Abstract
Universal principles underlying network science, and their ever-increasing applications in biomedicine, underscore the unprecedented capacity of systems biology based strategies to synthesize and resolve massive high throughput generated datasets. Enabling previously unattainable comprehension of biological complexity, systems approaches have accelerated progress in elucidating disease prediction, progression, and outcome. Applied to the spectrum of states spanning health and disease, network proteomics establishes a collation, integration, and prioritization algorithm to guide mapping and decoding of proteome landscapes from large-scale raw data. Providing unparalleled deconvolution of protein lists into global interactomes, integrative systems proteomics enables objective, multi-modal interpretation at molecular, pathway, and network scales, merging individual molecular components, their plurality of interactions, and functional contributions for systems comprehension. As such, network systems approaches are increasingly exploited for objective interpretation of cardiovascular proteomics studies. Here, we highlight network systems proteomic analysis pipelines for integration and biological interpretation through protein cartography, ontological categorization, pathway and functional enrichment and complex network analysis.
Keywords: ATP-sensitive K+ channel, bioinformatics, complex network analysis, KATP channel, Kir6.2, genetics, heart disease, metabolism, network biology, proteome, regenerative medicine, SUR2A, stem cells, systems biology
Systems biology approaches are accelerating elucidation of disease prediction, progression, and outcome, enabling previously unattainable comprehension of biological complexity. Complex network analysis offers an integrative multi-modal approach by which to achieve inclusivity and maximize systems comprehension of high throughput datasets at molecular, pathway, and network scales, incorporating full complements of identified molecular components together with their recognized interrelationships and functional contributions. Here, we describe integration and biological interpretation of proteomic data, encompassing protein cartography, ontology, pathway and network analysis, highlighting salient aspects in recent cardiovascular applications.
Proteomic cartography
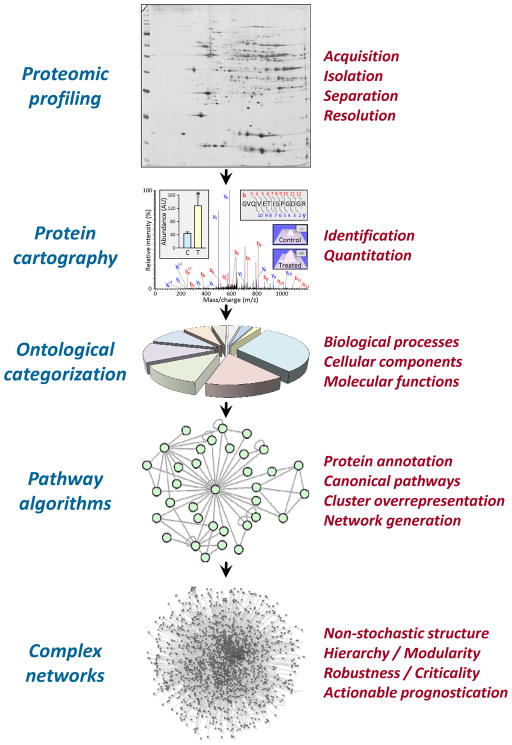
Proteomics encompasses diverse methods by which to study proteins, their abundance, structure, post-translational modifications (PTMs), and physical or functional interacting partners to map the proteome, the protein complement of a genome. While such methods can be applied to individual proteins, proteomics facilitates large-scale analysis of complete proteomes or targeted subproteomes (Table 1). Moreover, proteomics enables comparisons between distinct conditions/states, from defined biological sources, across discrete timelines. In general, a proteomic cartography pipeline involves sample acquisition, followed by protein isolation, separation, resolution, and identification (Figure). Along this continuum, each successive module harbors a multi-step process whereby proteomic methodologies can be implemented.
Table 1.
Systems proteomics lexicon
| Subproteome | Proteome subset, corresponding to particular intra- or extra-cellular compartments, involvement in specific functions, or modulated by or dependent upon particular biological contexts |
| Metaboproteome | Subproteome involved in and supporting cellular metabolic function |
| Systems biology | Analytical approach to investigate and model relationships among system components to understand and/or predict emergent properties |
| Network/Interactome | Mathematical representation of pairwise collections of proteins connected by interdependent interactions and/or relationships, the structure of which conveys emergent properties |
| Network medicine/biology | Analysis of molecular (e.g., protein, gene, metabolite) interaction networks through complex network theory, comprising topological features characterizing symmetric or asymmetric relationships amongst discrete network objects and their known or established interactions to guide medical or biological interpretation |
| Node/Vertex | Fundamental unit of a network, in network biology including one or more of the following: genes, proteins, metabolites, drugs, and endogenous small molecules |
| Edge | In network biology, a relationship between two nodes or node self-interaction, involving physical, regulatory, or genetic interactions which may be undirected or directed (uni- or bi-directional). Edges can include information regarding relationship confidence (e.g., node weighting) or function (e.g., interaction/pathway kinetics) |
| Degree distribution | Node degree defines number of connections between a network node and other nodes, with degree distribution being the probability distribution of the degrees of all nodes |
| Clustering coefficient | Node clustering coefficient is determined by proportion of nodes connected to it that also connect to one another, providing an indication of neighborhood relatedness, the degree to which nodes within a network cluster together |
| Scale-free topology | Biological networks are postulated to exhibit power law distributions of node connectivity, which influences emergent network properties, such as robustness |
| Path length | Shortest distance required to connect two nodes. Scale-free networks have extremely small average path lengths |
| Network diameter | Maximum path length necessary to connect any two network nodes |
| Network motif | Patterns of interconnections in complex networks occurring at significantly higher rates than expected in randomly assembled networks |
| Network module | Collection of nodes with high internal connectivity but few connections with remainder of the network, typically comprising nodes sharing a particular function (e.g., complex, signaling cascade) |
| Hub | Node or small number of nodes much more highly connected to other network constituents than expected randomly |
| Bridging node | Node bridging the shortest path between two other nodes or modules in a network, which can be critical bottlenecks in network functionality |
Figure 1.
Network systems proteomic pipeline. Strategy extending proteomic profiling from mapping and quantitation of protein identities to ontological categorization, pathway analysis and network generation for inclusive systems interpretation.
Once proteins are isolated from the source of interest, two major separation strategies have evolved to address proteome complexity.1,2 Traditional gel-based approaches, particularly two-dimensional gel electrophoresis (2-DE) involving isoelectric focusing followed orthogonally by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, were the earliest methods adopted for protein separation and resolution, comparative assessment of relative abundance, and initial reduction of protein complexity prior to mass spectrometry (MS). Although 2-DE remains common, the advent of quantitative MS techniques has led to increased application of various gel-based and gel-free alternatives, whereby protein and/or peptide complexity is addressed by separation and resolution prior to or in conjunction with MS, while quantitation is subsequently measured from MS precursor or fragment ion spectra. Choice of separation strategy is influenced by study objectives and resources, often guided by advantages and limitations to each approach. Quantitative MS methods tend to be less time- and labor-intensive while offering greater potential throughput and data acquisition, yet in instances where 2-DE approaches are applied in parallel, use of duplicate approaches can provide independent confirmation3,4 or yield complementary, non-overlapping information.5,6 Regardless of separation strategy, bottom-up or top-down MS analysis7,8 facilitates protein assignment, generating extensive lists that, depending on experimental design, typically include relative abundance between conditions or states (Figure). A number of reviews provide greater technical detail on proteomic methodologies and their broad-based application to cardiovascular research.1,2,9–11
A decade ago, an initial survey of the cardiovascular proteomics domain emphasized that successful incorporation of existing and new technologies would help to unravel intricacies of proteome dynamics.1 Indeed, introduction of several enabling technologies has accelerated this emerging field. The utility of 2-DE was enhanced by improved gel reproducibility attained with immobilized pH gradient isoelectric focusing,12 and fluorescent-based difference gel electrophoresis allowing co-resolution of paired samples.13 Numerous MS advances included algorithms for automatic spectral interpretation, and quantitative MS strategies such as isotope-code affinity tag peptide labeling14 or stable isotope labeling of amino acids.15 Subsequent instrumentation advances, such as development of high resolution, high mass accuracy Fourier transform-ion cyclotron resonance MS and introduction of Orbitrap MS technology for proteomic applications,16,17 expansion of quantitative MS techniques such as isobaric tag and label-free methods,18–20 development of selective PTM analysis strategies, e.g. phosphopeptide enrichment,21 plus continued growth and extensive curation of protein databases for comparison and identification of peptides and proteins from acquired MS data, all contributed to expansion of proteomic studies.
An important corollary of these developments is the increasing magnitude of acquired information. Individual proteomic datasets are not simply more numerous; they are growing ever larger. Regardless of whether gel-based or gel-free methodology is applied, if surveys are quantitative comparisons or qualitative cataloguing, or if they provide coverage of single or multiple time-point/phenotype dynamics, proteomic studies now regularly yield lists comprising hundreds of identified proteins. Thus, a critical consideration for continued maturation of proteomic research is defining appropriate measures by which to collate, prioritize, and integrate expansive protein lists to maximize biologically meaningful data extraction. When confronted with overwhelming protein numbers, where traditional reductionist analysis of each no longer remains feasible, it becomes tempting to focus on a particular subset. A fraction of proteins exhibiting greatest differential expression, those with established links to the topic of interest, or another delimiter to focus interpretation and narrow downstream analysis.22 As a consequence, mechanistically relevant proteins, complexes, and/or functional modules may be inadvertently marginalized or ignored. In this context, analysis of proteomic and other high throughput data acquired using objectively defined statistical criteria23 should instead be inclusive.22
Systems algorithms for proteome comprehension
Network systems biology offers an integrative approach to achieve inclusivity and maximize comprehension. Network platforms incorporate a complement of identified proteins together with their interactions and, where available, input of other high throughput data, to better comprehend the system in its entirety.22,24–29 Various network analysis approaches are becoming increasingly prevalent in genomic,30–34 microRNA,35,36 proteomic,3,37–41 and metabolomic42 cardiovascular research.43–45
Proteins function within intricate webs of interactions. Accordingly, derivation of biological interaction networks provides a rational means of assembling proteomic data in functional contexts. As an example, the minimal network required to incorporate all identified proteins via established interactions yields an immediate focal point of proteomic measurement within an experimental system, regardless of extent of dataset size or breadth of constituent protein functional variability. Derived networks are amenable to compositional assessment, based on function/ontology overrepresentation and enrichment analysis, and to structural examination, such as architectural topology or positional relevance of components, e.g. hubs or bridging nodes (Table 1).46,47 Moreover, network composition and structure may be exploited for mechanistic interpretation, hypothesis generation, and functional validation, enabling refinement of assembled networks.22,48 Network systems biology thus offers a logical, inclusive approach by which to process, prioritize, and interpret complex systems behavior from high throughput datasets of theoretically unlimited size.
Network biology approaches to proteomic data assessment and interpretation typically involve data reduction, clustering or grouping acquired data through ontological categorization, and a combination of pathway and complex network analysis (Figure).29,37–40 Functional ontology classification provides initial biological context for individual proteins, whereas pathway and network analysis integrate composition and architecture of associated interrelationships from defined proteomic observations. Both biological context and network architecture serve as tools to scrutinize acquired proteomes, and to develop informed follow-up investigation and/or validation of insights predicted from high throughput data.3,29,37–40
Ontological categorization
Proteomic studies are proficient at detecting changes, yet biological interpretation is often limited by data complexity. When lists remain small, relevance can be anticipated and/or functionally validated on a protein-by-protein basis. For large-scale studies, this approach is untenable, and there is a need to define appropriate methodology for bioinformatic assessment of high throughput datasets. Data comprehension is facilitated through clustering of proteins by biological processes, cellular components, or molecular functions (Figure).29,37–40,49 Accurate categorization requires the ability to cross-reference protein database information with complementary sources such as the Gene Ontology (GO) or pathway analysis applications. Despite progress, lack of conformity in nomenclature and in computational systems biology data and language formats can limit cross-platform interoperability.
For spectral assignment during tandem MS (MS/MS) and initial protein characterization, the UniProt Knowledge Base (UniProtKB) is an established reference. Alternatives include or have included IPI (International Protein Index), PIR (Protein Information Resource), NCBInr (National Center for Biotechnology Information, non-redundant), Swiss-Prot (from the Swiss Institute of Bioinformatics), and TrEMBL (Translated European Molecular Biology Laboratory Nucleotide Sequence Database, from the European Bioinformatics Institute). The UniProt Consortium initially combined PIR, Swiss-Prot and TrEMBL into the UniProtKB, and now also includes the IPI database. UniProtKB is curated and continuously updated to incorporate new entries and minimize redundancies, while maintaining a history of entry changes, such as consolidation, replacement, or protein/gene re-naming.
UniProtKB entries often include multiple identifiers, increasing the ability to acquire complementary data from other repositories during subsequent analyses. They also contain a wealth of related information, such as interacting partners and residency in protein complexes or functional modules, GO information regarding associated molecular functions or biological processes, and hyperlinks to additional function- and interaction-related databases. Of the protein identifiers used in UniProtKB, the Human Genome Organization (HUGO) gene symbol ID is prioritized and universally applied across databases. Therefore, it serves as a pragmatic choice for cross-database and downstream querying of UniProtKB-derived proteomic information. Protein complexity, however, still occasionally leads to HUGO ID nomenclature issues during bioinformatic data assessment. This may occur when: (i) a single ID represents multiple variants of one protein, all of which may be subsumed by MS/MS data and thus indistinguishable from one another, e.g. TNNT2 represents 10 human cardiac troponin T splice variants; (ii) other highly homologous proteins are subsumed by spectral data, e.g. actin isoforms; (iii) MS/MS peptide matches are subsumed by protein isoforms of identical sequence arising through gene duplication, e.g. calmodulins 1, 2 and 3; and (iv) synonyms exist for a single protein, e.g. histone 4 has 14 human and 12 mouse HUGO IDs. From each scenario, however, only one ID is assigned, in accordance with accepted parsimony standards.50 While indistinguishable isoforms and ID redundancy might be considered confounding issues during ontological classification and pathway/network analysis, their impact is negligible since they are variants of the same protein and their occurrence is infrequent in the context of hundreds of additional proteins.
Most cardiovascular proteomic studies now include ontological categorization, clustering identified proteins by GO biological process, cellular component or molecular function (Figure), with or without segregation into expression cohorts, i.e. upregulated, downregulated, or unchanged. While providing initial biological context, assignment of a single function to each protein is often an oversimplification, since many proteins participate in multiple physiological processes. For instance, promiscuous enzymes influencing an array of other proteins and their activity, e.g. oxidoreductases, kinases, and phosphatases, may carry out one specific function, such as phosphate transfer, yet this may impact numerous biological processes, confounding comprehensive protein categorization. In this regard, proteins with multiple GO associations are more appropriately accommodated, and more effectively accounted for, by calculating overrepresentation and/or enrichment significance during subsequent pathway and network analysis.
Pathway analysis algorithms
Pathway analysis maps high throughput data to, and assesses enrichment of, canonical pathways. Pathway algorithms facilitate data annotation, containing detailed descriptions of individual proteins and links to current knowledge. Pathway analysis software can also be used to calculate functional enrichment, by assessment of algorithm-specific functional terminology or GO molecular functions and biological processes. Some pathway algorithms enable biologically-oriented network analysis, where subsets of proteins, represented as nodes or vertices, are connected by edges via established relationships, generating networks that may be further scrutinized for function or pathway over-representation. Thus, pathway analysis extends functional categorization by evaluating collective consequences of proteomic findings in the context of established pathways, functions, and interactions (Figure).
Pathway analysis is an expanding field, with a diversity of bioinformatic applications designed to address molecular pathways and interactions. A comprehensive list of applications available at Pathguide51 outlines over 300 existing pathway and/or molecular relationship resources. Collectively, they encompass 8 subcategories, including protein-protein interactions, metabolic pathways, signaling pathways, pathway diagrams, transcription factor/gene regulatory networks, protein-compound interactions, genetic interaction networks, protein sequence focused databases, and 14 repositories related to topics not covered by the 8 major categories. Pathguide provides information about each resource, facilitating informed decisions regarding algorithm selection.
Deconvolution of broad-based proteomic data is most readily accomplished with pathway resources that maximize information retrieval, such as Ingenuity Pathways Knowledge Base (IPKB), archived in 5 Pathguide categories – protein-protein interactions, metabolic pathways, signaling pathways, transcription factors and gene regulatory networks, and protein-compound interactions. IPKB is comprised of data manually curated or automatically extracted from the literature, and incorporates reviewed content from over 20 additional repositories. In turn, data is accessed and interrogated using the complementary bioinformatics tool, Ingenuity Pathways Analysis (IPA). Since IPKB is a compilation of existing data, it is important to emphasize that IPA analysis is a pathway-oriented interpretation of current knowledge rather than inference of novel outcomes and interactions. As data acquisition increases, gaps in knowledge and limitations in scope will be reduced, expanding and refining pathway analysis interpretation. Together, IPA and IPKB offer broad applicability and, as such, have become increasingly popular for cardiovascular proteomic studies,3,5,37–40,52–66 as have various alternative applications for ontology/enrichment analysis (e.g. Gene Set Enrichment Analysis,67 Ontologizer,68 and the Database for Annotation, Visualization, and Integrated Discovery,68) and dedicated pathway and network analysis (e.g. MetaCore’s GeneGo69).
Pertinent considerations for implementing pathway analysis can be garnered from an overview of IPA search parameters. IPA accepts 21 different identifiers to process and interpret high throughput datasets, from a number of gene/protein ID formats suitable for proteomic data, several mRNA microarray platforms, plus additional formats to facilitate microRNA or metabolomic studies, and more recently, RNA-Seq and NGS experiments. Thus, pathway analysis can be carried out using proteomic data alone, or integrated with other high throughput data. Protein IDs uploaded for individual or multiple observations may include quantitative values such as fold change, p-value, or log ratio. Additional user-specified variables include output dataset size (e.g. optional settings for maximum size and number of generated networks), desired stringency and scope (e.g. species specific information; consideration of direct and/or indirect relationships; inclusion of observed and/or predicted interactions; inclusion/exclusion of endogenous chemicals together with genes/proteins as network nodes), whether or not to focus on a subset in isolation (e.g. fold change and/or p-value cutoffs to focus on up-, down-, or both up- and down-regulated proteins simultaneously; inclusion/exclusion of unaltered proteins from assessment), and selection of a baseline or standard reference set against which proteins of interest are compared for pathway and function enrichment and/or overrepresentation. To cast as wide a net as possible for detection of pathway and network interactions, both direct and indirect relationships across a variety of tissues and species should be included,37–40 as this will obviate annotation limitations. For instance, tissue specific limits may be confounded by gene/protein annotation being based primarily on healthy tissues, which may not reflect observed expression under conditions of stress or disease, e.g. fetal isoform expression in adult tissue, whereas setting species limits may overlook relevant knowledge of a particular protein that is more extensively documented in a species other than the one under investigation. Search stringency for proteomic data is better attained by limiting interpretation to include proteins only, by limiting interactions to those that are experimentally observed, and by defining network generation parameters to minimize the number of networked molecules required to incorporate all submitted proteins.37–40
As an evolving component of proteomic research, pathway analysis is best exploited as an interpretation tool facilitating hypothesis generation for subsequent follow-up, rather than as a confirmatory/validation step within the data analysis pipeline. The utility and potential of pathway analysis software is tempered by the fact that documented relationships only comprise partial biological information. Moreover, reliability of data supporting these interactions is variable, and it is impractical to assess interactions for quality control on a protein-by-protein basis when thousands of connections are generated. Finally, substantial effects on pathway software have been demonstrated by annotation changes, updates, additions, and errors, all of which impact prediction and significance outcomes.70 Thus, pathway analysis searches run the risk of being incomplete or potentially misleading, and are best validated by experimental follow-up.
Overrepresentation of normal and adverse biological functions assessed in IPA can be useful in guiding follow-up. In a model of imposed stress in the context of ATP-sensitive K+ (KATP) channel deficiency, IPA analysis of proteomic data exclusively predicted cardiac adverse effects, which were subsequently validated by in vivo assessment in the KATP channel deficient cohort.38 Adverse effect screening was also employed in a study examining proteomic consequences of cardiomyopathy and structure/function remodeling mediated by stem cell therapy.40 In this regard, cardiomyopathic proteome changes predicted several cardiac adverse effects and over-representation of cardiac disease, which were diminished or no longer evident when assessing proteomic remodeling of stem cell-treated cardiomyopathic hearts. Validation of the predicted spectrum of structural and functional benefits by stem cell therapy was demonstrated by multi-modal imaging, indicating restoration of pre-stress cardiac functional levels and normalization of heart structure.40 Thus, proteome-specified functional targets can be identified by pathway analysis for experimental follow-up and validation.
Networks generated during pathway analysis also yield biologically relevant contexts for functional prioritization, hypothesis generation, and experimental validation (Figure). IPA networks are ranked by calculated over-representation of the proportion of input proteins within each network relative to expected numbers if proteins were instead randomly selected from a background reference set. Included with these prioritized networks is predicted intra-network functional enrichment, which can further guide hypothesis generation. For example, the top prioritized network generated from proteomic analysis of endoderm secretome-mediated cardiopoiesis was associated with cardiovascular growth and development.37 Validation was carried out in silico by removal of the most highly connected network node from the list of input proteins in a parallel IPA assessment, which demoted cardiovascular development prioritization. Validation was independently conducted in vitro by targeted pharmacological inhibition, which abolished the potentiation of cardiac differentiation, thereby confirming network contributions to cardiopoiesis.37
Default IPA constraints typically double the number of proteins from initial upload to network output,3,37–40 thereby increasing complexity, but this also provides additional targets for hypothesis generation by incorporating potentially relevant proteins not detected during initial proteomic screening that can be targeted, assessed and validated.37 Another case in point is an IPA-derived network assessing crosstalk between endothelial and vascular smooth muscle cells in response to differential shear stress.64 Here, the primary proteomic network included a number of secretory proteins involved in cellular communication and function modulation. Some of these proteins were not detected during proteomic analysis, yet their presence in the network suggested criticality for intercellular crosstalk underlying observed vascular remodeling, which was subsequently assessed and validated.64 Thus, pathway analysis provides actionable predictions for validation via network functional prioritization, including addition of target proteins networked with, but not initially observed among, acquired proteomic data.
Pathway algorithms are advantageous for enrichment analysis or function and ontology representation, but lack tools necessary to characterize network architectural topology and positional relevance of network components. Pathway analysis networks emphasize biological relationships over structure, and are designed to be sufficiently large to provide biological interactions, yet small enough for rapid network inference and construction, and to readily visualize all nodes simultaneously. IPA network settings, for instance, range from 35–140 nodes, and although multiple networks can be combined, merges may comprise no more than 500 nodes at a time. Moreover, no provisions are included to assess functional enrichment within merged composite interactomes. To fully comprehend network structure and composition for inclusive systems interpretation of proteomic data, particularly as datasets continue increasing in magnitude, it is necessary to move beyond pathway software to applications dedicated to network visualization and interpretation.
Complex network analysis
Complex network analysis enables integrative proteome study across the spectrum of properties inherent within assembled protein-protein interrelationships. These mathematical representations comprise individual protein constituents, termed nodes or vertices in a derived network, held together by physical or regulatory interconnections, defined as edges (Table 1). Nodes and edges can be assessed for architectural connectivity, superimposed with qualitative or quantitative information, and, for some network algorithms, linked to external annotation or ontological information sources. Networks can also be modified or updated in an iterative fashion. While popular for proteomic and other systems interpretation, network approaches are limited with respect to input data quality, output variability from differing algorithms, and lack of flexibility,22,71 with limited capacity to account for post-translational or spatiotemporal dynamics, or when networking data acquired from multiple high throughput sources. Metabolic networks, for example, may comprise protein nodes with metabolite edges, metabolite nodes with protein edges, or both proteins and metabolites as nodes connected by various interrelationships. Applied judiciously in light of such caveats, networks remain versatile platforms upon which to integrate proteomic and other high throughput data for systems comprehension.22
Current understanding of the non-stochastic nature of networks arose from the demonstration that supposedly random networks from a variety of sources instead possess non-random connectivity.72 In these networks, the vast majority of nodes possess one or very few edges, whereas an exceedingly small proportion are more highly connected. Node connectivity is defined as degree, the number of edges a node possesses, i.e. how many other nodes it connects to in the network. Rather than exhibiting Gaussian node degree distributions, expected if randomly connected, networks approximate power law distributions which form characteristic scale-free topologies. Debate continues regarding whether network structures are scale-free, exponential or adhere to more esoteric mathematical constraints, yet it is clear that their organization is not random. Non-stochasticity of network architecture is now known to be universally applicable, and this imparts emergent structural properties of biological relevance, such as network motifs, modularity, and functional robustness (Table 1).46,47 Proteome networks, too, follow principles of non-stochasticity, reflecting non-random, ordered structure, consistently observed regardless of network scale, which may be exploited on the basis of these properties in a variety of ways not apparent when viewed simply as a list of protein identities.
To assess such properties for proteomic experiments, interaction networks are typically derived by impartial assembly from pathway analysis resources or by manual screening of the literature, and can be generated de novo by assessing protein-protein interaction data. Alternative criteria may be used to delineate network connectivity, such as statistically on the basis of co-expression matrices and Pearson correlation thresholds,33 or by ab initio reverse engineering network models from gene expression dynamics.73 Statistical approaches may yield novel relationships based on gene co-regulation, but require confirmation to determine whether or not connections are functionally meaningful.74 Moreover, extensive expression profiling over numerous perturbations or time points is necessary to provide sufficient data for reliable network modeling. Until such time as proteomic data is produced in quantities or at scales similar to transcript profiling, proteomic network generation will rely primarily upon current biological knowledge to establish node connectivity.
Just as Pathguide serves as a portal into pathway analysis, Graph Visualization Software References (GVSR) provides an online resource detailing over 80 network analysis algorithms to enlighten and guide selection of network software, including additional links to other sites and resources.75 As with pathway analysis, those interested in network generation may avail themselves of specialized or generalized algorithms through GVSR. A comprehensive GVSR-listed application is Cytoscape, an open source platform for visualization of proteomic molecular interaction networks and their integration with other state data.76 At the core of Cytoscape is a network visualization tool, for which both developers and users alike can create and exploit specific peripheral tools, termed plug-ins, to facilitate network interpretation, including network and molecular analyses, additional layout options and file format support, and connections to external databases and annotation.
Network generation in Cytoscape requires no specialized knowledge of network theory or computer programming. In their simplest form, networks consist of a collection of pairwise interactions. Thus, Cytoscape generates networks from spreadsheets or simple interaction files (.sif extension) for which each row expresses an interaction and the nature of that interaction, e.g. undirected, uni-directional, or bi-directional. Once pairwise interactions are compiled, e.g. from pathway analysis sources, and uploaded, additional files containing expression attributes may be included and applied to network layouts and node or edge attributes for network visualization. For instance, node and edge shape, size, and color can be used to convey direction and extent of expression, node functional category, edge confidence, and so on, and layouts can be used to emphasize or highlight particular attributes, such as the most highly connected or functionally related nodes.37–40 For instance, network information can be accentuated by clustering nodes of common function via layout co-localization,38–40 or by presenting common elements of adjacent networks by locating them in the same relative location for both networks.40 Unlike standard pathway analysis network settings that limit network size and scope, dedicated complex network analysis software can generate networks comprising hundreds of thousands of nodes, accommodating the largest proteomic interactomes.
To complement network assembly, Cytoscape users can presently select from 155 plug-ins to suit particular needs, including analysis of existing networks, import of networks and network attributes, network inference, functional enrichment, communication and scripting applications, and a miscellany of additional applications. Download statistics indicate that the most popular Cytoscape plug-in is BiNGO, for Biological Network Gene Ontology.77 BiNGO assesses biological network overrepresentation for various gene ontology categories – biological process, cellular component, and molecular function – by comparison to a species specific reference, similar to enrichment analysis tools in pathway algorithms. Other top downloads include plug-ins for assessment of network structural attributes, such as highly clustered or activated network modules (MCODE and jActive Modules, respectively), and those facilitating access to other databases (NCBI Entrez Gene User Interface and NCBI Client) and manuscript searches (Agilent Literature Search) for network annotation (Table 2). Another popular plug-in is Network Analyzer,78 a useful tool for assessing network topology, the benefits of which become more apparent as network size increases. This application computes and displays a wide array of topological parameters for both directed and undirected networks. These include number of nodes, edges, connected components, network diameter, degree distribution, clustering coefficient, and shortest path lengths, among others (Tables 1, 2).78 Due to the versatility of Cytoscape, this network visualization and interpretation tool has become increasingly useful for complex network analysis in cardiovascular proteomic studies.3,37–40,67
Table 2.
Examples of popular and useful network support applications
| Application | Network function addressed |
|---|---|
| Network Analyzer | Calculates an extensive array of network topological parameters to facilitate identification of nodes of interest and relevant network structural characteristics |
| MCODE | Detects and prioritizes highly interconnected regions within a network |
| jActive Modules | Identifies expression activated subnetworks, connected network regions that exhibit significant changes in expression |
| ReOrient Plugin | Facilitates visual comparison of adjacent networks by locating common nodes in the same relative location |
| BiNGO | Determines GO category statistical over- or under-representation from a set of genes/proteins within a network or subnetwork of interest, visualized as a GO hierarchy network to map significant themes by biological process, cellular component, or molecular function |
| Enrichment Map | Visualizes networks of gene set enrichment analysis results, where nodes represent gene sets and edges their mutual overlap (rather than connecting nodes by nested GO classification, as done in BiNGO), clustering highly redundant gene sets to improve enrichment analysis navigation and interpretation |
| NCBI Entrez Gene User Interface/NCBI Client | Imports gene/protein interactions, annotations, and attributes from NCBI Entrez Gene for network derivation |
| Agilent Literature Search | Queries multiple text-based search engines to obviate the need for manual search and extraction of gene/protein interactions for network generation |
Complex network analysis ontological overrepresentation extends observations available during pathway analysis by providing enrichment assessment for entire network neighborhoods rather than partial subnetworks. Although functional enrichment analysis of merged composite networks is not available within IPA proper, they can be assembled and scrutinized elsewhere, such as within Cytoscape. For example, widespread metabolic remodeling in response to KATP channel deficiency was apparent by IPA functional ontology classification, but was reinforced and expanded by BiNGO analysis of the derived composite network with Cytoscape.39 BiNGO assesses protein or gene networks for ontological representation, generating a hierarchical network of GO terms as nodes to visualize overrepresented functions, with connecting edges comprising nested connections within the GO hierarchy (Table 2). Specifically, BiNGO analysis extended the KATP channel-dependent metaboproteome (Table 1) by generating an ontology network from the proteomic network, demonstrating that from all 962 network-associated biological processes, the 55 that were overrepresented were all metabolic processes, associated with protein catabolism, glycolysis, the tricarboxylic acid cycle, fatty acid, and other substrate metabolism.39 Enrichment Map offers an alternative plug-in for Cytoscape functional interpretation.79 This tool creates an ontological network with nodes again comprising overrepresented functions, but with connecting edges showing a weighted intensity based on extent of proteomic dataset overlap between nodes, and an automated layout situating connected nodes closer to one another to facilitate comprehension of closely related enrichment terms (Table 2).79 By including all detected protein changes rather than a portion of those most differentially expressed, Enrichment Map was combined with Gene Set Enrichment Analysis to expand biological consequences evident from cardiomyopathy proteomic profiling.67
Network topological analysis provides yet another advantage of moving beyond pathway analysis enrichment, facilitating actionable prognostication of emergent structural properties (Figure). Since most nodes in scale-free networks possess only one or at most a few connections, and a very small number are highly connected hubs, non-stochastic networks are functionally robust.80 Thus, random node removal would result in a high probability of targeting low connectivity nodes and a small likelihood of radically disrupting network connectivity and functionality by removing a critical network hub.80 Therefore, assessing functional integrity in the context of specific hub inhibition could provide understanding of network biological relevance. This approach was used to confirm the potentiating effect of an endodermal secretome network on cardiac differentiation by targeted inhibition of either the primary or secondary hub of the derived network.37 Thus, understanding network mathematical architecture enables interrogation of constituent node positional relevance. Together, these examples demonstrate how network-oriented analysis algorithms extend pathway analysis data interpretation, apply alternative sources of network assembly to comprehend function (i.e. GO terms rather than proteins as nodes), make use of all protein IDs rather than a small proportion of the most extensively observed differences for data interpretation (i.e. inclusivity), and extract testable hypotheses from emergent structural properties of complex networks.
Future prospects
As the science of proteomics advances, network-based systems analysis is becoming increasingly integral to high throughput protein deconvolution with the ultimate purpose of furthering fundamental knowledge and translational ramifications. Rigor in proteomic experimental design and stringency in protein-protein interaction studies will be paramount for confidence in data interpretation and network protein interaction quality. Network systems medicine approaches offer unprecedented opportunity for integration of proteomic data with other high throughput sources to better comprehend function in health and disease. Accurate systems interpretation ultimately requires assessment of function or activity, such as biochemical rate constants or metabolic flux analysis, to facilitate prognostic modeling of prioritized pathways and cellular functions, requiring proteomic studies to transition from static state analyses examining change in protein abundance toward dynamical systems analysis. This necessitates incorporation of modalities facilitating state space comprehension to track both deterministic and stochastic effectors influencing complex cellular outcomes. The established capacity of network based systems templates to account for and incorporate a diversity of input data is well suited for this evolutionary transition at the forefront of emerging fields of translational network medicine.
Acknowledgments
The authors thank colleagues for helpful discussions.
Funding Sources: The authors are supported by National Institutes of Health, Marriott Foundation, and Mayo Clinic.
Footnotes
Conflict of Interest Disclosures: None
References
- 1.Arrell DK, Neverova I, Van Eyk JE. Cardiovascular proteomics: evolution and potential. Circ Res. 2001;88:763–773. doi: 10.1161/hh0801.090193. [DOI] [PubMed] [Google Scholar]
- 2.McGregor E, Dunn MJ. Proteomics of the heart: unraveling disease. Circ Res. 2006;98:309–321. doi: 10.1161/01.RES.0000201280.20709.26. [DOI] [PubMed] [Google Scholar]
- 3.Behfar A, Perez-Terzic C, Faustino RS, Arrell DK, Hodgson DM, Yamada S, et al. Cardiopoietic programming of embryonic stem cells for tumor-free heart repair. J Exp Med. 2007;204:405–420. doi: 10.1084/jem.20061916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Folmes CD, Nelson TJ, Martinez-Fernandez A, Arrell DK, Lindor JZ, Dzeja PP, et al. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 2011;14:264–271. doi: 10.1016/j.cmet.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu Z, Wang M, Gucek M, Zhang J, Wu J, Jiang L, et al. Milk fat globule protein epidermal growth factor-8: a pivotal relay element within the angiotensin II and monocyte chemoattractant protein-1 signaling cascade mediating vascular smooth muscle cells invasion. Circ Res. 2009;104:1337–1346. doi: 10.1161/CIRCRESAHA.108.187088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gundry RL, Tchernyshyov I, Sheng S, Tarasova Y, Raginski K, Boheler KR, et al. Expanding the mouse embryonic stem cell proteome: combining three proteomic approaches. Proteomics. 2010;10:2728–2732. doi: 10.1002/pmic.201000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chait BT. Mass spectrometry: bottom-up or top-down? Science. 2006;314:65–66. doi: 10.1126/science.1133987. [DOI] [PubMed] [Google Scholar]
- 8.Zhang H, Ge Y. Comprehensive Analysis of Protein Modifications by Top-Down Mass Spectrometry. Circ Cardiovasc Genet. 2011;4:711. doi: 10.1161/CIRCGENETICS.110.957829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Macri J, Rapundalo ST. Application of proteomics to the study of cardiovascular biology. Trends Cardiovasc Med. 2001;11:66–75. doi: 10.1016/s1050-1738(01)00088-3. [DOI] [PubMed] [Google Scholar]
- 10.Arab S, Gramolini AO, Ping P, Kislinger T, Stanley B, van Eyk J, et al. Cardiovascular proteomics: tools to develop novel biomarkers and potential applications. J Am Coll Cardiol. 2006;48:1733–1741. doi: 10.1016/j.jacc.2006.06.063. [DOI] [PubMed] [Google Scholar]
- 11.Van Eyk JE. Overview: the maturing of proteomics in cardiovascular research. Circ Res. 2011;108:490–498. doi: 10.1161/CIRCRESAHA.110.226894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanchez JC, Rouge V, Pisteur M, Ravier F, Tonella L, Moosmayer M, et al. Improved and simplified in-gel sample application using reswelling of dry immobilized pH gradients. Electrophoresis. 1997;18:324–327. doi: 10.1002/elps.1150180305. [DOI] [PubMed] [Google Scholar]
- 13.Unlü M, Morgan ME, Minden JS. Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis. 1997;18:2071–2077. doi: 10.1002/elps.1150181133. [DOI] [PubMed] [Google Scholar]
- 14.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 15.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 16.Håkansson K, Emmett MR, Hendrickson CL, Marshall AG. High-sensitivity electron capture dissociation tandem FTICR mass spectrometry of microelectrosprayed peptides. Anal Chem. 2001;73:3605–3610. doi: 10.1021/ac010141z. [DOI] [PubMed] [Google Scholar]
- 17.Hu Q, Noll RJ, Li H, Makarov A, Hardman M, Cooks GR. The Orbitrap: a new mass spectrometer. J Mass Spectrom. 2005;40:430–443. doi: 10.1002/jms.856. [DOI] [PubMed] [Google Scholar]
- 18.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Dayon L, Hainard A, Licker V, Turck N, Kuhn K, Hochstrasser DF, et al. Relative quantification of proteins in human cerebrospinal fluids by MS/MS using 6-plex isobaric tags. Anal Chem. 2008;80:2921–2931. doi: 10.1021/ac702422x. [DOI] [PubMed] [Google Scholar]
- 20.Fang R, Elias DA, Monroe ME, Shen Y, McIntosh M, Wang P, et al. Differential label-free quantitative proteomic analysis of Shewanella oneidensis cultured under aerobic and suboxic conditions by accurate mass and time tag approach. Mol Cell Proteomics. 2006;5:714–725. doi: 10.1074/mcp.M500301-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Edwards AV, Cordwell SJ, White MY. Phosphoproteomic profiling of the myocyte. Circ Cardiovasc Genet. 2011;4:575. doi: 10.1161/CIRCGENETICS.110.957787. [DOI] [PubMed] [Google Scholar]
- 22.Arrell DK, Terzic A. Network systems biology for drug discovery. Clin Pharmacol Ther. 2010;88:120–125. doi: 10.1038/clpt.2010.91. [DOI] [PubMed] [Google Scholar]
- 23.Levin Y. The role of statistical power analysis in quantitative proteomics. Proteomics. 2011;11:2565–2567. doi: 10.1002/pmic.201100033. [DOI] [PubMed] [Google Scholar]
- 24.Hood L, Heath JR, Phelps ME, Lin B. Systems biology and new technologies enable predictive and preventative medicine. Science. 2004;306:640–643. doi: 10.1126/science.1104635. [DOI] [PubMed] [Google Scholar]
- 25.Weston AD, Hood L. Systems biology, proteomics, and the future of health care: toward predictive, preventative, and personalized medicine. J Proteome Res. 2004;3:179–196. doi: 10.1021/pr0499693. [DOI] [PubMed] [Google Scholar]
- 26.Hood L, Perlmutter RM. The impact of systems approaches on biological problems in drug discovery. Nat Biotechnol. 2004;22:1215–1217. doi: 10.1038/nbt1004-1215. [DOI] [PubMed] [Google Scholar]
- 27.Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabási A-L. The human disease network. Proc Natl Acad Sci USA. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohl P, Crampin EJ, Quinn TA, Noble D. Systems biology: an approach. Clin Pharmacol Ther. 2010;88:25–33. doi: 10.1038/clpt.2010.92. [DOI] [PubMed] [Google Scholar]
- 29.Arrell DK, Zlatkovic Lindor J, Yamada S, Terzic A. KATP channel-dependent metaboproteome decoded: systems approaches to heart failure prediction, diagnosis, and therapy. Cardiovasc Res. 2011;90:258–266. doi: 10.1093/cvr/cvr046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Faustino RS, Behfar A, Perez-Terzic CM, Terzic A. Genomic chart guiding embryonic stem cell cardiopoiesis. Genome Biol. 2008;9:R6. doi: 10.1186/gb-2008-9-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faustino RS, Chiriac A, Niederlander NJ, Nelson TJ, Behfar A, Mishra PK, et al. Decoded calreticulin-deficient embryonic stem cell transcriptome resolves latent cardiophenotype. Stem Cells. 2010;28:1281–1291. doi: 10.1002/stem.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiriac A, Nelson TJ, Faustino RS, Behfar A, Terzic A. Cardiogenic induction of pluripotent stem cells streamlined through a conserved SDF-1/VEGF/BMP2 integrated network. PLoS One. 2010;5:e9943. doi: 10.1371/journal.pone.0009943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dewey FE, Perez MV, Wheeler MT, Watt C, Spin J, Langfelder P, et al. Gene coexpression network topology of cardiac development, hypertrophy, and failure. Circ Cardiovasc Genet. 2011;4:26–35. doi: 10.1161/CIRCGENETICS.110.941757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Civelek M, Manduchi E, Riley RJ, Stoeckert CJ, Jr, Davies PF. Coronary artery endothelial transcriptome in vivo: identification of endoplasmic reticulum stress and enhanced reactive oxygen species by gene connectivity network analysis. Circ Cardiovasc Genet. 2011;4:243–252. doi: 10.1161/CIRCGENETICS.110.958926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parikh VN, Jin RC, Rabello S, Gulbahce N, White K, Hale A, et al. MicroRNA-21 integrates pathogenic signaling to control pulmonary hypertension: Results of a network bioinformatics approach. Circulation. 2012;125:1520–1532. doi: 10.1161/CIRCULATIONAHA.111.060269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zampetaki A, Willeit P, Drozdov I, Kiechl S, Mayr M. Profiling of circulating microRNAs: from single biomarkers to re-wired networks. Cardiovasc Res. 2012;93:555–562. doi: 10.1093/cvr/cvr266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arrell DK, Niederländer NJ, Faustino RS, Behfar A, Terzic A. Cardioinductive network guiding stem cell differentiation revealed by proteomic cartography of tumor necrosis factor alpha-primed endodermal secretome. Stem Cells. 2008;26:387–400. doi: 10.1634/stemcells.2007-0599. [DOI] [PubMed] [Google Scholar]
- 38.Zlatkovic J, Arrell DK, Kane GC, Miki T, Seino S, Terzic A. Proteomic profiling of KATP channel-deficient hypertensive heart maps risk for maladaptive cardiomyopathic outcome. Proteomics. 2009;9:1314–1325. doi: 10.1002/pmic.200800718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arrell DK, Zlatkovic J, Kane GC, Yamada S, Terzic A. ATP-sensitive K+ channel knockout induces cardiac proteome remodeling predictive of heart disease susceptibility. J Proteome Res. 2009;8:4823–4834. doi: 10.1021/pr900561g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zlatkovic Lindor J, Arrell DK, Yamada S, Nelson TJ, Terzic A. ATP-sensitive K+ channel-deficient dilated cardiomyopathy proteome remodeled by embryonic stem cell therapy. Stem Cells. 2010;28:1355–1367. doi: 10.1002/stem.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barallobre-Barreiro J, Didangelos A, Schoendube FA, Drozdov I, Yin X, Fernández-Caggiano M, et al. Proteomics analysis of cardiac extracellular matrix remodeling in a porcine model of ischemia/reperfusion injury. Circulation. 2012;125:789–802. doi: 10.1161/CIRCULATIONAHA.111.056952. [DOI] [PubMed] [Google Scholar]
- 42.Stegemann C, Drozdov I, Shalhoub J, Humphries J, Ladroue C, Didangelos A, et al. Comparative lipidomics profiling of human atherosclerotic plaques. Circ Cardiovasc Genet. 2011;4:232–242. doi: 10.1161/CIRCGENETICS.110.959098. [DOI] [PubMed] [Google Scholar]
- 43.Lusis AJ, Weiss JN. Cardiovascular networks: systems-based approaches to cardiovascular disease. Circulation. 2010;121:157–170. doi: 10.1161/CIRCULATIONAHA.108.847699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sperling SR. Systems biology approaches to heart development and congenital heart disease. Cardiovasc Res. 2011;91:269–278. doi: 10.1093/cvr/cvr126. [DOI] [PubMed] [Google Scholar]
- 45.MacLellan WR, Wang Y, Lusis AJ. Systems-based approaches to cardiovascular disease. Nat Rev Cardiol. 2012;9:172–184. doi: 10.1038/nrcardio.2011.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barabási A-L, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 47.Newman MEJ. The structure and function of complex networks. SIAM Rev. 2003;45:167–256. [Google Scholar]
- 48.Pawson T, Linding R. Network medicine. FEBS Lett. 2008;582:1266–1270. doi: 10.1016/j.febslet.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 49.Arrell DK, Elliott ST, Kane LA, Guo Y, Ko YH, Pedersen PL, et al. Proteomic analysis of pharmacological preconditioning: Novel protein targets converge to mitochondrial metabolism pathways. Circ Res. 2006;99:706–714. doi: 10.1161/01.RES.0000243995.74395.f8. [DOI] [PubMed] [Google Scholar]
- 50.Nesvizhskii AI, Aebersold R. Interpretation of shotgun proteomic data: The protein inference problem. Mol Cell Proteomics. 2005;4:1419–1440. doi: 10.1074/mcp.R500012-MCP200. [DOI] [PubMed] [Google Scholar]
- 51.Bader GD, Cary MP, Sander C. Pathguide: a pathway resource list. Nucleic Acids Res. 2006;34:D504–D506. doi: 10.1093/nar/gkj126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mayr M, Zampetaki A, Sidibe A, Mayr U, Yin X, De Souza AI, et al. Proteomic and metabolomic analysis of smooth muscle cells derived from the arterial media and adventitial progenitors of apolipoprotein E-deficient mice. Circ Res. 2008;102:1046–1056. doi: 10.1161/CIRCRESAHA.108.174623. [DOI] [PubMed] [Google Scholar]
- 53.Grant JE, Bradshaw AD, Schwacke JH, Baicu CF, Zile MR, Schey KL. Quantification of protein expression changes in the aging left ventricle of Rattus norvegicus. J Proteome Res. 2009;8:4252–4263. doi: 10.1021/pr900297f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Z, Liu N, Zhang L, Gong K, Cai Y, Gao W, et al. Proteomic profiling reveals comprehensive insights into adrenergic receptor-mediated hypertrophy in neonatal rat cardiomyocytes. Proteomics Clin Appl. 2009;3:1407–1421. doi: 10.1002/prca.200900029. [DOI] [PubMed] [Google Scholar]
- 55.Mayr M, Liem D, Zhang J, Li X, Avliyakulov NK, Yang JI, et al. Proteomic and metabolomic analysis of cardioprotection: Interplay between protein kinase C epsilon and delta in regulating glucose metabolism of murine hearts. J Mol Cell Cardiol. 2009;46:268–277. doi: 10.1016/j.yjmcc.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Agnetti G, Kaludercic N, Kane LA, Elliott ST, Guo Y, Chakir K, et al. Modulation of mitochondrial proteome and improved mitochondrial function by biventricular pacing of dyssynchronous failing hearts. Circ Cardiovasc Genet. 2010;3:78–87. doi: 10.1161/CIRCGENETICS.109.871236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bugger H, Schwarzer M, Chen D, Schrepper A, Amorim PA, Schoepe M, et al. Proteomic remodelling of mitochondrial oxidative pathways in pressure overload-induced heart failure. Cardiovasc Res. 2010;85:376–384. doi: 10.1093/cvr/cvp344. [DOI] [PubMed] [Google Scholar]
- 58.Farina A, Chambery A, Esposito S, Agozzino L, Cotrufo M, Della Corte A, et al. Proteomic profiling of medial degeneration in human ascending aorta. Clin Biochem. 2010;43:387–396. doi: 10.1016/j.clinbiochem.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 59.Parguiña AF, Grigorian-Shamajian L, Agra RM, Teijeira-Fernández E, Rosa I, Alonso J, et al. Proteins involved in platelet signaling are differentially regulated in acute coronary syndrome: A proteomic study. PLoS One. 2010;5:e13404. doi: 10.1371/journal.pone.0013404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Angel PM, Nusinow D, Brown CB, Violette K, Barnett JV, Zhang B, et al. Networked-based characterization of extracellular matrix proteins from adult mouse pulmonary and aortic valves. J Proteome Res. 2011;10:812–823. doi: 10.1021/pr1009806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fogle RL, Hollenbeak CS, Stanley BA, Vary TC, Kimball SR, Lynch CJ. Functional proteomic analysis reveals sex-dependent differences in structural and energy-producing myocardial proteins in rat model of alcoholic cardiomyopathy. Physiol Genomics. 2011;43:346–356. doi: 10.1152/physiolgenomics.00203.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hammer E, Goritzka M, Ameling S, Darm K, Steil L, Klingel K, et al. Characterization of the human myocardial proteome in inflammatory dilated cardiomyopathy by label-free quantitative shotgun proteomics of heart biopsies. J Proteome Res. 2011;10:2161–2171. doi: 10.1021/pr1008042. [DOI] [PubMed] [Google Scholar]
- 63.Hanzu FA, Palomo M, Kalko SG, Parrizas M, Garaulet M, Escolar G, et al. Translational evidence of endothelial damage in obese individuals: inflammatory and prothrombotic responses. J Thromb Haemost. 2011;9:1236–1245. doi: 10.1111/j.1538-7836.2011.04285.x. [DOI] [PubMed] [Google Scholar]
- 64.Qi YX, Jiang J, Jiang XH, Wang XD, Ji SY, Han Y, et al. PDGF-BB and TGF-β1 on crosstalk between endothelial and smooth muscle cells in vascular remodeling induced by low shear stress. Proc Natl Acad Sci USA. 2011;108:1908–1913. doi: 10.1073/pnas.1019219108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li W, Rong R, Zhao S, Zhu X, Zhang K, Xiong X, et al. Proteomic analysis of metabolic, cytoskeletal and stress response proteins in human heart failure. J Cell Mol Med. 2012;16:59–71. doi: 10.1111/j.1582-4934.2011.01336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.de la Cuesta F, Barderas MG, Calvo E, Zubiri I, Maroto AS, Darde VM, et al. Secretome analysis of atherosclerotic and non-atherosclerotic arteries reveals dynamic extracellular remodeling during pathogenesis. J Proteomics. 2012;75:2960–2971. doi: 10.1016/j.jprot.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 67.Isserlin R, Merico D, Alikhani-Koupaei R, Gramolini A, Bader GD, Emili A. Pathway analysis of dilated cardiomyopathy using global proteomic profiling and enrichment maps. Proteomics. 2010;10:1316–1327. doi: 10.1002/pmic.200900412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cammarato A, Ahrens CH, Alayari NN, Qeli E, Rucker J, Reedy MC, et al. A mighty small heart: the cardiac proteome of adult Drosophila melanogaster. PLoS One. 2011;6:e18497. doi: 10.1371/journal.pone.0018497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun Z, Biela LM, Hamilton KL, Reardon KF. Concentration-dependent effects of the soy phytoestrogen genistein on the proteome of cultured cardiomyocytes. J Proteomics. 2012;75:3592–3604. doi: 10.1016/j.jprot.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 70.Henderson-MacLennan NK, Papp JC, Talbot CC, Jr, McCabe ERB, Presson AP. Pathway analysis software: annotation errors and solutions. Mol Genet Metab. 2010;101:134–140. doi: 10.1016/j.ymgme.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Merico D, Gfeller D, Bader GD. How to visually interpret biological data using networks. Nat Biotechnol. 2009;27:921–924. doi: 10.1038/nbt.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barabasi A-L, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- 73.Khalil I, Brewer MA, Neyarapally T, Runowicz CD. The potential of biologic network models in understanding the etiopathogenesis of ovarian cancer. Gynecol Oncol. 2010;116:282–285. doi: 10.1016/j.ygyno.2009.10.085. [DOI] [PubMed] [Google Scholar]
- 74.MacCannell KA, Shohet RV. Toward a holistic view of transcriptional regulation. Circ Cardiovasc Genet. 2011;4:2–3. doi: 10.1161/CIRCGENETICS.111.959452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pinaud B, Kuntz P. GVSR: an On-Line Guide for Choosing a Graph Visualization Software. In: Brandes U, Cornelsen S, editors. Graph Drawing: 18th Int Symp, GD 2010, Lecture Notes in Computer Science. Berlin: Springer; 2011. pp. 400–401. [Google Scholar]
- 76.Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 78.Assenov Y, Ramírez F, Schelhorn SE, Lengauer T, Albrecht M. Computing topological parameters of biological networks. Bioinformatics. 2008;24:282–284. doi: 10.1093/bioinformatics/btm554. [DOI] [PubMed] [Google Scholar]
- 79.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Albert R, Jeong H, Barabási A-L. Error and attack tolerance of complex networks. Nature. 2000;406:378–382. doi: 10.1038/35019019. [DOI] [PubMed] [Google Scholar]