Abstract
Myristoyl-CoA (CoA):protein N-myristoyltransferase (NMT) catalyzes protein modification through covalent attachment of a C14 fatty acid (myristic acid) to the N-terminal glycine of proteins, thus promoting protein-protein and protein-membrane interactions. NMT is essential for the viability of numerous human pathogens and is also up-regulated in several tumors. Here we describe a new, nonradioactive, ELISA-based method for measuring NMT activity. After the NMT-catalyzed reaction between a FLAG-tagged peptide and azido-dodecanoyl-CoA (analog of myristoyl-CoA), the resulting azido-dodecanoyl-peptide-FLAG was coupled to phosphine-biotin by Staudinger ligation, captured by plate-bound anti-FLAG antibodies and detected by streptavidin-peroxidase. The assay was validated with negative controls (including inhibitors), corroborated by HPLC analysis, and demonstrated to function with fresh or frozen tissues. Recombinant murine NMT1 and NMT2 were characterized using this new method. This versatile assay is applicable for exploring recombinant NMTs with regard to their activity, substrate specificity, and possible inhibitors as well as for measuring NMT-activity in tissues.
Keywords: azido-fatty acid, N-myristoyl transferase, protein acylation, Staudinger ligation
N-myristoylation refers to an irreversible protein modification involving the covalent attachment of n-tetradecanoic acid (myristic acid) to the N-terminal glycine residue of proteins (1, 2). This process can occur co- or post-translationally (3–6) and helps to promote protein interactions with membranes or with hydrophobic domains of other proteins (7, 8). Myristoyl-CoA:protein N-myristoyltransferase (NMT; EC 2.3.1.97) is the enzyme catalyzing this modification. It exerts its activity after cleavage of the initiator methionine residue or after exposure of an internal glycine (3, 5, 9). Several mammalian and plant species (Bos taurus, Homo sapiens, Mus musculus, Rattus norvegicus, and Arabidopsis thaliana) were demonstrated to express two isoenzymes of NMT (NMT1 and NMT2) localized in the cytosol (10–14). NMT1 and NMT2 have divergent peptide substrate specificities but share a high specificity for myristoyl-CoA (2, 15). An increase or decrease in fatty acid chain length significantly reduces the incorporation rate of the fatty acid into the peptide (16).
N-terminally myristoylated proteins are involved in a variety of cellular processes, including proliferation, differentiation, and apoptosis, and have been shown to be essential for embryogenesis (17, 18). Increased expression and activity of NMT has also been demonstrated in human tumors (e.g., colorectal carcinoma, gallbladder carcinoma, oral squamous cell carcinoma, and brain tumors) (19–23).
More importantly, several human protozoan pathogens, such as Leishmania major, Leishmania donovani, Plasmodium falciparum, and Trypanosoma brucei, that are responsible for debilitating tropical infections possess a single-copy NMT gene and require NMT activity for their viability (24). Similarly, prominent fungal pathogens (Candida albicans, Cryptococcus neoformans, and Histoplasma capsulatum) encode for their own NMT, the activity of which is essential for their survival (25, 26). Recently, several NMT inhibitors with the potency to kill these parasites have been developed (25, 27–29). N-myristoylation is also a prerequisite for the proper function of viral proteins (e.g., negative regulatory factor [Nef] and Gag of HIV-1) that are crucial for virus assembly and propagation (30, 31). Finally, N-myristoylation of bacterial and plant proteins has been shown to be involved in development and host defense (e.g., SnRK1 of Arabidopsis thaliana or Fen of Solanum lycopersicum) (13, 32, 33).
The vital dependency on protein myristoylation in the aforementioned human pathogens and tumors emphasizes the importance of further insight into myristoylation-related processes and makes NMT a potential target for antimicrobial and antineoplastic chemotherapy (23, 25, 34).
A central tool for accomplishing these goals is a robust, reliable, and user-friendly assay to measure NMT activity. Methods for detecting NMT activity have involved radioactive and nonradioactive assays, such as use of reverse-phase HPLC, and the so-called “continuous assays” in which enzymatic activity is detected while the reaction is progressing. Radioactive methods are based on the quantification of [3H]myristate incorporation into peptides, whereas continuous assays depend on the spectrophotometric or fluorometric detection of products released during myristoylation (e.g., HS-CoA) (1, 4, 35–40). A nonradioactive assay based on ELISA has been described using an antibody against N-myristoyl-glycine (41). Continuous and ELISA-based assays are more suitable for inhibitor screening. In contrast, radioactive and HPLC-based approaches are distinguished by a higher sensitivity and substrate specificity; however, they are expensive and time consuming and require complex technical skills.
Here we describe a new, versatile, nonradioactive ELISA-based method for measuring N-myristoyltransferase activity in vitro. We validate this assay using various negative controls and HPLC analysis. We demonstrate that it is applicable to exploring recombinant NMTs with regard to their activity, substrate specificity, and possible inhibitors. In addition, we show that the assay is suitable for measuring endogenous NMT activity in fresh or frozen tissues.
MATERIALS AND METHODS
Synthesis of azido-dodecanoyl-CoA
Azido-dodecanoyl-CoA (CoA) was synthesized according to Kostiuk et al. (42): 20 mM Tris-HCl (pH 7.4), 1 mM dithiothreitol (DTT), 0.1 M EGTA, 5 mM ATP, 1 mM CoA, 0.15 U/ml of Pseudomonas acyl-CoA synthetase (all Sigma, Schnelldorf, Germany), 0.25% Triton X-100, and 100 mM azido-dodecanoic-acid (dissolved in DMSO) were mixed and incubated for 1 h in the dark at room temperature (RT). The final azido-dodecanoyl-CoA concentration was measured to be 1 mM. The product was stored at −20°C until use.
RNA isolation, cloning, expression, and purification of recombinant Nmt1 and Nmt2
Total RNA was isolated from the brain of a male C57BL/6 mouse (Charles River Wiga, Germany) as described previously (43). After reverse transcription, full-length Nmt1 and Nmt2 were amplified using the following primers: 5′-GTAGGAATTCATGGCGGATGAGAGTGAGA-3′, 5′-CCCAAGCTTCTGTAACACCAACCCAACC-3′ for Nmt1, and 5′-ACGGAATTCATGGCGGAGGACAGCGA-3′, 5′-CGGGATCCCTGTAGAACAGTCCAACC-3′ for Nmt2. The products for Nmt1 and Nmt2 were cloned into pQC-His bacterial expression vector (kindly provided by Hanswalter Zentgraf, DKFZ Heidelberg, Germany), resulting in the generation of a C-terminal histidine-tagged fusion protein. Escherichia coli SG13009 (Qiagen, Hilden, Germany) was used for transformation and synthesis of the recombinant proteins according to manufacturer's instructions. Recombinant Nmt1 and Nmt2 were purified under native conditions using Ni-NTA agarose columns according to the specifications of the producer (Qiagen). To detect the fractions containing the enzyme, protein samples were separated by a discontinuous SDS-PAGE. The gels were either stained with Coomassie Brilliant Blue (Merck, Darmstadt, Germany) or blotted onto nitrocellulose membranes (GE Healthcare, Freiburg, Germany). The membranes were incubated with anti-(His)6-tag antibody (kindly provided by Hanswalter Zentgraf) followed by incubation with horseradish peroxidase conjugated anti-mouse secondary antibody (1: 10,000 dilution; Santa Cruz, Heidelberg, Germany). For development of the blots, an enhanced chemiluminescence system (GE Healthcare) was used according to manufacturer's instructions.
Cell culture and siRNA transfection
The human ovarian adenocarcinoma cell line SK-OV-3 (ATCC number: HTB-77) was cultured in DMEM supplemented with 10% FBS (Life Technologies, Darmstadt, Germany). SK-OV-3 cells were transfected with siRNAs (Ambion, Life Technologies) to knockdown NMT1 (5′-ATGAGGAGGACAACAGCTAC-3′) and/or NMT2 (5′-AAAAGGTTGGACTAGTACTAC-3′) at a final concentration of 10 nM (11). When transfecting NMT1 and NMT2 siRNAs simultaneously, each of them was applied at 10 nM. As nonsilencing siRNA, Negative control 1 (Ambion, Life Technologies) was used at a concentration of 20 nM. The transfection was done by using LipofectAMINE 2000 (Invitrogen, Life Technologies) according to manufacturer's recommendations. Cells were harvested 48 h after transfection and used for RNA isolation and for the NMT-azido-ELISA. The transfection efficiency was measured by flow cytometry (FACSCalibur, Becton Dickinson, Heidelberg, Germany) using alexa-fluor (488 nm) conjugated nonsilencing siRNA (Qiagen) (44) and was >95%.
qPCR analysis
Quantitative PCR was performed by LightCycler (Roche, Mannheim, Germany) as described previously (44). The following primers were used (BioSpring, Frankfurt am Main, Germany): 5′-CAACTACATGGTTTACATGTTC-3′ and 5′-GCCAGTGGACTCCACGAC-3′ for GAPDH, 5′-TTTTATACGCTGCCCTCCAC-3′ and 5′-TCCCCTATGCCAAACTTGAG-3′ for NMT1, and 5′-GGCTCCAGTGATGGATGAAG-3′ and 5′-GGCTTTGAGGCTCTTGTGAG-3′ for NMT2.
Preparation of mouse tissue extracts
C57BL/6 male mice were euthanized by cervical dislocation. Tissues were homogenized in digitonin lysis buffer (20 mM HEPES-NaOH buffer [pH 7.4] containing 25 mM KCl, 250 mM sucrose, 2 mM MgCl2, with freshly added 1% digitonin [Sigma], complete protease inhibitors [Roche], and 0.5 mM DTT) using Ultra Turrax T25 (Ika, Koenigswinter, Germany). The homogenate was centrifuged at 2,000 g for 20 min at 4°C, and the supernatant was further centrifuged at 39,000 g for 1 h at 4°C. The cytosolic fraction (supernatant) obtained was used immediately. Protein concentrations were determined by Bradford assay (Invitrogen, Karlsruhe, Germany) using BSA as protein standards. Animal procedures were approved by local authorities.
NMT enzymatic reaction
Synthetic peptides representing the N-terminal myristoylation sequence of Lck (GCVCSSNPE-DYKDDDDK), HIV-1 Nef (GGKWSKRSM-DYKDDDDK), and Trypanosoma brucei ADP-ribosylation factor (Arf) (GQWLASAFK-DYKDDDDK) linked to FLAG sequence (DYKDDDDK) were provided by the Peptide Synthesis Unit at DKFZ Heidelberg, Germany. As negative controls, mutated Lck (ACVCSSNPE-DYKDDDDK) or azido-tetradecanoyl-CoA (Invitrogen) instead of azido-dodecanoyl-CoA were used. Reactions (50 µl) containing the purified enzyme or 50 µg of cytosolic fraction obtained from organ homogenates, azido-dodecanoyl-CoA, and peptide-FLAG at different concentrations in 50 mM HEPES (pH 7.4) were allowed to proceed at 37°C for 1 h. In case of kinetic studies, the reaction was allowed to proceed for 2 min and terminated by incubation at 95°C for 5 min.
NMT-azido-ELISA
Ninety-six-well plates (Nunc, Langenselbold, Germany) were coated overnight with 100 µl of anti-FLAG antibody (10 µg/ml; F1804, Sigma) in coating buffer (0.05 M carbonate-bicarbonate [pH 9.6]) at 4°C and blocked with 200 µl of 5% BSA (w/v) in PBS (pH 7.4) for 2 h at RT. After three washings (washing buffer: 25 mM Tris, 150 mM NaCl, 0.1% BSA [w/v] and 0.05% Tween [v/v] [pH 7.2]), 50 µl of the NMT reaction products (see above) were placed into the wells and diluted with 50 µl of washing buffer. Plates were incubated for 2 h at RT. The wells were filled with 100 µl of phosphine-biotin (50 µM in DMSO; Fisher Scientific, Schwerte, Germany), and the Staudinger ligation was allowed to proceed at 4°C overnight. Wells were washed three times, followed by the addition of 100 µl streptavidin-HRP (1:10,000 in blocking buffer; BD Biosciences, Heidelberg, Germany) and incubation for 1 h at RT. Bound streptavidin-HRP was detected by adding 100 µl of the substrate tetramethylbenzidine (TMB) (BD Bioscience) for 20–30 min in the dark at RT. The reaction was terminated by the addition of 50 µl of 2 M H2SO4, and the absorbance was measured at 450 nm using ELISA reader (Tecan, Mainz, Germany). For the inhibitor study, Tris(dibenzylideneacetone)dipalladium (Tris DBA) (Sigma) and 5-(3-carboxy-4-nitrophenyl)disulfanyl-2-nitrobenzoic acid (DTNB) (Sigma) were used at concentrations indicated in the Results section. Absorbance values were defined as the absorbance of each sample well minus the mean absorbance of background controls in wells containing anti-FLAG antibody and phosphine biotin. A standard curve was created using biotin-FLAG (Peptide Synthesis Unit, DKFZ) as standard. Calculations in kinetic and inhibitor studies were performed using GraphPad Prism 5 (GraphPad Software, La Jolla, CA).
HPLC
Analytical HPLC-ESI-MS was conducted on a Hewlett-Packard 1090 liquid chromatograph (Agilent Technologies, Waldbronn, Germany) fitted with a reverse-phase C18 Gemini column (250 mm × 4 mm i.d., 5 µm; (Phenomenex Ltd, Aschaffenberg, Germany). The mobile phase consisted of 15% acetonitrile in double distilled water (solvent A) and 10% double distilled water in acetonitrile (solvent B) with the following gradient profile: 100% A for 5 min; reduced to 50% A over 14 min; 0% A over 1 min, continuing at 0% A until completion of the run. The flow rate of the mobile phase was constantly 1.0 ml/min. The injection volume was 1.0 µl and myristoylated compounds in the eluent were detected at 214 nm with a diode-array UV detector (Hewlett-Packard 1040M).
Mass spectra in the negative-ion mode were generated under the following conditions: fragmenter voltage = 100 V, capillary voltage = 2500 V, nebulizer pressure = 30 psi, drying gas temperature = 350°C, m/z = 100–1,500. Instrument control and data handling was by means of a Hewlett-Packard Chemstation operating in the Microsoft Windows software environment.
RESULTS
The principle of the NMT-azido-ELISA assay for measurement of NMT activity
Our aim was to develop an assay that would allow measurement of NMT activity in tissues and cell lysates as well as in experiments dealing with recombinant NMTs, enzyme kinetics, substrate specificity, and inhibitors. The assay should be easy to establish and suitable for high-throughput applications. To address these issues, an ELISA-like approach (Fig. 1) was established. As acyl donor, azido-dodecanoyl-CoA, which represents a broadly applied bio-orthogonal analog of myristoyl-CoA, was used (45). Three paradigmatic nonapeptides were selected as Nmt substrates. They represent the N-termini of proteins previously demonstrated to be myristoylated and of therapeutic interest. The peptides were derived from three different species: Lck from mouse, Nef from HIV-1, and Arf from Trypanosoma brucei (34, 46, 47). The peptides were each C-terminally linked to a FLAG-tag and are referred to as Lck-FLAG, Nef-FLAG, and Arf-FLAG.
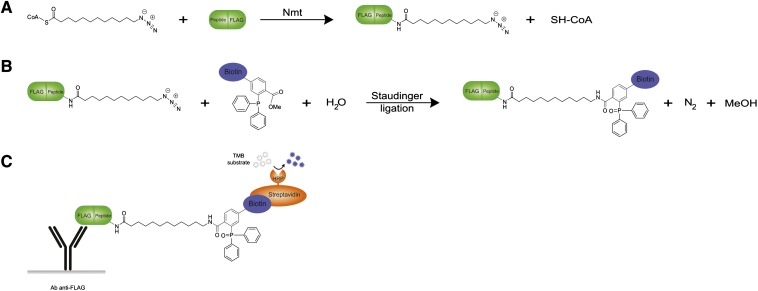
Fig. 1.
Scheme of the NMT-azido-ELISA. a: N-myristoyl transferase reaction between a FLAG-tagged peptide and azido-dodecanoyl-CoA, a bio-orthogonal azido-analog of myristoyl-CoA. b: Incorporation of biotin via Staudinger ligation between the azido group and phosphine. c: Detection of the azido-dodecanoylated-peptide-FLAG via ELISA using plate-bound anti-FLAG antibodies and streptavidin-linked HRP. TMB, tetramethylbenzidine.
After the Nmt reaction (Fig. 1a), the azido-dodecanoylated nonapeptide-FLAG was coupled to phosphine-biotin via Staudinger ligation (Fig. 1b). The products of the reaction, coupled to biotin, could be detected and quantified by an ELISA, in which it was captured on an immunoplate precoated with anti-FLAG antibody and detected by the addition of streptavidin-peroxidase (Fig. 1c).
Cloning and expression of murine Nmt1 and Nmt2
Murine Nmt1 and Nmt2 were cloned from the brain of a C57BL/6 male mouse and inserted into the pQC-His expression vector. Insert sequencing revealed the canonical form of Nmt1. In the case of Nmt2, repetitive sequencing of different clones demonstrated a previously unknown splice variant of Nmt2 lacking exons 3 and 4 (Nmt2_OTTMUST00000025234). Both recombinant (His)6-tagged enzymes were expressed in E. coli and purified under native conditions. The concentration of Nmt1 or Nmt2 used to perform each experiment, except where otherwise specified, was 30 nM.
Assay validation
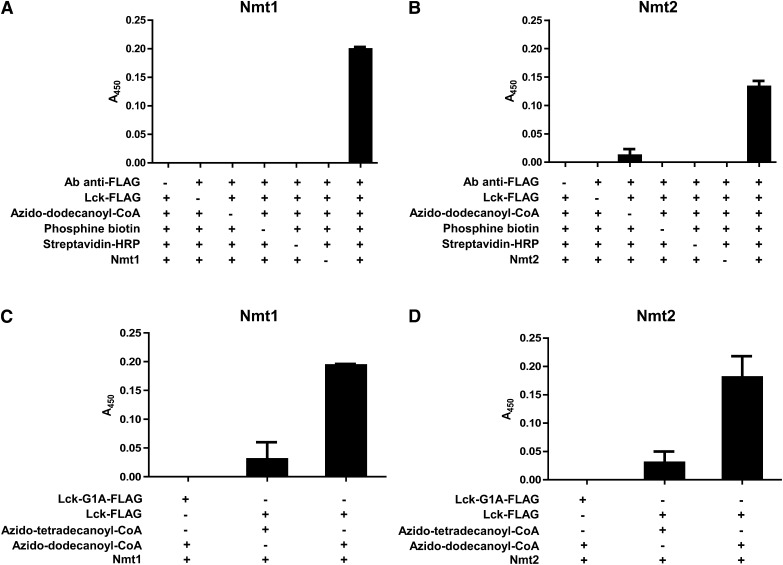
Numerous controls were used to validate the assay. Signals from both enzymes were observed exclusively in the presence of all assay components (Fig. 2a, b). In contrast, omitting each of the assay components interfered with the development of a signal (Fig. 2a, b).
Fig. 2.
Validation of the NMT-azido-ELISA using recombinant Nmt1 and Nmt2 and a set of negative controls. The assay was performed for Nmt1 (a) and Nmt2 (b) in the presence of all reaction and detection components (positive control, last columns) or omitting successively single components as indicated. With the exception of Nmt2 and azido-dodecanoyl-CoA, where a low signal was detected, the absence of each component interfered with signal development. c and d: Standard reactions (last columns) were compared with reactions with false substrates. To this end, Lck-FLAG was replaced by a peptide with N-terminal glycine to alanine substitution (Lck-G1A-FLAG), which was not recognized by the enzyme. Replacing azido-dodecanoyl-CoA for a two-carbon longer azido-tetradecanoyl-CoA diminished the signals. Values shown are the averages of two independent experiments, which were performed in duplicates. Error bars represent SD.
Myristoylation can occur only at the N-terminal glycine (1). To demonstrate that the reaction assayed here was specific for the N-terminal glycine, Lck-FLAG was replaced by a mutated peptide with N-terminal glycine to alanine substitution (Lck-G1A-FLAG). As expected, this mutated peptide elicited no signals for either Nmt1 or Nmt2 (Fig. 2c, d).
It has been reported that NMT can use, albeit with a lower affinity, palmitoyl-CoA as a substrate (4). To test this in our assay, azido-dodecanoyl-CoA was substituted by azido-tetradecanoyl-CoA, an azido-analog of palmitoyl-CoA, which led to low but detectible signals for both enzymes (Fig. 2c, d).
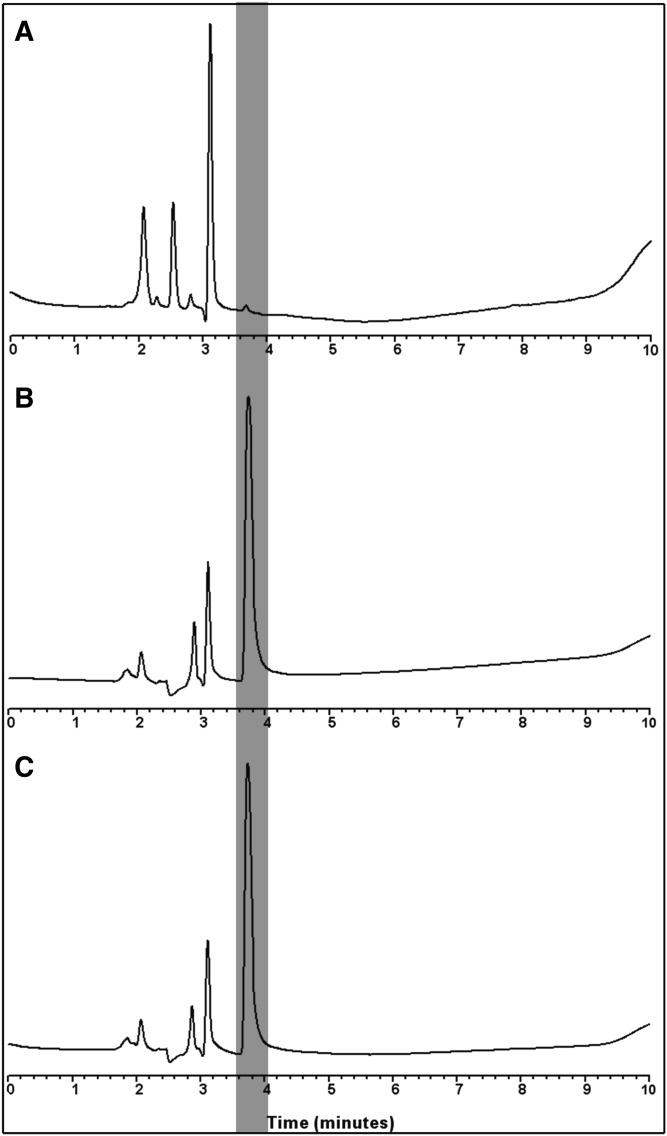
To confirm that the Nmt reactions yielded azido-dodecanoyl-Lck-FLAG, the reaction products for both enzymes were separated by HPLC. As compared with a negative control that lacked enzyme (Fig. 3a), the addition of Nmt1 or Nmt2 to the substrates led to the emergence of a new peak (Fig. 3b, c, gray line).
Fig. 3.
Formation of the azido-dodecanoyl-Lck-FLAG was confirmed via reverse-phase HPLC. Chromatograms of reactants without addition of the enzyme (a) and after the addition of Nmt1 (b) or Nmt2 (c). As compared with the analysis of reactants alone (a), the addition of the enzyme (b and c) led to the emergence of a new peak (gray line), which corresponds to azido-dodecanoyl-Lck-FLAG.
The sensitivity of the NMT-azido-ELISA, defined as the slope of the calibration curve, (48) was 0.112 liters/µg. The limit of detection (LOD), calculated as xbl + 3sbl, where xbl is the mean of the blank measures and sbl is the standard deviation of the blank measures (48), had a value of 10 ng/l. Linear and dynamic ranges were two and three orders of magnitude, respectively. Signal to noise ratio, calculated as (xsignal − xbl)/sbl, where xsignal is the mean signal and xbl and sbl are as defined above (49), had a value of 385 for recombinant Nmt1 (applied at a concentration of 0.9 µg/l), 146 for brain (using 50 µg of lysate), and 15 for liver (using 50 µg of lysate).
NMT-azido-ELISA can be used for comparative substrate studies
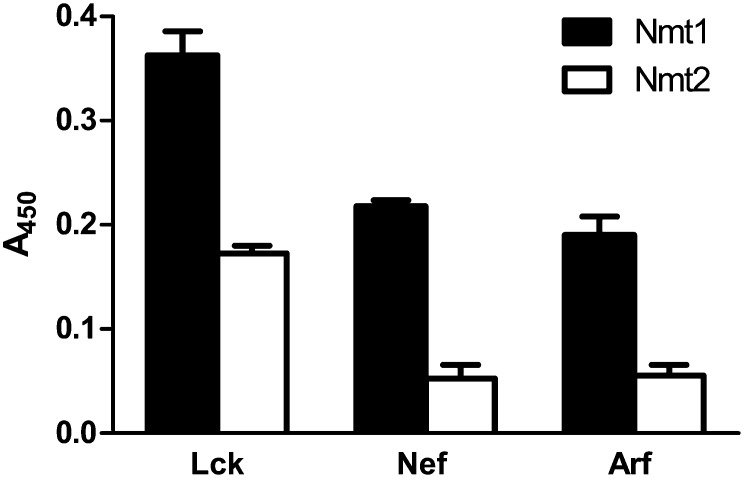
We sought to develop an assay that would allow testing a variety of substrates without changing the setting of the assay. To validate this, Lck-FLAG was tested in parallel with other paradigmatic peptides (Nef-FLAG and Arf-FLAG). Nmt1 and Nmt2 generated different signals according to the specific substrates-FLAG, demonstrating that each isoform has different peptide specificities (Fig. 4). Finally, as described above for azido-dodecanoyl-Lck-FLAG, the formation of azido-dodecanoylated Nef-FLAG and azido-dodecanoylated Arf-FLAG was confirmed by HPLC analysis (data not shown).
Fig. 4.
Comparison between different peptide-FLAG substrates in the presence of Nmt1 or Nmt2. The reactions were carried out using 100 μM of the following peptide-FLAGs: Lck-FLAG (from mouse), Nef-FLAG (from HIV-1), and Arf-FLAG (from Trypanosoma brucei). Values shown are the average of duplicate measurements of two independent experiments. Error bars represent SD.
Kinetic and inhibitor studies using NMT-azido-ELISA
Before implementing the NMT-azido-ELISA in the investigation of enzyme kinetics, we generated a standard curve that allowed correlating the absorbance values to the numbers of molecules captured on the plate. To this end, the assay was performed with increasing concentrations of biotinylated FLAG (data not shown).
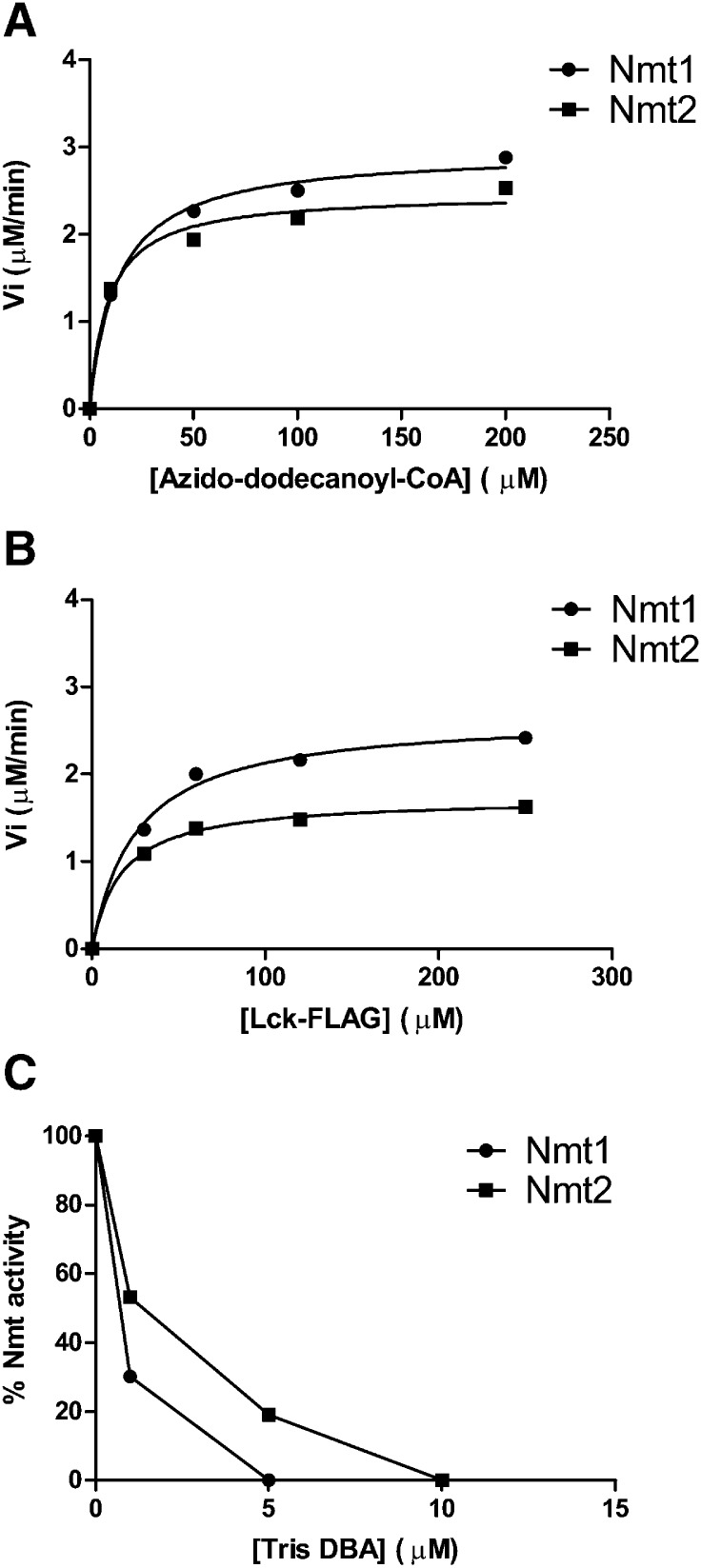
Using the NMT-azido-ELISA, Km values of the azido-dodecanoyl-CoA and of one of the paradigmatic nonapeptides (Lck-FLAG) were determined for Nmt1 and Nmt2. After verifying that the rate of myristoylation remained linear for the first 2 min, the initial velocity (Vi) was calculated over the first 2 min of the reaction. The Km value of azido-dodecanoyl-CoA was determined using increasing concentrations of azido-dodecanoyl-CoA, and a saturating concentration (200 µM) of Lck-FLAG. Similarly, the Km value of Lck-FLAG was calculated using increasing concentrations of Lck-FLAG, and a saturating concentration (200 µM) of azido-dodecanoyl-CoA. Azido-dodecanoyl-CoA showed a Km of 14 ± 2 (SD) µM for Nmt1 and 9 ± 3 µM for Nmt2 (Fig. 5a). Lck-FLAG had a Km of 26 ± 5 µM and 17 ± 2 µM for Nmt1 and Nmt2, respectively (Fig. 5b).
Fig. 5.
Kinetic and inhibitor studies using NMT-azido-ELISA. a and b: Kinetic parameters of azido-dodecanoyl-CoA and Lck-FLAG determined with the NMT-azido-ELISA using recombinant Nmt1 and Nmt2. Shown is the initial velocity (Vi) as a function of the concentrations of azido-dodecanoyl-CoA (a) and Lck-FLAG (b). The Km of azido-dodecanoyl-CoA was 14 ± 2 (SD) µM for Nmt1 and 9 ± 3 µM for Nmt2, whereas Lck-FLAG had a Km of 26 ± 5 µM for Nmt1 and 17 ± 2 µM for Nmt2. c: Inhibition of murine Nmt1 and Nmt2 by Tris DBA. Enzymes were incubated with increasing concentrations of Tris DBA using azido-dodecanoyl-CoA and Lck-FLAG as substrates. The calculated values of IC50 were 0.5 ± 0.1 µM for Nmt1 and 1.3 ± 0.1 µM for Nmt2. Values shown are the average of duplicate measurements.
For the investigation of enzyme inhibition, a previously described NMT1 inhibitor Tris DBA was used (50). Nmt1 and Nmt2 were inhibited by increasing Tris DBA concentrations (Fig. 5c). The calculated half-maximal inhibition IC50 was 0.5 ± 0.1 µM for Nmt1 and 1.3 ± 0.1 µM for Nmt2.
Repression of cellular NMT1 and NMT2 using siRNA
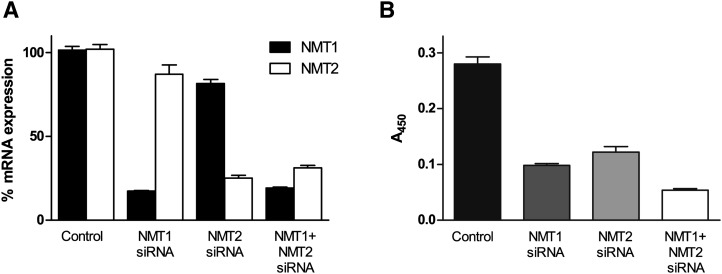
All aforementioned experiments were performed with recombinant enzymes. To assess that myristoylation activity can also be measured in cell lysates and that the detected signals reflect NMT activity, we selectively repressed NMT1 and/or NMT2 in the human ovarian adenocarcinoma cell line SK-OV-3 using previously described NMT1 and NMT2 siRNAs (11). The repression of NMT1 and NMT2 was quantified by qPCR. As compared with control siRNA, the treatment with NMT1 or NMT2 siRNA decreased their mRNA levels by 83% and 76%, respectively. The simultaneous treatment with both NMT siRNAs reduced the NMT1 and NMT2 mRNA levels by 81% and 70%, respectively (Fig. 6a).
Fig. 6.
Repression of NMT1 and NMT2 using siRNA. SK-OV-3 cells were transfected with nonsilencing control siRNA (20 nM), NMT1 siRNA (10 nM), NMT2 siRNA (10 nM), and both NMT1 and NMT2 siRNAs (20 nM). After 48 h, the mRNA expression of both enzymes was analyzed (a), and NMT activity was measured using NMT-azido-ELISA (b). Values are the averages of duplicate measurements.
The diminished expression of NMT1 and/or NMT2 was paralleled by a diminished myristoylation activity, which decreased by 80% when the expression of NMT1 and NMT2 was repressed (Fig. 6b). This implicates that the signal generated by the NMT-azido-ELISA specifically reflects the NMT activity in cell lysates.
NMT-azido-ELISA is suitable for the measurement of myristoylation activity in tissues
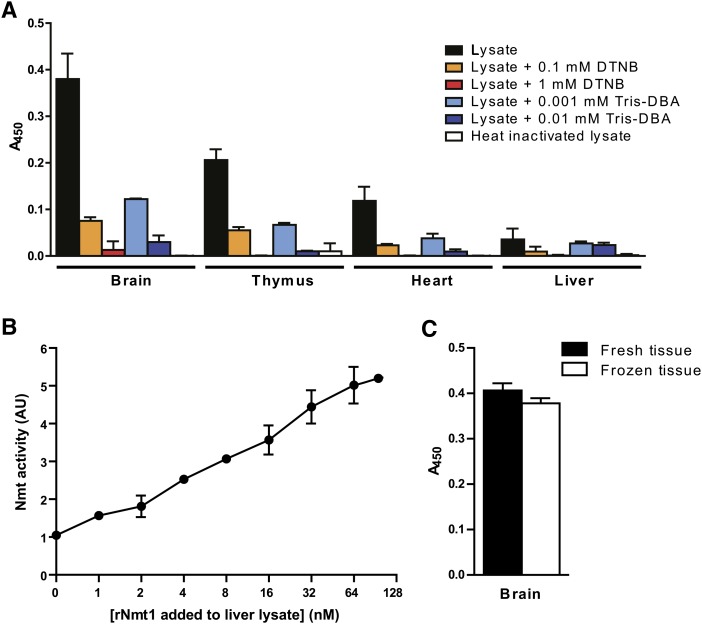
To document that NMT-azido-ELISA is suitable for the measurement of NMT activity in tissues, different murine organs were assayed for Nmt activity. Organs from adult C57BL/6 male mice were lysed, and cytosolic proteins were enriched. To demonstrate that the signals detected were a result of enzymatic activity, an aliquot of the preparation was thermally inactivated and treated under the same conditions as detailed above. Moreover, in parallel, two Nmt inhibitors were used: Tris DBA and DTNB, each of them at two different concentrations (Fig. 7a) (50, 51).
Fig. 7.
Detection of NMT activity in murine organs. a: Tissue enzyme activity was determined using NMT-azido-ELISA and Lck-FLAG as acyl acceptor. To demonstrate that the detected signal was a result of enzyme activity, aliquots of organ preparation were thermally inactivated at 95°C for 10 min or treated with two inhibitors (Tris DBA and DTNB) at different concentrations as indicated. All samples were processed in parallel. b: The liver lysate was spiked with increased amounts of recombinant Nmt1, and the Nmt activity was measured. Enzyme activity was normalized for the activity in the liver lysate without enzyme addition. The x axis has a log2 scale. AU, arbitrary unit. c: Measurement of enzymatic activity in brain lysates prepared freshly in comparison to frozen brains stored at −80°C. Values shown are the average of duplicate measurements of two independent experiments. Error bars are SD.
Murine brain was found to contain the highest activity of Nmt, followed by thymus, heart, and liver (Fig. 7a). Heat inactivation of the organ extracts abolished the signals. In samples treated with Nmt inhibitors Tris DBA or DTNB, the myristoylation activity decreased in a dose-dependent manner (Fig. 7a). The presence of the azido-dodecanoyl-Lck-FLAG in organ lysates was confirmed by HPLC (supplemental Fig. I and data not shown). In addition, the liver preparation that showed the lowest Nmt activity (Fig. 7a) was spiked with increasing concentrations of recombinant Nmt1, which led to a linear increase of the signal (Fig. 7b).
To test whether this assay could be applied to frozen tissues (e.g., samples from a tissue bank), activity measurement was performed in murine brain, which had been stored at −80°C for 1 week. In parallel, a freshly isolated brain was analyzed. The results demonstrated that Nmt activity could be assayed in frozen tissues without a pronounced decline in activity (Fig. 7c).
DISCUSSION
Protein myristoylation has been attracting increasing attention because it has been demonstrated to be of importance in embryogenesis, neoplasia, and plant biology and in the propagation of protozoal, fungal, and viral infections. It is anticipated that future research will focus on the development of small molecule inhibitors of NMT with the aim of suppressing infectious and malignant diseases. This goal requires a robust and easy to use assay for the measurement of NMT activity. Most of the methods in use are radioactive (1, 4, 36, 37, 40). Nonradioactive HPLC-based methods represent a sensitive alternative to radioactive assays but are not suitable for high-throughput analysis (41). Several continuous spectrophotometric or fluorescent procedures have been developed (35, 38, 39). Although they are relatively easy to perform and applicable for high throughput analysis, continuous assays depend strongly on the milieu in which the reaction occurs. The purity of enzymes, or the presence of particular contaminants in the samples (e.g., derived from a tissue preparation), may influence the assay outcome (35). Similarly, certain groups of substances (e.g., strong nucleophilic inhibitors) in these assays are excluded from testing because they interfere with the signal detection used (38). Takamune et al. (41) described a potential method in which antibodies directed against myristoyl-glycine were used in an ELISA. This approach has not been used according to published reports since its description.
The assay described here represents a new method for myristoylation measurement that uses affordable, commercially available substances in combination with an ELISA-like system. It has the advantage of using the myristoylation reaction that occurs between a FLAG-tagged peptide and azido-dodecanoyl-CoA, a bio-orthogonal analog of myristoyl-CoA (45). After subsequent coupling to phosphine-biotin via Staudinger ligation, the product is detected by ELISA using plate-bound anti-FLAG antibodies and avidin-conjugated HRP (Fig. 1). The combination of Staudinger ligation and ELISA has also been described to be useful for the detection of glycosyltransferase activity, although the assay arrangement was different (52). Furthermore, it has been shown that the azido-myristoylation can be used to tag proteins for proteomic analysis (53).
The presented assay was validated using recombinant murine Nmt1 and Nmt2, azido-dodecanoyl-CoA, and three paradigmatic FLAG-tagged substrate peptides (Lck, Nef, and Arf). Successive omission of each single component of the reaction and of the detection system abolished the signals, implicating a high specificity of this approach (Fig. 2a, b). Similarly, substitution of the indispensable N-terminal glycine for alanine interfered with substrate recognition (Fig. 2c, d). The identity of the reaction products was verified by HPLC-MS when using recombinant Nmt1, Nmt2, or tissue lysates as the source of Nmt activity (Fig. 3, supplemental Fig. I, and data not shown). Offering a substrate with a two-carbon longer acyl chain (i.e., azido-tetradecanoyl-CoA), low signals were detected (Fig. 2c, d); this is in line with a previous observation that NMT can also use palmitoyl-CoA, albeit inefficiently (4).
The presence of certain substances can potentially modify the assay readout. In particular, reducing thiol compounds such as DTT (400 mM) and 2-mercaptoethanol (2 mM) can enhance NMT activity up to 6-fold (51). Similarly, various types of detergents (Triton 770 and deoxycholate) can increase NMT activity (54). Conversely, certain oxidizing agents can have an inhibitory effect on NMT (51). Reducing reagents might interfere with the stability of the azido-group, and oxidation of the phosphine can hinder the Staudinger ligation (55).
A general myristoylation consensus sequence is already known, but the substrate specificity of NMT differs among species and among different isoenzymes (15, 56, 57). For this reason, myristoylation predictions based on this consensus sequence are not fail safe (58). We believe that the method presented here would allow easy screening of peptide libraries (e.g., based on certain protein motifs or on the proteome of a particular cell of interest) for potential myristoylation. We tested this possibility using three different peptides (Fig. 4).
In addition, the assay can be used for enzyme kinetic studies (Fig. 5a, b). We have now obtained kinetic parameters of murine recombinant NMT. Comparison of our data to the human Km for myristoyl-CoA showed similar values; it has been given to be 8.24 µM for NMT1 and 7.24 µM for NMT2 (38), as compared with our results of 14 µM and 9 µM for murine NMT1 and NMT2, respectively.
Inhibitor studies are of particular interest because several pathogens encode for their own NMT (25, 26). We demonstrated that the NMT-azido-ELISA is in principle also suitable for this application (Figs. 5c and 7a). Moreover, IC50 values for the inhibitor Tris DBA calculated by this approach for recombinant Nmt1 (0.5 ± 0.1 µM) and Nmt2 (1.3 ± 0.1 µM) were comparable to IC50 values previously published for this inhibitor (1.0 ± 0.26 µM for NMT1) (50).
Measurement of NMT activity is of interest in cancer research. An important aim of the present study was to demonstrate that the assay could be used without further adaptations in the measurement of enzymatic activity in organ lysates. To this end, Nmt activity was measured in different murine organs: brain, thymus, heart, and liver (Fig. 7a, b). The Nmt activity has not been previously reported in these murine organs; however, its distribution among these tissues is consistent with previous observations in rat and cattle (4, 14). The possibility of measuring Nmt activity in frozen organs (Fig. 7c) constitutes a significant advantage in clinical research and therapy trials.
We are convinced that a major advantage of this assay is its versatility. We show that it can be applied for investigations on recombinant Nmt enzymes and their substrates and inhibitors as well as for assessing Nmt activity in fresh or frozen tissues. The assay is user friendly, does not require complex equipment such as HPLC or radioactive material, and can be adopted for high-throughput analysis. Moreover, with appropriate modifications, other enzymes involved in protein acylation (e.g., palmitoylation) might be tested using this system.
Supplementary Material
Acknowledgments
The authors thank Mariona Rabionet Roig and Alexander Feuerborn for helpful consultations and Mahnaz Bonrouhi for technical assistance.
Footnotes
Abbreviations:
- Arf
- ADP-ribosylation factor
- DTNB
- 5-(3-carboxy-4-nitrophenyl)disulfanyl-2-nitrobenzoic acid
- Lck
- lymphocyte-specific protein tyrosine kinase
- Nef
- negative regulatory factor
- NMT
- N-myristoyltransferase
- RT
- room temperature
- Tris DBA
- Tris(dibenzylideneacetone)dipalladium
This work was supported by grant SFB 938 from the Deutsche Forschungsgemeinschaft (H.-J.G., S.P.).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the one figure.
REFERENCES
- 1.Towler D., Glaser L. 1986. Protein fatty acid acylation: enzymatic synthesis of an N-myristoylglycyl peptide. Proc. Natl. Acad. Sci. USA. 83: 2812–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farazi T. A., Waksman G., Gordon J. I. 2001. The biology and enzymology of protein N-myristoylation. J. Biol. Chem. 276: 39501–39504 [DOI] [PubMed] [Google Scholar]
- 3.Wilcox C., Hu J. S., Olson E. N. 1987. Acylation of proteins with myristic acid occurs cotranslationally. Science. 238: 1275–1278 [DOI] [PubMed] [Google Scholar]
- 4.King M. J., Sharma R. K. 1991. N-myristoyl transferase assay using phosphocellulose paper binding. Anal. Biochem. 199: 149–153 [DOI] [PubMed] [Google Scholar]
- 5.Zha J., Weiler S., Oh K. J., Wei M. C., Korsmeyer S. J. 2000. Posttranslational N-myristoylation of BID as a molecular switch for targeting mitochondria and apoptosis. Science. 290: 1761–1765 [DOI] [PubMed] [Google Scholar]
- 6.Pillai S., Baltimore D. 1987. Myristoylation and the post-translational acquisition of hydrophobicity by the membrane immunoglobulin heavy-chain polypeptide in B lymphocytes. Proc. Natl. Acad. Sci. USA. 84: 7654–7658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Resh M. D. 1999. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta. 1451: 1–16 [DOI] [PubMed] [Google Scholar]
- 8.Pellman D., Garber E. A., Cross F. R., Hanafusa H. 1985. An N-terminal peptide from p60src can direct myristylation and plasma membrane localization when fused to heterologous proteins. Nature. 314: 374–377 [DOI] [PubMed] [Google Scholar]
- 9.Towler D. A., Adams S. P., Eubanks S. R., Towery D. S., Jackson-Machelski E., Glaser L., Gordon J. I. 1987. Purification and characterization of yeast myristoyl CoA:protein N-myristoyltransferase. Proc. Natl. Acad. Sci. USA. 84: 2708–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giang D. K., Cravatt B. F. 1998. A second mammalian N-myristoyltransferase. J. Biol. Chem. 273: 6595–6598 [DOI] [PubMed] [Google Scholar]
- 11.Ducker C. E., Upson J. J., French K. J., Smith C. D. 2005. Two N-myristoyltransferase isozymes play unique roles in protein myristoylation, proliferation, and apoptosis. Mol. Cancer Res. 3: 463–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qi Q., Rajala R. V., Anderson W., Jiang C., Rozwadowski K., Selvaraj G., Sharma R., Datla R. 2000. Molecular cloning, genomic organization, and biochemical characterization of myristoyl-CoA:protein N-myristoyltransferase from Arabidopsis thaliana. J. Biol. Chem. 275: 9673–9683 [DOI] [PubMed] [Google Scholar]
- 13.Boisson B., Giglione C., Meinnel T. 2003. Unexpected protein families including cell defense components feature in the N-myristoylome of a higher eukaryote. J. Biol. Chem. 278: 43418–43429 [DOI] [PubMed] [Google Scholar]
- 14.Glover C. J., Goddard C., Felsted R. L. 1988. N-myristoylation of p60src. Identification of a myristoyl-CoA:glycylpeptide N-myristoyltransferase in rat tissues. Biochem. J. 250: 485–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Towler D. A., Adams S. P., Eubanks S. R., Towery D. S., Jackson-Machelski E., Glaser L., Gordon J. I. 1988. Myristoyl CoA:protein N-myristoyltransferase activities from rat liver and yeast possess overlapping yet distinct peptide substrate specificities. J. Biol. Chem. 263: 1784–1790 [PubMed] [Google Scholar]
- 16.Towler D. A., Eubanks S. R., Towery D. S., Adams S. P., Glaser L. 1987. Amino-terminal processing of proteins by N-myristoylation. Substrate specificity of N-myristoyl transferase. J. Biol. Chem. 262: 1030–1036 [PubMed] [Google Scholar]
- 17.Wright M. H., Heal W. P., Mann D. J., Tate E. W. 2010. Protein myristoylation in health and disease. J. Chem. Biol. 3: 19–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang S. H., Shrivastav A., Kosinski C., Sharma R. K., Chen M. H., Berthiaume L. G., Peters L. L., Chuang P. T., Young S. G., Bergo M. O. 2005. N-myristoyltransferase 1 is essential in early mouse development. J. Biol. Chem. 280: 18990–18995 [DOI] [PubMed] [Google Scholar]
- 19.Magnuson B. A., Raju R. V., Moyana T. N., Sharma R. K. 1995. Increased N-myristoyltransferase activity observed in rat and human colonic tumors. J. Natl. Cancer Inst. 87: 1630–1635 [DOI] [PubMed] [Google Scholar]
- 20.Rajala R. V., Radhi J. M., Kakkar R., Datla R. S., Sharma R. K. 2000. Increased expression of N-myristoyltransferase in gallbladder carcinomas. Cancer. 88: 1992–1999 [PubMed] [Google Scholar]
- 21.Lu Y., Selvakumar P., Ali K., Shrivastav A., Bajaj G., Resch L., Griebel R., Fourney D., Meguro K., Sharma R. K. 2005. Expression of N-myristoyltransferase in human brain tumors. Neurochem. Res. 30: 9–13 [DOI] [PubMed] [Google Scholar]
- 22.Shrivastav A., Sharma A. R., Bajaj G., Charavaryamath C., Ezzat W., Spafford P., Gore-Hickman R., Singh B., Copete M. A., Sharma R. K. 2007. Elevated N-myristoyltransferase activity and expression in oral squamous cell carcinoma. Oncol. Rep. 18: 93–97 [PubMed] [Google Scholar]
- 23.Selvakumar P., Lakshmikuttyamma A., Shrivastav A., Das S. B., Dimmock J. R., Sharma R. K. 2007. Potential role of N-myristoyltransferase in cancer. Prog. Lipid Res. 46: 1–36 [DOI] [PubMed] [Google Scholar]
- 24.Price H. P., Menon M. R., Panethymitaki C., Goulding D., McKean P. G., Smith D. F. 2003. Myristoyl-CoA:protein N-myristoyltransferase, an essential enzyme and potential drug target in kinetoplastid parasites. J. Biol. Chem. 278: 7206–7214 [DOI] [PubMed] [Google Scholar]
- 25.Georgopapadakou N. H. 2002. Antifungals targeted to protein modification: focus on protein N-myristoyltransferase. Expert Opin. Investig. Drugs. 11: 1117–1125 [DOI] [PubMed] [Google Scholar]
- 26.Langner C. A., Lodge J. K., Travis S. J., Caldwell J. E., Lu T., Li Q., Bryant M. L., Devadas B., Gokel G. W., Kobayashi G. S., et al. 1992. 4-oxatetradecanoic acid is fungicidal for Cryptococcus neoformans and inhibits replication of human immunodeficiency virus I. J. Biol. Chem. 267: 17159–17169 [PubMed] [Google Scholar]
- 27.Brannigan J. A., Smith B. A., Yu Z., Brzozowski A. M., Hodgkinson M. R., Maroof A., Price H. P., Meier F., Leatherbarrow R. J., Tate E. W., et al. 2010. N-myristoyltransferase from Leishmania donovani: structural and functional characterisation of a potential drug target for visceral leishmaniasis. J. Mol. Biol. 396: 985–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowyer P. W., Tate E. W., Leatherbarrow R. J., Holder A. A., Smith D. F., Brown K. A. 2008. N-myristoyltransferase: a prospective drug target for protozoan parasites. ChemMedChem. 3: 402–408 [DOI] [PubMed] [Google Scholar]
- 29.Frearson J. A., Brand S., McElroy S. P., Cleghorn L. A., Smid O., Stojanovski L., Price H. P., Guther M. L., Torrie L. S., Robinson D. A., et al. 2010. N-myristoyltransferase inhibitors as new leads to treat sleeping sickness. Nature. 464: 728–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bryant M., Ratner L. 1990. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc. Natl. Acad. Sci. USA. 87: 523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maurer-Stroh S., Eisenhaber F. 2004. Myristoylation of viral and bacterial proteins. Trends Microbiol. 12: 178–185 [DOI] [PubMed] [Google Scholar]
- 32.Pierre M., Traverso J. A., Boisson B., Domenichini S., Bouchez D., Giglione C., Meinnel T. 2007. N-myristoylation regulates the SnRK1 pathway in Arabidopsis. Plant Cell. 19: 2804–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nimchuk Z., Marois E., Kjemtrup S., Leister R. T., Katagiri F., Dangl J. L. 2000. Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell. 101: 353–363 [DOI] [PubMed] [Google Scholar]
- 34.Seaton K. E., Smith C. D. 2008. N-Myristoyltransferase isozymes exhibit differential specificity for human immunodeficiency virus type 1 Gag and Nef. J. Gen. Virol. 89: 288–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boisson B., Meinnel T. 2003. A continuous assay of myristoyl-CoA:protein N-myristoyltransferase for proteomic analysis. Anal. Biochem. 322: 116–123 [DOI] [PubMed] [Google Scholar]
- 36.French K. J., Zhuang Y., Schrecengost R. S., Copper J. E., Xia Z., Smith C. D. 2004. Cyclohexyl-octahydro-pyrrolo[1,2-a]pyrazine-based inhibitors of human N-myristoyltransferase-1. J. Pharmacol. Exp. Ther. 309: 340–347 [DOI] [PubMed] [Google Scholar]
- 37.French S. A., Christakis H., O'Neill R. R., Miller S. P. 1994. An assay for myristoyl-CoA: protein N-myristoyltransferase activity based on ion-exchange exclusion of [3H]myristoyl peptide. Anal. Biochem. 220: 115–121 [DOI] [PubMed] [Google Scholar]
- 38.Goncalves V., Brannigan J. A., Thinon E., Olaleye T. O., Serwa R., Lanzarone S., Wilkinson A. J., Tate E. W., Leatherbarrow R. J. 2012. A fluorescence-based assay for N-myristoyltransferase activity. Anal. Biochem. 421: 342–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pennise C. R., Georgopapadakou N. H., Collins R. D., Graciani N. R., Pompliano D. L. 2002. A continuous fluorometric assay of myristoyl-coenzyme A:protein N-myristoyltransferase. Anal. Biochem. 300: 275–277 [DOI] [PubMed] [Google Scholar]
- 40.McIlhinney R. A., McGlone K. 1989. A simplified assay for the enzyme responsible for the attachment of myristic acid to the N-terminal glycine residue of proteins, myristoyl-CoA: glycylpeptide N-myristoyltransferase. Biochem. J. 263: 387–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takamune N., Hamada H., Sugawara H., Misumi S., Shoji S. 2002. Development of an enzyme-linked immunosorbent assay for measurement of activity of myristoyl-coenzyme A:protein N-myristoyltransferase. Anal. Biochem. 309: 137–142 [DOI] [PubMed] [Google Scholar]
- 42.Kostiuk M. A., Corvi M. M., Keller B. O., Plummer G., Prescher J. A., Hangauer M. J., Bertozzi C. R., Rajaiah G., Falck J. R., Berthiaume L. G. 2008. Identification of palmitoylated mitochondrial proteins using a bio-orthogonal azido-palmitate analogue. FASEB J 22: 721–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chomczynski P. 1993. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques. 15: 532–534, 536–537 [PubMed] [Google Scholar]
- 44.Feuerborn A., Srivastava P. K., Kuffer S., Grandy W. A., Sijmonsma T. P., Gretz N., Brors B., Grone H. J. 2011. The Forkhead factor FoxQ1 influences epithelial differentiation. J. Cell. Physiol. 226: 710–719 [DOI] [PubMed] [Google Scholar]
- 45.Martin D. D., Vilas G. L., Prescher J. A., Rajaiah G., Falck J. R., Bertozzi C. R., Berthiaume L. G. 2008. Rapid detection, discovery, and identification of post-translationally myristoylated proteins during apoptosis using a bio-orthogonal azidomyristate analog. FASEB J. 22: 797–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Price H. P., Stark M., Smith D. F. 2007. Trypanosoma brucei ARF1 plays a central role in endocytosis and golgi-lysosome trafficking. Mol. Biol. Cell. 18: 864–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marchildon G. A., Casnellie J. E., Walsh K. A., Krebs E. G. 1984. Covalently bound myristate in a lymphoma tyrosine protein kinase. Proc. Natl. Acad. Sci. USA. 81: 7679–7682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McNaught A. D., Wilkinson A. 1997. IUPAC. Compendium of chemical terminology, 2nd ed. Blackwell Scientific Publications, Oxford, UK [Google Scholar]
- 49.Zhang J. H., Chung T. D., Oldenburg K. R. 1999. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4: 67–73 [DOI] [PubMed] [Google Scholar]
- 50.Bhandarkar S. S., Bromberg J., Carrillo C., Selvakumar P., Sharma R. K., Perry B. N., Govindarajan B., Fried L., Sohn A., Reddy K., Arbiser J. L. 2008. Tris (dibenzylideneacetone) dipalladium, a N-myristoyltransferase-1 inhibitor, is effective against melanoma growth in vitro and in vivo. Clin. Cancer Res. 14: 5743–5748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raju R. V., Sharma R. K. 1996. Coenzyme A dependent myristoylation and demyristoylation in the regulation of bovine spleen N-myristoyltransferase. Mol. Cell. Biochem. 158: 107–113 [DOI] [PubMed] [Google Scholar]
- 52.Hang H. C., Yu C., Pratt M. R., Bertozzi C. R. 2004. Probing glycosyltransferase activities with the Staudinger ligation. J. Am. Chem. Soc. 126: 6–7 [DOI] [PubMed] [Google Scholar]
- 53.Heal W. P., Wickramasinghe S. R., Bowyer P. W., Holder A. A., Smith D. F., Leatherbarrow R. J., Tate E. W. 2008. Site-specific N-terminal labelling of proteins in vitro and in vivo using N-myristoyl transferase and bioorthogonal ligation chemistry. Chem. Commun. (Camb.). 480–482 [DOI] [PubMed] [Google Scholar]
- 54.Boutin J. A., Clarenc J. P., Ferry G., Ernould A. P., Remond G., Vincent M., Atassi G. 1991. N-myristoyl-transferase activity in cancer cells. Solubilization, specificity and enzymatic inhibition of a N-myristoyl transferase from L1210 microsomes. Eur. J. Biochem. 201: 257–263 [DOI] [PubMed] [Google Scholar]
- 55.Schilling C. I., Jung N., Biskup M., Schepers U., Brase S. 2011. Bioconjugation via azide-Staudinger ligation: an overview. Chem. Soc. Rev. 40: 4840–4871 [DOI] [PubMed] [Google Scholar]
- 56.Raju R. V., Anderson J. W., Datla R. S., Sharma R. K. 1997. Molecular cloning and biochemical characterization of bovine spleen myristoyl CoA:protein N-myristoyltransferase. Arch. Biochem. Biophys. 348: 134–142 [DOI] [PubMed] [Google Scholar]
- 57.Maurer-Stroh S., Eisenhaber B., Eisenhaber F. 2002. N-terminal N-myristoylation of proteins: prediction of substrate proteins from amino acid sequence. J. Mol. Biol. 317: 541–557 [DOI] [PubMed] [Google Scholar]
- 58.Ashrafi K., Farazi T. A., Gordon J. I. 1998. A role for Saccharomyces cerevisiae fatty acid activation protein 4 in regulating protein N-myristoylation during entry into stationary phase. J. Biol. Chem. 273: 25864–25874 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.