerol)
- HDSF
- hexadecane-1-sulfonyl fluoride
- IDFP
- isopropyl dodecylfluorophosphonate
- LPA
- 1-oleoyl-sn-glycero-3-phosphate (lysophosphatidic acid)
- MAFP
- methylarachidonoylfluorophosphonate
- MAG
- monoacylglycerol
- MAGL
- monoacylglycerol lipase (monoglyceride lipase)
- SHD
- Ser-His-Asp
- TBS-T
- Tris-buffered saline containing 0.1% Tween
- THL
- tetrahydrolipstatin (orlistat)
- WT
- wild type
This study was supported by Academy of Finland grant 139620 (J.T.L.).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of five figures and one table.
REFERENCES
- 1.Simon G. M., Cravatt B. F. 2010. Activity-based proteomics of enzyme superfamilies: serine hydrolases as a case study. J. Biol. Chem. 285: 11051–11055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Long J. Z., Cravatt B. F. 2011. The metabolic serine hydrolases and their functions in mammalian physiology and disease. Chem. Rev. 111: 6022–6063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bisogno T., Howell F., Williams G., Minassi A., Cascio M. G., Ligresti A., Matias I., Schiano-Moriello A., Paul P., Williams E. J., et al. 2003. Cloning of the first sn1-DAG lipases points to the spatial and temporal regulation of endocannabinoid signaling in the brain. J. Cell Biol. 163: 463–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Piomelli D. 2003. The molecular logic of endocannabinoid signalling. Nat. Rev. Neurosci. 4: 873–884 [DOI] [PubMed] [Google Scholar]
- 5.Di Marzo V., Bisogno T., De Petrocellis L. 2007. Endocannabinoids and related compounds: walking back and forth between plant natural products and animal physiology. Chem. Biol. 14: 741–756 [DOI] [PubMed] [Google Scholar]
- 6.Kano M., Ohno-Shosaku T., Hashimotodani Y., Uchigashima M., Watanabe M. 2009. Endocannabinoid-mediated control of synaptic transmission. Physiol. Rev. 89: 309–380 [DOI] [PubMed] [Google Scholar]
- 7.Blankman J. L., Simon G. M., Cravatt B. F. 2007. A comprehensive profile of brain enzymes that hydrolyze the endocannabinoid 2-arachidonoylglycerol. Chem. Biol. 14: 1347–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Savinainen J. R., Saario S. M., Laitinen J. T. 2012. The serine hydrolases MAGL, ABHD6 and ABHD12 as guardians of 2-arachidonoylglycerol signalling through cannabinoid receptors. Acta Physiol. (Oxf.). 204: 267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karlsson M., Contreras J. A., Hellman U., Tornqvist H., Holm C. 1997. cDNA cloning, tissue distribution, and identification of the catalytic triad of monoglyceride lipase. Evolutionary relationship to esterases, lysophospholipases, and haloperoxidases. J. Biol. Chem. 272: 27218–27223 [DOI] [PubMed] [Google Scholar]
- 10.Zechner R., Zimmermann R., Eichmann T. O., Kohlwein S. D., Haemmerle G., Lass A., Madeo F. 2012. FAT SIGNALS: lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 15: 279–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dinh T. P., Freund T. F., Piomelli D. 2002. A role for monoglyceride lipase in 2-arachidonoylglycerol inactivation. Chem. Phys. Lipids. 121: 149–158 [DOI] [PubMed] [Google Scholar]
- 12.Nomura D. K., Long J. Z., Niessen S., Hoover H. S., Ng S. W., Cravatt B. F. 2010. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell. 140: 49–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nomura D. K., Morrison B. E., Blankman J. L., Long J. Z., Kinsey S. G., Marcondes M. C., Ward A. M., Hahn Y. K., Lichtman A. H., Conti B., et al. 2011. Endocannabinoid hydrolysis generates brain prostaglandins that promote neuroinflammation. Science. 334: 809–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goparaju S. K., Ueda N., Taniguchi K., Yamamoto S. 1999. Enzymes of porcine brain hydrolyzing 2-arachidonoylglycerol, an endogenous ligand of cannabinoid receptors. Biochem. Pharmacol. 57: 417–423 [DOI] [PubMed] [Google Scholar]
- 15.Saario S. M., Savinainen J. R., Laitinen J. T., Järvinen T., Niemi R. 2004. Monoglyceride lipase-like enzymatic activity is responsible for hydrolysis of 2-arachidonoylglycerol in rat cerebellar membranes. Biochem. Pharmacol. 67: 1381–1387 [DOI] [PubMed] [Google Scholar]
- 16.Savinainen J. R., Yoshino M., Minkkilä A., Nevalainen T., Laitinen J. T. 2010. Characterization of binding properties of monoglyceride lipase inhibitors by a versatile fluorescence-based technique. Anal. Biochem. 399: 132–134 [DOI] [PubMed] [Google Scholar]
- 17.Li F., Fei X., Xu J., Ji C. 2009. An unannotated alpha/beta hydrolase superfamily member, ABHD6 differentially expressed among cancer cell lines. Mol. Biol. Rep. 36: 691–696 [DOI] [PubMed] [Google Scholar]
- 18.Max D., Hesse M., Volkmer I., Staege M. S. 2009. High expression of the evolutionarily conserved alpha/beta hydrolase domain containing 6 (ABHD6) in Ewing tumors. Cancer Sci. 100: 2383–2389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marrs W. R., Blankman J. L., Horne E. A., Thomazeau A., Lin Y. H., Coy J., Bodor A. L., Muccioli G. G., Hu S. S., Woodruff G., et al. 2010. The serine hydrolase ABHD6 controls the accumulation and efficacy of 2-AG at cannabinoid receptors. Nat. Neurosci. 13: 951–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marrs W. R., Horne E. A., Ortega-Gutierrez S., Cisneros J. A., Xu C., Lin Y. H., Muccioli G. G., Lopez-Rodriguez M. L., Stella N. 2011. Dual inhibition of alpha/beta-hydrolase domain 6 and fatty acid amide hydrolase increases endocannabinoid levels in neurons. J. Biol. Chem. 286: 28723–28728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fiskerstrand T., H'mida-Ben Brahim D., Johansson S., M'zahem A., Haukanes B. I., Drouot N., Zimmermann J., Cole A. J., Vedeler C., Bredrup C., et al. 2010. Mutations in ABHD12 cause the neurodegenerative disease PHARC: an inborn error of endocannabinoid metabolism. Am. J. Hum. Genet. 87: 410–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Savinainen J. R., Järvinen T., Laine K., Laitinen J. T. 2001. Despite substantial degradation, 2-arachidonoylglycerol is a potent full efficacy agonist mediating CB1 receptor-dependent G-protein activation in rat cerebellar membranes. Br. J. Pharmacol. 134: 664–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakurada T., Noma A. 1981. Subcellular localization and some properties of monoacylglycerol lipase in rat adipocytes. J. Biochem. 90: 1413–1419 [DOI] [PubMed] [Google Scholar]
- 24.Somma-Delpéro C., Valette A., Lepetit-Thévenin J., Nobili O., Boyer J., Vérine A. 1995. Purification and properties of a monoacylglycerol lipase in human erythrocytes. Biochem. J. 312: 519–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karlsson M., Tornqvist H., Holm C. 2000. Expression, purification, and characterization of histidine-tagged mouse monoglyceride lipase from baculovirus-infected insect cells. Protein Expr. Purif. 18: 286–292 [DOI] [PubMed] [Google Scholar]
- 26.Imamura S., Kitaura S. 2000. Purification and characterization of a monoacylglycerol lipase from the moderately thermophilic Bacillus sp. H-257. J. Biochem. 127: 419–425 [DOI] [PubMed] [Google Scholar]
- 27.Taschler U., Radner F. P., Heier C., Schreiber R., Schweiger M., Schoiswohl G., Preiss-Landl K., Jaeger D., Reiter B., Koefeler H. C., et al. 2011. Monoglyceride lipase deficiency in mice impairs lipolysis and attenuates diet-induced insulin resistance. J. Biol. Chem. 286: 17467–17477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aaltonen N., Lehtonen M., Varonen K., Arrufat Goterris G., Laitinen J. T. 2012. Lipid phosphate phosphatase inhibitors locally amplify lysophosphatidic acid LPA1 receptor signalling in rat brain cryosections without affecting global LPA degradation. BMC Pharmacol. 12: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bertrand T., Augé F., Houtmann J., Rak A., Vallée F., Mikol V., Berne P. F., Michot N., Cheuret D., Hoornaert C., et al. 2010. Structural basis for human monoglyceride lipase inhibition. J. Mol. Biol. 396: 663–673 [DOI] [PubMed] [Google Scholar]
- 30.Okazaki T., Sagawa N., Okita J. R., Bleasdale J. E., MacDonald P. C., Johnston J. M. 1981. Diacylglycerol metabolism and arachidonic acid release in human fetal membranes and decidua vera. J. Biol. Chem. 256: 7316–7321 [PubMed] [Google Scholar]
- 31.Prescott S. M., Majerus P. W. 1983. Characterization of 1,2-diacylglycerol hydrolysis in human platelets. Demonstration of an arachidonoyl-monoacylglycerol intermediate. J. Biol. Chem. 258: 764–769 [PubMed] [Google Scholar]
- 32.Rindlisbacher B., Reist M., Zahler P. 1987. Diacylglycerol breakdown in plasma membranes of bovine chromaffin cells is a two-step mechanism mediated by a diacylglycerol lipase and a monoacylglycerol lipase. Biochim. Biophys. Acta. 905: 349–357 [DOI] [PubMed] [Google Scholar]
- 33.Chanda P. K., Gao Y., Mark L., Btesh J., Strassle B. W., Lu P., Piesla M. J., Zhang M. Y., Bingham B., Uveges A., et al. 2010. Monoacylglycerol lipase activity is a critical modulator of the tone and integrity of the endocannabinoid system. Mol. Pharmacol. 78: 996–1003 [DOI] [PubMed] [Google Scholar]
- 34.Schlosburg J. E., Blankman J. L., Long J. Z., Nomura D. K., Pan B., Kinsey S. G., Nguyen P. T., Ramesh D., Booker L., Burston J. J., et al. 2010. Chronic monoacylglycerol lipase blockade causes functional antagonism of the endocannabinoid system. Nat. Neurosci. 13: 1113–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jung K. M., Clapper J. R., Fu J., D'Agostino G., Guijarro A., Thongkham D., Avanesian A., Astarita G., Dipatrizio N. V., Frontini A., et al. 2012. 2-arachidonoylglycerol signaling in forebrain regulates systemic energy metabolism. Cell Metab. 15: 299–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rengachari S., Bezerra G. A., Riegler-Berket L., Gruber C. C., Sturm C., Taschler U., Boeszoermenyi A., Dreveny I., Zimmermann R., Gruber K., et al. 2012. The structure of monoacylglycerol lipase from Bacillus sp. H257 reveals unexpected conservation of the cap architecture between bacterial and human enzymes. Biochim. Biophys. Acta. 1821: 1012–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cabral G. A., Raborn E. S., Griffin L., Dennis J., Marciano-Cabral F. 2008. CB2 receptors in the brain: role in central immune function. Br. J. Pharmacol. 153: 240–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Labar G., Bauvois C., Muccioli G. G., Wouters J., Lambert D. M. 2007. Disulfiram is an inhibitor of human purified monoacylglycerol lipase, the enzyme regulating 2-arachidonoylglycerol signaling. ChemBioChem. 8: 1293–1297 [DOI] [PubMed] [Google Scholar]
- 39.Nomura D. K., Blankman J. L., Simon G. M., Fujioka K., Issa R. S., Ward A. M., Cravatt B. F., Casida J. E. 2008. Activation of the endocannabinoid system by organophosphorus nerve agents. Nat. Chem. Biol. 4: 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoover H. S., Blankman J. L., Niessen S., Cravatt B. F. 2008. Selectivity of inhibitors of endocannabinoid biosynthesis evaluated by activity-based protein profiling. Bioorg. Med. Chem. Lett. 18: 5838–5841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li W., Blankman J. L., Cravatt B. F. 2007. A functional proteomic strategy to discover inhibitors for uncharacterized hydrolases. J. Am. Chem. Soc. 129: 9594–9595 [DOI] [PubMed] [Google Scholar]
- 42.King A. R., Dotsey E. Y., Lodola A., Jung K. M., Ghomian A., Qiu Y., Fu J., Mor M., Piomelli D. 2009. Discovery of potent and reversible monoacylglycerol lipase inhibitors. Chem. Biol. 16: 1045–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chicca A., Marazzi J., Gertsch J. 2012. The antinociceptive triterpene β-amyrin inhibits 2-arachidonoylglycerol (2-AG) hydrolysis without directly targeting CB receptors. Br. J. Pharmacol. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.